Summary
Retrotrapezoid nucleus (RTN) neurons in the brainstem regulate the ventilatory response to hypercarbia. It is unclear how PHOX2B-polyalanine repeat mutations (PHOX2B-PARMs) alter the function of PHOX2B and perturb the formation of RTN neurons. Here, we generated human brainstem organoids (HBSOs) with RTN-like neurons from human pluripotent stem cells. Single-cell transcriptomics revealed that expression of PHOX2B+7Ala PARM alters the differentiation trajectories of the hindbrain neurons and hampers the formation of the RTN-like neurons in HBSOs. With the unguided cerebral organoids (HCOs), PHOX2B+7Ala PARM interrupted the patterning of PHOX2B+ neurons with dysregulation of Hedgehog pathway and HOX genes. With complementary use of HBSOs and HCOs with a patient and two mutant induced pluripotent stem cell lines carrying different polyalanine repetition in PHOX2B, we further defined the association between the length of polyalanine repetition and malformation of RTN-respiratory center and demonstrated the potential toxic gain of function of PHOX2B-PARMs, highlighting the uniqueness of these organoid models for disease modeling.
Keywords: PHOX2B, PHOX2B-PARMs, Human brainstem organoids, Human cerebral organoids, Human pluripotent stem cells, Retrotrapezoid nucleus (RTN) neurons, Congenital central hypoventilation syndrome, Breathing disorder, carbon dioxide (CO2)-sensitive glutamatergic neurons
Highlights
-
•
Newly established brainstem organoids containing hypercapnia-responsive neurons
-
•
PHOX2B-7Ala alters the differentiation trajectories of the hindbrain neurons
-
•
Dysregulation of Hedgehog and HOX genes in PHOX2B+ neurons with patterning defect
-
•
Longer polyalanine repetition leads to an additional loss of C1 neurons
E. Ngan and colleagues established a new brainstem organoid model for studying the breathing disorders and highlighted the advantage of the complementary uses of the region-specific and the unguided neural organoids to study the congenital neurogenic disorder.
Introduction
Ventilatory responses to hypoxia and hypercapnia are regulated by a specific set of chemoreceptors and motor neurons in the hindbrain. Retrotrapezoid nucleus (RTN) is a major center of chemoreceptors in the ventrolateral medulla that provides excitatory drives to the central respiratory pattern generator. It comprises a bilateral cluster of carbon dioxide (CO2)-sensitive glutamatergic neurons that express vesicular-glutamate transporter-2 (VGLUT2) and homeobox transcription factor PHOX2B (Goridis et al., 2010; Guyenet et al., 2019). These glutamatergic RTN neurons not only detect the CO2 level in the brain but also integrate chemosensory information from chemoreceptors, where innervating serotonin (5-HT) neurons enhance their chemosensitivity (Moreira et al., 2016). In response to elevated CO2 levels, RTN neurons intermingle with C1 neurons to excite A5 neurons and stimulate breathing and increase the rhythmic pattern of respiratory activity (Guyenet et al., 2005). This RTN-mediated central respiratory chemoreflex provides an important respiratory stimulus during apnea and dyspnea.
Congenital central hypoventilation syndrome (CCHS) is a life-threatening disease caused by the loss of RTN neurons in the brainstem. It is characterized by inadequate autonomic control of breathing due to impaired ventilatory response to hypercarbia and hypoxemia (Dubreuil et al., 2008). Polyalanine repeat expansion mutations (PARMs), resulting in +5 to +13 additional alanine (Ala) residues in the 20-residue polyalanine tract of PHOX2B, are highly associated with CCHS, where longer polyalanine repetition is associated with more severe disease phenotypes (Amiel et al., 2003; Bachetti and Ceccherini, 2020; Matera et al., 2004).
Mouse mutants carrying +7Ala-PARM in Phox2b lack Phox2b+ RTN neurons but have enriched Phox2b+ glutamatergic neurons on the dorsal side of facial nucleus (nVII). These mutants exhibit CCHS-like phenotypes including breathing defects, absence of response to hypercapnia, and death from central apnea soon after birth (Dubreuil et al., 2008, 2009). Several attempts have been made to establish hindbrain (Butts et al., 2019) or brainstem (Eura et al., 2020) organoids using human pluripotent stem cells (hPSCs). Nevertheless, these protocols did not provide cues for the generation of functional RTN neurons and thus hampered their application in disease modeling for CCHS. It remains unclear how PARMs affect the functions of PHOX2B and the formation of RTN neurons in the brainstem and whether longer PARMs lead to more severe CNS defects.
In this project, we generated hPSC-based brainstem (HBSO) and cerebral (HCO) organoids with cytoarchitectures resembling the RTN-respiratory center. We then unveiled the molecular mechanisms underlying PHOX2B-PARMs on the development of hindbrain neurons through analyzing the single-cell transcriptomes of HBSOs and HCOs derived from a PHOX2B +7Ala-PARM hPSC line. Proof-of-concept studies were also conducted with additional mutant hPSC lines carrying different lengths of PARMs and a CCHS-patient-specific hPSC line to illustrate the potential applications of these organoid models for recapitulating the PARM-associated RTN defects in vitro.
Results
Generation and characterization of human brainstem organoids from hPSCs
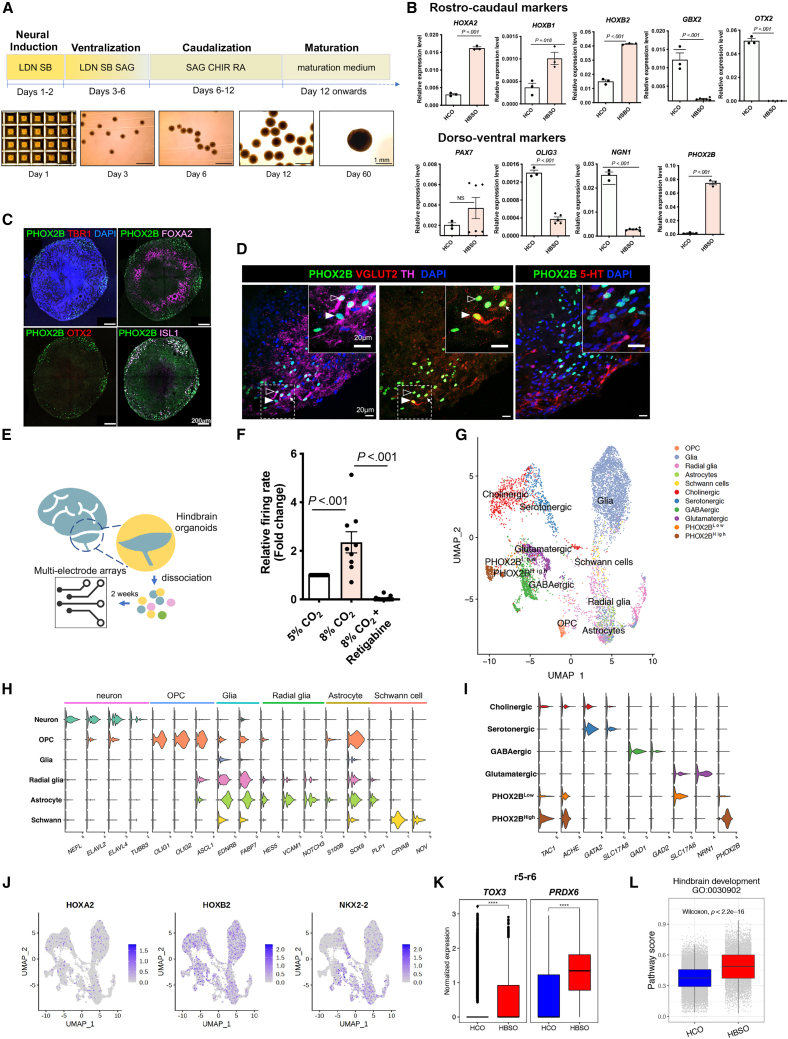
To develop an in vitro human model of CCHS, we first used a control hPSC cell line (UE02306) to establish a differentiation protocol for the generation of HBSOs with enriched PHOX2B+ and RTN neurons. RTN neurons originate from the rhombomere 5 (r5) and dB2 domains, which require WNT and retinoic acid (RA) signaling during development (Guyenet et al., 2019). Therefore, after directing the hPSCs to neural cell fate by dual-SMAD inhibitors (LDN193189 and SB431542) and ventralization by SHH agonist (SAG), the spheroids were caudalized to hindbrain cell fate with WNT agonist (CHIR99021) and RA (Figure 1A). Compared with HCOs comprising different subregions of the brain generated from an unguided differentiation protocol (Giandomenico et al., 2021; Lancaster and Knoblich, 2014; Lancaster et al., 2013), HBSOs expressed significantly higher levels of r4-5 markers (HOXA2, HOXB1, and HOXB2) but lower levels of r1 marker (GBX2) and undetectable levels of midbrain marker (OTX2) by day 20 of differentiation (Figure 1B). Regarding the dorsal-ventral specification, HBSOs expressed a much higher level of PHOX2B but lower levels of OLIG3 and NGN1 than HCOs while retaining the PAX7 expression (Figure 1B). This implies HBSOs have a spatial identity similar to the dB2 domain but not the other two PHOX2B+ domains (i.e., dA3 and MNv).
Figure 1.
Generation and characterization of brainstem organoids (HBSOs)
(A) Schematic showing the derivation of HBSOs from hPSCs. Representative phase-contrast images showing the HBSOs derived from the control hPSCs at different stages of differentiation.
(B) qRT-PCR analyses show the relative expressions of rostro-caudal (HOXA2, HOXB1, HOXB2, GBX2, and OTX2) and dorsoventral (PAX7, OLIG3, NGN, and PHOX2B) markers in the day-20 control HBSOs in comparison to the day-60 HCOs. Data are represented as mean ± SEM of three independent pools of ≥4 organoids. Unpaired t test; NS, non-significant.
(C) Immunostaining of PHOX2B and markers of the forebrain (TBR1), midbrain (FOXA2, OTX2), and hindbrain (ISL1) in the day-60 control HBSOs.
(D) Immunostaining of PHOX2B, TH, VGLUT2, and 5-HT in the day-60 control HBSOs. The inset on the top-right corner is an enlargement of a square region in the image. RTN (PHOX2B+ VGLUT2+ TH−), A5-like (PHOX2B+; TH+; VGLUT2–), and C1-like (PHOX2B+; TH+; VGLUT2+) neurons are marked by a filled arrowhead, open arrowhead, and an arrow, respectively.
(E) Schematic showing MEA analysis of day-60 control HBSOs.
(F) Firing rate of HBSO neurons in response to different CO2 levels and RTN blocker (KCNQ channel activator, retigabine). The mean spike firing rate of each MEA recording was normalized with the first recording undertaken in 5% CO2 condition to calculate the fold change ± SEM from ≥9 independent experiments per group. One-way ANOVA.
(G) UMAP projection of all 23,088 individual cells from day-60 HBSOs, colored by cell types.
(H) Violin plots showing the key markers of six main cell types. OPC, oligodendrocyte progenitor cell.
(I) Violin plots showing the key markers of six neuronal subtypes.
(J) Expression of hindbrain markers (HOXA2, HOXB2, and NKX2-2) in the HBSO cells.
(K) Relative expression of regional markers TOX3 (r5) and PRDX6 (r6) in HCOs and HBSOs.
(L) Pathway scores of hindbrain development in HCOs and HBSOs. p value (non-parametric Wilcoxon rank-sum test) from comparing HBSOs and HCOs is shown. ∗∗∗∗p value < 0.0001. See also Figure S1 and Data S1.
By day 60, most of the PHOX2B+ cells in the HBSOs retained specific position codes for hindbrain neurons, co-expressing the hindbrain marker (ISL1) but not the midbrain (OTX2/FOXA2) or the forebrain (TBR1) markers (Figure 1C). In addition, PHOX2B+ glutamatergic RTN-like (PHOX2B+; VGLUT2+; TH−) neurons were residing near to A5-(PHOX2B+; TH+; VGLUT2-), C1- (PHOX2B+; TH+; VGLUT2+) and 5-HT+ neurons, resembling the respiratory center of the brainstem (Figure 1D). We then assessed the potential hypercapnia response of HBSO neurons by performing multielectrode array (MEA) analyses. Dissociated HBSO neurons were plated onto an MEA plate for 1–2 weeks for network re-establishment (Figure 1E), and the neural activity was then measured under different CO2 levels (5% or 8%). Increased firings were detected in response to the elevated CO2 level, and that was blocked by an RTN-specific blocker (retigabine) (Hawryluk et al., 2012) (Figure 1F), suggesting the presence of hypercapnia-responsive RTN neurons in the HBSOs.
scRNA-seq analyses reveal cell type diversity and hindbrain molecular signatures of HBSOs
To further investigate the cellular composition and molecular signatures of HBSOs, we performed single-cell transcriptomic analyses of day-60 HBSOs and compared with the HCOs derived from the same control hPSC line. A total of 11,347 cells from a pool of >20 HBSOs and 42,970 cells from a pool of eight HCOs were sequenced using 10X Genomics, with 2,319 median genes and 24,389 mean unique molecular identifiers detected per cell (Figure S1A). All single cells were projected onto Uniform Manifold Approximation and Projection (UMAP) plots, and single-cell RNA-sequencing (scRNA-seq) datasets of hindbrain cells from embryonic (La Manno et al., 2021) and adolescent (Zeisel et al., 2018) mouse brains were used as references to annotate the HBSO cells (Figure S1B). 11 main cell types were found in the HBSOs (Figure 1G) with approximately 40% expressing pan-neuronal markers (NEFL, ELAVL2, ELAVL4, and TUBB3), 45% expressing glial markers (EDNRB and FABP7), and the remaining 5% exhibiting molecular signatures resembling radial glia, astrocytes, Schwann cells, or oligodendrocyte progenitor cells (OPCs) (Figure 1H and Data S1).
Using an unsupervised clustering analysis (Liu et al., 2020), HBSO neurons were then classified into four subclasses: cholinergic (TAC1+, ACHE+), serotonergic (GATA2+, SLC17A8+), GABAergic (GAD1+, GAD2+), and glutamatergic (SLC17A6+, NRN1+). In addition, there were two distinct groups of PHOX2B+ neurons with differential expression profiles (PHOX2BLow and PHOX2BHigh) (Figures 1I and 1G, Data S1). PHOX2BLow group also expressed the glutamatergic marker (SLC17A6, which encodes VGLUT2), representing RTN-like neurons, while PHOX2BHigh neurons would be the cholinergic population of PHOX2B+ hindbrain neurons.
The day-60 HBSOs expressed various caudal hindbrain markers (HOXA2, HOXB2, and NKX2-2) and region-specific markers of r5 (TOX3) and r6 (PRDX6) (Figures 1J and 1K). Gene Ontology (GO) term enrichment analysis of global gene expression profiles revealed that pathways associated with hindbrain development (GO: 0030902) were highly enriched in the HBSOs in comparison to the HCOs (Figure 1L). In sum, HBSOs exhibited unique hindbrain-specific molecular signatures and comprised the major neuronal subtypes found in the mouse hindbrain/brainstem (La Manno et al., 2021; Liu et al., 2020), including glutamatergic PHOX2B+ neurons.
PHOX2B-7Ala hampers the generation of hindbrain/RTN neurons as revealed by the HBSO model
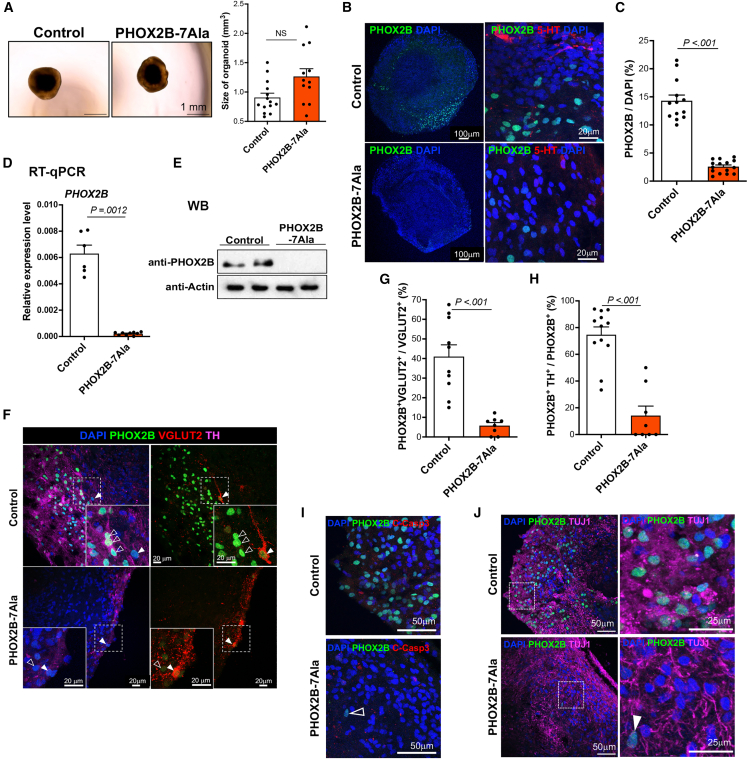
Using the newly established HBSO model, we examined the effect of PHOX2B-PARMs on the development of RTN neurons, particularly focusing on PHOX2B+7Ala PARM, the most common form of PARMs. Using CRISPR-Cas9-mediated homology-directed repair (HDR) platform, seven additional trinucleotide repeats (+7Ala) were introduced into the polyalanine tract of PHOX2B (Figure S2). An in-frame insertion of an enhanced green fluorescent protein with a 2A self-cleaving peptide was added immediately after the stop codon of PHOX2B to allow subsequent detection of the PARM-expressing mutant cells (Figure S2). At day 60, the HBSOs generated from PHOX2B-7Ala-hPSCs had a comparable size with the control HBSOs (Figure 2A). However, compared with the control, fewer PHOX2B+ cells were observed, and only a few were found adjacent to 5-HT+ neurons in the PHOX2B-7Ala HBSOs (Figures 2B and 2C). The loss of PHOX2B-expressing cells in PHOX2B-7Ala HBSOs was further confirmed by reverse transcription quantitative PCR (qRT-PCR) (Figure 2D) and western blot (Figure 2E) analyses. Consistently, PHOX2B+ TH+ neurons and PHOX2B+VGLUT2+TH- RTN neurons were barely found in the PHOX2B-7Ala HBSOs (Figure 2F). The percentages of PHOX2B+VGLUT2+ (Figure 2G) and PHOX2B+TH+ (Figure 2H) neurons drastically decreased in the PHOX2B-7Ala HBSOs. In both the control and PHOX2B-7Ala HBSOs, all PHOX2B+ neurons were negative for apoptotic markers (cleaved caspase-3, C-Casp3–) (Figure 2I) and expressed pan-neuronal marker (TUJ1) (Figure 2J), suggesting that the loss of PHOX2B+ neurons in the mutant HBSOs is unlikely due to the poor survival or neuronal differentiation defect of PHOX2B-7Ala-expressing cells.
Figure 2.
Detrimental effect of PHOX2B-7Ala in the generation of RTN-like neurons in HBSOs
(A) Representative phase-contrast images of the day-60 control and PHOX2B-7Ala HBSOs. Bar chart showing the sizes of the day-60 control and PHOX2B-7Ala HBSOs (mean ± SEM). n ≥ 9 per group from three independent experiments. Unpaired t test; NS, non-significant.
(B) Immunostaining of PHOX2B, 5-HT in the day-60 control and PHOX2B-7Ala HBSOs.
(C) Bar charts showing the percentages of PHOX2B+ cells in the day-60 HBSOs derived from the control and PHOX2B-PARM mutants (mean ± SEM). n ≥ 8 per group from three independent experiments.
(D) qRT-PCR and (E) western blot analyses of PHOX2B expression in the day-60 control and PHOX2B-7Ala HBSOs. Data are represented as mean ± SEM. n ≥ 9 per group from three independent experiments. Unpaired t test.
(F) Immunostaining of PHOX2B, TH, and VGLUT2 in the day-60 control and PHOX2B-7Ala HBSOs. The inset on the bottom is an enlargement of a square region in the image. Filled and open arrow heads mark the RTN-like (PHOX2B+ VGLUT2+ TH–) and PHOX2B+ TH+ neurons, respectively.
(G and H) Bar charts showing the percentages of (G) PHOX2B+VGLUT2+ and (H) PHOX2B+TH+ cells in the day-60 control and PHOX2B-7Ala HBSOs (mean ± SEM). n ≥ 8 per group from three independent experiments. Unpaired t test.
(I and J) Immunostaining of PHOX2B with (I) C-Casp3 and (J) TUJ1 (an enlargement of a square is shown in the right panel) in the day-60 control and PHOX2B-7Ala HBSOs. Mutant PHOX2B+ neurons that were viable (C-Casp3–) and expressed pan-neuronal marker (TUJ1+) are marked by open and filled arrowheads, respectively. See also Figure S2.
PHOX2B-7Ala HBSOs exhibit aberrant neuronal differentiation paths and hindbrain signatures
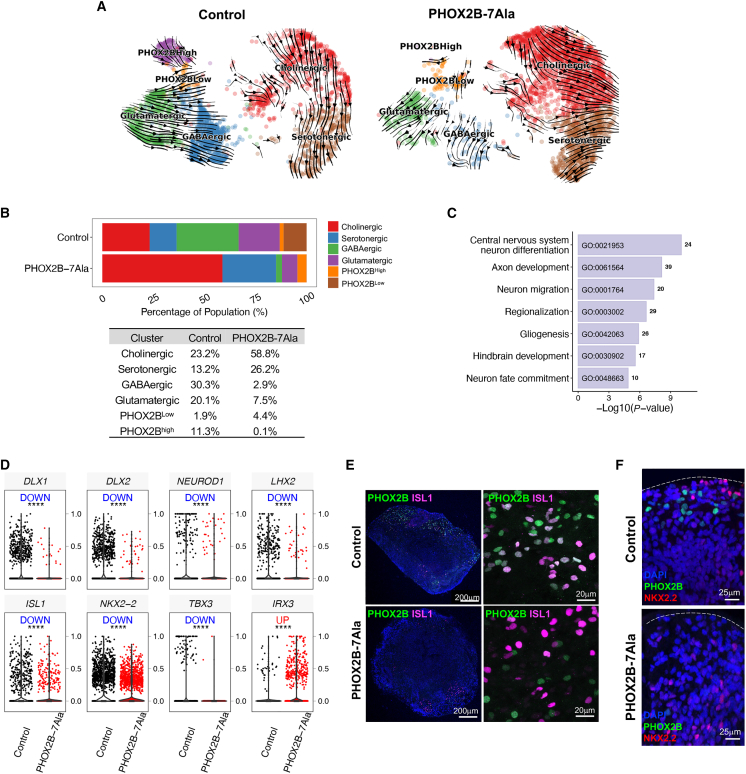
To delineate how PHOX2B-7Ala interferes the formation of the respiratory unit, another 11,741 cells from day-60 PHOX2B-7Ala HBSOs were sequenced. Despite the presence of the main neuronal and glial cell subtypes in the PHOX2B-7Ala HBSOs, an obvious change in the cellular composition of the neuronal population was observed (Figures S3A and S3B). RNA velocity analyses revealed that neuronal differentiation paths for both glutamatergic and GABAergic lineages were disrupted (Figure 3A), accompanied by increased proportions of cholinergic and serotonergic-like cells in the PHOX2B-7Ala HBSOs (Figure 3B). In addition, the PHOX2BHigh population was almost completely diminished, and the PHOX2B-expressing cells only expressed minimal levels of PHOX2B in the PHOX2B-7Ala HBSOs (Figure 3B). Thus, the loss of PHOX2B+ neurons in PHOX2B-7Ala HBSOs likely interferes with the pattern and/or development of the hindbrain and respiratory unit. A total of 642 genes (differentially expressed genes, DEGs) were found differentially expressed in PHOX2B-7Ala HBSOs. They were enriched in pathways such as “neuron differentiation/migration/fate commitment,” “regionalization,” and “hindbrain development” (Figure 3C). Genes associated with neuronal differentiation (DLX1, DLX2, NEUROD1, and LHX2) were consistently downregulated in PHOX2B-7Ala neurons, and genes implicated in hindbrain morphogenesis (ISL1, NKX2-2, and TBX3) and the midbrain-hindbrain boundary development (IRX3) were also dysregulated (Figure 3D). Subsequent immunostaining of the hindbrain marker (ISL1) with PHOX2B demonstrated that most PHOX2B+ cells in the PHOX2B-7Ala HBSOs did not co-express ISL1, suggesting a potential loss of hindbrain identity in the PHOX2B-7Ala-expressing cells (Figure 3E). Consistent with this observation, the NKX2.2+ domain (the inferred mantle layer of the hindbrain) shifted from the outer layer in the control HBSOs to the inner layer in the PHOX2B-7Ala HBSOs (Figure 3F), indicating that PHOX2B-7Ala perturbs the patterning of HBSOs.
Figure 3.
Aberrant differentiation and hindbrain signatures in the PHOX2B-7Ala HBSOs
(A) RNA velocity analyses revealed the differentiation paths of various neuronal subtypes were perturbed in the PHOX2B-7Ala HBSOs.
(B) Barplot showing the population size difference of six neuronal subtypes between the control and PHOX2B-7Ala mutant, and the sizes of each population are listed in the bottom table.
(C) Barplot showing the disrupted pathways in pooled neuronal cells in PHOX2B-7Ala HBSOs.
(D) Violin plots showing the key DEGs involved in neuronal differentiation (upper panel) and hindbrain regionalization (lower panel) in PHOX2B-7Ala mutant.
(E and F) Immunostaining of (E) PHOX2B and ISL1 and (F) NKX2-2 in the day-60 control and PHOX2B-7Ala HBSOs. Dotted lines mark the outer surface of the HBSOs. See also Figure S3.
Depletion of PHOX2B+ glutamatergic neurons in the PHOX2B-PARM HCOs
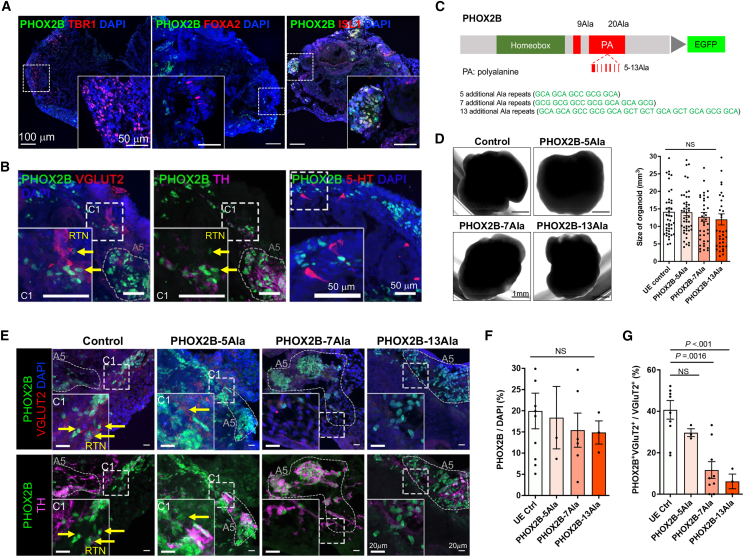
We next sought to address the hindbrain patterning defects associated with the PHOX2B-PARMs unbiasedly using unguided HCOs (Figure S4A). The control unguided HCOs comprised the TBR1+ forebrain, FOXA2+ midbrain, and ISL1+ hindbrain regions (Figure 4A). Importantly, PHOX2B+ cells were mainly co-localized in the ISL1+ hindbrain region where the A5, C1 and RTN-like neurons were found near to the 5-HT+ neurons (Figure 4B), forming a respiratory center-like region. In parallel, we generated two additional PHOX2B mutant hPSC lines carrying +5Ala- and +13Ala-PARM in PHOX2B for the subsequent comparative studies (Figures 4C and S2). The general morphology, size, and regional diversity of day-60 HCOs derived from all three PHOX2B-PARM hPSC lines were highly comparable to the control (Figures 4D and S4B). Unlike the HBSO model, we detected more cells expressing high levels of PHOX2B in the mutant HCOs. In both the control and mutants, PHOX2B+ cells were largely clustered near 5-HT+ neurons to form the respiratory center-like regions of comparable sizes (Figure S4C). The A5-like regions were identified in the control and all PHOX2B-PARM HCOs (Figure 4E). However, the C1-like region was absent in the PHOX2B-7Ala and PHOX2B-13Ala HCOs, whereas the PHOX2B-5Ala HCOs contained a slightly lower percentage of RTN-like neurons than the control HCOs in the respiratory center-like regions (Figure 4E). Although the number of PHOX2B-expressing cells was comparable between the control and all the PHOX2B-PARMs mutant HCOs (Figure 4F), the percentages of PHOX2B+; VGLUT2+ cells were significantly lower in the PHOX2B-7Ala and PHOX2B-13Ala HCOs than in the control HCOs (Figure 4G). These results demonstrated that PHOX2B-PARMs disrupt the formation of PHOX2B+; VGLUT2+ RTN neurons, as observed in the PHOX2B-7Ala-HBSOs. Moreover, using the HCO model, we demonstrated the direct correlation between the severity of the defect and the length of PARMs on the formation of PHOX2B+ domains during the hindbrain development.
Figure 4.
PHOX2B-PARMs disrupt the formation of respiratory center-like region
(A) Immunostaining of PHOX2B and markers of forebrain (TBR1), midbrain (FOXA2), and hindbrain (ISL1) in the day-60 control HCOs.
(B) Immunostaining of different markers of the respiratory center-like region in the day-60 control HCOs. Yellow arrows mark the RTN-like (PHOX2B+ VGLUT2+ TH−) neurons.
(C) Schematic showing the additional Ala repeats added to the PHOX2B-PARM mutant hPSC lines.
(D) Representative phase-contrast images of day-60 HCOs derived from the control and the PHOX2B-PARM mutants. Bar chart showing the sizes of day-60 HCOs derived from the control and the PHOX2B-PARM mutants (mean ± SEM). n ≥ 36 per group from four independent experiments. One-way ANOVA; NS, non-significant.
(E) Immunostaining of markers of A5 domain (PHOX2B+; TH+; VGLUT2−), C1 domain (PHOX2B+; TH+; VGLUT2+), and RTN neurons (PHOX2B+; TH−; VGLUT2+) in the day-60 HCOs derived from the control and the PHOX2B-PARM mutants. Yellow arrows mark the RTN-like (PHOX2B+ VGLUT2+ TH−) neurons.
(F and G) Bar charts showing the percentages of (F) PHOX2B+ and (G) PHOX2B+; VGLUT2+ double-positive cells in respiratory center-like region of the day-60 HCOs derived from the control and PHOX2B-PARM mutants. n ≥ 3 per group from three independent experiments. One-way ANOVA; NS, non-significant. All the insets on the bottom of the immunostaining image are the enlargement of the respective square regions in the images. See also Figure S4.
PARM interrupts the progenitor-to-neuron transition of PHOX2B+ cells in HCOs
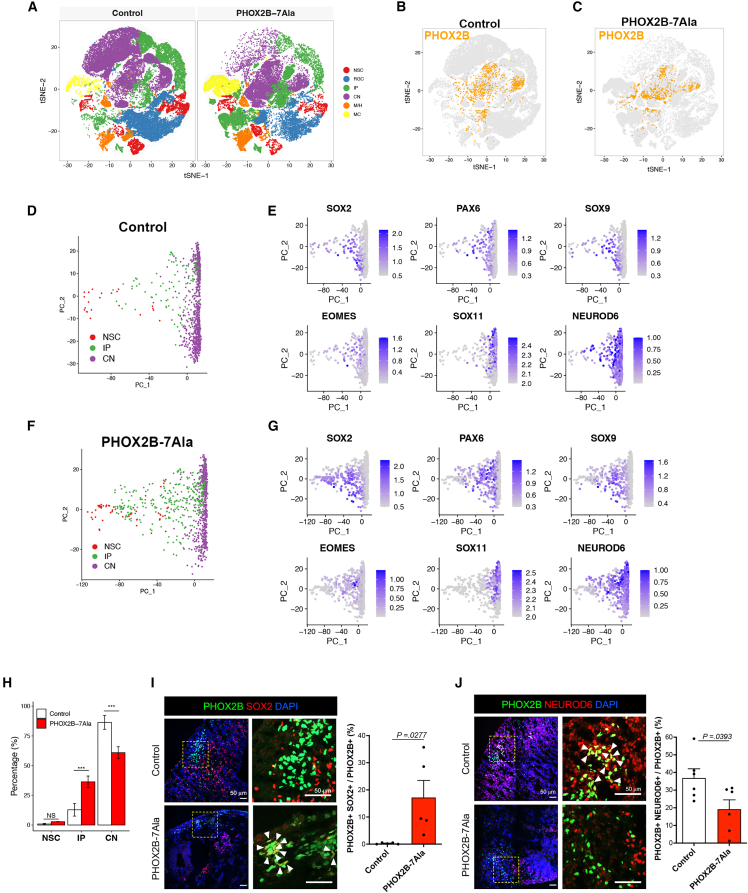
PHOX2B+ cells could be found in the PHOX2B-7Ala HCOs, so we then sequenced PHOX2B-7Ala day-60 HCOs at high resolution (Figure S5A). We projected all single cells from the control and PHOX2B-7-Ala PARM HCOs on a t-distributed stochastic neighbor embedding (t-SNE) plot (Figures S5B–S5D) and identified 16 transcriptionally distinct clusters (Data S2). After processing with Seurat software (Hao et al., 2021), the 16 unbiased clusters were further classified into six cell types, including neural stem cells (NSCs), radial glia cells (RGCs), intermediate progenitors (IPs), cortical neurons (CNs), midbrain/hindbrain (M/H), and mesenchymal-like cells (MCs) (Figures 5A, S6A, and S6B and Data S3) based on the expression of canonical markers and cell annotations from a public scRNA-seq dataset of 2-month-old HCOs (Kanton et al., 2019).
Figure 5.
scRNA-seq analysis of HCOs reveals that PHOX2B-PARM interrupts the progenitor-to-neuronal transition of PHOX2B+ cells
(A) t-SNE projection of all 72,800 individual cells from day-60 HCOs into six main cell types, colored by cell types. PHOX2B+ cells in (B) control and (C) PHOX2B-7Ala HCOs after imputation analysis.
(D) Trajectory constructed by PCA analysis shows the lineage progression of PHOX2B+ cells from NSC to IP and CN states in the control HCOs.
(E) Canonical markers expressed in PHOX2B+ cells at NSC, IP, and CN states in the control HCOs.
(F) Trajectory constructed by PCA analysis shows the lineage progression of PHOX2B+ cells from NSC to IP and CN states in the PHOX2B-7Ala HCOs.
(G) Canonical markers expressed in PHOX2B+ cells at NSC, IP, and CN states in the PHOX2B-7Ala HCOs.
(H) Barplot showing the population sizes of PHOX2B+ cells at NSC, IP, and CN states in the control and PHOX2B-7Ala HCOs (mean ± SEM). ∗∗p < 0.05 and ∗∗∗p < 0.001; NS, non-significant.
(I and J) Immunostaining of (I) PHOX2B+; SOX2+ and (J) PHOX2B+; NEUROD6+ cells in the control and PHOX2B-7Ala mutant HCOs as marked by the arrowheads. An enlargement of a square region in the image is shown in the right panel. Bar charts showing the quantitative data represented as mean ± SEM. n ≥ 3 per group from three independent experiments. Unpaired t test; two-sided. NSC, neural stem cells; RGC, radial glial cells; IP, intermediate progenitors; CN, cortical neurons; M/H, mid/hindbrain cells; MC, mesenchymal-like cells. See also Figures S5–S7 and Data S2.
Unexpectedly, the expression of PHOX2B was extremely low in HCO cells (Figure S7C). This may be due to the high cell heterogeneity in the HCOs, leading to a high dropout rate of rare cells in the 10X Genomics platform (Kim et al., 2020), which makes most of the PHOX2B of relatively short transcript length undetectable. Thus, we applied Markov affinity-based graph imputation of cells to denoise the cell count matrix and fill in the missing transcripts (van Dijk et al., 2018). The expression of PHOX2B could be recovered well and comparable numbers of PHOX2B+ cells were identified in the control and PHOX2B-7Ala HCOs (Figure S6D). Most of the PHOX2B+ cells were found in cell clusters inferred to be less mature progenitors (IPs) or more mature neurons (CNs), and a few were categorized into NSCs (Figures 5B and 5C). We next inferred the lineage relationship among PHOX2B+ cells in the control HCOs using principal component analysis (PCA). The transition states that linked NSCs through IPs to CNs could be well represented by PC1, while the diversity of CNs was reflected on PC2 (Figure 5D). In the control HCOs, only a small portion of PHOX2B+ cells remained as NSC/IPs, resided at the beginning of the PC1 axis, and expressed NSC (SOX2, PAX6, SOX9) or IP (EOMES) markers, while the majority of PHOX2B+ cells were found at the end of the PC1 axis with higher expression of mature neuronal markers (SOX11 and NEUROD6), suggesting that most PHOX2B+ cells were mature neurons (CNs) (Figures 5D and 5E). Although a similar differentiation trajectory (Figures 5F and 5G) was detected in PHOX2B-7Ala HCOs, the percentage of PHOX2B+ cells in the CN state was significantly reduced, and more PHOX2B+ cells remained in the progenitor (IP) state in the PHOX2B-7Ala HCOs (Figure 5H). Subsequent immunofluorescence analyses of day-60 HCOs further confirmed the higher percentage of PHOX2B+ cells in the NSC/IP state (PHOX2B+; SOX2+) (Figure 5I), accompanied by a reduced percentage of mature PHOX2B+ neurons (PHOX2B+; NEUROD6+) (Figure 5J) in the PHOX2B-7Ala HCOs when compared with the control. Thus, PHOX2B-7Ala interrupted the progenitor-to-neuron transition of the PHOX2B+ neurons.
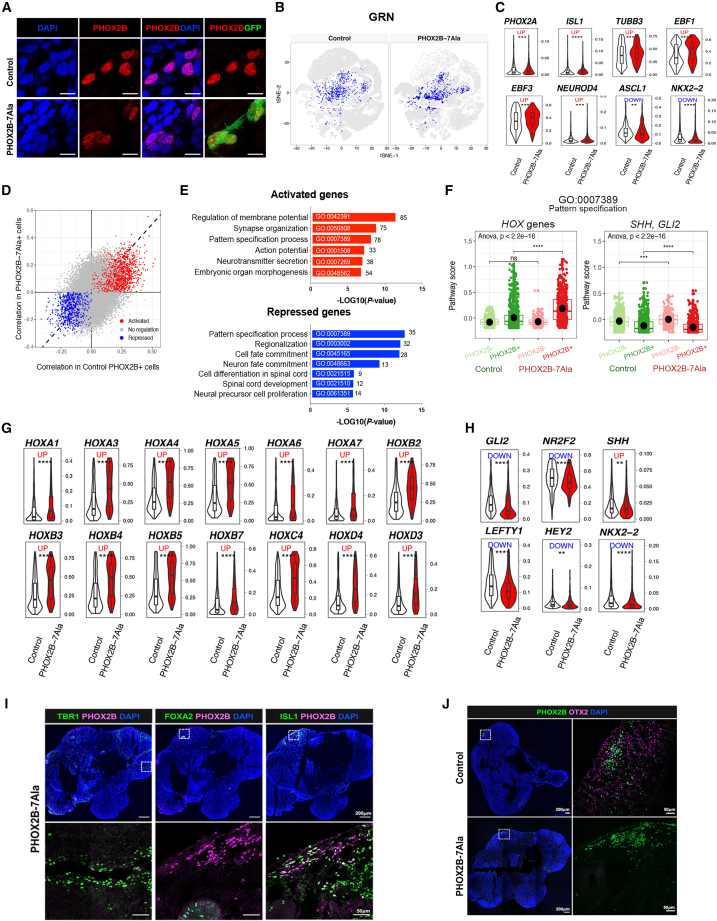
Dysregulation of pattern specification pathway genes and loss of PHOX2B+ neurons in OTX2+ domain of PHOX2B-7Ala HCOs
The mutant PHOX2B proteins were largely localized at the nuclei of the HCO neurons, where the expression of the PHOX2B-7Ala mutant allele was confirmed by the co-expression of GFP signals (Figure 6A). Thus, we performed single-cell regulatory network inference and clustering analysis (Aibar et al., 2017) to establish a gene regulatory network (GRN) of PHOX2B in the control and mutant HCOs. PHOX2B-7Ala cells showed a different regulatory architecture from that of the control HCOs (Figure 6B). In mice, Phox2b upregulates Phox2a, Isl1, Tubb3, and Ebf1-3 and downregulates Ascl1 and Nkx2.2 during the hindbrain development (Pattyn et al., 2000). We observed a similar expression pattern of these genes in the cells expressing PHOX2B-7Ala but with the greater changes when compared with the wild-type cells (Figure 6C). Therefore, it is conceivable that PHOX2B-7Ala does not simply lead to loss of function or function in a dominant-negative manner but may cause aberrant expression of different gene sets. To address this point, whole-transcriptome correlation analysis was performed to identify PHOX2B-7Ala-mediated (activated or repressed) genes and pathways. 2,393 and 640 genes were predicted as PHOX2B-7Ala activated (red dots) and repressed (blue dots) genes, respectively, where the activated genes represent the direct targets of PHOX2B-7Ala with binding motif(s) in their promoter regions (Figure 6D). Based on the GO database, the putative PHOX2B-7Ala activated/repressed genes were mainly implicated in pattern specification or neurogenesis (Figure 6E). Intriguingly, HOX- and Hedgehog pathway (SHH, GLI2)-associated genes belonging to GO: 0007389 (pattern specification process) were found to be significantly upregulated and downregulated, respectively, in the PHOX2B-7Ala-expressing cells, suggesting that this pathway was severely interrupted in the mutant cells. The overall changes in these two pathway gene sets in PHOX2B+ and PHOX2B− cells were then scored using an additive model. The upregulation of HOX- and the downregulation of Hedgehog pathway-associated genes implicated in pattern specification were found in all the PHOX2B+ cells expressing both wild-type and mutant forms of PHOX2B, but their log2 fold changes (PHOX2B+/PHOX2B−) were much higher in the cells expressing mutant PHOX2B (Figure 6F), including upregulation of HOX genes (Figure 6G) and downregulation of multiple SHH downstream effectors (Figure 6H). Noteworthy, the Hedgehog pathway-associated genes were slightly affected in PHOX2B− cells in the PHOX2B-7Ala mutant HCOs (Figure 6F), suggesting that the patterning defects in PHOX2B+ may also influence neighboring PHOX2B− cells and their development. These analyses suggest that PARMs in PHOX2B likely alter its transcriptional activity, leading to either the overactivation or downregulation of its target genes.
Figure 6.
PHOX2B-PARM interrupts the pattern specification of PHOX2B+ neurons in HCOs
(A) Immunostaining showing the nuclear localization of PHOX2B proteins in the day-60 control and PHOX2B-7Ala mutant HCOs.
(B) GRN activity of PHOX2B in the control and mutant HCOs.
(C) Violin plots showing the expression of PHOX2B hindbrain targets in the control and mutant CNs. ∗∗p < 0.05, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
(D) Scatterplot showing the putative PHOX2B-7Ala activated (red) and repressed (blue) targeted genes.
(E) Gene Ontology (GO) enrichment analyses of PHOX2B activated and repressed targeted genes.
(F) Overall pathway scores of embryonic organ morphogenesis and pattern specification. ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001; ns, non-significant.
(G) Violin plots showing the expression of HOX genes in the control and mutant CNs. ∗∗∗∗p < 0.0001.
(H) Violin plots showing the expression of Hedgehog pathway genes in the control and mutant CNs. ∗∗p < 0.05 and ∗∗∗∗p < 0.0001.
(I) Immunostaining of PHOX2B and markers of forebrain, midbrain, and hindbrain (TBR1, FOXA2, and ISL1) in the day-60 PHOX2B-7Ala HCOs.
(J) Immunostaining of PHOX2B and OTX2 in the day-60 control and PHOX2B-7Ala HCOs. An enlargement of a square region in the image is shown in the (I) bottom and (J) right panel.
Although some of the mutant PHOX2B+ cells still retained hindbrain identity and were localized largely in the ISL1+ hindbrain region and not in the FOXA2+ midbrain region or the TBR1+ forebrain region of the HCOs (Figure 6I), they might have lost some patterning information. For instance, in control HCOs, some PHOX2B+ cells were found in the OTX2+ domain of the mid-hindbrain boundary (MHB), but this was not observed in mutant HCOs (Figure 6J), implying that the genetic program for the positioning of PHOX2B+ cells may be altered in mutant HCOs. Phox2b has been suggested to regulate dopaminergic neuron development in the embryonic mouse midbrain (Hoekstra et al., 2012), so PHOX2B-7Ala may also interrupt the formation of dopaminergic neurons in the MHB. Taken together, we found that the pattern specification genes were severely interrupted in the mutant PHOX2B+ cells, with a concomitant loss of PHOX2B+ cells in the respiratory center and OTX2+ domain in the PHOX2B-7Ala HCOs.
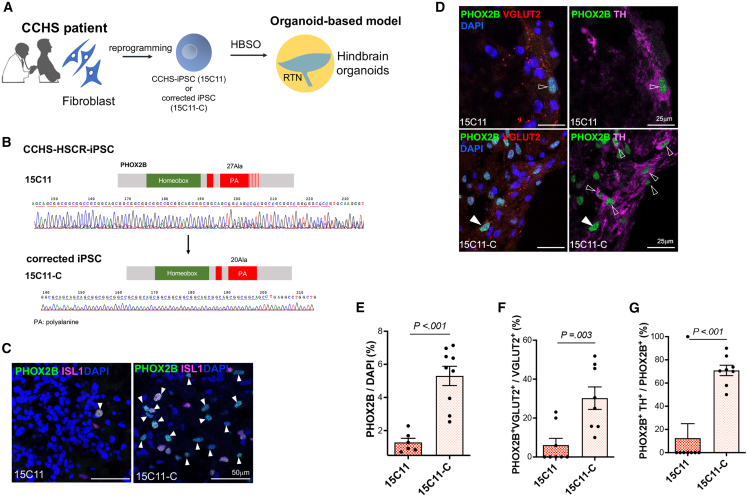
An HBSO model of CCHS
We next aimed to use the HBSOs to model the CCHS. A patient-specific hPSC line (15C11) was established from skin biopsy of a CCHS patient, who carried a heterozygous +7Ala repeat mutation in the PHOX2B (Figures 7A and S7A–S7D). The impact of the +7Ala-PARM was then demonstrated by comparing to the isogenic “corrected” line (15C11-C) where the heterozygous +7Ala repeat mutation in the PHOX2B locus was “corrected” using CRISPR-Cas9-mediated HDR (Figures 7B and S2). The 15C11 HBSOs had defects similar to those observed in the PHOX2B-7Ala HBSOs, including the loss of PHOX2B+ (Figure 7C), PHOX2B+ TH+, and PHOX2B+ VGLUT2+ neurons (Figure 7D). These defects were significantly rescued in the 15C11-C HBSOs (Figures 7E–7G), suggesting that the PHOX2B-associated CCHS phenotypes can be nicely recapitulated by the HBSOs.
Figure 7.
A HBSO model of CCHS
(A) Schematic showing the establishment of hPSC-based model of CCHS.
(B) Electrographs of Sanger sequencing of PHOX2B exon 3 showing the “correction” of +7Ala-PARM in PHOX2B locus.
(C) Immunostaining of PHOX2B and ISL1 in the day-60 15C11 and 15C11-C HBSOs. Arrowheads mark the PHOX2B+ISL1+ neurons.
(D) Immunostaining of PHOX2B, TH, and VGLUT2 in the day-60 15C11 and 15C11-C HBSOs. The putative RTN (PHOX2B+VGLUT2+TH−) and PHOX2B+ TH+ neurons are marked by filled and open arrowheads, respectively.
(E–G) Bar charts showing the percentages of (E) PHOX2B+, (F) PHOX2B+VGLUT2+, and (G) PHOX2B+TH+ cells in the day-60 15C11 and 15C11-C HBSOs. Data are represented as mean ± SEM. n ≥ 6 per group from three independent experiments. Unpaired t test, two-sided. See also Figure S7.
Discussion
In this study, we used hPSC-derived regionalized (HBSOs) and unguided (HCOs) brain organoids to study the molecular controls underpinning the formation of the hypercapnia-responsive domain of the respiratory center and the molecular basis of CCHS. We presented a robust differentiation protocol to generate HBSOs that contain a diversity of CNS neurons with hindbrain signatures. The HBSOs comprise the respiratory center-like region and RTN-like neurons of a functional response to hypercapnia. Subsequent analysis of the single-cell transcriptomes of the control HBSOs confirmed the presence of the key populations of neurons and glia and the expression of hindbrain markers, resembling the cellular composition and molecular features of the developing mouse hindbrains. More importantly, our data demonstrated PHOX2B-PARMs not only deterred the formation of glutamatergic RTN neurons but also the differentiation of other lineages of hindbrain neurons, perturbing the cellular composition of HBSOs.
The use of self-patterned HCOs further allowed us to unbiasedly reveal the molecular changes associated with PHOX2B-PARMs. PHOX2B-PARMs not only interfered with the progenitor-to-neuronal transition of PHOX2B+ neurons during neuronal differentiation but also with their morphogenesis and pattern specification. In particular, many HOX genes were dysregulated in the PHOX2B-PARM HCOs. Since HOX genes regulate the effectors of SHH signaling as well as ASCL1/NGN signaling during rostro-caudal and dorsoventral patterning specification (Gaufo et al., 2000), PHOX2B-PARMs may disrupt hindbrain development by dysregulating SHH signaling. The defective patterning of PHOX2B+ neurons may account for their absence in the OTX+ domain and RTN-respiratory center in the PHOX2B-PARM-HCOs and/or HBSOs. In addition, SHH is a key factor for the generation of HSBOs, particularly for ventralizing the organoids to enrich hindbrain neurons, so the loss of response to SHH signaling in PHOX2B-PARMs expressing neurons may lead to the depletion of PHOX2B+ neurons in mutant HBSOs. Noteworthy, a delayed maturation of the arcuate nucleus was observed in a syndromic CCHS patient (Tomycz et al., 2010), highlighting the possible association between the delayed neuronal maturation and PHOX2B-PARMs in human.
As demonstrated in this study, the complementary uses of HCO and HBSO models will allow us to address a board range of scientific questions. For instance, the HCOs are generated at a close-to-physiological condition and driven by the self-patterning signals, allowing the study of the interactions among various functional domains as well as genes implicated in the patterning of the brain, while the HBSOs are enriched with hindbrain neurons with high reproducibility, and that makes it most suitable for functional analyses or drug screening. The hypercapnia-responsive neurons in HBSOs can be easily detected not only based on the expression of the molecular markers but also their electroactivities in response to the change in CO2 levels, ion channel mediators, or neurotransmitters. This greatly expedites its potential applications for the high-throughput drug screenings.
PHOX2B-PARM proteins, when expressed at a normal endogenous level, still localized in the nuclei of mutant neurons. Our data suggest that PHOX2B-PARMs may have a toxic gain of function that alters their GRNs. Regardless of the length, PHOX2B-PARMs disrupted the formation of glutamatergic RTN neurons, while PARMs with longer alanine repeats also perturbed the formation of PHOX2B+VGLUT2+TH+ neurons in C1. The C1 neurons innervating the A5 domain are important for sympathetic and respiratory outputs (Malheiros-Lima et al., 2022); therefore, the additional loss of C1 neurons associated with the longer polyalanine repetition in PARMs probably leads to more severe breathing defects. Additional proof-of-concept study using a CCHS patient-specific and the “corrected” isogenic hPSC lines further suggests the potential application of HBSOs for disease modeling. Using the experimental paradigm established in this study, HBSOs derived from multiple patient lines can be used in conjunction with the scRNA-seq platform to further delineate the molecular mechanisms underlying PHOX2B-PARMs in CCHS pathogenesis. In addition, CCHS patients are frequently complicated with Hirschsprung disease (absence of enteric neurons in the distal colon) (Bachetti and Ceccherini, 2020). Therefore, HBSOs and enteric neurons can be derived from the same hPSC lines with identical genetic backgrounds and used for a side-by-side comparison to unveil the causal link between these closely related diseases in the future. In sum, HBSOs and HCOs comprise enriched hindbrain neurons and self-assembled multicomponent structures, respectively, and complementary uses of these two models allow us to unambiguously reveal the detrimental effects of PHOX2B-PARMs in hindbrain development.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Elly Ngan (engan@hku.hk).
Materials availability
All unique/stable reagents generated in this study are available from the corresponding author without restriction.
Generation and maintenance of hPSC lines
UE02302 hPSC line was used as the control (Xue et al., 2013). To generate PHOX2B-PARM mutant hPSC line, UE02302 hPSCs were transfected with the mutant PHOX2B-PARM donor plasmids together with gRNA constructs targeting PHOX2B and a Cas9 nickase expression plasmid by Nucleofection (Lonza). The transfected cells were selected with 0.2 μg/mL puromycin, and the PARM insertions were confirmed with Sanger sequencing. The patient hPSC line (15C11) was generated from skin fibroblast of a CCHS-HSCR patient (Figure S7) with episomal reprogramming vectors as described previously (Lai et al., 2017; Takahashi et al., 2007). The CRISPR-Cas9 homologous recombination strategy was used for the correction of PHOX2B-7Ala mutation in the patient hPSC line with a wild-type PHOX2B donor plasmid (15C11-C) (supplemental experimental procedures). All the hPSC lines were tested Mycoplasma-free and maintained in Matrigel-coated plates with mTeSR1 medium.
Generation of HSBOs and HCOs from hPSCs
To generate HSBOs, 2.7 × 106 hPSCs were seeded per well in an AggreWell800 24-well plate with mTeSR1 containing 10 μM Y-27632. 1 day later, the spheroids were transferred to an ultra-low-attachment plate and cultured with Neural Induction Medium. From days 3–6, the organoids were incubated in Neural Differentiation Medium (NDM) with 1 μM LDN-193189, 10 μM SB431542, and 1 μM SAG and kept rotating from day 3 onward. From days 6–12, the organoids were maintained in NDM with 1 μM SAG, 3 μM CHIR99021, and 1 μM RA, and the medium was changed every 2–3 days. From day 12 onward, the organoids were maintained in Neural Maturation Medium with 20 ng/mL BDNF, 20 ng/mL GDNF, 50 nM dibutyryl-cAMP, and 200 μM ascorbic acid. The medium was changed every 3–4 days.
HCOs were generated from hPSCs using a commercially optimized kit, STEMdiff Cerebral Organoid Kit (STEMCELL Technologies), following the manufacturer’s protocol.
Details of the cell culture reagents are listed in supplemental experimental procedures and Table S1.
Multielectrode array analysis
Day 60 HBSOs were dissociated with Papain Dissociation System and seeded into a 24-well CytoView MEA plate pre-coated with 0.1% polyethylenimine and maintained for 2 weeks before MEA analysis. The electric signals were measured at 37°C with 5%–8% CO2 using Maestro Edge MEA System (Axion Biosystems) in absence or presence of retigabine (10 μM). The recordings were analyzed using AxIS Navigator 1.5 and Neural Metric Tool software.
Reverse transcription quantitative PCR
Total RNA from the organoids were extracted and reverse-transcribed to cDNA. The cDNA samples were amplified with specific primers (Table S2). Each individual sample was assayed in triplicate and normalized with GAPDH expression.
Western blot
Protein lysates from HBSOs were separated by SDS-PAGE and then transferred to PVDF membrane. After blocking, the membrane was incubated with primary antibodies, followed by HRP-conjugated secondary antibodies (Table S3). The HRP signals were detected using WesternBright ECL kit.
Immunostaining
Organoids samples were fixed with 4% paraformaldehyde, dehydrated, and embedded in O.C.T. Compound. After blocking, the frozen sections (10–20 μm) were incubated with primary antibodies, followed by the respective secondary antibodies (Table S3).
Droplet-based single-cell RNA-sequencing
Day-60 HBSOs and HCOs were dissociated into single-cell suspension. Viable cells negative for 7-AAD were sorted with BD FACSAria III Cell Sorter and then subjected to 10X Genomics’ scRNA-seq with Illumina NextSeq 500. The details of data analyses are available in the supplemental experimental procedures.
Image analysis and statistical analysis
All the fluorescence images were acquired by Carl Zeiss LSM800 or LSM900. Cell numbers were quantified using Fiji ImageJ software (Schindelin et al., 2012; Schneider et al., 2012). Statistical analysis was performed with the software Prism 9.0 (GraphPad). Unless otherwise noted, the data were expressed as mean ± SEM from at least three independent experiments. Two-tailed unpaired Student’s t test was used for two-group comparisons. One-way ANOVA, followed by Turkey’s multiple comparison test, was used for comparisons involving more than two groups. Probability values p < 0.05 were considered statistically significant.
Author contributions
K.N.C.L., S.T.L., and F.P.L.L. established hPSC lines and performed in vitro functional assays. Z.L. performed the bioinformatics analyses. K.N.C.L., Z.L., and E.S.W.N. prepared the manuscript. E.S.W.N. supervised the project.
Acknowledgments
The authors acknowledge the assistance of Center of PanorOmic Science (CPOS), Li Ka Shing Faculty of Medicine, The University of Hong Kong. The work described in this paper was substantially supported by the HMRF grants (Project nos. 06173306, 07181926, 08192786) from the Health Department, China Hong Kong and the General Research Fund (GRF: HKU 17108019, 17101320) from the Research Grant Council of Hong Kong Special Administrative Region, China Hong Kong. The study was approved by the Institutional Review Board of The University of Hong Kong and the Hospital Authority (UW 13–419).
Conflict of interests
The authors are filing a patent related to the method of generating HBSOs as described in this manuscript.
Published: June 22, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2023.05.020.
Supplemental information
Data and code availability
Raw sequencing data are available in the Sequence Read Archive at the NCBI Center with the accession number PRJNA891126. The processed scRNA-seq datasets are available at https://doi.org/10.5281/zenodo.7215153.
The detailed protocols are available in supplemental experimental procedures.
References
- Aibar S., Gonzalez-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel J., Laudier B., Attie-Bitach T., Trang H., de Pontual L., Gener B., Trochet D., Etchevers H., Ray P., Simonneau M., et al. Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat. Genet. 2003;33:459–461. doi: 10.1038/ng1130. [DOI] [PubMed] [Google Scholar]
- Bachetti T., Ceccherini I. Causative and common PHOX2B variants define a broad phenotypic spectrum. Clin. Genet. 2020;97:103–113. doi: 10.1111/cge.13633. [DOI] [PubMed] [Google Scholar]
- Butts J.C., Iyer N., White N., Thompson R., Sakiyama-Elbert S., McDevitt T.C. V2a interneuron differentiation from mouse and human pluripotent stem cells. Nat. Protoc. 2019;14:3033–3058. doi: 10.1038/s41596-019-0203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V., Ramanantsoa N., Trochet D., Vaubourg V., Amiel J., Gallego J., Brunet J.F., Goridis C. A human mutation in Phox2b causes lack of CO2 chemosensitivity, fatal central apnea, and specific loss of parafacial neurons. Proc. Natl. Acad. Sci. USA. 2008;105:1067–1072. doi: 10.1073/pnas.0709115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V., Thoby-Brisson M., Rallu M., Persson K., Pattyn A., Birchmeier C., Brunet J.F., Fortin G., Goridis C. Defective respiratory rhythmogenesis and loss of central chemosensitivity in Phox2b mutants targeting retrotrapezoid nucleus neurons. J. Neurosci. 2009;29:14836–14846. doi: 10.1523/JNEUROSCI.2623-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eura N., Matsui T.K., Luginbühl J., Matsubayashi M., Nanaura H., Shiota T., Kinugawa K., Iguchi N., Kiriyama T., Zheng C., et al. Brainstem organoids from human pluripotent stem cells. Front. Neurosci. 2020;14:538. doi: 10.3389/fnins.2020.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaufo G.O., Flodby P., Capecchi M.R. Hoxb1 controls effectors of sonic hedgehog and Mash1 signaling pathways. Development. 2000;127:5343–5354. doi: 10.1242/dev.127.24.5343. [DOI] [PubMed] [Google Scholar]
- Giandomenico S.L., Sutcliffe M., Lancaster M.A. Generation and long-term culture of advanced cerebral organoids for studying later stages of neural development. Nat. Protoc. 2021;16:579–602. doi: 10.1038/s41596-020-00433-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goridis C., Dubreuil V., Thoby-Brisson M., Fortin G., Brunet J.F. Phox2b, congenital central hypoventilation syndrome and the control of respiration. Semin. Cell Dev. Biol. 2010;21:814–822. doi: 10.1016/j.semcdb.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Guyenet P.G., Mulkey D.K., Stornetta R.L., Bayliss D.A. Regulation of ventral surface chemoreceptors by the central respiratory pattern generator. J. Neurosci. 2005;25:8938–8947. doi: 10.1523/JNEUROSCI.2415-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyenet P.G., Stornetta R.L., Souza G.M.P.R., Abbott S.B.G., Shi Y., Bayliss D.A. The retrotrapezoid nucleus: central chemoreceptor and regulator of breathing automaticity. Trends Neurosci. 2019;42:807–824. doi: 10.1016/j.tins.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk J.M., Moreira T.S., Takakura A.C., Wenker I.C., Tzingounis A.V., Mulkey D.K. KCNQ channels determine serotonergic modulation of ventral surface chemoreceptors and respiratory drive. J. Neurosci. 2012;32:16943–16952. doi: 10.1523/JNEUROSCI.3043-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra E.J., Von Oerthel L., Van Der Linden A.J.A., Smidt M.P. Phox2b influences the development of a caudal dopaminergic subset. PLoS One. 2012;7:e52118. doi: 10.1371/journal.pone.0052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanton S., Boyle M.J., He Z., Santel M., Weigert A., Sanchís-Calleja F., Guijarro P., Sidow L., Fleck J.S., Han D., et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Zhou X., Chen M. Demystifying "drop-outs" in single-cell UMI data. Genome Biol. 2020;21:196. doi: 10.1186/s13059-020-02096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G., Siletti K., Furlan A., Gyllborg D., Vinsland E., Mossi Albiach A., Mattsson Langseth C., Khven I., Lederer A.R., Dratva L.M., et al. Molecular architecture of the developing mouse brain. Nature. 2021;596:92–96. doi: 10.1038/s41586-021-03775-x. [DOI] [PubMed] [Google Scholar]
- Lai F.P.L., Lau S.T., Wong J.K.L., Gui H., Wang R.X., Zhou T., Lai W.H., Tse H.F., Tam P.K.H., Garcia-Barcelo M.M., Ngan E.S.W. Correction of hirschsprung-associated mutations in human induced pluripotent stem cells via clustered regularly interspaced short palindromic repeats/Cas9, restores neural crest cell function. Gastroenterology. 2017;153:139–153.e8. doi: 10.1053/j.gastro.2017.03.014. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Venugopal S., Majid S., Ahn I.S., Diamante G., Hong J., Yang X., Chandler S.H. Single-cell RNA-seq analysis of the brainstem of mutant SOD1 mice reveals perturbed cell types and pathways of amyotrophic lateral sclerosis. Neurobiol. Dis. 2020;141:104877. doi: 10.1016/j.nbd.2020.104877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malheiros-Lima M.R., Silva T.M., Takakura A.C., Moreira T.S. A5 noradrenergic-projecting C1 neurons activate sympathetic and breathing outputs in anaesthetized rats. Exp. Physiol. 2022;107:147–160. doi: 10.1113/EP089691. [DOI] [PubMed] [Google Scholar]
- Matera I., Bachetti T., Puppo F., Di Duca M., Morandi F., Casiraghi G.M., Cilio M.R., Hennekam R., Hofstra R., Schöber J.G., et al. PHOX2B mutations and polyalanine expansions correlate with the severity of the respiratory phenotype and associated symptoms in both congenital and late onset Central Hypoventilation syndrome. J. Med. Genet. 2004;41:373–380. doi: 10.1136/jmg.2003.015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira T.S., Takakura A.C., Czeisler C., Otero J.J. Respiratory and autonomic dysfunction in congenital central hypoventilation syndrome. J. Neurophysiol. 2016;116:742–752. doi: 10.1152/jn.00026.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A., Hirsch M., Goridis C., Brunet J.F. Control of hindbrain motor neuron differentiation by the homeobox gene Phox2b. Development. 2000;127:1349–1358. doi: 10.1242/dev.127.7.1349. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Okita K., Nakagawa M., Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- Tomycz N.D., Haynes R.L., Schmidt E.F., Ackerson K., Kinney H.C. Novel neuropathologic findings in the Haddad syndrome. Acta Neuropathol. 2010;119:261–269. doi: 10.1007/s00401-009-0599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A.J., Burdziak C., Moon K.R., Chaffer C.L., Pattabiraman D., et al. Recovering gene interactions from single-cell data using data diffusion. Cell. 2018;174:716–729.e27. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Cai X., Wang L., Liao B., Zhang H., Shan Y., Chen Q., Zhou T., Li X., Hou J., et al. Generating a non-integrating human induced pluripotent stem cell bank from urine-derived cells. PLoS One. 2013;8:e70573. doi: 10.1371/journal.pone.0070573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L.E., La Manno G., et al. Molecular architecture of the mouse nervous System. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data are available in the Sequence Read Archive at the NCBI Center with the accession number PRJNA891126. The processed scRNA-seq datasets are available at https://doi.org/10.5281/zenodo.7215153.
The detailed protocols are available in supplemental experimental procedures.