Abstract
Lung cancer as the leading cause of cancer related mortality is always one of the main global health challenges. Despite the recent progresses in therapeutic methods, the mortality rate is still significantly high among lung cancer patients. A wide range of therapeutic methods including chemotherapy, radiotherapy, and surgery are used to treat lung cancer. Doxorubicin (DOX) and Paclitaxel (TXL) are widely used as the first-line chemotherapeutic drugs in lung cancer. However, there is a significant high percentage of DOX/TXL resistance in lung cancer patients, which leads to tumor recurrence and metastasis. Considering, the side effects of these drugs in normal tissues, it is required to clarify the molecular mechanisms of DOX/TXL resistance to introduce the efficient prognostic and therapeutic markers in lung cancer. MicroRNAs (miRNAs) have key roles in regulation of different pathophysiological processes including cell division, apoptosis, migration, and drug resistance. MiRNA deregulations are widely associated with chemo resistance in various cancers. Therefore, considering the importance of miRNAs in chemotherapy response, in the present review, we discussed the role of miRNAs in regulation of DOX/TXL response in lung cancer patients. It has been reported that miRNAs mainly induced DOX/TXL sensitivity in lung tumor cells by the regulation of signaling pathways, autophagy, transcription factors, and apoptosis. This review can be an effective step in introducing miRNAs as the non-invasive prognostic markers to predict DOX/TXL response in lung cancer patients.
Keywords: Lung cancer, Paclitaxel, Doxorubicin, Chemo resistance, microRNA
Background
Lung cancer is the most frequent cancer and is responsible for the highest number of cancer-related deaths in the world [1]. Despite the recent progresses in molecular targeted therapies and surgical techniques, the overall 5-years survival rate is still around 15% for lung cancer patients [2]. Non-small cell lung cancer (NSCLC) is the most frequent lung tumor type accounting for 85% of all cases [1, 3]. Targeted drugs have efficient therapeutic benefits for NSCLC patients; however, drug resistance is a frequent challenge that is finally observed among a large proportion of NSCLC patients [4, 5]. Cisplatin, carboplatin, paclitaxel, docetaxel, gemcitabine, and pemetrexed are the frequently used chemotherapeutic options for NSCLC patients [6–8]. DNA-damaging factors are the most commonly used types of chemotherapeutic drugs [9, 10]. They prevent cell proliferation while induce cell death by the suppression of the double-strand breaks rejoining [11]. Microtubule targeting agents (MTAs) are also conventional chemotherapeutic drugs for NSCLC patients. They bind to microtubules at various sites to disrupt their dynamics and structure, resulting in cell cycle arrest and subsequent cell death [12, 13]. Doxorubicin is considered as an inhibitor of DNA synthesis and transcription by targeting topoisomerase II that results in cell cycle arrest and apoptosis [14, 15]. The combination of doxorubicin with other chemotherapeutic drugs is a standard therapeutic regimen for lung carcinoma [16]. Nevertheless, the emergence of drug resistance has impaired its efficacy as a therapeutic agent [17]. Doxorubicin as an anthracycline is frequently used to treat SCLC patients. Despite more than half of the patient’s response to drug, the median survival of SCLC patients is approximately 10–12 months in primary tumor stage [18–20]. Paclitaxel (PTX) is a critical therapeutic option for the advanced stage NSCLC [21]. It functions by binding to the β subunit of tubulin to inhibit the establishment of microtubules that results in cell cycle disruption and apoptosis [22]. However, the development of resistance to paclitaxel leads to treatment failure and reduced survival rates for patients. As the poor prognosis is associated with advanced stages and drug resistance in lung cancer, it is required to introduce the novel diagnostic and prognostic biomarkers to improve the therapeutic strategies in these patients. MiRNAs are small non-coding RNAs that are found in all eukaryotic cells and play a vital role in post-transcriptional inhibition of target mRNAs [23, 24]. MiRNAs have pivotal roles in lung tumor progression by regulation of various cellular processes such as cell proliferation, angiogenesis, and epithelial-mesenchymal transition (EMT) [25, 26]. They are implicated in drug resistance by affecting various cellular processes such as cell survival, apoptosis, angiogenesis, and migration [27]. MiRNAs deregulations are associated with chemo resistance in various cancers [28, 29]. Therefore, in the present review we discussed the role of miRNAs in DOX/TXL responses in lung tumor cells to introduce them as the probable non-invasive prognostic markers in lung cancer patients (Table 1). Web of Science, Embase, PubMed, Cochrane Library, and Google scholar were searched and assessed until the May 2023 without language limitations. The reference lists were also manually searched for the relevant publications including the review articles and original researches. The search strategy was based on “microRNA”, “Doxorubicin”, “Paclitaxel”, “Drug resistance”, and “Lung cancer” keywords.
Table 1.
Role of miRNAs in regulation of DOX and TXL responses in lung tumor cells
| miRNA | Target | Samples | Results | Clinical application | Study | Year |
|---|---|---|---|---|---|---|
| Signaling pathways | ||||||
| miR-7 | EGFR |
20 T 20N* A549, H1395, 95C and 95D cell lines |
Increased Paclitaxel sensitivity | Diagnosis | Liu [39] | 2014 |
| miR-4262 | PTEN |
20 T 20N A549, H1299, A549/PTX and H1299/PTX cell lines Xenograft model |
Increased Paclitaxel resistance | Diagnosis | Sun [42] | 2019 |
| miR-181a | PTEN | A549, A549/PTX, and A549/DDP cell lines | Increased Paclitaxel resistance | Diagnosis | Li [43] | 2015 |
| miR-4715-5p | RAC1 |
25 T 25N A549, Calu1, H1299, and HOP62 cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis | Yang [52] | 2019 |
| miR-9600 | STAT3 |
144 T 20N A549, SPC-A-1, H1299, SK-MES-1, NCI-H520, 95D and 16HBE cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis and prognosis | Sun [57] | 2016 |
| miR-1247-3p | STAT5A |
162 T 162N NCI-H1299, NCI-H1395, A549, NCL-H460, PG49, NCI-H1993 cell lines Xenograft model |
Increased Doxorubicin sensitivity | Diagnosis and prognosis | Lin [59] | 2022 |
| miR-337-3p | RAP1A | H1155, H1299, H1819, H1993, HCC2935, and HCC515 cell lines | Increased Paclitaxel sensitivity | Diagnosis | Du [67] | 2012 |
| miR-34c | NOTCH1 |
30 T 30N A549, H1299, and 293 T cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis | Yang [69] | 2020 |
| Transcription factors and DNA binding proteins | ||||||
| miR-138 | ZEB2 | A549, NCI-H23, A549/ADM and NCI-H23/ADM cell lines | Increased Doxorubicin sensitivity | Diagnosis | Jin [73] | 2016 |
| miR-194-5p | HIF-1 | H460 and A549 cell lines | Increased Doxorubicin sensitivity | Diagnosis | Xia [75] | 2021 |
| mR-608 | TFAP4 |
37 T 37N 96 T serum 136N serum A549 and HCC4006 cell lines |
Increased Doxorubicin sensitivity | Diagnosis | Wang [81] | 2019 |
| miR-935 | SOX7 |
30 T 30N A549 cell line |
Increased Paclitaxel resistance | Diagnosis | Peng [88] | 2018 |
| miR-30c | MTA1 | A549 and H460 cell lines | Increased Paclitaxel sensitivity | Diagnosis | Lu [97] | 2017 |
| miR- 137 | NUCKS1 |
50 T 50N A549, A549/PTX and A549/CDDP cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis and prognosis | Shen [104] | 2016 |
| Structural factors | ||||||
| miR-200c | CTSL | A549 and A549/TAX cell lines | Increased Paclitaxel sensitivity | Diagnosis | Zhao [114] | 2018 |
| miR-421 | KEAP1 |
129 T 129N 10 T serum 10N serum A549, H358, H1650, H460, and H1975 cell lines Xenograft model |
Increased Paclitaxel resistance | Diagnosis and prognosis | Duan [119] | 2019 |
| miR-223 | FBW7 | A549, NCI-H358, NCI-H1299 and HCC827 cell lines | Increased Doxorubicin resistance | Diagnosis | Li [126] | 2016 |
| miR-490-5p | UBE2T |
50 T (20R 30S) 50N H1299 and A549 cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis | Wang [129] | 2023 |
| miR-558 | MMP1/MMP17 |
46 T 46N A549, H1299, H358, and PC9 cell lines |
Increased Paclitaxel resistance | Diagnosis | Li [131] | 2021 |
| miR-197-3p | p120-ctn |
326 T 326N A549, H1299, H460 and SPC-A-1 cell lines Xenograft model |
Increased Paclitaxel and Doxorubicin sensitivity | Diagnosis and prognosis | Yang [137] | 2019 |
| miR-708-5p | COX-2/mPGES-1 | A549, A549-ER, and A549-PR cell lines | Increased Paclitaxel sensitivity | Diagnosis | Monteleone [143] | 2020 |
| miR-486-3p | CRABP2 |
65 T (30R 35S) 65N A549 and H1299 cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis | Wu [144] | 2022 |
| miR-526b-5p | GRK5 |
65 T 65N A549, H3122, H1975, and H2342 cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis | Liu [146] | 2021 |
| mR-299-3p | ABCE1 |
20 T 20N NCI-H69 cell line |
Increased Doxorubicin sensitivity | Diagnosis | Zheng [149] | 2015 |
| Apoptosis and DNA repair | ||||||
| miR-1273f | MDM2 |
20 T 20N A549 and A549/Taxol cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis and prognosis | Xu [153] | 2021 |
| miR-107 | Bcl-w |
A549 and HEK 293 T cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis | Lu [161] | 2017 |
| miR-30a-5p | BCL-2 |
94 T 94N A549, H460, A549/PR, and H460/PR cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis and prognosis | Xu [162] | 2017 |
| miR-7-5p | PARP1 | H69, H69AR, and H446AR | Increased Doxorubicin sensitivity | Diagnosis | Lai [167] | 2019 |
| miR-195 | CHEK1 |
57 T 57N H1155, H1993 and H358 cell lines Xenograft model |
Increased Paclitaxel sensitivity | Diagnosis and prognosis | Yu [169] | 2018 |
| miR-433-3p | CHEK1 |
41 T 41N A549, H1299, A549/PTX and H1299/PTX cell lines |
Increased Paclitaxel sensitivity | Diagnosis | Jin [170] | 2022 |
| Autophagy and drug efflux | ||||||
| miR-17-5p | Beclin1 | A549, H596, A549-T24, and H596-TxR cell lines | Increased Paclitaxel sensitivity | Diagnosis | Chatterjee [180] | 2014 |
| miR-199a-5p | ATG5 | A549, H1299, H661, H522, H1944, and A549/T cell lines | Increased Paclitaxel resistance | Diagnosis | Zeng [183] | 2021 |
| miR-155 | AKT/ERK | A549 and A549/dox cell lines | Increased Doxorubicin resistance | Diagnosis | Lv [189] | 2016 |
*Tumor (T) and normal (N) tissues
Signaling pathways
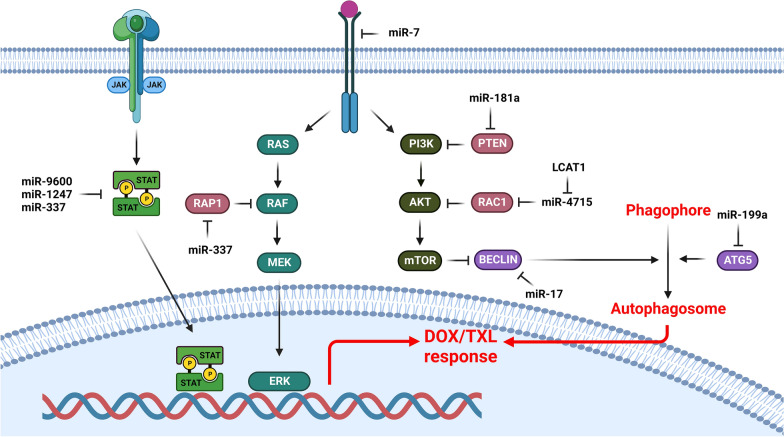
MiRNAs are involved in DOX/TXL response of lung tumor cells via the regulation of signaling pathways (Fig. 1). PI3K/AKT is one of the main oncogenic signaling pathways that is directly associated with the extracellular growth factors. It is mainly triggered by the activation of receptor tyrosine kinases (RTKs) that subsequently activates PI3K/AKT/mTOR axis [30]. EGFR belongs to the RTK protein family that has a key role in cell proliferation by activation of PI3K/AKT and MAPK signaling pathways. MiR-7 attenuated NSCLC progression via targeting several oncogenes, such as PAK1, EGFR, RAF1, IRS1, and IRS2 that resulted in inhibition of the EGFR/AKT axis [31–34]. Activation of EGFR downstream pathways, including STAT, PI3K/AKT, and MAKP intensifies the chemo resistance of tumor cells [32, 34–38]. It has been reported that miR-7 increased the PTX sensitivity via EGFR targeting in NSCLC cells [39]. PTEN is a negative regulator of the PI3K/Akt axis and is frequently down regulated or mutated in lung cancer [40, 41]. MiR-4262 promoted PTX resistance through PTEN targeting and subsequent PI3K/AKT activation in NSCLC cells [42]. MiRNA-181a has been identified as a contributor to the acquisition of EMT, as well as increased invasion and migration in lung adenocarcinoma cells through PTEN targeting. MiR-181a also increased the sensitivity of cancer cells to paclitaxel treatment [43]. Reactive oxygen species (ROS) is involved in VEGF induced activation of the PI3K/AKT axis [44, 45]. Rac1 belongs to the Rho protein family that regulates growth factors and cytokines [46]. P21-activated kinase (PAK1) is a ser/thr kinase that interacts with Rac1 and Cdc42 [47]. EGF promotes tumor cell migration by Rac1 mediated activation of PI3K/Akt and PAK1 [48]. Long noncoding RNAs (lncRNAs) are a class of non-coding RNAs that have pivotal roles in regulation of cell growth, angiogenesis, survival, and motility [49–51]. The significant up regulation of LCAT1 has been reported in lung cancer tissues that were associated with unfavorable prognosis. LCAT1 enhanced the lung tumor growth through the miR-4715-5p/RAC1 axis. The reduction of RAC1 activity hindered the cell proliferation and mobility and its function was regulated by PAK1. Both RAC1 and PAK1 were found to be reduced in cells with elevated levels of miR-4715-5p and in cells where LCAT1 was silenced. EHop-016 as a Rac GTPase inhibitor reduced the viability of lung tumor cells. The efficacy of EHop-016 and paclitaxel in treating lung cancer cells was improved when they were used in combination. EHop-016 as an adjuvant therapy enhanced the paclitaxel efficacy in lung cancer patients who had LCAT1 up regulation [52].
Fig. 1.
Role of miRNAs in DOX/TXL responses via regulation of signaling pathways and autophagy in lung tumor cells. (Created with BioRender.com)
JAK/STAT pathway has a critical role in cell proliferation, inflammation, and apoptosis. IL-6 activates the JAK2 that promotes the STAT3 dimerization and nuclear transportation to regulate the JAK/STAT target genes [53]. STAT3 is a key regulator of cancer-related inflammation and tumor progression [54]. It promotes tumor cell growth, invasion, immunosuppression, angiogenesis, and drug resistance [55]. STAT3 also promotes tumorigenesis by inhibiting cell death via Bcl-xL and Bcl-2 up regulations [56]. MiR-9600 enhanced paclitaxel sensitivity of NSCLC through targeting STAT3 that resulted in CDK2, CCND1, cyclin E, and p-RB down regulations [57]. STAT5A is a transcription factor that participates in cell proliferation, migration, and aggressiveness [58]. MiR-1247-3p has been reported to be down regulated in lung adenocarcinoma tissues that were associated with advanced stages and metastatic tumors. It suppressed Doxorubicin resistance in lung tumor cells via STAT5A targeting [59]. RAP1A as one of the RAP1 isoforms is involved in regulation of microtubule dynamics. RAP1 triggers the MAPK/ERK axis and phosphorylates microtubule-associated proteins such as MAP2 and MAP4 [60–64]. It can also regulate the paclitaxel sensitivity of tumor cells via extracellular matrix and cell interactions [65, 66]. MiR-337-3p increased the paclitaxel sensitivity of lung tumor cells via STAT3 and RAP1A targeting. STAT3 antagonized microtubule depolymerization by binding to stathmin, while RAP1A suppressed microtubule polymerization via triggering ERK/MAPK and MAP2 and MAP4 phosphorylations. Depletion of RAP1A or STAT3 disrupted normal microtubule dynamics that sensitized tumor cells toward the microtubule-targeting agents. Therefore, paclitaxel treatment and RAP1A/STAT3 down regulation synergistically disrupted microtubule function, resulting in G2/M arrest and cell death [67].
NOTCH is a developmental signaling pathway that has critical roles in embryogenesis and tumor progression. It can be triggered by activation of NOTCH receptors that releases the NICD into the cytoplasm. Subsequently, NICD enters into the nucleus and regulate the NOTCH target genes by MAML/CSL transcriptional machinery [68]. There was significant miR-34c down regulation in NSCLC tissues. MiR-34c sensitized the NSCLC cells to paclitaxel and cisplatin through the NOTCH1 targeting [69].
Transcription factors and DNA binding proteins
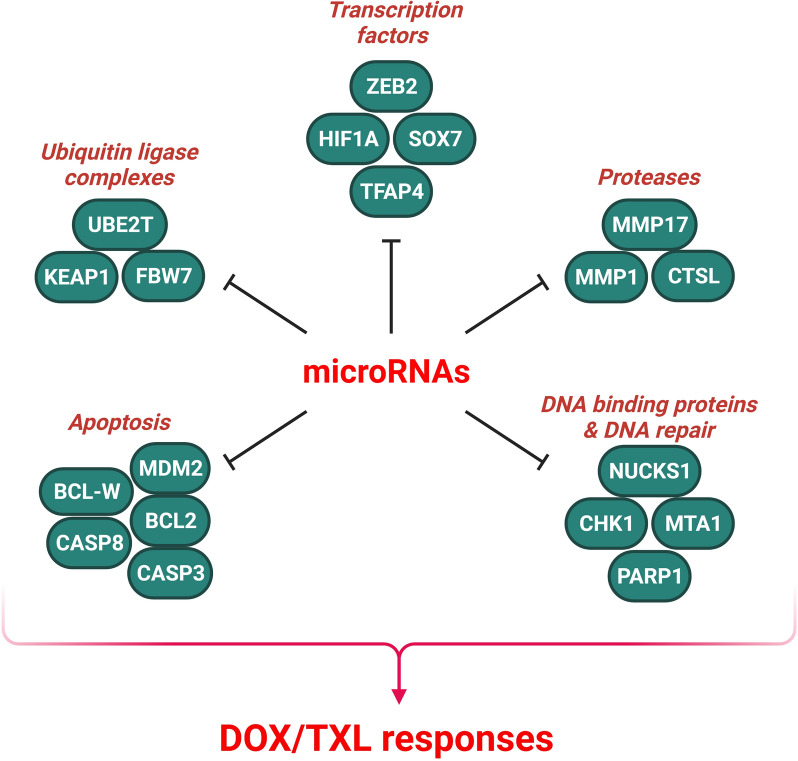
MiRNAs are involved in DOX/TXL response of lung tumor cells via the regulation of transcription factors and DNA binding proteins (Fig. 2). EMT has a key role in NSCLC progression and chemotherapy response, by which tumor cells lose their epithelial features and acquire a mesenchymal and aggressive phenotype [70]. ZEB2 belongs to the zinc finger homeobox protein family that regulates the tumor progression and chemotherapy response [71]. ZEB2 suppresses the CDH1 to promote tumor cell invasion and chemo resistance. However, the inhibition of ZEB2 by several miRNAs can effectively reverse this effect and lead to the suppression chemo resistance [72]. There was miR-138 down regulation in chemo resistant NSCLC cells. MiR-138 up regulated the E-cadherin while down regulated the Vimentin to sensitize NSCLC cells to DOX via ZEB2 targeting [73]. HIF1A as a basic helix-loop-helix protein is the master regulator of hypoxia response that mediates drug resistance via up regulation of P-glycoprotein (P-gp) [74]. There was significant down regulation of miR-194-5p in yypoxia-induced DOX-resistant NSCLC cells. MiR-194-5p directly targeted HIF-1, which subsequently impaired the expression of downstream proteins, such as P-gp, to enhance the sensitivity of NSCLC cells to DOX. In addition, miR-194-5p regulated the expression of several apoptotic proteins such as PARP and BAX that increased DOX-mediated apoptosis of NSCLC cells [75]. TFAP4 is a transcription factor that is involved in progression of various human cancers [76–79]. It promotes tumor cell proliferation and metastasis, while represses the cell death [79, 80]. It has been shown to activate the Wnt/β-catenin pathway to enhance hepatocellular carcinoma progression [80]. There was miR-608 down regulation in NSCLC samples. MiR-608 facilitated doxorubicin mediated apoptosis in NSCLC cells by targeting TFAP4 [81]. SOX7 is a transcription factor that regulates the cell differentiation, proliferation, migration, and apoptosis and acts as a tumor suppressor in different cancers [82, 83]. In lung cancer, reduced expression of SOX7 is associated with an unfavorable prognosis [84]. Additionally, SOX7 physically interacts with β-catenin and transcription factor 4 to inhibit the Wnt pathway and stemness [85]. Down regulation of SOX7 promotes tumor cell stemness and chemo-resistance [86]. PI3K/Akt axis is a key regulator of cell migration, growth, death, and blood vessel formation [87]. MiR-935 silencing increased paclitaxel mediated apoptosis in NSCLC cells by SOX7 targeting. This intervention down regulated Bcl-2 and p-AKT while up regulated Bax [88]. MTA1 is a member of chromatin remodeling complexes that has key roles in nucleosome remodeling and transcriptional regulation [89]. Curcumin inhibits the tumor cell growth while promotes the programmed cell death [90–92]. It has been shown that Curcumin functions as an anti-tumor drug by modulating signaling pathways, transcription factors, and miRNAs [93–96]. According to a recent investigation, Curcumin enhanced the response of NSCLC cells to Paclitaxel by MTA1 down regulation following the miR-30c-5p up regulation [97]. NUCKS1 is a DNA-binding protein that is a nuclear substrate for DNA-activated Kinase, CDK1, and CK2 [98–101]. It has a key role in regulation of cell cycle progression and transcription during rapid cell growth [102, 103]. MiR-137 promoted PTX sensitivity through NUCKS1 targeting in lung tumor cells [104].
Fig. 2.
Role of miRNAs in DOX/TXL responses via regulation of transcription factors, ubiquitination, proteases, and DNA repair in lung tumor cells. (Created with BioRender.com)
Structural factors
Cathepsin L (CTSL) belongs to the papain-like cysteine protease family that is associated with the tumor progression [105–107]. It has a crucial role in the various activities of tumor cells, including cell proliferation, migration, viability, invasion, and drug resistance [108–110]. CTSL as an EMT regulator alters the aggressiveness and migration of tumor cells [111]. CTSL also affects drug resistance via EMT-associated transcription factors, such as ZEB1, ZEB2, Slug, and Snail [112]. EMT is regulated by several transcription factors such as Twist, ZEB1, ZEB2, and Snail/Slug [113]. MiRNA-200c suppression reduced paclitaxel sensitivity in lung tumor cells via the up regulation of EMT-related transcription factors. MiRNA-200c inhibited EMT and subsequently improved the response to paclitaxel in lung tumor cells through CTSL targeting [114].
MiRNAs are involved in DOX/TXL response of lung tumor cells via the regulation of protein ubiquitination (Fig. 2). ROS is implicated in both targeted-therapy resistance and chemical resistance that introduce the redox pathway as a reliable tumor therapeutic target [21–25]. KEAP1 acts as an adaptor for substrates by attaching to the CuI3-containing E3 ubiquitin ligase and destructing them through the proteasome pathway [115]. KEAP1 is modified due to ROS-induced oxidative stress, which releases Nrf2 from the KEAP1-Cul3 E3 ligase complex [116]. Subsequently, Nrf2 moves into the nucleus and binds to the antioxidant response element along with a small-Maf binding partner [117, 118]. KEAP1 down regulation was contributed to paclitaxel resistance in NSCLC through the up regulation of miR-421. β-catenin mediated transcription also up regulated the miR-421 [119]. EMT is a multifaceted and reversible process that induces a mesenchymal morphology while reduces the epithelial cell adhesion [120]. FBW7 functions as the substrate recognition component in SCF E3 ligase complex [121]. FBW7 modulates several oncoproteins, including c-Myc, c-Jun, Notch, and CCNE1 [122, 123]. F-box proteins are involved in EMT by modulating inducers and transcription factors [124, 125]. The miR-223/FBW7 axis has been found to enhance doxorubicin sensitivity by regulating EMT in NSCLC cells. Doxorubicin treatment induced EMT in NSCLC cells, but knockdown of Twist hindered this transition through CDH1 up regulation and Vimentin down regulation. Moreover, hypoxia-induced EMT and increased resistance to doxorubicin was accompanied by the reduced levels of FBW7 and E-cadherin while increased Vimentin expression [126]. UBE2T functions as a E2 ubiquitin-conjugating enzyme that catalyzes the ubiquitination of FANCD2 in DNA damage response [127]. Circular RNAs (circRNAs) are a type of non-coding RNA with a stable covalently closed-loop structure [128]. Circ_0092887 inhibiting decreased cell growth and migration, while increased apoptosis in NSCLC cells treated with PTX. Circ_0092887 regulated the PTX resistance via miR-490-5p/UBE2T axis in NSCLC [129]. Matrix metalloproteinases (MMPs) are the key enzymes that break down extracellular matrix (ECM) and collagen to promote tumor angiogenesis and metastasis [130]. Circ_0030998 reduced Taxol resistance by miR-558/MMP1 and MMP17 axes in lung tumor cells [131]. The p120-catenin (p120-ctn) interacts with EMT marker E-cadherin to enhance the lung tumor cell proliferation [132]. It has a pivotal role in modification of the intercellular adhesion and EMT process by interaction with E-cadherin [133–135]. It also bound to cellular structures such as microtubules and cytocentrum to suppress the cell proliferation [136]. There was MALAT1 up regulation in NSCLC tissues that was correlated with poor survival. MALAT1 was associated with resistance to chemotherapeutic drugs such as TXL, gefitinib, DOX, and CDDP. It promoted the cell growth and survival while induced the EMT process in NSCLC cells via miR-197-3p/p120-ctn pathway [137]. ITGB8 is a fibronectin receptor that is involved in cell-cell interactions. There were circDNER up regulations in tumor tissues and plasma exosomes of lung cancer patients. It also promoted the paclitaxel resistance through the miR-139-5p sponging and subsequent ITGB8 up regulation in lung tumor cells [138].
Arachidonic Acid (AA) pathway regulates the cell proliferation, immunity, and homeostasis [139]. COX-1 or COX-2 convert free cytosolic AA to PGH2 [140]. PGE2 has a critical oncogenic role via activation of PI3K/AKT, MAPK, β-catenin, and NF-kB signaling pathways [141, 142]. CHOP is a member of the C/EBP transcription factors involved in adipogenesis and erythropoiesis. Chemotherapy up regulated miR-708-5p while down regulated the AA pathway in lung tumor cells. CHOP and p53 were the transcription factors involved in regulation of chemotherapeutic-mediated miR-708-5p expression. MiR-708-5p also up regulated the p53 and CHOP via a positive feedback loop. There was COX-2 up regulation while miR-708-5p down regulation in paclitaxel resistant lung tumor cells. MiR-708-5p played a tumor suppressive role by COX-2, mPGES-1, and Survivin targeting that resulted in immune evasion [143]. CRABP2 is a retinoic acid binding protein that functions as a cytosol-to-nuclear shuttle to facilitate RA nucleus transfer. Circ_0011298 promoted Taxol resistance via miR-486-3p/CRABP2 axis in NSCLC cells [144].
G protein-coupled receptor kinase 5 (GRK5) belongs to the serine/threonine kinase protein family that is involved in sensing various internal stimuli and regulation of the subsequent signaling pathways [145]. There was circ_0001821 up regulation in NSCLC tissues that was correlated with poor prognosis. Circ_0001821 blocking inhibited the TAX resistance, colony formation, and tumor proliferation via miR-526b-5p/GRK5 axis in NSCLC cells [146]. ABCE1 is a protein that belongs to the ATP-binding cassette (ABC) family and suppresses the RNase L as and interferon-induced nuclease in mammalian cells. ABCE1 is a potential tumor suppressor that is involved in regulation of cell proliferation and apoptosis [147, 148]. It has been indicated that miRNA-299-3p enhanced the doxorubicin-sensitivity in lung cancer via targeting ABCE1. There was miR-299-3p down regulation in doxorubicin-resistant lung tumor tissues compared with the sensitive tissues [149].
Apoptosis and DNA repair
Tumor cells develop paclitaxel resistance through the various processes such as increased DNA repair, cell cycle regulation, and anti-apoptotic pathways [150–152]. MiRNAs are involved in DOX/TXL response of lung tumor cells via the regulation of apoptosis (Fig. 2). Mouse double minute 2 homolog (MDM2) is an E3 ubiquitin ligase that has a key role in p53 inhibition. There was circ_0002874 up regulation in NSCLC tissues that was correlated with higher stages. Although, there was MDM2 down regulation in NSCLC tissues compared with normal counterparts, increased expression of MDM2 was associated with TNM staging. Circ_0002874 induced paclitaxel resistance by miR-1273f/MDM2 axis in NSCLC cells [153]. Bcl-w belongs to the BCL2 family that blocks apoptosis and promotes cell proliferation [154, 155]. Bcl-w enhances tumor progression by targeting pro-apoptotic factors such as Bax and Bak [156, 157]. Bcl-w deregulation is significantly associated with various types of cancers [158–160]. MiR-107 down regulation was associated with paclitaxel resistance in NSCLC. MiR-107 reduced the levels of p-Akt and p-GSK3β, which were restored by Bcl-w. MiR-107/Bcl-w axis regulated paclitaxel resistance via the PI3K-Akt pathway. MiR-107 increased paclitaxel sensitivity by regulation of Bcl-w expression and PI3K/Akt pathway in NSCLC cells [161]. MiR-30a-5p increased the sensitivity of NSCLC cells to paclitaxel by suppressing BCL-2 and promoting apoptosis. There was a correlation between the miR-30a-5p up regulation and a positive response to paclitaxel treatment in NSCLC patients [162].
MiRNAs are involved in DOX/TXL response of lung tumor cells via the regulation of DNA repair factors (Fig. 2). PARP1 has a key role in DNA repair and gene transcription [163, 164]. DNA damage activates PARP1, which polymerizes ADP-ribose units to recruit the DNA repair proteins in DNA damage location [165]. Homologous recombination (HR) is essential to preserve the genomic stability and chemotherapy response that can be regulated by PARP1 [166]. It has been shown that SCLC cells utilized the miR-7-5p-mediated HR repair by PARP1 targeting to increase the doxorubicin resistance. MiR-7-5p down regulated the BRCA1 and Rad51 in DOX-resistant SCLC cells via PARP1 targeting [167]. Checkpoint kinase 1 (CHK1) is a ser/thr kinase that is involved in regulation of DNA damage and cell cycle response [168]. It promotes cell cycle arrest, DNA repair, and apoptosis. MiR-195 sensitized NSCLC cells to paclitaxel and targeted CHEK1 to modulate the effectiveness of MTAs [169]. Circ_0011292 was up regulated in PTX-resistant NSCLC cells. Depletion of circ_0011292 increased the sensitivity to PTX, suppressed cell growth, aggressiveness, and migration, while induced apoptosis in PTX-resistant NSCLC cells. Circ_0011292 was also contributed to PTX resistance via targeting the miR-433-3p/CHEK1 axis [170].
Autophagy and drug efflux
Multidrug resistance (MDR) is the ability of tumor cell to resist against the chemotherapy drugs [171]. MDR is acquired through several mechanisms, such as up regulation of ABC transports, inhibition of apoptosis, hypoxia, autophagy, DNA repair, miRNA regulation, and epigenetic changes [172]. Autophagy is a defensive mechanism in tumor cells toward the chemotherapeutic treatment. Chemotherapy mediated autophagy supports the tumor cell metabolism through the recycling of damaged organelles and proteins to prevent DNA damage [173, 174]. Autophagy breaks down damaged cellular components using a lysosomal degradation pathway [175, 176]. This process improves the tumor cell resistance toward apoptosis, hypoxia, and other stress responses, which is essential for MDR [177, 178]. MiRNAs are involved in DOX/TXL response of lung tumor cells via the regulation of autophagy (Fig. 1). Beclin1 is one of the components of autophagy process that facilitates the autophagosomal membrane formation [179]. MiR-17-5p was down regulated in paclitaxel-resistant lung cancer cells, and its up regulation enhanced the paclitaxel response. Inhibition of miR-17-5p ameliorated Beclin1 levels and autophagy, which protected cells against paclitaxel-induced apoptosis. MiR-17-5p-mediated autophagy and paclitaxel treatment also triggered ROS and induced apoptosis in A549-T24 cells [180]. Autophagy-related (ATG) proteins as the main components of the autophagy process are involved in regulation of the autophagy initiation, autophagosomal maturation, lysosomal fusion, and autophagolyosomal degradation [181]. There was LINC01296 up regulation in NSCLC samples. LINC01296 promoted the NSCLC progression and paclitaxel resistance through miR-143-3p/ATG2B axis [182]. MiR-199a-5p inhibited autophagy in MDR lung tumor cells by activating the PI3K/Akt/mTOR axis, eEF2K expression, and decreasing ATG5 expression. There was miR-199a-5p up regulation in PTX resistant lung tumor cells [183]. Tumor cells can develop chemo resistance by decreasing drug absorption and facilitating drug efflux [184]. MRP1, MDR1, and BCRP belong to the ABC protein family involved in drug efflux [185]. GST-π reduces drug toxicity by binding to the hydrophobic and electrophilic compounds via glutathione reduction that result in chemo resistance [186–188]. Inhibition of miR-155 down regulated the MRP1, MDR1, GST-π, and BCRP in A549/Dox cells. MiR-155 repressing also down regulated Bcl-2 and Survivin, while up regulated CASP8 and CASP3 that enhanced apoptosis in lung tumor cells. MiR-155 inhibition also reduced AKT and ERK phosphorylation to inhibit PI3K/AKT and MAPK signaling pathways that reversed DOX resistance in lung tumor cells [189].
Conclusions
Doxorubicin and Paclitaxel are widely used as the first line chemotherapeutic drugs in lung cancer patients. However, a significant percentage of patients show resistance to these drugs. Therefore, considering the DOX/TXL side effects in normal body tissues, it is required to introduce the novel prognostic markers to predict the Doxorubicin and Paclitaxel responses in lung cancer. The present review is an effective step towards introducing miRNAs as the non-invasive markers to predict DOX/TXL response in lung cancer which improves the therapeutic strategies to prolong the survival rates in these patients. However, the introduction of miRNAs as the non-invasive prognostic markers in lung cancer patients requires more clinical studies. In this context, it is required to assess the circulating levels of miRNAs in body fluids to clinically use them as the non-invasive markers in screening programs among lung cancer patients and healthy people with a positive familial history. Considering that the miRNAs mainly promote the sensitivity of lung tumor cells to Paclitaxel and Doxorubicin, microRNA mimics strategies can have the promising therapeutic effects in these patients. However, more animal studies and clinical trials are needed to be able to clinically use the microRNA mimics to treat the DOX/TXL-resistant lung cancer patients.
Acknowledgements
None.
Abbreviations
- AA
Arachidonic acid
- ATG
Autophagy-related
- CTSL
Cathepsin L
- CHK1
Checkpoint kinase 1
- circRNAs
Circular RNAs
- DOX
Doxorubicin
- EMT
Epithelial-mesenchymal transition
- ECM
Extracellular matrix
- GRK5
G protein-coupled receptor kinase 5
- HR
Homologous recombination
- lncRNAs
Long noncoding RNAs
- MMPs
Matrix metalloproteinases
- miRNAs
MicroRNAs
- MTAs
Microtubule targeting agents
- MDM2
Mouse double minute 2 homolog
- MDR
Multidrug resistance
- NSCLC
Non-small cell lung cancer
- PAK1
P21-activated kinase
- TXL
Paclitaxel
- P-gp
P-glycoprotein
- ROS
Reactive oxygen species
- RTKs
Receptor tyrosine kinases
Author contributions
AM was involved in search strategy and drafting. MM designed, revised, structured, and edited the manuscript. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Martin P, Leighl NB. Review of the use of pretest probability for molecular testing in non-small cell lung cancer and overview of new mutations that may affect clinical practice. Therapeutic Adv Med Oncol. 2017;9(6):405–14. doi: 10.1177/1758834017704329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reck M, Rabe KF. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017;377(9):849–61. doi: 10.1056/NEJMra1703413. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16(9):e447–e59. doi: 10.1016/S1470-2045(15)00246-6. [DOI] [PubMed] [Google Scholar]
- 6.Rowinsky EK, Donehower RC. Paclitaxel (taxol) N Engl J Med. 1995;332(15):1004–14. doi: 10.1056/NEJM199504133321507. [DOI] [PubMed] [Google Scholar]
- 7.Gelmon K. The taxoids: paclitaxel and docetaxel. Lancet (London England) 1994;344(8932):1267–72. doi: 10.1016/S0140-6736(94)90754-4. [DOI] [PubMed] [Google Scholar]
- 8.Maharati A, Zanguei AS, Khalili-Tanha G, Moghbeli M. MicroRNAs as the critical regulators of tyrosine kinase inhibitors resistance in lung tumor cells. Cell Commun Signal. 2022;20(1):27. doi: 10.1186/s12964-022-00840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung-Ong K, Giaever G, Nislow C. DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem Biol. 2013;20(5):648–59. doi: 10.1016/j.chembiol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Zangouei AS, Moghbeli M. MicroRNAs as the critical regulators of cisplatin resistance in gastric tumor cells. Genes Environ. 2021;43(1):21. doi: 10.1186/s41021-021-00192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moyal L, Goldfeiz N, Gorovitz B, Rephaeli A, Tal E, Tarasenko N, et al. AN-7, a butyric acid prodrug, sensitizes cutaneous T-cell lymphoma cell lines to doxorubicin via inhibition of DNA double strand breaks repair. Investig New Drugs. 2018;36(1):1–9. doi: 10.1007/s10637-017-0500-x. [DOI] [PubMed] [Google Scholar]
- 12.Yvon AM, Wadsworth P, Jordan MA. Taxol suppresses dynamics of individual microtubules in living human tumor cells. Mol Biol Cell. 1999;10(4):947–59. doi: 10.1091/mbc.10.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: mitotic catastrophe. Cell Death Differ. 2008;15(7):1153–62. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- 14.Müller I, Niethammer D, Bruchelt G. Anthracycline-derived chemotherapeutics in apoptosis and free radical cytotoxicity (review) Int J Mol Med. 1998;1(2):491–4. doi: 10.3892/ijmm.1.2.491. [DOI] [PubMed] [Google Scholar]
- 15.Khalili-Tanha G, Moghbeli M. Long non-coding RNAs as the critical regulators of doxorubicin resistance in tumor cells. Cell Mol Biol Lett. 2021;26(1):39. doi: 10.1186/s11658-021-00282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu T, Li LL, Xiao GF, Luo QZ, Liu QZ, Yao KT, et al. Berberine increases Doxorubicin Sensitivity by suppressing STAT3 in Lung Cancer. Am J Chin Med. 2015;43(7):1487–502. doi: 10.1142/S0192415X15500846. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Feng C, He Y, Ding W, Sheng J, Arshad M, et al. Phosphorylation of apoptosis repressor with caspase recruitment domain by protein kinase CK2 contributes to chemotherapy resistance by inhibiting doxorubicin induced apoptosis. Oncotarget. 2015;6(29):27700–13. doi: 10.18632/oncotarget.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackman DM, Johnson BE. Small-cell lung cancer. Lancet (London England) 2005;366(9494):1385–96. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- 19.Schmittel A. Second-line therapy for small-cell lung cancer. Expert Rev Anticancer Ther. 2011;11(4):631–7. doi: 10.1586/era.11.7. [DOI] [PubMed] [Google Scholar]
- 20.Lally BE, Urbanic JJ, Blackstock AW, Miller AA, Perry MC. Small cell lung cancer: have we made any progress over the last 25 years? Oncologist. 2007;12(9):1096–104. doi: 10.1634/theoncologist.12-9-1096. [DOI] [PubMed] [Google Scholar]
- 21.Teow HM, Zhou Z, Najlah M, Yusof SR, Abbott NJ, D’Emanuele A. Delivery of paclitaxel across cellular barriers using a dendrimer-based nanocarrier. Int J Pharm. 2013;441(1–2):701–11. doi: 10.1016/j.ijpharm.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277(5698):665–7. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 23.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Moghbeli M. Molecular interactions of miR-338 during tumor progression and metastasis. Cell Mol Biol Lett. 2021;26(1):13. doi: 10.1186/s11658-021-00257-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doghish AS, Ismail A, Elrebehy MA, Elbadry AMM, Mahmoud HH, Farouk SM, et al. A study of miRNAs as cornerstone in lung cancer pathogenesis and therapeutic resistance: a focus on signaling pathways interplay. Pathol Res Pract. 2022;237:154053. doi: 10.1016/j.prp.2022.154053. [DOI] [PubMed] [Google Scholar]
- 26.Elkady MA, Doghish AS, Elshafei A, Elshafey MM. MicroRNA-567 inhibits cell proliferation and induces cell apoptosis in A549 NSCLC cells by regulating cyclin-dependent kinase 8. Saudi J Biol Sci. 2021;28(4):2581–90. doi: 10.1016/j.sjbs.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamidi AA, Zangoue M, Kashani D, Zangouei AS, Rahimi HR, Abbaszadegan MR, et al. MicroRNA-217: a therapeutic and diagnostic tumor marker. Expert Rev Mol Diagn. 2022;22(1):61–76. doi: 10.1080/14737159.2022.2017284. [DOI] [PubMed] [Google Scholar]
- 28.Fan Z, Cui H, Yu H, Ji Q, Kang L, Han B, et al. MiR-125a promotes paclitaxel sensitivity in cervical cancer through altering STAT3 expression. Oncogenesis. 2016;5(2):e197. doi: 10.1038/oncsis.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zangouei AS, Rahimi HR, Mojarrad M, Moghbeli M. Non coding RNAs as the critical factors in chemo resistance of bladder tumor cells. Diagn Pathol. 2020;15(1):136. doi: 10.1186/s13000-020-01054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navaei ZN, Khalili-Tanha G, Zangouei AS, Abbaszadegan MR, Moghbeli M. PI3K/AKT signaling pathway as a critical regulator of cisplatin response in tumor cells. Oncol Res. 2021;29(4):235–50. doi: 10.32604/or.2022.025323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rai K, Takigawa N, Ito S, Kashihara H, Ichihara E, Yasuda T, et al. Liposomal delivery of MicroRNA-7-expressing plasmid overcomes epidermal growth factor receptor tyrosine kinase inhibitor-resistance in lung cancer cells. Mol Cancer Ther. 2011;10(9):1720–7. doi: 10.1158/1535-7163.MCT-11-0220. [DOI] [PubMed] [Google Scholar]
- 32.Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, et al. microRNA-7 inhibits the epidermal growth factor receptor and the akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68(10):3566–72. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 33.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284(9):5731–41. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 34.Chou YT, Lin HH, Lien YC, Wang YH, Hong CF, Kao YR, et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010;70(21):8822–31. doi: 10.1158/0008-5472.CAN-10-0638. [DOI] [PubMed] [Google Scholar]
- 35.Petitprez A, Larsen AK. Irinotecan resistance is accompanied by upregulation of EGFR and src signaling in human cancer models. Curr Pharm Design. 2013;19(5):958–64. doi: 10.2174/138161213804547204. [DOI] [PubMed] [Google Scholar]
- 36.Rekhtman N, Paik PK, Arcila ME, Tafe LJ, Oxnard GR, Moreira AL, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res. 2012;18(4):1167–76. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, Gini B, et al. Oncogenic EGFR signaling activates an mTORC2-NF-κB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1(6):524–38. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee KM, Choi EJ, Kim IA. microRNA-7 increases radiosensitivity of human cancer cells with activated EGFR-associated signaling. Radiother Oncol. 2011;101(1):171–6. doi: 10.1016/j.radonc.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 39.Liu R, Liu X, Zheng Y, Gu J, Xiong S, Jiang P, et al. MicroRNA-7 sensitizes non-small cell lung cancer cells to paclitaxel. Oncol Lett. 2014;8(5):2193–200. doi: 10.3892/ol.2014.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowen KA, Doan HQ, Zhou BP, Wang Q, Zhou Y, Rychahou PG, et al. PTEN loss induces epithelial–mesenchymal transition in human colon cancer cells. Anticancer Res. 2009;29(11):4439–49. [PMC free article] [PubMed] [Google Scholar]
- 41.Soria JC, Lee HY, Lee JI, Wang L, Issa JP, Kemp BL, et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res. 2002;8(5):1178–84. [PubMed] [Google Scholar]
- 42.Sun H, Zhou X, Bao Y, Xiong G, Cui Y, Zhou H. Involvement of miR-4262 in paclitaxel resistance through the regulation of PTEN in non-small cell lung cancer. Open Biol. 2019;9(7):180227. doi: 10.1098/rsob.180227. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Li H, Zhang P, Sun X, Sun Y, Shi C, Liu H, et al. MicroRNA-181a regulates epithelial-mesenchymal transition by targeting PTEN in drug-resistant lung adenocarcinoma cells. Int J Oncol. 2015;47(4):1379–92. doi: 10.3892/ijo.2015.3144. [DOI] [PubMed] [Google Scholar]
- 44.Liu LZ, Hu XW, Xia C, He J, Zhou Q, Shi X, et al. Reactive oxygen species regulate epidermal growth factor-induced vascular endothelial growth factor and hypoxia-inducible factor-1alpha expression through activation of AKT and P70S6K1 in human ovarian cancer cells. Free Radic Biol Med. 2006;41(10):1521–33. doi: 10.1016/j.freeradbiomed.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Weber DS, Taniyama Y, Rocic P, Seshiah PN, Dechert MA, Gerthoffer WT, et al. Phosphoinositide-dependent kinase 1 and p21-activated protein kinase mediate reactive oxygen species-dependent regulation of platelet-derived growth factor-induced smooth muscle cell migration. Circul Res. 2004;94(9):1219–26. doi: 10.1161/01.RES.0000126848.54740.4A. [DOI] [PubMed] [Google Scholar]
- 46.Sundaresan M, Yu ZX, Ferrans VJ, Sulciner DJ, Gutkind JS, Irani K, et al. Regulation of reactive-oxygen-species generation in fibroblasts by Rac1. Biochem J. 1996;318(Pt 2):379–82. doi: 10.1042/bj3180379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao ZS, Manser E. PAK and other rho-associated kinases–effectors with surprisingly diverse mechanisms of regulation. Biochem J. 2005;386(Pt 2):201–14. doi: 10.1042/BJ20041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Du J, Hu Z, Liu J, Tian Y, Zhu Y, et al. Activation of Rac1-PI3K/Akt is required for epidermal growth factor-induced PAK1 activation and cell migration in MDA-MB-231 breast cancer cells. J biomedical Res. 2011;25(4):237–45. doi: 10.1016/S1674-8301(11)60032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lorenzi L, Avila Cobos F, Decock A, Everaert C, Helsmoortel H, Lefever S, et al. Long noncoding RNA expression profiling in cancer: challenges and opportunities. Genes Chromosom Cancer. 2019;58(4):191–9. doi: 10.1002/gcc.22709. [DOI] [PubMed] [Google Scholar]
- 50.Rahmani Z, Mojarrad M, Moghbeli M. Long non-coding RNAs as the critical factors during tumor progressions among iranian population: an overview. Cell Biosci. 2020;10:6. doi: 10.1186/s13578-020-0373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamidi AA, Khalili-Tanha G, Nasrpour Navaei Z, Moghbeli M. Long non-coding RNAs as the critical regulators of epithelial mesenchymal transition in colorectal tumor cells: an overview. Cancer Cell Int. 2022;22(1):71. doi: 10.1186/s12935-022-02501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, Qiu Q, Qian X, Yi J, Jiao Y, Yu M, et al. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging mir-4715-5p in lung cancer. Mol Cancer. 2019;18(1):171. doi: 10.1186/s12943-019-1107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin W. Role of JAK/STAT3 signaling in the regulation of metastasis, the transition of cancer stem cells, and chemoresistance of cancer by epithelial-mesenchymal transition. Cells. 2020;9(1):217. doi: 10.3390/cells9010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–46. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 56.Lee JH, Kim C, Sethi G, Ahn KS. Brassinin inhibits STAT3 signaling pathway through modulation of PIAS-3 and SOCS-3 expression and sensitizes human lung cancer xenograft in nude mice to paclitaxel. Oncotarget. 2015;6(8):6386–405. doi: 10.18632/oncotarget.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun CC, Li SJ, Zhang F, Zhang YD, Zuo ZY, Xi YY, et al. The Novel miR-9600 suppresses tumor progression and promotes paclitaxel sensitivity in non-small-cell lung cancer through altering STAT3 expression. Mol Ther Nucleic acids. 2016;5(11):e387. doi: 10.1038/mtna.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang T, Zhong H, Zhang W, Wen J, Yi Z, Li P, et al. STAT5a induces endotoxin tolerance by alleviating pyroptosis in kupffer cells. Mol Immunol. 2020;122:28–37. doi: 10.1016/j.molimm.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 59.Lin J, Zheng X, Tian X, Guan J, Shi H. Mir-1247-3p targets STAT5A to inhibit lung adenocarcinoma cell migration and chemotherapy resistance. J Cancer. 2022;13(7):2040–9. doi: 10.7150/jca.65167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fujita H, Fukuhara S, Sakurai A, Yamagishi A, Kamioka Y, Nakaoka Y, et al. Local activation of Rap1 contributes to directional vascular endothelial cell migration accompanied by extension of microtubules on which RAPL, a Rap1-associating molecule, localizes. J Biol Chem. 2005;280(6):5022–31. doi: 10.1074/jbc.M409701200. [DOI] [PubMed] [Google Scholar]
- 61.Pizon V, Baldacci G. Rap1A protein interferes with various MAP kinase activating pathways in skeletal myogenic cells. Oncogene. 2000;19(52):6074–81. doi: 10.1038/sj.onc.1203984. [DOI] [PubMed] [Google Scholar]
- 62.Vossler MR, Yao H, York RD, Pan MG, Rim CS, Stork PJ. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89(1):73–82. doi: 10.1016/S0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 63.Holmfeldt P, Brattsand G, Gullberg M. MAP4 counteracts microtubule catastrophe promotion but not tubulin-sequestering activity in intact cells. Curr biology: CB. 2002;12(12):1034–9. doi: 10.1016/S0960-9822(02)00897-7. [DOI] [PubMed] [Google Scholar]
- 64.Hoshi M, Ohta K, Gotoh Y, Mori A, Murofushi H, Sakai H, et al. Mitogen-activated-protein-kinase-catalyzed phosphorylation of microtubule-associated proteins, microtubule-associated protein 2 and microtubule-associated protein 4, induces an alteration in their function. Eur J Biochem. 1992;203(1–2):43–52. doi: 10.1111/j.1432-1033.1992.tb19825.x. [DOI] [PubMed] [Google Scholar]
- 65.Kinbara K, Goldfinger LE, Hansen M, Chou FL, Ginsberg MH. Ras GTPases: integrins’ friends or foes? Nat Rev Mol Cell Biol. 2003;4(10):767–76. doi: 10.1038/nrm1229. [DOI] [PubMed] [Google Scholar]
- 66.Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, et al. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12(6):514–27. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Du L, Subauste MC, DeSevo C, Zhao Z, Baker M, Borkowski R, et al. Mir-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PLoS ONE. 2012;7(6):e39167. doi: 10.1371/journal.pone.0039167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moghbeli M, Mosannen Mozaffari H, Memar B, Forghanifard MM, Gholamin M, Abbaszadegan MR. Role of MAML1 in targeted therapy against the esophageal cancer stem cells. J Transl Med. 2019;17(1):126. doi: 10.1186/s12967-019-1876-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang LZ, Lei CC, Zhao YP, Sun HW, Yu QH, Yang EJ, et al. MicroRNA-34c-3p target inhibiting NOTCH1 suppresses chemosensitivity and metastasis of non-small cell lung cancer. J Int Med Res. 2020;48(3):300060520904847. doi: 10.1177/0300060520904847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Jiao M, Wu K, Li L, Zhu G, Wang X, et al. TNF-α induced epithelial mesenchymal transition increases stemness properties in renal cell carcinoma cells. Int J Clin Exp Med. 2014;7(12):4951–8. [PMC free article] [PubMed] [Google Scholar]
- 71.Peinado H, Olmeda D, Cano A, Snail Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 72.Liu X, Wang C, Chen Z, Jin Y, Wang Y, Kolokythas A, et al. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem J. 2011;440(1):23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin Z, Guan L, Song Y, Xiang GM, Chen SX, Gao B. MicroRNA-138 regulates chemoresistance in human non-small cell lung cancer via epithelial mesenchymal transition. Eur Rev Med Pharmacol Sci. 2016;20(6):1080–6. [PubMed] [Google Scholar]
- 74.Kim HG, Hien TT, Han EH, Hwang YP, Choi JH, Kang KW, et al. Metformin inhibits P-glycoprotein expression via the NF-κB pathway and CRE transcriptional activity through AMPK activation. Br J Pharmacol. 2011;162(5):1096–108. doi: 10.1111/j.1476-5381.2010.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xia M, Sheng L, Qu W, Xue X, Chen H, Zheng G, et al. MiR-194-5p enhances the sensitivity of nonsmall-cell lung cancer to doxorubicin through targeted inhibition of hypoxia-inducible factor-1. World J Surg Oncol. 2021;19(1):174. doi: 10.1186/s12957-021-02278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Butter F, Davison L, Viturawong T, Scheibe M, Vermeulen M, Todd JA, et al. Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding. PLoS Genet. 2012;8(9):e1002982. doi: 10.1371/journal.pgen.1002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma W, Liu B, Li J, Jiang J, Zhou R, Huang L, et al. MicroRNA-302c represses epithelial-mesenchymal transition and metastasis by targeting transcription factor AP-4 in colorectal cancer. Biomed Pharmacother. 2018;105:670–6. doi: 10.1016/j.biopha.2018.06.025. [DOI] [PubMed] [Google Scholar]
- 78.Wu H, Liu X, Gong P, Song W, Zhou M, Li Y, et al. Elevated TFAP4 regulates lncRNA TRERNA1 to promote cell migration and invasion in gastric cancer. Oncol Rep. 2018;40(2):923–31. doi: 10.3892/or.2018.6466. [DOI] [PubMed] [Google Scholar]
- 79.Jackstadt R, Röh S, Neumann J, Jung P, Hoffmann R, Horst D, et al. AP4 is a mediator of epithelial-mesenchymal transition and metastasis in colorectal cancer. J Exp Med. 2013;210(7):1331–50. doi: 10.1084/jem.20120812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song J, Xie C, Jiang L, Wu G, Zhu J, Zhang S, et al. Transcription factor AP-4 promotes tumorigenic capability and activates the Wnt/β-catenin pathway in hepatocellular carcinoma. Theranostics. 2018;8(13):3571–83. doi: 10.7150/thno.25194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang YF, Ao X, Liu Y, Ding D, Jiao WJ, Yu Z, et al. MicroRNA-608 promotes apoptosis in Non-Small Cell Lung Cancer cells treated with Doxorubicin through the inhibition of TFAP4. Front Genet. 2019;10:809. doi: 10.3389/fgene.2019.00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stovall DB, Cao P, Sui G. SOX7: from a developmental regulator to an emerging tumor suppressor. Histol Histopathol. 2014;29(4):439–45. doi: 10.14670/hh-29.10.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cui J, Xi H, Cai A, Bian S, Wei B, Chen L. Decreased expression of Sox7 correlates with the upregulation of the Wnt/β-catenin signaling pathway and the poor survival of gastric cancer patients. Int J Mol Med. 2014;34(1):197–204. doi: 10.3892/ijmm.2014.1759. [DOI] [PubMed] [Google Scholar]
- 84.Li B, Ge Z, Song S, Zhang S, Yan H, Huang B, et al. Decreased expression of SOX7 is correlated with poor prognosis in lung adenocarcinoma patients. Pathol Oncol research: POR. 2012;18(4):1039–45. doi: 10.1007/s12253-012-9542-8. [DOI] [PubMed] [Google Scholar]
- 85.Guo L, Zhong D, Lau S, Liu X, Dong XY, Sun X, et al. Sox7 is an independent checkpoint for beta-catenin function in prostate and colon epithelial cells. Mol cancer research: MCR. 2008;6(9):1421–30. doi: 10.1158/1541-7786.MCR-07-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng Z, Liu J, Yang Z, Wu L, Xie H, Jiang C, et al. MicroRNA-452 promotes stem-like cells of hepatocellular carcinoma by inhibiting Sox7 involving Wnt/β-catenin signaling pathway. Oncotarget. 2016;7(19):28000–12. doi: 10.18632/oncotarget.8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baselga J. Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. Oncologist. 2011;16(Suppl 1):12–9. doi: 10.1634/theoncologist.2011-S1-12. [DOI] [PubMed] [Google Scholar]
- 88.Peng B, Li C, Cai P, Yu L, Zhao B, Chen G. Knockdown of miR–935 increases paclitaxel sensitivity via regulation of SOX7 in non–small–cell lung cancer. Mol Med Rep. 2018;18(3):3397–402. doi: 10.3892/mmr.2018.9330. [DOI] [PubMed] [Google Scholar]
- 89.Li DQ, Kumar R. Unravelling the complexity and functions of MTA coregulators in Human Cancer. Adv Cancer Res. 2015;127:1–47. doi: 10.1016/bs.acr.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 90.Wen X, Cheng X, Hu D, Li W, Ha J, Kang Z, et al. Combination of Curcumin with an anti-transferrin receptor antibody suppressed the growth of malignant gliomas in vitro. Turkish Neurosurg. 2016;26(2):209–14. doi: 10.5137/1019-5149.JTN.7966-13.1. [DOI] [PubMed] [Google Scholar]
- 91.Mosieniak G, Sliwinska MA, Przybylska D, Grabowska W, Sunderland P, Bielak-Zmijewska A, et al. Curcumin-treated cancer cells show mitotic disturbances leading to growth arrest and induction of senescence phenotype. Int J Biochem Cell Biol. 2016;74:33–43. doi: 10.1016/j.biocel.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 92.Fan Z, Duan X, Cai H, Wang L, Li M, Qu J, et al. Curcumin inhibits the invasion of lung cancer cells by modulating the PKCα/Nox-2/ROS/ATF-2/MMP-9 signaling pathway. Oncol Rep. 2015;34(2):691–8. doi: 10.3892/or.2015.4044. [DOI] [PubMed] [Google Scholar]
- 93.Mishra A, Kumar R, Tyagi A, Kohaar I, Hedau S, Bharti AC, et al. Curcumin modulates cellular AP-1, NF-kB, and HPV16 E6 proteins in oral cancer. Ecancermedicalscience. 2015;9:525. doi: 10.3332/ecancer.2015.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pandey A, Vishnoi K, Mahata S, Tripathi SC, Misra SP, Misra V, et al. Berberine and curcumin target survivin and STAT3 in gastric cancer cells and synergize actions of standard chemotherapeutic 5-Fluorouracil. Nutr Cancer. 2015;67(8):1293–304. doi: 10.1080/01635581.2015.1085581. [DOI] [PubMed] [Google Scholar]
- 95.Zhao Z, Li C, Xi H, Gao Y, Xu D. Curcumin induces apoptosis in pancreatic cancer cells through the induction of forkhead box O1 and inhibition of the PI3K/Akt pathway. Mol Med Rep. 2015;12(4):5415–22. doi: 10.3892/mmr.2015.4060. [DOI] [PubMed] [Google Scholar]
- 96.Chen J, Xu T, Chen C. The critical roles of miR-21 in anti-cancer effects of curcumin. Ann Transl Med. 2015;3(21):330. doi: 10.3978/j.issn.2305-5839.2015.09.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu Y, Wang J, Liu L, Yu L, Zhao N, Zhou X, et al. Curcumin increases the sensitivity of Paclitaxel-resistant NSCLC cells to Paclitaxel through microRNA-30c-mediated MTA1 reduction. Tumour Biol. 2017;39(4):1010428317698353. doi: 10.1177/1010428317698353. [DOI] [PubMed] [Google Scholar]
- 98.Walaas SI, Ostvold AC, Laland SG. Phosphorylation of P1, a high mobility group-like protein, catalyzed by casein kinase II, protein kinase C, cyclic AMP-dependent protein kinase and calcium/calmodulin-dependent protein kinase II. FEBS Lett. 1989;258(1):106–8. doi: 10.1016/0014-5793(89)81626-6. [DOI] [PubMed] [Google Scholar]
- 99.Maelandsmo GM, Ostvold AC, Laland SG. Phosphorylation of the high-mobility-group-like protein P1 by casein kinase-2. Eur J Biochem. 1989;184(3):529–34. doi: 10.1111/j.1432-1033.1989.tb15046.x. [DOI] [PubMed] [Google Scholar]
- 100.Meijer L, Ostvold AC, Walass SI, Lund T, Laland SG. High-mobility-group proteins P1, I and Y as substrates of the M-phase-specific p34cdc2/cyclincdc13 kinase. Eur J Biochem. 1991;196(3):557–67. doi: 10.1111/j.1432-1033.1991.tb15850.x. [DOI] [PubMed] [Google Scholar]
- 101.Ostvold AC, Norum JH, Mathiesen S, Wanvik B, Sefland I, Grundt K. Molecular cloning of a mammalian nuclear phosphoprotein NUCKS, which serves as a substrate for Cdk1 in vivo. Eur J Biochem. 2001;268(8):2430–40. doi: 10.1046/j.1432-1327.2001.02120.x. [DOI] [PubMed] [Google Scholar]
- 102.Wisniewski JR, Schwanbeck R. High mobility group I/Y: multifunctional chromosomal proteins causally involved in tumor progression and malignant transformation (review) Int J Mol Med. 2000;6(4):409–19. doi: 10.3892/ijmm.6.4.409. [DOI] [PubMed] [Google Scholar]
- 103.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14(11):4376–86. doi: 10.1091/mbc.e03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shen H, Wang L, Ge X, Jiang CF, Shi ZM, Li DM, et al. MicroRNA-137 inhibits tumor growth and sensitizes chemosensitivity to paclitaxel and cisplatin in lung cancer. Oncotarget. 2016;7(15):20728–42. doi: 10.18632/oncotarget.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bylaite M, Moussali H, Marciukaitiene I, Ruzicka T, Walz M. Expression of cathepsin L and its inhibitor hurpin in inflammatory and neoplastic skin diseases. Exp Dermatol. 2006;15(2):110–8. doi: 10.1111/j.1600-0625.2005.00389.x. [DOI] [PubMed] [Google Scholar]
- 106.Tyagi C, Grover S, Dhanjal J, Goyal S, Goyal M, Grover A. Mechanistic insights into mode of action of novel natural cathepsin L inhibitors. BMC Genomics. 2013;14(Suppl 8):10. doi: 10.1186/1471-2164-14-S8-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tholen M, Wolanski J, Stolze B, Chiabudini M, Gajda M, Bronsert P, et al. Stress-resistant translation of cathepsin L mRNA in breast Cancer progression. J Biol Chem. 2015;290(25):15758–69. doi: 10.1074/jbc.M114.624353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fei Y, Xiong Y, Zhao Y, Wang W, Han M, Wang L, et al. Cathepsin L knockdown enhances curcumin-mediated inhibition of growth, migration, and invasion of glioma cells. Brain Res. 2016;1646:580–8. doi: 10.1016/j.brainres.2016.06.046. [DOI] [PubMed] [Google Scholar]
- 109.Zheng X, Chou PM, Mirkin BL, Rebbaa A. Senescence-initiated reversal of drug resistance: specific role of cathepsin L. Cancer Res. 2004;64(5):1773–80. doi: 10.1158/0008-5472.CAN-03-0820. [DOI] [PubMed] [Google Scholar]
- 110.Zheng X, Chu F, Chou PM, Gallati C, Dier U, Mirkin BL, et al. Cathepsin L inhibition suppresses drug resistance in vitro and in vivo: a putative mechanism. Am J Physiol Cell Physiol. 2009;296(1):C65–74. doi: 10.1152/ajpcell.00082.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang Q, Han M, Wang W, Song Y, Chen G, Wang Z, et al. Downregulation of cathepsin L suppresses cancer invasion and migration by inhibiting transforming growth factor–β–mediated epithelial–mesenchymal transition. Oncol Rep. 2015;33(4):1851–9. doi: 10.3892/or.2015.3754. [DOI] [PubMed] [Google Scholar]
- 112.Han ML, Zhao YF, Tan CH, Xiong YJ, Wang WJ, Wu F, et al. Cathepsin L upregulation-induced EMT phenotype is associated with the acquisition of cisplatin or paclitaxel resistance in A549 cells. Acta Pharmacol Sin. 2016;37(12):1606–22. doi: 10.1038/aps.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–94. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 114.Zhao YF, Han ML, Xiong YJ, Wang L, Fei Y, Shen X, et al. A miRNA-200c/cathepsin L feedback loop determines paclitaxel resistance in human lung cancer A549 cells in vitro through regulating epithelial-mesenchymal transition. Acta Pharmacol Sin. 2018;39(6):1034–47. doi: 10.1038/aps.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16(2):123–40. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 116.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23(22):8137–51. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci USA. 2004;101(17):6379–84. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fan Z, Wirth AK, Chen D, Wruck CJ, Rauh M, Buchfelder M, et al. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis. 2017;6(8):e371. doi: 10.1038/oncsis.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Duan FG, Wang MF, Cao YB, Dan L, Li RZ, Fan XX, et al. MicroRNA-421 confers paclitaxel resistance by binding to the KEAP1 3’UTR and predicts poor survival in non-small cell lung cancer. Cell Death Dis. 2019;10(11):821. doi: 10.1038/s41419-019-2031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Investig. 2009;119(6):1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science (New York, NY) 2008;321:1499–502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Teixeira LK, Reed SI. Ubiquitin ligases and cell cycle control. Annu Rev Biochem. 2013;82:387–414. doi: 10.1146/annurev-biochem-060410-105307. [DOI] [PubMed] [Google Scholar]
- 123.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 124.Díaz VM, de Herreros AG. F-box proteins: keeping the epithelial-to-mesenchymal transition (EMT) in check. Sem Cancer Biol. 2016;36:71–9. doi: 10.1016/j.semcancer.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 125.Su J, Yin X, Zhou X, Wei W, Wang Z. The functions of F-box proteins in regulating the epithelial to mesenchymal transition. Curr Pharm Design. 2015;21(10):1311–7. doi: 10.2174/1381612821666141211144203. [DOI] [PubMed] [Google Scholar]
- 126.Li R, Wu S, Chen X, Xu H, Teng P, Li W. miR-223/FBW7 axis regulates doxorubicin sensitivity through epithelial mesenchymal transition in non-small cell lung cancer. Am J translational Res. 2016;8(6):2512–24. [PMC free article] [PubMed] [Google Scholar]
- 127.Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D’Andrea AD, et al. UBE2T is the E2 in the fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23(4):589–96. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 128.Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang L, Zhang Z, Tian H. Hsa_circ_0092887 targeting miR-490-5p/UBE2T promotes paclitaxel resistance in non-small cell lung cancer. J Clin Lab Anal. 2023;37(1):e24781. doi: 10.1002/jcla.24781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yadav L, Puri N, Rastogi V, Satpute P, Ahmad R, Kaur G. Matrix metalloproteinases and cancer - roles in threat and therapy. Asian Pac J Cancer Prev. 2014;15(3):1085–91. doi: 10.7314/APJCP.2014.15.3.1085. [DOI] [PubMed] [Google Scholar]
- 131.Li X, Feng Y, Yang B, Xiao T, Ren H, Yu X, et al. A novel circular RNA, hsa_circ_0030998 suppresses lung cancer tumorigenesis and taxol resistance by sponging miR-558. Mol Oncol. 2021;15(8):2235–48. doi: 10.1002/1878-0261.12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jiang G, Wang Y, Dai S, Liu Y, Stoecker M, Wang E, et al. P120-catenin isoforms 1 and 3 regulate proliferation and cell cycle of lung cancer cells via β-catenin and Kaiso respectively. PLoS ONE. 2012;7(1):e30303. doi: 10.1371/journal.pone.0030303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163(3):525–34. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Davis MA, Reynolds AB. Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Dev Cell. 2006;10(1):21–31. doi: 10.1016/j.devcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 135.Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, et al. A novel role for p120 catenin in E-cadherin function. J Cell Biol. 2002;159(3):465–76. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chartier NT, Oddou CI, Lainé MG, Ducarouge B, Marie CA, Block MR, et al. Cyclin-dependent kinase 2/cyclin E complex is involved in p120 catenin (p120ctn)-dependent cell growth control: a new role for p120ctn in cancer. Cancer Res. 2007;67(20):9781–90. doi: 10.1158/0008-5472.CAN-07-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang T, Li H, Chen T, Ren H, Shi P, Chen M. LncRNA MALAT1 Depressed Chemo-Sensitivity of NSCLC cells through directly functioning on miR-197-3p/p120 catenin Axis. Mol Cells. 2019;42(3):270–83. doi: 10.14348/molcells.2019.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li J, Zhu T, Weng Y, Cheng F, Sun Q, Yang K, et al. Exosomal circDNER enhances paclitaxel resistance and tumorigenicity of lung cancer via targeting miR-139-5p/ITGB8. Thorac cancer. 2022;13(9):1381–90. doi: 10.1111/1759-7714.14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tallima H, El Ridi R. Arachidonic acid: physiological roles and potential health benefits - a review. J Adv Res. 2018;11:33–41. doi: 10.1016/j.jare.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fitzpatrick FA. Cyclooxygenase enzymes: regulation and function. Curr Pharm Design. 2004;10(6):577–88. doi: 10.2174/1381612043453144. [DOI] [PubMed] [Google Scholar]
- 141.Sonoshita M, Takaku K, Sasaki N, Sugimoto Y, Ushikubi F, Narumiya S, et al. Acceleration of intestinal polyposis through prostaglandin receptor EP2 in apc(Delta 716) knockout mice. Nat Med. 2001;7(9):1048–51. doi: 10.1038/nm0901-1048. [DOI] [PubMed] [Google Scholar]
- 142.Salcedo R, Zhang X, Young HA, Michael N, Wasserman K, Ma WH, et al. Angiogenic effects of prostaglandin E2 are mediated by up-regulation of CXCR4 on human microvascular endothelial cells. Blood. 2003;102(6):1966–77. doi: 10.1182/blood-2002-11-3400. [DOI] [PubMed] [Google Scholar]
- 143.Monteleone NJ, Lutz CS. Mir-708-5p enhances erlotinib/paclitaxel efficacy and overcomes chemoresistance in lung cancer cells. Oncotarget. 2020;11(51):4699–721. doi: 10.18632/oncotarget.27840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu Y, Xie J, Wang H, Hou S, Feng J. Circular RNA hsa_circ_0011298 enhances taxol resistance of non-small cell lung cancer by regulating miR-486-3p/CRABP2 axis. J Clin Lab Anal. 2022;36(5):e24408. doi: 10.1002/jcla.24408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gambardella J, Franco A, Giudice CD, Fiordelisi A, Cipolletta E, Ciccarelli M, et al. Dual role of GRK5 in cancer development and progression. Translational Med @ UniSa. 2016;14:28–37. [PMC free article] [PubMed] [Google Scholar]
- 146.Liu Y, Li C, Liu H, Wang J. Circ_0001821 knockdown suppresses growth, metastasis, and TAX resistance of non-small-cell lung cancer cells by regulating the miR-526b-5p/GRK5 axis. Pharmacol Res Perspect. 2021;9(4):e00812. doi: 10.1002/prp2.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huang B, Gong X, Zhou H, Xiong F, Wang S. Depleting ABCE1 expression induces apoptosis and inhibits the ability of proliferation and migration of human esophageal carcinoma cells. Int J Clin Exp Pathol. 2014;7(2):584–92. [PMC free article] [PubMed] [Google Scholar]
- 148.Wang L, Zhang M, Liu DX. Knock-down of ABCE1 gene induces G1/S arrest in human oral cancer cells. Int J Clin Exp Pathol. 2014;7(9):5495–504. [PMC free article] [PubMed] [Google Scholar]
- 149.Zheng D, Dai Y, Wang S, Xing X. MicroRNA-299-3p promotes the sensibility of lung cancer to doxorubicin through directly targeting ABCE1. Int J Clin Exp Pathol. 2015;8(9):10072–81. [PMC free article] [PubMed] [Google Scholar]
- 150.Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer. 2010;10(3):194–204. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- 151.Zheng H, Liu Z, Liu T, Cai Y, Wang Y, Lin S, et al. Fas signaling promotes chemoresistance in gastrointestinal cancer by up-regulating P-glycoprotein. Oncotarget. 2014;5(21):10763–77. doi: 10.18632/oncotarget.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vargas JR, Stanzl EG, Teng NN, Wender PA. Cell-penetrating, guanidinium-rich molecular transporters for overcoming efflux-mediated multidrug resistance. Mol Pharm. 2014;11(8):2553–65. doi: 10.1021/mp500161z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xu J, Ni L, Zhao F, Dai X, Tao J, Pan J, et al. Overexpression of hsa_circ_0002874 promotes resistance of non-small cell lung cancer to paclitaxel by modulating miR-1273f/MDM2/p53 pathway. Aging. 2021;13(4):5986–6009. doi: 10.18632/aging.202521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tang X, Tang J, Liu X, Zeng L, Cheng C, Luo Y, et al. Downregulation of mir-129-2 by promoter hypermethylation regulates breast cancer cell proliferation and apoptosis. Oncol Rep. 2016;35(5):2963–9. doi: 10.3892/or.2016.4647. [DOI] [PubMed] [Google Scholar]
- 155.Datta S, Ray A, Singh R, Mondal P, Basu A, De Sarkar N, et al. Sequence and expression variations in 23 genes involved in mitochondrial and non-mitochondrial apoptotic pathways and risk of oral leukoplakia and cancer. Mitochondrion. 2015;25:28–33. doi: 10.1016/j.mito.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 156.Bae IH, Yoon SH, Lee SB, Park JK, Ho JN, Um HD. Signaling components involved in Bcl-w-induced migration of gastric cancer cells. Cancer Lett. 2009;277(1):22–8. doi: 10.1016/j.canlet.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 157.Um HD. Bcl-2 family proteins as regulators of cancer cell invasion and metastasis: a review focusing on mitochondrial respiration and reactive oxygen species. Oncotarget. 2016;7(5):5193–203. doi: 10.18632/oncotarget.6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zubor P, Hatok J, Moricova P, Kajo K, Kapustova I, Mendelova A, et al. Gene expression abnormalities in histologically normal breast epithelium from patients with luminal type of breast cancer. Mol Biol Rep. 2015;42(5):977–88. doi: 10.1007/s11033-014-3834-x. [DOI] [PubMed] [Google Scholar]
- 159.Zhang Y, Yu J, Liu H, Ma W, Yan L, Wang J, et al. Novel epigenetic CREB-miR-630 signaling axis regulates radiosensitivity in colorectal cancer. PLoS ONE. 2015;10(8):e0133870. doi: 10.1371/journal.pone.0133870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Katoh M. Cardio-miRNAs and onco-miRNAs: circulating miRNA-based diagnostics for non-cancerous and cancerous diseases. Front cell Dev biology. 2014;2:61. doi: 10.3389/fcell.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lu C, Xie Z, Peng Q. MiRNA-107 enhances chemosensitivity to paclitaxel by targeting antiapoptotic factor Bcl-w in non small cell lung cancer. Am J cancer Res. 2017;7(9):1863–73. [PMC free article] [PubMed] [Google Scholar]
- 162.Xu X, Jin S, Ma Y, Fan Z, Yan Z, Li W, et al. miR-30a-5p enhances paclitaxel sensitivity in non-small cell lung cancer through targeting BCL-2 expression. J Mol Med. 2017;95(8):861–71. doi: 10.1007/s00109-017-1539-z. [DOI] [PubMed] [Google Scholar]
- 163.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113(6):677–83. doi: 10.1016/S0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 164.Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18(10):610–21. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Teloni F, Altmeyer M. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016;44(3):993–1006. doi: 10.1093/nar/gkv1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11(3):196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lai J, Yang H, Zhu Y, Ruan M, Huang Y, Zhang Q. MiR-7-5p-mediated downregulation of PARP1 impacts DNA homologous recombination repair and resistance to doxorubicin in small cell lung cancer. BMC Cancer. 2019;19(1):602. doi: 10.1186/s12885-019-5798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol Ther. 2014;142(1):1–10. doi: 10.1016/j.pharmthera.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 169.Yu X, Zhang Y, Ma X, Pertsemlidis A. miR-195 potentiates the efficacy of microtubule-targeting agents in non-small cell lung cancer. Cancer Lett. 2018;427:85–93. doi: 10.1016/j.canlet.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jin M, Zhang F, Li Q, Xu R, Liu Y, Zhang Y. Circ_0011292 knockdown mitigates progression and drug resistance in PTX-resistant non-small-cell lung cancer cells by regulating miR-433-3p/CHEK1 axis. Thorac cancer. 2022;13(9):1276–88. doi: 10.1111/1759-7714.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Alfarouk KO, Stock CM, Taylor S, Walsh M, Muddathir AK, Verduzco D, et al. Resistance to cancer chemotherapy: failure in drug response from ADME to P-gp. Cancer Cell Int. 2015;15:71. doi: 10.1186/s12935-015-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wu Q, Yang Z, Nie Y, Shi Y, Fan D. Multi-drug resistance in cancer chemotherapeutics: mechanisms and lab approaches. Cancer Lett. 2014;347(2):159–66. doi: 10.1016/j.canlet.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 173.Chang H, Zou Z. Targeting autophagy to overcome drug resistance: further developments. J Hematol Oncol. 2020;13(1):159. doi: 10.1186/s13045-020-01000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.White E. Autophagy and p53. Cold Spring Harb Perspect Med. 2016;6(4):a026120. doi: 10.1101/cshperspect.a026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mishima Y, Terui Y, Mishima Y, Taniyama A, Kuniyoshi R, Takizawa T, et al. Autophagy and autophagic cell death are next targets for elimination of the resistance to tyrosine kinase inhibitors. Cancer Sci. 2008;99(11):2200–8. doi: 10.1111/j.1349-7006.2008.00932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hu YL, Jahangiri A, Delay M, Aghi MK. Tumor cell autophagy as an adaptive response mediating resistance to treatments such as antiangiogenic therapy. Cancer Res. 2012;72(17):4294–9. doi: 10.1158/0008-5472.CAN-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pavlinov I, Salkovski M, Aldrich LN. Beclin 1-ATG14L protein-protein Interaction inhibitor selectively inhibits autophagy through disruption of VPS34 complex I. J Am Chem Soc. 2020;142(18):8174–82. doi: 10.1021/jacs.9b12705. [DOI] [PubMed] [Google Scholar]
- 180.Chatterjee A, Chattopadhyay D, Chakrabarti G. Mir-17-5p downregulation contributes to paclitaxel resistance of lung cancer cells through altering beclin1 expression. PLoS ONE. 2014;9(4):e95716. doi: 10.1371/journal.pone.0095716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19(1):12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li Y, Zhang H, Guo J, Li W, Wang X, Zhang C, et al. Downregulation of LINC01296 suppresses non-small-cell lung cancer via targeting miR-143-3p/ATG2B. Acta Biochim Biophys Sin. 2021;53(12):1681–90. doi: 10.1093/abbs/gmab149. [DOI] [PubMed] [Google Scholar]
- 183.Zeng T, Xu M, Zhang W, Gu X, Zhao F, Liu X, et al. Autophagy inhibition and microRNA–199a–5p upregulation in paclitaxel–resistant A549/T lung cancer cells. Oncol Rep. 2021;46(1):149. doi: 10.3892/or.2021.8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhao YY, Yu L, Liu BL, He XJ, Zhang BY. Downregulation of P-gp, ras and p-ERK1/2 contributes to the arsenic trioxide-induced reduction in drug resistance towards doxorubicin in gastric cancer cell lines. Mol Med Rep. 2015;12(5):7335–43. doi: 10.3892/mmr.2015.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Keppler D. Multidrug resistance proteins (MRPs, ABCCs): importance for pathophysiology and drug therapy. Handb Exp Pharmacol. 2011;201:299–323. doi: 10.1007/978-3-642-14541-4_8. [DOI] [PubMed] [Google Scholar]
- 186.Chen S, Jiao JW, Sun KX, Zong ZH, Zhao Y. MicroRNA-133b targets glutathione S-transferase π expression to increase ovarian cancer cell sensitivity to chemotherapy drugs. Drug Des Devel Ther. 2015;9:5225–35. doi: 10.2147/DDDT.S87526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Surowiak P, Materna V, Kaplenko I, Spaczyński M, Dietel M, Lage H, et al. Augmented expression of metallothionein and glutathione S-transferase pi as unfavourable prognostic factors in cisplatin-treated ovarian cancer patients. Virchows Archiv. 2005;447(3):626–33. doi: 10.1007/s00428-005-1228-0. [DOI] [PubMed] [Google Scholar]
- 188.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol/Hematol. 2007;63(1):12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 189.Lv L, An X, Li H, Ma L. Effect of miR-155 knockdown on the reversal of doxorubicin resistance in human lung cancer A549/dox cells. Oncol Lett. 2016;11(2):1161–6. doi: 10.3892/ol.2015.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.