Abstract
Burkholderia cepacia MBA4 has been shown to produce a single dehalogenase batch culture. Moreover, other cryptic dehalogenases were also detected when the cells were grown in continuous culture. In this paper, we report the cloning and characterization of one of the cryptic dehalogenases in MBA4. This cryptic haloacid dehalogenase, designated Chd1, was expressed constitutively in Escherichia coli. This recombinant Chd1 had a relative molecular weight of 58,000 and existed predominantly as a dimer. The subunits had a relative molecular weight of 27,000. Chd1 exhibited isomer specificity, being active towards the l-isomer of 2-monochloropropionic acid only. The structural gene, chd1, was isolated on a 1.7-kb PstI fragment. This fragment contains a functional promoter, because expression of chd1 in E. coli is orientation independent. The nucleotide sequence of this fragment was determined and characterized. An open reading frame of 840 bp encoding a putative peptide of 280 amino acids was identified. This corresponds closely with the size of the subunit. The nucleotide sequence of chd1 did not show any homology with those of other dehalogenase genes. Comparison of the predicted amino acid sequence, however, shows significant homology, ranging from 42 to 50%, with the amino acid sequences of many other dehalogenases. Chd1 is unusual in having a long leader sequence, a property of periplasmic enzymes.
2-Haloacid dehalogenases, or halidohydrolases, are hydrolytic enzymes that cleave the halogen-carbon bond(s) in halogenated aliphatic acids, yielding hydroxy- or oxoalkanoic acids from a substrate with a mono- or disubstitution, respectively (14, 39). Burkholderia cepacia MBA4 (formerly considered a Pseudomonas species) was isolated from soil by batch enrichment culture with monobromoacetic acid (MBA) as the sole carbon and energy source. MBA4 is able to grow on MBA, monochloroacetate (MCA), 2-mono-chloropropionate (2MCPA), and 2-monobromopropionate (2MBPA) (43). This bacterium produces a single dehalogenase (DehIVa) in batch culture, and this enzyme has been purified and characterized (43). The active enzyme is a dimeric protein of 45 kDa. The structural gene for DehIVa, hdlIVa, was isolated from a genomic library (42), and analysis of the DNA sequence revealed an open reading frame (ORF) for 231 amino acids and a putative protein of 25.9 kDa (32).
Cryptic genes are silent DNA sequences; i.e., they are not normally expressed in an individual (13). Previous studies suggest the presence of cryptic dehalogenases in the gene pools of some dehalogenase-producing bacteria (15, 19, 38, 40, 45). During the course of studying the physiology of MBA4 in continuous culture, other dehalogenases were also detected by activity-stained gel electrophoresis and by a change in specific dehalogenase activity. These cells grew in a fraction of the maximum specific growth rate (35a, 41). However, because of their lack of expression in normal batch culture, the availability of these cryptic dehalogenases is scarce. It is therefore difficult to characterize these cryptic dehalogenases unless sufficient amounts of the enzymes can be obtained, for example, by means of heterologous expression.
Producing an enzyme in a heterologous host requires the isolation of the corresponding structural gene. Cloning and expression of the MBA4 hdlIVa gene in Pseudomonas putida and in Escherichia coli suggested that the promoter of the Burkholderia gene is not regulated in these foreign hosts (42). These results provide the basis for the hypothesis that genes normally silent in Burkholderia could be expressed constitutively, although at a basal level, in E. coli. In this study, we report the cloning and characterization of one of the cryptic dehalogenases found in B. cepacia MBA4.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
B. cepacia MBA4 (43) was used as the source of chromosomal DNA. E. coli XL1-Blue MR (Stratagene) was used for library construction, and strain TOP10F′ (Invitrogen) was used as a host strain for gene cloning and plasmid construction. Plasmid pSP73 (Promega) was used as a cloning vector. B. cepacia was grown in an MBA medium as described previously (43). Recombinant E. coli cells were grown at 37°C in Luria broth (1% tryptone, 0.5% yeast extract) with or without 0.5% NaCl and supplemented with 50 μg of ampicillin per ml. E. coli IT41 (20) is temperature sensitive and was grown at 30°C unless otherwise specified.
Enzymes and chemicals.
Restriction endonucleases, T4 DNA ligase, and sequencing oligonucleotide primers were obtained from Gibco-BRL and Advanced Biotechnologies. Calf intestine alkaline phosphate was purchased from Boehringer Mannheim. Deoxynucleotides and a sequencing kit including Cy5-labelled fluorescent DNA were obtained from Pharmacia. MCA was obtained from Sigma. All other chemicals were of analytical grade.
DNA manipulation and transformation.
Restriction endonuclease treatments, alkaline phosphatase treatments, and ligations were performed according to the suppliers’ recommended protocols. Plasmid DNA was prepared with a Qiagen plasmid kit. For analytical purposes, the boiling method of Holmes and Quigley (17) was used. DNA fragments were purified with a GeneClean kit (Bio 101) after agarose gel electrophoresis. Transformation of E. coli TOP10F′ with plasmid DNA was performed by the CaCl2 method (8).
Construction of a genomic DNA library and screening of dehalogenase-producing clones.
A genomic DNA library of B. cepacia MBA4 was constructed in the cosmid vector SuperCos1 according to the manufacturer’s protocol (Stratagene). The library DNA was packaged in vitro with Gigapack II XL packaging extract and transfected into E. coli XL1-Blue MR. Putative dehalogenase-producing transfectants were screened with a 96-well microtiter plate with MCA as the reaction substrate as described previously (32, 42). MCA was used because MBA is toxic to E. coli cells and medium containing MBA inhibited the growth of the bacterium (32, 42).
Preparation of total protein extract and activity-stained PAGE.
Total protein extracts of the cells were prepared by sonication (MSE Soniprep 150) or by two passages through a French press (SLM Aminco) at 40,000 lb/in2. The remaining whole cells and cell debris were removed by centrifugation at 48,400 × g for 45 min. The relative mobilities of the dehalogenases were analyzed by nondenaturing activity-stained polyacrylamide gel electrophoresis (PAGE) (43) with MCA as the reaction substrate. Crude protein extracts prepared from B. cepacia MBA4 were used as positive controls to determine the relative mobilities of the tested enzymes.
Dehalogenase and protein assays.
The dehalogenase assay was routinely carried out with MCA as the substrate, owing to its better stability than that of MBA. A standard assay mixture (1 ml) consisted of 50 mM MCA, 20 mM Tris-sulfate buffer (pH 7.9), and enzyme and was incubated at 30°C. The halide ions released were measured with a Chloride Analyzer 925 (Corning). When the substrate concentration was below 1 mM, chloride ions were determined spectrophotometrically (3). Protein concentrations were determined with Bio-Rad protein assay reagent.
Enzyme purification.
All steps were carried out at 0 to 5°C unless otherwise specified. All buffers contained 1 mM phenylmethylsulfonyl fluoride and 1 mM EDTA to prevent inactivation of the enzyme.
Step 1. Preparation of crude cell extracts.
Total protein extracts were prepared as described above.
Step 2. Protamine sulfate precipitation.
Nucleic acids were removed from the crude extract by protamine sulfate precipitation. Protamine sulfate (0.4% [wt/vol] final concentration) was added to the cell extracts, with stirring, for 20 min, and the precipitated nucleic acids were removed by centrifugation at 48,400 × g for 45 min. The supernatant was dialyzed overnight against 20 mM Tris-sulfate (pH 7.5).
Step 3. Ammonium sulfate precipitation.
The dialyzed enzyme preparation was fractionated by stepwise addition of solid ammonium sulfate to 10 to 60% saturation. At each step, the precipitate was collected by centrifugation at 9,000 × g and dissolved in 20 mM Tris-sulfate (pH 7.5). The active fractions at 10 to 30% were dialyzed overnight against Tris-sulfate.
Step 4. Anion-exchange chromatography.
The dialyzed enzyme was applied to a column (2.6 by 10 cm) of Sepharose Q Fast Flow (Pharmacia), equilibrated with 20 mM Tris (pH 8.2). The column was eluted at a flow rate of 7 ml/min with 1 liter of a linear gradient of 0 to 1 M NaCl in the same Tris buffer. Dehalogenase was eluted at 0.33 to 0.41 M NaCl. The active fractions were combined and dialyzed against 20 mM Tris-sulfate (pH 7.5).
Step 5. Hydroxylapatite chromatography.
The enzyme was applied to a column (10 ml) of Hydroxylapatite Fast Flow (Calbiochem) equilibrated with 20 mM Tris-sulfate (pH 7.5). The column was washed with 100 ml of the same buffer and stepwise elution was carried out with a linear gradient of 5 to 90 mM trisodium phosphate in 20 mM Tris-sulfate (pH 7.5). The active fractions, which were eluted at 10 to 15 mM buffer concentrations, were pooled, dialyzed, and concentrated with Centricon 30 (Amicon). The purified enzyme was stored at −20°C, with a negligible loss of activity, for a period of over 1 month.
Molecular weight determination.
The native molecular weight of the protein was determined by gel filtration on a High Prep 26/60 Sephacryl S-200 high-resolution column (Pharmacia). The medium was equilibrated with 20 mM Tris-sulfate, pH 7.9. Cell extracts were applied to the column and eluted with the same buffer at a flow rate of 1 ml/min. The column was calibrated with blue dextran (2,000 kDa), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and ribonuclease A (13.7 kDa). The molecular weight of the partially purified denatured dehalogenase was determined by sodium dodecyl sulfate (SDS)-PAGE on a 12% gel (26). Rainbow molecular weight markers (Amersham) were used as standards, and the gels were visualized with Coomassie blue R-250.
N-terminal amino acid sequence determination.
Protein samples were transferred onto a Hybond-P polyvinylidene difluoride membrane (Amersham) from a denaturing gel by using a Bio-Rad electroblotting system. Electroblotting was carried out in 10 mM (cyclohexylamino)-1-propanesulfonic acid (CAPS) buffer, pH 11, containing 10% (vol/vol) methanol for 1 h at 75 V. The amino acid sequence was determined with a G1000 protein sequencer (Hewlett-Packard).
Nucleotide sequence determination and alignment.
An automated DNA sequencer (ALF-express; Pharmacia) was used to determine the DNA sequences of the inserts, using Cy5-labelled nucleotides according to the protocol provided by the supplier (Pharmacia). The 1.7-kb PstI fragment in pHKU104 was initially sequenced with primers SP6 and T7. New oligonucleotide primers were then designed according to the obtained sequencing data. The designed primers used for sequencing in this study were as follows: S1, GGACA GCAGG GAAGC CGTCA C; S2, CCATC TATCT GCGTG TTTGA; S3, TATGG GACAC AATTG GTGCG; S4, TTTGC CAGGA GTCGC TTCCC; T1, GACTT TTGCG CCTGT CACGA; T2, GAGTA CAAGA TAAGC TGATT; T3, ATATC CATAT CATTA CGGTC; and T4, TTTTT GTTAG TTAGA TATCC. The fragment was sequenced in both directions. The nucleotide sequences and the predicted amino acid sequences were analyzed by University of Wisconsin Genetics Computer Group (GCG) programs and by National Center for Biotechnology Information programs. Nucleotide sequences were compared with those in the EMBL nucleotide sequence data library. Amino acid sequences were compared with those in the SWISS-PROT protein database. Alignment of the sequence of Chd1 with those of other dehalogenases was performed with the GCG Pileup program.
Primer extension.
The transcriptional start site was mapped by primer extension analysis (4). Total RNAs were isolated from cultures of HKU103 with the High Pure RNA isolation kit (Boehringer Mannheim). The Cy5-labelled primer (5′-GTCTCCGAACACACGCTGA-3′) was complementary to the sequence at positions 822 to 804. The RNA-primer mixture, containing 5 μg of total RNA and a 2 μM concentration of the primer, was incubated at 70°C for 10 min and quenched on ice for 1 min. The reverse transcription reaction was performed by first-strand cDNA synthesis with the SuperScript preamplification system (Gibco-BRL) according to the manufacturer’s protocol. The size of the primer extension product was estimated by parallel sequencing reactions on pHKU103 by using the same primer. The transcriptional start site was determined with the ALF-express DNA sequencer.
Southern blot analysis.
The enhanced chemiluminescence method (Amersham) was used for Southern hybridization. DNA fragments were labelled according to the manufacturer’s protocol and hybridized to DNA-containing Hybond-N membranes.
Nucleotide sequence accession number.
The nucleotide sequences presented here have been submitted to the EMBL database under accession no. AJ005843.
RESULTS
Cloning of the structural gene (chd1) for the cryptic haloacid dehalogenase from MBA4.
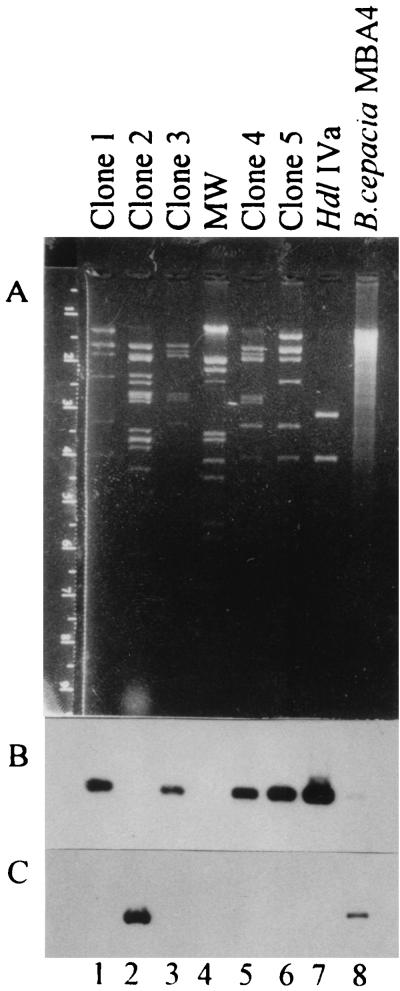
Among 1,200 ampicillin-resistant transfectants, five putative dehalogenase-positive clones, which produced white precipitates upon addition of silver nitrate, were obtained. Total protein extracts were prepared from these putative dehalogenase-producing clones and analyzed by activity-stained PAGE. Clones 1, 3, 4, and 5 produced a dehalogenase migrating similarly to DehIVa. On the other hand, clone 2 produced a novel dehalogenase migrating slower than DehIVa (data not shown). This dehalogenase was tentatively named Chd1 to indicate its cryptic nature.
The corresponding plasmids of the clones were isolated and cut with EcoRI restriction endonuclease. Figure 1A shows that they give different restriction patterns, indicating their independence in origin (lanes 1 to 3, 5, and 6). In order to eliminate DehIVa-producing clones, Southern blot analysis was carried out. Figure 1B shows that clones 1, 3, 4, and 5 (lanes 1, 3, 5, and 6, respectively) exhibited positive signals when hybridized with a probe containing the hdlIVa gene. This result suggested that four out of the five dehalogenase-producing clones contained DehIVa. Only clone 2 (lane 2) did not exhibit any hybridization signal. The structural gene for Chd1 was cloned (see below) and used as a probe. Figure 1C shows the specific hybridization of this chd1-containing probe with DNA of the various clones and with the total DNA of MBA4.
FIG. 1.
Agarose gel electrophoresis of plasmids isolated from various clones. (A) Ethidium bromide-stained gel. (B) Southern blot hybridized with probes containing the hdlIVa gene. (C) Southern blot hybridized with probes containing the chd1 gene. Lanes 1 to 3, 5, and 6 contain plasmid DNAs isolated from various clones and digested with EcoRI. Lane 7 contains a plasmid bearing the hdlIVa gene in a 1.6-kb EcoRI fragment. Lane 4 contains a molecular weight (MW) ladder (lambda DNA cut with HindIII and EcoRI). Lane 8 contains an MBA4 total DNA digested with EcoRI.
Subcloning of the DNA fragment for sequence analysis.
The plasmid pHKU100, isolated from clone 2, was ca. 50 kb in size. In order to identify the location for the structural gene, chd1, of the cryptic dehalogenase, the plasmid was subjected to further subcloning. Plasmid DNA was partially digested with Sau3AI and ligated to the BamHI site of pSP73 (Promega). The ligation products were transformed into E. coli TOP10F′, and transformants were screened for dehalogenase production. This produced plasmid pHKU101, which contains chd1 in a 10-kb fragment. Plasmid pHKU101 was then subjected to EcoRI digestion, and a 3-kb fragment conferring dehalogenase activity was cloned into pSP73 to form pHKU102. This plasmid was then cleaved with PstI, and a 1.7-kb fragment was cloned into pSP73 in both orientations to form pHKU103 and pHKU104. Subcloning of chd1 was confirmed by the hybridization of the 1.7-kb PstI fragment to DNA of all the subclones (data not shown). Cells harboring either pHKU103 or pHKU104 were dehalogenase positive (data not shown). This suggested that the 1.7-kb PstI fragment contains a promoter that is functional in E. coli.
Nucleotide sequence of the chd1 gene and flanking regions.
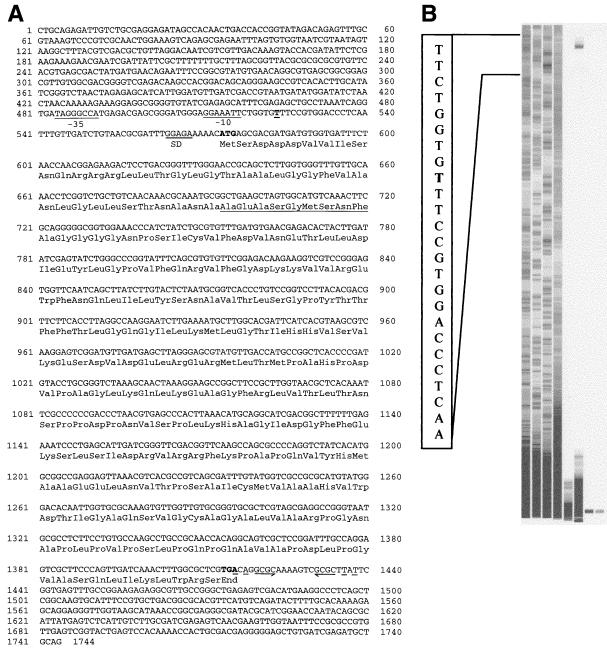
The nucleotide sequence of the 1,744-bp insert in pHKU103 was determined on plasmid DNA templates by an automated DNA sequencer (Pharmacia). Primers SP6 and T7, annealing to the nucleotide sequences flanking the insert, were used. Other primers corresponding to sequences found within the inserts were also used. The nucleotide sequence is given in Fig. 2A. An ORF consisting of 840 nucleotides started from the ATG beginning at position 574 and ended with codon TGA beginning at position 1414. This sequence would encode a protein of 280 amino acid residues with a predicted molecular weight of 29,939.
FIG. 2.
Structure of the chd1 gene. (A) Nucleotide sequence. The 1.7-kb PstI fragment contains an ORF of 840 nucleotides putatively encoding a protein of 280 amino acids. The deduced amino acid sequence is shown below the nucleotide sequence. Possible −10 and −35 consensus sequences of E. coli and a possible Shine-Dalgarno (SD) sequence are indicated. The transcriptional start site is in boldface and underlined. The underlined portion of the deduced protein sequence was confirmed by direct N-terminal sequencing. (B) Mapping of the transcriptional start site of chd1. The products of primer extension and a parallel sequencing reaction were analyzed by the ALF-express DNA sequencer. The gel image was generated by the sequencer. The sequence illustrated represents positions 516 to 540. The end of the extension product is determined to be the T (in boldface) at position 524.
Within the same reading frame another ATG codon starting at position 709 could give rise to a protein with a predicted molecular weight of 25,439. In order to define the initiation codon for Chd1, the N-terminal amino acid sequence of the recombinant protein was determined. The results showed that the nucleotide sequence corresponding to the N terminus started between the first and the second ATG codons (Fig. 2A).
A postulated Shine-Dalgarno (GGAGA) region was found close to the first ATG of the gene, beginning at position 564. Figure 2B is an image of the primer extension result showing the start of the transcript at the T at position 524. Possible −35 and −10 consensus sequences beginning at positions 484 (TAGGGCCA) and 512 (GGAAATT) were detected. Downstream of the structural gene, a putative stem-loop structure was detected between nucleotides 1416 and 1438, followed by a stretch of T residues.
Enzyme purification and biochemical characteristics.
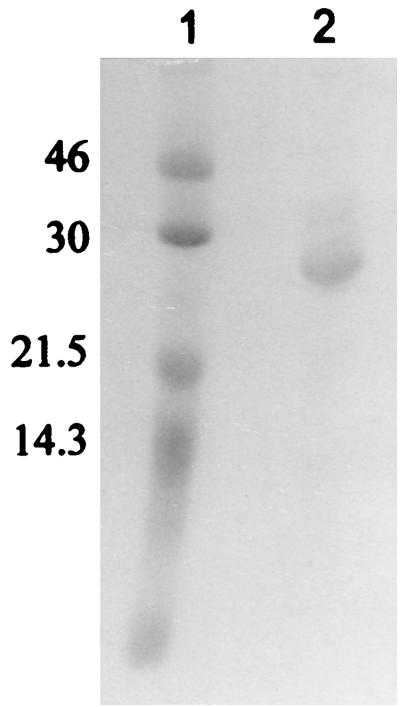
Total protein extracts were prepared from cells harboring pHKU103. The enzyme was purified 18.5-fold, with an overall yield of 15%, and the dehalogenase accounted for 5.5% of the total soluble cellular protein (Table 1). The purified enzyme preparation was considered to be 90% homogeneous when electrophoresed by denaturing PAGE (Fig. 3). The molecular mass of the dehalogenase was estimated to be 58 kDa by gel filtration chromatography on Sephacryl S-200, while in SDS–12% PAGE the apparent molecular mass was 27 kDa.
TABLE 1.
Purification of Chd1
| Purification step | Total protein (mg) | Total activity (U) | Sp act (μmol of Cl−/min/mg of protein) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 2,880 | 63,360 | 22 | 100 | |
| Protamine sulfate | 2,323 | 58,075 | 25 | 92 | 1.1 |
| (NH4)2SO4 | 1,211 | 44,807 | 37 | 71 | 1.7 |
| Sepharose Q | 96 | 19,968 | 208 | 32 | 9.5 |
| Hydroxylapatite | 24 | 9,840 | 410 | 16 | 18.5 |
FIG. 3.
SDS-PAGE of the purified dehalogenase on a 12% gel. Lane 1, molecular mass standards. Lane 2, 15 μg of the purified enzyme preparation. The gel was stained with Coomassie blue R-250. Numbers on the left are molecular masses, in kilodaltons.
The purified enzyme was most active towards MBA, with specific activities of 5.44, 1.5, 4.56, and 1.95 μmol of halide released/min/mg of protein for MBA, MCA, 2MBPA, and 2MCPA, respectively. When activities towards d- and l-2MCPA were considered individually, the enzyme was found to be active towards the l-isomer only. The enzyme was not active towards dichloroacetate or chlorobutane. The Michaelis constant (Km) for MBA at 25°C over the substrate range of 0.1 to 3.0 mM was 1.13 mM, and a Vmax of 30 μmol/min/mg was calculated.
The pH curve was bell shaped, with optimum activity at pH 6.5. The enzyme was also sensitive to heat treatment. A preincubation at 35°C for 15 min abolished 20 to 25% of the enzyme activity. The enzyme was not inactivated by incubation with 1 mM EDTA, CaCl2, MnCl2, NiCl2, ZnCl2, hydroxylamine, iodoacetate, N-ethylmaleimide, or FeSO4 but was effectively inhibited by HgCl2. The enzyme was partially inhibited by CuSO4 (41%), RbCl2 (70%), and ρ-mercuric chlorobenzoate (87%).
DISCUSSION
In this paper, we have reported the cloning and characterization of a cryptic dehalogenase, Chd1, from B. cepacia MBA4. This dehalogenase was not detected previously in MBA4 as visualized by activity-stained PAGE, which can differentiate dehalogenases according to their electrophoretic mobilities. In previous experiments using continuous-culture techniques, at least two other dehalogenases were detected (41). The genes for these previously identified dehalogenases were not isolated in the present study. It is likely that many of the bacterial promoters did not function properly in E. coli, leading to a lack of expression. It is common for dehalogenases expressed in soil isolates to not be expressed in E. coli (16, 25). Moreover, many natural isolates do contain unexpressed cryptic dehalogenases (16, 19, 25).
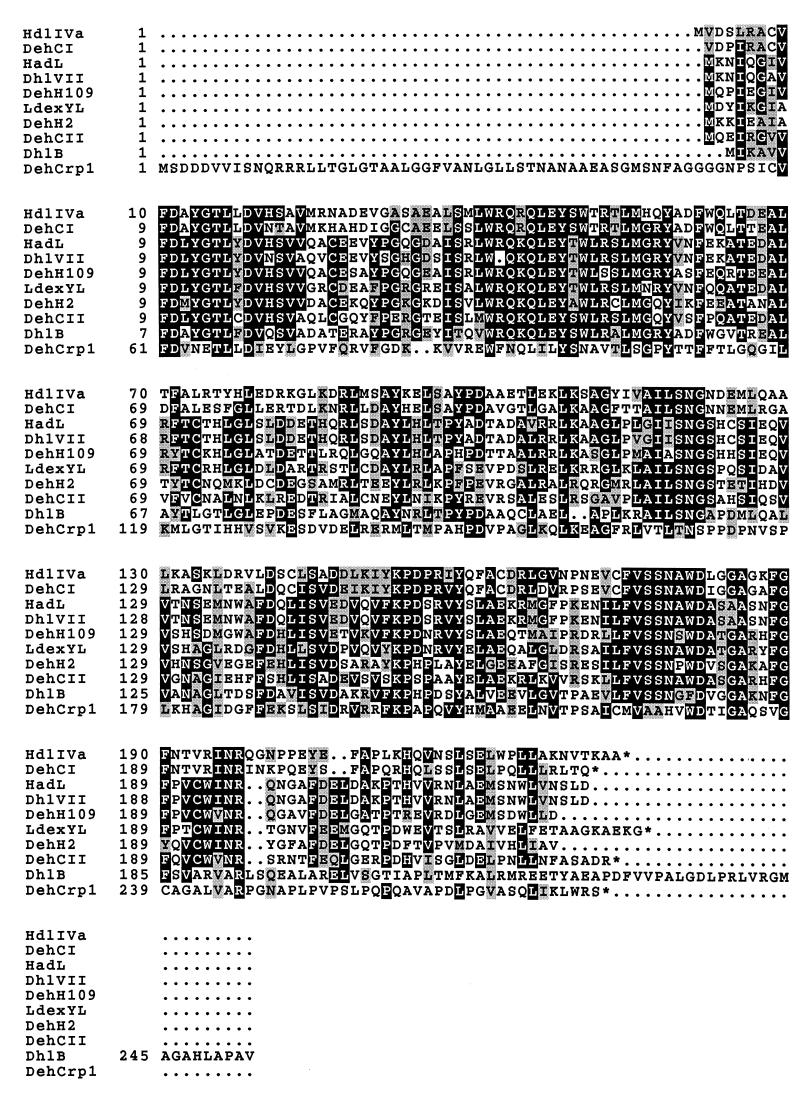
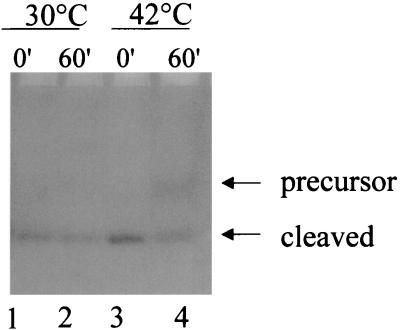
We have purified and characterized Chd1. It is a 2-haloacid dehalogenase with substrate specificity towards the l-isomer only. This protein, however, has some unique properties. Unlike previously isolated dehalogenases for which the optimal pH is alkaline (9, 11, 12, 24, 27, 30, 31, 46), the present enzyme is most active at pH 6.5. Comparison of the ORF for Chd1 with those for other dehalogenases (Fig. 4) shows that Chd1 possesses a long N-terminal peptide not found in other dehalogenases. Primer extension results confirmed that the transcript is sufficient to provide a polypeptide to start at the first AUG of the ORF. N-terminal amino acid sequence determination also confirmed that the protein starts upstream of the second in-frame methionine. The putative N-terminal sequence also exhibited the signal peptide property of periplasmic enzymes at Ser-Thr-Asn-Ala-Asn-Ala-↓-Ala-Glu-Ala, where the arrow indicates the postulated cleavage site (34). This fits well with the N-terminal amino acid sequence of the purified protein, which starts at Ala-Glu-Ala. Periplasmic proteins were isolated from cells harboring pHKU103 by a quantitative method (1). This protein fraction was shown to contain dehalogenase activity (data not shown). Plasmid pHKU103 was also transformed into E. coli strain IT41 (20), which contains a temperature-sensitive leader peptidase gene. Figure 5 shows that precursor molecules of Chd1 were detected, with a concomitant decrease in the cleaved protein, in cells grown at a nonpermissive temperature (compare lanes 3 and 4). These data strongly suggested that Chd1 is a periplasmic protein.
FIG. 4.
Comparison of haloacid dehalogenases. The amino acid sequences of haloacid dehalogenases from B. cepacia MBA4 (DehIVa and Chd1 [DehCrp1]) (accession no. Q51645 [29; also this study]), Pseudomonas sp. strains CBS3 (DehCI and DehCII) (accession no. P24069 and P24070 [33]) and YL (LdexYL) (accession no. Q53464 [30]), P. putida AJ1 (HadL) (accession no. A44830 [18]) and 109 (DehH109) (accession no. Q59728 [19]), X. autotrophicus GJ10 (DhlB) (accession no. Q60099 [41]), P. fluorescens (DhlVII) (accession no. Q59666 [16]), and Moraxella sp. strain B (DehH2) (accession no. Q01399 [20]) are aligned. The numbers on the left are the residue numbers of each amino acid sequence. The conserved residues are boxed and shaded.
FIG. 5.
Accumulation of precursor proteins in a temperature-sensitive leader peptidase mutant. IT41(pHKU103) cells were grown at 30°C until an optical density at 600 nm of 0.55 was reached and then were divided into two aliquots. The two cultures were then incubated at 30 or 42°C. Total protein extracts were prepared from samples taken at 0 and 60 min and analyzed on an activity-stained gel. The substrate used for visualization of the dehalogenase was MBA. “Cleaved” refers to cleaved dehalogenase.
Activity-stained PAGE analysis indicated that Chd1 has a smaller relative mobility value than does DehIVa of MBA4. DehIVa is a dimeric protein of 45 kDa with a subunit size of 23 kDa (43). SDS-PAGE analysis of purified Chd1 suggested that it has a subunit size of ca. 27 kDa. Its shorter migration distance in activity-stained PAGE and the gel filtration chromatography results indicate that Chd1 is most likely a dimer consisting of two identical subunits. Thus far, most of the dehalogenases isolated are dimers (21, 22, 29, 30, 43), although monomeric and tetrameric dehalogenases have also been detected in nature (21, 24, 28, 44).
The GCG program FASTA has been used to search for sequences similar to the chd1 gene sequence. No significant homology was discovered among sequences in the EMBL database. However, when the National Center for Biotechnology Information program BLASTP was used to search for similarity with the predicted sequence of Chd1, a few dehalogenases showing significant amino acid sequence homology were obtained. The observed lack of DNA sequence homology among the dehalogenases is consistent with the hypothesis that the enzymes resulted from specialization of existing hydrolases (23, 25, 43, 46).
The protein sequences of these dehalogenases were then compared with the predicted Chd1 amino acid sequences by using the GCG program BESTFIT. The gap creation penalty and the gap extension penalty set for the BESTFIT program were 3 and 1, respectively. The similarities with the other dehalogenases are as follows: B. cepacia MBA4 (32), 49% (HdlIVa); Pseudomonas sp. strain CBS3 (36), 50% (DehCI) and 42% (DehCII); Pseudomonas sp. strain YL (33), 48%; P. putida AJ1 (21), 47% (HadL); P. putida 109 (22), 45% (DehH109); Xanthobacter autotrophicus GJ10 (44), 49% (DhlB); Pseudomonas fluorescens (18), 47% (DhlVII); and Moraxella sp. strain B (23), 45% (DehH2). Phylogenetic analysis was performed by using the PHYLIP package (version 3.5). A phylogenetic tree of haloacid dehalogenases was constructed by the neighbor-joining method (35), with 100 bootstrapped replicates (10). All the dehalogenases analyzed were closely related to each other, with Chd1 most closely related to DehCI of Pseudomonas sp. strain CBS3 and to DehIVa (data not shown).
Haloacid dehalogenases have also been isolated in other types of bacteria, such as P. putida (2), Rhizobium (7), Alcaligenes xylosoxidans subsp. denitrificans (6), and Agrobacterium tumefaciens (37). However, no significant homologies were detected by comparing the nucleotide or the deduced amino acid sequences of these enzymes with those in the EMBL database (5, 7). The molecular characterization of dehalogenases awaits future investigation.
ACKNOWLEDGMENTS
We thank the Department of Zoology for determining the N-terminal amino acid sequence of the purified protein. We also thank J. Felsenstein for the PHYLIP package and Y. Nakamura for E. coli IT41.
This work was supported by grant from the Hong Kong Research Grants Council. L.S. thanks the University of Hong Kong for a studentship.
REFERENCES
- 1.Ames G F-L, Prody C, Kustu S. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J Bacteriol. 1984;160:1181–1183. doi: 10.1128/jb.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth P T, Bolton L, Thomson J C. Cloning and partial sequencing of an operon encoding two Pseudomonas putida haloalkanoate dehalogenases of opposite stereospecificity. J Bacteriol. 1992;174:2612–2619. doi: 10.1128/jb.174.8.2612-2619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann J G, Sanik J. Determination of trace amounts of chlorine in naphtha. Anal Biochem. 1957;29:241–243. [Google Scholar]
- 4.Boorstein W R, Craig E A. Primer extension analysis of RNA. Methods Enzymol. 1989;180:347–369. doi: 10.1016/0076-6879(89)80111-9. [DOI] [PubMed] [Google Scholar]
- 5.Brokamp A, Schmidt F R J. Survival of Alcaligenes xylosoxidans degrading 2,2-dichloropropionate and horizontal transfer of its halidohydrolase gene in a soil microcosm. Curr Microbiol. 1991;22:299–306. [Google Scholar]
- 6.Brokamp A, Happe B, Schmidt F R J. Cloning and nucleotide sequence of a d,l-haloalkanoic acid dehalogenase encoding gene from Alcaligenes xylosoxidans ssp. denitrificans ABIV. Biodegradation. 1997;7:383–396. doi: 10.1007/BF00056422. [DOI] [PubMed] [Google Scholar]
- 7.Cairns S S, Cornish A, Cooper R A. Cloning, sequencing and expression in Escherichia coli of two Rhizobium sp. genes encoding haloalkanoate dehalogenases of opposite stereospecificity. Eur J Biochem. 1996;235:744–749. doi: 10.1111/j.1432-1033.1996.t01-1-00744.x. [DOI] [PubMed] [Google Scholar]
- 8.Cohen S N, Chang A C Y, Shu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of E. coli by R-factor DNA. Proc Natl Acad Sci USA. 1972;69:2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies J I, Evans W C. The elimination of halide ions from aliphatic halogen-substituted organic acids by an enzyme preparation from Pseudomonas dehalogenans. Biochem J. 1962;82:50–51. [Google Scholar]
- 10.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Goldman P. The enzymatic cleavage of the carbon-fluorine bond in fluoroacetate. J Biol Chem. 1965;240:3434–3438. [PubMed] [Google Scholar]
- 12.Goldman P, Milne G W A, Keister D B. Carbon-halogen bond cleavage. III. Studies on bacterial halidohydrolases. J Biol Chem. 1968;243:428–434. [PubMed] [Google Scholar]
- 13.Hall B G, Yokoyama S, Calhoun D H. Role of cryptic genes in microbial evolution. Mol Biol Evol. 1983;1:109–124. doi: 10.1093/oxfordjournals.molbev.a040300. [DOI] [PubMed] [Google Scholar]
- 14.Hardman D J. Biotransformation of halogenated compounds. Crit Rev Biotechnol. 1991;11:1–40. doi: 10.3109/07388559109069182. [DOI] [PubMed] [Google Scholar]
- 15.Hardman D J, Slater J H. The dehalogenase complement of a soil pseudomonad grown in closed and open cultures on haloalkanoic acids. J Gen Microbiol. 1981;127:399–405. [Google Scholar]
- 16.Hill K E, Marchesi J R, Weightman A J. Investigation of two evolutionarily unrelated halocarboxylic acid dehalogenase gene families. J Bacteriol. 1999;181:2535–2547. doi: 10.1128/jb.181.8.2535-2547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 18.Honnens, E., R. Reiting, A. Brokamp, and F. J. R. Schmidt. 1995. EMBL accession no. X94147.
- 19.Hope S J, Slater J H. Cryptic dehalogenase and chloroamidase genes in Pseudomonas putida and the influence of environmental conditions on their expression. Arch Microbiol. 1995;163:57–64. doi: 10.1007/BF00262204. [DOI] [PubMed] [Google Scholar]
- 20.Inada T, Court D L, Ito K, Nakamura Y. Conditionally lethal amber mutations in the leader peptidase gene of Escherichia coli. J Bacteriol. 1989;171:585–587. doi: 10.1128/jb.171.1.585-587.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones D H A, Barth P T, Byrom D, Thomas C M. Nucleotide sequence of the structural gene encoding a 2-haloalkanoic acid dehalogenase of Pseudomonas putida AJ1 and purification of the encoded protein. J Gen Microbiol. 1992;138:675–683. doi: 10.1099/00221287-138-4-675. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki H, Toyama T, Maeda T, Nishino H, Tonomura K. Cloning and sequence analysis of a plasmid encoded 2-haloacid dehalogenase gene from Pseudomonas putida no. 109. Biosci Biotechnol Biochem. 1994;58:160–163. doi: 10.1271/bbb.58.160. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki H, Tsuda K, Matsushita I, Tonomura K. Lack of homology between two haloacetate dehalogenase genes encoded on a plasmid from Moraxella sp. strain B. J Gen Microbiol. 1992;138:1317–1323. doi: 10.1099/00221287-138-7-1317. [DOI] [PubMed] [Google Scholar]
- 24.Klages U, Krauss S, Lingens F. 2-Haloacid dehalogenase from a 4-chlorobenzoate-degrading Pseudomonas spec. CBS3. Hoppe-Seyler’s Z Physiol Chem. 1983;364:529–535. doi: 10.1515/bchm2.1983.364.1.529. [DOI] [PubMed] [Google Scholar]
- 25.Köhler R, Brokamp A, Schwarze R, Reiting R H, Schmidt F R J. Characteristics and DNA-sequence of a cryptic haloalkanoic acid dehalogenase from Agrobacterium tumefaciens RS5. Curr Microbiol. 1998;36:96–101. doi: 10.1007/s002849900286. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Little M, Williams P A. A bacterial halidohydrolase: its purification, some properties and its modification by specific amino acid reagents. Eur J Biochem. 1971;21:99–109. doi: 10.1111/j.1432-1033.1971.tb01445.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu J-Q, Kurihara T, Quamrul Hasan A K M, Nardi-Dei V, Koshikawa H, Esaki N, Soda K. Purification and characterization of thermostable and nonthermostable 2-haloacid dehalogenases with different stereospecificities from Pseudomonas sp. strain YL. Appl Environ Microbiol. 1994;60:2389–2393. doi: 10.1128/aem.60.7.2389-2393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mörsberger F-M, Müller R, Otto M K, Lingens F, Kulbe K D. Purification and characterization of 2-halocarboxylic acid dehalogenase II from Pseudomonas spec. CBS3. Biol Chem Hoppe-Seyler. 1991;372:915–922. doi: 10.1515/bchm3.1991.372.2.915. [DOI] [PubMed] [Google Scholar]
- 30.Motosugi K, Esaki N, Soda K. Purification and properties of 2-haloacid dehalogenase from Pseudomonas putida. Agric Biol Chem. 1982;46:837–838. doi: 10.1128/jb.150.2.522-527.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motosugi K, Esaki N, Soda K. Purification and properties of a new enzyme, dl-2-haloacid dehalogenase, from Pseudomonas sp. J Bacteriol. 1982;150:522–527. doi: 10.1128/jb.150.2.522-527.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murdiyatmo U, Asmara W, Tsang J S H, Baines A J, Bull A T, Hardman D J. Molecular biology of the 2-haloacid halidohydrolase IVa from Pseudomonas cepacia MBA4. Biochem J. 1992;284:87–93. doi: 10.1042/bj2840087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nardi-Dei V, Kurihara T, Okamura T, Liu J-Q, Koshikawa H, Ozaki H, Terashima Y, Esaki N, Soda K. Comparative studies of genes encoding thermostable l-2-halo acid dehalogenase from Pseudomonas sp. strain YL, other dehalogenases, and two related hypothetical proteins from Escherichia coli. Appl Environ Microbiol. 1994;60:3375–3380. doi: 10.1128/aem.60.9.3375-3380.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35a.Sam, L., and J. S. H. Tsang. Unpublished results.
- 36.Schneider B, Müller R, Frank R, Lingens F. Complete nucleotide sequences and comparison of the structural genes of two 2-haloalkanoic acid dehalogenases from Pseudomonas sp. strain CBS3. J Bacteriol. 1991;173:1530–1535. doi: 10.1128/jb.173.4.1530-1535.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarze R, Brokamp A, Schmidt F R J. Isolation and characterization of dehalogenases from 2,2-dichloropropionate-degrading soil bacterium. Curr Microbiol. 1997;34:103–109. doi: 10.1007/s002849900152. [DOI] [PubMed] [Google Scholar]
- 38.Senior E, Bull A T, Slater J H. Enzyme evolution in a microbial community growing on the herbicide Dalapon. Nature. 1976;263:476–479. doi: 10.1038/263476a0. [DOI] [PubMed] [Google Scholar]
- 39.Slater J H, Bull A T, Hardman D J. Microbial dehalogenation of halogenated alkanoic acids, alcohols and alkanes. Adv Microb Physiol. 1997;38:133–176. doi: 10.1016/s0065-2911(08)60157-5. [DOI] [PubMed] [Google Scholar]
- 40.Slater J H, Hope S J. Interactions between populations of Pseudomonas putida leading to the expression of a cryptic dehalogenase gene (dehII) World J Microbiol Biotechnol. 1995;11:186–192. doi: 10.1007/BF00704646. [DOI] [PubMed] [Google Scholar]
- 41.Tsang J S H. Ph.D. thesis. Canterbury, United Kingdom: University of Kent; 1987. [Google Scholar]
- 42.Tsang J S H, Hardman D J, Bull A T. Cloning and expression of 2-haloalkanoic acid dehalogenase of Pseudomonas cepacia MBA4 in Escherichia coli and in Pseudomonas putida. In: Chang S T, Chan K-Y, Woo N Y S, editors. Recent advances in biotechnology and applied biology. Hong Kong: Chinese University Press; 1988. pp. 231–239. [Google Scholar]
- 43.Tsang J S H, Sallis P J, Bull A T, Hardman D J. A monobromoacetate dehalogenase from Pseudomonas cepacia MBA4. Arch Microbiol. 1988;150:441–446. [Google Scholar]
- 44.Van der Ploeg J, van Hall G, Janssen D B. Characterization of the haloacid dehalogenase from Xanthobacter autotrophicus GJ10 and sequencing of the dhlB gene. J Bacteriol. 1991;173:7925–7933. doi: 10.1128/jb.173.24.7925-7933.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weightman A J, Slater J H. Selection of Pseudomonas putida strains with elevated dehalogenase activities by continuous culture growth on chlorinated alkanoic acids. J Gen Microbiol. 1980;121:187–193. [Google Scholar]
- 46.Weightman A J, Weightman A L, Slater J H. Stereospecificity of 2-monochloropropionate dehalogenation by the two dehalogenases of Pseudomonas putida PP3: evidence for two different dehalogenation mechanisms. J Gen Microbiol. 1982;128:1755–1762. doi: 10.1099/00221287-128-8-1755. [DOI] [PubMed] [Google Scholar]