Abstract
The parCBA operon of the 3.2-kb stabilization region of plasmid RK2 encodes three cotranslated proteins. ParA mediates site-specific recombination to resolve plasmid multimers, ParB has been shown to be a nuclease, and the function of ParC is unknown. In this study ParB was overexpressed by cotranslation with ParC in Escherichia coli by using a plasmid construct that contained the parC and parB genes under the control of the T7 promoter. Purification was achieved by treatment of extracts with Polymin P, followed by ammonium sulfate precipitation and heparin and ion-exchange chromatography. Sizing-column analysis indicated that ParB exists as a monomer in solution. Analysis of the enzymatic properties of purified ParB indicated that the protein preferentially cleaves single-stranded DNA. ParB also nicks supercoiled plasmid DNA preferably at sites with potential single-stranded character, like AT-rich regions and sequences that can form cruciform structures. ParB also exhibits 5′→3′ exonuclease activity. This ParB activity on a 5′-end-labeled, double-stranded DNA substrate produces a 3′,5′-phosphorylated dinucleotide which is further cleaved to a 3′,5′-phosphorylated mononucleotide. The role of the ParB endonuclease and exonuclease activities in plasmid RK2 stabilization remains to be determined.
The broad-host-range IncPα plasmid RK2 (identical to RP4) is capable of being stably maintained in a wide variety of gram-negative hosts (reviewed in reference 42). Several regions of RK2 have been identified which ensure stable maintenance of the plasmid despite a relatively low copy number of five to eight copies per chromosome in Escherichia coli (13). The psa (postsegregational arrest) locus causes the growth inhibition of host cells from which the plasmid is lost (21). The IncC and KorB proteins, encoded by the central control region (Ctl) of the IncPα plasmid RK2 and related IncPβ plasmids (25), have been proposed to play a role in plasmid partitioning (27). Perhaps the most important contributor to RK2 stability, however, is the 3.2-kb par region, which has been shown to stabilize plasmids in a host- and vector-independent manner (31, 36). It encodes five proteins on two divergent operons (16, 30, 36), parCBA and parDE, which are autoregulated by ParA and ParD, respectively (8, 11). The parDE operon (8, 20, 40) encodes an effective proteic postsegregational killing system similar to the ccd locus of plasmid F (4, 19), the parD/pem locus of R1/R100 (5, 32, 43), and the phd-doc system of prophage P1 (24). In these systems, the plasmid expresses two proteins, a toxin and an antidote protein. It has been proposed in each case that the antidote protein is less stable than the toxin protein, resulting in a release of toxin activity and the death of cells that have lost the plasmid.
It has been proposed that the parCBA operon of RK2 encodes a partitioning region (16, 30), as has been shown for par of P1 (3, 47), sop of F (26), and parA of R1/R100 (7, 15). While the parCBA operon is effective in stabilizing the plasmid, particularly in certain bacterial strains (9, 10, 39), there is no direct evidence to support the involvement of this region in a physical partitioning process. The ParA protein, encoded by the parCBA operon, is part of a multimer resolution system which is related to the Tn3 family of resolvases (12, 16). ParA catalyzes the site-specific recombination of plasmid multimers, acting at a specific res site which is situated between the parCBA and parDE operons. The res site is similar in structure and exhibits limited DNA sequence identity to in cis sites found in Tn3 resolvase systems. In in vitro assays in which ParA acts on artificial dimer molecules containing two res sites, the product is two monomers linked in the form of a catenane. The ParA protein and its res site alone will provide a substantial level of stabilization of plasmid RK2, but in certain E. coli hosts the products of the parB and parC genes enhance this stabilizing activity (9). Grohmann et al. have reported that ParB is a Ca2+-dependent nuclease, based on work with partially purified preparations (17). Little is known about the function of the ParC protein. The expression of the ParC, ParB, and ParA proteins is translationally coupled (16).
The amino acid sequence of the ParB protein (16) shows significant similarity to several nucleases, including the chromosomally encoded nuclease RuvC from E. coli (see reference 45 for a review), and the staphylococcal nuclease (38), as well as nucleases encoded by the plasmids pKM101 (28), pSa (6), and R100 (46). While these proteins all show some structural similarities to ParB, their diverse roles in vivo provide little insight into a role for ParB in the stabilization of RK2. Two of the ParB-homologous proteins (encoded by nuc of pKM101 and orfA of R100) are encoded by genes located in tra regions, suggesting an involvement in conjugal transfer. Two others, the nucleases of Staphylococcus aureus and plasmid pSa, are involved in extracellular degradation of nucleic acids. RuvC, which has the least homology with ParB (17), is involved in the processing of DNA molecules joined as a result of recombination events.
The aim of this study was to purify ParB and characterize in further detail the biochemical properties of the ParB nuclease. Here we report the extensive purification of ParB and present data demonstrating that this protein is a monomer in solution, prefers single-stranded DNA as a substrate, and cleaves supercoiled DNA in regions of high potential single strandedness. ParB also possesses a 5′→3′ exonuclease activity, generating di- and mononucleotides as reaction products.
MATERIALS AND METHODS
Materials.
Restriction enzymes, Klenow fragment of E. coli DNA polymerase, shrimp alkaline phosphatase, T4 DNA ligase, T4 DNA polymerase, T4 polynucleotide kinase, Taq polymerase, Sequenase 2.0, exonuclease III, and T7 gene 6 exonuclease were obtained from commercial suppliers and used according to the manufacturers’ instructions. Antibiotics were obtained from Sigma Chemical Co. (St. Louis, Mo.).
Bacterial strains and plasmids.
E. coli XLI Blue MRF′, pBluescript II SK(−), and bacteriophage VCS-M13 (Stratagene, La Jolla, Calif.) were used for single-stranded DNA production. E. coli BL21(DE3) (41) was used for overexpression of ParB from plasmid pEJ18. E. coli DH5 (18) was used for plasmid constructions. E. coli strains were grown in Lennox L broth (Gibco-BRL, Gaithersburg, Md.), and selection for plasmids was performed with 250 μg of penicillin per ml when necessary.
DNA manipulations were performed by standard procedures (34). Plasmid pRR71 was used for the construction of plasmid pEJ16. pRR71 (30), which contains the 3.2-kb stabilization region inserted into the HindIII and KpnI sites of pUC19, was digested with AvaI to remove a 422-bp fragment of the parA gene and treated with Klenow fragment to generate blunt ends. The plasmid was then religated with an 8-bp BamHI linker, generating pEJ16. Plasmid pEJ17 was produced by digesting pEJ16 with BamHI, which cleaves at the linker site and at a BamHI site within the multiple cloning site. The fragment containing the par region was then ligated into the BamHI site of pET11c (Novagen, Madison, Wis.) oriented with the parB gene closest to the terminator sequence. For the construction of pEJ18, pEJ17 was partially digested with StyI, followed by gel purification of singly cut molecules and redigestion with NdeI. When the desired StyI site is cleaved, the NdeI digestion removes the parDE operon and the sequence encoding the N terminus of the parC product. A modified N terminus was introduced by ligating a DNA linker consisting of annealed oligonucleotides with the sequences 5′-TATGGGTATACCCAATTTGAC-3′ and 5′-ACCCATATGGGTTAAACTGGTTC-3′ (synthesized by OPERON, Alameda, Calif.), respectively, to produce pEJ18, which contains parCB and a truncated parA gene downstream of the T7 promoter. For expression purposes, pEJ18 was then transformed into BL21(DE3), which contains the T7 RNA polymerase gene under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter.
Purification of ParB.
One liter of L broth was inoculated with a 30-ml overnight culture of BL21(DE3)(pEJ18) and grown at 30°C with penicillin and shaking until an optical density at 600 nm of 0.8 was reached. The culture was then induced by the addition of IPTG to a final concentration of 1 mM, and growth was continued for an additional 3 h. Growth and induction were performed in nonbaffled flasks, as a lower aeration aided the expression of ParB. After induction, cells were harvested and washed once in 50 ml of 20 mM sodium phosphate (pH 7.6), and the pellet was resuspended in buffer A (20 mM Tris-HCl [pH 7.4], 10% glycerol, 25 mM KCl). After lysis by sonication at 4°C and centrifugation at 10,000 × g for 30 min, the supernatant was mixed with Polymin P (Sigma) to a final concentration of 0.75%. The mixture was then recentrifuged at 8,000 × g for 10 min, and the supernatant was retained. Polymin P was then removed by performing two successive 80% ammonium sulfate precipitations at 4°C, each followed by resuspension in buffer B (20 mM HEPES [pH 7.4], 10% glycerol, 50 mM KCl). After overnight dialysis against buffer B at 4°C, the protein solution was then filtered (Acrodisc [0.2-μm pore size]; Gelman Sciences, Ann Arbor, Mich.) and loaded onto a 7-ml heparin column equilibrated with buffer B. Proteins were eluted at 4°C with a linear 50 to 500 mM KCl gradient in buffer B, and the ParB-containing fractions were pooled and dialyzed against buffer B. The proteins were again filtered and loaded onto a Mono-S cation-exchange column (HR 5/5; Pharmacia, Uppsala, Sweden). The proteins were then eluted by using fast protein liquid chromatography (FPLC) and a 50 to 500 mM KCl gradient in buffer B at 4°C. The ParB-containing fractions were pooled. With this procedure, 1 liter of culture typically yielded about 0.5 mg of ParB which was greater than 98% pure. The procedure was monitored by polyacrylamide gel electrophoresis (PAGE) (14% Tris-glycine gel; Novex, San Diego, Calif.) and staining with Coomassie brilliant blue R. Protein concentrations were determined by using the Bio-Rad (Hercules, Calif.) protein assay with the supplied protocol.
Sizing column.
Four hundred micrograms of purified ParB was loaded onto a fast protein liquid chromatography sizing column (Superose-12; Pharmacia) equilibrated with buffer B along with 400 μg each of cytochrome c (12.5 kDa), carbonic anhydrase (29 kDa), and ovalbumin (45 kDa). Fractions were then collected and visualized by PAGE (14% Tris-glycine gel; Novex) and Coomassie blue staining.
Nuclease assay.
Nuclease assays were carried out by using procedures modified from those described previously (17). Time course nuclease assays were performed by treating 200 ng of supercoiled pUC19 with 12.5 ng of purified ParB in ParB nuclease buffer (50 mM Tris base [pH 8.0], 3 mM β-mercaptoethanol, 5 mM CaCl2) in a final volume of 30 μl. Reaction mixtures were incubated at 37°C for 0.5, 1, 2, 4, and 8 min, and reactions were stopped with 2 μl of 500 mM EDTA. Reaction products were electrophoresed in a 0.8% agarose gel and visualized by staining with ethidium bromide (EtBr).
Single-stranded pBluescript II SK(−) DNA was produced by using the strain XLI Blue [pBluescript II SK(−)] and helper phage VCS-M13 (Stratagene) by a protocol provided by Stratagene. Nuclease assays with double-stranded and single-stranded pBluescript II SK(−) were carried out by reacting 2 μg of the double- or single-stranded DNA in 30 μl of ParB nuclease buffer for 5 min at 37°C.
Restriction endonuclease assay.
Supercoiled pUC19 DNA was treated with ParB as described above for a sufficient time to produce the linear form of the DNA, which was then purified by gel electrophoresis and electroelution as described previously (34). Undigested pUC19, ParB-linearized pUC19, and pUC19 digested with EcoRI were then cleaved with SspI and DraI. Undigested pUC19 and ParB-linearized pUC19 were also digested with EcoRI. Reaction products were then analyzed by 0.8% agarose gel electrophoresis and visualized by EtBr staining. All digestions were performed by using the specifications of the manufacturer.
Primer extension.
pUC19 was treated with ParB as described above for sufficient time to produce the linear and open circular forms of DNA or was digested to completion with FspI and AlwNI. Linear and open circular DNA forms, as well as the 702-bp fragment from the FspI-AlwNI digest, were then gel purified, using the UltraClean system (Mo Bio Laboratories, Solana Beach, Calif.) for DNA purification. Four synthetic primers were then 32P labeled by treatment with T7 polynucleotide kinase and [γ-32P]ATP for use in sequencing reactions (Sequenase 2.0; Amersham, Cleveland, Ohio) and for primer extension. Primers 1428TSpUC (5′-GCAAGCAGCAGATTACG-3′) and 1293TSpUC (5′-CGGCTACACTAGAAGGACAGTATT-3′) were used in the analysis of the top strand of pUC19. Primers 1720BSpUC (5′-TCGTAGTTATCTACACG-3′) and 1484BSpUC (5′-GACCCCGTAGAAAAGATCAAAGG-3′) were used to analyze the bottom strand. Primer extension reactions were performed as previously described (23). Three hundred nanograms of ParB-treated linear, open circular, or FspI-AlwNI-digested DNA was treated with Taq polymerase in a final volume of 25 μl of a PCR buffer (100 mM Tris-HCl, 500 mM KCl 0.25 mM deoxynucleoside triphosphates, 1.5 mM MgCl2 [pH 8.3]) with 0.5 pmol of the appropriate primer. Reactions were performed for 15 cycles (1 min at 94°C, followed by 1 min at 42°C and 2 min at 72°C) except when primer 1484BSpUC was used, in which case the annealing temperature was increased to 60°C. Reaction products were then mixed with 12.5 μl of 3× formamide buffer (55 mM Tris-HCl [pH 8.0], 2 mM EDTA, 0.2% sodium dodecyl sulfate [SDS], 0.02% bromophenol blue, and 0.02% xylene cyanol in 100% deionized N′,N′-dimethyl formamide) and analyzed by denaturing PAGE, with visualization by autoradiography.
Internal stability determination.
The internal stability of pUC19 sequences was determined with the primer analysis application Oligo 5.0. The entire pUC19 sequence was analyzed in 10-bp windows.
Exonuclease assay.
The exonuclease activity of ParB was assayed by using a previously published procedure (35). A 5′-radiolabeled substrate to be used in the assay was made by first digesting pUC19 with BamHI and end labeling by treatment with T7 polynucleotide kinase and [γ-32P]ATP. The recessed 3′ end was then filled by treatment with Klenow fragment, and the DNA was further digested with EcoRI. A 20-bp labeled fragment was then purified by standard means (34). This procedure resulted in labeling of just one 5′ end of the double-stranded 20-bp molecule. A 1.2-pg portion of the labeled substrate was then treated with increasing amounts of purified ParB in ParB nuclease buffer in a final volume of 10 μl. Reactions were performed at 37°C for 5 min. Two other sets of reactions were performed by treating the substrate with increasing amounts of exonuclease III or T7 gene 6 exonuclease. Both of these reactions were carried out at 37°C for 5 min with buffers provided by the manufacturers. Reactions were stopped by adding 5 μl of formamide buffer, and the products were analyzed by electrophoresis through a 15% Tris-borate-EDTA gel under denaturing conditions followed by visualization by autoradiography.
For further characterization of reaction products, a substrate was generated by annealing synthetic oligonucleotides with the sequences 5′-AATTCGAGCTCGGTACCCGGGGATC-3′ (top) and 5′-GATCCCCGGGTACCGAGCTCGAATT-3′ (bottom) (synthesized by IDT). The oligonucleotides were purified by thin-layer chromatography (2), and the bottom oligonucleotide was 5′ radiolabeled by treatment with T7 polynucleotide kinase and [γ-32P]ATP. The oligonucleotides were then annealed and purified by acrylamide gel electrophoreses (34). This substrate was then treated with ParB as described above. The reaction products were subjected to thin-layer chromatography as previously described (29) by using polyethyleneimine cellulose and resolved with 1 M lithium chloride. Plates were not prerun and were obtained from EM Science (Gibstown, N.J.). Products were visualized by autoradiography, and the standards were visualized by short-wavelength UV shadowing.
Synthesis of pGp.
N-2-Isobutyryl-2′-deoxyguanidine (0.5 g, 1.48 mmol) was dissolved in 50 ml of anhydrous pyridine (Aldrich, Milwaukee, Wis.), dried, and then left under vacuum overnight. The nucleoside was redissolved in 50 ml of anhydrous pyridine, and phosphorous oxychloride (0.30 ml, 3.2 mmol; Aldrich) was added dropwise under argon. After 30 min, the solution was cooled to 0°C, and 50 ml of water was added dropwise with rapid stirring. After 30 min, the solution was dried to a yellow oil. Two hundred milliliters of concentrated ammonia was added, and the slurry was heated at 65°C for 16 h with rapid stirring. The final solution was dried to a yellow oil, which was dissolved in 100 ml of water and filtered. The solution was cooled to 0°C, and 10 ml of a 20% (wt/vol) barium acetate solution and 200 ml of absolute ethanol were added sequentially. The precipitate was collected by centrifugation and resuspended in 100 ml of water by adding approximately 5 g of 50W-X2 resin (hydrogen form; Bio-Rad). Ten milliliters of the resulting solution was added to a 2.5- by 10-cm DEAE column. 2′-Deoxyguanosine 3′,5′-phosphate (pGp) was resolved by using a linear gradient of ammonium acetate (0.06 to 6 M; 2.5 ml/min; 10-ml fractions). Peak fractions were identified by polyethyleneimine thin-layer chromatography (1 M LiCl) and combined. Barium acetate was added to make a final 2% (wt/vol) solution, and an equal volume of absolute ethanol was added. The precipitate was collected by centrifugation, dried, and redissolved in 2 ml of water by adding approximately 0.5 g of 50W-X2 resin (hydrogen form). The solution was filtered (0.1-μm pore size), and the final solution was reduced to 1 ml under reduced pressure.
RESULTS
Purification of ParB.
Several constructs were made to facilitate the overexpression of ParB. The parB gene alone was placed under the control of both T7 and tac promoters and also was modified by the addition of sequence encoding six histidine residues or by fusion with the glutathione S transferase gene at the end corresponding to the N terminus. In each of these cases, the level of expression of ParB after induction was poor (data not shown). Satisfactory expression, however, was achieved by the cotranslation of ParB and ParC, driven by the T7 promoter, using the plasmid pEJ18 in E. coli BL21(DE3). Under most growth conditions, BL21(DE3)(pEJ18) expresses a large quantity of ParC and very little ParB. Roughly equal amounts of ParB and ParC are produced, however, when the cells are grown in a nonbaffled flask to ensure low aeration and at 30°C before induction. Under these conditions, ParB is largely in the soluble fraction after cell lysis (Fig. 1). When overexpression was attempted with a plasmid construct with parB alone (17), a lower level of soluble ParB was produced, since most of the protein is in the insoluble fraction. Invariably, the ParC protein produced by any of the vectors and under any of the conditions tested so far was found in fast-sedimenting material, presumably inclusion bodies. After induction of ParC expression in the presence or absence of ParB, inclusion-like bodies are detectable in the host cell by light microscopy.
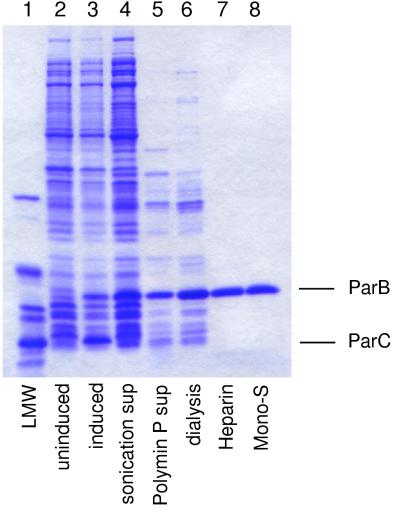
FIG. 1.
Purification of the ParB protein. Fractions of material from the purification steps described in Materials and Methods were analyzed by SDS-PAGE. The gels were stained with Coomassie brilliant blue R. The positions of ParB and ParC are indicated. Lane 1, molecular weight standards (LMW); lanes 2 and 3, total SDS-soluble protein of uninduced and induced cultures, respectively; lane 4, supernatant after sonication; lane 5, supernatant after treatment with Polymin P; lane 6, material after dialysis of the Polymin P supernatant; lanes 7 and 8, peak fractions after chromatography on heparin and Mono-S columns, respectively.
Early attempts to remove DNA from crude extracts by using a DEAE anion-exchange column were unsuccessful because of heavy losses due to the binding of ParB (calculated pI of 11.0) to DNA in the crude extract. For this reason, Polymin P was used to separate ParB from DNA in the early stages of purification. The nonspecific DNA binding activity of ParB was confirmed by gel mobility shift assays, which indicated that ParB bound to all of the nine linear DNA fragments generated from the par region by restriction enzyme cleavage (data not shown). The results of the purification procedure developed in this study are shown in Fig. 1. The various purification steps resulted in ParB preparations that were greater than 98% pure (Fig. 1).
ParB is a monomer.
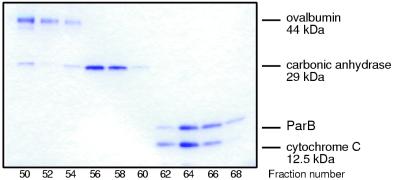
A sizing column was employed to determine if ParB exists as a monomer or as a multimer in solution. ParB was loaded onto a Superose-12 column along with samples of cytochrome c (12.5 kDa), carbonic anhydrase (29 kDa), and ovalbumin (45 kDa). ParB, with a calculated molecular mass of 20 kDa on the basis of the nucleotide sequence of the ParB gene (16), clearly exhibits exclusion properties between those of carbonic anhydrase and cytochrome c, having a molecular mass of approximately 17 kDa with respect to the referenced proteins (Fig. 2). These data indicate that ParB is a monomer in solution, a conclusion supported by the failure to detect higher forms of ParB by SDS-PAGE after treatment with glutaraldehyde (data not shown) and by the results of light-scattering studies carried out with this protein (22a).
FIG. 2.
Superose-12 gel filtration of purified ParB. ParB, ovalbumin (44 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.5 kDa) were mixed and subjected to Superose-12 gel filtration. The column fractions are indicated at the bottom, and the position of each protein is indicated on the right.
ParB enzymatic activity.
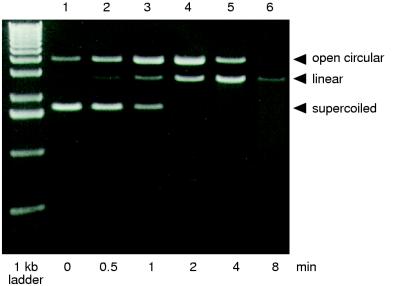
Several approaches were used to characterize the enzymatic activity of purified ParB. First, pUC19 DNA was treated with purified ParB for various time periods (Fig. 3). The pUC19 substrate consisted primarily of supercoiled DNA, with a small amount of the open circular form. When the DNA is treated with ParB, the amount of the supercoiled form is reduced, while the proportion of open circular DNA is increased, suggesting that ParB first acts to nick supercoiled DNA. With increased time, the linear form of pUC19 DNA appears, with a concomitant reduction of the open circular form. This is followed by the degradation of the linear DNA. These results are consistent with those seen previously with partially purified ParB (17) and indicate that ParB has endonuclease activity. Similar experiments were conducted with pUC19 plasmid constructs containing the 3.2-kb par region of RK2, which contains the parB gene. ParB displayed no significantly increased activity as a result of insertion of the par sequence into pUC19 (data not shown). As shown previously (17) the activity of the ParB protein is Ca2+ dependent. We also found that this activity is inhibited by Co2+ (data not shown).
FIG. 3.
Time course treatment of pUC19 DNA by ParB. Supercoiled pUC19 DNA was treated with ParB for 0, 0.5, 1, 2, 4, or 8 min (lanes 2 to 6, respectively) as described in Materials and Methods. The positions of the various forms of pUC19 are indicated.
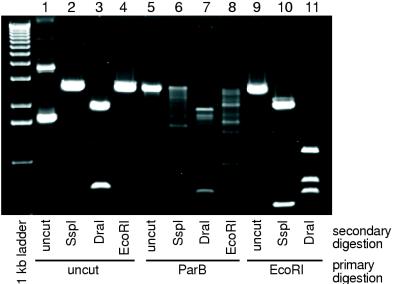
Experiments were also carried out to determine if purified ParB cleaved plasmid pUC19 at specific sites or randomly. Plasmid pUC19 DNA was first treated with ParB for a sufficient time to convert the supercoiled form to the linear form. The linear DNA was then gel purified and subjected to secondary digestion as described in Materials and Methods. When supercoiled pUC19 is digested with SspI, DraI, or EcoRI, either without prior ParB treatment or after digestion with EcoRI, the reaction products are of the expected size as determined by agarose gel electrophoresis (Fig. 4). When supercoiled pUC19 first is linearized by ParB treatment and then is digested with restriction endonucleases, however, only a small preference for any specific ParB site is observed. This is in contrast to previous experiments with plasmid pGMA30, a pUC derivative containing the RK2 par region (17), which suggested that ParB cleaves plasmid pGMA30 at a preferred site within the pUC sequence. The difference in cleavage properties exhibited by ParB with pUC19 and pGMA30 may be attributed to differences in topology between the two plasmids because of the dissimilarities in DNA sequence. As reported by Grohmann et al. (17), the number of preferred cleavage sites varied with different plasmids, but no specific influence on the site preference of ParB activity by the presence or absence of par DNA sequences was observed.
FIG. 4.
Restriction enzyme cleavage of ParB-linearized pUC19 DNA. Supercoiled DNA was untreated (lanes 1 to 4) or treated with ParB (lanes 5 to 8) or with EcoRI (lanes 9 to 11). Samples were then analyzed without an additional digestion (lanes 1 and 5) or after digestion with SspI (lanes 2, 6, and 10), DraI (lanes 3, 7, and 11), or EcoRI (lanes 4 and 8). Agarose gel electrophoresis was carried out as described in Materials and Methods.
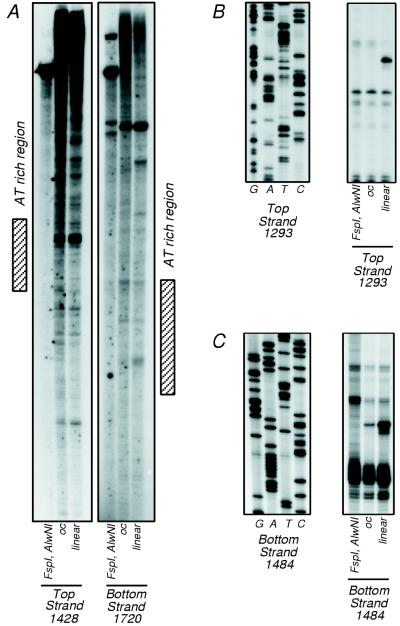
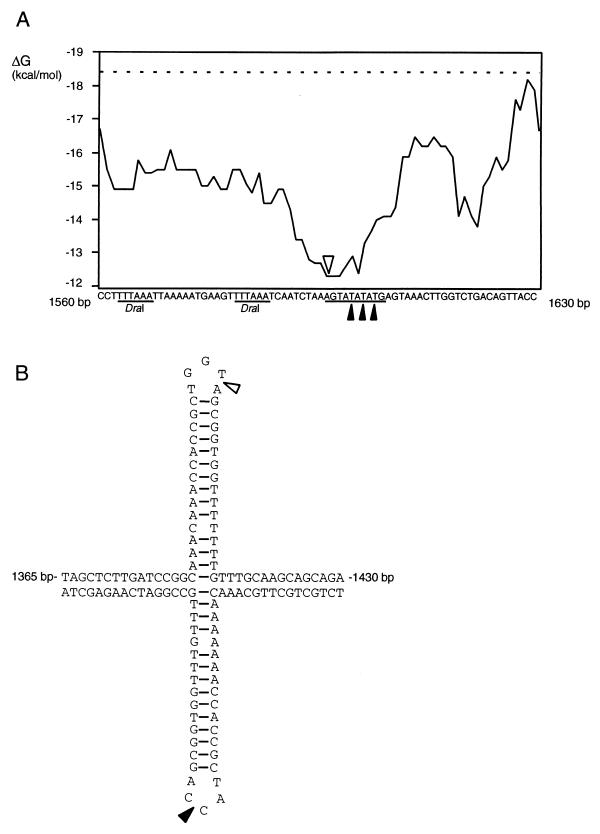
To further address the question of site preference, primer extension was used to localize sites of cleavage within pUC19 treated with ParB. Plasmid pUC19 DNA was treated with ParB for a sufficient time to produce both open circular and linear DNAs. Each of these products was purified separately from agarose gels and subjected to primer extension analysis. The results are shown in Fig. 5A. While one major band is visible, as expected, when a restriction enzyme fragment derived from pUC19 untreated with ParB was analyzed, a ladder of bands is visible when ParB-treated DNA was used as a template (Fig. 5A). This strongly suggests that ParB cleaves at many sites. Some increased ParB activity is visible, however, in areas of high A+T content (Fig. 5A). Analysis of the internal stability of 10-bp windows of the pUC19 sequence revealed that the preferred cleavage sites were in a very A+T-rich region, with a ΔG of −12.3 kcal/mol (Fig. 6A). Stability analysis showed that of the entire pUC19 sequence, which has an average stability of −18.4 kcal/mol for a 10-bp window, this region has the lowest stability and is the most likely to exist at some time in a single-stranded form. A second preferred site was discovered downstream of the A+T-rich region (Fig. 5B and C). While not in a region of A+T richness, this site lies between two indirect repeats with a high probability of forming a cruciform structure (Fig. 6B). This observation suggests that the preferential cleavage of this region by ParB is due to the potential single strandedness of the loop region of the cruciform.
FIG. 5.
Primer extension of ParB-treated supercoiled DNA. (A) pUC19 DNA was treated with FspI and AlwNI (lanes 1 and 4) or with ParB to produce open circular (oc) (lanes 2 and 5) or linear (lanes 3 and 6) DNA. Primer extension was then performed with radiolabeled primers against the top strand (lanes 1 to 3) and against the bottom strand (lanes 4 to 6) of the treated pUC19. The hatched boxes indicate the position of the AT-rich region. (B) Results of primer extension performed on the top strand of the FspI-AlwNI fragment of pUC19 and on ParB-treated open circular and linear DNAs with primer 1293TSpUC (right panel) and results of sequencing with the same primer (left panel). (C) Results of primer extension performed on the bottom strand of the FspI-AlwNI fragment of pUC19 and on ParB-treated open circular and linear DNAs with primer 1484BSpUC (right panel) and sequencing of this region with the same primer (left panel).
FIG. 6.
Preferred ParB-nicked sites in pUC19 DNA. (A) Sites of preferred ParB nicking adjacent to the DraI restriction enzyme sites. By using Oligo 5.0, the internal binding stabilities of 10-bp windows of base pairs within the pUC19 sequence were calculated. The nucleotide sequence of the DraI region is shown at the bottom. A point along the x axis of the graph corresponds to the stability of a 10-bp window beginning with the nucleotide shown directly below it. The position on the graph indicated with an open arrowhead corresponds to the 10-bp sequence underlined. The filled arrowheads indicate sites of preferred cleavage by ParB suggested by analysis of the top strand. The dashed line indicates the average stability of 10-bp windows of the entire pUC19 sequence. The positions of the DraI restriction sites are also indicated. (B) Site of preferred ParB nicking within the bla gene of pUC19. The sequences making up the cruciform structure and the positions on the pUC19 map are shown. The open arrowhead indicates the position of preferred ParB cleavage on the top strand. The filled arrowhead indicates the position of preferred ParB cleavage on the bottom strand.
Substrate specificity.
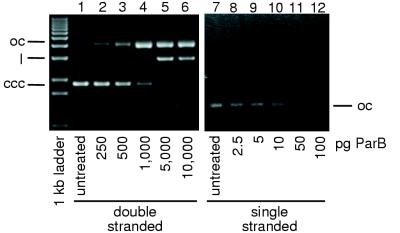
To test the idea that ParB preferentially cleaves single-stranded DNA, ParB activities on double- and single-stranded pBluescript II SK(−) substrates were compared. The DNA substrates were digested for 5 min with increasing amounts of ParB protein, and the reaction products were subjected to agarose gel electrophoresis followed by staining with EtBr. It should be noted that single-stranded DNA is detected poorly with EtBr on an agarose gel, requiring roughly 10-fold more single-stranded DNA to be visualized with an intensity similar to that of double-stranded DNA. For the purpose of this experiment, the relaxation of the supercoiled double-stranded substrate and the linearization of the single-stranded circular substrate were considered analogous, as both reactions require a single-strand cleavage event. Approximately 1 ng of ParB protein was required to nick the majority of the supercoiled double-stranded substrate (Fig. 7), while only 50 pg of ParB was required to substantially reduce the level of single-stranded circular DNA. Similarly, double-stranded DNA was essentially completely converted to the open circular form by the addition of between 1 and 5 ng of ParB, while single-stranded circular DNA is completely linearized by between 50 and 100 pg of ParB. These data suggest that ParB cleaves single-stranded DNA roughly 20- to 50-fold more effectively than double-stranded substrate.
FIG. 7.
ParB activity on double-stranded and single-stranded DNAs. Double-stranded and single-stranded pBluescript II SK(−) DNAs were treated with ParB for 5 min. Double-stranded DNA was treated with 0, 250, 500, 1,000, 5,000, or 10,000 pg of ParB (lanes 1 to 6), while single-stranded DNA was treated with 0, 2.5, 5, 10, 50, and 100 pg of ParB (lanes 7 to 12). The positions of the open circular (oc), linear (l), and supercoiled (ccc) DNA forms are shown.
ParB exonuclease activity.
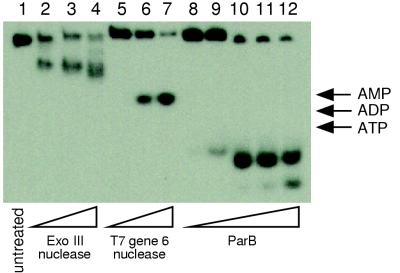
It has been suggested that ParB possesses an exonuclease activity, in addition to the protein’s role as an endonuclease (17). To investigate the exonuclease activity of ParB, a 20-bp DNA fragment, radiolabeled on one of its 5′ ends, was treated with ParB. Exonuclease III, which cleaves mononucleotides from the 3′ end of a double-stranded substrate, and T7 gene 6 exonuclease, which cleaves mononucleotides from the 5′ end, were also employed as controls. As expected, cleavage of the 20-bp fragment by exonuclease III produced a ladder of reaction products, with smaller molecules being generated as the reaction time was increased (Fig. 8). Also as expected, cleavage of the substrate by T7 gene 6 exonuclease generated a single band of mononucleotides which increased in intensity with time. In the case of treatment with ParB (Fig. 8), two products were generated, both running faster on the gel than a trinucleotide but more slowly than inorganic phosphate (not shown). The high mobility of the labeled products during electrophoresis suggests that they are more highly charged or possess a different geometry than a mononucleotide. Also, the slower-migrating product appears after 5 min of digestion with 1 ng of ParB (Fig. 8, lane 9), while the faster product appears at a later time (after exposure to 40 ng of ParB) (lane 12). These reaction products were further characterized by thin-layer chromatography with lithium chloride as a solvent and visualized by autoradiography. The mobilities of the reaction products were compared to those of standard molecules considered likely to be similar in structure and charge to the reaction products. pGp, pGpA, and pGpAp were chosen as standards, since the terminal guanine and the adjacent adenine are the first two nucleotides present at the labeled end of the substrate DNA strand. Standards were visualized by UV shadowing. As shown in Table 1, the reaction products of ParB digestion appear to comigrate with pGpAp and pGp, respectively. Comigration was also observed when potassium phosphate was used as a solvent instead of lithium chloride. These results suggest that ParB has an exonuclease activity, producing from the 5′ end of the double-stranded substrate a dinucleotide that is 3′ and 5′ phosphorylated (pGpAp). The dinucleotide is then further cleaved to a 3′,5′-diphosphomononucleotide (pGp).
FIG. 8.
Exonuclease assay. A 20-bp double-stranded DNA fragment was 5′ radiolabeled on one end and then treated with 0.01, 0.1, or 1 U of exonuclease III (lanes 2 to 4, respectively); with 0.1, 1, or 2 U of the T7 gene 6 exonuclease (lanes 5 to 7, respectively); or with 0.1, 1, 10, 20, or 40 ng of purified ParB (lanes 8 to 12, respectively). The positions of AMP, ADP, and ATP are indicated.
TABLE 1.
Resolution of ParB reaction products by thin-layer chromatographya
| Sample | Rf(s) |
|---|---|
| ParB products | 0.19, 0.30 |
| pGpAp | 0.27b |
| pGpA | 0.38b |
| pGp | 0.21b |
| ParB products + pGpAp | 0.19, 0.29c |
| ParB products + pGpA | 0.19, 0.30, 0.38 |
| ParB products + pGp | 0.22d, 0.30 |
The solvent used was 1 M lithium chloride.
Standard.
When samples were cospotted, comigration was observed at an Rf of 0.29.
When samples were cospotted, comigration was observed at an Rf of 0.22.
DISCUSSION
The ParB protein encoded by plasmid RK2 was highly purified and shown to possess both endonuclease and 5′-exonuclease activities. Overexpression was accomplished by using pEJ18, a vector which contains both the parC and parB genes driven by the T7 promoter. Induction of E. coli BL21(DE3)(pEJ18) resulted in the cotranslation of ParB and ParC. After induction, ParC was invariably found in fast-sedimenting material, presumably in inclusion bodies. Under conditions of relatively slow growth of the cells at 30°C with low aeration, ParB and ParC were produced in roughly equal amounts. While most of the ParB has been reported previously to reside primarily in the insoluble fraction (17), ParB produced in BL21(DE3) from pEJ18 is found almost exclusively in the soluble fraction. This may be due in part to the coproduction of ParB and ParC by using pEJ18. The amino acid sequence of ParB contains a putative transmembrane region and a signal sequence cleavage site at the N terminus of the ParB protein (16, 17), suggesting the localization of the protein in the periplasmic space. Several other nucleases with sequences homologous to that of ParB have been shown to be exported from the cytosol, but none have been suggested to have a role in plasmid stability. The nucleases of plasmid pSa (6) and S. aureus (44) are transported across the cell membrane, while nuc of pKM101 encodes a protein likely to be involved in conjugal transfer (28). One model of RK2 par region function proposes that ParB aids in the partition of plasmid monomers by anchoring them to the cell wall during cell division. A second possible role of ParB is in the conversion of the catenane product of the action of ParA on dimers to monomeric forms (9, 17). As parB is not essential in all hosts for establishing full plasmid stabilization, host-encoded nucleases may in certain strains partly substitute for ParB activity (9, 10, 16, 17). Clearly, additional study will be required to definitively localize ParB and to clarify its role in plasmid stabilization.
Our studies with highly purified ParB confirm the previously demonstrated progression of reaction products from supercoiled DNA to open circular DNA and finally to the linear form (17). In studies with pUC19, however, ParB seemed to display only weak sequence specificity (Fig. 4 through 6). Digestion of ParB-linearized DNA by specific restriction enzymes (Fig. 4), as well as primer extension with ParB-treated templates (Fig. 6), indicates that ParB cleaves pUC19 at many more sites than reported for the par region containing pGMA30 in experiments with partially purified ParB. Our primer extension studies, however, do suggest some preference for cleavage at the site adjacent to one of the two DraI sites on pUC19 DNA that was previously found to be a preferential site in early studies with partially purified ParB (17). The adjacent and newly detected sites of preferential nicking are located in an AT-rich region just downstream of those previously detected (Fig. 5A and 6A). An additional site of preferential cleavage is located within the bla gene of pUC19. While this additional site is not located in a region of high AT content, it appears to lie within the loop region of a potential cruciform structure (Fig. 5A and B and 6B). This is consistent with the observation that ParB is 20- to 50-fold more active on a single-stranded DNA substrate than on a double-stranded one. It therefore seems likely that ParB cleaves supercoiled DNA substrates in localized regions in which strand separation due to breathing is more frequent, such as in AT-rich regions in a supercoiled DNA substrate. It is possible that a supercoiled molecule would be nicked only once by ParB and that linearization follows due to ParB-mediated nicking of the opposite strand which is made possible by greater access to the phosphodiester bond. This would be similar to the function of the S1 nuclease from Aspergillus oryzae, which nicks supercoiled DNA but prefers to cleave open circular double-stranded DNA opposite a nicked site (14, 37). Linearization may also result from random nicks at many sites on both strands of the double-stranded DNA molecule.
Our studies on the exonuclease activity of ParB provide direct evidence that ParB also has 5′→3′ exonuclease activity, cleaving double-stranded DNA to 3′-phospho di- and mononucleotides. Preliminary experiments with a 3′-labeled substrate appear to confirm the 5′→3′ exonuclease activity. Several of the proteins shown to have sequence similarity to ParB display enzymatic properties similar to those of ParB. The most similar to ParB with respect to amino acid sequence and in vitro activity seems to be staphylococcal nuclease, which is Ca2+ dependent and prefers a single-stranded DNA substrate but will also nick supercoiled DNA substrates (44). The staphylococcal nuclease also exhibits exonuclease activity, cleaving double-stranded DNA and generating 3′-phosphorylated mononucleotides and dinucleotides. Unlike that of ParB, however, the exonuclease activity of the staphylococcal nuclease proceeds in a 3′→5′ orientation. In addition, the S. aureus nuclease, and possibly the nuclease of plasmid pSa (6), appears likely to function in the extracellular digestion of nucleic acids, which is an unlikely function of ParB.
Other exonucleases similar to ParB in enzymatic activity investigated so far generate 5′-phosphorylated nucleotides from 5′→3′ activity or 3′-phosphorylated nucleotides from 3′→5′ exonuclease activity. However, ParB cleaves DNA with 5′→3′ exonuclease activity to generate 3′-phosphonucleotides. Thus, the enzymatic activity of ParB is highly unusual among nucleases but is not unprecedented. The endo-exonuclease nuclease β of Ustilago maydis possesses 5′→3′ activity but also generates 3′-phosphorylated nucleotides (33). This unique enzyme functions independently of divalent cations and is not inhibited by the presence of 10 mM EDTA. A biological role for this protein is unknown.
While the endonuclease and exonuclease activities of ParB bear resemblance to the endo-exonuclease class of nucleases (for a review, see reference 14), none have yet been found to have amino acid sequence similarity to ParB. Like ParB, members of this class of enzymes prefer to cleave single-stranded DNA, exhibit 5′→3′ exonuclease activity, and are capable of nicking supercoiled DNA. Identified in fungi and mitochondria, several appear to have a role in either recombination or DNA repair. The similarities in properties of ParB and the endo-exonuclease class of proteins raise the possibility that ParB plays a similar role in vivo as well. It is of interest to speculate that ParB serves a role in the resolution of a Holliday structure which may be an intermediate in the decatenation of the products of the ParA resolution activity on plasmid dimers. It has been proposed that bacterial partitioning mechanisms involve as a crucial step the resolution of chromosomal dimers that are formed during the course of DNA replication (1, 22). A putative signal sequence and cleavage site found in the amino acid sequence of ParB may suggest localization of the protein in the inner membrane or periplasmic space, consistent with the putative membrane location of a partitioning apparatus. Additionally, preliminary studies suggest that partially purified preparations of ParC protein bind specifically to the intercistronic region of the 3.2-kb par region (unpublished results). This leaves open the possibility that during the resolution process a ParC-B-A complex may be involved in anchoring a plasmid dimer to the inner membrane, followed by resolution of the dimer during cell division. The availability of highly purified ParB and ParA and progress in the purification of the ParC protein should allow the testing in vitro of the various possible interactions between the three RK2 par proteins.
ACKNOWLEDGMENTS
We thank Aresa Toukdarian and George A. Kassavetis for helpful discussions and Aresa Toukdarian for critical reading of the manuscript.
This work was supported by NIH grant AI-07194. E.P.J. was supported by NIH training grant 5T32GM07317.
REFERENCES
- 1.Adams D E, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 2.Alvarado-Urbina G, Sathe G M, Liu W C, Gillen M F, Duck P D, Bender R, Ogilvie K K. Automated synthesis of gene fragments. Science. 1981;214:270–274. doi: 10.1126/science.6169150. [DOI] [PubMed] [Google Scholar]
- 3.Austin S, Abeles A. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983;169:373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- 4.Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-Topoisomerase II complexes. J Mol Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 5.Bravo A, de Torrontegui G, Diaz R. Identification of components of a new stability system of R1, ParD, that is close to the origin of replication of the plasmid. Mol Gen Genet. 1987;210:101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- 6.Close S M, Kado C I. A gene near the plasmid pSa origin of replication encodes a nuclease. Mol Microbiol. 1992;6:521–527. doi: 10.1111/j.1365-2958.1992.tb01497.x. [DOI] [PubMed] [Google Scholar]
- 7.Dam M, Gerdes K. Partitioning of plasmid R1. Ten direct repeats flanking the parA promoter constitute a centromere-like partition site parC, that expresses incompatibility. J Mol Biol. 1994;236:1289–1298. doi: 10.1016/0022-2836(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 8.Davis T L, Helinski D R, Roberts R C. Transcription and autoregulation of the stabilizing functions of broad-host-range plasmid RK2 in Escherichia coli, Agrobacterium tumefaciens and Pseudomonas aeruginosa. Mol Microbiol. 1992;6:1981–1994. doi: 10.1111/j.1365-2958.1992.tb01371.x. [DOI] [PubMed] [Google Scholar]
- 9.Easter C L, Schwab H, Helinski D R. Role of the parCBA operon of the broad-host-range plasmid RK2 in stable plasmid maintenance. J Bacteriol. 1998;180:6023–6030. doi: 10.1128/jb.180.22.6023-6030.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Easter C L, Sobecky P A, Helinski D R. Contribution of different segments of the par region to stable maintenance of the broad-host-range plasmid RK2. J Bacteriol. 1997;179:6472–6479. doi: 10.1128/jb.179.20.6472-6479.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberl L, Givskov M, Schwab H. The divergent promoters mediating transcription of the par locus of plasmid RP4 are subject to autoregulation. Mol Microbiol. 1992;6:1969–1979. doi: 10.1111/j.1365-2958.1992.tb01370.x. [DOI] [PubMed] [Google Scholar]
- 12.Eberl L, Kristensen C, Givskov M, Grohmann E, Gerlitz M, Schwab H. Analysis of the multimer resolution system encoded by the parCBA operon of broad-host-range plasmid RP4. Mol Microbiol. 1994;12:131–141. doi: 10.1111/j.1365-2958.1994.tb01002.x. [DOI] [PubMed] [Google Scholar]
- 13.Figurski D H, Meyer R, Helinski D R. Suppression of ColE1 replication properties by the IncP-1 plasmid RK2 in hybrid plasmids constructed in vitro. J Mol Biol. 1979;133:295–318. doi: 10.1016/0022-2836(79)90395-4. [DOI] [PubMed] [Google Scholar]
- 14.Fraser M J, Low R L. Fungal and mitochondrial nucleases. In: Linn S M, Lloyd R S, Roberts R J, editors. Nucleases. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 171–208. [Google Scholar]
- 15.Gerdes K, Molin S. Partitioning of plasmid R1—structural and functional analysis of the parA locus. J Mol Biol. 1986;190:269–279. doi: 10.1016/0022-2836(86)90001-x. [DOI] [PubMed] [Google Scholar]
- 16.Gerlitz M, Hrabak O, Schwab H. Partitioning of broad-host-range plasmid RP4 is a complex system involving site-specific recombination. J Bacteriol. 1990;172:6194–6203. doi: 10.1128/jb.172.11.6194-6203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grohmann E, Stanzer T, Schwab H. The ParB protein encoded by the RP4 par region is a Ca(2+)-dependent nuclease linearizing circular DNA substrates. Microbiology. 1997;143:3889–3898. doi: 10.1099/00221287-143-12-3889. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning: a practical approach. Vol. 1. McLean, Va: IRL Press; 1985. p. 109. [Google Scholar]
- 19.Jaffé A, Ogura T, Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985;163:841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson E P, Ström A R, Helinski D R. Plasmid RK2 toxin protein ParE: purification and interaction with ParD antitoxin protein. J Bacteriol. 1996;178:1420–1429. doi: 10.1128/jb.178.5.1420-1429.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jovanovic O S, Ayres E K, Figurski D H. Host-inhibitory functions encoded by promiscuous plasmids; transient arrest of Escherichia coli segregants that fail to inherit plasmid RK2. J Mol Biol. 1994;237:52–64. doi: 10.1006/jmbi.1994.1208. [DOI] [PubMed] [Google Scholar]
- 22.Kato J-I, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 22a.Keller, W. Personal communication.
- 23.Konieczny I, Doran K S, Helinski D R, Blasina A. Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J Biol Chem. 1997;272:20173–20178. doi: 10.1074/jbc.272.32.20173. [DOI] [PubMed] [Google Scholar]
- 24.Lehnherr H, Maguin E, Jafri S, Yarmolinsky M B. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol. 1993;233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- 25.Macartney D P, Williams D R, Stafford T, Thomas C M. Divergence and conservation of the partitioning and global regulation functions in the central control region of the IncP plasmids RK2 and R751. Microbiology. 1997;143:2167–2177. doi: 10.1099/00221287-143-7-2167. [DOI] [PubMed] [Google Scholar]
- 26.Mori H, Mori Y, Ichinose C, Niki H, Ogura T, Kato A, Hiraga S. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J Biol Chem. 1989;264:15535–15541. [PubMed] [Google Scholar]
- 27.Motallebi-Veshareh M, Rouch D A, Thomas C M. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol Microbiol. 1990;4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- 28.Pohlman R F, Liu F, Wang L, Moré M I, Winans S C. Genetic and biochemical analysis of an endonuclease encoded by the IncN plasmid pKM101. Nucleic Acids Res. 1993;21:4867–4872. doi: 10.1093/nar/21.21.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Randerath E, Randerath K. Ion-exchange thin-layer chromatography. XII. Quantitative elution and microdetermination of nucleoside monophosphates, ATP, and other nucleotide coenzymes. Anal Biochem. 1965;12:83–93. doi: 10.1016/0003-2697(65)90145-4. [DOI] [PubMed] [Google Scholar]
- 30.Roberts R C, Burioni R, Helinski D R. Genetic characterization of the stabilizing functions of broad-host-range plasmid RK2. J Bacteriol. 1990;172:6204–6216. doi: 10.1128/jb.172.11.6204-6216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts R C, Ström A, Helinski D R. The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J Mol Biol. 1994;237:35–51. doi: 10.1006/jmbi.1994.1207. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz-Echevarría M J, Giménez-Gallego G, Sabariegos-Jareño R, Díaz-Orejas R. Kid, a small protein of the parD stability system of plasmid R1, is an inhibitor of DNA replication acting at the initiation of DNA synthesis. J Mol Biol. 1995;247:568–577. doi: 10.1006/jmbi.1995.0163. [DOI] [PubMed] [Google Scholar]
- 33.Rusche J R, Rowe T C, Holloman W K. Purification and characterization of nuclease β from Ustilago maydis. J Bacteriol. 1980;255:9117–9123. [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sander M, Benhaim D. Drosophila Rrp1 3′-exonuclease: demonstration of DNA sequence dependence and DNA strand specificity. Nucleic Acids Res. 1996;24:3926–3933. doi: 10.1093/nar/24.20.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saurugger P N, Hrabak O, Schwab H, Lafferty R M. Mapping and cloning of the par-region of broad-host-range plasmid RP4. J Biotechnol. 1986;4:333–343. [Google Scholar]
- 37.Shishido K, Ando T. Single-strand-specific nucleases. In: Linn S M, Roberts R J, editors. Nucleases. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 155–185. [Google Scholar]
- 38.Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983;22:181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- 39.Sia E A, Roberts R C, Easter C, Helinski D R, Figurski D H. Different relative importances of the par operons and the effect of conjugal transfer on the maintenance of intact promiscous plasmid RK2. J Bacteriol. 1995;177:2789–2797. doi: 10.1128/jb.177.10.2789-2797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobecky P A, Easter C, Bear P D, Helinski D R. Characterization of the stable maintenance properties of the par region of broad-host-range plasmid RK2. J Bacteriol. 1996;178:2086–2093. doi: 10.1128/jb.178.7.2086-2093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 42.Thomas C M, Helinski D R. Vegetative replication and stable inheritance of IncP plasmids. In: Thomas C M, editor. Promiscous plasmids of gram-negative bacteria. London, United Kingdom: Academic Press Ltd.; 1989. pp. 1–25. [Google Scholar]
- 43.Tsuchimoto S, Nishimura Y, Ohtsubo E. The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J Bacteriol. 1992;174:4205–4211. doi: 10.1128/jb.174.13.4205-4211.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tucker P W, Hazen J E E, Cotton F A. Staphylococcal nuclease reviewed: a prototypic study in contemporary enzymology. I. Isolation; physical and enzymatic properties. Mol Cell Biochem. 1978;22:67–77. doi: 10.1007/BF00496235. [DOI] [PubMed] [Google Scholar]
- 45.West S C. The RuvABC proteins and Holliday junction processing in Escherichia coli. J Bacteriol. 1996;178:1237–1241. doi: 10.1128/jb.178.5.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshioka Y, Fujita Y, Ohtsubo E. Nucleotide sequence of the promoter-distal region of the tra operon of plasmid R100, including traI (DNA helicase I) and traD genes. J Mol Biol. 1990;214:39–53. doi: 10.1016/0022-2836(90)90145-C. [DOI] [PubMed] [Google Scholar]
- 47.Youngren B, Austin S. Altered ParA partition proteins of plasmid P1 act via the partition site to block plasmid propagation. Mol Microbiol. 1997;25:1023–1030. doi: 10.1046/j.1365-2958.1997.4761842.x. [DOI] [PubMed] [Google Scholar]