Abstract
A Tn917 insertion mutant of an M49 serotype, opacity factor-positive Streptococcus pyogenes, was isolated. It had the following phenotypes: decreased β-hemolysis mediated by streptolysin S, reduction in the activity of a secreted cysteine protease and streptokinase, and an altered immunoglobulin and fibrinogen-binding phenotype. The site of insertion of Tn917 into the chromosome and the surrounding sequence, the pel region (pleiotropic effect locus), was determined. Phage A25 transduction confirmed that the pleiotropic changes in phenotype could be cotransduced with Tn917. The pel region was cloned and sequenced, and the transposon was found to be inserted upstream of a single open reading frame which led to a failure to transcribe a 500-base mRNA. The loss of this transcript decreased the transcription of emm and speB genes and reduced the secretion of streptokinase. Enhanced Pel expression from a nisin-inducible plasmid resulted in increased message levels for emm in a wild-type organism. Characterization of the pel mutant provides evidence for the coordinated regulation of secreted and surface proteins and suggests the existence of a new global regulatory factor in S. pyogenes.
Group A streptococci (GAS) are capable of causing a variety of human diseases ranging from superficial or deep tissue infections to noninfectious poststreptococcal infection sequelae (7, 13, 31, 53, 61, 64). Many GAS products, either located on the bacterial cell surface or secreted by the bacteria, have been implicated in pathogenesis (8, 22, 39, 48). Recent studies have demonstrated that GAS can coordinately regulate the expression and production of certain virulence factors in response to environmental signals (11, 20, 32, 63).
The most intensively studied regulatory mechanism in GAS involves the mga gene product (10, 33, 34, 41, 42, 44, 46, 57). Mga is a transacting positive regulatory protein, which is required for the full expression of several important GAS virulence factors including M protein, M-like proteins, C5a peptidase, and opacity factor. It was proposed, based on sequence comparison, that Mga is a transducer protein involved in a two-component regulatory system. However, the putative sensor protein that responds to environmental signals and activates Mga remains unidentified.
A second GAS regulator, RofA, which regulates the expression of protein F, is also a positive transacting protein (18). Protein F is a fibronectin-binding protein associated with the attachment of bacteria to the host extracellular matrix. RofA was identified as a transcriptional activator for the expression of prtF, which encodes the GAS fibronectin-binding protein, protein F. Recent data suggested that prtF might be regulated in response to superoxide levels (20); however, the precise mechanisms by which environmental signals influence RofA are still unknown.
Recent studies have also identified a novel negative regulatory activity of GAS present in a variety of opacity factor-positive isolates (47). This negative regulator controls both its own expression and the regulation of two adjacent operons. The regulator also influences the expression of mga and demonstrates its effects in a growth phase-dependent manner. Similar regulatory effects of growth phase have been reported for the expression of capsule, SpeB, and other putative virulence factors in different GAS isolates (14, 26, 33, 56). Interestingly, Nra and RofA have 62% homology (47); however, these two genes are not present in all GAS isolates studied. rofA is present in both opacity factor-positive and -negative isolates, while nra appears to be preferentially associated with opacity factor-positive isolates.
With the exception of mga and rofA, little is known about the environmental regulation of virulence gene expression in GAS, and only a few potential sensors for virulence gene regulation have been identified. For example, the oligopeptide and dipeptide permeases have been associated with the regulation of cysteine protease SpeB production (30, 43, 45), and csrS and csrR form a two-component regulatory system involved in capsule expression (2, 5, 27). More recently, studies by Federle et al. suggest that this system may regulate the expression of other virulence factors, including strepokinase and streptolysin S (SLS) in addition to capsule (17a). These investigators suggest that the CovR-CovS (csrR-csrS) system acts as a multiple-gene repression system that functions independently of growth phase regulators in GAS (17a).
Even less is known about molecules in GAS that mediate signal transduction in control of virulence factor expression. To date, most differences in gene expression in GAS are associated with physical or physicochemical conditions, e.g., temperature, oxygen, carbon dioxide tension, pH, or the concentration of iron in the media (20, 32). No peptide signaling molecules or RNA regulatory mechanisms have been identified in GAS. Precedents for each of these types of regulation in gram-positive cocci have been reported, including Com peptides in Streptococcus gordonii and pneumococci (35), sex pheromones in enterococci (4, 19, 25), and regulatory RNA in Streptococcus aureus (1, 23, 38, 50). Based on these models and some recent observations of changes in phenotype of GAS M and M-related proteins expressed by an mga isogenic mutant following selection for virulence with a skin infection model (8), we predicted that GAS might express global regulators that in turn control the expression of mga.
This hypothesis was further supported by analysis of a randomly mutagenized Tn917 library of an M49 serotype GAS isolate, CS101. During screening of this library, a mutant was identified that exhibited a pleiotropic defect in expression of several secreted proteins as well as changes in surface-associated M and M-related proteins. This mutant contained a single Tn917 insertion and was stable after repeated subculturing in selective media. The phenotype of this mutant suggested that a key gene involved in transcription, translation, or posttranslation regulation, similar to those previously identified in S. aureus and other organisms (1, 9, 23, 38, 40, 49, 50), was mutated.
MATERIALS AND METHODS
Bacteria, plasmids, and chemicals.
The bacteria used in this study and their characteristics are described in Table 1. Todd-Hewitt broth (THB) was obtained from BBL (Becton Dickinson, Cockeysville, Md.), hyaluronidase was purchased from Sigma (St. Louis, Mo.), and human plasma was obtained from the American Red Cross (Toledo, Ohio).
TABLE 1.
Bacterial strains, plasmids, and bacteriophage used in this study
| Strains, plasmids, or bacteriophage | Descriptiona | Reference or source |
|---|---|---|
| S. pyogenes | ||
| CS101 | OF(+); emm49 mrp49 sk49 speB sls slo (wild type) | 42 |
| CS101:ZQ6 | CS101; pel:Tn917 (pel-1; Mrp− SK− SLS− Slo+) | This work |
| 1881 | OF(−); emm1 ska1 speB (wild type) | 48 |
| NZ131 | OF(+); emm49 mrp49 sk49 speB (wild type) | 55 |
| SF370 | OF(−); emm1 ska1 speB (wild type) | 60 |
| MGAS 166s | OF(−); emm1 ska1 speB (wild type) | 6 |
| E. coli | ||
| HB101 | hsd-520 recA13 supE44 ara-14 galK2 lacY1 proA2 rpsL20 xyl-5 leu mtl-11 mcrB | 52 |
| MC1061 | Δ(araA-leu)7697 araD139 Δ(codB-lac)3 galE15 galK16 mcroAO relAI rpsL150 spT1 mcrB9999 hsdR2 | 15 |
| Plasmids | ||
| pTV1-OK | repA(Ts)-pWVO1Ts aphA3 Tn917 (erm) | 21 |
| pUC18 | lacZα bla | Pharmacia, Piscataway, N.J. |
| Bacteriophage | ||
| A25 | GAS-transducing bacteriophage | 12 |
OF, opacity factor.
Tn917-mediated transposon mutagenesis.
The Tn917-mediated transposon mutagenesis was performed as previously described (21, 28). In brief, the temperature-sensitive shuttle vector pTV1-OK was electroporated into GAS isolate CS101. Transformants were selected based on a temperature-dependent resistance to kanamycin. A single transformed colony was inoculated into 5 ml of THB containing 0.3% yeast (THY) and erythromycin (5 μg/ml) and grown overnight at 30°C. This culture was diluted 1:100 in THY, prewarmed to 39°C, and grown at that temperature for 10 h. An aliquot (100 μl) of the culture was plated onto THY agar containing 5% heat-inactivated human plasma and either kanamycin (50 μg/ml) or erythromycin (5 μg/ml). All colonies that were sensitive to kanamycin but resistant to erythromycin were considered potential Tn917 integrants. Southern blot analysis using biotinylated pTV1-OK as a probe confirmed the insertion of a single copy of Tn917 into the chromosome.
Phenotypic characterization.
The mutagenized library was screened for a number of secreted and surface expressed proteins as described below.
β-Hemolysis assay.
Initially, individual colonies of mutants were screened for β-hemolysis on sheep blood agar plates. Potential β-hemolysis-negative colonies were selected and restreaked onto sheep blood agar plates (erythromycin, 5 μg/ml) and screened for β-hemolysis following overnight growth at 37°C. Analysis of the contribution of streptolysin O (SLO) and SLS was carried out by the methods described in reference 29.
Streptokinase assay.
Colonies were initially screened for products of streptokinase by using a plate overlay assay described previously (28). Potential streptokinase-negative colonies were expanded and analyzed by using a sensitive functional assay in which the activation of human plasminogen was measured with a plasmin-selective synthetic substrate as described previously (16). Briefly, the culture supernatants of wild-type CS101 and selected Tn917 mutants were precipitated by adding ammonium sulfate to 80% saturation at 4°C and stirring them overnight. The suspension was then centrifuged at 9,000 × g for 30 min at 4°C, and the resulting pellet was resuspended in 1 ml of 0.01 M phosphate-buffered saline (pH 7.6) and dialyzed extensively against the same buffer. An aliquot of the dialysate (100 μl) was mixed with either 1 μg of purified human plasminogen or buffer and a plasmin-selective synthetic substrate, S-2251 (H-d-Val-Leu-Lys-paranitroanilide), was added to a final concentration of 400 μM. Plasmin generation was monitored by reading product generation at 405 nm.
Cysteine endopeptidase assay.
Cysteine protease activity present in ammonium sulfate-precipitated culture supernatants was assayed according to the method of North (37). Briefly, 50 μl of ammonium sulfate-concentrated culture supernatant, without or with 0.1 μM dithiothreitol, was added to the wells of a microtiter plate. Following incubation for 30 min at 37°C to allow reduction, 150 μl of the substrate-buffer solution (3.2 ml of 2.5 mM Bz-Pro-Phe-Arg-paranitroanilide [Sigma]) dissolved in distilled water at pH 4.0 plus 4.8 ml of 100 mM sodium phosphate, pH 6.0, was added to each well. Cleavage of the substrate was monitored by measuring the A405 over time in a microtiter plate reader (Biotek, Winooska, Vt.). The cleavage of substrate and generation of product were determined to be linear with time to an A405 of 1.5. The cysteine protease-specific inhibitor, E64 (Sigma), was included in parallel assays at a concentration of 1 μM to determine if all of the enzymatic activity being measured could be attributed to the presence of a cysteine protease.
Analysis of immunoglobulin-binding proteins.
Mutants were extracted with CNBr as described previously (48). Solubilized proteins were separated in sodium dodecyl sulfate–10% polyacrylamide gels under reducing conditions and screened for immunoglobulin G (IgG)-binding activity by Western blotting as described previously (48).
Direct binding assay for fibrinogen or fibronectin.
The ability of bacteria to bind fibrinogen or fibronectin was determined by their ability to bind the specific radiolabeled ligand. Human fibrinogen and fibronectin were radiolabeled with 125I (Amersham, Chicago, Ill.) by using a lactoperoxidase method (62). Different numbers of bacteria were incubated with 20,000 cpm of either 125I-labeled fibrinogen or fibronectin for 60 min at 37°C. The bacteria were pelleted by centrifugation at 5,000 × g for 20 min and washed twice with 2 ml of 0.15 M Veronal buffered saline, pH 7.35, containing 0.1% gelatin. The radiolabel associated with the bacterial pellet was quantified with a Beckman automatic gamma counter.
Determination of hyaluronic acid capsule level.
The level of capsular hyaluronic acid was determined by a chemical method as described previously (64).
Southern blot analysis.
Analysis of chromosomal DNA for the insertion of Tn917 was carried out as described previously (59). Briefly, chromosomal DNA was isolated from wild-type CS101 isolates as described previously (31). Chromosomal DNA was digested to completion with either EcoRI or HindIII. DNA fragments were separated in a 0.8% agarose gel and transferred to nylon membranes by blotting. The membrane was probed with biotinylated pTV1-OK probe and developed with a PhotoGene kit (Life Technologies, Inc., Gaithersburg, Md.).
DNA cloning.
A shotgun cloning strategy was used to recover the gene containing the Tn917 insertion by standard protocols (52). In brief, chromosomal DNA was isolated from the selected mutant and digested to completion with HindIII. A sample of pUC18 was also treated with HindIII, dephosphorylated with bacteria alkaline phosphatase (Life Technologies, Inc.), and then ligated with the HindIII-digested chromosomal DNA. The ligation mixture was used to transform Escherichia coli MC1061 by standard methods (15). Transformants were selected by growing bacteria on Luria-Bertani agar containing erythromycin (300 μg/ml) and ampicillin (100 μg/ml). Transformants were further analyzed by determining the size of the insert and by Southern blot analysis with biotinylated pTV1-OK probe as described previously. Plasmid was recovered from the selected transformants and purified with the Plasmid Maxi-Prep kit (Qiagen, Santa Clara, Calif.).
DNA sequencing.
The isolated plasmid was subjected to DNA sequencing by an automated DNA sequencer (Ana-Gen, Inc., Palo Alto, Calif.) with a specific sequencing primer corresponding to the 5′ end of Tn917 and extended into the adjoining streptococcus-derived DNA. DNA sequence analysis was compared to the GAS DNA sequence database (51) for homology. Potential open reading frames of the DNA sequence were identified with MacVector (Oxford Molecular, Oxford, England) computer software.
PCR and XL PCR.
PCR was used to amplify the genes from wild-type CS101, its Tn917 mutants, and transductants. To amplify a potential DNA fragment that contains the entire Tn917 transposon and flanking sequences, an extra-long PCR (XL PCR) procedure was performed (3). In this procedure the forward and reverse primers for specific amplification were designed on the basis of our sequence data generated during the study and a recently published sequence for a GAS streptolysin S-associated gene (sagA) (6).
The sequence of the two primers was as follows: forward primer, 5′-ACC TAA ATA TCA CTA CCA CAT-3′, and reverse primer, 5′-ATG TCT CTT CTT TTA GGT ATG-3′. The optimal conditions for XL PCR were determined empirically. Optimal product was obtained by using 1 μg of chromosomal DNA, 4 mM Mg++, 20 pmol of each primer, and 1 U of rtTh polymerase for each 100 μl of PCR reaction mixture. The PCR cycling parameters were 94°C for 30 s, followed by 16 cycles of 94°C for 30 s, 50°C for 1 min, and 68°C for 6.5 min followed by 18 cycles of 94°C for 30 s, 50°C for 1 min, and 68°C for 6.5 min with 15 s of segment extension at 68°C, followed by 72°C for 10 min. The PCR product was gel purified and subjected to DNA sequencing by an automated DNA sequencer with two primers specific for either the 5′ or 3′ end of Tn917.
Transduction.
A GAS-virulent bacteriophage, A25, was used to transduce Tn917 mutations into CS101 and other GAS isolates. The following procedure was modified from a previously published transduction protocol (12). A lysate of A25 phage from a Tn917 mutant was prepared by growing the Tn917 mutant of interest in 10 ml of transduction broth (THY supplemented immediately before use with 1% heat-inactivated horse serum, 50 mM CaCl2, and 340 μg of hyaluronidase per ml) and incubated at 30°C overnight. This culture was diluted 1:50 in transduction broth (without Ca++) and grown to an optical density at 600 nm (OD600) of 0.7. Then approximately 109 PFU of phage A25 was added to the broth and incubated at 30°C for another 4 h with gentle shaking. The culture was cleared by the addition of 100 μl of CHCl3 followed by centrifugation, and the supernatant was stored at 4°C for later use.
To perform the transduction, the recipient strain was grown in transduction broth at 30°C overnight. This culture was then diluted in 5 ml of fresh transduction broth and grown at 30°C to an OD600 of approximately 0.6. Ten microliters of an A25 lysate, generated as described above, was added to the culture and incubated at 30°C for 30 min. The cells were then pelleted by centrifugation, resuspended in 5 ml of THY broth, and incubated for 2 h at 30°C, at which time the bacteria were harvested by centrifugation. The bacterial pellet was resuspended in 0.5 ml of THY, and aliquots containing 0.1 ml of the suspension were plated onto selective THY agar plates supplemented with 5% human plasma and 5 μg of erythromycin per ml. Potential transductants were selected on the basis of erythromycin resistance phenotypes and confirmed to contain Tn917 by XL PCR and sequencing as described above.
RNA techniques.
For RNA preparations, serotype M49 GAS strains were grown aerobically to an OD600 of 0.9, which corresponded to late logarithmic growth phase. Prior to preparation, cells were sedimented by 2 min of centrifugation at 4°C, suspended in ice-cold 20 mM Tris (pH 7.5) containing 5 mM MgCl2, 20 mM Na azide, 400 mg of chloramphenicol per liter. RNA preparation was done according to the protocol of Shaw and Clewell (54). Denaturing agarose gel electrophoresis and quantitative Northern blot hybridizations with digoxigenin-dUTP-labeled probes were performed as previously described (43). Probes were generated by asymmetric PCR, with only 10−2 to 10−3 of the normal amounts of the appropriate upstream primers. The sequences of primers used for asymmetric PCR were described previously in reference 47.
Generation of GAS mutant carrying an inducible pel gene.
A 186-bp fragment containing the pel open reading frame of GAS CS101 was amplified by PCR, with oligonucleotides pel FOR (5′-AGG AGG TAA ACC TTA TGT T-3′) and pel REV (5′-GCT AAA TAG ATT ATT TAC CTG-3′) as primers. The product was cloned into the PstI and XbaI sites or plasmid pNZ8048 (17, 24). Plasmid pNZ8048 harbors a nisin-inducible promoter. The recombinant plasmid was coestablished with plasmid pNZ9531 in the serotype M49 CS101 wild-type strain. Plasmid pNZ9531 carries the genes for a nisin-responsive sensor-regulator pair. The optimum amount of nisin leading to maximum stimulation of the inducible promoter, while not affecting the growth rate of the bacteria, was 10 μg/liter. For studying the effects of pel hyperexpression, the recombinant and wild-type isolates of strain CS101 were exposed to 10 μg of nisin per liter and grown to an OD600 of 0.9. RNA was prepared from these bacteria and analyzed as described above. These conditions were critical, since the ability to hyperexpress pel in this system is influenced by as yet unexplained cell cycle-dependent effects.
RESULTS
Identification of Tn917-generated pleiotropic mutants.
A Tn917-mutagenized library of GAS isolate CS101 was screened for a variety of phenotypic characteristics. These characteristics included the ability to bind fibrinogen, fibronectin, expression of hyaluronic acid capsule, and expression of surface Ig-binding M and M-related proteins. In addition, analysis of β-hemolysis, streptokinase production, and secretion of a cysteine protease were included in the initial screening assays. The focus of these studies was to identify any mutation that affected both surface and secreted products. The initial screening was carried out with the assays described in Materials and Methods. Approximately 104 individual colonies were screened by plate assays, and any mutant colony which demonstrated an apparent change in β-hemolysis as well as a decrease in SpeB and streptokinase activity was screened for changes in surface Ig-binding proteins and a fibrinogen-binding or fibronectin-binding activity. Emr mutants that had changes in surface and secreted proteins were characterized further by more quantitative functional assays.
After this procedure one mutant (designated CS101:ZQ6) was identified that demonstrated an altered expression of several surface (IgG-binding and fibrinogen-binding proteins) and secreted proteins (Ska and SpeB). The results presented in Table 2 and Fig. 1 summarize the characteristics of the wild-type CS101 organism and mutant CS101:ZQ6. Although it was noted that the expression of selected streptococcal products can vary with the growth phase (14, 33, 56), the differences between wild-type CS101 and mutant CS101:ZQ6 could not be attributed to growth phase variation (data not shown).
TABLE 2.
Phenotypic comparison of wild-type CS101 and Tn917 mutant CS101:ZQ6
| Phenotype | Wild type | CS101:ZQ6 |
|---|---|---|
| β-Hemolysis | ||
| SLS | Positive | Negative |
| SLO | Positive | Positive |
| SK activity | Positive | Negative |
| Cysteine protease | ||
| Enzymatic activity | High | Low |
| E-64 inhibitable | Yes | No |
| Fibrinogen binding (%) | 100 | 33 |
| Fibronectin binding (%) | 100 | 100 |
| Capsular hyaluronic acid | Positive | Positive |
| Ig bindinga | ||
| Emm (IgG3) | Positive | Negative |
| Mrp (IgG1, IgG2, IgG4) | Positive | Negative |
| Enn (IgA) | Positive | Negative |
Determined in CNBr extracts of bacteria by Western blotting as described in reference 48.
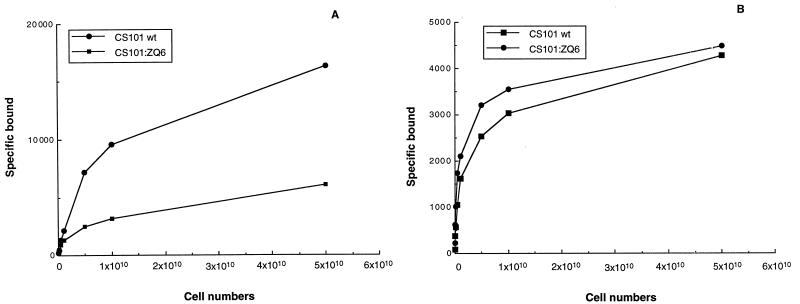
FIG. 1.
Direct binding (counts per minute) of human fibrinogen (A) and fibronectin (B) by GAS isolate CS101 and Tn917 mutant CS101:ZQ6. After growth in THB at 37°C overnight, bacteria were harvested and washed with 0.15 M Veronal buffered saline, pH 7.35, containing 0.1% gelatin. Different numbers of cells were then incubated with 125I-human fibrinogen or 125I-human fibronectin, as described in Materials and Methods. Following three washings binding was measured in a gamma counter. All estimations were carried out in duplicate, and <5% variation was observed between samples.
Characterization of the Tn917 insertion in CS101:ZQ6.
We next determined the nature of the Tn917 insertion associated with mutant CS101:ZQ6. Southern blot analysis was performed as described in Materials and Methods and fragments screened for the presence of the transposon by using biotinylated pTV1-OK as a probe (Fig. 3). There was no binding of the biotinylated pTV1-OK probe to digested chromosomal DNA from wild-type GAS isolate CS101. An EcoRI digest of chromosomal DNA from mutant CS101:ZQ6 contained a single reactive band, while for the HindIII digest three reactive bands were identified (data not shown). This was consistent with a single Tn917 insertion since EcoRI does not cut within the transposon, while there are two HindIII sites present in Tn917 (54). This locus, disrupted by Tn917, was designated pel (pleiotropic effect locus).
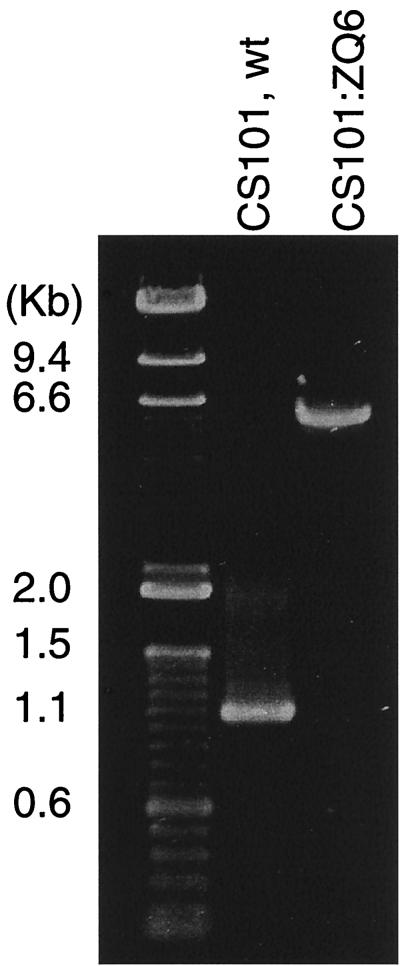
FIG. 3.
PCR amplification of the locus of Tn917 insertion from chromosomal DNA of wild-type (wt) CS101 and Tn917 mutant CS101:ZQ6. The locus containing the Tn917 insertion was amplified either from chromosomal DNA of wild-type CS101 by a conventional PCR protocol or from chromosomal DNA of the mutant CS101:ZQ6 by an XL PCR protocol as described in Materials and Methods. A primer designed specifically for the sagA gene and a second primer specifically complementary to a sequence about 700 bp downstream of the Tn917 insertion site were used to determine if the proposed insertion site of Tn917 was correct. The PCR products were separated in a 1% agarose gel and visualized under UV light, after ethidium bromide staining.
The next series of experiments were designed to map the position of pel in the streptococcal chromosome. Chromosomal DNA from CS101:ZQ6 was digested with HindIII, which cut twice inside the Tn917 sequence, leaving the 5′ end sequence containing the erythromycin resistance cassette intact. This digest was cloned into pUC18. The plasmid library was used to transform E. coli, and colonies resistant to ampicillin and erythromycin were selected. Plasmid DNA from one of the erythromycin-resistant clones was isolated and shown to contain Tn917 by Southern blot analysis (data not shown). The plasmid was sequenced by using a primer that would hybridize to the 5′ end of Tn917 and extend into the flanking GAS sequence (21). Sequence analysis confirmed that the plasmid contained the expected sequence of Tn917 and led to the identification of an additional 240 bp of flanking streptococcal DNA.
Mapping of pel.
Recently, a putative streptolysin-associated gene, called sagA, was cloned from an M1 GAS isolate and proposed to be responsible for the SLS-deficient phenotype of a Tn917 mutant (6). The first 32 nucleotides of sagA were 100% complementary to the first 32 nucleotides of the 5′ sequence of pel after the Tn917 insertion (Fig. 2B). This result suggested that a sagA-like sequence might be the sequence immediately flanking the 3′ end of the Tn917 insertion in pel. Based on this prediction, a pair of specific primers were designed for PCR analysis based on the sagA sequence and the 5′ flanking sequence shown in Fig. 2A. Assuming that the two sequences flanked both ends of the Tn917, a single 6.3-kb fragment should be amplified from the CS101:ZQ6 mutant chromosomal DNA and an approximately 1.1-kb product should be amplified from the wild-type strain.
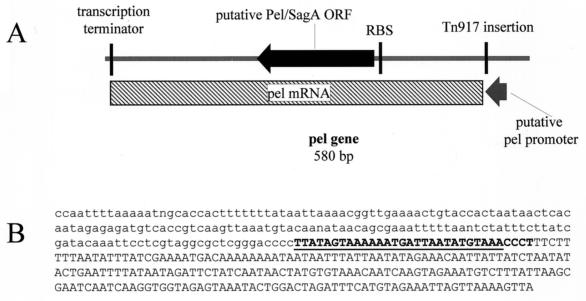
FIG. 2.
Schematic representation of the pleiotropic effect locus (pel) (A) and nucleotide sequence of part of pClone2 (B). The filled arrow represents the putative pel-sagA protein-coding region. The gray arrow below the line represents the putative pel promoter. The vertical lines indicate the insertion site of Tn917 and potential ribosome binding site (RBS) and transcription terminator. The hatched box represents the pel mRNA. The size of this mRNA is consistent with the Northern blot analysis of pel (Fig. 4). The lowercase region is Tn917 sequence derived from pClone2. The 32 nucleotides in bold are 100% homologous to the published sagA sequence (6). The underlined sequence is part of the putative pel promoter. The remainder of the streptococcal sequence was 93% identical to the corresponding sequence in the M1 streptococcal genome database. The sequence shown is upstream of the pel gene.
For these experiments an XL PCR strategy was utilized (3). A 1.1-kb fragment and a 6.3-kb fragment were observed when the chromosomal DNA from the wild-type isolate and mutant CS101:ZQ6, respectively, were used as templates (Fig. 3). This result was consistent with pel and sagA being proximal or overlapping. The XL PCR product was sequenced with two sequencing primers specific for Tn917 (21). Sequencing data confirmed that the XL PCR products contained the Tn917 sequence with our cloned DNA sequence flanking the 5′ end and a sagA-like sequence flanking the 3′ end (data not shown). Thus, we were able to locate both ends of the Tn917 insertion.
Based on our cloned partial sequence and the streptococcal genome sequence of an M1 isolate, we were able to generate a genetic map of the pel locus. The Tn917 insertion was present within the promotor region of sagA, and this gene was the only open reading frame in the region that appeared to have a promoter, a Shine-Dalgarno sequence, and a ribosome binding site to enable transcription. This result assumes that the CS101:ZQ6 mutant contains only a single mutation and that the entire change in phenotype could be attributed to the single Tn917 insertion.
To determine whether the pleiotropic phenotypic changes were due entirely to the disruption of pel or could be attributed in part to a second spontaneous mutation, a phage transduction strategy was used. The Tn917-associated mutation in pel, pel-1, was transduced to the wild-type CS101 strain by using phage A25 as described in Materials and Methods. Potential transductants were selected on erythromycin-containing medium and analyzed for phenotypic changes. After conditions were optimized, numerous transductants were recovered and five of them were analyzed. Chromosomal DNA was isolated from each transductant, and XL PCR was performed with the same primer pairs described previously to amplify the 6.3-kb pel-1 fragment. This procedure resulted in the amplification of an identical molecular size fragment (6.3 kb) when DNA from each transductant was used as a template (data not shown).
A similar strategy was used to transduce the mutation into three M1 serotype isolates, 1881, MGAS 166s, and SF370, as well as into an additional M49 serotype isolate, NZ131. Erythromycin-resistant transductants were obtained for all isolates, and PCR analysis and Southern blotting demonstrated that they contained the expected Tn917 insertion. This result indicates that the pel region was present in both opacity factor-positive and -negative isolates. The NZ131 pel mutant strain had a phenotype identical to that of CS101:ZQ6 (Table 3). The opacity factor-negative wild-type M1 isolates differed in their phenotypes from CS101 in that they failed to bind significant levels of fibronectin, and thus this property could not be used to monitor changes in the transductants (Table 3).
TABLE 3.
Analysis of the phenotypes of representative opacity factor-positive and -negative GAS isolates transduced with phage A25 containing a disrupted pel regiona
| Phenotype | % Expression in:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CS101 (M49)
|
NZ131 (M49)
|
1881 (M1)
|
SF370 (M1)
|
MGAS 166s (M1)
|
||||||
| Wild type | Transductant | Wild type | Transductant | Wild type | Transductant | Wild type | Transductant | Wild type | Transductant | |
| SK | 100 | <10 | 100 | <10 | 100 | <10 | 100 | <10 | 100 | <10 |
| SpeB | 100 | <10 | 100 | <10 | 100 | <10 | 100 | <10 | 100 | <10 |
| β-Hemolysis | + | − | + | − | + | − | + | − | + | − |
| Fibrinogen binding | 100 | 36 | 100 | 35 | 100 | 42 | 100 | 38 | 100 | 37 |
All transductants were Emr and contained the expected 6.6-kb fragment containing Tn917 (Fig. 4). Expression of each surface or secreted virulence factor in the transductant was compared to the level of expression of the wild-type isogenic pair, which was designated as 100%. +, detectable; −, nondetectable.
Transductants of either M1 isolate 1881, MGAS 166s, or SF370 demonstrated a loss of hemolyzing activity on blood agar plates, significant reduction in streptokinase and SpeB activity, and a down-regulation in fibrinogen-binding activity (Table 3). The latter property is particularly interesting, since the major fibrinogen-binding activity of opacity factor-negative M1 isolates is the M protein while in opacity factor-positive isolates like CS101 this activity is mediated by the mrp gene product (16), a gene that is not normally present in opacity factor-negative isolates.
Similar transduction studies were carried out with the Tn916 mutation (sagA) described by Betschtel et al. (6). The resulting sagA mutants were phenotypically identical to the isogenic pel-1 mutants (data not shown). The phenotypic differences reported here for pel and by Betschtel et al. for the sagA mutant (6) most probably represent differences in the phenotypic screening assays performed in the two studies.
Transcription analysis of pel and overexpression of pel.
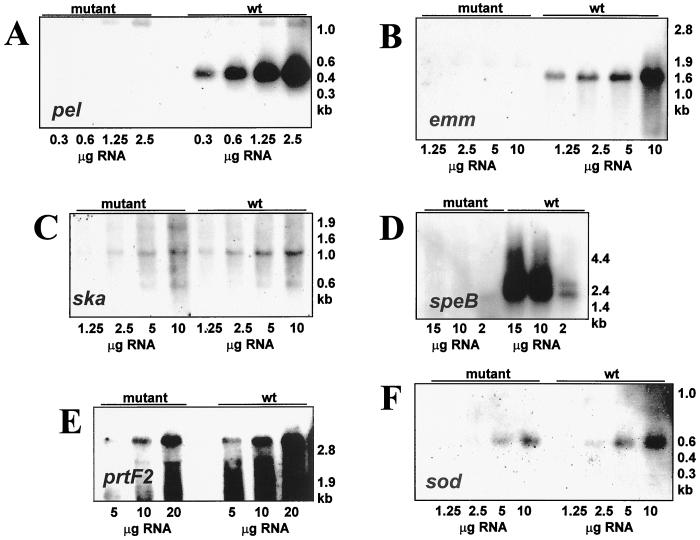
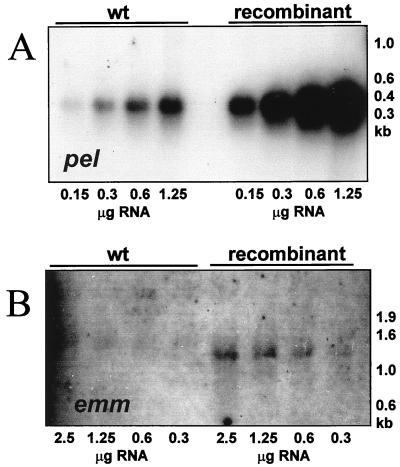
The results described above indicate that a genetic disruption in the pel region of either opacity factor-positive or -negative isolates resulted in a markedly changed phenotype (Table 3). This involved both surface proteins associated with virulence, e.g., M and M-related proteins, and putative secreted virulence factors, e.g., SpeB and Ska. To analyze whether these effects were occurring at the transcriptional, translational, or posttranslational level, a series of quantitative Northern blots were performed, comparing the M49 wild-type strain CS101 and the corresponding pel mutant. The gene expression monitored included that of the pel gene, prtF2, speB, emm, and ska. The housekeeping gene sod (superoxide dismutase) was used as an internal standard. In wild-type cells an mRNA of approximately 500 bases was detected with a pel probe (Fig. 4A). The size of this mRNA is consistent with the predicted size of the pel transcript (Fig. 2A). This mRNA was not detected in the pel mutant strain (Fig. 4A). No transcription from the speB or emm genes was detected in the pel mutant strain (Fig. 4B and D). The levels of sod transcripts were decreased approximately twofold, while the ska gene was reduced two- to fourfold (Fig. 4F and C, respectively). The prtF2 transcript was unchanged (Fig. 4E). These results suggested that the pel gene product regulated the transcription of the emm and speB genes but regulated streptokinase at the translational or posttranslational level.
FIG. 4.
Northern blot transcript analysis of the serotype M49 GAS strain CS101 (wild type [wt]) and its isogenic pel mutant (mutant). mRNA probes used were pel (A), emm, M protein (B), ska, streptokinase (C), speB, pyrogenic exotoxin B (D), prtF2, protein F (E), and sod, superoxide dismutase (F). Total cellular RNA was isolated from each strain in late logarithmic growth phase. RNAs were separated by MOPS (morpholinepropanesulfonic acid)-formaldehyde agarose gel electrophoresis. The source of the RNA is shown above the lanes, and the amount of RNA loaded is shown below the corresponding lanes. Uniform loading of the gel was confirmed after electrophoresis by visual inspection of the ethidium bromide-stained RNA. The approximate sizes of the molecular weight markers are shown on the right. The results shown are representative of those from at least two independent RNA isolations.
In order to test whether increased amounts of the pel message would have an effect opposite to that of the gene disruption, the pel gene was cloned downstream of a nisin-inducible promoter in plasmid pNZ8048. The recombinant plasmid together with plasmid pNZ9531 harboring the genes for a pair of nisin sensor-regulators were transformed into serotype M49 GAS strain CS101. The wild-type strain and the strain carrying both plasmids were exposed to nisin, and RNA from both strains was analyzed. In the plasmid-containing strain the pel and emm messages were clearly increased compared to wild-type levels (Fig. 5).
FIG. 5.
Northern blot transcript analysis of the serotype M49 GAS strain CS101 (wild type [wt]) and an isogenic strain expressing a nisin-inducible pel gene. mRNA probes used were pel (A) and emm, M protein (B). Total cellular RNA was isolated from each strain in late logarithmic growth phase after exposure to nisin. RNAs were separated by MOPS-formaldehyde agarose gel electrophoresis. The source of the RNA is shown above the lanes, and the amount of RNA loaded is shown below the corresponding lanes. Uniform loading of the gel was confirmed after electrophoresis by visual inspection of the ethidium bromide-stained RNA. The approximate sizes of the molecular weight markers are shown on the right. The results shown are representative of those from at least two independent RNA isolations.
DISCUSSION
In this study we described a pleiotropic Tn917 insertion mutant in an opacity factor-positive M49 Streptococcus pyogenes isolate, CS101. A single Tn917 insertion was found to decrease the expression of a number of secreted products, including the plasminogen activator streptokinase and a secreted cysteine protease, as well as totally inhibiting the activity of the hemolysin SLS. In addition, decreased expression of surface fibrinogen-binding and IgG-binding proteins was also observed in the absence of a change in surface fibronectin binding or a significant change in the level of hyaluronic acid capsule.
The Tn917 insertion in CS101 disrupts transcription of the 500-base pel RNA. Since no other RNAs were detected in this region of the chromosome and the overexpression of pel increases expression of one pel-regulated gene, we conclude that the phenotypes associated with the Tn917 insertion are due to the loss of pel transcription.
In related studies, three groups have reported SLS-negative mutants after Tn916 mutagenesis. Nida and Cleary demonstrated a Tn916 mutation in an M12 serotype isolate that lacked SLS activity (36), and Liu et al., studying an M3 serotype isolate, found that the inactivation of SLS was associated with a requirement for riboflavin for growth of the mutant (29). The site of the mutation in these mutants was, however, not mapped. Betschel et al. also described an SLS-negative Tn916 mutant in an M1 serotype isolate (6). This phenotype negative mutant was associated with disruption of the sagA gene. The Tn917 mutation in the opacity factor-positive M49 isolate CS101 used in this study and the Tn916 mutation in the opacity factor-negative M1 isolate used in the study of Betschel et al. (6) mapped to within 5 bp of each other, and both were in the promoter region of the gene. Transduction studies using pel-1 containing Tn917 or sagA containing Tn916 resulted in similar phenotypic changes in CS101 and M1 strains.
The results presented in this paper suggest that pel has an important effect on the expression of key virulence factors in GAS and that this gene region is associated with expression of only the hemolysin SLS. The genes regulated by pel are not clustered in a single region of the chromosome, e.g., a pathogenicity island. Analysis of transcription in the pel mutant supports a key role for Pel in the global regulation of gene expression in S. pyogenes. The disruption of expression of the pel gene results in the absence or reduction of emm message and speB but had minor effects on the expression of ska, ptrf, or sod message.
Phenotypically the mutant also demonstrates the absence of expression of other M-related gene products (Mrp and Enn), as well as the absence of streptokinase and β-hemolysin activity. The discrepancy between the discrete reduction of ska message and the almost complete loss of streptokinase activity could be attributed to a change in translation efficiency processing and/or secretion of functional streptokinase.
The message from the pel gene shown in Fig. 4 is consistent with a monocystronic transcript that begins upstream of the pel open reading frame and ends before the predicted start of the next downstream open reading frame. This suggests that either the RNA or protein encoded by this gene is a direct global positive regulator or interacts directly or indirectly with a global positive or negative regulatory system in S. pyogenes. The fact that both a loss of pel expression and increased pel expression influence the expression of several virulence genes indicates that the pel-associated regulatory mechanism does not exclusively react to a threshold level of the pel gene product but quantitatively responds to different levels of pel expression.
Although sequence analysis of Pel suggests a potentially secreted peptide with a signal peptidase 1-associated leader sequence (data not shown), it is not currently clear whether pel encodes a protein that acts as a transcriptional regulator or if the untranslated Pel mRNA could be involved. Precedents for each of these types of regulator signals in controlling the expression of genes in bacteria exists, e.g., RNA regulators in S. aureus (1, 23, 38, 50), Enterococcus faecalis (4, 19), and E. coli (58). Further analysis of the nature of pel regulation is now required to address these questions and to determine the relationship of this regulatory activity to other regulatory circuits recently described in GAS (2, 5, 17a, 47).
ACKNOWLEDGMENTS
We thank June Scott, Emory University, Atlanta, Ga., for providing the A25 phage and helpful advice on the transduction strategy, Joyce de Azavedo, Mt. Sinai Hospital, Toronto, Ontario, Canada, for providing us with M1 isolate MGAS 166s and the corresponding β-hemolytic Tn916 mutant described in reference 6, A. Flosdorff, for expert technical assistance, and Carol Hepner for typing the manuscript.
This work was supported by grants from the National Institutes of Health (M.D.P.B.) A1 43474 and by DFG Sch877/1-2 (A.P.).
REFERENCES
- 1.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 2.Ashbaugh C D, Albertí S, Wessels M R. Molecular analysis of the capsule gene region of group A Streptococcus: the hasAB genes are sufficient for capsule expression. J Bacteriol. 1998;180:4955–4959. doi: 10.1128/jb.180.18.4955-4959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes W M. PCR amplification of up to 35 kb DNA with high fidelity and high yield from lambda templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bensing B A, Dunny G M. Pheromone-inducible expression of an aggregation protein in Enterococcus faecalis requires interaction of a plasmid-encoded RNA with components of the ribosome. Mol Microbiol. 1997;24:295–308. doi: 10.1046/j.1365-2958.1997.3311709.x. [DOI] [PubMed] [Google Scholar]
- 5.Bernish B, van de Rijn I. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J Biol Chem. 1999;274:4786–4793. doi: 10.1074/jbc.274.8.4786. [DOI] [PubMed] [Google Scholar]
- 6.Betschel S D, Borgia S M, Barg N L, Low D E, De Azavedo J C S. Reduced virulence of group A streptococcal Tn916 mutants not producing streptolysin S. Infect Immun. 1998;66:1671–1679. doi: 10.1128/iai.66.4.1671-1679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisno A L, Stevens D L. Streptococcal infections of skin and soft tissues. N Engl J Med. 1996;334:240–245. doi: 10.1056/NEJM199601253340407. [DOI] [PubMed] [Google Scholar]
- 8.Boyle M D P, Raeder R, Flosdorff A, Podbielski A. Role of emm and mrp genes in the virulence of group A streptococcal isolate 64/14 in a mouse model of skin infection. J Infect Dis. 1998;177:991–997. doi: 10.1086/515241. [DOI] [PubMed] [Google Scholar]
- 9.Bruberg M B, Nes I F, Eijsink V G H. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol Microbiol. 1997;26:347–360. doi: 10.1046/j.1365-2958.1997.5821951.x. [DOI] [PubMed] [Google Scholar]
- 10.Caparon M, Scott J. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caparon M G, Geist R T, Perez-Casal J, Scott J. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J Bacteriol. 1992;174:5693–5701. doi: 10.1128/jb.174.17.5693-5701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caparon M G, Scott J R. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 1991;204:556–586. doi: 10.1016/0076-6879(91)04028-m. [DOI] [PubMed] [Google Scholar]
- 13.Chaussee M S, Liu J, Stevens D L, Ferretti J J. Genetic and phenotypic diversity among isolates of Streptococcus pyogenes from invasive infections. J Infect Dis. 1996;173:901–908. doi: 10.1093/infdis/173.4.901. [DOI] [PubMed] [Google Scholar]
- 14.Chaussee M S, Phillips E R, Ferretti J J. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect Immun. 1997;65:1956–1959. doi: 10.1128/iai.65.5.1956-1959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung C T, Miller R H. A rapid convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988;16:3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christner R, Li Z, Raeder R, Podbielski A, Boyle M D P. Identification of key gene products required for acquisition of plasmin-like enzymatic activity by group A streptococci. J Infect Dis. 1998;175:1115–1120. doi: 10.1086/516450. [DOI] [PubMed] [Google Scholar]
- 17.Eichenbaum Z, Federle M J, Marra D, de Vos W M, Kuipers O P, Kleerebezem M, Scott J R. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl Environ Microbiol. 1998;64:2763–2769. doi: 10.1128/aem.64.8.2763-2769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Federle M J, McIver K S, Scott J R. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fogg G C, Gibson C M, Caparon M G. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an mγδ-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol Microbiol. 1994;11:671–684. doi: 10.1111/j.1365-2958.1994.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 19.Fujimoto S, Clewell D B. Regulation of the pAD1 sex pheromone response of Enterococcus faecalis by direct interaction between the cAD1 peptide mating signal and the negatively regulating, DNA-binding TraA protein. Proc Natl Acad Sci USA. 1998;95:6430–6435. doi: 10.1073/pnas.95.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson C M, Caparon M G. Insertional inactivation of Streptococcus pyogenes sod suggests that prtF is regulated in response to superoxide signal. J Bacteriol. 1996;178:4688–4695. doi: 10.1128/jb.178.15.4688-4695.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez J A, Crowley P L, Brown D P, Hillman J D, Youngman P, Bleiweis A S. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J Bacteriol. 1996;178:4166–4175. doi: 10.1128/jb.178.14.4166-4175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanski E, Caparon M G. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus (Streptococcus pyogenes) Proc Natl Acad Sci USA. 1992;89:6172–6176. doi: 10.1073/pnas.89.13.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji G, Beavis R C, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 24.Kleerebezem M, Beerthuyzen M M, Vaughan E E, de Vos W M, Kuipers O P. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol. 1997;63:4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleerebezem M, Quadri L E N, Kuipers O P, de Vos W M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 26.Leonard B A B, Woischnik M, Podbielski A. Production of stabilized virulence factor-negative variants by group A streptococci during stationary phase. Infect Immun. 1998;66:3841–3847. doi: 10.1128/iai.66.8.3841-3847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin J C, Wessels M R. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 28.Li Z Q, Gutierrez J A, Bleiweis A S, Boyle M D P. Efficient insertional mutagenesis in group A streptococci mediated by Tn917 transposon. J Microbiol Methods. 1997;30:203–215. [Google Scholar]
- 29.Liu S, Shlomo S, Cohen G, Jadoun J, Cheung A, Ofek I. Insertional inactivation of streptolysin S expression is associated with altered riboflavin metabolism in Streptococcus pyogenes. Microb Pathog. 1997;22:227–234. doi: 10.1006/mpat.1996.0107. [DOI] [PubMed] [Google Scholar]
- 30.Lyon W R, Gibson C M, Caparon M G. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin D R, Single L A. Molecular epidemiology of group A streptococcus M type 1 infections. J Infect Dis. 1993;167:1112–1117. doi: 10.1093/infdis/167.5.1112. [DOI] [PubMed] [Google Scholar]
- 32.McIver K S, Heath A S, Scott J R. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation in emm transcription. Infect Immun. 1997;63:4540–4542. doi: 10.1128/iai.63.11.4540-4542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McIver K S, Scott J R. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLandsborough L A, Cleary P P. Insertional inactivation of virR in Streptococcus pyogenes M49 demonstrates that VirR functions as a positive regulator of ScpA, FcRA, OF, and M protein. FEMS Microbiol Lett. 1995;128:45–52. doi: 10.1111/j.1574-6968.1995.tb07498.x. [DOI] [PubMed] [Google Scholar]
- 35.Morrison D A. Streptococcal competence for genetic transformation: regulation by peptide pheromones. Microb Drug Resist. 1997;3:27–37. doi: 10.1089/mdr.1997.3.27. [DOI] [PubMed] [Google Scholar]
- 36.Nida K, Cleary P P. Insertional inactivation of streptolysin S expression in Streptococcus pyogenes. J Bacteriol. 1983;155:1156–1161. doi: 10.1128/jb.155.3.1156-1161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.North M J. Cysteine endopeptidase of parasitic protozoa. Methods Enzymol. 1994;244:523–539. doi: 10.1016/0076-6879(94)44038-7. [DOI] [PubMed] [Google Scholar]
- 38.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada N, Pentland A P, Falk P, Caparon M G. M protein and protein F act as important determinants of cell-specific tropism of Streptococcus pyogenes in skin tissue. J Clin Investig. 1994;94:965–977. doi: 10.1172/JCI117463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perego M. Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Micro. 1998;6:366–370. doi: 10.1016/s0966-842x(98)01350-x. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Casal J, Caparon M, Scott J. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulator systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Podbielski A, Leonard B A B. The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol Microbiol. 1998;28:1323–1334. doi: 10.1046/j.1365-2958.1998.00898.x. [DOI] [PubMed] [Google Scholar]
- 44.Podbielski A, Peterson J A, Cleary P. Surface protein-CAT reporter fusions demonstrate differential gene expression in the vir regulon of Streptococcus pyogenes. Mol Microbiol. 1992;6:2253–2265. doi: 10.1111/j.1365-2958.1992.tb01401.x. [DOI] [PubMed] [Google Scholar]
- 45.Podbielski A, Pohl B, Woischnik M, Körner C, Schmidt K H, Rozdzinski E, Leonard B A B. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (Opp) and its effect on cysteine protease production. Mol Microbiol. 1996;21:1087–1099. doi: 10.1046/j.1365-2958.1996.661421.x. [DOI] [PubMed] [Google Scholar]
- 46.Podbielski A, Woischnik M, Pohl B, Schmidt K H. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med Microbiol Immunol. 1996;185:171–181. doi: 10.1007/s004300050028. [DOI] [PubMed] [Google Scholar]
- 47.Podbielski A, Woischnik M, Leonard B A B, Schmidt K-H. Characterization of nra, a global negative regulator gene in group A streptococci. Mol Microbiol. 1999;26:347–360. doi: 10.1046/j.1365-2958.1999.01241.x. [DOI] [PubMed] [Google Scholar]
- 48.Raeder R, Boyle M D P. Distinct profiles of immunoglobulin G binding protein expression by invasive serotype M1 isolates of Streptococcus pyogenes. Clin Diagn Lab Immunol. 1995;2:478–483. doi: 10.1128/cdli.2.4.478-483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rather P N, Parojcic M M, Paradise M R. An extracellular factor regulating expression of the chromosomal aminoglycoside 2′-N-acetyltransferase of Providencia stuartii. Antimicrob Agents Chemother. 1997;41:1749–1754. doi: 10.1128/aac.41.8.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gruss A, Novick R P. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 51.Roe B A, Linn S P, Song L, Yuan X, Clifton S, McShan M, Ferretti J J. posting date. [Online.] GAS genome present project supported by NIH grant A1 S8406. 27 September 1998. http://dna1.chem.uoknor.edu http://dna1.chem.uoknor.edu. [1 July 1999, last date accessed.] . [1 July 1999, last date accessed.] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 53.Schlievert P M, Assimacopoulos A P, Cleary P P. Severe invasive group A streptococcal disease: clinical description and mechanisms of pathogenesis. J Lab Clin Med. 1996;127:13–22. doi: 10.1016/s0022-2143(96)90161-4. [DOI] [PubMed] [Google Scholar]
- 54.Shaw J H, Clewell D B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985;164:782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon D, Ferretti J J. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol Lett. 1991;82:219–224. doi: 10.1016/0378-1097(91)90336-9. [DOI] [PubMed] [Google Scholar]
- 56.Simpson W J, Cleary P P. Expression of M type 12 protein by a group A streptococcus exhibits phaselike variation: evidence for coregulation of colony opacity determinants and M protein. Infect Immun. 1987;55:2448–2455. doi: 10.1128/iai.55.10.2448-2455.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simpson W J, LaPenta D, Chen C, Cleary P P. Coregulation of type 12 M protein and streptococcal C5a peptidase genes in group A streptococci: evidence for a virulence regulon controlled by the virR locus. J Bacteriol. 1990;172:696–700. doi: 10.1128/jb.172.2.696-700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 60.Suvorov A N, Ferretti J J. Physical and genetic chromosomal map of an M type 1 strain of Streptococcus pyogenes. J Bacteriol. 1996;178:5546–5549. doi: 10.1128/jb.178.18.5546-5549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Talkington D F, Schwartz B, Black C M, Todd J K, Elliot J, Breiman R F, Facklam R R. Association of phenotypic and genotypic characteristics of invasive Streptococcus pyogenes isolates with clinical components of streptococcal toxic shock syndrome. Infect Immun. 1993;61:3369–3374. doi: 10.1128/iai.61.8.3369-3374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thorell J I, Johnson B G. Enzymatic iodination of polypeptides with 125I to high specific activity. Biochim Biophys Acta. 1971;151:363–368. doi: 10.1016/0005-2795(71)90123-1. [DOI] [PubMed] [Google Scholar]
- 63.Van-Heyningen T, Fogg G, Yates D, Hanski E, Caparon M. Adherence and fibronectin binding are environmentally regulated in the group A streptococci. Mol Microbiol. 1993;9:1213–1222. doi: 10.1111/j.1365-2958.1993.tb01250.x. [DOI] [PubMed] [Google Scholar]
- 64.Wessels M R, Moses A E, Goldberg J B, DiCesare T J. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc Natl Acad Sci USA. 1991;88:8317–8321. doi: 10.1073/pnas.88.19.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]