Abstract
Cyclohexanone monooxygenases (CHMO) consume molecular oxygen and NADPH to catalyze the valuable oxidation of cyclic ketones. However, CHMO usage is restricted by poor stability and stringent specificity for NADPH. Efforts to engineer CHMO have been limited by the sensitivity of the enzyme to perturbations in conformational dynamics and long-range interactions that cannot be predicted. We demonstrate an aerobic, high-throughput growth selection platform in Escherichia coli for oxygenase evolution based on NADH redox balance. We applied this NADH-dependent selection to alter the cofactor specificity of CHMO to accept NADH, a less expensive cofactor than NADPH. We first identified the variant CHMO DTNP (S208D-K326T-K349N-L143P) with a ~1200-fold relative cofactor specificity switch from NADPH to NADH compared to the wild type through semirational design. Molecular modeling suggests CHMO DTNP activity is driven by cooperative fine-tuning of cofactor contacts. Additional evolution of CHMO DTNP through random mutagenesis yielded the variant CHMO DTNPY with a ~2900-fold relative specificity switch compared to the wild type afforded by an additional distal mutation, H163Y. These results highlight the difficulty in engineering functionally innovative variants from static models and rational designs, and the need for high throughput selection methods. Our introduced tools for oxygenase engineering accelerate the advancements of characteristics essential for industrial feasibility.
Keywords: cyclohexanone monooxygenase, Baeyer–Villiger monooxygenase, directed evolution, cofactor specificity, redox balance, high-throughput selection
Graphical Abstract
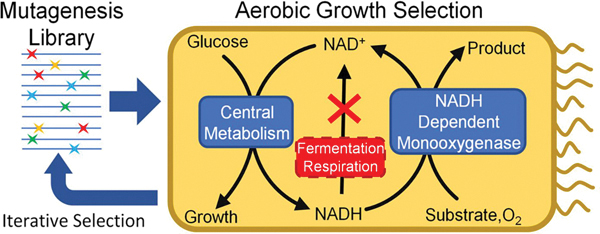
High-throughput screening methods are essential to the enzyme engineering workflow. Compared to other methods using 96 well plates,1,2 microfluidics,3,4 and fluorescence sorting5 based assays, cell growth-based selections are more facile and scalable because they utilize cell survival as an easy readout. Furthermore, the selected enzyme variants are directly compatible with in vivo applications. Implementing growth selection requires building a specific and robust connection between the desired enzymatic activity and cell growth. This is especially challenging for biomanufacturing applications, where non-native enzyme functions that do not contribute to cell fitness are targeted. Interestingly, there exists a unique opportunity to link the function of virtually any NAD(P)/H-dependent oxidoreductase to cell survival via employing the redox balance principle: Microbes must maintain supply of two universal cofactors at appropriate redox states. Microbial hosts with disrupted NAD or NADP recycling pathways become redox cofactor auxotrophs and can only grow when complemented by heterologous redox activity.
Several redox cofactor auxotrophs have been built on Escherichia coli. These include the anaerobic NAD+ 6,7 and NADP+ 8 auxotrophs unable to regenerate the oxidized form of the cofactors in the absence of oxygen, and the NADH9 and NADPH10,11 auxotrophs unable to produce the reduced form of the cofactors. While these strains have facilitated the identification of dehydrogenases for cofactor regeneration applications9,11 and provided insights into the extremes of cellular redox states,12,13 none of these platforms are suitable for directed evolution of oxygenases, a superfamily of versatile catalysts14–16 that consume NAD(P)H and oxygen. As a result, aerobic NAD(P)+ auxotrophs, which over accumulate NAD-(P)H even with the presence of oxygen, are needed.
Recently, we described the construction and application of an aerobic NADP+ auxotroph for engineering NADPH-dependent oxygenases.17,18 This system has also been used to engineer carboxylic acid reductases.19 Using this selection platform, we identified a thermostable variant of the NADPH-dependent Baeyer–Villiger monooxygenase (BVMO),20 Acinetobacter sp. cyclohexanone monooxygenase (Ac CHMO), a promising catalyst with broad applications.21–23 In addition to the poor thermostability, another limitation of Ac CHMO is that it strictly prefers NADPH.24 At large scale in vitro, NADPH is less stable and more costly than NADH,25 and in vivo, the rate of NADPH regeneration relative to NADH is lower in ubiquitous microbial chassis such as Escherichia coli and Bacillus subtilis.25,26 The high cost of cofactors is also a challenge for the development of efficient bioproduction processes. Switching cofactor preference from NADPH to NADH can result in a 15-fold reduction in cofactor cost and has been previously demonstrated as a strategy in the production of a variety of chemical commodities.27–30 CHMO has been shown to be notoriously averse to cofactor specificity switching with existing methods24,27,28 and a high-throughput selection is advantageous for exploring alternative engineering strategies.
Here we present the development and utilization of an aerobic, NADH-dependent selection platform to obtain variants of Ac CHMO with improved NADH activity. This selection platform, E. coli strain MX304, features disrupted fermentation, respiration, and NADH-to-NADPH transhydrogenation. As a result, it cannot use oxygen as an electron sink and must rely on a heterologous NADH-consuming enzyme to grow. Through a single round of NADH-dependent selection, we obtained a variant, CHMO DTNP (S208D-K326T-K349N-L143P), with ~1200-fold improvement in catalytic efficiency (kcat/KM) with NADH over NADPH in comparison to CHMO WT. Further directed evolution of this variant yielded DNTPY (DTNP + H163Y) which displays a ~2-fold improvement in catalytic efficiency over DTNP and an overall ~2900-fold cofactor specificity switch compared to the wild type. Molecular dynamics (MD) simulation suggest that the selected mutations may function by tuning the conformational dynamics of the protein and the cofactors, which would not be readily predicted in structure-guided protein design. For example, the key mutation L143P in CHMO DTNP emerged as a spontaneous mutation outside the three rationally picked positions (S208, K326, K349) for site-saturated mutagenesis. Although L143P does not directly interact with NADH, MD analysis suggests that it tunes the conformation of the flavin adenine dinucleotide (FAD) in CHMO DTNP, allowing more efficient hydride transfer from NADH. Similarly, the additional mutation in DTNPY, H163Y, is a surface residue that improves catalysis despite being distal to the catalytic pocket, likely due to subtle changes in enzyme dynamics. These effects would not be evident through analysis of static models, highlighting the difficulty of engineering functionally innovative variants through rational design and the critical role of high-throughput selections.
This work completes the full scope of redox-balance-based growth selections. Oxygenases are promising catalysts, but difficult to engineer due to their highly complex behavior during catalysis.31–34 We envision that the tool developed here will open new opportunities for engineering NADH-dependent oxygenases. While NADPH-dependent oxygenases such as most P450s naturally function in anabolism, the NADH-dependent oxygenases involved in catabolism have untapped potential as catalysts converting recalcitrant substrates such as xylene35 and plastics.36
RESULTS AND DISCUSSION
Development of an Aerobic, NADH-Dependent Selection Platform.
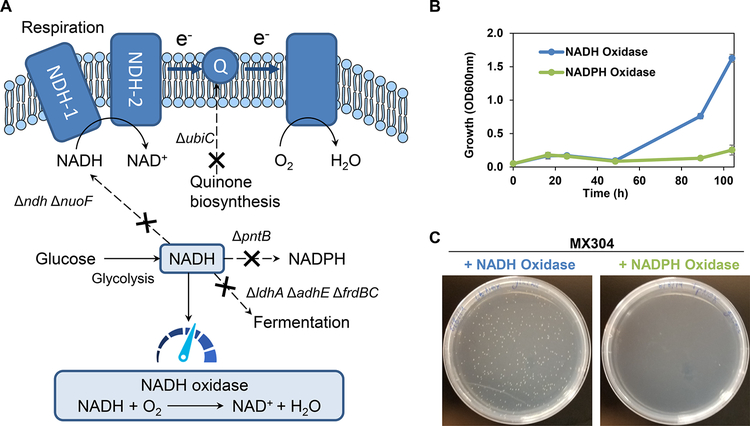
The NADH-dependent selection platform utilizes the principle of cofactor redox balance, where the inability to recycle NADH results in an elevated NADH/NAD+ ratio which greatly inhibits growth in the engineered E. coli strain MX304. NAD+ and NADH play key roles in regulating cellular metabolism and the accumulation of NADH is tied to the inhibition of glycolysis and the tricarboxylic acid (TCA) cycle.37,38 Growth of the selection strain MX304 is restored by expressing a heterologous oxygenase mutant with the desired activity8,17 to alleviate imbalance (Figure 1). Previously, anaerobic NADH accumulation in strain JCL166 (Table S1) was achieved by eliminating native fermentation pathways adhE, ldhA, and frdBC. When cultured anaerobically this strain experiences inhibitory levels of NADH, while microaerobically it is able to grow and demonstrates a roughly 4-fold increase in NADH levels while cultured as a production strain.39 Accumulating NADH in aerobic conditions represents a different set of challenges, since numerous metabolic pathways funnel NADH through the respiration chain. These challenges were tackled here by disrupting multiple respiratory chain components: ndh (encoding NADH:quinone oxidoreductase II), nuoF (encoding subunit of NADH:ubiquinone oxidoreductase I), and ubiC (encoding chorismate lyase, which catalyzes the first committed step in the biosynthesis of ubiquinone, an electron carrier in the respiration chain). In addition, the conversion of NADH to NADPH was diminished by disrupting pntAB (Figure 1A). Notably, a viable strain containing the above-mentioned knockouts could not be made without simultaneously expressing a water-producing NADH-oxidase from Lactobacillus brevis (Lb Nox),40 indicating the successful establishment of high NADH stress to a growth-inhibitory level. Lb Nox was expressed under an arabinose-inducible promoter (on plasmid pLS101, Table S1), so that its expression can be turned off during selection of desired oxygenases. The final NADH accumulation strain MX304 (Table S1) harbors a tightly controllable Lb Nox to support strain stability.
Figure 1.
Redox balance-based selection platform for NADH-dependent oxygenases. (A) Deletion of mixed acid fermentation pathways (ldhA, adhE, and frdBC), respiration associated genes (nuoF, ndh, and ubiC), and transhydrogenase component (pntB) greatly diminished native NADH oxidation. The aerobic accumulation of NADH severely inhibited growth without an alternative means of cofactor consumption. NADH-consuming oxidase Lb Nox was expressed on an arabinose inducible plasmid to alleviate NADH stress during nonselection conditions. (B, C) Growth of the NADH selection strain (MX304) is rescued by an NADH oxidase, but not by an NADPH oxidase in liquid media and on agar plates. Values are an average of at least three replicates, and the error bars represent one standard deviation.
We used Lb Nox and its NADPH-dependent counterpart, TP Nox,17,41 introduced on plasmids pLS401 and pLS402, respectively (Table S1), to further validate the newly developed NADH-dependent selection platform. As expected, only Lb Nox, but not TP Nox rescued the growth of MX304 (with its arabinose-inducible copy of Lb Nox turned off) in both liquid and solid media (Figure 1B, C, Figure S1). Uninduced Lb Nox under both the arabinose-inducible promoter and the IPTG-inducible promoter (pLS401) was insufficient to restore growth (Figure S2). These results confirm that the selection platform exhibits the desired cofactor specificity and can be used with flexible experimental setup. Evaluation of the growth response of MX304 with the NADPH-dependent wild type Ac CHMO (CHMO WT, on plasmid pLS201) confirmed CHMO WT fails to restore the growth of the NADH-dependent selection strain (MX304, with its arabinose-inducible copy of Lb Nox turned off) in selection conditions (Figure S1). MX304’s cofactor-dependent growth suggests that this strain can function as a high-throughput selection platform for identification of oxygenases with altered cofactor specificity toward NADH.
Directed Evolution of CHMO for NADH-Dependent Activity.
BVMOs cycle between the “open” and “closed” conformations during catalysis, with the cofactor NADP(H) not only participating in the reaction, but also playing an important role in coordinating the conformational change.34 Therefore, we hypothesized that to switch the cofactor specificity of Ac CHMO, it would be crucial to target sites that guide the conformational changes accompanying catalytic cycles, in addition to the ones that directly discriminate the 2′-phosphate of NADPH. Specifically, we chose four positions, K326, K349, S208, and T378. These sites were also subjected to extensive rational design efforts by Beier et al. to improve NADH-dependent activity.24 However, only a fraction of the possible sequence space (204 = 1.5 × 105) was searched by rational engineering strategies.
We incorporated NNK degenerate codons at the four sites mentioned above to yield a CHMO library (pLS403, Table S1). The library size constructed was estimated to be 2.5 × 107 independent transformants. Once the diversity of the library was confirmed by sequencing, pLS403 was transformed into the selection strain MX304 and the selection was conducted on M9 selection agar plates containing 2 g/L D-glucose and 2 g/L cyclohexanone at 30 °C for up to 120 h. Colonies were selected based on improved initial time of observable growth, growth rate, and colony size, compared to CHMO WT. Interestingly, many colonies harbored the same variant, CHMO DTNP (S208D-K326T-K349N-L143P). This variant retained the wild type residue at position T378 that is typically conserved across the BVMO family,24 and contained an unanticipated mutation L143P which possibly originates from spontaneous error in PCR or in vivo plasmid replication.
We characterized the kinetic parameters of the CHMO variants toward different cofactors (Table 1). Compared to wild type, CHMO DTNP exhibited ~5-fold increase in catalytic efficiency (kcat/KM) for NADH and ~255-fold decrease for NADPH, which collectively gives a ~1200-fold relative cofactor specificity switch. The improvement we achieved on NADH-dependent activity is substantial, given BVMOs well-documented recalcitrance to efforts aiming to increase NADH activity.25 Furthermore, the kinetic characterization shows that the spontaneous mutation L143P is critical to increased NADH-dependent activity (Table 1), which is consistent with the fact that CHMO DTN (S208D-K326T-K349N) cannot support growth (Figure S1) and was never discovered in growth selection. Contrastingly, the catalytic efficiency for NADPH is only moderately impacted by this spontaneous mutation (Table 1, comparing CHMO DTNP and DTN), suggesting that L143P is specifically involved in NADH-dependent catalysis.
Table 1.
Kinetic Characterization of CHMO Variantsa
| NADH |
NADPH |
|||||
|---|---|---|---|---|---|---|
| variant | kcat (s−1) | KM (mM) | kcat/KM (mM−1 s−1) | kcat (s−1) | KM (mM) | kcat/KM (mM−1 s−1) |
| WT | n.d. | n.d. | 0.18 ± 0.01 | 13.5 ± 0.55b | 0.037 ± 0.01b | 370 ± 85b |
| DTN | n.d. | n.d. | 0.21 ± 0.03 | n.d. | n.d. | 3.39 ± 0.03 |
| DTNP | 2.46 ± 0.36 | 2.79 ± 0.43 | 0.88 ± 0.01 | n.d. | n.d. | 1.45 ± 0.12 |
| DTNPY | 5.08 ± 0.48 | 2.81 ± 0.21 | 1.81 ± 0.03 | n.d. | n.d. | 1.28 ± 0.04 |
n.d. Individual kcat or KM were not determined because the enzyme cannot be saturated with the cofactor concentrations tested (up to 4 mM). In which case, the catalytic efficiency (kcat/KM) was determined by fitting the modified Michaelis–Menten equation ν0 = Vmax [S]/KM
Values taken from Maxel et al.20
The Effect of Cofactor Specificity-Switching Mutations on Protein and Cofactor Dynamics.
Rosetta modeling suggests that S208D and K349N function cooperatively to recognize NADH, with K349N supporting the S208D loop through backbone hydrogen bond and S208D forming a novel bidentate hydrogen bond to the NADH adenosine ribose that resembles interactions seen in native NADH specific proteins (Figure 2A, B). The contribution of K326T and L143P are not evident from static structural analysis since they do not directly contact NADH (Figure 2A, C, D), which motivated further analysis through MD simulations.
Figure 2.
Homology model of CHMO DTNP. (A) Overview of the cofactor binding pocket, NADH (in green) is pressed on one end by the control loop (pink). FAD (yellow) is held opposite of the NADH, and positions of the selected mutations are represented as spheres. (B) K349N supports the loop that S208D sits on, and S208D makes a bidentate hydrogen bond to the NADH adenosine ribose. (C) K326T initiates a hydrogen bond to the control loop at the backbone carbonyl of A487. The additional hydrogen bond may cause the control loop to favor maintaining the catalytically relevant, closed conformation. (D) L143P impacts the conformations that FAD can adopt.
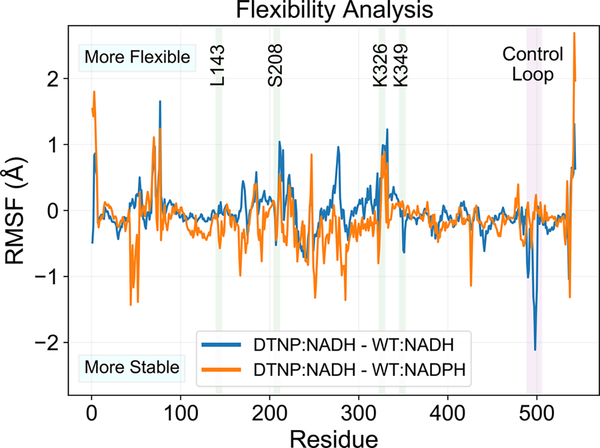
K326T contacts the “control loop” through A487 (Figure 2A, C). The control loop is observed to be ordered in some CHMO crystal structures but disordered in others,32,34,42 suggesting that its flexibility varies depending on the enzyme’s position in catalytic cycle. Specifically, it is hypothesized that the control loop must become rigid to hold the NADPH cofactor and cyclohexanone during the hydride transfer stages.32 We analyzed the flexibility of the control loop through α-carbon root-mean-square fluctuation (RMSF), and plot the difference between CHMO DTNP (bound with NADH) and CHMO WT (bound with NADH or NADPH, respectively) (Figure 3). The results show that CHMO DTNP with NADH bound maintains greater rigidity over the control loop in comparison to the WT with NADH bound and the level of control loop rigidity in CHMO DTNP with NADH bound is nearly identical with that of the CHMO WT binding NADPH. These results support the role of K326T in stabilizing the control loop which clamps on the otherwise loosely bound cofactor NADH.
Figure 3.
Root mean square fluctuation (RMSF) analysis of CHMO DTNP. Results are plotted as the difference in RMSF between DTNP (with NADH bound) and WT (with NADH or NADPH bound, respectively). The control loop is found to have greater stability with DTNP:NADH, comparable to the native condition of WT:NADPH. The inactive pairing of WT:NADH exhibits high flexibility at the control loop.
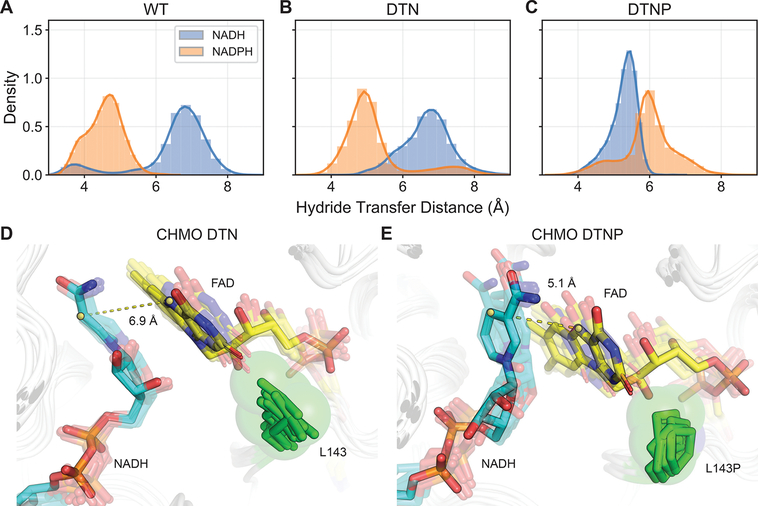
L143P contacts FAD and not NADH (Figure 2D, 4). Therefore, we hypothesized that this mutation is involved in positioning FAD and in turn affects hydride transfer from the nicotinamide cofactors to FAD. We first compared the hydride transfer distances (the distance between nicotinamide C4 and FAD N5) when CHMO WT utilizes different cofactors, and confirmed that higher activity is linked to a shorter hydride transfer distance (4.6 ± 0.5 Å for NADPH versus 6.5 ± 1.0 Å for NADH, Figure 4A),34 in line with the logic that enzymes capable of sampling catalytic geometry more frequently are more active.
Figure 4.
Evaluation of hydride transfer efficiency in CHMO variants. Shorter distances between NAD(P)H C4 and FAD N5 contribute to enhanced catalysis. (A, B) WT and DTN are both marked by NADPH sampling distances ~5 Å, while the less active NADH samples distances are >6 Å and are too remote to engage in hydride transfer. (C) DTNP displays the opposite arrangement, with NADH sampling closer distances than NADPH. (D) L143 in DTN firmly packs under the flavin ring, blocking the flavin from moving closer to nicotinamide, resulting in suboptimal hydride transfer distance. (E) L143P presses against the FAD ribitol rather than contacting the flavin. This anchors the FAD core and allows the flavin head to rotate in response to changes in the nicotinamide positioning to sustain closer contact. The dashed lines show representative distances between the nicotinamide C4 and FAD N5.
Interestingly, the hydride transfer distance when using NADH as the cofactor was much shorter in CHMO DTNP compared to in CHMO DTN (Figure 4B, C), which is consistent with our hypothesis that L143P facilitates more efficient hydride transfer from NADH.
To understand the role of L143P in influencing hydride transfer, we identified metastable FAD conformations in CHMO DTN (with NADH bound) and CHMO DTNP (with NADH bound) based on minimum heavy atom distance to residues within 4 Å by performing PCA and K-means clustering (Figure S3). Conformations from the most populated cluster of each sample were compared and indicate that the native leucine packs underneath the flavin (Figure 4D), limiting flavin’s flexibility to move closer to the nicotinamide. This limited flexibility is likely advantageous under the native condition of NADPH binding to CHMO WT, resulting in precise organization of the flavin for hydride transfer. However, upon mutation at S208D and K326T which contact the adenosine end of the cofactor, the binding pose of the cofactor is altered, which requires the flavin to adjust its position accordingly. The L143P proline in CHMO DTNP appears to pack along the FAD ribitol instead of directly against flavin (Figure 4E). This may support twisting motion of FAD to optimally orient the flavin toward the NADH nicotinamide ring.
Iterative Selection Improves NADH Dependent Conversion.
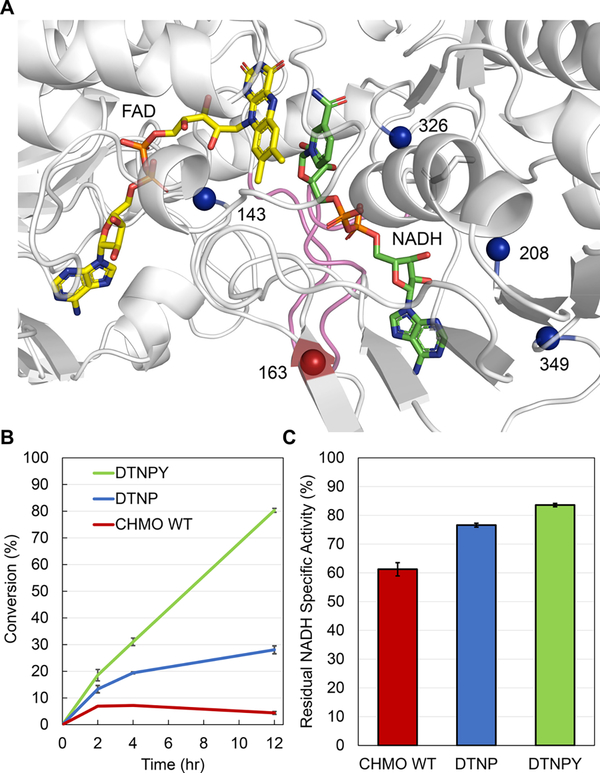
To further improve the NADH dependent activity of CHMO DTNP, a random mutagenesis library was built on top of this variant through error-prone PCR. By leveraging the high-throughput of growth-based selection we sought to explore a large sequence space and identify further mutations outside the catalytic pocket. A library of roughly 1.9 × 106 transformants was subjected to selection in strain MX304 on agar plates with 2 g/L D-glucose in M9 minimal medium and 2 g/L cyclohexanone at 30 °C. After incubation for ~120 h we identified 2 colonies with improved growth phenotypes compared to CHMO WT and CHMO DTNP (Figure S1). These two colonies carried the same variant, CHMO DTNPY (DTNP + H163Y) (Figure 5A).
Figure 5.
Assessment of the improved CHMO variant DTNPY (S208D-K326T-K349N-L143P-H163Y). (A) H163 (red) is positioned on the surface of CHMO, away from the control loop (pink), and preceding mutations (blue). (B) Conversion of cyclohexanone to caprolactone by CHMO variants. The conversion was performed with 5 mM NADH, 5 mM cyclohexanone, 50 mM Tris-Cl buffer pH 9, and 25 μg/mL purified protein, at 25 °C. CHMO WT did not achieve more than 10% conversion, whereas CHMO DTNP and CHMO DTNPY achieved ~30% and ~80% conversion, respectively. (C) Residual NADH specific activity of the characterized CHMO variants after incubation at 25 °C for one hour. CHMO WT retained only 61% of its activity, whereas CHMO DTNP and CHMO DTNPY retained 77% and 84% activity, respectively. The activity was measured with 1 mM NADH, 5 mM cyclohexanone, 50 mM Tris-Cl buffer pH 9, and 20–40 μg/mL purified protein. The structure is predicted based on homology modeling. Values are an average of two or three replicates for residual activity and conversion respectively, and the error bars represent one standard deviation.
We examined the potential application of our CHMO variants as industrial biocatalysts by performing a conversion assay with 5 mM cyclohexanone as the substrate and 5 mM NADH as the cofactor at 25 °C. Whereas CHMO WT did not achieve more than 10% conversion after 12 h incubation, CHMO DTNP achieved ~30% conversion, and CHMO DTNPY achieved ~80% conversion (Figure 5B). Analysis of the kinetic parameters for DTNPY shows that the addition of H163Y results in a slight decrease in the catalytic efficiency (kcat/KM) for NADPH compared to DTNP, but increases the catalytic efficiency for NADH by ~2-fold which is primarily driven by the increased kcat (Table 1). Overall, DTNPY showed ~10-fold increase in catalytic efficiency for NADH and ~290-fold decrease for NADPH, which collectively results in a ~2900-fold relative cofactor specificity switch from NADPH to NADH compared to the wild type. Moreover, CHMO wild type retained only 61% of its activity after incubation at 25 °C for an hour, whereas CHMO DTNP and CHMO DTNPY retained 77% and 84% activity, respectively (Figure 5C). This suggests that H163Y may also play a role in improving stability. CHMO DTNPY also shows improved cofactor selectivity compared to previously reported CHMO variants. Beier et al. reported a rationally designed CHMO mutant (S186P-S208E-K326H-K349R) with a ~2-fold higher conversion with NADH than with NADPH.24 The best variant reported in this work, CHMO DTNPY, shows ~8-fold higher NADH dependent conversion than NADPH (Figure S5).
It has been well documented that small structural changes of noncatalytic residues can have strong effects on the activity and stability of enzymes.43–45 Random mutagenesis opens the possibility of discovering these distal mutations that are difficult to predict through rational design. For this to be effective though, a high throughput screening method is needed. Here we present the use of an NADH-specific selection platform for screening a large error prone library which identified a mutation, H163Y, that improves on our semirationally designed variant CHMO DTNP.
The kinetic analysis shows that DTNPY shares the same KM as DTNP but has a doubled kcat with NADH. This suggests that H163Y may have little effect on cofactor binding but plays a role in improving turnover. In addition to activity, conversion with this variant is enhanced by DTNPY’s high coupling efficiency of ~90% with NADH (Figure S4), resulting in a 25% improvement in caprolactone formation per NADH oxidized compared to DTNP. Engineered oxygenase variants are susceptible to incomplete reaction cycles which can result in formation of the reactive oxygen species (ROS) hydrogen peroxide and superoxide.24,46 The generation of these compounds is detrimental to cell growth, and previous growth-based selections have demonstrated concomitant improvement in both activity and coupling efficiency18 as a result of strain sensitivity to ROS. Future work is needed to evaluate whether MX304 growth is similarly regulated by the additional oxidative stress from oxygenase uncoupling.
The H163 residue is located on the surface of the enzyme on a β sheet that participates in conformational changes during catalysis (Figure 5A). Conformational changes have been shown to be particularly important for the catalytic activity and coupling efficiency of oxygenases, and the cofactor plays a key role in coordinating these conformational changes in CHMO.21,23 Therefore, we hypothesize that small changes in hydrogen bond stabilization, secondary structure compaction, or orbital steering can have significant effects on CHMO’s ability to function with NADH. To elucidate the exact mechanism of CHMO DTNPY’s improved NADH activity and stability, further mutations at this position should be investigated and its crystal structure resolved.
CONCLUSION
The growth-based, high-throughput selection platform described in this work can facilitate the directed evolution of NADH-dependent oxygenases, and complements the NADPH-dependent systems developed previously.17,19 We used this selection platform to engineer a monooxygenase, CHMO, into a more viable catalyst for large-scale processes by switching its cofactor specificity toward NADH and increasing its stability. Despite engineering efforts to tackle both limitations previously,24,47–49 we hypothesized that the combination of semirational design and directed evolution, coupled with high throughput screening, would enable the discovery of innovative variants. First, selection of a four-residue, site-saturated mutagenesis library obtained CHMO DTNP with ~1200-fold relative cofactor specificity switch from NADPH to NADH. Second, selection from a random mutagenesis library built on top of CHMO DTNP yielded the additional mutation H163Y. This new variant CHMO DTNPY exhibited a ~2900-fold relative cofactor specificity switch from NADPH to NADH compared to the wild type.
Interestingly, the contributions of the selected mutations are not immediately evident from inspection of static structure. For example, CHMO DTNP harbors a spontaneous mutation L143P which falls outside the targeted sites deemed important in cofactor recognition. Likewise, DTNPY’s improved activity is caused by a mutation on the surface of the protein. MD simulations suggest that most of the mutations play a role in fine-tuning the conformational dynamics of enzyme and cofactor, which are subtle effects that are challenging to design without a high-throughput method. Conformational dynamics are particularly critical for catalysis in oxygenases,31–34 and the establishment of this selection will support the directed evolution of these complex and powerful enzymes.
METHODS
Media and Growth Conditions.
Cloning was carried out with E. coli XL-1 blue and protein expression was performed with E. coli BL21 (DE3). All E. coli were cultured in 2xYT unless otherwise noted. M9 Wash Buffer consisted of 1 mM MgSO4, 0.1 mM CaCl2, trace metal mix A5 with Co (H3BO3 2860 μg/L, MnCl2·4H2O 1810 μg/L, ZnSO4·7H2O 222 μg/L, Na2MoO4·2H2O 390 μg/L, CuSO4·5H2O 79 μg/L, Co-(NO3)2·6H2O (49 μg/L), and BD Difco M9 salts (Na2HPO4 6.78 g/L, KH2PO4 3g/L, NaCl 0.5 g/L, NH4Cl 1 g/L). M9 Selection Media shared the same composition of M9 Wash Buffer with the inclusion of 2 g/L D-glucose, 0.01 g/L thiamine, 0.04 g/L FeSO4·7H2O. For solid media M9 Selection Plates, 15 g/L agar was added in addition to M9 Selection Media composition. Concentrations for antibiotic selection were 100 mg/L for ampicillin, 50 mg/L for kanamycin, 50 mg/L for spectinomycin, and 10 mg/L for tetracycline. All strains were cultured at 37 °C with 250 rpm agitation unless otherwise noted. Induction in growth experiments and overnights was initiated with final concentrations of 0.05 mM IPTG for strains with Plac promoter. Nonselection media for the NADH strain MX304 was supplemented with 0.1% arabinose and 5 g/L acetoin to provide NADH consuming outlets by the heterologous NADH oxidase (pLS101) and endogenous gldA activity,7 respectively.
Preliminary characterization of strain growth in the presence of substrates was necessary to establish substrate availability and characterize basal effects of substrate addition. Moderate toxicity for the substrate cyclohexanone has been reported previously.20,50 During selections and growth experiments evaluating CHMO variants, substrate cyclohexanone was added at a concentration of 2 g/L to ensure substrate availability and minimize growth inhibition. Because presence of 2 g/L cyclohexanone was found to moderately extend lag phase in positive control (Lb Nox), overnight cultures substrate cyclohexanone were supplemented with 1 g/L cyclohexanone to minimize time needed for cellular adaptation to the substrate presence in selection experiments. In addition, plate cultures were parafilmed to minimize evaporation of substrate and media.
Strain Construction.
E. coli strain JCL166 served as starting strain for NADH strain MX304 construction.51 Plasmid pCP20 was used to eliminate kanamycin resistance in between knockouts performed.52 Knockouts ΔnuoF, Δndh, ΔubiC, and ΔpntB on JCL166 were generated using the P1 phage transduction method.53 Keio collection strains52 JW229–3 (ΔnuoF::kan), JW1095–1 (Δndh::kan), JW5713–1 (ΔubiC::kan), and JW1594–1 (ΔpntB::kan) served as donors for the generation of P1 lysate with gene knockout cassettes containing a kanamycin resistance marker. Construction of strain MX301 (JCL166 ΔnuoF Δndh::kan) resulted in a strain that demonstrated poor growth in even rich media. Outside of expressing heterologous genes to restore growth, strains will evolve under stressful conditions; therefore, suitable growth conditions were necessary to maintain the strain in nonselection conditions. Strictly regulated NADH consumption strategies were achieved via expression of Lb Nox on a tightly controlled arabinose inducible plasmid (pLS101). Supplementation of pLS101 and 0.1% arabinose was maintained in all further strain variants to provide NADH sink in nonselection culturing to preserve growth phenotype of the strains. All knockouts were confirmed by MyTaq Colony PCR (Bioline).
Characterization of MX304 Growth Behavior with Oxidases.
The growth rescue condition used for Figure 1B, 1C, and S2 are as follows: Briefly, the strains were first cultured in 2xYT under aerobic conditions at 30 °C overnight with appropriate antibiotics, inducers, and 2 g/L glucose. Next, overnight cultures were washed 3 times and resuspended in M9 Wash Buffer. For liquid growth, a 0.1% (v/v) volume of washed culture was used to inoculate to an OD600nm of ~0.05 in 0.3 mL of M9 Selection Media, with appropriate antibiotics and inducers. Culture tubes were incubated at 30 °C in a rotary shaker, and OD600nm was measured using a 96-well plate reader (Figure 1B). For solid growth of Figure 1C, ~104 cells/mL were prepared in M9 Wash and 100 μL aliquots were dispensed on an agar plate of M9 Selection Media, with appropriate antibiotics and inducers. For solid growth of Figure S2, targeted serial dilutions of 106, 105, and 104 cells/mL were prepared in M9 Wash and 2 μL aliquots were dispensed in series on an agar plate of M9 Selection Media, with appropriate antibiotics and inducers. Plates were grown at designated temperatures and photos were taken to document growth progress (Figure 1C, S2).
Plasmid Construction.
All PCR fragments were generated using PrimeSTAR Max DNA Polymerase (TaKaRa) unless otherwise noted. Splicing-by-overlap extension (SOE) PCR and degenerate codon PCR was conducted using KOD Xtreme Hot Start DNA Polymerase (Novagen).
The Lactobacillus brevis nox gene was amplified from previously constructed plasmid pSM101 originally cloned from genomic DNA from Lactobacillus brevis strain 118–8 [ATCC 367]. After PCR and gel extraction, the Lb Nox gene fragment was inserted into the pQElac vector backbone which contains a 6 × His tag at the N-terminus (ColE1 ori, Ampr) using Gibson isothermal DNA assembly method,54 resulting in plasmid pLS401. Plasmid pLS402 carrying the TP Nox gene was generated using previously constructed plasmid pLS102 as a template which carries the following mutations G159A-D177A-A178R-M179S-P184R.
The Acinetobacter sp. NCIMB 9871 chnB gene was previously cloned into the pQElac vector backbone using Gibson isothermal DNA assembly method, resulting in plasmid pLS201. Plasmid pLS405 was generated by the reverting the L143P mutation to the wildtype residue using pLS404 as a template.
Generation of S208-K326-K349-T378 NNK Library.
The Ac CHMO combinatorial site-saturation mutagenesis library was cloned using primers containing NNK or MNN degenerate codons at targeted sites. PCR was used to amplify a DNA fragment from pLS201 between a forward primer beginning at position A209 and a reverse primer beginning at position A325. An additional fragment was generated by PCR of the same template using a forward primer with NNK codon at position K326 and a reverse primer with MNN codon at position K349. A final PCR fragment was amplified from the same template using primers containing a forward primer beginning at position A350 and a reverse primer containing an MNN codon at position T378. The fragments of corresponding length were purified by gel electrophoresis and three purified fragments were used as templates in an SOE PCR reaction to amplify a single fragment containing mutations at sites K326, K349, and T378. PCR was used to amplify a DNA fragment from pLS208 between a forward primer beginning at position G379 and a reverse primer containing an MNN codon at position S208. Both fragments were purified by gel electrophoresis, assembled using Gibson Isothermal Assembly, and then transformed into ElectroMAX DH10β competent cells (Invitrogen) by electroporation. Transformed cells were rescued in 600 μL SOC medium by shaking for 1 h at 37 °C. After rescue, cells were added to 20 mL 2xYT medium in a 250 mL baffled shake flask and 2, 20, and 200 μL was plated on 2xYT agar plates with appropriate antibiotics. Colonies formed on these plates were counted to estimate library size. The liquid culture was incubated at 37 °C for 10 h before plasmid extraction to generate the plasmid library, pLS403. Ten colonies from the library estimation plates were cultured in liquid medium for plasmid extraction and sequencing to confirm diverse mutations at all four targeted sites.
DNTP Error-Prone Library Construction.
The Ac CHMO Error-Prone PCR Library was cloned using the GeneMorph II Random Mutagenesis Kit (Agilent) to generate 1 to 2 random mutations on gene Ac chnB (L143P-S208D-K326T-K349N). Using 175 ng of template DNA (provided by plasmid pLS404), the gene insert was amplified via error prone PCR with 25 reaction cycles according to manufacturer’s instructions. A panel of starting template material (150 ng to 250 ng) was initially evaluated to tune range of mutations observed. To construct the Ac chnB error-prone insert library, a forward primer containing a BamHI-HF digestion site was used in a PCR reaction with a reverse primer containing an XbaI digestion site to generate the full-length gene insert with random PCR errors. The gel purified insert fragment and plasmid pLS201 (backbone) were separately digested with BamHI-HF and XbaI (NEB). Both digestion products were again gel purified and the appropriate length fragments were assembled by ligation, purified, and transformed into Electro-MAX DH10β cells (Invitrogen) using electroporation. Then cells were rescued with SOC medium at 37 °C for 1 h and then added to 20 mL 2xYT medium with appropriate antibiotics. Then 2, 20, and 200 μL of culture was sampled from the culture and plated on an 2xYT agar plates with appropriate antibiotics. The remaining liquid culture was incubated at 37 °C with 250 rpm agitation for 10 h, before extraction of the library DNA (pLS406). The plates were incubated at 37 °C overnight, and colonies formed were counted to estimate the library size (1.2 × 107). Six single colonies were also cultured individually to extract plasmids, which were sequenced to sample the sequence space of the library.
Transformation of Libraries for Selections with NADH Strains (MX304).
To generate electrocompetent E. coli, selection strain cells were cultured in 200 mL SOB medium with appropriate antibiotics at 30 °C with shaking at 250 rpm until OD600nm reached 0.4–0.6. The culture was chilled on ice for 15 min and the cells were pelleted at 4 °C, 4000g. The cells were washed at 4 °C three times with 40 mL 10% glycerol solution (ice cold). After, cells were finally resuspended with 500 μL 10% glycerol solution (ice cold), and aliquoted for transformation.
The transformation was performed as follows: After electrocompetent cells were prepared, 20 μL of library DNA was added to 200 μL competent cells. Cell-DNA mixture was added to four ice chilled 1 mm gap electroporation cuvettes (55 μL of per electroporation cuvette). Cells were electroporated at 2 kV, 129 Ω, 50 μF, resistance 2.5 kV; 200 μL of SOC medium was immediately added and transferred to a microcentrifuge tube at room temperature. This step was repeated twice more. Cells were rescued at 37 °C with shaking for 1 h. Serial dilution of the cells was performed and then plated on 2xYT agar plates with appropriate antibiotics and addition of acetoin and arabinose as needed. After incubation at 37 °C overnight, colonies formed were counted to estimate transformation efficiency.
Selection of Ac CHMO NADH Library.
E. coli MX304 cells were transformed with the Ac CHMO NNK NADH library pLS403 or Ac CHMO DTNP Error Prone Library pLS406 via electroporation. After rescue in SOC medium for 1 h, cultures were combined in 20 mL 2xYT with appropriate antibiotics, 0.1% arabinose, 5 g/L acetoin, and 1 g/L cyclohexanone in a 250 mL baffled shake flask. A dilution series from the culture was plated to determine transformation of individual variants: 2.47 × 107 for the NNK library and 1.9 × 106 for the error prone library. Cultures with CHMO WT (pLS201) negative control and NADH oxidase (pLS401) positive control were added to 5 mL of the same induction media described above in a 50 mL conical tube (cap loose to allow increased aeration). All cultures were grown at 30 °C for ~8 h or until OD600nm = 0.6 was reached. Subsequently, controls and the library were induced by addition of IPTG. Cultures were grown for an additional 4 h or until OD600nm = ~1.
To prepare cells for the selection condition, 1 mL of each culture was pelleted in 2 mL microcentrifuge tubes and washed three times with M9 Wash Buffer. After wash, cells were finally resuspended in 1 mL M9 Wash Buffer. Cells were diluted with M9 Wash Buffer to a final cell concentration of ~107 cells/mL for the NNK selection, and ~105 for the error prone selection. Ten 100 μL aliquots of this cell suspension were plated on separate M9 Selection Plates supplemented with 2 g/L cyclohexanone and incubated at 30 °C for 48–120 h. The colony growth was monitored periodically. Selected colonies were restreaked onto fresh selection media and again incubated to obtain single colonies. Colonies were favored based on their initial time of observable growth, speed of growth, and the size of the colonies produced. In the case of our initial selections, growth rescue was compared to CHMO WT, whereas in iterative selections growth was compared to the best available variant. From NNK and error prone selections, 26 and 3 single colonies, respectively, were cultured in liquid media to extract plasmids using QIAprep Spin Miniprep kit (Qiagen). Sequencing results highlighted variant on plasmid pLS404 (DTNP) which appeared four times in the NNK library selection, as well as the error prone mutant DTNPY which appeared twice in contrast to majority of variants which appeared once. To confirm growth phenotype conferred, isolated variant plasmids were transformed individually into MX304 and subjected to identical selection protocol. Due to the superior growth conferred by DTNP and DTNPY compared to other variants and preliminary activity assays, only variant encoding the CHMO DTNP and DTNPY were extensively characterized.
Characterization of MX304 Growth Behavior with CHMO Variants.
The growth rescue condition used are as follows: Briefly, the strains tested were first cultured in 2xYT under aerobic conditions at 30 °C overnight with appropriate antibiotics, inducers, and 2 g/L glucose. Next, overnight cultures were washed 3 times and resuspended in M9 Wash Buffer. For solid growth, targeted serial dilutions of 107 cells/mL, 106 cells/mL, 105 cells/mL, and 104 cells/mL were prepared in M9 Wash and 2 μL aliquots were dispensed in series on an agar plate of M9 Selection Media, with appropriate antibiotics and inducers. Plates were grown at designated temperatures and photos were taken to document growth progress (Figure S1). Plates were parafilmed to minimize evaporation of substrate and media.
Expression and Purification of CHMO Wild Type and Variants.
Enzyme expression of pQElac based plasmids used E. coli BL21 (DE3) containing the plasmids encoding for the CHMO wild type or variants. Expression was carried out by inoculation of 50 mL 2xYT media supplied with 200 mg/L ampicillin and an overnight culture. Cells were grown at 37 °C in baffled shaking flasks and were induced at an OD600nm of 0.6–0.8 with 0.25 mM IPTG. The 50 mL culture was then incubated at 25 °C and 250 rpm for 24 h. After 24 h cells were harvested for protein extraction (centrifugation at 4 °C, 4000g for 15 min). Cell pellets were resuspended in HisPur Ni-NTA equilibration buffer (Thermo Fisher Scientific). Resuspended cell mixture was added to 0.5 mL of 0.1 mm glass beads (Biospec) and homogenized using a benchtop homogenizer (FastPrep-24, MP Biomedicals). Cell debris was separated from the crude extract by centrifugation at 4 °C, 15 000 rcf for 15 min. Protein purification was performed using HisPur Ni-NTA Protein Miniprep (Thermo Fisher Scientific) according to the manufacturer’s instructions. The histidine-tagged protein was eluted in HisPur elution buffer (50 mM sodium phosphate buffer pH 7.5, 300 mM NaCl, 300 mM imidazole). The concentrations of purified protein were quantified by Bradford assay using BSA as standards. Purified protein was stored with 20% (v/v) glycerol at −80 °C.
Steady State Kinetic Analyses for Cyclohexanone and NAD(P)H.
Catalytic efficiencies (kcat/KM) for NAD(P)H (Table 1) were measured according to previously described methods.24 The reaction mixture containing 50 mM Tris-Cl buffer (pH 9.0), 1 mM cyclohexanone, and varied NAD(P)H concentrations from 0.02 mM to 4 mM was incubated at 25 °C. All the reactions were initiated by addition of an appropriate amount of the enzyme (purified protein from −80 °C stock) and the kinetic parameters were measured by monitoring the NAD(P)H consumption at 340 nm in 96-well plate. The rate of NAD(P)H consumption was determined upon stabilization of linear region, after approximately ~200 s. Protein concentrations used were 0.7–0.15 μM for NADH, and 0.04–0.015 μM for NADPH.
Cyclohexanone Conversion Assay.
Purified and diluted CHMO variants (25 μg/mL, purified protein from −80 °C, 20% glycerol stock) were incubated in Tris-HCl buffer (50 mM, pH 9) with 5 mM NADH and 5 mM cyclohexanone (1 M stock dissolved in ethanol) at incubation temperature (25 °C) in sealed glass vials. At 2, 4, and 12 h, the reaction mixture was sampled. Caprolactone and cyclohexanone were extracted with an equal volume of ethyl acetate with 200 mg/L octanol as an internal standard. To confirm productive NADH oxidation, cyclohexanone conversion to caprolactone was determined for CHMO variants via gas chromatography–flame ionization detection (GC–FID). Samples were analyzed using previously described GC methods.20 To monitor the amount of remaining cofactor, the aqueous phase of the extraction was measured at 340 nm in 96 well plate.
GC–FID Analytical Methods.
All GC analysis was performed on an Agilent 6850 (Agilent Technologies) equipped with an FID. An Agilent DB-WAXetr capillary column (30 m × 0.25 mm × 0.5 μm) was used for separation. The inlet and detector were held at 250 and 260 °C, respectively. The GC was operated in constant flow mode with a flow of 2.7 mL min−1. Helium was used as the carrier gas. Air and hydrogen were supplied to the FID at 350 and 40 mL min−1, respectively. All gases were purchased from Airgas and 1 μL of sample was injected with a split ratio of 5:1. For analysis of cyclohexanone and its oxidation product caprolactone, the oven was initially held at 170 °C for 2 min and then ramped at a rate of 5 °C min−1 to 190 °C. Cyclohexanone and caprolactone eluted at 1.2 and 3.9 min, respectively. Octanol was used as an internal standard.
Residual Specific Activity Analysis with NADH.
Stability of CHMO variants was assessed by measuring the residual specific activity for each variant with NADH (Figure 5C). Specific activity assays were performed according to previously described methods.48 The reaction mixture containing 50 mM Tris-Cl buffer (pH 9.0), 5 mM cyclohexanone, and 0.02 mg/mL (DTNP, DTNPY) or 0.04 mg/mL protein (WT CHMO) protein (purified protein from −80 °C stock) was incubated either on ice or at 25 °C for 1 h prior to specific activity measurements. All reactions were initiated by addition of 1 mM NADH, and activity was measured by monitoring the NADH consumption at 340 nm in a 96-well plate at 25 °C. The residual specific activity was determined by calculating the ratio of the activity of the 25 °C incubated reaction mixture to the activity of the reaction mixture incubated on ice.
Ac CHMO Homology Modeling.
The model of WT Ac CHMO with cofactors FAD and NADPH bound was generated with Rosetta CM.55,56 Threading templates with high sequence identity and cocrystallized cofactors were identified through BLASTP search of the Protein Data Bank with Ac CHMO as the query.57,58 Crystal structures of CHMO from Rhodococcus sp. (PDB: 4RG3, 3GWD, 3GWF, 3UCL), which shares 57.8% sequence identity to Ac CHMO, were selected as input models.34,42,59 The Rosetta CM protocol consisted of repeated rounds where the target Ac CHMO sequence was threaded onto the template structure based on MAFFT sequence alignment, segments of the protein structure were constructed through insertion of fragments drawn from the library provided by the templates through Monte Carlo evaluation, followed by minimization to relax the final output.60 1500 homology modeling trajectories were completed, the output structure with the most favorable total Rosetta energy was selected as the representative model for all further analysis. Point mutations for the Ac CHMO variants were produced through 1000 further Rosetta docking simulations on the homology model with backbone flexibility, side chain repacking, and ligand minimizations; final models were selected by lowest total Rosetta energies. The NADH binding poses were prepared by deleting atoms of the ribose 2′ phosphate group on the existing NADPH models prior to the Rosetta Design trials.
The Ac CHMO model shows that NADPH binds in an extended conformation with the nicotinamide ring tucked into a small binding pocket against the FAD flavin. The NADPH binding mode is characterized by the conserved Rossman fold with β-α-β secondary structure motifs enclosing the cofactor. The primary interactions predicted to hold the NADPH in place include: Q190 side-chain amide which contacts the NADPH carboxamide oxygen, W490 indole which forms a hydrogen bond to the nicotinamide ribose hydroxyl, backbone polar interactions at the N-terminus of the Rossman α-helix and hydrogen bonding from the T189 hydroxyl to the pyrophosphate, a salt-bridge from K326 extending from a loop across the substrate channel to the pyrophosphate, and R207 guanidinium forming a salt-bridge to the ribose 2′ phosphate group and packing against the adenosine ring. The side of the NADPH facing away from the Rossman fold is marginally exposed, with the control loop defined as residues 489–505 lightly packing against the cofactor. Since NADH differs from NADPH only in the absence of the 2′ phosphate group and is capable of establishing the same set of binding interactions along the pyrophosphate and nicotinamide ring, we postulate that Ac CHMO’s strict specificity for NADPH is driven by the R207 contact spanning from the end of the second Rossman β-strand stabilizing the adenosine tail of the cofactor for optimal packing dynamics with the control loop. The FAD is held opposite of the nicotinamide cofactor with the flavin group facing the nicotinamide ring and is tightly bound through polar contacts throughout the adenosine tail, ribose, pyrophosphate, ribitol, and flavin carbonyls. The numerous hydrogen bonds restrict FAD mobility, position the flavin for efficient hydride transfer with the nicotinamide cofactor or substrate, and prevent the FAD release. The cyclohexanone binding pocket is adjacent to the nicotinamide ring, it is proposed that some degree of protein and NADPH flexibility is required to allow the substrate to move close enough to the FAD for electron transfer
Molecular Dynamics Simulations.
MD simulations of the Ac CHMO variants were completed with PMEMD from the AMBER 18 package utilizing the ff14sb force field and 8 Å Particle Mesh Ewald real space cutoff.61–64 Cofactor parameters were obtained from the AMBER parameter database and protonation states of titratable residues were determined with the H++ Web server.65–67 The TLEAP program was utilized to solvate the complexes with TIP3P water molecules in a truncated octahedron with 10 Å buffer and neutralizing Na+/Cl− counterions. The Ac CHMO systems were minimized in two stages, first with 2500 steps of steepest decent and 2500 steps of conjugate gradient where all nonhydrogen solute atoms were restrained with a 20 kcal mol−1 Å−2 force to relieve solvent clash. The second stage minimization to remove solute steric clashes was run with the same cycle settings and restraints removed. Heating from 0 to 300 K was performed over 0.5 ns with 10 kcal mol−1 Å −2 restraints on all non-hydrogen solute atoms under NPT conditions at 1 atm pressure with Langevin thermostat and 1 fs time step. Solvent density equilibration over 5 ns with 5 kcal mol−1 Å −2 restraints on all solute atoms and an unrestrained 10 ns equilibration using 2 fs time step to clear remaining structural artifacts followed the heating stage. Production MD trajectories were each carried out for 400 ns with 2 fs time step, SHAKE restraints on hydrogens, NVT ensemble, Langevin thermostat with collision frequency 1.0 ps−1, and periodic boundary conditions.
Cofactor Binding Analysis.
Cofactor binding comparisons were completed between WT Ac CHMO, DTN CHMO, and DTNP CHMO with either NADH or NADPH bound. Protein backbone flexibility over the trajectories was measured through alpha-carbon RMSF. Hydride transfer potential was recorded as the distance between the nicotinamide C4 to FAD N5. Metastable FAD binding conformations were established through featurization on minimum heavy atom distance to residues with any atom within 4 Å contact and PCA dimensionality reduction with K-means clustering. The distance array was standardized to zero mean and unit variance and transformed to lower dimensional space with components maintaining maximal variance through PCA. K-means clustering was performed to discretize the sampled conformations projected onto the free energy landscape of the first two PCA components into metastable states. The optimal number of clusters was selected by the elbow heuristic where clustering over a range of K values, from one to nine here, is completed and the sum of squared distances from the sample points to their assigned cluster center is computed. The value of K where the sum of squared distances decrease becomes linear is selected as optimal and indicates that increasing K further will result in overfitting. The residue positions neighboring FAD include: 12, 13, 15, 16, 17, 36, 37, 44, 45, 46, 48, 49, 51, 56, 57, 58, 63, 109, 110, 140, 141, 142, 390, 426, 434, 435, 436, and 439. MDTraj and cpptraj were utilized for trajectory analysis, data processing was completed with the NumPy and scikit-learn packages.68–71 PyMol was used to illustrate the structures (Schrödinger LLC, 2020).
Supplementary Material
ACKNOWLEDGMENTS
H.L. acknowledges support from University of California, Irvine, the National Science Foundation (NSF) (Award No. 1847705), and the National Institutes of Health (NIH) (Award No. DP2 GM137427). S.M. acknowledges support from the NSF Graduate Research Fellowship Program (Grant No. DGE-1839285). D.A. acknowledges support from the NSF Graduate Research Fellowship Program (Grant No. DGE-1839285).
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.1c00258.
Plasmids and strains used in this study (Table S1); Growth phenotypes of MX304 with complementation by different CHMO variants (Figure S1); Induction of NADH oxidase is necessary for growth restoration of MX304 (Figure S2); Free energy landscapes for CHMO DTN, DTNP, and WT with NADH or NADPH (Figure S3); Coupling efficiency of CHMO DTNPY, DTNP, and wild type (Figure S4); NADPH-dependent cyclohexanone to caprolactone conversion by CHMO variants (Figure S5) (PDF)
Contributor Information
Sarah Maxel, Departments of Chemical and Biomolecular Engineering, University of California, Irvine, California 92697, United States.
Samer Saleh, Departments of Chemical and Biomolecular Engineering, University of California, Irvine, California 92697, United States.
Edward King, Molecular Biology and Biochemistry, University of California, Irvine, California 92697, United States.
Derek Aspacio, Departments of Chemical and Biomolecular Engineering, University of California, Irvine, California 92697, United States.
Linyue Zhang, Departments of Chemical and Biomolecular Engineering, University of California, Irvine, California 92697, United States.
Ray Luo, Molecular Biology and Biochemistry, Departments of Chemical and Biomolecular Engineering, and Biomedical Engineering, University of California, Irvine, California 92697, United States.
Han Li, Departments of Chemical and Biomolecular Engineering and Biomedical Engineering, University of California, Irvine, California 92697, United States.
REFERENCES
- (1).Rousseau O; Ebert MCCJC; Quaglia D; Fendri A; Parisien AH; Besna JN; Iyathurai S; Pelletier JN Indigo Formation and Rapid NADPH Consumption Provide Robust Prediction of Raspberry Ketone Synthesis by Engineered Cytochrome P450 BM3. ChemCatChem 2020, 12, 837–845. [Google Scholar]
- (2).Weingartner AM; Sauer DF; Dhoke GV; Davari MD; Ruff AJ; Schwaneberg U A hydroquinone-specific screening system for directed P450 evolution. Appl. Microbiol. Biotechnol. 2018, 102, 9657–9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gielen F; Hours R; Emond S; Fischlechner M; Schell U; Hollfelder F Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS). Proc. Natl. Acad. Sci. U. S. A. 2016, 113, E7383–E7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Debon A; Pott M; Obexer R; Green AP; Friedrich L; Griffiths AD; Hilvert D Ultrahigh-throughput screening enables efficient single-round oxidase remodelling. Nat. Catal. 2019, 2, 740–747. [Google Scholar]
- (5).Spielmann A; Brack Y; van Beek H; Flachbart L; Sundermeyer L; Baumgart M; Bott M NADPH biosensor-based identification of an alcohol dehydrogenase variant with improved catalytic properties caused by a single charge reversal at the protein surface. AMB Express 2020, 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Machado HB; Dekishima Y; Luo H; Lan EI; Liao JC A selection platform for carbon chain elongation using the CoA-dependent pathway to produce linear higher alcohols. Metab. Eng. 2012, 14, 504–511. [DOI] [PubMed] [Google Scholar]
- (7).Liang K; Shen CR Selection of an endogenous 2,3-butanediol pathway in Escherichia coli by fermentative redox balance. Metab. Eng. 2017, 39, 181–191. [DOI] [PubMed] [Google Scholar]
- (8).Zhang L; King E; Luo R; Li H Development of a High-Throughput, in Vivo Selection Platform for NADPH-Dependent Reactions Based on Redox Balance Principles. ACS Synth. Biol 2018, 7, 1715–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wenk S; Schann K; He H; Rainaldi V; Kim S; Lindner SN; Bar-Even A An “energy-auxotroph” Escherichia coli provides an in vivo platform for assessing NADH regeneration systems. Biotechnol. Bioeng. 2020, 117, 3422–3434. [DOI] [PubMed] [Google Scholar]
- (10).Lindner SN; Ramirez LC; Krüsemann JL; Yishai O; Belkhelfa S; He H; Bouzon M; Döring V; Bar-Even A NADPH-Auxotrophic E. coli: A Sensor Strain for Testing in Vivo Regeneration of NADPH. ACS Synth. Biol. 2018, 7, 2742–2749. [DOI] [PubMed] [Google Scholar]
- (11).Calzadiaz-Ramirez L; Calvó-Tusell C; Stoffel GMM; Lindner SN; Osuna S; Erb TJ; Garcia-Borràs M; Bar-Even A; Acevedo-Rocha CG In Vivo Selection for Formate Dehydrogenases with High Efficiency and Specificity toward NADP+. ACS Catal. 2020, 10, 7512–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Auriol C; Bestel-Corre G; Claude J; Soucaille P; Meynial-Salles I Stress-induced evolution of Escherichia coli points to original concepts in respiratory cofactor selectivity. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Zhou Y; Wang L; Yang F; Lin X; Zhang S; Zhao ZK Determining the extremes of the cellular NAD(H) level by using an Escherichia coli NAD + -auxotrophic mutant. Appl. Environ. Microbiol. 2011, 77, 6133–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lin Y; Yan Y Biotechnological production of plant-specific hydroxylated phenylpropanoids. Biotechnol. Bioeng. 2014, 111, 1895–1899. [DOI] [PubMed] [Google Scholar]
- (15).Urlacher VB; Girhard M Cytochrome P450 Monooxygenases in Biotechnology and Synthetic Biology. Trends Biotechnol. 2019, 37, 882–897. [DOI] [PubMed] [Google Scholar]
- (16).Furuya T; Kino K Catalytic activity of the two-component flavin-dependent monooxygenase from Pseudomonas aeruginosa toward cinnamic acid derivatives. Appl. Microbiol. Biotechnol. 2014, 98, 1145–1154. [DOI] [PubMed] [Google Scholar]
- (17).Maxel S; Aspacio D; King E; Zhang L; Acosta AP; Li H A Growth-Based, High-Throughput Selection Platform Enables Remodeling of 4-Hydroxybenzoate Hydroxylase Active Site. ACS Catal. 2020, 10, 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Maxel S; King E; Zhang Y; Luo R; Li H Leveraging oxidative stress to regulate redox balance-based, in vivo growth selections for oxygenase engineering. ACS Synth. Biol. 2020, 9, 3124–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kramer L; Le X; Rodriguez M; Wilson MA; Guo J; Niu W Engineering Carboxylic Acid Reductase (CAR) through a Whole-Cell Growth-Coupled NADPH Recycling Strategy. ACS Synth. Biol. 2020, 9, 1632–1637. [DOI] [PubMed] [Google Scholar]
- (20).Maxel S; Zhang L; King E; Acosta AP; Luo R; Li H In Vivo, High-Throughput Selection of Thermostable Cyclohexanone Monooxygenase (CHMO). Catalysts 2020, 10, 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zhang Y; Wu YQ; Xu N; Zhao Q; Yu HL; Xu JH Engineering of Cyclohexanone Monooxygenase for the Enantioselective Synthesis of (S)-Omeprazole. ACS Sustainable Chem. Eng. 2019, 7, 7218–7226. [Google Scholar]
- (22).Xu N; Zhu J; Wu YQ; Zhang Y; Xia JY; Zhao Q; Lin GQ; Yu HL; Xu JH Enzymatic Preparation of the Chiral (S)-Sulfoxide Drug Esomeprazole at Pilot-Scale Levels. Org. Process Res. Dev. 2020, 24, 1124–1130. [Google Scholar]
- (23).de Gonzalo G; Alcantara AR Multienzymatic Processes Involving Baeyer-Villiger Monooxygenases. Catalysts 2021, 11, 605. [Google Scholar]
- (24).Beier A; Bordewick S; Genz M; Schmidt S; van den Bergh T; Peters C; Joosten HJ; Bornscheuer UT Switch in Cofactor Specificity of a Baeyer-Villiger Monooxygenase. ChemBioChem 2016, 17, 2312–2315. [DOI] [PubMed] [Google Scholar]
- (25).Chánique AM; Parra LP Protein engineering for nicotinamide coenzyme specificity in oxidoreductases: Attempts and challenges. Front. Microbiol. 2018, DOI: 10.3389/fmicb.2018.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Cahn JKB; Werlang CA; Baumschlager A; Brinkmann-Chen S; Mayo SL; Arnold FH A General Tool for Engineering the NAD/NADP Cofactor Preference of Oxidoreductases. ACS Synth. Biol. 2017, 6, 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Mordhorst S; Andexer JN Round, round we go-strategies for enzymatic cofactor regeneration. Nat. Prod. Rep. 2020, 37, 1316–1333. [DOI] [PubMed] [Google Scholar]
- (28).Pick A; Ott W; Howe T; Schmid J; Sieber V Improving the NADH-cofactor specificity of the highly active AdhZ3 and AdhZ2 from Escherichia coli K-12. J. Biotechnol. 2014, 189, 157–165. [DOI] [PubMed] [Google Scholar]
- (29).You Z-N; Chen Q; Shi S-C; Zheng M-M; Pan J; Qian X-L; Li C-X; Xu J-H Switching cofactor dependence of 7β-hydroxysteroid dehydrogenase for cost-effective production of ursodeoxycholic acid. ACS Catal. 2019, 9, 466–473. [Google Scholar]
- (30).Wu W; Zhang Y; Liu D; Chen Z Efficient mining of natural NADH-utilizing dehydrogenases enables systematic cofactor engineering of lysine synthesis pathway of Corynebacterium glutamicum. Metab. Eng. 2019, 52, 77–86. [DOI] [PubMed] [Google Scholar]
- (31).Acevedo-Rocha CG; Li A; D’Amore L; Hoebenreich S; Sanchis J; Lubrano P; Ferla MP; Garcia-Borràs M; Osuna S; Reetz MT Pervasive cooperative mutational effects on multiple catalytic enzyme traits emerge via long-range conformational dynamics. Nat. Commun. 2021, DOI: 10.1038/s41467-021-21833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Yachnin BJ; Lau PCK; Berghuis AM The role of conformational flexibility in Baeyer-Villiger monooxygenase catalysis and structure. Biochim. Biophys. Acta, Proteins Proteomics 2016, 1864, 1641–1648. [DOI] [PubMed] [Google Scholar]
- (33).Butler CF; Peet C; Mason AE; Voice MW; Leys D; Munro AW Key mutations alter the cytochrome P450 BM3 conformational landscape and remove inherent substrate bias. J. Biol. Chem. 2013, 288, 25387–25399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Mirza IA; Yachnin BJ; Wang S; Grosse S; Bergeron H; Imura A; Iwaki H; Hasegawa Y; Lau PCK; Berghuis AM Crystal Structures of Cyclohexanone Monooxygenase Reveal Complex Domain Movements and a Sliding Cofactor. J. Am. Chem. Soc. 2009, 131, 8848–8854. [DOI] [PubMed] [Google Scholar]
- (35).Luo ZW; Lee SY Biotransformation of p-xylene into terephthalic acid by engineered Escherichia coli. Nat. Commun. 2017, 8, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jeon HJ; Kim MN Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int. Biodeterior. Biodegrad. 2015, 103, 141–146. [Google Scholar]
- (37).Kim Y; Ingram LO; Shanmugam KT Dihydrolipoamide Dehydrogenase Mutation Alters the NADH Sensitivity of Pyruvate Dehydrogenase Complex of Escherichia coli K-12. J. Bacteriol. 2008, 190, 3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wang X; Wang A; Zhu L; Hua D; Qin J Altering the sensitivity of Escherichia coli pyruvate dehydrogenase complex to NADH inhibition by structure-guided design. Enzyme Microb. Technol. 2018, 119, 52–57. [DOI] [PubMed] [Google Scholar]
- (39).Atsumi S; Cann AF; Connor MR; Shen CR; Smith KM; Brynildsen MP; Chou KJY; Hanai T; Liao JC Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 2008, 10, 305–311. [DOI] [PubMed] [Google Scholar]
- (40).Titov DV; Cracan V; Goodman RP; Peng J; Grabarek Z; Mootha VK Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science 2016, 352, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Cracan V; Titov DV; Shen H; Grabarek Z; Mootha VK A genetically encoded tool for manipulation of NADP+/NADPH in living cells. Nat. Chem. Biol. 2017, 13, 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Yachnin BJ; McEvoy MB; Maccuish RJD; Morley KL; Lau PCK; Berghuis AM Lactone-bound structures of cyclohexanone monooxygenase provide insight into the stereochemistry of catalysis. ACS Chem. Biol. 2014, 9, 2843–2851. [DOI] [PubMed] [Google Scholar]
- (43).Xi L; Cho KW; Herndon ME; Tu SC; et al. Elicitation of an oxidase activity in bacterial luciferase by site-directed mutation of a noncatalytic residue. J. Biol. Chem. 1990, 265, 4200–4203. [PubMed] [Google Scholar]
- (44).Wong KF; Selzer T; Benkovic SJ; Hammes-Schiffer S; et al. Impact of distal mutations on the network of coupled motions correlated to hydride transfer in dihydrofolate reductase. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 6807–6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Kennedy EJ; Yang J; Pillus L; Taylor SS; Ghosh G; et al. Identifying critical non-catalytic residues that modulate protein kinase A activity. PLoS One 2009, 4, 4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Morlock LK; Böttcher D; Bornscheuer UT Simultaneous detection of NADPH consumption and H 2 O 2 production using the AmplifluTM Red assay for screening of P450 activities and uncoupling. Appl. Microbiol. Biotechnol. 2018, 102, 985–994. [DOI] [PubMed] [Google Scholar]
- (47).van Beek HL; de Gonzalo G; Fraaije MW Blending Baeyer-Villiger monooxygenases: Using a robust BVMO as a scaffold for creating chimeric enzymes with novel catalytic properties. Chem. Commun. 2012, 48, 3288–3290. [DOI] [PubMed] [Google Scholar]
- (48).Schmidt S; Genz M; Balke K; Bornscheuer UT The effect of disulfide bond introduction and related Cys/Ser mutations on the stability of a cyclohexanone monooxygenase. J. Biotechnol. 2015, 214, 199–211. [DOI] [PubMed] [Google Scholar]
- (49).van Beek HL; Wijma HJ; Fromont L; Janssen DB; Fraaije MW Stabilization of cyclohexanone monooxygenase by a computationally designed disulfide bond spanning only one residue. FEBS Open Bio 2014, 4, 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Lee W; Park J; Park K; Kim M; Seo J Enhanced production of ε-caprolactone by overexpression of NADPH-regenerating glucose 6-phosphate dehydrogenase in recombinant Escherichia coli harboring cyclohexanone monooxygenase gene. Appl. Microbiol. Biotechnol. 2007, 76, 329–338. [DOI] [PubMed] [Google Scholar]
- (51).Shen CR; et al. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Datsenko KA; Wanner BL One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Thomason LC; Costantino N; Court DLE coli Genome Manipulation by P1 Transduction. Curr. Protoc. Mol. Biol. 2007, 1.17.1–1.17.8. [DOI] [PubMed] [Google Scholar]
- (54).Gibson DG; et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [DOI] [PubMed] [Google Scholar]
- (55).Fleishman SJ; Leaver-Fay A; Corn JE; Strauch E-M; Khare SD; Koga N; Ashworth J; Murphy P; Richter F; Lemmon G; Meiler J; Baker D Rosettascripts: A scripting language interface to the Rosetta Macromolecular modeling suite. PLoS One 2011, 6, e20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Song Y; et al. High-resolution comparative modeling with RosettaCM. Structure 2013, 21, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Madden T Chapter 16: The BLAST Sequence Analysis Tool. In The NCBI Handbook; National Center for Biotechnology Information, 2002. [Google Scholar]
- (58).Berman HM The Protein Data Bank/Biopython. Presentation 2000, 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Yachnin BJ; Sprules T; McEvoy MB; Lau PCK; Berghuis AM The substrate-bound crystal structure of a baeyer-villiger monooxygenase exhibits a criegee-like conformation. J. Am. Chem. Soc. 2012, 134, 7788–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Katoh K MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Essmann U; et al. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar]
- (62).Case DA; et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Salomon-Ferrer R; Götz AW; Poole D; Le Grand S; Walker RC Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- (64).Maier JA; et al. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Antony J; Medvedev DM; Stuchebrukhov AA Theoretical study of electron transfer between the photolyase catalytic cofactor FADH- and DNA thymine dimer. J. Am. Chem. Soc. 2000, 122, 1057–1065. [Google Scholar]
- (66).Walker RC; De Souza MM; Mercer IP; Gould IR; Klug DR Large and fast relaxations inside a protein: Calculation and measurement of reorganization energies in alcohol dehydrogenase. J. Phys. Chem. B 2002, 106, 11658–11665. [Google Scholar]
- (67).Anandakrishnan R; Aguilar B; Onufriev AV H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012, 40, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Pedregosa F; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12 (85), 2825–2830. http://scikit-learn.sourceforge.net. [Google Scholar]
- (69).Van Der Walt S; Colbert SC; Varoquaux G The NumPy array: A structure for efficient numerical computation. Comput. Sci. Eng. 2011, 13, 22–30. [Google Scholar]
- (70).Roe DR; Cheatham TE PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- (71).McGibbon RT; et al. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109, 1528–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.