Abstract
Sleep-disordered breathing is an important health issue for children. The objective of this study was to develop a machine learning classifier model for the identification of sleep apnea events taken exclusively from nasal air pressure (NAP) measurements acquired during overnight polysomnography (PSG) for pediatric patients. A secondary objective of this study was to differentiate site of obstruction exclusively from hypopnea event data using the model.
Computer vision classifiers were developed via transfer learning to either normal breathing while asleep, obstructive hypopnea, obstructive apnea, or central apnea. A separate model was trained to identify site of obstruction as either adeno-tonsillar or tongue base. In addition, a survey of board-certified and board-eligible sleep physicians was completed to compare clinician versus model classification performance of sleep events, and indicated very good performance of our model relative to human raters. The NAP sample database available for modeling comprised 417 normal, 266 obstructive hypopnea, 122 obstructive apnea, and 131 central apnea events derived from 28 pediatric patients. The four-way classifier achieved a mean prediction accuracy of 70.0% (95% confidence interval (CI): 67.1–72.9). Clinician raters correctly identified sleep events from NAP tracings 53.8% of the time, whereas the local model was 77.5% accurate. The site of obstruction classifier achieved a mean prediction accuracy of 75.0% (CI 95: 68.7–81.3). Machine learning applied to NAP tracings is feasible and may exceed the diagnostic performance of expert clinicians. NAP tracings of obstructive hypopneas may “encode” information regarding the site of obstruction which may only be discernable by machine learning.
Keywords: sleep apnea, polysomnography, machine learning, nasal air pressure
INTRODUCTION
Sleep-disordered breathing is an important health issue for children and adolescents, with current estimates of the prevalence of obstructive sleep apnea (OSA) between 6% and 9% in the pediatric population (Marcus et al., 2012; Tsukada et al., 2018). A steadily building body of evidence suggests that untreated OSA in children and adolescents has important potential impacts on health and well-being, contributing not only to cardiovascular, metabolic, and pulmonary disease, but also to behavioral, learning, neuropsychiatric, and developmental problems (Bhatt et al., 2021; Hunter et al., 2016; Teo & Mitchell, 2013; Tzeng et al., 2019). Children with specific genetic syndromes (e.g., trisomy 21) are at higher risk for OSA, and may be particularly susceptible to the neurodevelopmental impacts of untreated OSA (Grieco et al., 2021; Lee et al., 2018).
In-laboratory polysomnography (PSG) remains the gold standard in the diagnosis of pediatric sleep disordered breathing and provides important information to enable risk stratification for pediatric patients pre-operatively (Kirk et al., 2017; Owens et al., 2012). However, access to this gold standard test is limited by insufficient testing capacity in the United States of America despite an increasing recognition of the importance of early detection and treatment of pediatric OSA (Owens et al., 2012). Due in part to the access problem, investigators have developed other diagnostic approaches to OSA. Prior studies have used machine learning models to classify OSA based on patient-reported questionnaires(Ahmed et al., 2018), pulse oximetry(Hornero et al., 2017; Vaquerizo-Villar et al., 2021), and heart-rate variability(Uçar et al., 2018) among others. Machine learning approaches have been shown to enhance the diagnostic accuracy of OSA (Gutiérrez-Tobal et al., 2021; Gutiérrez-Tobal et al., 2022). Though in-lab PSG is a sensitive and specific test for sleep-disordered breathing, , it does not provide clinicians with actionable information regarding site of obstruction, for which invasive procedures such as drug-induced sleep endoscopy (DISE) may still be required (Baldassari et al., 2021).
In this work, we describe the development of a machine learning classifier model for the identification of sleep apnea events taken exclusively from nasal air pressure (NAP) measurements acquired during clinical in-lab PSG. While others have described using single-channel NAP and expert-based rules as a means for diagnosing OSA via AHI enumeration (Erman et al., 2007), our contribution uses machine learning to classify specific sleep apnea events. Using the same data type, we also describe the development of a classifier that can differentiate site of obstruction exclusively from hypopnea event data. Accurately differentiating the site of obstruction exclusively from hypopnea event data may be informative in the formulation of specific therapeutic interventions for a given case of OSA. Ultimately, these models serve as proof-of-concept for the future development of novel lightweight diagnostic technology for pediatric sleep apnea detection using fewer data channels, as well as to provide non-invasive means for characterizing the anatomic site of obstruction.
METHODS
This research was conducted under Mass General Brigham (MGB) Institutional Review Board (IRB) approval and oversight (MGB IRB protocol numbers 2021P001529 and 2021P003186). The development and reporting of this predictive model was completed in accordance with published guidelines from a multidisciplinary panel (Luo et al., 2016).
Clinical Setting.
The patient population was comprised of 28 children and adolescents, ages 1–16 years old, who underwent a standard overnight in-laboratory clinical polysomnography (PSG) test battery as ordered by a clinician (e.g., family physician, pediatrician, pulmonologist or otolaryngologist) to evaluate sleep disordered breathing. The standard PSG test battery includes nasal and oral airflow sensors, snoring microphone, respiratory impedance plethysmography, pulse oximetry, electrocardiography, carbon dioxide monitors, electroencephalography, and body position monitoring sensors. All PSGs were obtained as part of the usual medical management of the patient, and were scored and interpreted using American Academy of Sleep Medicine Pediatric criteria (Berry, 2020). All studies were obtained in the same lab, which adheres to AASM-specified inter-rater scoring standards, including >85% correlation between scorers and regular testing of scorer performance against Gold Standard studies. For our analysis, training and testing datasets were generated from the NAP exclusively. NAP was recorded, as part of the broader polysomnogram, using a Nihon Kohden PSG 1100 system (headbox model JB-110A) via Salter-brand cannulas (SL-5052–7-7–25 and SL-5044–7-7–25). The sampling rate for the NAP transducer signal was 25Hz, which is within AASM-outlined parameters (American Academy of Sleep Medicine, 2020).
Diagnostic Prediction Task.
We structured our analysis as a multi-classification prediction task using computer vision deep learning. The primary diagnostic prediction task of this study was to classify NAP signals as representing either i) normal breathing while asleep (during rapid eye movement (REM) or non-rapid eye movement (NREM) sleep), ii) obstructive hypopnea, iii) obstructive apnea, or iv) central apnea. Central hypopneas were not scored on the included polysomnograms, and so were not included in this study. Model success was primarily determined by the classification accuracy in assigning the correct label (%-correct). Precision, recall, and F-1 scores were also computed.
We performed a secondary diagnostic prediction experiment to determine the predictive value of NAP signals for site of obstruction associated with the sleep apnea phenotype. Hypopnea events were isolated in a subset of our patients who underwent a successful adenotonsillectomy (AT) versus hypoglossal nerve (HGN) stimulation after their PSG study. Model success was primarily determined by the binary classification accuracy in assigning the correct surgery label (%-correct) as a proxy label for anatomic site of obstruction. Sensitivity, specificity, and precision were also computed.
Data Preparation.
Data were collected retrospectively from a database of pediatric patients who had successfully completed a full overnight PSG. Patient-level samples were included if the patient completed a full overnight PSG with data quality sufficient for routine clinical interpretation. Patient samples were excluded if there was disruption in the PSG testing protocols or poor data quality (e.g., limited continuous data available due to signal disruption).
For the neural network model development, training and testing datasets were comprised of 30-second samples of NAP signal, which were extracted from sleep study epochs containing either an obstructive apnea, obstructive hypopnea, central apnea, or a period of normal breathing as previously scored by our sleep lab technologists. To extract features from the NAP time series data, we performed continuous wavelet transformations to each NAP sample to produce a scalogram image representation of each NAP sample (Figure 1). Our scalogram image database was split with 80% of images used for training, and the remaining 20% used for validation to compute of out-of-training-set performance.
Figure 1.
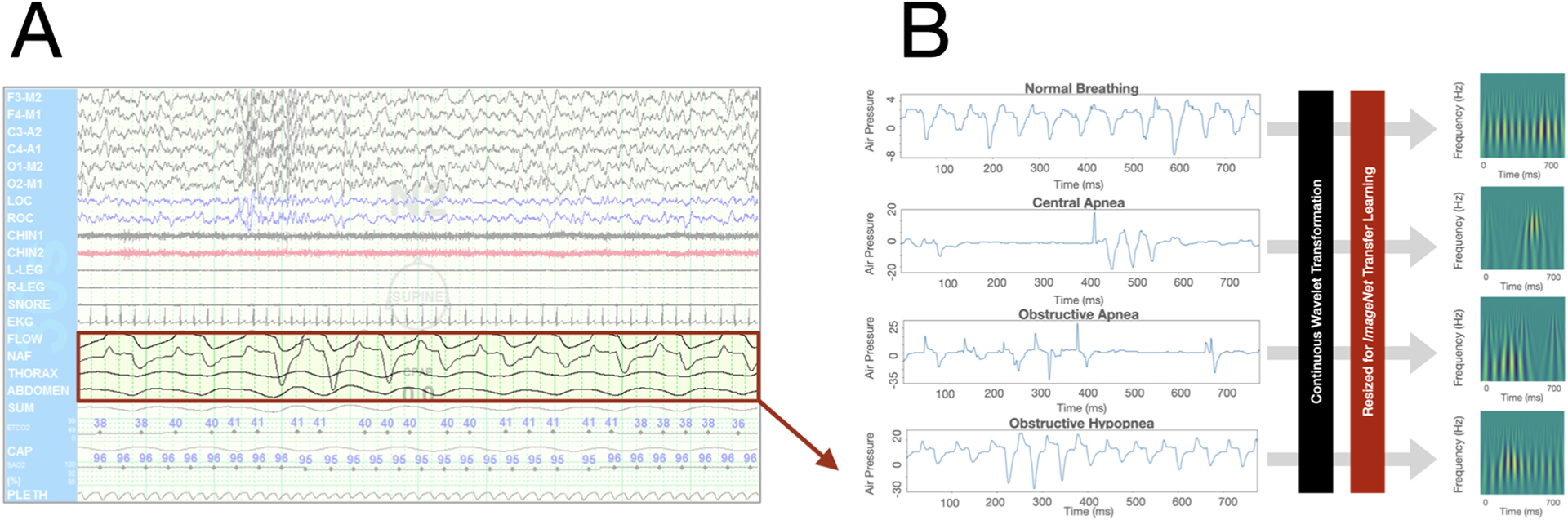
Nasal air pressure (NAP) feature engineering and processing prior to computer vision transfer learning modeling. A. NAP tracings were extracted from the patient-level clinical polysomnography dataset. B. Continuous wavelet transformations were performed on NAP samples to produce scalogram image representations.
Model Development.
We performed transfer learning using convolutional neural network architectures optimized for computer vision. We trained neural networks using the NAP scalogram images as the input data. The prediction output of the model was one label out of either i) normal breathing while asleep (during rapid eye movement (REM) or non-rapid eye movement (NREM) sleep), ii) obstructive hypopnea, iii) obstructive apnea, or iv) central apnea. We surveyed several different architectures with different layer depths including ResNet (ResNet-18, -34, -50, -101, and -152), DenseNet (DenseNet-121 and -201), and Visual Geometry Group (vgg-16, vgg-19) models previously trained on the ImageNet database comprised of millions of high-resolution images within 22,000 categories.(Deng et al.) Prior to training, images were resized to 512 by 512 pixels and normalized for ImageNet.
To produce an accurate estimate of the model performance, we used Monte Carlo resampling with five separate experiments using random training-validation data splits to generate a 95% confidence interval (CI-95) of the model classification accuracy performance. Each random repetition was trained with 25 epochs and a progressive-regressive learning rate strategy.(Smith, 2018) Confusion matrices were also generated to evaluate model performance within the classification labels strata. The modeling was completed with Python v3.9, PyTorch v1.9, and fast.ai v2.5 (available at: https://github.com/fastai/fastai).
‘Human vs. Machine’ Validation.
We performed an IRB-approved survey of board-certified and board-eligible sleep medicine physicians to assess the performance of an expert rater at identifying respiratory events (i.e., normal breathing, obstructive apneas, obstructive hypopneas, and central apneas) using only NAP tracings. Using an anonymized online survey platform (REDCap), physician respondents who affirmed their status as sleep medicine board-certified or board-eligible were shown a set of 30-second tracings of isolated NAP, taken from clinical in-lab PSGs. Each physician was shown a total of 40 unlabeled NAP tracings, which either represented normal breathing, an obstructive apnea, and obstructive hypopnea, or a central apnea. The ground truth labels were determined by a board-certified sleep medicine clinician who had access to the entire multi-channel PSG. Physician respondents were asked to give their best impression of each 30-second tracing, using pediatric rules for event duration (i.e. at least the duration of 2 breaths during baseline breathing) and for hypopnea scoring (i.e. peak signal excursions drop by ≥30% of pre-event baseline)(Berry, 2020). The respondents were instructed to choose among i) normal breathing while asleep (during rapid eye movement (REM) or non-rapid eye movement (NREM) sleep), ii) obstructive hypopnea, iii) obstructive apnea, or iv) central apnea. The survey invitation was distributed to a convenience sample of board-certified and board-eligible pediatric sleep clinicians through a pediatric sleep clinician mailing list (PedSleep2.0 via Google Groups) and through the American Academy of Sleep Medicine (AASM) Engage online forum. The classification performance of the clinicians was compared directly against the performance of our local model.
RESULTS
Patient Cohort.
The final patient cohort comprised 28 pediatric patients with sleep disordered breathing evaluated in our pediatric sleep lab between 2016 and 2022. This small cohort was selected based on polysomnographic or strong clinical evidence of significant improvement in OSA following a single-site operative intervention. The average age of the patient at time of PSG was 8.75 years (range 1.59 – 16.4 years, standard deviation (SD) 4.81 years). The study population was 32% female. The population was ethnically and racially diverse, with 53.6% self-reporting as non-hispanic white, 21.4% self-reporting as Hispanic, 10.7% self-reporting as non-hispanic Asian, and 3.6% self-reporting as non-hispanic African American. The average BMI was 19.74 kg/m2 (range 13.33 – 31.33 kg/m2, SD 4.04 kg/m2), and the BMI percentile was 69.5% (SD 28.3%). The average apnea hypopnea index (AHI) of the 29 polysomnograms (1 patient contributed 2 separate PSGs at clinically discrete timepoints) included in our training and testing datasets was 15 events per hour (range 1.4 – 33.8/hour, SD 7.7/ hour), with an average obstructive apnea and hypopnea (OAHI) of 13.7/ hour (range 1.4 – 30.4/ hour, SD 7.3/ hour).
Of the 28 patients in this cohort, 27 underwent a surgical intervention and had either post-operative PSG studies or hypoglossal nerve stimulator (HGNS) titration studies available. The prevalence of trisomy 21 (T21) in this cohort was 36%, with most of this subset of patients undergoing hypoglossal nerve stimulator placement. Of those patients who underwent a surgical intervention for their OSA, 46% of the patients underwent tonsillectomy-adenoidectomy, 36% underwent hypoglossal nerve stimulator placement, and 7% underwent adenoidectomy only. One patient underwent nasal turbinate ablation, one underwent lingual tonsillectomy, and one underwent supraglottoplasty. Following either surgical intervention or hypoglossal nerve stimulator activation and titration, the average residual AHI was 3.1/ hour (range 0–10.9/ hour, SD 2.5/ hour), with a residual OAHI of 2.3/ hour (range 0–8.7/ hour, SD 2.3/ hour).
Classifier Model Performance.
Our aggregate NAP sample database available for modeling comprised 417 normal, 266 obstructive hypopnea, 122 obstructive apnea, and 131 central apnea events derived from 29 patients (28 polysomnograms, with 1 patient contributing 2 separate studies to the dataset). Using Monte Carlo resampling with five separate random repetitions, our four-way classifier using the ResNet-50 architecture achieved a mean prediction accuracy of 70.0% (95% confidence interval (CI): 67.1–72.9) on held-out validation data. The classifier performed best in identifying ‘Normal’ images and had notable difficulty in differentiating actual ‘hypopnea’ events from ‘normal’ (Table 1; Figure 2).
Table 1.
Performance statistics of the four-way multiclass model on held-out data
| Target Class | Precision | Recall | F1-Score |
|---|---|---|---|
| Apnea | 0.60 (0.53–0.67) | 0.59 (0.50–0.67) | 0.59 (0.52–0.66) |
| Central | 0.61 (0.58–0.64) | 0.63 (0.61–0.66) | 0.62 (0.59–0.64) |
| Hypopnea | 0.69 (0.66–0.71) | 0.55 (0.48–0.62) | 0.61 (0.56–0.65) |
| Normal | 0.80 (0.80–0.80) | 0.84 (0.83–0.86) | 0.82 (0.81–0.83) |
CI-95: 95% confidence interval
Figure 2.
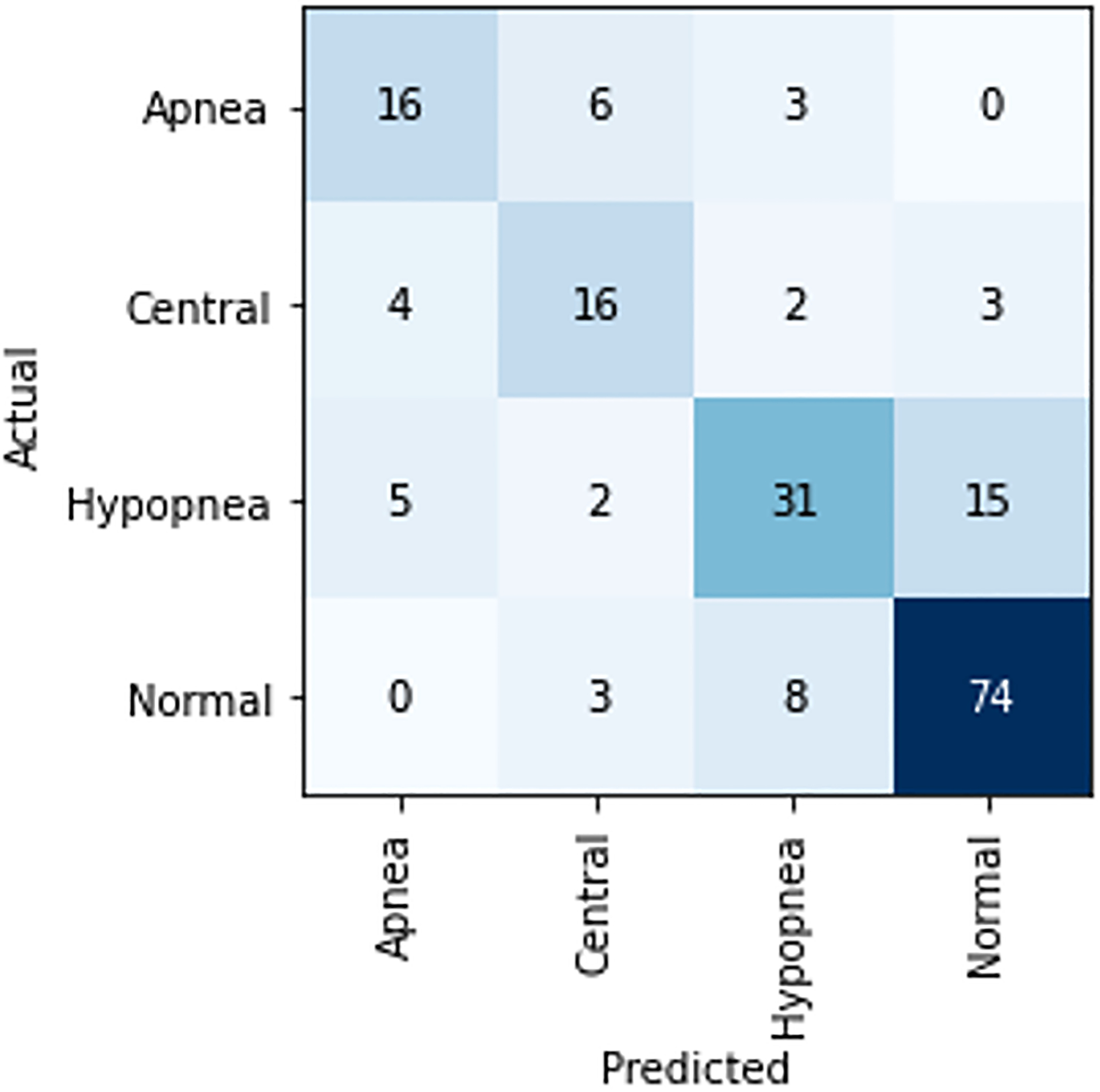
Confusion matrix for first replicate of the four-way classifier.
For our site of obstruction analysis, our NAP database available for modeling comprised 118 and 121 hypopnea events from patients who subsequently underwent adenoidectomy-tonsillectomy and hypoglossal nerve stimulation, respectively. Using Monte Carlo resampling with five separate random repetitions, our site of obstruction classifier achieved a mean prediction accuracy of 75.0% (CI 95: 68.7–81.3) on held-out validation data with a sensitivity of 78.1% (CI 95: 69.6–86.6%), specificity of 72.1% (CI 95: 66.4–77.7%), and a precision of 70.6% (CI 95: 61.3–80.0%)
‘Human vs. Machine’ Survey.
25 sleep medicine board-certified or board-eligible physicians participated in the survey. Overall, physician raters correctly identified NAP tracings 53.8% of the time on average (Table 2). Physician raters were most consistently able to discriminate normal breathing correctly, with an average identification rate of 84.4%. Physicians had relatively poor performance in identifying obstructive apneas (an average of 48.8% correctly identified), obstructive hypopneas (an average of 47.9% correctly identified), and central apneas (an average of 34.2% correctly identified). On the same dataset, our validated model achieved an average classification accuracy of 77.5% across all event types.
Table 2.
Human vs. machine performance on forty nasal air pressure classification images. Percentages represent classification accuracy.
| Clinicians | |
|---|---|
| 84.4% (SD 19.8) | 90% |
| 48.8% (SD 28.4) | 70% |
| 47.9% (SD 17.4) | 80% |
| 34.2% (SD 29.8) | 70% |
| 53.8% (SD 30.5) | 77.5% |
NAP: nasal air pressure
DISCUSSION
To the best of our knowledge, our study is the first to combine the concepts of computer vision transfer learning and single channel nasal air pressure measurements to classify sleep disordered breathing events during in-laboratory PSG in children. Our best-performing proof-of-concept classifier model, which was developed using a relatively small dataset of sleep disordered breathing events, demonstrated robust performance in identifying specific breathing events. When compared to clinician experts, our proof-of-concept model exceeded human classification performance in aggregate. Our secondary objective was to explore the predictive ability of NAP measurements for classifying anatomic subsite of obstruction based on post-PSG surgery as the ground truth label. We found that NAP measurements alone had a surprisingly accurate ability to correctly classify site of obstruction. Our results illustrate the potential for computer vision machine learning approaches based on limited data streams for the development of simpler, less-invasive PSG testing modalities for pediatric patients, as well as obtaining additional clinically useful information such as anatomic obstruction subsite through the application of principles associated with machine learning.
In-laboratory PSG remains the gold standard in the diagnosis of sleep-related breathing disorders in children and adolescents, however the scoring of these studies remains largely a manual and labor-intensive process. While quality assurance is performed continuously in accredited sleep laboratories, inter-rater reliability variance between scorers and interpreting physicians remains a concern. An automated system for accurately identifying respiratory events would be a cost and time-saving tool for busy sleep laboratories and could ensure consistency across scorers and laboratories and better access to PSG for pediatric patients. To that end, considerable interest in applying machine learning models to various aspects of PSG has emerged. Investigators have developed machine learning models based on machine learning to predict the presence and severity of sleep-disordered breathing using oximetry, actigraphy, and clinical variables (Bertoni et al., 2020; Calderón et al., 2020; Combs & Parthasarathy, 2017; Gutiérrez-Tobal et al., 2021; Jiménez-García et al., 2020; Vaquerizo-Villar et al., 2021; Vaquerizo-Villar et al., 2020). These efforts have demonstrated that machine learning models to predict the presence and severity of sleep-disordered breathing in any population may be a viable approach with acceptable accuracy. Our result adds to this growing body of literature in highlighting the predictive utility of using a non-invasive single-channel data source – such as nasal air pressure – in classifying sleep events observed during in-laboratory clinical PSG. Our approach and result also supports recent evidence that has demonstrated that wavelet transformation of single-channel data, such as oronasal air flow, data can produce accurate diagnostic accuracy for pediatric OSA (Barroso-García, Gutiérrez-Tobal, Gozal, et al., 2021; Barroso-García, Gutiérrez-Tobal, Kheirandish-Gozal, et al., 2021).
Previously published data indicates that expert inter-rater performance in identifying respiratory events with the benefit of a full standard polysomnographic montage exceeds 93% agreement overall, with very good performance on identifying epochs with normal breathing (97.4% agreement), and relatively weaker agreement between expert raters when scoring obstructive apneas (77.1%), obstructive hypopneas (65.4%), and central apneas (52.4%)(Rosenberg & van Hout, 2014). In our survey study, our clinicians’ performance in classifying NAP measurements exclusively tracked the general trends in clinician rating of PSG data in that hypopneas and central apneas are difficult to reliably classify. However, our clinicians were provided with an artificial environment of only having NAP measurements to consider. In clinical practice, clinicians rely upon multi-channel data in making their interpretations. However, in our case, we found the machine learning model trained on just a single channel of PSG data outperformed human clinicians on the same task.
Our study is not without limitations. Some potential pitfalls are inherent in interpreting the model. Our site of obstruction analysis uses surgery as a proxy measure for an anatomic subsite (i.e., adenotonsillar versus tongue base). We assumed that the correct surgery was selected and thus the proxy measure serves as a reasonably accurate assumption for obstruction at either the adenotonsillar or tongue base subsites. There exists potential bias of the data used in our modeling. Our sleep laboratory is located at a tertiary level academic medical center and may yield a different distribution of type and/or severity of pediatric sleep disordered breathing. As a result, the generalizability of our model may be impacted if applied to data from other patient populations. Our small sample size was selected based on the presence of a clinically well-delineated site of obstruction which responded to surgical intervention. In this way, we were able to train the model on site of obstruction with some degree of clinical confidence. Patients with T21 comprise a significant proportion of our dataset and were included as a source of tongue base obstruction data in the context of successful pediatric HGNS procedures performed at the study site. The NAP signature of tongue base obstruction from a patient with T21 may not be generalizable to patient without T21, and we will explore this further in subsequent work.
While in-laboratory PSG remains the gold standard in the diagnosis of sleep-related breathing disorders in children, the manual scoring and interpretation of PSG data is labor-intensive and subject to variance in inter-rater reliability. Our study suggests that machine learning applied to NAP tracings may exceed the diagnostic performance of expert clinician raters. Further, our site of obstruction experiment suggests that the NAP tracings of obstructive hypopneas may “encode” information regarding the site of obstruction which may only be discernable by a machine learning model. Significant further work, including confirmatory work by other labs, will be needed before clinical use of such a model would be appropriate in surgical planning. Ultimately, we provide support to the growing body of work that applying machine learning to clinical PSG data may help push for the development of scalable tools for simpler, more consistent, and time-saving analyses for clinical pediatric PSG. Future work is needed to explore incorporating other non-invasive data channels, externally validate these models and increase dataset sizes to enhance model classification accuracy.
ACKNOWLEDGEMENTS
Dr. Crowson’s effort is supported in part by an NIH grant (Biomedical Informatics and Data Science Research Training Program; T15LM007092-30; PI Nils Gehlenborg).
Funding Source:
No funding was secured for this study.
ABBREVIATIONS
- AASM
American Academy of Sleep Medicine
- AT
Adenotonsillectomy
- AHI
Apnea hypopnea index
- CI
Confidence interval
- HGN
Hypoglossal nerve
- REM
Rapid eye movement
- NREM
Non-rapid eye movement
- NAP
Nasal air pressure
- OSA
Obstructive sleep apnea
- OAHI
Obstructive apnea and hypopnea
- PSG
Polysomnography
- SD
Standard deviation
Footnotes
Institution where work was performed: Mass Eye & Ear and Massachusetts General Hospital
Financial Disclosure: No financial disclosures to report by the above authors of the study.
Conflict of Interest: The above authors have no conflicts of interest to disclose that might raise the question of bias in the work reported or the conclusions, implications, or opinions stated.
REFERENCES
- Ahmed S, Hasani S, Koone M, Thirumuruganathan S, Diaz-Abad M, Mitchell R, Isaiah A, & Das G (2018). An Empirical Study of Questionnaires for the Diagnosis of Pediatric Obstructive Sleep Apnea. Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual International Conference, 2018, 4097–4100. 10.1109/embc.2018.8513389 [DOI] [PubMed] [Google Scholar]
- Baldassari CM, Lam DJ, Ishman SL, Chernobilsky B, Friedman NR, Giordano T, Lawlor C, Mitchell RB, Nardone H, Ruda J, Zalzal H, Deneal A, Dhepyasuwan N, & Rosenfeld RM (2021). Expert Consensus Statement: Pediatric Drug-Induced Sleep Endoscopy. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery, 165(4), 578–591. 10.1177/0194599820985000 [DOI] [PubMed] [Google Scholar]
- Barroso-García V, Gutiérrez-Tobal GC, Gozal D, Vaquerizo-Villar F, Álvarez D, Del Campo F, Kheirandish-Gozal L, & Hornero R (2021). Wavelet Analysis of Overnight Airflow to Detect Obstructive Sleep Apnea in Children. Sensors (Basel, Switzerland), 21(4). 10.3390/s21041491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-García V, Gutiérrez-Tobal GC, Kheirandish-Gozal L, Vaquerizo-Villar F, Álvarez D, Del Campo F, Gozal D, & Hornero R (2021). Bispectral analysis of overnight airflow to improve the pediatric sleep apnea diagnosis. Computers in biology and medicine, 129, 104167. 10.1016/j.compbiomed.2020.104167 [DOI] [PubMed] [Google Scholar]
- Berry R, Quan SF, Abreau AR, et al. (2020). The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications. Version 2.6. American Academy of Sleep Medicine. [Google Scholar]
- Bertoni D, Sterni LM, Pereira KD, Das G, & Isaiah A (2020). Predicting polysomnographic severity thresholds in children using machine learning. Pediatric research, 88(3), 404–411. 10.1038/s41390-020-0944-0 [DOI] [PubMed] [Google Scholar]
- Bhatt SP, Guleria R, & Kabra SK (2021). Metabolic alterations and systemic inflammation in overweight/obese children with obstructive sleep apnea. PloS one, 16(6), e0252353. 10.1371/journal.pone.0252353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón JM, Álvarez-Pitti J, Cuenca I, Ponce F, & Redon P (2020). Development of a Minimally Invasive Screening Tool to Identify Obese Pediatric Population at Risk of Obstructive Sleep Apnea/Hypopnea Syndrome. Bioengineering (Basel, Switzerland), 7(4). 10.3390/bioengineering7040131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs D, & Parthasarathy S (2017). Machines Learning to Detect Obstructive Sleep Apnea in Children. Are We There Yet? American journal of respiratory and critical care medicine, 196(12), 1506–1507. 10.1164/rccm.201708-1688ED [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Dong W, Socher R, Li L-J, Li K, & Fei-Fei L ImageNet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition (pp. 248–255). IEEE. 10.1109/cvpr.2009.5206848 [DOI] [Google Scholar]
- Erman MK, Stewart D, Einhorn D, Gordon N, & Casal E (2007). Validation of the ApneaLink for the screening of sleep apnea: a novel and simple single-channel recording device. Journal of clinical sleep medicine : JCSM : official publication of the American Academy of Sleep Medicine, 3(4), 387–392. [PMC free article] [PubMed] [Google Scholar]
- Grieco JA, Hartnick CJ, Skotko BG, Yu PK, & Pulsifer MB (2021). Preliminary Neurocognitive Results Post Hypoglossal Nerve Stimulation in Patients With Down Syndrome. The Laryngoscope, 131(12), 2830–2833. 10.1002/lary.29808 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Tobal GC, Álvarez D, Kheirandish-Gozal L, Del Campo F, Gozal D, & Hornero R (2021). Reliability of machine learning to diagnose pediatric obstructive sleep apnea: Systematic review and meta-analysis. Pediatric pulmonology. 10.1002/ppul.25423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Tobal GC, Álvarez D, Vaquerizo-Villar F, Barroso-García V, Gómez-Pilar J, Del Campo F, & Hornero R (2022). Conventional Machine Learning Methods Applied to the Automatic Diagnosis of Sleep Apnea. Advances in experimental medicine and biology, 1384, 131–146. 10.1007/978-3-031-06413-5_8 [DOI] [PubMed] [Google Scholar]
- Hornero R, Kheirandish-Gozal L, Gutiérrez-Tobal GC, Philby MF, Alonso-Álvarez ML, Álvarez D, Dayyat EA, Xu Z, Huang Y-S, Tamae Kakazu M, Li AM, van Eyck A, Brockmann PE, Ehsan Z, Simakajornboon N, Kaditis AG, Vaquerizo-Villar F, Crespo Sedano A, Sans Capdevila O, … Gozal D (2017). Nocturnal Oximetry-based Evaluation of Habitually Snoring Children. American journal of respiratory and critical care medicine, 196(12), 1591–1598. 10.1164/rccm.201705-0930OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter SJ, Gozal D, Smith DL, Philby MF, Kaylegian J, & Kheirandish-Gozal L (2016). Effect of Sleep-disordered Breathing Severity on Cognitive Performance Measures in a Large Community Cohort of Young School-aged Children. American journal of respiratory and critical care medicine, 194(6), 739–747. 10.1164/rccm.201510-2099OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-García J, Gutiérrez-Tobal GC, García M, Kheirandish-Gozal L, Martín-Montero A, Álvarez D, Del Campo F, Gozal D, & Hornero R (2020). Assessment of Airflow and Oximetry Signals to Detect Pediatric Sleep Apnea-Hypopnea Syndrome Using AdaBoost. Entropy (Basel, Switzerland), 22(6). 10.3390/e22060670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk V, Baughn J, D’Andrea L, Friedman N, Galion A, Garetz S, Hassan F, Wrede J, Harrod CG, & Malhotra RK (2017). American Academy of Sleep Medicine Position Paper for the Use of a Home Sleep Apnea Test for the Diagnosis of OSA in Children. Journal of clinical sleep medicine : JCSM : official publication of the American Academy of Sleep Medicine, 13(10), 1199–1203. 10.5664/jcsm.6772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-F, Lee C-H, Hsueh W-Y, Lin M-T, & Kang K-T (2018). Prevalence of Obstructive Sleep Apnea in Children With Down Syndrome: A Meta-Analysis. Journal of clinical sleep medicine : JCSM : official publication of the American Academy of Sleep Medicine, 14(5), 867–875. 10.5664/jcsm.7126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Phung D, Tran T, Gupta S, Rana S, Karmakar C, Shilton A, Yearwood J, Dimitrova N, Ho TB, Venkatesh S, & Berk M (2016). Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research: A Multidisciplinary View. Journal of medical Internet research, 18(12), e323. 10.2196/jmir.5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus CL, Brooks LJ, Draper KA, Gozal D, Halbower AC, Jones J, Schechter MS, Ward SD, Sheldon SH, Shiffman RN, Lehmann C, & Spruyt K (2012). Diagnosis and management of childhood obstructive sleep apnea syndrome. Pediatrics, 130(3), e714–755. 10.1542/peds.2012-1672 [DOI] [PubMed] [Google Scholar]
- Owens J, Kothare S, & Sheldon S (2012). PRO: “Not just little adults”: AASM should require pediatric accreditation for integrated sleep medicine programs serving both children (0–16 years) and adults. Journal of clinical sleep medicine : JCSM : official publication of the American Academy of Sleep Medicine, 8(5), 473–476. 10.5664/jcsm.2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo DT, & Mitchell RB (2013). Systematic review of effects of adenotonsillectomy on cardiovascular parameters in children with obstructive sleep apnea. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery, 148(1), 21–28. 10.1177/0194599812463193 [DOI] [PubMed] [Google Scholar]
- Tsukada E, Kitamura S, Enomoto M, Moriwaki A, Kamio Y, Asada T, Arai T, & Mishima K (2018). Prevalence of childhood obstructive sleep apnea syndrome and its role in daytime sleepiness. PloS one, 13(10), e0204409. 10.1371/journal.pone.0204409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng N-S, Chung C-H, Chang H-A, Chang C-C, Lu R-B, Yeh H-W, Chiang W-S, Kao Y-C, Chang S-Y, & Chien W-C (2019). Obstructive Sleep Apnea in Children and Adolescents and the Risk of Major Adverse Cardiovascular Events: A Nationwide Cohort Study in Taiwan. Journal of clinical sleep medicine : JCSM : official publication of the American Academy of Sleep Medicine, 15(2), 275–283. 10.5664/jcsm.7632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uçar MK, Bozkurt MR, Bilgin C, & Polat K (2018). Automatic sleep staging in obstructive sleep apnea patients using photoplethysmography, heart rate variability signal and machine learning techniques. Neural Computing and Applications, 29(8), 1–16. 10.1007/s00521-016-2365-x [DOI] [Google Scholar]
- Vaquerizo-Villar F, Alvarez D, Kheirandish-Gozal L, Gutierrez-Tobal GC, Barroso-Garcia V, Santamaria-Vazquez E, Del Campo F, Gozal D, & Hornero R (2021). A Convolutional Neural Network Architecture to Enhance Oximetry Ability to Diagnose Pediatric Obstructive Sleep Apnea. IEEE journal of biomedical and health informatics, 25(8), 2906–2916. 10.1109/jbhi.2020.3048901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizo-Villar F, Alvarez D, Kheirandish-Gozal L, Gutierrez-Tobal GC, Gomez-Pilar J, Crespo A, Del Campo F, Gozal D, & Hornero R (2020). Automatic Assessment of Pediatric Sleep Apnea Severity Using Overnight Oximetry and Convolutional Neural Networks. Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual International Conference, 2020, 633–636. 10.1109/embc44109.2020.9176342 [DOI] [PubMed] [Google Scholar]


