Abstract
RNA methylation normally inhibits self-recognition and immunogenicity of RNA. As such, it is likely an important inhibitor of cancer immune recognition in the tumor microenvironment (TME), but how N6-methyladenosine (m6A) affects prognosis and treatment response remains unknown. In eight independent melanoma cohorts (1564 patients), the modification patterns of 21 m6A gene signatures were systematically correlated with the immune cell infiltration of melanoma TME. m6A modification patterns for each patient were quantified using the principal component analysis (PCA) method, yielding an m6Ascore that reflects the abundance of m6A RNA modifications. Two different m6A modification patterns were observed in melanoma patients, separated into high and low m6Ascores that correlated with survival and treatment response. Low m6Ascores were characterized by an immune-inflamed phenotype, with 61.1% five-year survival. High m6Ascores were characterized by an immune-excluded phenotype, with 52.2% five-year survival. Importantly, lower m6Ascores correlated with more sensitive anti-PD1 and anti-CTLA4 treatment responses, with 90% of patients with low m6Ascore responding while 10% of those with high m6Ascore non-responding (in cohort GSE63557). At single-cell and spatial transcriptome resolution, m6Ascore reflects melanoma malignant progression, immune exhaustion, and resistance to ICB therapy. Hence, the m6Ascore correlates to an important facet of tumor immune escape as a tool for personalized medicine to guide immunotherapy in melanoma patients.
INTRODUCTION
The effects of genomic and protein modification on tumors are well studied and diverse (Rodriguez-Paredes and Lyko, 2019). RNA methylation also regulates tumor development, specifically 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), N3-methylcytosine (m3C), and N6-methyladenosine (m6A) (Chen et al., 2019). Of these, N6-methyladenosine (m6A) methylation is the most prominent and regulates mRNA, lncRNA, and siRNA in eukaryotes, comprising 0.1–0.4% of all adenosine residues (Zhao et al., 2017). Increasingly, it has been shown that the dysregulation of m6A correlates with malignant progression, vigorous cell proliferation, impaired self-control ability of tumor cells, and invalid immune regulation (Wang et al., 2017). Thus, understanding the modification mechanism of m6A on melanoma may aid melanoma diagnosis, patient management, treatment, and prognosis.
Briefly, m6A modification is a dynamic and reversible process regulated by specific enzymes and binding proteins called writers, erasers, and readers, which work in concert to selectively arrange m6A on RNA. This process regulates RNA nuclear transcription, export, cytoplasmic stability, translation, and localized regulation (Fu et al., 2014). Writers (methyltransferases), composed of METTL3, METTL4, RMB15/15B, and a set of writer complexes, first catalyze m6A modifications in the nucleus. Erasers (demethylases), including ALKBH5 and FTO, concurrently remove m6A, refinining m6A positioning (Liu et al., 2016). Readers, consisting of YTHDC1/2, then regulate splicing and RNA output in the nucleus. In the cytoplasm, YTHDF1/2/3 enhances translation by accelerating the degradation of RNA methylation and recruiting ribosomes. Together, the expression of these enzymes resgulates m6A modifications.
A central function of RNA methylation is to decrease the immunogenicity and self-recognition of RNA by the innate immune system(Kariko et al., 2005). In this manner, host RNA is methylated and labeled to distinguish it from viral RNA, for example. This suggests that increased RNA methylation might also be used by cancers to evade immune detection. Immune checkpoint inhibitors or blockades (ICBs) have shown outstanding efficacy in the treatment of advanced tumors, especially for metastatic melanoma, achieving 6–11% objective response rates (Hodi et al., 2010). However, 60%–70% of melanoma patients cannot benefit from ICB treatment, so it is urgent to understand the basis of ICB resistance. Increased RNA methylation might be one such mechanism.
Increasingly, research has focused on melanoma tumor microenvironment (TME), which has been proven foundational for melanoma behavior and its predictive value for ICB treatment (Zhang et al., 2017). Immune cells inhibit melanoma development and regulates tumor progression by directly killing tumor cells, inducing apoptosis, controlling proliferation, inhibiting angiogenesis, and mediating hypoxia (Zhang et al., 2017).
The relationship between m6A modification and TME immune cells is gradually being discovered (Tong et al., 2018) and not as simple as more m6A RNA methylation causing less immune recognition. Li et al. found that in Tregs, one m6A mechanism enhances the effective signal of IL2 by methylating SOCS mRNA, thereby strengthening the immunosuppressive effect of Tregs (Li et al., 2017). In in vivo and in vitro experiments, the lack of the m6A reader YTHDF1 can slow down the rate of antigen degradation, allowing dendritic cells in melanoma TMEs to deliver cross-antigens to CD8+ T cells more effectively, further bolstering the efficacy of anti-PDL1 therapy (Han et al., 2019). However, prevoius m6A research is predominantly limited to a specific m6A pathway protein or highly basic models, whereas the regulation of cancer and melanoma is determined by the coordination of multiple m6A factors. Therefore, a comprehensive understanding of the effect of m6A’s overall landscape on the TME of melanoma patients is imperative for helping us more deeply understand the impact of RNA epigenetics on melanoma and broadening new perspectives on melanoma immunotherapy.
In this study, we included 1564 melanoma patients from eight independent cohorts, including ICB therapy cohorts. We found two m6A modification patterns in melanoma patients that correlate to the immune cell infiltration pattern of the melanoma TME. Therefore, we quantified the m6A modification (m6Ascore) of each patient and found that the m6Ascore could significantly distinguish patients with good and poor prognoses and predict the efficacy of ICB therapy. At the single-cell and spatial transcriptome levels, we demonstrated that a high m6Ascore (with more m6A methyltransferases) is positively correlated with tumor malignant progression, Cancer-associated fibroblast (CAF) infiltration, T cell depletion, immune exclusion, and ICB resistance, whereas a low m6Ascore indicates the opposite with good prognosis, and immunotherapy benefit.
RESULTS
The modification pattern of 21 m6A regulatory genes corresponds to distinct immune microenvironments in melanoma.
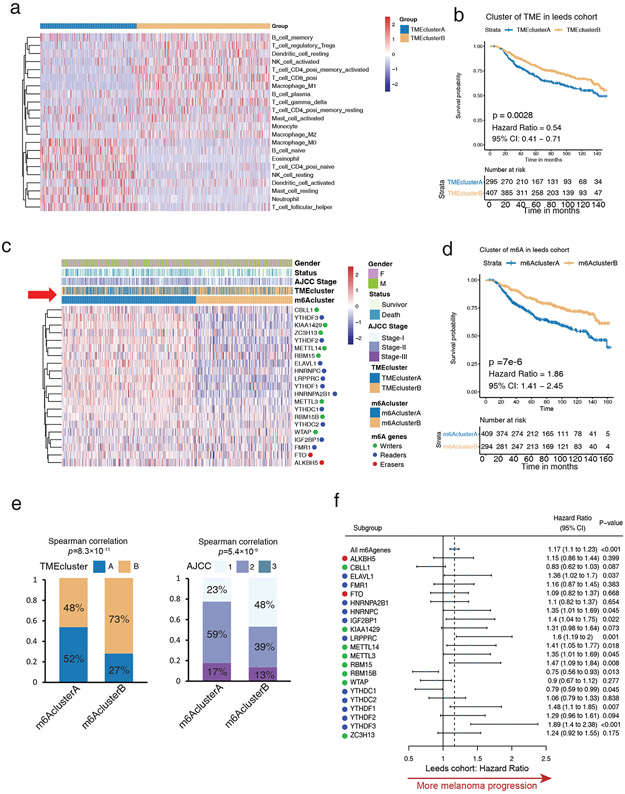
To study the association between m6A regulators and tumor-infiltrating immune cells within melanoma TME, CIBERSORT was used with the Leeds cohort that includes the largest number of melanoma patients (733) as a training cohort. Unsupervised clustering separated melanoma patients into two clusters (Figure 1a, Figure S1a; Table S1) based on different TME patterns. TMEclusterA displayed an immune-excluded phenotype, with increased resting mast cells, resting NK cells, naive B cells, and naive CD4 T cells (Figure 1a and S1b). Conversely, TMEclusterB exhibited an immune-inflamed phenotype with increased memory B cells, CD4 T cells, CD8 T cells, activated NK cells, and activated mast cells (Figure 1a and S1b). Consistent with these profiles, TMEclusterB had a significantly prominent survival advantage (HR, 0.54; 95% CI, 0.41–0.71; p = 0.0028) (Figure 1b).
Figure 1: High m6A modifications correlate to inactive immune microenvironments in the training cohort (LMC).
(a) Heatmap of unsupervised clustering based on 22 TME immune cells of 703 patients in the Leeds Melanoma Cohort (LMC) showing two distinct immune cell infiltration patterns: TMEclusterA (blue) and TMEclusterB (yellow). The rows of the heatmap show the value of immune cells (z-score) calculated by CIBERSORT. Naive B cells, neutrophils, and other resting type immune cells are highly expressed in TMEclusterA, while T cells, mature B cells, and other activated type immune cells are highly expressed in TMEclusterB. (b) Survival analyses based on TMEclusterA and B of 703 patients in LMC show that TMEclusterB has a significant survival advantage (HR, 0.54; 95% CI, 0.41–0.71; p = 0.0028). (c) Heatmap of unsupervised clustering based on 21 m6A regulators of 703 patients in LMC show two distinct m6A modification patterns, namely m6AclusterA (blue) and m6AclusterB (yellow). The overall expression of m6A regulators is higher in m6AclusterA and lower in m6AclusterB. The gender, survival status, AJCC stage, TMEcluster, and m6Acluster are presented as annotations. From the row of annotation (red arrow), most of TMEclusterA gathered with m6AclusterA and most of TMEclusterB gathered with m6AclusterB, reflecting the positive relationship with TME and m6A patterns. m6AclusterB was positively associated with favorable survival status and earlier AJCC stage. (d) Survival analyses based on m6AclusterA and B of 703 patients in Leeds melanoma cohorts show that m6AclusterB has a significant survival advantage (HR, 1.86; 95% CI, 1.41–2.45; p = 7X10-6). (e) The patients in m6AclusterB were associated with significantly higher proportion of TMEclusterB and earlier AJCC stage in the Leeds melanoma cohort. (f) Forest plot estimating the clinical prognostic value of each m6A regulator. The horizontal lines represent the hazard ratio (95% CI) of each m6A regulator. The dotted vertical line represents all m6A regulators’ hazard ratio which is >1, indicating an m6A signature is an unfavorable prognostic biomarker.
Twenty-one of the most important m6A methylation regulatory genes, including writers, erasers, and readers, were selected (see detailed information in the Methods section) and sorted. Two distinct m6A modification patterns emerged, termed m6AclusterA and B (Figure 1c, Figure S1c, Table S2). The expression of most writers and readers was high in m6AclusterA and low in m6AclusterB (Figure 1c), but no overall differences were observed between groups for erasers (FTO and ALKBH5) (Figure 1c). This suggests that melanomas in m6AclusterA had greater RNA methylation than in B. Previous studies have correlated increased m6A modification patterns with poorer prognoses in numerous cancers, and consistent with these studies, m6AclusterB exhibited a significant survival advantage over A (HR, 1.86; 95% CI, 1.41–2.45; p = 7 × 10−6) (Figure 1d) (Zhang et al., 2020). Interestingly, most melanomas in m6AclusterB also overlapped with the melanomas in TMEclusterB and presented more benign American Joint Committee on Cancer (AJCC) stages and greater survival (Figures 1c-e red arrow, Figure S1d). In all patients, most m6A genes correlated with survival for melanoma (Figure 1f and S1e, Table S3).
Transcriptomic traits in distinct m6A modification patterns
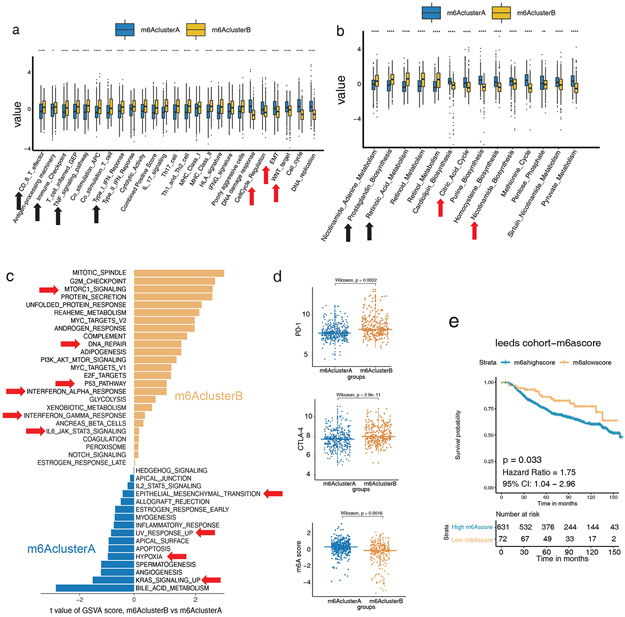
We next queried the biological features involved in distinct m6Aclusters. The transcriptome analysis showed that tumor progression-related functions such as cell cycle, WNT signaling, and epithelial-mesenchymal transition (EMT) were highly expressed in m6AclusterA, while anti-tumor functions such as CD8+ T cell effector, immune checkpoint, TNF, and IFNG signaling were significantly highly expressed in m6AclusterB (Figure 2a, Table S10). In terms of metabolic function, m6AclusterA showed higher oxidative phosphorylation and nicotinamide energy metabolism (Fania et al., 2019) (Figure 2b), while pro-inflammatory and immune-activating factors such as prostaglandin and retinoic acid were highly expressed in m6AclusterB (Figure 2b).
Figure 2: Low m6A RNA modifications correlate to an inflammatory transcriptome and better survival in the training cohort (LMC).
(a) Relative gene signature intensities enrich distinct categories in each m6Acluster: immune signal activation for clusterA; cell active proliferation, transformation, and DNA mismatch for clusterB. (b) Related metabolic functions enrich distinct categories in each m6Acluster: oxidative phosphorylation and nicotinamide biosynthesis for clusterA and prostaglandin and retinoic acid metabolism for clusterB. (c) Functional enrichment analysis of m6AclusterA was significantly correlated with EMT, UV response, hypoxia, tissue neogenesis, angiogenesis, and KRAS signaling, while m6AclusterB was significantly correlated with sufficient oxygen reaction, DNA repair, P53 signaling, IFNA, and IFNG signaling. (d) Differences in the expression of PD-L1, CTLA4, and m6Ascore among m6AclusterA and B (p = 0.0022, p = 2.9e-11, p = 0.0016, respectively, Wilcoxon test). (e) Survival analyses based on high and low m6Ascore of 703 patients in Leeds melanoma cohorts. Low m6Ascore has a significant survival advantage (HR, 1.75; 95% CI, 1.04–2.96; p = 0.033).
The gene set variation analysis (GSVA) between m6Aclusters showed that the pathways related to melanoma progression were higher in m6AclusterA, while pathways related to melanoma suppression were higher in m6AclusterB (Figure 2c, Table S4). Of note, both PD-1 and CTLA4 expression were significantly higher in m6AclusterB, suggesting a better immune checkpoint inhibitor treatment response (Figure 2d).
The above m6Aclusters is a classification based on the biological basis of m6A biologically focused genes expression but this is not convenient for clinical quantification and clinical use. For a more simple numerical scale appriopriate for clinical use, we converted the results of m6Aclusters into an m6Ascore for quantification (see Methods section for details ). m6Ascores were significantly higher in m6AclusterA (Figure 2d). Survival analysis showed that high m6Ascores correlated with significantly worse survival, demonstrating the effectiveness and utility of the m6Ascore in evaluating melanoma TME (HR, 1.75; 95% CI, 1.04–2.96; p = 0.033) (Figure 2e and S1f, Table S5).
Landscape of TME and clinicopathological characteristics of TME subtypes in melanoma
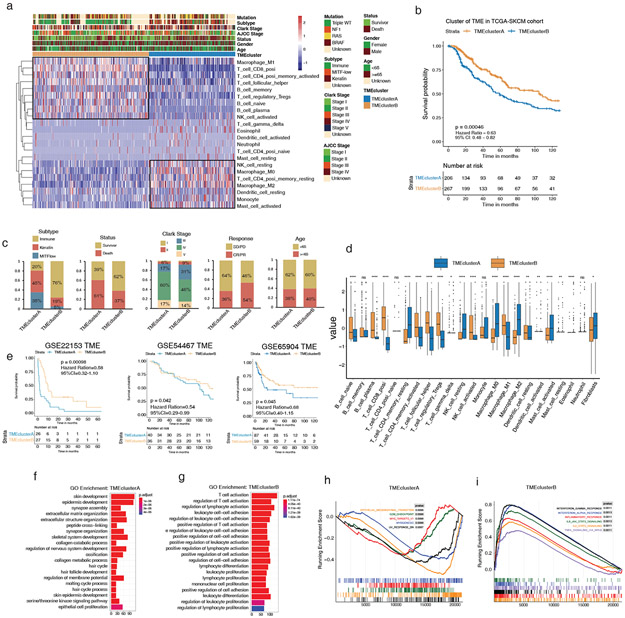
We next defined the melanoma TME landscapes in external non-overlapping validation datasets TCGA-SKCM, GSE22153, GSE54467, and GSE65904, which are independent and distinct from the training dataset, before testing the m6Ascore utility. Unsupervised clustering established two distinct immune cell infiltration patterns in TCGA-SKCM (Figure 3a, Figure S2a). Consistent with the training cohort, TMEclusterA and B exhibited immune-excluded and -inflamed phenotypes, respectively, with associated prognostic outcomes and cell types (HR, 0.63; 95% CI, 0.48–0.82; p = 0.00046) (Figures 3a-b; Tables S2, S6). TMEclusterB also had a higher proportion of immune subtypes and a lower proportion of keratin and MITFlow subtypes (Figure 3c, Table S2). In line with this sorting, the survivor ratio of TMEclusterB was significantly higher than that of A, and in Clark’s level staging system, TMEclusterB patients had better prognostic traits (Levels I to III) than A (Figure 3c). In terms of treatment, TMEclusterB also had a higher proportion of complete response and partial response (CR/PR) patients than TMEclusterA (Figure 3c). Comparing all 22 previously measured immune cells, 18 were significantly different between the groups (Figure 3d). Unsupervised clustering was also performed in the validation datasets GSE22153, GSE54456, and GSE65904, yielding similar infiltration patterns (Figure S2a, S3a-c). Survival analysis showed that melanomas in TMEclusterB correlated with better prognoses in GSE22153 (HR, 0.58; 95% CI, 0.32–1.10; p = 0.00098), GSE54467 (HR, 0.54; 95% CI, 0.29–0.99; p = 0.042), and GSE65904 (HR, 0.68; 95% CI, 0.40–1.15; p = 0.045) (Figure 3e).
Figure 3: The landscape and clinicopathological characteristics of TME in melanoma validation cohorts.
(a) Heatmap of unsupervised clustering based on 22 TME immune cells of 473 patients in TCGA-SKCM, showing two immune cell infiltration patterns: TMEclusterA (blue) and TMEclusterB (yellow). The rows of the heatmap show the value of immune cells (z-score) calculated by CIBERSORT. Macrophage, neutrophil, and other resting type immune cells are highly expressed in TMEclusterA while T cells and B cells are highly expressed in TMEclusterB. The mutation, subtype, Clark stage, AJCC stage, survival status, gender, age, and TMEcluster are presented as annotations. (b) Survival analyses based on TMEclusterA and B of 473 patients in TCGA cohorts show that TMEclusterB has a significant survival advantage (HR, 0.54; 95% CI, 0.41–0.71; p = 0.0028). (c) The clinicopathological parameters in TME clusterB were associated with better therapy response, more immune subtype, better survival status, and earlier Clark stage. Age is allocated evenly. CR/PR: complete response and partial response; SD/PD: stable and progressive disease. (d) The fraction of immune cells in TMEclusterA and B in TCGA-SKCM. The upper and lower ends of the vertical line represent the maximum and minimum values. The three horizontal lines of the boxplot represent the first quartile, median, and third quartile. Outliers are expressed in scattered dots. The Kruskal–Wallis test was used to calculate the statistical difference (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). “NS” indicates no significant difference. (e) Survival analyses based on TMEclusterA and B in GSE22153 (HR, 0.58; 95% CI, 0.32–1.10; p = 0.00098), in GSE54467 (HR, 0.54; 95% CI, 0.29–0.99; p = 0.042), and in GSE65904 (HR, 0.68; 95% CI, 0.40–1.15; p = 0.045), indicating that TMEclusterB has a significant survival advantage. (f–g) Gene ontology (GO) analysis of differentially expressed genes (DEG) presented higher immune cell activation in TMEclusterB (g) and higher skin and skin appendage development in TMEclusterA (f) in TCGA-SKCM. (h–i), GSEA enrichment analysis of DEG presented higher EMT, myogenesis, and UV response in TMEclusterA (h), and higher IFNG, IFNA response, inflammatory, and TNFA signaling in TMEclusterB (i).
We next parsed the transcriptome signature of melanoma TME in TCGA-SKCM, which showed that TMEclusterA had significantly increased skin development whereas TMEclusterB had significantly increased T cell activation and proliferation (Figures 3f-g, Table S7). Similar gene signatures were also presented in the distinct TME clusters of other validation cohorts (Figures S4a-f). The gene set enrichment analysis (GSEA) enrichment analysis confirmed that the traits related to poor prognosis of melanoma were highly expressed in TMEclusterA (Figure 3h). Conversely, anti-tumor immune pathways were significantly higher in TMEclusterB (Figure 3i). These multicohort results confirmed that melanoma TME and m6A status correlate to immune activity and survival.
m6Ascore associates with TME immune cells, clinical characteristics, and prognosis
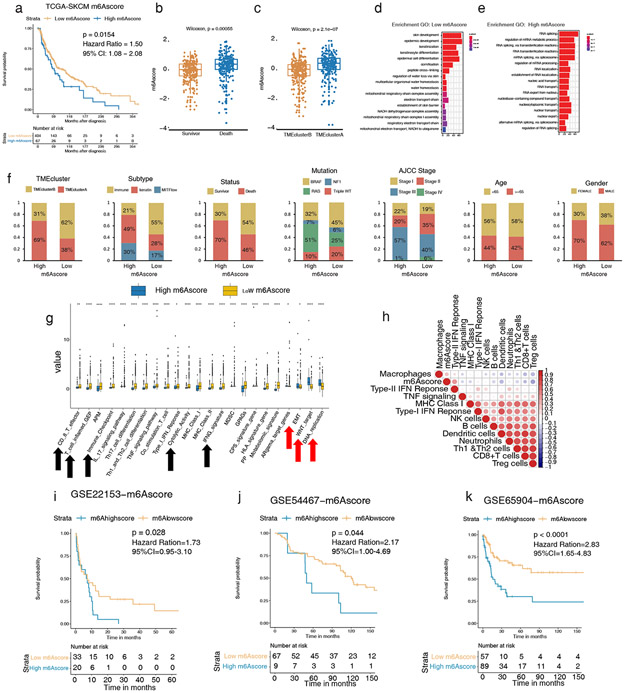
We next sought to validate the association found in the training cohort between m6A modification patterns and melanoma TME. Indeed, patients with low m6Ascores in TCGA-SKCM exhibited a significant survival advantage (HR, 1.50; 95% CI, 1.08–2.08; p = 0.0154) (Figure 4a). Melanomas captured in TME cluster A showed high m6A scores, consistent with their more malignant character as opposed to melanomas of TME cluster B (Figure 4b and 4c). Enrichment analysis further showed that patients with low m6Ascores had higher transcript expressions of keratinocyte differentiation (Figure 4d), while patients with high m6Ascores had significantly higher expressions of RNA related transcripts (Figure 4e). Similar gene signatures were also present in an analogous analysis of other validation cohorts (Figures S5a-f).
Figure 4: High m6A modifications correlate to worse tumor phenotypes and survival in melanoma validation cohorts.
(a) Survival analyses based on high and low m6Ascores of 473 patients in TCGA-SKCM show low m6Ascore has a significant survival advantage (HR, 1.50; 95% CI, 1.08–2.08; p = 0.0154). (b–c) Differences of m6Ascore and survival status among TMEclusters in TCGA-SKCM (p = 2.1e-07 and p = 5.5e-4 respectively, Wilcoxon test). (d–e) GO analysis of DEG presented higher skin cell differentiation and sufficient oxygen reaction in low m6Ascore (d) and higher active RNA modification and transcription in high m6Ascore (e) in TCGA-SKCM. (f) The clinicopathological characteristics in low m6Ascore are associated with more TMEclusterB, more immune subtype, better survival status, more triple WT, and earlier AJCC stage. Age and gender were allocated evenly. (g) Relative gene signature intensities enrich distinct categories in high and low m6Ascore: immune signal activation for low m6Ascore and cell active proliferation, transformation, and DNA mismatch for high m6Ascore. (h) Correlation of m6Ascore and infiltrating immune cells and immune response by Spearman correlation analysis. m6Ascore is negatively correlated with active immune singling in the TCGA-SKCM cohort. (i–k) Survival analyses based on high and low m6Ascore in GSE22153 (HR, 1.73; 95% CI, 0.95–3.10; p = 0.028), in GSE54456 (HR, 2.17; 95% CI, 1.00–4.69; p = 0.044), and in GSE65904 (HR, 2.83; 95% CI, 1.65–4.83; p < 0.0001), indicating that low m6Ascore has a significant survival advantage.
The m6Ascore also correlates to clinical differences. Low m6Ascore melanomas overlaped with TMEclusterB melanomas and possessed greater immune infiltration, survival, absence of BRAF/RAS/NF1 mutations, and more benign AJCC stages; no age or gender differences were noted (Figure 4f).
We found that patients with low m6Ascores had significantly higher expression of anti-tumor pathways and significantly lower expressions of tumor-promoting pathways (Figure 4g, Tables S8, S10). Correlation analysis showed that the m6Ascore is negatively correlated with an immune cell anti-tumor response (Figure 4h).
Multicohort survival analysis showed that the prognosis of patients with low m6Ascores was significantly better than those with high m6Ascores in GSE22153 (HR, 1.73; 95% CI, 0.95–3.10; p = 0.028), GSE54456 (HR, 2.17; 95% CI, 1.00–4.69; p = 0.044), and GSE65904 (HR, 2.83; 95% CI, 1.65–4.83; p < 0.0001) (Figures 4i-k, Table S5).
Single-cell and spatial RNA-seq reveals m6A modification correlates to lower immune infiltration and malignant progression in melanoma
We analyzed single-cell RNA-seq (GSE72056) and spatial transcriptomic data (GSE159709) of melanoma to measure m6A levels. First, we divided the single-cell data of 18 patients into malignant cells and subtypes of nonmalignant cells:immune cells, endothelial cells, and CAFs. In a PCA anlysis using the 500 m6A-related genes trained in the Leeds dataset (see Methods section for details), we found that malignant cells from different patients were clustered and segregated in distinct areas corresponding to individual(Figure 5a), which indicated that the expression patterns of m6A varied greatly among the malignant cells of different patients. In immune cells, endothelial cells, and CAFs cells, the aggregation was by cell type and not by individual (Figure 5b), which indicated that different cell types, especially immune cells, have highly distinct m6A modification patterns. We then quantified m6Ascores and found that malignant cells had significantly higher m6Ascores than other cells, followed by vascular endothelial cells and CAFs, which contributed to melanoma development (Figure 5c). The m6Ascore of malignant-suppressing B cells, T cells, macrophages, and NK cells was significantly lower (Figure 5c). We further calculated the m6Ascore of malignant cells of different patients, and divided the patients into low m6Ascore patients and high m6Ascore patients for further investigation (Figure 5d).
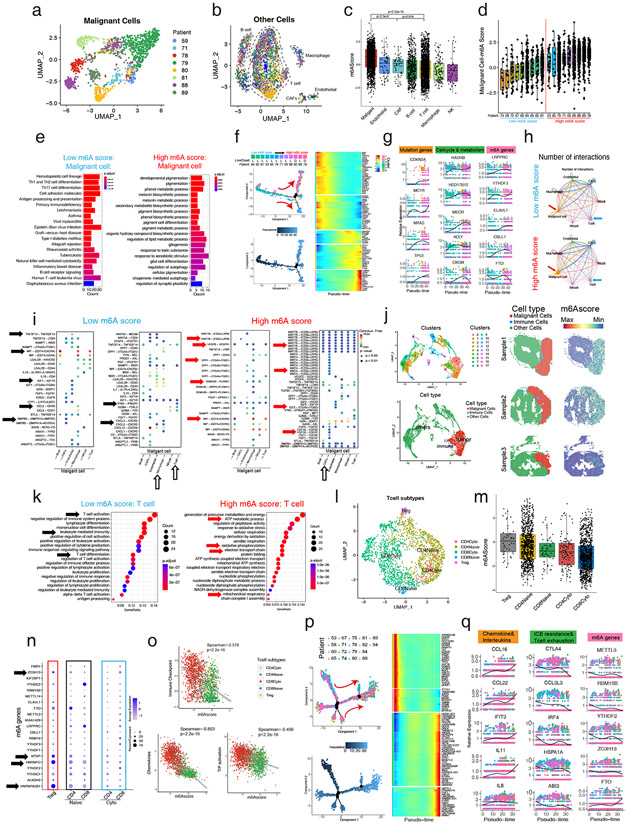
Figure 5: Single-cell and spatial RNA-seq reveals high m6A modification promotes malignant progression and reduces immune infiltration in melanoma.
(a) UMAP plot of 500 m6A relative gene expression profiles of malignant cells in melanoma patients. Malignant cells from different patients gather together. Different patients with malignant cells are shown in different colors. (b) UMAP plot of 500 m6A relative gene expression profiles of other cells of melanoma patients. Different types of cells gather together. Different patients with malignant cells are shown in different colors. (c) The m6Ascores of malignant cells are significantly higher than other cells, especially immune cells. (d) The m6Ascores of different patients. Melanoma patients were divided into high m6Ascore and low m6Ascore groups equally. (e) KEGG enrichment analysis of the DEGs illustrates antigen presentation, T helper cells, NK cells, and B cell signaling highly expressing in malignant cells of low m6Ascore patients, while pigmentation signals, melanosome metabolic signals, and energy metabolism signals highly expressing in malignant cells of high m6Ascore patients. (f) Pseudotime analysis showed that the malignant cell development pattern follows the patient’s m6Ascore ranking and patient mortality trends. (g) Melanoma mutation, cell cycle, metabolism, and m6A readers (LRPPRC, YTHDF3, ELAVL1) and writers (CBLL1) of malignant cells increase while m6A eraser (FTO) decreased with pseudotime. (h) The T cell and malignant cell interaction is greater, while CAFs and malignant cell interaction is lower in the low m6Ascore patients than in the high m6Ascore patients. The thicker line indicates a greater number of interactions. (i) The ligand-receptor pairs of different cells that originate from malignant cells (left) or act on malignant cells (right) of low m6Ascore patients or high m6Ascore patients (j) UMAP shows the clustering and cell classification of each locus of the spatial transcriptome (left), and the m6Ascore in the malignant cells are significantly higher than in the immune cells and other cells (right). (k) KEGG enrichment analysis of the DEGs illustrates T cell activation, immune cell activation, and T cell differentiation pathways highly expressed in T cells of low m6Ascore patients, and energy metabolism signals highly expressing in T cells of high m6Ascore patients. (l) UMAP plot of 500 m6A relative gene expression profiles of T cells of melanoma patients. Different types of T cells gather together and are shown in different colors. (m) Tregs have the highest m6Ascore, while cytotoxic T cells’ m6Ascores are significantly lower. (n) The expression of individual m6A genes in different T cell subtypes. (o) Spearman correlation analysis shows that the m6Ascore is negatively correlated with immune checkpoint, chemokines, and TIP activation in T cells. (p) Pseudotime analysis showed the T cell development pattern. (q) Chemokines and interleukins increase while ICB resistance, T cell exhaustion and m6A readers (YTHDF2, RBM15B) and writers (METTL3, ZC3H13) of T cells decreased but m6A eraser (FTO) increased with pseudotime.
We found that malignant cells from patients with low m6Ascores expressed higher immune cell infiltration signatures, while patients with high m6Ascores highly expressed higher hipigmentation signals and melanosome metabolic signals (Figure 5e). To further understand the expression trend of the m6A gene in the malignant progression of melanoma, we used pseudotime to simulate the developmental progression of melanoma. The malignancy of the patients varied significantly (Figure 5f). Interestingly, we found that maturing melanoma pseudotime evolution trajectory matched with increasing m6Ascore ranking and mortality (Figure 5f, left up). At an individual gene level we also discovered that progression of pseudotime correlated with increased expression of individual cell cycle and metabolism genes, genes mutated in melanoma, as well as m6A reader and writer but not eraser (FTO) genes (Figure 5g).
We next compared the interactions between malignant cells and other cells and found that in patients with low m6Ascores, malignant cells (red arrows) released significantly lower signals to cancer associated fibroblasts (CAFs) for example than high m6Ascore tumours. Meanwhile, the communication between macrophage cells and malignant cells was higher in low m6Ascore tumors (Figure 5h). Further observation of the specific signals of interaction showed that low m6Ascore malignant cells release chemokines, while high m6Ascore malignant cells release developmental signals (Figure 5i). In addition, other low m6Ascore cells, especially immune cells, release inflammatory signals, while other high m6Ascore cells, especially CAFs, release growth factors (Figure 5i).
To visualize m6A modification patterns in melanoma, we used melanoma spatial transcriptome data. Through cell clustering and marker gene expression, we identified malignant cells, immune cells, and other cells (Figure 5j). Projecting the cell classification into the spatial 2-D structure of melanomas, we found that immune cells infiltrated the melanoma periphery, consistent with the original research’s pathology (Figure 5j) (Hunter et al., 2021). Next, we found that the m6Ascore was significantly overexpressed at malignant cell locations, while cells at other cell locations were significantly underexpressed, indicating that the important role of m6Ascore in melanoma.
Next, we queried if m6A RNA methylation varies in immune cells during immune attack of melanoma. We found that the T cells of low m6Ascore patients had higher T cell activation and differentiation pathway expression, while the T cells of high m6Ascore patients had higher expression of energy metabolism, which is a prominent feature of T cell exhaustion (Figure 5k). We divided T cells into Tregs, naive (CD4, CD8), and cytotoxic (CD4, CD8) T cells (Figure 5l, S5g-h). Tregs had the highest m6Ascore, and the m6Ascore of cytotoxic T cells, especially CD8+ cytotoxic T cells, was significantly lower than that of naive T cells, indicated that high m6A suggested cold melanoma and poor prognosis (Figure 5m). By displaying 21 m6A genes in detail, we found that Tregs highly expressed WTAP(writer), HNRNPA2B1 (reader), HNRNPC(reader), and ZCH13(writer) (Figure 5n). We found that m6Ascore was negatively correlated with ICB, chemokines, and TIP activation in T cells, especially in CD8+ cytotoxic T cells (red), which had the lowest m6Ascore expression (Figure 5o).
Finally, we used pseudotime to analyze the developmental patterns of the T cells (Figure 5p). The expression of chemokine and interleukin genes significantly increased with T cell development, and interestingly, ICB resistance, T cell exhaustion, and m6A genes except for eraser (FTO) decreased significantly (Figure 5q).
m6Ascore is a candidate prognostic biomarker that correlates to the efficacy of immune checkpoint therapy.
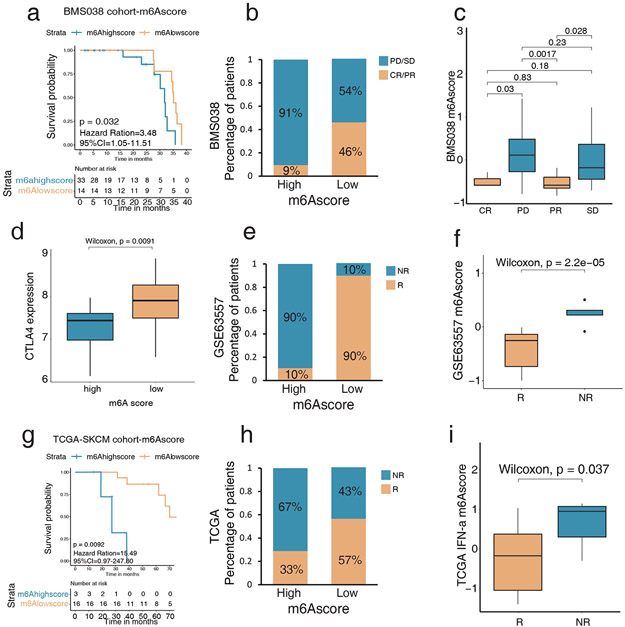
In the anti-PD-1 cohort, patients with low m6Ascores exhibited markedly prolonged survival (HR, 3.48; 95% CI, 1.05–11.51; p = 0.032) (Figure 6a-c, Tables S5, S9). For anti-CTLA4 therapy, patients with low m6Ascores also correlated with better treatment responses and clinical characteristics (Figures 6d-f). For IFN-α- therapy, patients with low m6Ascores had a significant survival benefit during treatment (HR, 15.49; 95% CI, 0.97–247.80; p = 0.0092) (Figures 6g-i, Tables S5 and S9).
Figure 6: Low m6Ascore predicts better efficacy of melanoma checkpoint immunotherapy.
(a) Survival analyses based on high and low m6A of 47 patients in the anti-PD-1 therapy cohort (BMS038) show low m6Ascore is significantly associated with better anti-PD-1 response (HR, 3.48; 95% CI, 1.05–11.51; p = 0.032). (b) The percentage of patients with anti-PD-1 response in the high and low m6Ascore in BMS038 show progressive disease, and stable disease (PD/SD) patients are greater in the high m6Ascore, and complete responder or partial responder (CR/PR) patients are greater in the low m6Ascore. (c) Difference of m6Ascore in distinct anti-PD-1 clinical response groups show PD/SD patients’ m6Ascores are significantly higher than CR/PR patients’. (d) Differences in the expression of CTLA4 among high and low m6Ascore in anti-CTLA4 cohorts (GSE63557) (p = 0.0091, Wilcoxon test). (e) The percentage of patients with anti-CTLA4 response in the high and low m6Ascore. NR: non-responder; R: responder. (f) Difference of m6Ascore in distinct anti-CTLA4 clinical response groups. NR patients’ m6Ascores are significantly higher than R patients’. (g) Survival analyses based on high and low m6Ascore in IFN-α therapy cohort (TCGA-SKCM). Low m6Ascore is significantly associated with a better IFN-α response (HR, 15.49; 95% CI, 0.97–247.80; p = 0.0092). (h) The percentage of patients with IFN-α response in the high and low m6Ascore. NR: non-responder; R: responder. (i) Difference of m6Ascore in distinct IFN-α clinical response groups. NR patients’ m6Ascores are significantly higher than R patients’.
In conclusion, we demonstrated that m6Ascore is positively correlated with malignant progression of melanomas, poor infiltration and function of T cells, high expression of CAFs, low expression of effective immune cells, and ICB resistance at the single-cell level, suggesting that m6Ascore plays an important role in the prognosis and immunotherapy of melanoma patients.
DISCUSSION
RNA post-transcriptional modifications are important steps to decrease self-recognition. Increasingly, studies have also found that N6-methyladenosine (m6A) is an important RNA methylation modification pattern in immune regulation and tumor development (Su et al., 2018). Most previous research, however, is limited to the regulation of a specific m6A gene or a specific tumor-infiltrating immune cell in melanoma.
Here, we systematically dissected the correlation of these two important tumorigenic regulators—m6A modification patterns and the melanoma TME—in a large sample of multicohort melanoma patients. To quantify the m6A modification pattern of melanoma in a patient-specific manner, given the high degree of m6A heterogeneity between individuals, we established a scoring system: the m6Ascore. A high m6Ascore represented greater m6A RNA additions and an immune-excluded phenotype, while a low m6Ascore revealed decreased m6A RNA additions and corresponded to an immune-inflamed phenotype. In this study, patients with high m6Ascores correlated with worse prognoses, clinical classifications, and molecular subtypes, while those with low m6Ascores correlated with better prognoses, clinical classifications, and molecular subtypes.
At the single-cell and spatial transcriptomic levels, we demonstrated that the m6Ascore positively correlated with tumor malignant progression, CAF infiltration, T cell depletion, immune exclusion, and ICB resistance, suggesting that a high m6Ascore was associated with cold tumors, poor prognosis, and no benefit from immunotherapy. These results are all consistent with greater innate immune recognition of unmodified RNA thus allowing greater anti-tumor immunity.
Since this work is a retrospective analysis showing only correlations, prospective clinical trials are needed to further validate the robustness of the m6Ascore in clinical practice. Besides predicting checkpoint inhibitor responders, the m6Ascore should also be tested as a predictor of sentinel node biopsy positivity, potentially reducing unnecessary procedures and resultant lymphedema risks.
Materials & Methods
Human Subjects Compliance
All data analyzed was downloaded from the listed publically available data consortiums. All data was de-identified and no further annotations used for the data than provided in public forums. The compliance of these studies with human subjects regulations including original informed consent of participants is as listed in their primary publications. No repeat informed consents are required for reuse of this publically available data since all data is entirely de-identified.
Inference of immune cell infiltration pattern in melanoma tumor microenvironment
To quantify tumor-infiltrating immune cells in melanoma, we used CIBERSORT (https://cibersort.stanford.edu), a deconvolution algorithm, and the LM22 gene signature matrix, which uses 574 immune cell markers to quantify the infiltration of 22 immune cells in tissues.
Unsupervised consensus clustering of immune cells and m6A regulators in melanoma tumor microenvironment
Twenty-one m6A regulator genes were selected, including eight writers (CBLL1, KIAA1429, METTL3, RBM15, ZC3H13, WTAP, RBM15B, METTL14), eleven readers (YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, IGF2BP1, HNRNPA2B1, HNRNPC, FMR1, LRPPRC, ELAVL1), and two erasers (FTO, ALKBH5). Based on distinct TME and m6A modification patterns, melanomas were organized through hierarchical agglomerative clustering (Ward’s method Euclidean distance). We also employed K-means unsupervised clustering methods to analyze the datasets after defining different infiltrating features.
Definition of m6Ascore in melanoma and classification of high and low m6Ascore
We first converted the gene expression of each m6A gene into a z-score to define transcript levels (RMA method normalization). Next, we separated entities using principal component analysis (PCA). Then, we separated component principle 1 (PC1 = Coefi*xi) to define the gene signature score. We applied the following method to calculate the m6Ascore of each patient:
In this equation, i is the signature score of the cluster whose Cox coefficient is positive. To distinguish between high and low m6Ascores, aggregate results were divided into two groups around the median as either high or low.
Pathway and functional enrichment analysis
We used the R package clusterprofile to perform gene annotation enrichment analysis on DEG defined above. Using the limits of false discovery rate (FDR) less than 0.05 and p < 0.01, we determined the term of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG).
Single-cell RNA-seq and bioinformatics analysis
The R program was used for single-cell RNA-seq analysis. The Seurat package was used for standardized analysis. Patients who have less than 50 malignant cells were excluded from partial analysis. Markers used to label cells were shown in the corresponding figures. The UMAP function was used to cluster the single cells into 6 groups based on known specific markers: Malignant, B, T, Endothelial cells, Macrophage, and CAFs. Then the T cells were clustered into 5 subtypes according to published T cell markers: Treg, CD4 Naïve, CD4 Cyto, CD8 Naïve, CD8Cyto (Tirosh et al., 2016). The interaction of different cell types was analyzed using the R package Cellchat. The immune checkpoint, cell cycle, EMT, chemokines, and TIP activation scores were calculated using the PCA method described above. Gene ontology enrichment analysis was performed using the R package Clusterprofile. Pseudotime analysis was performed using the R package Monocle.
Statistical analysis
All statistical analyses were performed using R (https://www.r-project.org/). For the comparison of the two groups, we used the unpaired student t-test to compare non-normally distributed variables. For the comparison of more than two groups, we used one-way analysis of variance as the parameter statistics. All p-values are two-sided, and p-values of less than 0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, part of the National Institutes of Health, under R01AR074846 01 and UG3 AR079376 to LAG as well as the National Natural Science Foundation of China (Grant No.82202468) to G.W. The authors the Chinese Skin Microbiome (CSM) Study Group at the Department of Plastic and Aesthetic Surgery, Nanfang Hospital, and Guangdong Provincial Key Laboratory of Construction and Detection in Tissue Engineering for technical support. For additional correspondence queries, please contact Gaofeng Wang (wanggaofeng@smu.edu.cn).
Abbreviations
- TME
tumor microenvironment
- m6A
N6-methyladenosine
- PCA
principal component analysis
- ICB
immune checkpoint blockades
- CAF
Cancer-associated fibroblast
- AJCC
American Joint Committee on Cancer
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LMC
Leeds Melanoma Cohort
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest.
Data availability statement
Datasets related to this article can be found at the following links with corresponded accession number. 703 melanoma patients’ data were kindly provided by Leeds cohort (https://ega-archive.org/studies/EGAS00001002922), 473 melanoma patients’ data came from TCGA-SKCM (https://portal.gdc.cancer.gov/projects/TCGA-SKCM), and 323 melanoma patients’ data were obtained from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) under GSE22153, GSE54467, GSE65904, and GSE63557. Forty-seven melanoma patients’ data were obtained from BMS038 (https://clinicaltrials.gov/ct2/show/NCT01621490). Single-cell RNA-seq and spatial transcriptomics data were obtained from GEO under GSE72056 and GSE159709.
References
- Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J, et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res 2019;47(5):2533–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fania L, Mazzanti C, Campione E, Candi E, Abeni D, Dellambra E. Role of Nicotinamide in Genomic Stability and Skin Cancer Chemoprevention. Int J Mol Sci 2019;20(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 2014;15(5):293–306. [DOI] [PubMed] [Google Scholar]
- Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 2019;566(7743):270–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363(8):711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MV, Moncada R, Weiss JM, Yanai I, White RM. Spatially resolved transcriptomics reveals the architecture of the tumor-microenvironment interface. Nat Commun 2021;12(1):6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005;23(2):165–75. [DOI] [PubMed] [Google Scholar]
- Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017;548(7667):338–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 2016;167(3):816–28 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Lyko F. The importance of non-histone protein methylation in cancer therapy. Nat Rev Mol Cell Biol 2019;20(10):569–70. [DOI] [PubMed] [Google Scholar]
- Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 2018;172(1–2):90–105 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd,, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352(6282):189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res 2018;28(2):253–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen H, Tian R, Zhang Y, Drutskaya MS, Wang C, et al. Macrophages induce AKT/beta-catenin-dependent Lgr5(+) stem cell activation and hair follicle regeneration through TNF. Nature communications 2017;8:14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer 2020;19(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kurupati R, Liu L, Zhou XY, Zhang G, Hudaihed A, et al. Enhancing CD8(+) T Cell Fatty Acid Catabolism within a Metabolically Challenging Tumor Microenvironment Increases the Efficacy of Melanoma Immunotherapy. Cancer Cell 2017;32(3):377–91 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 2017;18(1):31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets related to this article can be found at the following links with corresponded accession number. 703 melanoma patients’ data were kindly provided by Leeds cohort (https://ega-archive.org/studies/EGAS00001002922), 473 melanoma patients’ data came from TCGA-SKCM (https://portal.gdc.cancer.gov/projects/TCGA-SKCM), and 323 melanoma patients’ data were obtained from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) under GSE22153, GSE54467, GSE65904, and GSE63557. Forty-seven melanoma patients’ data were obtained from BMS038 (https://clinicaltrials.gov/ct2/show/NCT01621490). Single-cell RNA-seq and spatial transcriptomics data were obtained from GEO under GSE72056 and GSE159709.