Abstract
Previous studies have shown that the production of extracellular enzymes (pectate lyase [Pel], polygalacturonase [Peh], cellulase [Cel], and protease [Prt]) and harpinEcc (the elicitor of hypersensitive reaction) in Erwinia carotovora subsp. carotovora is regulated by RsmA, an RNA-binding protein, and rsmB, a regulatory RNA (Rsm stands for regulator of secondary metabolites) (Y. Liu et al., Mol. Microbiol. 29:219–234, 1998). We have cloned and characterized a novel regulatory gene, rsmC, that activates RsmA production and represses extracellular enzyme and harpinEcc production, rsmB transcription, and virulence in E. carotovora subsp. carotovora. In an rsmC knockout mutant of E. carotovora subsp. carotovora Ecc71 carrying the chromosomal copy of the wild-type rsmA+ allele, the basal levels of Pel, Peh, Cel, Prt, and harpinEcc as well as the amounts of rsmB, pel-1, peh-1, celV, and hrpNEcc transcripts are high, whereas the levels of rsmA transcripts and RsmA protein are low. Furthermore, the expression of an rsmA-lacZ gene fusion is lower in the RsmC− mutant than in the RsmC+ parent. Conversely, the expression of an rsmB-lacZ operon fusion is higher in the RsmC− mutant than in the RsmC+ parent. These observations establish that RsmC negatively regulates rsmB transcription but positively affects RsmA production. Indeed, comparative studies with an RsmC− mutant, an RsmA− mutant, and an RsmA− RsmC− double mutant have revealed that the negative effects on exoprotein production and virulence are due to the cumulative regulatory effects of RsmC on rsmA and rsmB. Exoprotein production by the RsmC− mutant is partially dependent on the quorum sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Southern blot data and analysis of PCR products disclosed the presence of rsmC sequences in E. carotovora subsp. atroseptica, E. carotovora subsp. betavasculorum, and E. carotovora subsp. carotovora. These findings collectively support the idea that rsmA and rsmB expression in these plant pathogenic Erwinia species is controlled by RsmC or a functional homolog of RsmC.
Erwinia species produce extracellular enzymes and proteins, polysaccharides, pigments, and small diffusible metabolites (2, 5, 43). The production of such substances is markedly stimulated during late exponential and early stationary growth phases when bacteria reach high cell density and experience nutrient limitation and other forms of stress (14, 31, 36, 39, 44). How bacteria perceive these conditions and then activate gene expression are issues that have lately attracted considerable attention. Consequently, at least four regulatory parameters are now recognized to be of critical importance in the activation of growth phase-dependent (secondary) metabolite production. One entails the production of RpoS, an alternate sigma factor responsible for the activation of many genes expressed mainly during the stationary phase (14, 44). The other factor is the cell density (quorum) sensing signal, N-acylated derivatives of homoserine lactone, that apparently accumulate during these growth conditions and activate the expression of an array of genes many of which are expressed during postexponential growth (11, 12, 36, 41). The other two parameters, RsmA and rsmB RNA, control the production of extracellular enzymes, phytohormones, antibiotics, pigments, and polysaccharides, the synthesis of flagella, and levels of the quorum sensing signal N-(3-oxohexanoyl)-l-homoserine lactone (OHL) in various Erwinia species; they also affect virulence and the production of Erwinia carotovora subsp. carotovora harpin (harpinEcc), the elicitor of the hypersensitive reaction (4, 6, 19, 26).
Recent studies have disclosed that RsmA is an RNA-binding protein and that it promotes message decay (6), although how this is brought about awaits clarification. rsmB (previously aepH [28]), on the other hand, specifies a unique RNA regulator that apparently neutralizes RsmA action by forming an inactive ribonucleoprotein complex (19). The current model postulates that RsmA and rsmB act antagonistically to modulate the expression of many genes, particularly those that are expressed in a growth phase-dependent manner. Romeo and associates have characterized a very similar system comprising CsrA (a RsmA homolog) and csrB (a rsmB homolog), which controls glycogen accumulation, cell surface properties, and cell size in Escherichia coli (reference 34 and references cited therein).
There is growing evidence that RsmA levels and RsmA activity are rigorously controlled by bacteria to prevent extensive decay of transcripts of genes for essential functions. Several lines of evidence support this view. First, overexpression of rsmA from high-copy-number plasmids or artificial strong promoters is generally detrimental to cell physiology and in certain hosts is even lethal (25). Second, rsmA expression in Erwinia and other enterobacteria (i.e., Salmonella typhimurium) is controlled by sigma-S as well as sigma-70 (27). Moreover, E. carotovora subsp. carotovora uses a novel regulatory mechanism involving KdgR, a global negative regulator of IclR family, to modulate the levels of rsmB RNA (20), which in turn controls RsmA action.
In the course of our search for regulatory mutants of E. carotovora subsp. carotovora, we discovered a class of transposon insertion mutants that produced very high basal levels of extracellular enzymes, as previously noted with RsmA− mutants (4, 6). Subsequent physical evidence, however, revealed that the phenotypes of the new mutants resulted from disruption of a previously unidentified locus, which we have designated, rsmC (for regulator of secondary metabolism). We describe here the structure and function of rsmC. Our data for the first time show that RsmC controls the production of RsmA and rsmB RNA and that the phenotypic changes in RsmC− mutants are due to these regulatory effects of RsmC. Physical evidence shows that homologs of E. carotovora subsp. carotovora Ecc71 rsmC occur in other E. carotovora subspecies, i.e., atroseptica and betavasculorum. Based on the data presented here and previously reported (4, 6, 19), we conclude that RsmA, rsmB, and RsmC are the major components of a global regulatory system that controls gene expression in several plant pathogenic enterobacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. The compositions of Luria-Bertani (LB) medium, minimal salts medium, minimal salts medium plus celery extract, nutrient gelatin agar have been previously described (4, 29). When required, antibiotics were added as follows: ampicillin, 100 μg/ml; gentamicin, 10 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 50 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 10 μg/ml. Media were solidified by the addition of 1.5% (wt/vol) agar. The compositions of agarose media for semiquantitative plate assay for extracellular pectate lyase (Pel), polygalacturonase (Peh), cellulase (Cel), and protease (Prt) have been described by Chatterjee et al. (4).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. carotovora subsp. carotovora | ||
| Ecc71 | Wild type | 45 |
| AC5047 | Nalr derivative of AC5006 | 4 |
| AC5050 | RsmC− derivative of AC5047 by mini-Tn5-Km mutagenesis | This work |
| AC5051 | Ohl− derivative of AC5050 by marker exchange with pAKC855 | This work |
| AC5053 | RsmC− derivative of Ecc71 by marker exchange with pAKC970 | This work |
| AC5054 | RsmC− derivative of AC5071 by marker exchange with pAKC980 | This work |
| AC5070 | RsmA− derivative of AC5047 | 4 |
| AC5071 | RsmA− derivative of Ecc71 | 24 |
| AC5091 | Ohl− derivative of AC5047 | 4 |
| SCRI193 | Wild type | 37 |
| E. carotovora subsp. atroseptica Eca12 | Wild type | 45 |
| E. carotovora subsp. betavasculorum Ecb11129 | Wild type | J. E. Loper |
| E. herbicola pv. gypsophilae PD713 | Wild type | 17 |
| Escherichia coli | ||
| DH5α | φ80lacZΔM15Δ(lacZYA-argF) U169 hsdR17 recA1 endA1 thi-1 | Gibco BRL |
| JM109(DE3) | endA1 recA1 gyrA96 hsdR17 supE44 relA1 thi Δ(lac-pro) F′ (traD36 proAB+lacIqlacZΔM15) λcI857 ind1 Sam7 lacU5-T7 gene 1 | Promega Biotec |
| MC4100 | araD139 Δ(lacIPOZYA)U169 recA1 thi-1 Strr | 18 |
| Plasmids | ||
| pBluescript SK(+) | Apr | Stratagene |
| pCL1920 | Spr, Smr | 16 |
| p34S-Gm | Gmr; source of Gmr cassette | 8 |
| pCL1920Gmr | Gmr, Spr, Smr, Gmr cassette inserted at the BamHI site of pCL1920 | This work |
| pET-28a(+) | Kmr | Novagen |
| pHP45Ω | Source of Spr omega fragment | 33 |
| pLARF5 | Tcr | 15 |
| pMP220 | Tcr; promoter-probe vector | 40 |
| pNM481 | Apr, promoter-probe vector | 23 |
| pNM481Spr | Spr, Spr omega fragment inserted at the ScaI site of pNM481 | This work |
| pRK415 | Tcr | 15 |
| pRK2013 | Mob+, Tra+, Kmr | 10 |
| pAKC781 | Apr, peh-1+ | 18 |
| pAKC783 | Apr, pel-1+ | 18 |
| pAKC882 | Apr, pT7-rsmA | 26 |
| pAKC855 | Spr, Tcr, Ohl− | 4 |
| pAKC887 | Spr, rsmAEcc-lacZ, 279-bp (245 bases upstream of translational start site to 34 bases downstream of translational start site) PCR product from Ecc71 in pNM481Spr | This work |
| pAKC888 | Spr, rsmAEa-lacZ, 317-bp (283 bases upstream of translational start site to 34 bases downstream of translational start site) PCR product from E9 in pNM481Spr | This work |
| pAKC889 | Spr, rsmAEhg-lacZ, 300-bp (266 bases upstream of translational start site to 34 bases downstream of translational start site) PCR product from PD713 in pNM481Spr | This work |
| pAKC890 | Spr, csrA-lacZ, 287 bp (252 bases upstream of translational start site to 34 bases downstream of translational start site) PCR product from MC4100 in pNM481Spr | This work |
| pAKC924 | Apr, hrpNEcc in pBluescript SK(+) | 7 |
| pAKC970 | Kmr, Tcr; pLARF5 containing mini-Tn5-Km and flanking chromosomal DNA from AC5050 | This work |
| pAKC971 | Tcr; pLARF5 containing rsmC from genomic library of Ecc71 | This work |
| pAKC972 | Tcr; pLARF5 containing rsmC from genomic library of Ecc71 | This work |
| pAKC973 | Apr, 4.0-kb ClaI fragment of pAKC970 containing mini-Tn5-Km and flanking DNA in pBluescript SK (+) | This work |
| pAKC974 | Apr; RsmC+, 2.0-kb ClaI fragment of pAKC971 in pBluescript SK (+) | This work |
| pAKC975 | Spr, RsmC+, 2.0-kb ClaI fragment of pAKC971 in pCL1920 | This work |
| pAKC976 | Apr, 1.5-kb ClaI-HindIII fragment of pAKC973 (flanking DNA of mini-Tn5-Km) in pBluescript SK (+) | This work |
| pAKC977 | Apr, 0.5-kb ClaI-HindIII fragment of pAKC973 (flanking DNA of mini-Tn5-Km) in pBluescript SK (+) | This work |
| pAKC978 | Kmr, 371-bp EcoRV-HindIII fragment of pAKC974 in pET28a(+) | This work |
| pAKC979 | Tcr, RsmC+, 2.0-kb ClaI fragment of pAKC971 in pRK415 | This work |
| pAKC980 | Tcr, Spr; RsmC was inactivated in pAKC979 by inserting an omega fragment at the EcoRV site | This work |
| pAKC1002 | Tcr, rsmB-lacZ in pMP220 | 19 |
| pAKC1004 | Spr, plac-rsmB′ in pCL1920 | 19 |
| pAKC1004Gmr | Gmr, Spr, Gmr cassette inserted at the BamHI site of pAKC1004 | This work |
| pAKC1034 | Apr, 200-bp DNA fragment of celV from Ecc71 in pGEM-T Easy | 19 |
Preparation of samples for enzyme assays and assay conditions.
The preparation of enzyme samples for extracellular Pel, Peh, Prt, and Cel, and the assay procedures were carried out according to Murata et al. (29). The semiquantitative agarose plate assays for extracellular Pel, Peh, Prt, and Cel were performed as described by Chatterjee et al. (4).
Isolation of RsmC− mutant by mini-Tn5 mutagenesis.
The Nalr strain AC5047 (Table 1) was mutagenized with mini-Tn5-Km as described by Chatterjee et al. (4). Transconjugants were selected on nutrient gelatin agar medium containing kanamycin and nalidixic acid. Protease-overproducing mutants, identified by the size of halo around the colony, were tested for a pleiotropic phenotype by semiquantitative agarose plate assays for Pel, Peh, and Cel.
DNA techniques.
Standard procedures were used in the isolation of plasmid and chromosomal DNAs, transformation, restriction endonuclease digests, gel electrophoresis, DNA ligation, and colony in situ hybridization (38). Southern blot hybridizations were carried out as described by Cui et al. (6). PCR was performed as described by Liu et al. (19). Restriction and modifying enzymes were obtained from Promega Biotec (Madison, Wis.).
Nucleotide sequence analysis of rsmC.
For nucleotide sequence analysis, the rsmC fragments flanking the mini-Tn5-Km sequence in pAKC973 (Table 1) were cloned into the ClaI-HindIII sites of pBluescript SK(+). The resulting plasmids, pAKC976 and pAKC977, were used for producing unidirectional deletions into the rsmC sequence with the Erase-a-Base system (Promega Biotec). Plasmids carrying deletions were used for sequence analysis using Sequenase version 2.0 (U.S. Biochemical, Cleveland, Ohio). Oligonucleotide primers were also used in nucleotide sequence determinations. DNA and protein sequence analyses were performed with the PC gene software (IntelliGenetics, Inc., Mountain View, Calif.).
RNA assays.
Total RNA was extracted by the method of Aiba et al. (1) from E. carotovora subsp. carotovora strains grown at 28°C in minimal salts medium plus sucrose (0.5% wt/vol) or in this medium supplemented with appropriate drugs.
Northern blot analyses were performed as described by Liu et al. (19). The probes used in this study were the 304-bp EcoRV-HindIII fragment of rsmC from pAKC975 (see Fig. 3A), the 314-bp EcoRV-KpnI fragment of pel-1 from pAKC783 (18), the 743-bp HindIII fragment of peh-1 from pAKC781 (18), the 200-bp EcoRI fragment of celV from pACK1034 (19), the 779-bp EcoRV-SmaI fragment of hrpNEcc from pAKC924 (7), the 183-bp NdeI-SalI fragment of rsmA from pAKC882, and the 321-bp BamHI-HindIII fragment of rsmB from pAKC1004. DNA probes were labeled with [α-32P]dATP by using the Prime-a-Gene labeling system (Promega Biotec) according to the manufacturer’s instructions.
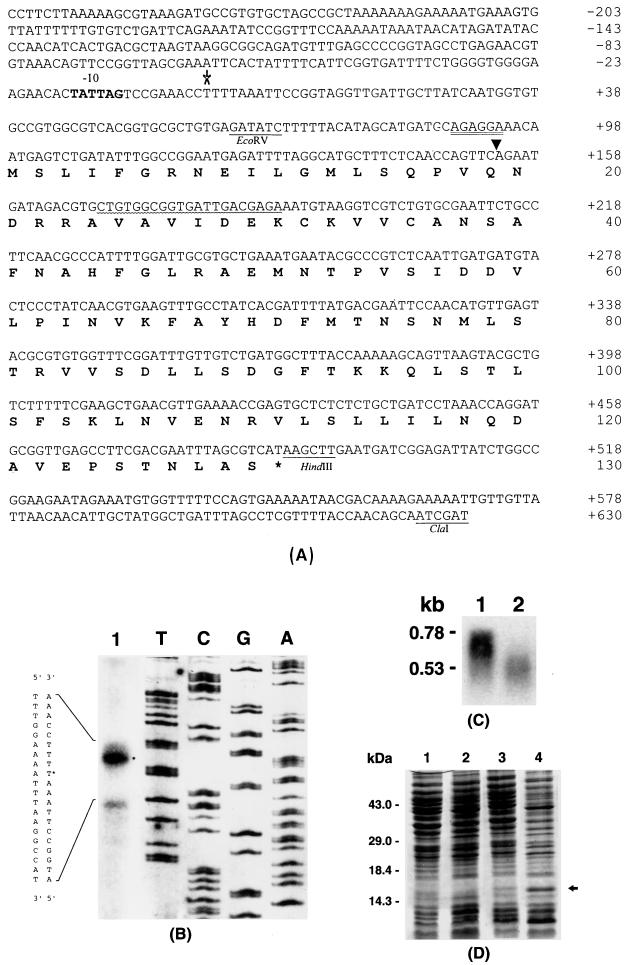
FIG. 3.
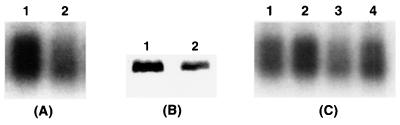
(A) Nucleotide and deduced amino acid sequences of E. carotovora subsp. carotovora rsmC. The putative ribosome binding site is double underlined. The transcriptional start site at the thymine residue is indicated with an asterisk. The −10 consensus sequences are shown in boldface. The nucleotide sequence used for the synthesis of a complementary oligonucleotide for the primer extension assay is underlined with a wavy line. The position of mini-Tn5-Km insertion in the RsmC− mutants AC5050 and AC5053 is indicated with an arrowhead. Some restriction endonuclease sites are also shown. Numbers on the right refer to positions of the nucleotides and the amino acid residues for each line. (B) Primer extension analysis of rsmC mRNA. Lane 1, 20 μg of total RNA sample from E. carotovora subsp. carotovora Ecc71 grown at 28°C in minimal salts medium plus sucrose to an A600 of 2.0. The products of the primer extension reaction were run alongside a regular sequencing gel. Nucleotides on the left refer to the nucleotide sequence beyond the transcriptional start site; the asterisk denotes the thymine residue at which transcription was initiated. (C) Northern blot analysis of rsmC mRNA in E. carotovora subsp. carotovora Ecc71 (RmsC+; lane 1) and its RsmC− derivative, AC5053 (lane 2). Total RNAs were isolated from bacteria grown at 28°C in minimal salts medium plus sucrose to an A600 of 2.0. Each lane contained 20 μg of total RNA. (D) Overproduction of RsmC in E. coli strain JM109(DE3). Bacteria carrying the cloning vector pET28a(+) or the rsmC+ plasmid pAKC978 were grown in LB medium plus kanamycin with or without IPTG as described in Materials and Methods. Each lane contained 10 μg of total bacterial protein. The arrow indicates the 15-kDa overproduced RsmC protein. Lane 1, pET28(+), no IPTG; lane 2, pET28(+), with IPTG (final concentration, 1 mM); lane 3, pAKC978, no IPTG; lane 4, pAKC978 with IPTG (final concentration, 1 mM).
Primer extension assay was performed as instructed by the manufacturer (Promega Biotec) with primer rsmC1 (see Fig. 3A) and 20 μg of RNA.
Identification of the rsmC product.
E. coli JM109(DE3) carrying the cloning vector, pET28a(+), or pAKC978 (Table 1), which contains the coding region of rsmC in the expression vector pET28a(+), were grown at 37°C in LB medium containing kanamycin. When the cultures reached an A600 of 0.7, each culture was divided into two parts; isopropyl-β-d-thiogalactopyranoside (IPTG) was added to one part to yield a final concentration of 1.0 mM, and the other part served as the control. Following an additional 3 h of incubation, cells were collected by centrifugation. Double-strength sodium dodecyl sulfate (SDS) loading buffer (100 mM Tris-HCl [pH 6.8], 200 mM dithiothreitol, 4% [wt/vol] SDS, 0.2% [wt/vol] bromophenol blue, 20% [vol/vol] glycerol) was added, and the samples were boiled for 5 min. Proteins were fractionated by 0.1% (wt/vol) SDS–15% (wt/vol) polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie brilliant blue.
Western blot analysis.
E. carotovora subsp. carotovora Ecc71, AC5053, AC5054, and AC5071 were grown at 28°C in minimal salts medium plus sucrose to an A600 value of 2.3. Total bacterial protein was precipitated with trichloroacetic acid at a final concentration of 10% (vol/vol) and resuspended in 1× SDS loading buffer. The protein concentrations were determined with a bicinchoninic acid protein assay kit (Pierce Corp., Rockford, Ill.) according to the manufacturer’s specifications. Western blot analysis of the total bacterial protein was carried out as described by Mukherjee et al. (24). The antibody raised against the harpin from strain E. chrysanthemi (3) was used as the probe for harpinEcc. The anti-RsmA antiserum produced against a synthesized peptide from amino acids 48 to 61 of RsmA (6) in rabbit by Genemed Biotechnologies Inc. (San Francisco, Calif.) was used as the probe for RsmA.
Construction of rsmA-lacZ and csrA-lacZ fusions and β-galactosidase assay.
To construct lacZ fusions of the rsmA genes of E. amylovora, E. carotovora subsp. carotovora, and E. herbicola pv. gypsophilae and the csrA gene of E. coli, the promoter regions of these genes were amplified by PCR from the chromosomal DNAs of E. amylovora E9, E. carotovora subsp. carotovora Ecc71, E. herbicola pv. gypsophilae PD713, and E. coli MC4100 with primers designed from the nucleotide sequences of these genes (6, 25, 35). PCR products were digested with EcoRI and BamHI and cloned into the promoter-probe vector pNM481Spr to yield pAKC887, pAKC888, pAKC889, and pAKC890 (Table 1). E. carotovora subsp. carotovora AC5047 and AC5050 carrying these constructs were grown at 28°C in minimal salts medium plus sucrose and spectinomycin to an A600 of 2.0, and culture samples were assayed for β-galactosidase activity as described by Miller (22).
Construction of RsmA− RsmC− and RsmC− Ohl− double mutants.
To inactivate rsmC in the RsmC+ plasmid pAKC979, the omega Spr DNA fragment was inserted at the EcoRV site (see Fig. 3A) to yield pAKC980. To confirm that rsmC was inactivated in pAKC980, this plasmid was transformed into Ecc71 and AC5053, and the exoenzyme levels were checked. The assay data showed that pAKC980 did not suppress levels of Pel, Peh, Cel, and Prt, indicating that rsmC was inactivated. To construct an RsmA− RsmC− double mutant, pAKC980 was transferred into the RsmA− strain AC5071 by using the helper plasmid pRK2013. To construct an RsmC− Ohl− double mutant, pAKC855 (Table 1) was transferred into the RsmC− strain AC5050 by using the helper plasmid pRK2013. Transconjugants were selected on minimal salts agar plus sucrose (0.2%, wt/vol) and spectinomycin, and Spr Tcs isolates were obtained. The inactivation of rsmC in AC5054 was confirmed by Northern analysis. The inactivation of ohl in AC5051 was determined by assaying for OHL production (4).
Plant tissue maceration.
The celery petiole assays were previously described (29). The extent of tissue maceration was estimated visually.
Nucleotide sequence accession number.
The GenBank accession number for rsmC is AF178852.
RESULTS
Isolation of mini-Tn5-Km insertion RsmC− mutants.
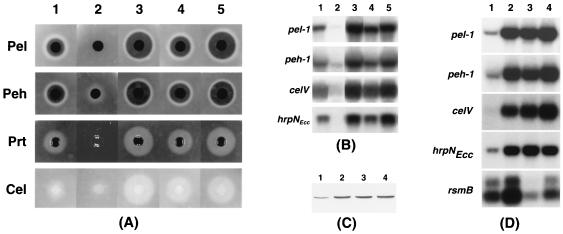
E. carotovora subsp. carotovora AC5047 was mutagenized with mini-Tn5-Km, and the Kmr transconjugants were screened for increased protease activity on nutrient gelatin agar medium. Colonies with high protease activity were subsequently tested for the levels of pectinases and cellulase. The characteristics of one class of mutants represented by AC5070 and AC5071 and designated as RsmA− have been described elsewhere (4, 6, 24). Here we report the characteristics of another class of derepressed mutant and the corresponding gene, rsmC. The mutant strain, AC5050, and its parent strain, AC5047, were grown in minimal salts medium supplemented with sucrose, and culture samples were assayed for the levels of extracellular enzymes. The data in Fig. 1A show that the levels of Pel, Peh, Cel, and Prt activities in AC5050 are markedly higher than those in the parent strain, AC5047. Moreover, the results of Northern hybridization analysis using pel-1, peh-1, celV, and hrpNEcc as probes revealed that the levels of transcripts of these genes are considerably higher in the RsmC− mutant than in the parent strain (Fig. 1B). These findings demonstrate that the negative effect of rsmC on Pel, Peh, and Cel production is due to the modulation of transcript levels. Further support for this conclusion comes from the findings with an RsmC− mutant of E. carotovora subsp. carotovora Ecc71 constructed by marker exchange and the transdominant effects of the cloned rsmC+ DNA (see below).
FIG. 1.
(A) Agarose plate assays for Pel, Peh, Prt, and Cel activities of E. carotovora subsp. carotovora AC5047 (column 1), the Ohl− derivative of AC5047 (AC5091; column 2), the RsmC− mutant (AC5050; column 3), the Ohl− derivative of AC5050 (AC5051; column 4), and the RsmA− mutant of AC5047 (AC5070; column 5). Bacteria were grown at 28°C in minimal salts medium plus sucrose and harvested at an A600 value of 2.5. Culture supernatants (10 μl) were added to each well. After 18 h of incubation at 28°C, the Pel and Peh assay plates were developed with 4 N HCl, and the Cel assay plate was developed with Congo red and NaCl solutions. Halos around the wells in the Prt assay plate became visible within 24 h without any further treatment. (B) Northern blot analysis of pel-1, peh-1, celV, and hrpNECC mRNA in E. carotovora subsp. carotovora AC5047 (RsmA+ RsmC+ Ohl+; lane 1), AC5091 (RsmA+ RsmC+ Ohl−; lane 2), AC5050 (RsmA+ RsmC− Ohl+; lane 3), AC5051 (RsmA+ RsmC− Ohl−; lane 4), and AC5070 (RsmA− RsmC+ Ohl+; lane 5). Bacteria were grown at 28°C in minimal salts medium plus sucrose to an A600 of 2.0 for total RNA extraction. Each lane contained 10 μg of total RNA. (C) Western blot analysis of harpinEcc produced by E. carotovora subsp. carotovora Ecc71 (RsmA+ RsmC+; lane 1), AC5053 (RsmA+ RsmC−; lane 2), AC5071 (RsmA− RsmC+; lane 3), and AC5054 (RsmA− RsmC−; lane 4). Each lane contained 20 μg of total bacterial protein. (D) Northern blot analysis of pel-1, peh-1, celV, hrpNECC, and rsmB transcripts in E. carotovora subsp. carotovora Ecc71 (RsmA+ RsmC+; lane 1), AC5053 (RsmA+ RsmC−; lane 2), AC5071 (RsmA− RsmC+; lane 3), and AC5054 (RsmA− RsmC−; lane 4). Bacteria were grown at 28°C in minimal salts medium plus sucrose to an A600 of 2.0 for total RNA extraction. Each lane contained 10 μg of total RNA.
Since the phenotypes of AC5050 and AC5070 are very similar (Fig. 1A, columns 3 and 5; also see references 4, 6, and 7), it was initially deemed important to determine if the mutants were genetically different. For this, we first localized the sites of mini-Tn5-Km insertion in these mutants. Chromosomal DNAs of the mutants and the parent strain, AC5047, were digested with the restriction enzyme ClaI, which does not have a recognition site in the mini-Tn5-Km DNA. The fragments were resolved in an agarose gel, transferred to a nylon membrane, and hybridized with the labeled transposon DNA under stringent conditions. Results (data not shown) indicated that while there were no hybridizing signals with AC5047, the probe hybridized to DNA fragments of about 11 kb of AC5070 and 4 kb of AC5050. Since rsmA does not possess a ClaI site (4), these data indicated that the mutants have insertions in different genes. To determine if the disruption of a functional gene due to the insertion of the transposon in the chromosome of AC5050 was in fact responsible for the higher levels of enzymatic activities, we cloned the mini-Tn5-Km DNA along with flanking AC5050 DNA into the cosmid vector pLAFR5, yielding pAKC970 (Table 1). This plasmid was transferred to Ecc71, and a spontaneous Kmr Tcs derivative, AC5053, was obtained according to the procedures described by Chatterjee et al. (4). The absence of a functional rsmC+ allele in AC5053 due to its replacement by rsmC::mini-Tn5 DNA was confirmed by Northern analysis (data not shown). The parent strain, Ecc71, and its RsmC− derivative, AC5053, were grown in minimal salts medium supplemented with sucrose, and culture samples were assayed for Pel, Peh, Cel, and Prt activities as well as harpinEcc. The data (Table 2; Fig. 1C, lanes 1 and 2) show that the levels of Pel, Peh, Cel, Prt, and harpinEcc in AC5053 are markedly higher than those in the parent strain, Ecc71. Moreover, the results of Northern hybridization analysis (Fig. 1D, lanes 1 and 2) reveal that the levels of transcripts of pel-1, peh-1, celV and hrpNEcc are considerably higher in the RsmC− mutant than in the parent strain.
TABLE 2.
Levels of Pel, Peh, Prt, and Cel produced by E. carotovora subsp. carotovora strains
| Straina | Relevant phenotypeb | Sp act (U/ml/A600 unit; mean ± SD)c
|
|||
|---|---|---|---|---|---|
| Pel | Peh | Prt | Cel | ||
| Ecc71 | RsmA+ RsmC+ | 0.19 ± 0.03 | 66.2 ± 3.21 | 1.4 ± 0.28 | ND |
| AC5053 | RsmA+ RsmC− | 5.00 ± 0.19 | 441.8 ± 8.64 | 57.9 ± 2.32 | 1.32 ± 0.26 |
| AC5071 | RsmA− RsmC+ | 6.97 ± 0.23 | 391.8 ± 2.31 | 23.8 ± 2.74 | 0.90 ± 0.03 |
| AC5054 | RsmA− RsmC− | 15.77 ± 0.43 | 744.5 ± 18.69 | 77.8 ± 4.15 | 3.32 ± 0.09 |
| AC5047(pCL1920) | RsmA+ RsmC+ (none [vector]) | 0.17 ± 0.02 | 10.3 ± 0.26 | — | — |
| AC5047(pAKC975) | RsmA+ RsmC+ (RsmC+) | 0.06 ± 0.01 | ND | — | — |
| AC5050(pCL1920) | RsmA+ RsmC− (none [vector]) | 2.10 ± 0.11 | 341.0 ± 9.70 | — | — |
| AC5050(pAKC975) | RsmA+ RsmC− (RsmC+) | 0.11 ± 0.01 | 36.7 ± 1.70 | — | — |
Bacteria were grown at 28°C in minimal salts medium plus sucrose (0.5% [wt/vol]) for Pel, Peh, and Cel assays or in this medium plus celery extract for Prt assay. Strains carrying plasmids were grown in minimal salts medium plus sucrose and spectinomycin. Cultures were harvested at an A600 value of 2.5, and the culture supernatants were used for enzyme assays.
Relevant phenotypes conferred by the plasmids are indicated in parentheses.
ND, not detectable; —, not done.
Cloning of the wild-type rsmC+ allele.
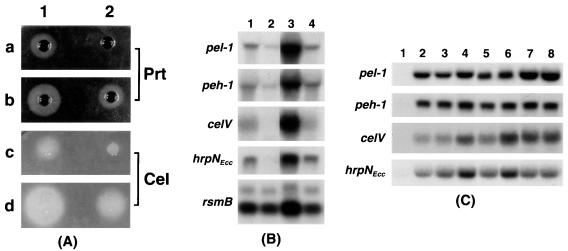
The 4-kb ClaI fragment from pAKC970 encompassing the transposon sequence and the flanking DNA (Table 1) was used as the probe in colony hybridization of Ecc71 genomic library, yielding two hybridizing plasmids, pAKC971 and pAKC972. The parent strain (AC5047) and its RsmC− mutant (AC5050) carrying these plasmids produced levels of extracellular enzymes much lower than the levels produced by these strains carrying the cloning vector pLAFR5 (data not shown). The 2-kb wild-type ClaI fragment of pAKC971 containing the rsmC+ DNA was subcloned into plasmid pCL1920, yielding pAKC975. E. carotovora subsp. carotovora AC5047 and AC5050 were transformed with pAKC975 or the cloning vector pCL1920, and the bacterial constructs were grown in minimal salts medium supplemented with sucrose and spectinomycin. The culture supernatants were assayed for Pel, Peh, Cel, and Prt activities. It is apparent that multiple copies of rsmC+ DNA caused the repression of Pel and Peh production (Table 2) as well as Prt and Cel levels (Fig. 2A) in both the parent strain (AC5047) and the RsmC− (AC5050). Moreover, these strains carrying multiple copies of the rsmC+ DNA produced significantly lower levels of transcripts of pel-1, peh-1, celV, and hrpNEcc than the strains carrying the vector (Fig. 2B). A very similar effect of multiple copies of the rsmC+ DNA was noted with Ecc71 and its RsmC− mutant, AC5053 (data not shown). This transdominant effect of the cloned DNA taken along with other findings described above indicated that the phenotypes of AC5050 and AC5053 resulted from the inactivation of a regulatory gene which negatively controls harpin and extracellular enzyme production.
FIG. 2.
(A) Agarose plate assays for Prt and Cel activities of E. carotovora subsp. carotovora AC5047 (a and c) and its RsmC− mutant (AC5050; b and d) carrying the cloning vector, pCL1920 (column 1) or the RsmC+ plasmid, pAKC975 (column 2). Bacteria were grown at 28°C in minimal salts medium plus sucrose and spectinomycin and harvested at an A600 of 2.5. Culture supernatants (10 μl) were added to each well. (B) Northern blot analysis of pel-1, peh-1, celV, hrpNEcc, and rsmB transcripts produced by E. carotovora subsp. carotovora AC5047 (RsmC+) and AC5050 (RsmC−) carrying the cloning vector pCL1920 or the RsmC+ plasmid pAKC975. Lane 1, AC5047 carrying pCL1920; lane 2, AC5047 carrying pAKC975; lane 3, AC5050 carrying pCL1920; lane 4, AC5050 carrying pAKC975. Total RNAs were isolated from bacteria grown at 28°C in minimal salts medium plus sucrose and spectinomycin to an A600 of 2.0. Each lane contained 10 μg of total RNA. (C) Northern blot analysis of pel-1, peh-1, celV, and hrpNEcc mRNA produced by E. carotovora subsp. carotovora Ecc71 (RsmA+ RsmC+), AC5053 (RsmA+ RsmC−), AC5071 (RsmA− RsmC+), and AC5054 (RsmA− RsmC−) carrying the cloning vector pCL1920Gmr or the rsmB+ plasmid pAKC1004Gmr. Lane 1, Ecc71/pCL1920Gmr; lane 2, Ecc71/pAKC1004Gmr; lane 3, AC5053/pCL1920Gmr; lane 4, AC5053/pAKC1004Gmr; lane 5, AC5071/pCL1920Gmr; lane 6, AC5071/pAKC1004Gmr; lane 7, AC5054/pCL1920Gmr; lane 8, AC5054/pAKC1004Gmr. Total RNAs were isolated from bacteria grown at 28°C in minimal salts medium plus sucrose and gentamicin to an A600 of 2.0. Each lane contained 10 μg of total RNA.
Analysis of the nucleotide sequence of rsmC and identification of the gene product.
Sequence analysis of the DNA regions flanking mini-Tn5-Km in pAKC973 revealed that the minitransposon had inserted into an open reading frame (ORF) (Fig. 3A) which could encode a protein of 130 amino acid residues. Ten bases upstream of the transcriptional start site (Fig. 3B), there is a typical −10 consensus sequence of a sigma-70-type promoter (Fig. 3A). Database (BLAST 2.0) search for sequences homologous to RsmC revealed no protein with significant homology. However, a segment of RsmC (amino acid residues 33 to 99) has 43% identity with a segment (amino acid residues 734 to 800) of a putative transcriptional adaptor of Caenorhabditis elegans (GenBank accession no. U20864). To establish that the ORF deduced from the sequence data is actually functional, we identified the transcripts and the corresponding protein product. Total RNA samples from the parent strain (Ecc71) and the RsmC− mutant (AC5053) grown in minimal medium were hybridized with rsmC. The results (Fig. 3C) show that Ecc71 produced an rsmC transcript of about 600 bases, while AC5053 produced a diffuse weak signal of an apparently truncated transcript. The size of the transcript produced by Ecc71 is consistent with the size of the rsmC ORF. To identify the product of rsmC, the 371-bp EcoRV-HindIII DNA fragment containing the coding region of rsmC (Fig. 3A) was cloned into the expression vector pET28a(+), making pAKC978, where rsmC is under the transcriptional control of T7 promoter. E. coli JM109(DE3) carrying pAKC978 and the cloning vector were grown at 37°C in LB medium containing kanamycin and induced by IPTG. Bacterial cells were collected and the total bacterial protein samples were assayed by SDS-PAGE in a 15% (wt/vol) polyacrylamide gel. After IPTG induction, a protein of ca. 15 kDa was produced by JM109(DE3) carrying pAKC978 (Fig. 3D, lane 4) but not by JM109(DE3) carrying the cloning vector pET28a(+) (Fig. 3D, lane 2). The apparent molecular mass of 15 kDa of the overproduced protein matches the mass of 14.5 kDa of the polypeptide deduced from the rsmC sequence, further indicating that this protein band is the product of rsmC. Without IPTG induction, this protein band was not detected with JM109(DE3) carrying pET28a(+) (Fig. 3D, lane 1), although a faint band was visible at the same position with JM109(DE3) carrying pAKC978 (Fig. 3D, lane 3). This may have resulted from a leaky RsmC production due to the activity of T7 promoter.
Exoenzyme production by the RsmC− mutant in the absence of a quorum sensing signal.
Our studies had shown that RsmA− mutants of E. carotovora subsp. carotovora do not require the cell density/quorum sensing signal OHL for enzyme and harpinEcc overproduction, pathogenicity, and elicitation of a hypersensitive reaction (4, 6). Since AC5050, like AC5070, overproduces enzymes and harpinEcc, we wanted to determine if this mutant also is OHL independent. For this, we made an Ohl− and RsmC− double mutant, AC5051, by replacing ohlI+ of AC5050 with ohlI::omega (Spr) of pAKC855 (Table 1). The results (Fig. 1A) show that AC5051 produced less Pel, Peh, Cel, and Prt than AC5050, although these levels were somewhat higher than the levels in the RsmC+ Ohl+ strain (Fig. 1A, column 1) and considerably higher than the levels in its RsmC+ Ohl− derivative (Fig. 1A, column 2; also see reference 4). We noted a similar effect of OHL deficiency on the levels of pel-1, peh-1, celV, and hrpNEcc transcripts in RsmC+ and RsmC− bacteria (Fig. 1B). These observations demonstrate that in the RsmC− mutant, the requirement for OHL is partially relieved.
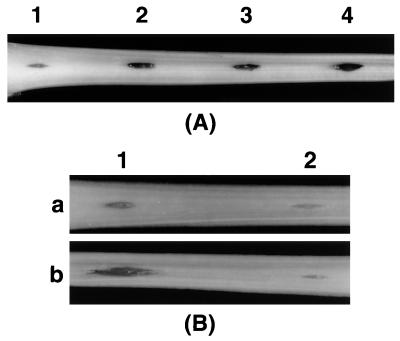
Pathogenicity assays.
Figure 4A shows that AC5053 caused more extensive maceration of celery petioles than Ecc71. This observation was expected since the mutant produced higher levels of extracellular enzymes and harpinEcc than the RsmA+ RsmC+ parent. Also, AC5053 carrying multiple copies of rsmC caused significantly less maceration than the same strains carrying the vector (Fig. 4B), most likely due to the inhibition of extracellular protein production (see above).
FIG. 4.
(A) Maceration of a celery petiole by E. carotovora subsp. carotovora Ecc71 (RsmA+ RsmC+; site 1), AC5053 (RsmA+ RsmC−; site 2), AC5071 (RsmA− RsmC+; site 3), and AC5054 (RsmA− RsmC−; site 4). (B) Maceration of celery petioles by E. carotovora subsp. carotovora AC5047 (RsmA+ RsmC+; a) and AC5050 (RsmA+ RsmC−; b) carrying the cloning vector pCL1920 (site 1) or the RsmC+ plasmid pAKC975 (site 2). About 2 × 108 cells were injected into the celery petiole at each inoculation site and covered with petroleum jelly. The inoculated petioles were incubated in a moist chamber at 25°C for 24 h.
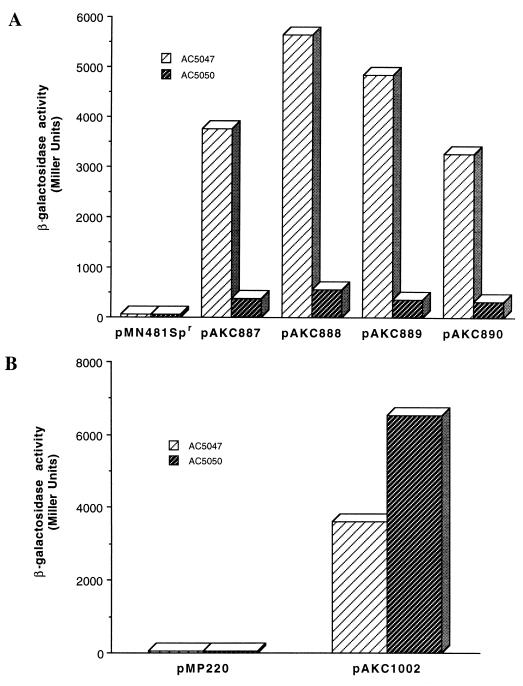
rsmC positively controls RsmA production.
The similarities in phenotypes of RsmA− and RsmC− mutants strongly suggested that the RsmC effect could partly manifest itself by modulating RsmA levels. To confirm this, we compared the levels of rsmA transcripts, the expression of a rsmA-lacZ gene fusion, and the levels of RsmA protein in RsmC+ and RsmC− E. carotovora subsp. carotovora strains. Unlike pel-1, peh-1, celV, and hrpNEcc transcripts (see above), the levels of rsmA transcript produced by AC5053 were lower than the levels produced by the parent strain Ecc71 (Fig. 5A). The data in Fig. 6A show that the level of β-galactosidase produced by the RsmC+ strain AC5047 carrying the fusion plasmid pAKC887 was 10- to 12-fold higher than the level produced by the RsmC− strain AC5050 carrying the fusion plasmid. Similarly, the results of Western blot analysis (Fig. 5B) also show that the level of RsmA polypeptide is higher in the RsmC+ strain than in the RsmC− mutant. Moreover, the data shown in Fig. 5C revealed that Ecc71 and AC5053 carrying multiple copies of rsmC produced higher levels of rsmA transcript than these bacteria carrying the cloning vector, pCL1920. These observations establish that RsmC has a positive effect on RsmA production and that the RsmC effect on exoprotein production may be attributed, at least in part, to this regulatory effect. To strengthen the latter conclusion, we constructed an RsmA− RsmC− double mutant and assayed for the levels of exoproteins and transcripts (Table 2; Fig. 1D). The levels of Pel, Peh, Cel, and Prt were even higher in the double mutant than in the RsmA− RsmC+ parent. Similarly, the levels of pel-1, peh-1, and celV transcripts were higher in the double mutant (Fig. 1D, column 4) than in the RsmA− RsmC+ parent (Fig. 1D, column 3). These findings raised the possibility that RsmC acted on a regulatory component in addition to RsmA. We should note that the levels of hrpNEcc transcripts (Fig. 1D) and harpinEcc protein (Fig. 1C) were comparable in RsmA− RsmC+ and RsmA− RsmC− mutants. These results suggested that the RsmC effect on hrpNEcc expression occurs primarily by its effect on RsmA, whereas the effect of RsmC on exoenzyme production is mediated via multiple regulators.
FIG. 5.
(A) Northern analysis of rsmA transcripts produced by E. carotovora subsp. carotovora Ecc71 (lane 1) and its RsmC− derivative, AC5053 (lane 2). Total RNAs were extracted from bacteria grown in minimal salts medium plus sucrose at 28°C to an A600 of 2.0. Each lane contained 10 μg of total RNA. (B) Western blot analysis of RsmA produced by E. carotovora subsp. carotovora Ecc71 (RsmC+; lane 1) and AC5053 (RsmC−; lane 2). Each lane contained 20 μg of total bacterial protein. (C) Levels of rsmA transcripts in E. carotovora subsp. carotovora Ecc71 (RsmC+) carrying the cloning vector pCL1920 (lane 1) or the rsmC+ plasmid pAKC975 (lane 2) and AC5053 (RsmC−) carrying the cloning vector pCL1920 (lane 3) or the rsmC+ plasmid pAKC975 (lane 4). Total RNAs were extracted from bacteria grown in minimal salts medium plus sucrose and spectinomycin at 28°C to an A600 of 2.0 and subjected to Northern analysis. Each lane contained 10 μg of total RNA.
FIG. 6.
(A) β-Galactosidase assays of E. carotovora subsp. carotovora AC5047 or its RsmC− mutant AC5050 carrying the cloning vector pNM481Spr, pAKC887 (rsmAEcc-lacZ fusion), pAKC888 (rsmAEa-lacZ fusion), pAKC889 (rsmAEhg-lacZ fusion), and pAKC890 (csrA-lacZ fusion). Bacteria were grown at 28°C in minimal salts medium plus sucrose and spectinomycin to an A600 of 2.0 and assayed for β-galactosidase activity. (B) β-Galactosidase assays of E. carotovora subsp. carotovora AC5047 or its RsmC− mutant AC5050 carrying the cloning vector pMP220 or the rsmB-lacZ plasmid pAKC1002. Bacteria were grown at 28°C in minimal salts medium plus sucrose and tetracycline to an A600 of 2.0 and assayed for β-galactosidase activity.
rsmA or rsmA-like genes have been cloned from several enterobacterial species, including E. amylovora (designated rsmAEa [25]), E. herbicola pv. gypsophilae (designated rsmAEhg [25]), and E. coli (designated csrA [35]). To test the effects of RsmC of E. carotovora subsp. carotovora Ecc71 on the expression of these heterologous rsmA (csrA) species, we made lacZ gene fusions of rsmAEa, rsmAEhg, and csrA. Plasmids carrying these fusions were transferred to RsmC+ strain AC5047 and RsmC− strain AC5050. These strains carrying the rsmAEcc-lacZ gene fusion served as the controls. The data shown in Fig. 6A clearly indicate that with each fusion, higher levels of β-galactosidase were produced in the RsmC+ strain than in the RsmC− strain. Although the degree of stimulation was variable depending on the source of the rsmA gene, these results demonstrate that RsmC stimulates the expression of rsmA homologs in E. carotovora subsp. carotovora.
Effect of RsmC on rsmB expression.
We have shown that rsmB specifies a regulatory RNA which positively controls exoprotein production and various secondary metabolites (19). The similarities in phenotypes due to RsmC deficiency (see above) and the dosage of rsmB RNA (19) suggested that the RsmC effect could be modulated via rsmB RNA. To test this idea, we examined the levels of rsmB transcripts in RsmC− and RsmC+ strains as well as in the RsmC− strain carrying multiple copies of rsmC+ DNA. The data in Fig. 1D and 2B show that (i) rsmB transcript levels are lower in the RsmC+ parent than in the RsmC− mutant and (ii) multiple copies of rsmC+ DNA inhibit the production of rsmB transcripts. The expression of a rsmB-lacZ transcriptional fusion in RsmC+ and RsmC− bacteria (Fig. 6B) also demonstrates a negative effect of RsmC on rsmB transcription.
We should note that the level of rsmB RNA in the RsmA− RsmC+ strain was about 10% of the level found in its RsmA+ RsmC+ parent strain (Fig. 1D). We have determined that this reduced level of rsmB RNA is primarily due to shorter half-life of the transcripts in RsmA− mutants than in the RsmA+ strains (25). Thus, it appears that RsmA contributes to the stability of rsmB RNA, perhaps by forming a ribonucleoprotein complex. This effect notwithstanding, it is evident that as in RsmA+ bacteria, the deficiency of RsmC in the RsmA− mutant stimulated the production of rsmB RNA.
We next examined the effects of the dosage of rsmB+ DNA in E. carotovora subsp. carotovora Ecc71 (RsmA+ RsmC+ RsmB+), AC5053 (RsmA+ RsmC− RsmB+), AC5071 (RsmA− RsmC+ RsmB+), and AC5054 (RsmA− RsmC− RsmB+). The data in Table 3 show that Pel levels were 20-fold higher in Ecc71 and two- to threefold higher in AC5053 and AC5054 carrying the rsmB+ plasmid than in bacteria carrying the cloning vector. Similar effects were noted with Peh, Cel, and Prt activities (data not shown) and the transcripts of pel-1, peh-1, and celV genes (Fig. 2C). A notable exception was the strain AC5054, which is deficient in both RsmA and RsmC, in that the enzyme and transcript levels were comparable in bacteria carrying a single copy of rsmB or multiple copies of rsmB (Table 3; Fig. 2C, columns 7 and 8).
TABLE 3.
Levels of Pel produced by E. carotovora subsp. carotovora strains carrying multiple copies of rsmB+ DNA
| Bacterial constructa | Relevant phenotypeb | Pel sp act (U/ml/A600 unit; mean ± SD) |
|---|---|---|
| Ecc71(pCL1920Gmr) | RsmA+ RsmB+ RsmC+ (none [vector]) | 0.2 ± 0.00 |
| Ecc71(pAKC1004Gmr) | RsmA+ RsmB+ RsmC+ (RsmB+) | 4.2 ± 0.08 |
| AC5053(pCL1920Gmr) | RsmA+ RsmB+ RsmC− (none [vector]) | 4.6 ± 0.04 |
| AC5053(pAKC1004Gmr) | RsmA+ RsmB+ RsmC− (RsmB+) | 10.6 ± 0.58 |
| AC5071(pCL1920Gmr) | RsmA− RsmB+ RsmC+ (none [vector]) | 7.8 ± 0.14 |
| AC5071(pAKC1004Gmr) | RsmA− RsmB+ RsmC+ (RsmB+) | 19.1 ± 0.08 |
| AC5054(pCL1920Gmr) | RsmA− RsmB+ RsmC− (none [vector]) | 22.9 ± 0.36 |
| AC5054(pAKC1004Gmr) | RsmA− RsmB+ RsmC− (RsmB+) | 21.3 ± 0.03 |
Bacteria were grown at 28°C in minimal salts medium plus sucrose (0.5% [wt/vol]) and gentamicin to an A600 value of 2.5, and culture supernatants were used for enzyme assays.
Relevant phenotypes conferred by the plasmids are given in parentheses.
rsmC homologs occur in E. carotovora subspecies.
Southern hybridization analysis using the rsmC DNA as the probe revealed that rsmC homologs exist in strains of E. carotovora subspecies carotovora, atroseptica, and betavasculorum (Fig. 7) but not in the other Erwinia or enterobacterial species (data not shown). In addition, using synthetic oligonucleotide primers specific to the internal sequences of rsmC, we conducted PCR analysis of DNA preparations of strains of E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, E. carotovora subsp. betavasculorum, E. amylovora, E. rhapontici, E. stewartii, E. chrysanthemi, and E. herbicola, as well as E. coli, Salmonella, Serratia, Yersinia, and Shigella strains. The PCR-amplified DNA segments similar in size to the expected product from Ecc71 were detected only with E. carotovora subspecies and not with the other bacteria. These data suggest that rsmC sequences have been conserved in the E. carotovora group of the soft-rotting Erwinia species.
FIG. 7.
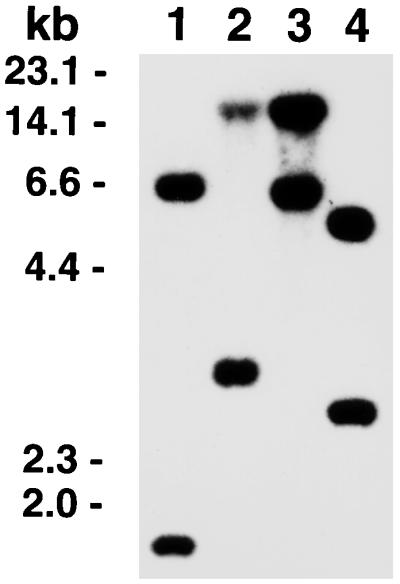
Southern blot analysis of EcoRI-digested chromosomal DNAs of E. carotovora subspecies with rsmC of E. carotovora subsp. carotovora Ecc71. Lane 1, E. carotovora subsp. atroseptica Eca12; lane 2, E. carotovora subsp. betavasculorum Ecb11129; lanes 3 and 4, E. carotovora subsp. carotovora strains Ecc71 and SCRI193.
DISCUSSION
The data presented here show that RsmC positively regulates the expression of rsmA, which encodes an RNA-binding protein (6) and negatively regulates rsmB, which specifies a regulatory RNA (19). For example, the RsmC− mutant produces low levels of rsmA transcripts and RsmA protein, but when provided with the rsmC+ DNA in trans it produces high levels of these transcripts and RsmA. The levels of rsmB transcripts, on the other hand, are higher in RsmC− bacteria than in RsmC+ strains. These observations were also confirmed by following the expression of rsmA-lacZ and rsmB-lacZ fusions. We do not yet know how RsmC interacts with the RNA polymerase holoenzyme to regulate gene expression. The absence of a DNA-binding motif in RsmC suggests that it probably does not function as a classical transcriptional factor by directly interacting with rsmA and rsmB sequences. It is perhaps significant that a stretch of RsmC has sequence homology with eukaryotic transcriptional adapters (9). Such adapters have been found to activate transcription by interacting with components of transcriptional machinery, i.e., via protein-protein interactions. By extrapolating from those observations, we postulate that RsmC interacts with RNA polymerase holoenzyme and this ternary complex modulates transcription. At this juncture we have to entertain the possibility that RsmC acts directly or indirectly both as a positive regulator of rsmA and a negative regulator of rsmB transcription. We have initiated in vitro transcription studies and mutational analysis of E. carotovora subsp. carotovora to identify the targets of RsmC and to elucidate its mode of action.
In a series of studies (4, 6, 19, 26), we have documented that RsmA and rsmB RNA control the production of exoproteins, polysaccharides, and an assortment of secondary metabolites as well as various virulence factors in Erwinia species. Those observations and the data presented here demonstrate that there is a correlation between expression of rsmA and rsmB and levels of exoenzymes, harpinEcc, and plant virulence. Indeed, the following findings establish that the pleiotropic effect of RsmC deficiency is mainly if not solely directed via the RsmA-rsmB regulatory pathway, i.e., due to the reduced levels of RsmA and high levels of rsmB RNA. (i) rsmC+ DNA stimulates RsmA production and concomitantly causes a severe repression of exoprotein production. (ii) The levels of exoenzymes are higher in the RsmA− RsmC− double mutant than in the RsmA− RsmC+ strain, indicating that a regulatory factor in addition to RsmA is also affected in the RsmC− mutant. (iii) The pleiotropic phenotypes of the RsmC− mutant and the RsmA− RsmC− double mutant are reminiscent of the effects of multiple copies of rsmB DNA (19). (iv) rsmB expression is much higher in the RsmC− mutant than in the RsmC+ parent. Furthermore, multiple copies of rsmC+ DNA lower the levels of rsmB RNA. (v) Multiple copies of rsmB+ DNA further stimulate the levels of exoproteins in RsmA− bacteria but not in the RsmA− RsmC− double mutant. (vi) The levels of rsmA and rsmB RNA, but not the transcripts of other known global regulator genes, such as kdgR (20), hexA (13), rpoS (27), hor (42), and ohlI (4), are affected in the RsmC− mutant (25).
Since RsmC positively regulates rsmA expression, it was expected that the phenotypes of RsmC− mutants would closely resemble those of the RsmA− strains. That this indeed is the case is evident from exoenzyme overproduction, hypervirulence, and overexpression of hrpNEcc. It was, therefore, surprising that the RsmC− mutant at least partially requires the quorum sensing signal, OHL, for exoenzyme overproduction (Fig. 1). This contrasts with the finding (4) that RsmA− mutants can overproduce exoenzymes in the absence of OHL. Since we do not yet know how OHL activates the expression of exoenzyme genes in E. carotovora subsp. carotovora, these findings with the RsmC− and RsmA− mutants are difficult to explain. We should, however, note that there are reports describing LuxR homologs that in conjunction with OHL could affect gene expression in soft-rotting Erwinia. For example, CarR (a regulator of carbapenem antibiotic production) together with OHL activates carbapenem biosynthetic genes, carA to carH, in E. carotovora subsp. carotovora GS101, but CarR does not affect exoenzyme production (21). Another luxR homolog, expR, has been detected in E. carotovora subsp. carotovora SCC3193 (32). However, ExpR− mutant does not have any effect on OHL or exoenzyme production. Moreover, a genetic homolog of expR has not been found in E. carotovora subsp. carotovora Ecc71, although OHL plays a key role in regulating exoenzymes and harpinEcc in this bacterium (4, 6, 25). Nasser et al. (30) reported that ExpR of E. chrysanthemi did bind sequences of a pel gene of E. carotovora subsp. carotovora SCRI193 and several pel genes of E. chrysanthemi 3193. The physiological significance of this physical interaction is not apparent since the ExpR− knockout mutants of E. chrysanthemi, like the ExpR− mutants of E. carotovora subsp. carotovora, were not affected in OHL and exoenzyme production. These uncertainties notwithstanding, the data presented here and in our previous publications (4, 19, 24) support the idea that the regulatory effects of RsmC and OHL are directed via common as well as nonoverlapping steps. As a prelude to understanding the basis for OHL independence in E. carotovora subsp. carotovora mutants, we have begun testing various strategies that would allow the identification of a regulator(s) which is responsible for the OHL effect and is affected by RsmA and RsmC.
Several lines of evidence have established that rsmA-like genes occur and are expressed in many enterobacteria (6, 26). In this report we have documented that RsmC is a positive regulator not only of Ecc71 rsmA but also of rsmA genes non-soft-rotting bacteria such as E. amylovora, E. herbicola pv. gypsophilae, and E. coli. For example, (i) the levels of Ecc71 rsmA transcript are higher in RsmC+ than in RsmC− bacteria and (ii) the expression of lacZ driven by promoters of rsmA genes of E. amylovora, E. herbicola pv. gypsophilae, E. coli, and E. carotovora subsp. carotovora is consistently higher in RsmC+ than in RsmC− E. carotovora subsp. carotovora strains. However, we were surprised to find that genetic homologs of rsmC are present in E. carotovora subsp. atroseptica, E. carotovora subsp. betavasculorum, and E. carotovora subsp. carotovora but not in other bacteria, including non-soft-rotting Erwinia species and E. coli. We have initiated a search for functional homologs of RsmC of E. coli and non-soft-rotting Erwinia species by testing for heterologous complementation.
ACKNOWLEDGMENTS
Our work was supported by the National Science Foundation (grant MCB-9728505) and the Food for the 21st Century program of the University of Missouri.
We thank David W. Emerich and Judy D. Wall for reviewing the manuscript.
Footnotes
Journal series 12,906 of the Missouri Agricultural Experiment Station.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Barras F, van Gijsegem F, Chatterjee A K. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- 3.Bauer D W, Wei Z-M, Beer S V, Collmer A. Erwinia chrysanthemi harpinEch: an elicitor of the hypersensitive response that contributes to soft-rot pathogenesis. Mol Plant-Microbe Interact. 1995;8:484–491. doi: 10.1094/mpmi-8-0484. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee A, Cui Y, Liu Y, Dumenyo C K, Chatterjee A K. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl Environ Microbiol. 1995;61:1959–1967. doi: 10.1128/aem.61.5.1959-1967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee A K, Vidaver A K. Genetics of pathogenicity factors: application to phytopathogenic bacteria. In: Ingram D S, Williams P H, editors. Advances in plant pathology. Vol. 4. London, England: Academic Press; 1986. pp. 1–224. [Google Scholar]
- 6.Cui Y, Chatterjee A, Liu Y, Dumenyo C K, Chatterjee A K. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J Bacteriol. 1995;177:5108–5115. doi: 10.1128/jb.177.17.5108-5115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui Y, Madi L, Mukherjee A, Dumenyo C K, Chatterjee A K. The RsmA− mutants of Erwinia carotovora subsp. carotovora strain Ecc71 overexpress hrpNEcc and elicit a hypersensitive reaction-like response in tobacco leaves. Mol Plant-Microbe Interact. 1996;9:565–573. doi: 10.1094/mpmi-9-0565. [DOI] [PubMed] [Google Scholar]
- 8.Dennis J J, Zylstra G J. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl Environ Microbiol. 1998;64:2710–2715. doi: 10.1128/aem.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edmondson D G, Roth S Y. Chromatin and transcription. FASEB J. 1996;10:1173–1182. doi: 10.1096/fasebj.10.10.8751719. [DOI] [PubMed] [Google Scholar]
- 10.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 depend on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua C, Greenberg E P. Self perception in bacteria—quorum sensing with acylated homoserine lactones. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 13.Harris S J, Shih Y-L, Bentley S D, Salmond G P C. The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence determinants. Mol Microbiol. 1998;28:705–717. doi: 10.1046/j.1365-2958.1998.00825.x. [DOI] [PubMed] [Google Scholar]
- 14.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 15.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 16.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lichter A, Barash I, Valinsky L, Manulis S. The genes involved in cytokinin biosynthesis in Erwinia herbicola pv. gypsophilae: characterization and role in gall formation. J Bacteriol. 1995;177:4457–4465. doi: 10.1128/jb.177.15.4457-4465.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Chatterjee A, Chatterjee A K. Nucleotide sequence and expression of a novel pectate lyase gene (pel-3) and a closely linked endopolygalacturonase gene (peh-1) of Erwinia carotovora subsp. carotovora 71. Appl Environ Microbiol. 1994;60:2545–2552. doi: 10.1128/aem.60.7.2545-2552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Cui Y, Mukherjee A, Chatterjee A K. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol Microbiol. 1998;29:219–234. doi: 10.1046/j.1365-2958.1998.00924.x. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Jiang G-Q, Cui Y, Mukherjee A, Ma W-L, Chatterjee A K. kdgREcc negatively regulates genes for pectinases, cellulase, protease, harpinEcc, and a global RNA regulator in Erwinia carotovora subsp. carotovora. J Bacteriol. 1999;181:2411–2422. doi: 10.1128/jb.181.8.2411-2421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGowan S J, Sebaihia M, Porter L E, Stewart G S A B, Williams P, Bycroft B W, Salmond G P C. Analysis of bacterial carbapenem antibiotic production genes reveals a novel β-lactam biosynthesis pathway. Mol Microbiol. 1996;22:415–426. doi: 10.1046/j.1365-2958.1996.00125.x. [DOI] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 23.Minton N P. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene. 1984;31:269–273. doi: 10.1016/0378-1119(84)90220-8. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee A, Cui Y, Liu Y, Chatterjee A K. Molecular characterization and expression of the Erwinia carotovora hrpNEcc gene, which encodes an elicitor of the hypersensitive reaction. Mol Plant-Microbe Interact. 1997;10:462–471. doi: 10.1094/MPMI.1997.10.4.462. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee, A., Y. Cui, Y. Liu, and A. K. Chatterjee. Unpublished data.
- 26.Mukherjee A, Cui Y, Liu Y, Dumenyo C K, Chatterjee A K. Global regulation in Erwinia species by Erwinia carotovora rsmA, a homologue of Escherichia coli csrA: repression of secondary metabolites, pathogenicity and hypersensitive reaction. Microbiology. 1996;142:427–434. doi: 10.1099/13500872-142-2-427. [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee A, Cui Y, Ma W-L, Liu Y, Ishihama A, Eisenstark A, Chatterjee A K. RpoS (sigma-S) controls expression of rsmA, a global regulator of secondary metabolites, harpin, and extracellular proteins in Erwinia carotovora. J Bacteriol. 1998;180:3629–3634. doi: 10.1128/jb.180.14.3629-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murata H, Chatterjee A, Liu Y, Chatterjee A K. Regulation of the production of extracellular pectinase, cellulase, and protease in the soft rot bacterium Erwinia carotovora subsp. carotovora: evidence that aepH of E. carotovora subsp. carotovora 71 activates gene expression in E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, and Escherichia coli. Appl Environ Microbiol. 1994;60:3150–3159. doi: 10.1128/aem.60.9.3150-3159.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murata H, McEvoy J L, Chatterjee A, Collmer A, Chatterjee A K. Molecular cloning of an aepA gene that activates production of extracellular pectolytic, cellulolytic, and proteolytic enzymes in Erwinia carotovora subsp. carotovora. Mol Plant-Microbe Interact. 1991;4:239–246. [Google Scholar]
- 30.Nasser W, Bouillant M L, Salmond G, Reverchon S. Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol Microbiol. 1998;29:1391–1405. doi: 10.1046/j.1365-2958.1998.01022.x. [DOI] [PubMed] [Google Scholar]
- 31.Pierson III L S, Wood D W, Pierson E A. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu Rev Phytophathol. 1998;36:207–225. doi: 10.1146/annurev.phyto.36.1.207. [DOI] [PubMed] [Google Scholar]
- 32.Pirhonen M, Flego D, Heikinheimo R, Palva E T. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 34.Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- 35.Romeo T, Gong M, Liu M-Y, Brun-Zinkernagel A-M. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol. 1993;175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. The bacterial ‘enigma’: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 37.Salmond G P C, Hinton J C D, Gill D R, Perombelon M C M. Transposon mutagenesis of Erwinia using phage λ vectors. Mol Gen Genet. 1986;203:524–528. [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Siegele D A, Kolter R. Life after log. J Bacteriol. 1992;174:345–348. doi: 10.1128/jb.174.2.345-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 41.Swift S, Throup J P, Williams P, Salmond G P C, Stewart G S A B. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem Sci. 1996;21:214–219. [PubMed] [Google Scholar]
- 42.Thomson N R, Cox A, Bycroft B W, Stewart G S A B, Williams P, Salmond G P C. The Rap and Hor proteins of Erwinia, Serratia and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol Microbiol. 1997;26:531–544. doi: 10.1046/j.1365-2958.1997.5981976.x. [DOI] [PubMed] [Google Scholar]
- 43.Vanneste J L. Erwinia amylovora. In: Singh U S, Singh R P, Kohmoto K, editors. Pathogenesis and host specificity in plant diseases: histopathological, biochemical, genetic and molecular bases. 1. Prokaryotes. Oxford, England: Pergamon Press; 1995. pp. 21–41. [Google Scholar]
- 44.Zambrano M M, Kolter R. Gasping for life in stationary phase. Cell. 1996;86:181–184. doi: 10.1016/s0092-8674(00)80089-6. [DOI] [PubMed] [Google Scholar]
- 45.Zink R T, Kemble R J, Chatterjee A K. Transposon Tn5 mutagenesis in Erwinia carotovora subsp. carotovora and Erwinia carotovora subsp. atroseptica. J Bacteriol. 1984;157:809–814. doi: 10.1128/jb.157.3.809-814.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]