Abstract
The Bacillus subtilis ripX gene encodes a protein that has 37 and 44% identity with the XerC and XerD site-specific recombinases of Escherichia coli. XerC and XerD are hypothesized to act in concert at the dif site to resolve dimeric chromosomes formed by recombination during replication. Cultures of ripX mutants contained a subpopulation of unequal-size cells held together in long chains. The chains included anucleate cells and cells with aberrantly dense or diffuse nucleoids, indicating a chromosome partitioning failure. This result is consistent with RipX having a role in the resolution of chromosome dimers in B. subtilis. Spores contain a single uninitiated chromosome, and analysis of germinated, outgrowing spores showed that the placement of FtsZ rings and septa is affected in ripX strains by the first division after the initiation of germination. The introduction of a recA mutation into ripX strains resulted in only slight modifications of the ripX phenotype, suggesting that chromosome dimers can form in a RecA-independent manner in B. subtilis. In addition to RipX, the CodV protein of B. subtilis shows extensive similarity to XerC and XerD. The RipX and CodV proteins were shown to bind in vitro to DNA containing the E. coli dif site. Together they functioned efficiently in vitro to catalyze site-specific cleavage of an artificial Holliday junction containing a dif site. Inactivation of codV alone did not cause a discernible change in phenotype, and it is speculated that RipX can substitute for CodV in vivo.
The reproduction of the circular chromosome during the cell cycles of both Escherichia coli and Bacillus subtilis begins with an initiation event at the origin of replication followed by bidirectional replication toward the terminus located approximately 180° away. Before chromosome replication is complete, recombination can occur between homologous regions of the replicated portion of the chromosome. Upon termination of replication, an odd number of crossovers would be expected to yield a dimeric chromosome that cannot be partitioned properly in the absence of a recombination-mediated resolution event. It has been proposed that in E. coli the XerC and XerD proteins act in concert at the dif site in order to resolve chromosome dimers. Thus, partitioning (Par) phenotypes are displayed by E. coli strains mutated at xerC, xerD, or dif (2, 3, 14). The absence of a Par phenotype when the mutations are combined with a recA mutation indicates that the formation of chromosome dimers requires RecA-mediated recombination during replication.
Homologues of XerC and XerD have been identified in a variety of species, and functional activity on reporter plasmids has been demonstrated in Pseudomonas aeruginosa, Salmonella typhimurium, and several Enterobacteriaceae (10, 11, 29). A function in chromosome dimer resolution is indicated by the ability of S. typhimurium xerC and xerD genes to complement E. coli xer mutations. In addition, in vitro recombination at dif has been demonstrated with XerC from Haemophilus influenzae acting with XerD from E. coli (20). Despite the many similarities between the various XerC and XerD homologues across species, such complementation is not guaranteed, as Proteus mirabilis xerD was unable to efficiently complement an E. coli xerD mutation (34).
B. subtilis is a gram-positive organism that has been widely studied and often juxtaposed with E. coli in comparative biology. The CodV and RipX proteins of B. subtilis have 35 and 44% identity with the XerC and XerD proteins, respectively, and 39% identity with each other. CodV and RipX also possess proper alignment of the six invariant catalytic residues found in all λ integrase family site-specific recombinases (27, 30). The strong amino acid similarity among the RipX and CodV proteins of B. subtilis and the XerC and XerD proteins of E. coli prompted us to investigate whether the RipX and CodV proteins have roles in resolving chromosome dimers and hence facilitate chromosome partitioning in B. subtilis.
Several details of the B. subtilis chromosome partitioning mechanism have recently been described. A critical event in the partitioning of circular chromosomes is the alleviation of catenates formed during replication. In B. subtilis, catenation nodes are removed from replicated chromosomes by the protein products of the parC and parE genes (12). ParC and ParE are subunits of topoisomerase IV. Conditional inactivation of either parC or parE results in a strong decrease in cell viability and yields a subpopulation of elongated cells containing large, asymmetrically located nucleoids (12). It is speculated that, as in E. coli, topoisomerase IV completely removes catenation nodes that may result from an even number of crossovers during replication. However, topoisomerases cannot effect the recombination events necessary to resolve chromosome dimers that result from an odd number of crossovers; RipX and CodV are hypothesized to have that role.
In this study, RipX is shown to be involved in chromosome partitioning during both vegetative growth and sporulation of B. subtilis. To our knowledge, this is the first report where a Xer homologue is shown to be required for chromosomal partitioning in a species other than E. coli. In sharp contrast to the chromosome partition phenotypes seen in E. coli xer mutants, those seen in a ripX mutant are not dependent on a functional RecA protein. The companion role of CodV in chromosome partitioning is presently circumspect because of the absence of phenotypes in mutant strains. However, we demonstrate that both CodV and RipX together are required for efficient recombination of dif substrates in vitro.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The parental strain used in this study was B. subtilis BR151 (trpC2 lys3 metB10). The YB886 strain (trpC2 metB10 xin-1 SPβ) that was used to evaluate prophage contribution to the phenotypes in ripX strains was obtained from the Bacillus Genetic Stock Center (Columbus, Ohio). Table 1 provides a complete listing of all B. subtilis strains used. The E. coli strain used for cloning knockout vectors was DH5α F− endA1 hsdR17(rK−mK+) supE44 thi-1 λ− recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 φ80dlacZΔM15 (Bethesda Research Laboratories, Bethesda, Md.). The E. coli strain used to clone maltose-binding protein (MBP) fusions was DS9009, a recF xerC::cm xerD::km derivative of AB1157.
TABLE 1.
B. subtilis strainsa
| Strain | Genotype | Source or constructionb |
|---|---|---|
| BR151 | trpC2 lys-3 metB10 | Standard lab strain |
| SL4257 | trpC2 metC3 rif-2 dinC::lacZ | MB24 × YB886c dinC::Tn917-lacZ |
| SL7131 | trpC2 lys-3 metB10 ripX::spc | This study |
| SL7224 | trpC2 lys-3 metB10 ripX::spc codV::neo | This study |
| SL7355 | trpC2 lys-3 metB10 codV::neo | This study |
| SL7360 | trpC2 lys-3 metB10 recA::neo | This study |
| SL7370 | trpC2 lys-3 metB10 ripX::spc recA::neo | SL7131 × SL7360 |
| SL7325 | trpC2 lys-3 metB10 ripX::spc dinC::lacZ | SL7131 × SL4257 |
| SL7326 | trpC2 lys-3 metB10 dinC::lacZ | BR151 × SL4257 |
| SL7375 | trpC2 lys-3 metB10 spoIIIE::spc | BR151 × PL412d |
| SL7377 | trpC2 lys-3 metB10 ripX::cm spoIIIE::spc | SL7131 × PL412d |
| SL7413 | trpC2 lys-3 metB10 ripX::cm spoIIIE::spc recA::neo | SL7377 × SL7360 |
All strains are isogenic with BR151 with the exception of SL4257.
The first strains listed in crosses are the recipient strains during transformation.
YB886 is a derivative of BR151 (37).
PL412 chromosomal DNA was kindly provided by Petra Levin.
Genetic manipulations.
B. subtilis transformations were performed as described previously (24) with selection on Luria-Bertani (LB) plates containing chloramphenicol (5 μg ml−1), neomycin (NEO) (12 μg ml−1), and spectinomycin (75 μg ml−1) where appropriate. Knockout mutations were constructed by ligating antibiotic resistance cassettes within flanking chromosomal DNA. Plasmids pSAS7-4, pSAS8-20, and pSAS12-4 are pBluescript (Stratagene) derivatives containing internal ripX and recA (pSAS12-4) gene fragments interrupted by spectinomycin, chloramphenicol, and NEO antibiotic resistance cassettes, respectively. The spc and cat cassettes were inserted into the EcoRV site at nucleotide 392 of the ripX open reading frame, and neo was inserted at nucleotide 530 of the recA open reading frame. Plasmids pSAS7-4, pSAS8-20, and pSAS12-4 were used as donors in transformation in order to obtain chromosomal knockout mutations. All knockout mutations were confirmed by PCR. The codV mutant has a neo cassette inserted codirectionally with the transcription of codV to permit transcription of downstream genes in the cod operon via readthrough from the neo promoter. This type of mutant has been defined for the purposes of this report as nonpolar. Transformation efficiencies in the ripX mutant were diminished ≈10-fold relative to the parent strain. To generate MBP fusions, codV and ripX were amplified by PCR with chromosomal DNA from B. subtilis followed by cloning into pMAL-C2. The MBP fusions were expressed in DS9009 and purified with amylose resin per the manufacturer’s instructions.
Gel retardation and in vitro recombination assays.
Methods and oligonucleotides used were those of Arciszewska et al. (1) and Blakely et al. (3). Binding reactions were performed in 50 mM NaCl–20 mM Tris (pH 8)–1 mM EDTA–10% glycerol–100 μg of poly(dI-dC) at 37°C for 10 min before electrophoresis through 6% polyacrylamide at 4°C. Holliday junction resolution reactions used binding buffer, as above, but were incubated at 37°C for 30 min before electrophoresis through 4% polyacrylamide containing 0.1% sodium dodecyl sulfate.
β-Galactosidase activity assay.
Cell samples were assayed for β-galactosidase activity by using o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate, as detailed by Nicholson and Setlow (21). Specific activity is expressed as nanomoles of ONPG hydrolyzed per minute per milligram (dry weight) of bacteria.
Spore preparation and germination of purified spores.
Spores were initially purified as previously described (21) by using modified Schaeffer’s sporulation medium (MSSM) to induce sporulation (23). Spores were further purified by ultracentrifugation at 75,000 × g for 16 h through a Urografin (Sigma) density gradient. Phase-bright spore fractions were pooled and counted in a Petroff-Hausser counting chamber to determine concentrations. The final preparations were 99.99% phase-bright. Spores were heat activated at 70°C for 15 min prior to germination and outgrowth in nutrient broth containing 0.5% glucose (28). Cells were outgrown at 30°C as a convenient means to slow cell cycle progression. Vegetative cultures of the parent and ripX strains grown at 30°C had proportionately longer generation times and were otherwise unaffected with respect to the phenotypes discussed in this report.
Immunofluorescence staining.
Cell fixation and staining were performed essentially as described by Harry et al. (9) and modified by Khvorova et al. (13). FtsZ antibodies were kind gifts of J. Lutkenhaus and P. Levin. Fluorescence observations were made with a Zeiss Axioskop fluorescent microscope with standard Cy3 and DAPI filter sets. Images were photographed with a Sony DKC-5000 digital camera and acquired with Adobe Photoshop Software, version 4.0. Software processing of photographs was restricted to brightness and contrast adjustments only.
DAPI staining.
Chromosome staining was performed with 4′,6-diamidino-2-phenylindole (DAPI). Cells were fixed prior to staining in 0.37% formalin. Twenty microliters of fixed cell samples were adsorbed onto 0.1% (wt/vol) poly-l-lysine-treated coverslips for 5 min before placing the coverslip onto a 20-μl pool of DAPI at a concentration of 1 μg ml−1 for 30 min. The coverslip was then placed upon a slide containing a single drop of Slow-Fade and sealed.
Cell measurements.
Random fields of view were photographed for each strain (exponential-phase samples) and measured by using Scion Image software, Release Beta 2.
Sequence analysis.
Sequence analyses were performed with the BestFit program in the Wisconsin Package Software (version 9.0) of the Genetics Computer Group. Accession numbers were BG10965 (for codV), BG11332 (for ripX), EG11069 (for xerC), and EG11071 (for xerD).
RESULTS
Disruption of ripX alters cell and nucleoid morphology.
The B. subtilis ripX locus is located immediately upstream of the drm pnp operon and encodes a protein having 37 and 44% amino acid identity with the XerC and XerD recombinases of E. coli, respectively. ripX is transcribed as a monocistronic message (25). In order to evaluate the physiological role of RipX, a ripX::spc knockout mutant was constructed in strain BR151. An initial indication of the importance of ripX for cellular growth was that mutant colonies on LB-spectinomycin plates had an altered colony morphology, compared to a BR151 derivative containing spc inserted in the thrC locus.
Phase-contrast microscopy examination of the ripX::spc strain grown in LB broth and in MSSM revealed that a portion of the population is present as chains of elongated cells connected with other cells of varying length. This heterogeneity in cell length within chains was seen in both exponential- and stationary-phase cultures (Fig. 1c and d). Concomitantly, a 10 to 20% increase (in different experiments) was observed in the generation time of the ripX mutant. Measurements of cell length for the parent strain and for the ripX mutant broadly describe the extent of cell length variability in the mutant population (Fig. 2). Introduction of the ripX::spc allele into a strain carrying a second intact copy of ripX inserted at the amyE locus did not result in a RipX phenotype. A nonpolar codV knockout mutant could not be distinguished from the parent strain, BR151. Similarly, a ripX codV double mutant was indistinguishable from the ripX strain (data not shown).
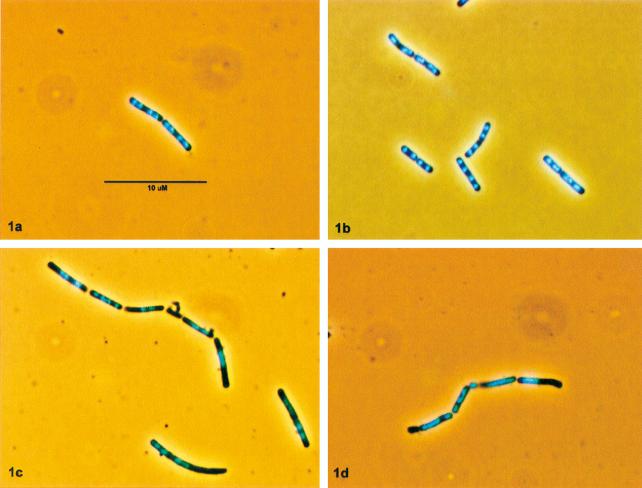
FIG. 1.
Micrographs of DAPI-stained cells of the parent strain (BR151) (a and b) and the ripX mutant (SL7131) (c and d) taken from exponential-phase (a and c) and stationary-phase (b and d) growth cultures. The scale bar in A applies to all images.
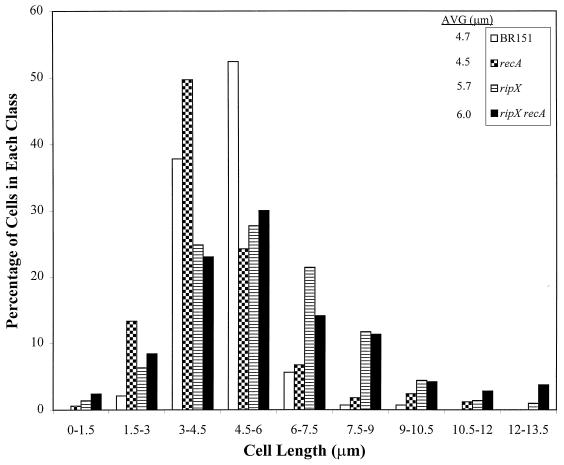
FIG. 2.
Frequency distribution of cell lengths of strains growing exponentially in LB medium. Open bar, BR151 (parent strain); checkered bar, SL7360 (recA::neo); horizontal lines in bar, SL7131 (ripX::spc); filled bar, SL7370 (recA::neo ripX::spc). At least 150 cells were measured per strain.
Nucleoids in mutated cells were visualized by using DAPI staining in conjunction with fluorescence microscopy. Nucleoid staining revealed a range of phenotypes indicative of partition failure. First, in larger cells within chains, a single, dense or diffuse nucleoid (Fig. 1c and d) was often observed. This nucleoid phenotype is very similar to those seen in E. coli xerC or xerD mutants and in B. subtilis parC or parE mutants (2, 12). A second indication of partitioning difficulties in ripX cells is the presence of small anucleate cells adjoining larger cell units containing a chromosome located close to the interfacing septum. A comparable situation has been described for E. coli parC (19, 33) and ftsK (16, 38) mutants. In addition, the ripX mutant cultures contained rare nucleated minicells and examples of what appeared to be nucleoids guillotined by a cell division. It has been reported that a small portion of cells in B. subtilis strains mutated at the spoIIIE locus experience difficulty completely clearing their DNA from the advance of the division septum when nucleoid partitioning or septum positioning is disturbed (26). However, the introduction of spoIIIE knockout and point mutations did not appear to appreciably alter the cell or nucleoid profiles of ripX mutants (data not shown).
ripX cells sporulate at a reduced frequency.
The indications that RipX may be involved in the resolution of dimeric chromosomes in B. subtilis led us to investigate the effect of ripX mutations on sporulation. Sporulation provides a separate opportunity to appraise chromosome partitioning failures apart from vegetative growth. After the start of sporulation, completely replicated chromosomes must be partitioned, with one chromosome occupying the larger mother cell compartment while the other advances into the developing spore compartment (22). Cultures of the parent strain and the ripX mutant were grown in MSSM in order to induce sporulation. Twenty hours after the cessation of exponential growth (t20), each culture was assessed for sporulation frequency by a heat-killing assay and by phase-contrast microscopy. The sporulation frequency varied little between the two scoring methods or as a function of time assayed (t19 to t31 [data not shown]). Our results show that the sporulation frequency of ripX cells is reduced to approximately half that of the parent strain (Table 2).
TABLE 2.
Sporulation frequency of parent and mutant strains
| Strain | % Phase-brighta | Relative sporulation frequency |
|---|---|---|
| BR151 (parent) | 80 | 1.0 |
| ripX | 45 | 0.56 |
| recA | 46 | 0.58 |
| ripX recA | 29 | 0.36 |
A minimum of 300 phase-bright spores and vegetative cells were counted in several fields of view at t20 by phase-contrast microscopy.
Analysis of ripX-associated events in outgrowing spores.
The analysis of cell division events in vegetatively growing and sporulating cultures is complicated by the interaction of several cell cycles within a single bacterium. Also, batch cultures are nonsynchronous, making progressive evaluations of cell populations problematic. To minimize these difficulties, outgrowing spore populations were employed in our assessment of cell division in ripX mutants. Spores of B. subtilis contain a single, completely replicated chromosome (6). The spores can be induced to germinate and return to vegetative growth (outgrow) by the addition of nutrients. Thus, outgrowing spores progress through the first cell cycle free from the influence of multiple ongoing rounds of chromosome replication. Also, highly purified spore populations can be germinated with a reasonable degree of synchrony, thereby providing a system for examining cell division processes in a relatively homogeneous population of cells with respect to the cell cycle (28, 35). Therefore, the use of outgrowing spores provides potential advantages over studies involving batch cultures of vegetatively growing bacteria.
Outgrowing spore populations were used to study chromosome partitioning during the first round of replication and cell division. DAPI-stained nucleoids of samples taken from outgrowing cultures were scored as being either bilobed (i.e., nucleoids have begun segregating but remain connected) or partitioned (i.e., there is identifiable space between nucleoids). We noted from these data an inverse relationship between the parent and ripX strains. Specifically, the ripX strain was more frequently found and spent a longer time in the bilobed state than the parent strain (Fig. 3). Conversely, the ripX strain was less frequently found in the partitioned state than the parent strain. This inverse relationship in nucleoid phenotype as cells outgrow is consistent with a role for RipX in the proper partitioning of chromosomes. It is suggested that the bilobed cells in the ripX mutant were of two types: those undergoing normal segregation as in the parent strain and those with chromosome dimers whose resolution is impaired because of the mutation. The bilobed class of nucleoid ultimately disappeared from the ripX mutant, although more slowly than for the parent strain, suggesting that there is some RipX-independent mechanism for slowly resolving chromosome dimers. Alternatively, it is also possible that there is some gradual lysis of bacteria with aberrant nucleoids and/or that some dimerized chromosomes replicate and individual dimers are partitioned.
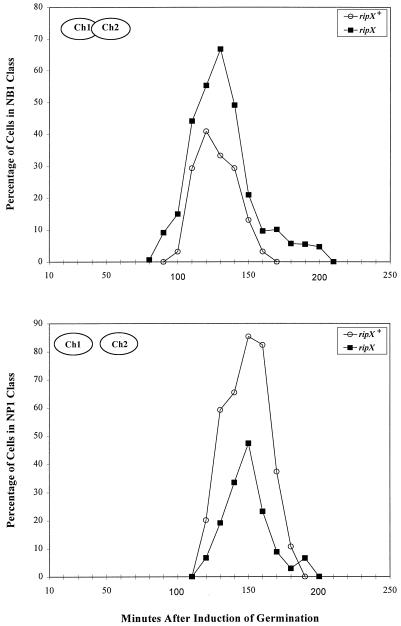
FIG. 3.
Change in nucleoid phenotype during outgrowth of germinated spores of the parent strain (BR151) (open circles) and the ripX mutant (SL7131) (filled squares). For each point, at least 200 cells were scored. The phenotypes scored are nucleoid-bilobed (NB1) and nucleoid-partitioned (NP1) (as described by Siccardi et al. [28]); they are illustrated schematically in the figure. Ch1, chromosome 1; Ch2, chromosome 2.
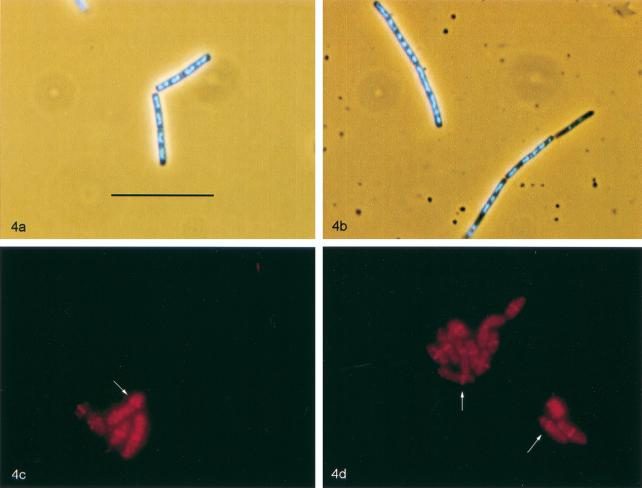
Outgrowing spores were also utilized in order to compare FtsZ ring (18) and septum formation in the parent and ripX strains. The touchstone for our scoring was that FtsZ rings or septa classified as asymmetric be no more than one-third of the way along the cell length from the closest pole. Counting was begun with the first samples that were positive for FtsZ rings or septa in >30% of the cells observed. Using our scoring system, we determined that ripX cells asymmetrically place FtsZ rings and septa at a rate that is, respectively, 6- and 10-fold higher than that for the parent strain (Table 3 and Fig. 4c and d). The frequency of FtsZ ring formation and of septation did not appear to be affected in either ripX or ripX codV strains (data not shown).
TABLE 3.
FtsZ ring and septum positions in outgrowing spores of parent and mutant strains during the primary cell cycle
| Strain | Position
|
% Asymmetryb
|
||||
|---|---|---|---|---|---|---|
| Midcell
|
Asymmetric
|
|||||
| FtsZ | Septum | FtsZ | Septum | FtsZ | Septum | |
| BR151 (parent)a | 5896 | 4956 | 32 | 18 | 0.54 | 0.36 |
| ripX | 2022 | 5112a | 68 | 183a | 3.3 | 3.5a |
| ripX codV | 5375 | 2181 | 138 | 53 | 2.6 | 2.4 |
Combined data from two separate experiments.
Level of significance between parent and mutant strains (P = 0.0001).
FIG. 4.
Collage illustrating phenotypes associated with the ripX::spc mutation. (a and b) Micrographs of DAPI-stained cells from exponential cultures of SL7630 (recA::neo) (a) and SL7370 (recA::neo ripX::spc) (b). (c and d) Immunolocalization of FtsZ in outgrowing spores of strain SL7131 (ripX::spc) (c) and SL7224 (ripX::spc codV::neo) (d). Visualization of FtsZ is with affinity-purified antibody against FtsZ and a secondary antibody coupled to Cy-3; arrows indicate cells with asymmetrically located FtsZ bands. Bar = 10 μm.
ripX phenotype is not abrogated by a recA mutation.
We sought to establish the connection between ripX phenotypes and RecA-mediated homologous recombination during the replication of the chromosome in B. subtilis. To this end, a recA insertion mutation, recA::neo, was introduced into the parent and ripX strains by transformation, selecting for Neor. The presence of the recA::neo mutation in both strains was confirmed by (i) PCR, (ii) sensitivity of the strains to mitomycin C (MMC), and (iii) inability of the strains to be transformed with homologous DNA (the frequency was <10−4 of that of the parental strain) (data not shown).
The phenotypes associated with the ripX mutation persisted in the recA::neo mutant. The cell elongation and aberrant nucleoid phenotypes associated with ripX were not abrogated but rather were slightly exaggerated in the ripX recA strain, compared to those in the ripX parent (Fig. 2 and 4b). Correspondingly, we noted in the ripX recA strain, in comparison to the ripX strain, that the amount of septation was somewhat reduced within longer chains bearing anomalous nucleoids. The slightly exaggerated phenotypes that we visualized in ripX recA cells were also reflected in further increases in generation time (data not shown) and in a decrease in sporulation frequency (Table 2). These data indicate that phenotypes associated with ripX strains can develop independently of RecA-mediated homologous recombination.
In addition to recA, mutations in the recB and recF loci have been shown to affect recombination frequency at dif in E. coli. Individual mutations in either the recB or recF locus diminish the frequency of recombination at dif by about 50%. When both the recB and recF loci of E. coli are mutated, there is almost no detectable recombination at dif (31). Therefore, we have also analyzed analogous rec loci in B. subtilis (addB and recF) (8), as well as less characterized genes which showed potential for recombinagenic activity based on homology to known recombination genes (yrrC, sms, and yshD). Neither individual mutations in these loci nor a variety of combination mutant constructs were able to suppress the effects of a ripX mutation (data not shown).
ripX mutation does not give rise to an SOS-like response.
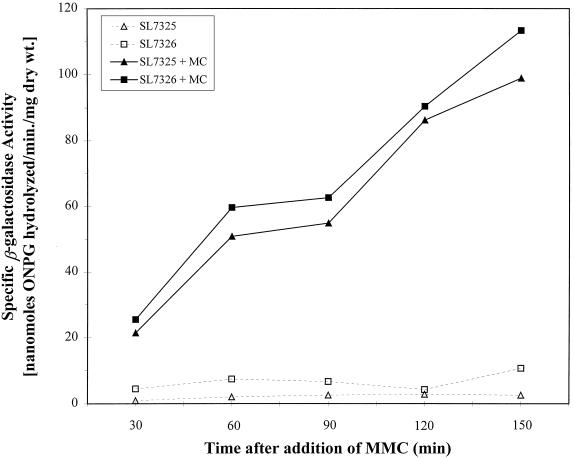
We utilized a dinC::lacZ fusion to monitor the SOS response in B. subtilis (36). Exponentially growing parent and ripX strains containing dinC::lacZ fusions were divided equally among two flasks. One of the two flasks for each strain received MMC in order to induce the SOS response, while the other was left untreated. Our results showed that β-galactosidase activity in MMC-treated cells of both strains steadily rose from the first analyzed sample onward, indicating that the dinC::lacZ fusions were responsive to MMC induction (Fig. 5). In contrast, the corresponding culture for each strain that was left untreated remained at the baseline level (less than 10% of the induced level) throughout the experiment. Thus, the ripX mutation by itself did not cause induction of dinC::lacZ, indicating that the RipX phenotypes do not result from a RecA-mediated SOS-like response. This interpretation is supported by the finding in the previous section which showed that RipX phenotypes persist in a recA background.
FIG. 5.
SOS response of a ripX mutant as indicated by the expression of a dinC::lacZ fusion. MMC was added to a final concentration of 500 ng ml−1 at time zero. Samples were taken from cultures with (filled symbols) and without (open symbols) MMC. Squares, SL7325 (ripX::spc); triangles, SL7326 (ripX+).
CodV and RipX bind dif DNA and interact with the E. coli recombinases XerC and XerD.
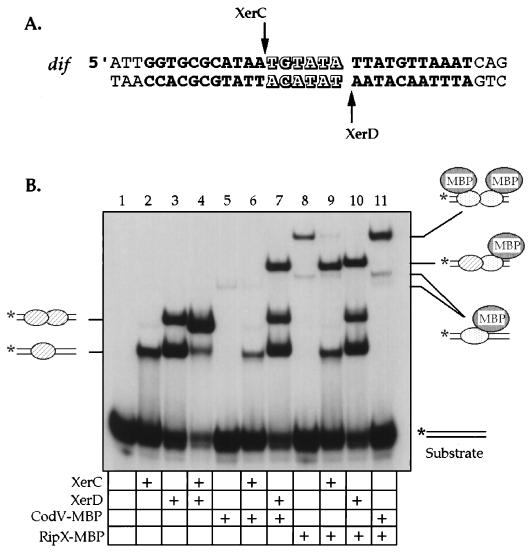
Homology between B. subtilis CodV and RipX and E. coli XerC and XerD, plus the similarity of the RipX and Xer phenotypes, suggests that there is a functional parallel between the two systems. Because we have been unsuccessful in identifying a B. subtilis chromosomal recombination site for CodV and RipX, we used the dif site from E. coli as a substrate to test DNA binding and catalytic activities of these recombinases. The dif site consists of two 12-bp recombinase binding sites containing limited dyad symmetry, separated by a 6-bp central region that delineates the points of strand cleavage and exchange (Fig. 6A) (5). To facilitate protein purification, we constructed MBP fusions of CodV and RipX; the increased molecular weights of the fusions were advantageous for identifying components within specific protein-DNA complexes in experiments where we tested possible interactions between E. coli and B. subtilis recombinases.
FIG. 6.
Binding of RipX and CodV to DNA containing the dif site. (A) The dif site from E. coli consists of two recombinase binding sites (bold lettering) separated by a central region (hollow lettering). Positions of recombinase-mediated strand cleavage and exchange are marked by arrows. (B) Autoradiogram of gel retardation analysis by using purified XerC, XerD, and MBP fusions of RipX and CodV. The dif-containing substrate was labeled with 32P (∗). The positions of complexes relating to the occupancy of either one or two monomers of recombinase are diagrammed at the sides of the gel. The inclusion of a specific protein in a reaction mixture is denoted by a plus in the grid below the gel.
In binding reaction mixtures containing radiolabeled dif DNA, both XerC and XerD were able to bind independently; at the concentration used in this experiment, a single monomer of XerC bound to the left half-site of dif, giving a single retarded complex in a gel shift assay. XerD bound preferentially to the right half-site of dif but is capable of binding both half-sites in a noncooperative manner (Fig. 6B, lanes 2 and 3). Together XerC and XerD show highly cooperative binding to dif, with an increase of apparent affinity of several 100-fold (Fig. 6B, lane 4) (3, 4).
CodV was able to bind dif with a low affinity, as demonstrated by a single faint complex that has a slower mobility than the XerC-XerD complex (Fig. 6B, lane 5); the small amount of binding was not due to low protein concentration. The addition of XerC to the reaction generated only one other complex that is equivalent to XerC alone bound to dif. When XerD was added to CodV plus dif, however, a complex with reduced mobility was formed that was consistent with cooperative interaction between a monomer of CodV and a monomer of XerD (Fig. 6B, lanes 6 and 7). In similar experiments, no detectable binding was observed to seven dif-like oligonucleotides (closest match, 23 of 30 bp) that were derived from a computer search of the B. subtilis genome for dif-like sequences (data not shown).
RipX was also able to bind dif, but it produced two protein-DNA complexes. Our interpretation is that the faster migrating complex represents a monomer of RipX bound to dif, whereas the slower migrating complex represents cooperative binding of two RipX monomers (Fig. 6B, lane 8). Mixing either XerC or XerD with RipX plus dif abolished the most retarded complex and gave rise to complexes consistent with a monomer of either XerC or XerD bound with RipX (Fig. 6B, lanes 9 and 10). In reaction mixtures containing both B. subtilis recombinases there was a fourfold increase in the amount of DNA found, compared to that found in reaction mixtures containing RipX alone (Fig. 6B, lane 11). From these data, we conclude that CodV and RipX can interact when bound to DNA; the apparently low level of cooperativity could be a consequence of the inappropriate substrate. Again, no binding was observed to the seven dif-like oligonucleotides derived from the B. subtilis genome.
These data clearly demonstrate that CodV and RipX are DNA-binding proteins that can interact with related recombinases from a gram-negative bacterium. This apparent conservation over such a great evolutionary distance argues that these two proteins play a crucial role in the bacterial life cycle.
Catalytic activity of CodV and RipX.
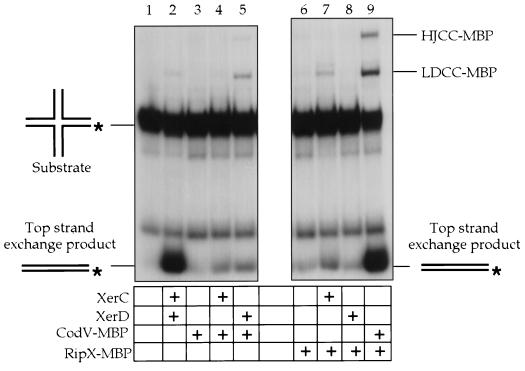
We had previously used various synthetic DNA substrates to study partial recombination reactions mediated by XerC and XerD at dif in vitro; these include linear suicide substrates and synthetic Holliday junctions (1, 5). To test the catalytic competence of CodV and RipX, we used a strand exchange assay based on an artificial Holliday junction (dif-HJ). Substrates are assembled by annealing four oligonucleotides to form a junction that can branch migrate within the 28 bp of the core recombination site. The arms of the junction are different lengths, thus allowing a distinction between the exchange of top or bottom strands. The presence of recombinase covalently attached to either the Holliday junction or a linear duplex recombination product is indicative of strand cleavage.
When XerC and XerD are incubated with the dif-HJ substrate, the major product is derived from an XerC-mediated exchange of top strands (by convention, the first strand exchanged in recombination at psi) (7). Approximately 40% of the substrate is converted to a linear recombinant product after 30 min at 37°C (Fig. 7, lane 2). XerD-mediated strand exchange does not usually occur at a significant level with this substrate, apparently because the DNA-protein complex adopts a conformation that is suitable for XerC but not XerD strand exchange (1). Neither XerC nor XerD alone are capable of catalyzing strand exchange on dif-HJ substrates (1).
FIG. 7.
Autoradiogram of in vitro strand exchange assay using artificial dif Holliday junction substrate labeled with 32P (∗). The exchange of top strands generated a 76-bp linear duplex product, while strand cleavages generated covalently bound recombinase-DNA molecules of either the dif-HJ substrate (HJCC-MBP) or a linear duplex product (LDCC-MBP). Note the high levels of product and covalent complexes in the reaction mixture containing CodV and RipX (lane 9). The inclusion of a specific protein in a reaction mixture is denoted by a plus in the grid below the gel.
Incubation of CodV alone with dif-HJ did not lead to detectable recombination; however, addition of XerC or XerD to the reaction mixture led to a low level of top-strand exchange, as indicated by the presence of a 76-bp linear product (Fig. 7, lanes 3 to 5). Reaction mixtures that contained RipX alone were capable of catalyzing low levels of top-strand exchange, a result that is consistent with cooperative binding of RipX to the left and right half-sites. Addition of XerC to the RipX plus dif-HJ reaction mixture also allowed top-strand exchange and covalent complex formation, whereas addition of XerD allowed a low level of top-strand exchange (Fig. 7, lanes 6 to 8).
The most compelling evidence which indicates that CodV and RipX function together is the high level of top-strand exchange that occurred on dif-HJ when both B. subtilis recombinases were present. Approximately 50% of the substrate was converted to a linear recombinant product after 30 min; the unusually high levels of covalent complexes also show that the recombinases are very active for strand cleavage (Fig. 7, lane 9). The linear duplex recombination product present could arise from two strand cleavages on a dif-HJ that fails to complete strand exchange, suggesting a partial uncoupling of the steps within the reaction. The presence of an excess of the Holliday junction also indicates that strand cleavages are not concerted.
The ability of CodV and RipX, when combined, to resolve a preformed Holliday junction indicates a requirement for appropriate protein-protein interactions for activation of catalysis. That both CodV and RipX are required for efficient resolution in vitro argues strongly that both could be required for in vivo recombination in B. subtilis. Sequence comparison between the B. subtilis and E. coli recombinases also suggests that CodV is the analogue of XerC and that RipX is the analogue of XerD (32). This suggestion is supported by the ability of CodV to interact with XerD but not XerC and the ability of RipX to cleave the bottom strand but not the top strand of a dif linear suicide substrate (data not shown).
DISCUSSION
We show here that a knockout mutation in the B. subtilis ripX gene results in a disturbance of chromosome partitioning that appears to be phenotypically similar to that observed in xer mutants of E. coli (2) and, more generally, to that observed in par mutants of both B. subtilis and E. coli (12, 19, 33). Specifically, in ripX mutants, we observed a subpopulation of elongated cells with abnormally dense or, in other cells, diffuse nucleoids. These elongated cells were usually connected with several other cells of varying length in a chain. In these abnormal chains, we occasionally noted small anucleate cells, and more rarely, DNA trapped in miniature cells or apparently guillotined between cells. We infer from these phenotypes that the primary defect in ripX cells is the inability to resolve dimeric chromosomes in advance of cellular division and that this inability ultimately results in minicell formation or guillotined DNA.
Analysis of outgrowing spores provided evidence that unpartitioned chromosomes can influence the positioning of the division septum in B. subtilis. By the first round of division, ripX mutants localize FtsZ rings and division septa away from the midcell at a 5- to 10-fold greater frequency than their parental strain. We suggest that when chromosomal partitioning is prevented, bulk DNA at the center of the cell may direct the formation of the division septum toward one of the cell poles (19). However, if the chromosome does have an influence on the positioning of FtsZ rings and septa in the ripX strain, it cannot be complete, as rare nucleated minicells and guillotined nucleoids were observed (Fig. 1d). This latter observation is consistent with the report by Wu et al. (35) which showed that completion of chromosomal replication is not required for medial division during outgrowth of B. subtilis.
Sporulation in B. subtilis offers a separate view of cellular partitioning difficulties apart from vegetative growth and division. Sporulation in B. subtilis is characterized by the positioning of a septum toward one of the two cell poles accompanied by the partitioning of a completely replicated chromosome into both the smaller prespore compartment and the larger mother cell compartment (6, 22). The ≈50% reduction in sporulation frequency for a ripX mutant is larger than what we would have expected based on our estimate of the penetration of the ripX phenotype among a vegetative cell population, which we placed at approximately 10 to 20%. A possible explanation for the difference between the observed sporulation defect and our estimate of the phenotype penetrance is that visual scoring by DAPI staining substantially underrecords the number of cells with unresolved chromosomes. It is also possible that there is a RipX-independent mechanism that allows some amount of resolution during vegetative growth but that is inadequate once sporulation has begun.
In E. coli, two separate recombinases, XerC and XerD, act in concert to execute cleavage and recombination reactions that resolve multimeric replicons (2, 3). Therefore, we sought to establish the presence of a sister recombinase to RipX. The closest potential partner recombinase for RipX is CodV, which, like RipX, has considerable amino acid identity with XerC and XerD. By analogy with E. coli, we suspected that RipX and CodV might function as a pair. This suspicion is supported by our in vitro assays which show that catalysis of strand exchange is most efficiently performed when both CodV and RipX are present in the reaction mixtures. However, a nonpolar knockout mutant of codV that we constructed had no discernible phenotype. This absence of phenotype could be the result of a redundancy in B. subtilis. That is, it is possible that RipX proteins acting by themselves are capable of functioning efficiently at the putative recombination site but that CodV does not possess the same flexibility and instead requires a RipX complement. This hypothesis is supported by the observation that RipX but not CodV was able to catalyze strand exchange in vitro by itself. Despite concerted efforts employing database searches and various genetic and biochemical strategies, we have been unable to identify a cognate dif site in B. subtilis.
The instigating event in the production of dimeric chromosomes in E. coli was determined to be RecA-mediated recombination (3, 14). E. coli xer phenotypes were abolished upon the introduction of recA mutations. This effect was not observed in B. subtilis. On the contrary, ripX phenotypes became slightly exaggerated in the absence of RecA. It should be noted that the recA mutant alone exhibits a broader range of cell length and has a more diminished capacity for sporulation than the parent strain. We suggest, therefore, that the exaggerated phenotypes seen in the ripX recA mutant are additive between ripX and recA. Analyses of several other known or suspected recombination genes (addB, recF, yrrC, sms, and yshF) in ripX backgrounds showed no suppression of the RipX phenotype. We have also examined the contribution of prophages as possible sources of recombination activity in the absence of RecA. The introduction of ripX and ripX recA mutations into a strain cured of the SPβ prophage and noninducible for the PBSX prophage, YB886 (37), had no obvious effect on the phenotypes associated with these mutations (data not shown). This result could mean (i) that some protein(s) other than RecA can catalyze the recombination that produces dimeric chromosomes or (ii) that RipX has a more general partitioning function in B. subtilis cell division than do the Xer proteins in E. coli.
We found no evidence of an SOS response in the B. subtilis ripX mutant as indicated by dinC::lacZ expression. Expression of dinC::lacZ is mediated by RecA and DinR, which are the homologues of RecA and LexA of E. coli. It is thus a good indicator of a DNA damage (SOS) response (36). However, although the general DNA damage response in B. subtilis is mediated by RecA, DNA damage-induced filamentation is not (17). Further, B. subtilis does not appear to have an SfiA homologue (15). Thus, although the ripX mutation does not induce a general DNA damage response, we cannot exclude the possibility that it might cause the little-studied RecA-independent DNA damage response that leads to filamentation. Such a response, however, would not explain the observed RipX phenotype which includes the presence of abnormally small cells (Fig. 1). We favor the explanation that the cell division defects resulting from the ripX mutation are a direct consequence of the defect in chromosome resolution and are not a consequence of an SOS-type response. It should be recalled that in E. coli dif mutants (and by inference, xerC and xerD mutants) SOS response activity is not the primary cause of filamentation (14).
ACKNOWLEDGMENTS
We thank Joe Lutkenhaus, Petra Levin, and Linc Sonenshein for provision of experimental materials and helpful advice.
This work was supported by Public Health Service grant GM43577 (to P.J.P.) and training grant T32 AI07101 (to S.A.S.). G.B. and D.J.S. were supported by the Wellcome Trust.
REFERENCES
- 1.Arciszewska L K, Grainge I, Sherratt D J. Action of site-specific recombinases XerC and XerD on tethered Holliday junctions. EMBO J. 1997;16:3731–3743. doi: 10.1093/emboj/16.12.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blakely G, Colloms S D, May G, Burke M, Sherratt D J. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991;3:789–798. [PubMed] [Google Scholar]
- 3.Blakely G, May G, McCulloch R, Arciszewska L K, Burke M, Lovett S T, Sherratt D J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993;75:351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- 4.Blakely G, Sherratt D. Determinants of selectivity in Xer site-specific recombination. Genes Dev. 1996;10:762–773. doi: 10.1101/gad.10.6.762. [DOI] [PubMed] [Google Scholar]
- 5.Blakely G W, Davidson A O, Sherratt D J. Binding and cleavage of nicked substrates by site-specific recombinases XerC and XerD. J Mol Biol. 1997;265:30–39. doi: 10.1006/jmbi.1996.0709. [DOI] [PubMed] [Google Scholar]
- 6.Callister H, Wake R G. Completed chromosomes in thymine-requiring Bacillus subtilis spores. J Bacteriol. 1974;120:579–582. doi: 10.1128/jb.120.2.579-582.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colloms S D, McCulloch R, Grant K, Neilson L, Sherratt D J. Xer-mediated site-specific recombination in vitro. EMBO J. 1996;15:1172–1181. [PMC free article] [PubMed] [Google Scholar]
- 8.Dubnau D. Genetic exchange and homologous recombination. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 555–584. [Google Scholar]
- 9.Harry E, Pogliano K, Losick R. Cell-specific gene expression in B. subtilis. J Bacteriol. 1995;177:3386–3393. doi: 10.1128/jb.177.12.3386-3393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes F, Lubetzki S A, Sherratt D J. Salmonella typhimurium specifies a circular chromosome dimer resolution system which is homologous to the Xer site-specific recombination system of Escherichia coli. Gene. 1997;198:105–110. doi: 10.1016/s0378-1119(97)00299-0. [DOI] [PubMed] [Google Scholar]
- 11.Hofte M, Dong Q, Kourambas S, Krishnapillai V, Sherratt D, Mergeay M. The sss gene product, which affects pyoverdin production in Pseudomonas aeruginosa 7NSK2, is a site-specific recombinase. Mol Microbiol. 1994;14:1011–1020. doi: 10.1111/j.1365-2958.1994.tb01335.x. [DOI] [PubMed] [Google Scholar]
- 12.Huang W M, Libbey J L, Van DerHoeven P, Yu S X. Bipolar localization of Bacillus subtilis topoisomerase IV, an enzyme required for chromosome segregation. Proc Natl Acad Sci USA. 1998;95:4652–4657. doi: 10.1073/pnas.95.8.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khvorova A, Zhang L, Higgins M L, Piggot P J. The spoIIE locus is involved in the SpoOA-dependent switch in the location of FtsZ rings in Bacillus subtilis. J Bacteriol. 1998;180:1256–1260. doi: 10.1128/jb.180.5.1256-1260.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuempel P L, Henson J M, Dircks L, Tecklenberg M, Lim D F. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 1991;3:799–811. [PubMed] [Google Scholar]
- 15.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, et al. The complete genome sequence of the gram positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 16.Liu G, Draper G C, Donachie W. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- 17.Love P E, Yasbin R E. Genetic characterization of the inducible SOS-like system of Bacillus subtilis. J Bacteriol. 1984;160:910–920. doi: 10.1128/jb.160.3.910-920.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 19.Mulder E, Woldringh C L. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989;171:4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neilson L, Blakely G, Sherratt D J. Site-specific recombination at dif by Haemophilus influenzae XerC. Mol Microbiol. 1999;31:915–926. doi: 10.1046/j.1365-2958.1999.01231.x. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson W L, Setlow P. Sporulation germination and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, West Sussex, England: John Wiley and Sons, Ltd.; 1990. pp. 391–429. [Google Scholar]
- 22.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piggot P J, Curtis C A M. Analysis of the regulation of gene expression during Bacillus subtilis sporulation by manipulation of the copy number of spo-lacZ fusions. J Bacteriol. 1987;169:1260–1266. doi: 10.1128/jb.169.3.1260-1266.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piggot P J, Curtis C A M, de Lencastre H. Use of integrational plasmid vectors to demonstrate the polycistronic nature of a transcriptional unit (spoIIA) required for sporulation of Bacillus subtilis. J Gen Microbiol. 1984;130:2123–2136. doi: 10.1099/00221287-130-8-2123. [DOI] [PubMed] [Google Scholar]
- 25.Schuch R. Ph.D. thesis. Philadelphia, Pa: Temple University School of Medicine; 1995. [Google Scholar]
- 26.Sharpe M E, Errington J. Postseptational chromosome partitioning in bacteria. Proc Natl Acad Sci USA. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherratt D J, Wigley D B. Conserved themes but novel activities in recombinases and topoisomerases. Cell. 1998;93:149–152. doi: 10.1016/s0092-8674(00)81566-4. [DOI] [PubMed] [Google Scholar]
- 28.Siccardi A G, Galizzi A, Mazza G, Clivio A, Albertini A. Synchronous germination and outgrowth of fractionated Bacillus subtilis spores: tool for the analysis of differentiation and division of bacterial cells. J Bacteriol. 1975;121:13–19. doi: 10.1128/jb.121.1.13-19.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sirois S, Szatmari G. Detection of XerC and XerD recombinases in gram-negative bacteria of the family Enterobacteriaceae. J Bacteriol. 1995;177:4183–4186. doi: 10.1128/jb.177.14.4183-4186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stark W M, Boocock M R, Sherratt D J. Catalysis by site-specific recombinases. Trends Genet. 1992;8:432–438. [PubMed] [Google Scholar]
- 31.Steiner W W, Kuempel P L. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J Bacteriol. 1998;180:6269–6275. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanya H S, Arciszewska L K, Baker R A, Bird L E, Sherratt D J, Wigley D B. Crystal structure of the site-specific recombinase, XerD. EMBO J. 1997;16:5178–5187. doi: 10.1093/emboj/16.17.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Q, Yu X, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 34.Villion M, Szatmari G. Cloning and characterisation of the Proteus mirabilis xerD gene. FEMS Microbiol Lett. 1998;164:83–90. doi: 10.1111/j.1574-6968.1998.tb13071.x. [DOI] [PubMed] [Google Scholar]
- 35.Wu L J, Franks A H, Wake R G. Replication through the terminus region of the Bacillus subtilis chromosome is not essential for the formation of a division septum that partitions the DNA. J Bacteriol. 1995;177:5711–5715. doi: 10.1128/jb.177.19.5711-5715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yasbin R E, Cheo D, Bol D. DNA repair systems. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 529–538. [Google Scholar]
- 37.Yasbin R E, Fields P I, Andersen B J. Properties of Bacillus subtilis 168 derivatives freed of their natural prophages. Gene. 1980;12:155–159. doi: 10.1016/0378-1119(80)90026-8. [DOI] [PubMed] [Google Scholar]
- 38.Yu X-C, Weihe E, Margolin W. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J Bacteriol. 1998;180:6424–6428. doi: 10.1128/jb.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]