Abstract
The abilities of two bacterial active heme transporters, HmbR of Neisseria meningitidis and HemR of Yersinia enterocolitica, to use different heme sources were compared. While HmbR-expressing cells used only hemoglobin (Hb) and heme, HemR-expressing bacteria were able to grow on Hb, heme, myoglobin, hemopexin, catalase, human and bovine serum albumin-heme, and haptoglobin-hemoglobin complexes as sources of iron. Expression of functional HemR allowed Escherichia coli cells to respond to heme-containing peptides, microperoxidases MP-8, MP-9, and MP-11, suggesting the ability of HemR to transport heme covalently linked to other molecules. Comparison of HemR with other heme receptors identified several highly conserved histidine residues as well as two conserved amino acid motifs, the FRAP and NPNL boxes. A site-directed mutagenesis approach was used to investigate the roles of His128, His192, His352, and His461 residues in HemR function. The HemR receptor with histidine changed to lysine at position 128 (HemRH128K), HemRH461L, HemRH461A, and HemRH128A,H461A mutant receptors were unable to use Hb, human serum albumin-heme, and myoglobin as sources of porphyrin and iron. Utilization of free heme was also severely affected, with some residual heme uptake in cells expressing HemRH128K, HemRH461A, and HemRH461L. Conversely, the HemRH192T, HemRH352A, HemRH352K, and HemRH192T,H352K mutant receptors were fully functional. All mutant HemR proteins were expressed in the outer membrane at levels similar to that of the wild-type HemR receptor. Nonfunctional HemRs were able to bind heme- and Hb-agarose. A hypothetical model of the HemR function in which two conserved histidine residues, His128 and His461, participate in the transport of heme through the receptor pore is postulated.
The heme molecule (iron protoporphyrin IX) is one of the most important enzyme cofactors. Bacteria devote up to 10% of the total cell iron content and a significant amount of cell energy to the biosynthesis of heme (3, 33). The high energy cost of heme biosynthesis coupled with the constant famine that bacteria face in their natural environments favored the evolution of heme-scavenging systems. The heme-scavenging systems developed in different bacteria are confronted with the following tasks: (i) binding a heme-containing compound, (ii) releasing heme from the compound, and (iii) transporting heme into the cell. Studies with Yersinia enterocolitica, Vibrio cholerae, and Haemophilus influenzae have begun to define models of heme utilization in gram-negative bacteria (6, 13, 17, 43, 44). These studies postulated that all heme-utilizing bacteria possess outer membrane (OM) receptors that bind heme or heme-containing proteins and transport heme across the otherwise impermeable OM.
Bacterial receptors that interact with heme-protein complexes, transferrin (Tf), and lactoferrin (Lf) as sources of iron are close relatives of siderophore and vitamin B12 receptors of gram-negative bacteria. However, while siderophore and vitamin B12 receptors bind their cognate ligands directly, receptors that use heme-protein complexes, Tf, or Lf must first release the ligand (i.e., heme or iron in the case of Tf and Lf receptors) from the protein complex. This is not a trivial task, since the bond between the heme and the globin in hemoglobin (Hb) has an association constant of 1012 to 1016 (5). Similarly, the constant of association of Tf and iron is 1020 (1). The mechanism and the protein domains involved in stripping heme or iron from protein complexes and transporting heme or iron through the receptor pore are currently unknown.
Heme and Hb are preferred substrates for the majority of bacterial heme acquisition systems (27). However, some bacteria use additional heme-protein complexes like hemopexin-heme, haptoglobin-Hb, heme-albumin, and myoglobin (8, 40, 48). Serratia marcescens responds to the challenge of multiple heme sources by producing a small protein, hemophore, that is able to extract heme from protein complexes and deliver it to the OM receptor (12, 28). Vibrio vulnificus and Porphyromonas gingivalis make heme available to their heme receptors by secreting proteases that degrade Hb and other heme-protein complexes (11, 38). Other heme-utilizing pathogens express multiple OM receptors that are specific for some but not all heme-protein complexes. H. influenzae, for example, expresses several distinct hemopexin and haptoglobin-hemoglobin utilization systems (6, 20, 32, 36). Conversely, when cloned into Escherichia coli, only one Yersinia pestis genetic locus allowed use of several heme-protein sources (18).
We have used the HemR protein of Y. enterocolitica as a model system to learn how heme and Hb receptors function. In this communication, we show that HemR of Y. enterocolitica is sufficient for the utilization of the largest variety of heme-containing compounds. HemR also used heme-peptide conjugates, which suggests that the receptor pore may accommodate molecules larger than heme itself. Amino acid comparisons of several heme and Hb receptors identified four conserved histidine residues and a conserved receptor domain that was specific for heme and Hb receptors. Results from site-directed mutagenesis experiments confirmed that two of four conserved histidine residues in HemR are essential for its function.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. Bacteria were routinely grown in Luria broth (LB). Iron-limiting conditions were accomplished by growing bacteria on nutrient broth plates supplemented with dipyridyl (NBD) (43). When necessary, supplementation with δ-aminolevulinic acid (final concentration, 50 μg/ml) was used. Chemicals were purchased from Sigma (St. Louis, Mo.), Aldrich Co. (Milwaukee, Wis.), and Porphyrin Products, Inc. (Logan, Utah).
TABLE 1.
Bacterial strains and plasmids used in the study
| Bacterial strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | Stratagene | |
| IR1532 | DH5α but heme biosynthesis mutant | 47 |
| IR1583 | As IR1532 but tonB exbB exbDNM | 47 |
| EB53 | E. coli hemA aroB Rifr | 43 |
| IR754 | EB53 but tonB::Kanr | 45 |
| Y. enterocolitica WA-C | ||
| H1883 | Plasmidless strain, hemPRSTVU+ | 43 |
| H5030 | hemR::phoA Ampr | 43 |
| PLASMIDS | ||
| pT76.91 | pT7-5, hemPR | 43 |
| pHEM101 | pBluescript hemPR | 43 |
| pT76.53 | hemPRST′ | 43 |
| pWKS130 | Low-copy-number vector, Neor | 49 |
| pWKS30 | Low-copy-number vector, Ampr | 49 |
| pAMP1173 | pWKS30 carrying hmbRWT | This study |
| pNEO76.91 | pWKS130, hemRWT | Laboratory collection |
| pNEO1187 | pWKS130, hemRH128K | This study |
| pNEO1188 | pWKS130, hemRH461K | This study |
| pNEO1194 | pWKS130, hemRH352K | This study |
| pWSH1332 | pWKS130, hemRH192T | This study |
| pNEO1391 | pWKS130, hemRSTOP | This study |
| pICE1403 | pWKS130, hemRH192T,H352K | This study |
| pNEO1505 | pWKS130, hemRH352A | This study |
| pNEO1507 | pWKS130, hemRH461A | This study |
| pCSB1707 | pWKS130, hemRH128A,H461A | This study |
hemRWT, wild-type hemR.
Growth promotion and inhibition assays.
The functions of mutant HemRs were tested by assaying the abilities of these receptors to supply an E. coli heme biosynthesis mutant with heme-iron and/or heme-porphyrin while growing in the presence of different heme sources. Assays done on LB plates tested the ability of HemRs to supply heme as a porphyrin source to cells. Assays done on nutrient broth plates supplemented with 0.25 mM dipyridyl [NBD(0.25)] tested the ability of HemRs to supply heme as both an iron source and a porphyrin source. Testing of HemR function on LB plates allows detection of very small functional defects, since under these growth conditions, HemR is expressed at lower levels than when grown on NBD(0.25) plates (approximately 12-fold ratio of induction) (43). Human serum albumin (HSA), bovine serum albumin (BSA), horse heart muscle and skeletal muscle myoglobin, cytochrome c, catalase, and human Hb A0 were purchased from Sigma and dissolved in 0.9% NaCl at a final concentration of 100 μM (except for catalase [25 μM]). One milligram of a lyophilized pooled human haptoglobin (Sigma) that binds between 0.5 and 0.9 mg of human Hb was dissolved in 100 μl of a 6.5-mg/ml solution of human Hb. Ten microliters of the haptoglobin-Hb complex was used in growth promotion assays. Twenty microliters of human hemopexin (2.5 mg/ml), a gift from E. Hansen, was used in growth promotion assays. Hemin was dissolved in 20 mM NaOH (final concentration, 10 mM). Albumin-heme solutions were prepared by mixing heme and HSA (or BSA) in a 1:1 molar ratio. Approximately 2 × 107 E. coli IR1532 cells expressing the wild-type or mutant HemRs or E. coli 1583 cells expressing HmbR were inoculated into 3 ml of 0.75% agarose in water and spread onto LB or NBD plates. The HemR and HmbR receptors were expressed from their own promoters. Paper discs (one to four discs, each 6 mm in diameter) were soaked with 10-μl amounts of different heme compounds and placed on plates. Zones of stimulation were measured after 18 to 24 h of incubation at 37°C. The zones of growth stimulation obtained with E. coli IR1532(pNEO76.91) were very similar to the zones of growth stimulation seen with E. coli EB53(pNEO76.91), E. coli EB53(pT76.53), and E. coli IR1532(pT76.53) (data not shown).
Microperoxidases were purchased from Sigma and dissolved in 0.9% NaCl to a final concentration of 5 mg/ml. Discs with 10-μl amounts of stock solutions were placed onto NBD(0.2) plates containing bacteria. δ-Aminolevulinic acid was added to the plates in experiments with E. coli EB53 to ensure bacterial growth. Due to the high bacterial inoculum and the presence of δ-aminolevulinic acid, E. coli EB53 could grow on NBD(0.2) (background growth visible after 1 to 2 days of incubation).
Site-directed mutagenesis of hemR.
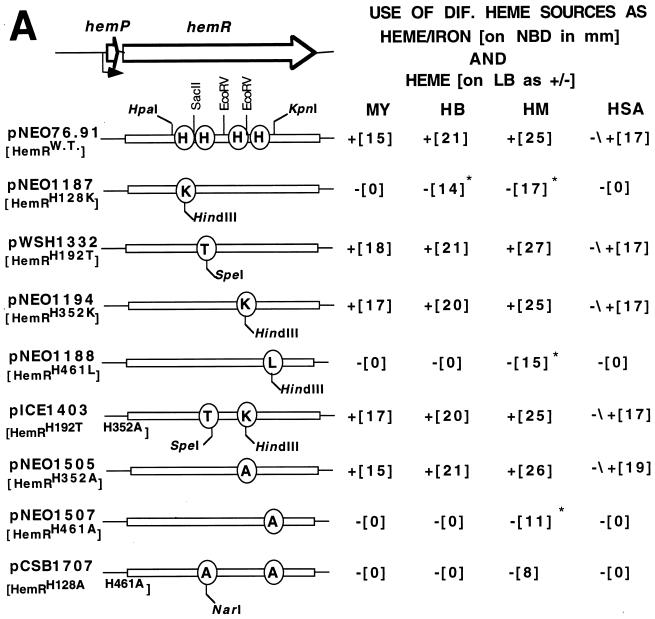
HemR mutants were constructed by using either PCR amplification or the QuickChange protocol (Stratagene). Plasmid pHEM101, which had been linearized with ScaI, was used as a template for mutagenesis. A unique restriction site (HindIII or SpeI) was introduced at the locations of His codons by PCR amplification with the PwoI DNA polymerase (Boehringer, Mannheim, Germany). His128 was replaced by a lysine residue, creating a unique HindIII restriction site. His192 was replaced by a threonine residue, creating a new SpeI restriction site. His352 was replaced by a lysine residue, creating a new HindIII restriction site. His461 was replaced by a leucine residue, creating a new HindIII restriction site. Other mutant HemRs (HemR receptor with H changed to A at position 352 [HemRH352A], HemRH461A, and HemRH128A,H461A) were constructed by the QuickChange protocol (Stratagene) with pT76.91 as a template for mutagenesis. The hemRH128A mutation can be detected by NarI digestion. Cartridge-purified PCR primers, 33 to 36 bp in length, were purchased from Gibco BRL. DNA fragments carrying the mutations (HpaI-KpnI, SacII-KpnI, or EcoRV-EcoRV [see Fig. 4]) were subcloned into pBluescript and recloned into the wild-type pT76.91 (43). All DNA fragments carrying the mutations were sequenced to rule out secondary mutations. Finally, the hemR gene, expressed from its own promoter, was subcloned onto a low-copy-number vector pWKS130 in the orientation opposite that of the lac promoter (49). The amount of HemR expressed from the low-copy-number plasmid approximated the amount of the receptor expressed in Y. enterocolitica cells (43).
FIG. 4.
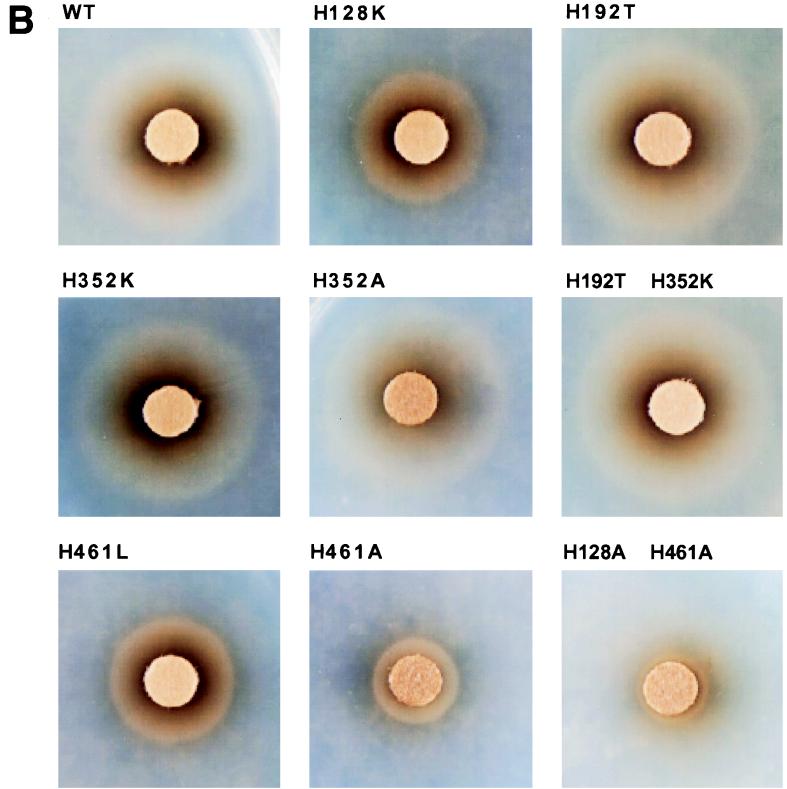
(A) A schematic representation of mutant HemRs and their phenotypes after expression in the E. coli heme biosynthesis mutant. The testing for growth stimulation was done on LB and NBD plates essentially as described in the legend to Fig. 1. Growth stimulation with four different (DIF.) heme sources, myoglobin (MY), hemoglobin (HB), heme (HM), and HSA (HSA-heme complex, is indicated as follows: +, stimulation of growth; −, no stimulation; ∗ and −/+, very light or small zone of stimulation. HemRW.T., wild-type HemR. (B) Growth zones of HemR histidine mutants around discs soaked in 10 μl of 10 mM heme (NBD plates). WT, wild type.
OM fraction preparation.
Triton X-100 OM fractions were prepared from E. coli IR1532, a DH5α heme biosynthesis mutant, expressing recombinant HemRs from low-copy-number (pWKS130) and medium-copy-number (pT7-5) plasmids. Briefly, 100-ml cultures were inoculated with 1-ml portions of cultures grown overnight and incubated at 37°C with shaking for 2.5 h. An iron chelator, dipyridyl (final concentration 0.3 mM) was added to induce the expression of HemRs, and the cultures were incubated for a further 2.5 h. Cells were harvested by centrifugation and resuspended in 1 ml of 200 mM Tris (pH 8.0), followed by addition of 2 ml of 1 M sucrose in 200 mM Tris (pH 8.0)–200 μl of 10 mM EDTA–200 μl of lysozyme at 2 mg/ml–6.4 ml of double-distilled water (ddH2O). Mixtures were left to stand at room temperature for 5 min to allow spheroplasts to form. Ten milliliters of a solution of 2% Triton X-100, 50 mM Tris (pH 8.0), 10 mM MgCl2, and 200 μl of DNase at 1 mg/ml was added, and the mixtures were spun at 18,000 rpm in a JA25.50 rotor for 60 min at 4°C. Triton X-100-insoluble OM fraction pellets were washed three times in 1 ml of ddH2O and resuspended in ddH2O to a final volume of 100 μl (14).
Affinity chromatography of HemR with heme and Hb-agarose.
OM preparations from bacteria expressing HemRs from medium-copy-number plasmids (pT7-5) were solubilized for affinity chromatography in Zwittergent 3-14 (9, 27). OM samples were diluted in 600 μl of phosphate-buffered saline (PBS) and then 600 μl of 1% Zwittergent 3-14 in PBS was added. Binding reactions were performed in Eppendorf tubes with 10 to 20 μl of hemin-agarose (Sigma H6390; 7.7 μmol of hemin per ml of packed gel) or 20 μl of Hb-agarose (Sigma H8756; 16.7 mg of Hb per ml of packed gel). Binding reaction mixtures were incubated on a rotator either for 2 h at room temperature or overnight at 4°C. Agarose was then washed four times with 1.2 ml of 0.5% Zwittergent 3-14 in PBS to remove unbound material. Agarose was boiled in Laemmli sample buffer for 3 min and then electrophoresed in a 10% polyacrylamide gel at constant voltage with Tris-glycine buffer (24). Proteins in the gel were visualized with Coomassie stain.
RESULTS
The HemR receptor uses a wide spectrum of heme compounds.
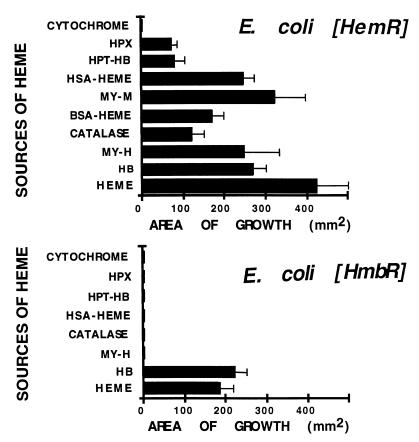
Use of different heme-containing compounds by an E. coli heme biosynthesis mutant expressing the Y. enterocolitica HemR and N. meningitidis HmbR was compared (45, 46). As can be seen from Fig. 1, the HemR-expressing strain was able to use heme, heart and skeletal muscle myoglobins (MY-M and MY-H), Hb, BSA- and HSA-heme complexes, hemopexin, and catalase as the sole sources of porphyrin and iron. The use of haptoglobin-Hb complexes (HPT-Hb) was also dependent on the expression of HemR, while both native and denatured cytochrome c could not support growth of HemR-expressing cells. Conversely, the strain expressing the HmbR receptor used only heme and Hb as sources of porphyrin and iron (Fig. 1). The spectrum of heme-containing compounds used by these receptors correlated with the spectrum of heme compounds used by yersiniae and neisseriae (8, 18, 40, 43, 45). The inability of HmbR-expressing cells to use some heme-protein complexes ruled out the possibility that heme released from denaturated complexes fed HemR-expressing bacteria. These results show that the expression of the HemR receptor is sufficient for utilization of heme from native heme-protein complexes.
FIG. 1.
Comparison of the abilities of heme and Hb receptors, HemR and HmbR, to use different heme-protein complexes as sources of iron and porphyrin in the E. coli heme biosynthesis mutant IR1532. HemR and HmbR receptors were expressed from low-copy-number plasmids pWKS130 and pWKS30, respectively (49). The expression of HmbR was done in the presence of N. meningitidis tonB exbB exbD genes (47). Bars represent the surface areas of bacterial growth around the discs on NBD plates (error bars show the standard deviations). Discs were soaked with equimolar amounts of heme-containing compounds (except for heme, see Material and Methods). Each measurement was repeated at least three times. Abbreviations: HB, hemoglobin; HPX, human hemopexin; HPT-HB, human haptoglobin-Hb complex; MY-M, skeletal muscle myoglobin; MY-H, heart muscle myoglobin.
The HemR receptor-expressing bacteria use a heme-peptide conjugate as a source of iron.
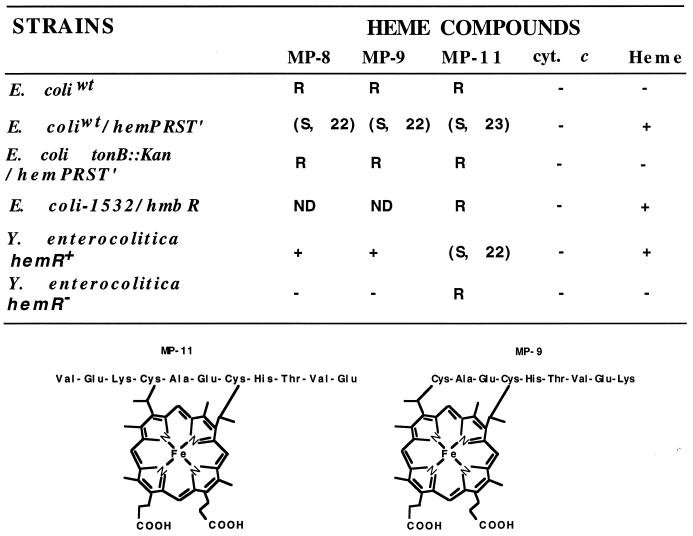
The inability of the native and denatured cytochrome c to serve as an iron source indicated that covalently attached heme cannot be taken up by the HemR receptor (Fig. 2). Interestingly, peptides derived from cytochrome c by proteolytic digestion, such as microperoxidases MP-8 and MP-9, stimulated growth, whereas MP-11 inhibited growth of Y. enterocolitica under iron-limiting growth conditions (Fig. 2). The hemR mutant of Y. enterocolitica was not stimulated by MP-8 and MP-9 or inhibited by MP-11. All three compounds were inhibitory for HemR-expressing E. coli. The heme moiety is covalently attached to a peptide in MP-11, MP-9, and MP-8, and these compounds have peroxidase activity (15). E. coli cells expressing the N. meningitidis HmbR protein were not sensitive to MP-11. The toxicity of microperoxidases depended on the genotype of the strain and growth conditions used in the experiments. Only HemR-expressing E. coli strains that possessed functional TonB protein were sensitive to these compounds. The presence of heme and Hb partially protected sensitive, HemR-expressing E. coli cells against MP-11 toxicity (data not shown). These results suggest that the HemR receptor recognizes the heme moiety of MP-11 and transports the whole compound into the bacterial periplasm. The mechanism by which microperoxidases inhibit bacterial growth and the reason for a difference in sensitivity of E. coli and Y. enterocolitica to MP-8 and MP-9 are not clear.
FIG. 2.
Growth-inhibitory or -stimulatory activity of microperoxidases on Y. enterocolitica and E. coli (top) and structures of microperoxidases MP-9 and MP-11 (bottom). Wild-type E. coli hemA aroB (E. coliwt) and E. coli IR1532 (E. coli-1532), a DH5α heme biosynthesis mutant (47), were two of the strains used. Growth-inhibitory or -stimulatory activity of microperoxidases MP-8, MP-9, and MP-11, cytochrome c (cyt. c), and heme are shown as follows: R, resistant to the inhibitory activity of microperoxidases; (S, 22), sensitive to inhibitory activity of microperoxidases, with a 22-mm-diameter inhibition zone; ND, not done; +, stimulation of growth under iron-limiting conditions; −, no stimulation of growth under iron-limiting conditions. Only bacteria that possess functional HemR can be stimulated or inhibited by microperoxidases. The same results were obtained with the native and denatured forms of cytochrome c. MP-8 is one amino acid (Lys) shorter than MP-9 (not shown). Note a covalent link between the peptide and the heme moiety.
Identification of conserved amino acid motifs present in different heme and Hb receptors.
The mechanism and protein domains by which heme receptors bind heme compounds, extract heme from these compounds, and transport heme into the cell are not known. Amino acid comparisons of different TonB-dependent siderophore receptors identified several conserved domains and amino acid residues (21). Similar analysis of bacterial heme receptors revealed two amino acid motifs that are highly conserved among different heme and Hb receptors but are not present or conserved in siderophore and vitamin B12 receptors (Fig. 3A). The FRAP and NPNL amino acid motifs and a conserved His residue between the motifs were found only in heme receptors and were not well conserved in siderophore and vitamin B12 receptors. The FRAP box and the His residue at position 461 were also present in some Tf and Lf receptors. The same analysis revealed that several heme receptors possess additional conserved histidine residues, corresponding to His128, His192, and His352 of HemR (Fig. 3B). Histidine 128 is located in the receptor domain that is conserved in all TonB-dependent receptors (21, 31). However, His residues were not found at the corresponding regions of FepA and FhuA receptors. The E. coli FepA receptor contains histidine residues at positions 194, 351, 473, and 474, but there is no significant homology between other parts of these histidine-containing domains (data not shown).
FIG. 3.
Amino acid comparisons of the conserved domains of the heme and Hb receptors. (A) A highly conserved receptor domain containing an invariant histidine residue, FRAP and NPNL amino acid boxes, present in all receptors that transport heme into the periplasm. Siderophore, vitamin B12, and some Tf and Lf receptors lack either the complete domain or have only the FRAP box and distal glutamic amino acid residues relatively well conserved. Highly conserved residues (indicated by asterisks), aromatic residue (Aro), and gaps introduced to maximize alignment (indicated by dashes) are shown. Hpt, haptoglobin. (B) Amino acid comparison of Hb and heme receptor domains around conserved histidine residues (conserved histidine residues are shown in bold). Y. enterocolitica HemR (HEMR), Y. pestis HmuR (HMUR), E. coli ChuA (CHUA), S. dysenteriae SduA (SHUA), H. influenzae HxuC (HXUC), E. coli FepA (FEPA), and E. coli FhuA (FHUA) are shown. Amino acid residues of all receptors except FepA and FhuA are numbered from the first methionine.
Comparison of the HemR model with crystal structures of FepA and FhuA.
The overall homology of E. coli FhuA and FepA and Y. enterocolitica HemR is relatively low: HemR shares 21% identical amino acid residues with FhuA and FepA. A two-dimensional model of the HemR receptor was designed in order to determine probable localization of the FRAP and NPNL boxes and conserved histidine residues on the receptor (42). Amino acid comparison analysis of HemR and its close relatives, heme receptors ChuA, SduA, HmuR, and PhuR, was used to assist in the determination of surface-exposed loops and transmembrane regions of the receptor. The model of the HemR receptor contains 22 transmembrane β-strands and 11 surface-exposed loops (data not shown). The first ∼150 amino acid residues together with the TonB box are localized in the periplasm, closing the receptor pore. This arrangement of the amino terminus of the receptor is based on the three-dimensional structures of FepA and FhuA receptors (4, 10, 30). The amino-terminal domain of HemR contains the invariable His128 residue. His128 aligns with arginine 105 of FepA and tyrosine 116 of FhuA. Arginine residues of the FepA amino terminus have been postulated to interact with the negatively charged enterobactin sideropore (4), while tyrosine 116 was found to make hydrogen bonds with the ferrichrome (10, 30). The HemR model predicted that the FRAP and NPNL boxes and His352 are localized in the transmembrane regions, while His192 and His461 are localized in the surface-exposed loops of the receptor. A limited homology between the FepA domain encompassing residues 485 to 532 and the HemR domain containing FRAP and NPNL motifs was found (Fig. 3A). Corresponding FepA residues participate in the formation of two β-sheets and an external loop, suggesting that the placement of FRAP and NPNL motifs is correct in the two-dimensional model of HemR (i.e., that the loop containing an invariant His461 is exposed on the surface). This prediction could not be independently tested with the FhuA model due to the lack of homology between HemR and FhuA receptors at corresponding regions.
Mutagenesis and phenotypic characterization of the HemR histidine mutants.
Conserved histidine residues of HemR may play an important role in binding of heme-protein complexes and/or transporting heme through the receptor pore. The role of conserved His residues in HemR function was tested by constructing site-directed HemR mutants (Fig. 4). Mutant receptor genes were expressed from the hemR iron-regulated promoter (43). All mutant receptors localized to the OM in approximately equal amounts (Fig. 5 and data not shown). The ability of the E. coli IR1532 heme biosynthesis mutant (47), transformed with different HemRs, to use 100 μM solutions of Hb, myoglobin, HSA-heme complexes, and heme (10 mM) as sources of heme and iron was assessed by the plate assay (see below). Plate assays were done on both NBD and LB plates in order to test the abilities of HemRs to use heme-protein complexes as sources of porphyrin and iron and porphyrin alone. In addition, testing mutants under iron-rich (LB plates) and iron-poor (NBD plates) growth conditions allows comparison of the receptor function at different levels of expression, since the expression of the hemR gene is regulated by the availability of iron (approximately 12-fold) (43). The mutant receptors HemRH128K (pNEO1187), HemRH461L (pNEO1188), HemRH461A (pNEO1507), and HemRH128A,H461A (pCSB1707) could not support growth of the E. coli heme biosynthesis mutant on any heme source when tested on LB plates (Fig. 4A). The HemR receptors HemRH192T (pWSH1332), HemRH352K (pNEO1194), HemRH352A (pNEO1505), and HemRH352K,H192T (pICE1403) were proficient in utilization of all heme sources on LB plates (Fig. 4A).
FIG. 5.
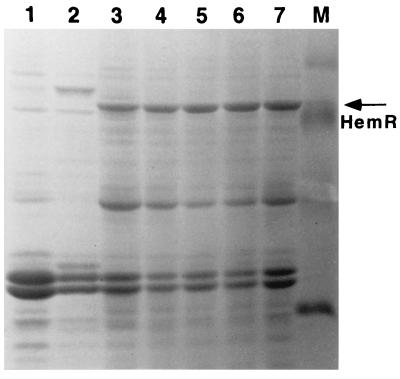
OMs of E. coli expressing different HemR histidine mutants from a pWKS130 plasmid. Lanes: 1, HemRSTOP (pNEO1391 [see Table 1]); 2, E. coli DH5α; 3, HemRH352K; 4, HemRH461L; 5, HemRH192T; 6, HemRH128K; 7, wild-type HemR; M, molecular mass markers (30, 46, 69, and 94 kDa).
Testing the mutant HemRs under iron-restricted growth conditions gave the following results: HemRH461L and HemRH461A were unable to use Hb, myoglobin, and HSA-heme, while free heme still had some growth-promoting effect. The hemRH461A allele had a much larger growth defect on heme than the hemRH461L allele. The hemRH128A,H461A double mutant was the most affected receptor allele; only a 1-mm-wide and weak growth zone could be seen around the heme disc (Fig. 4). HemRH128K was grossly affected in its ability to use three heme-protein complexes; however, a very small and weak zone of growth around the Hb disc was observed on the lawn of E. coli 1532(pNEO1187). The zone of growth around the heme disc was significantly smaller than the one observed with the wild-type HemR but larger than the zone of growth observed with the HemRH461L receptor (Fig. 4). Cells expressing the HemRH352K receptor grew slower and needed longer to form growth zones around different heme-containing discs, although the diameters of the zones were identical to the ones seen with the wild-type cells. The growth of cells expressing HemRH352A and the wild-type receptor were indistinguishable. Growth zones of HemRH192T-expressing cells around myoglobin and heme and were larger than those found in the wild-type strain (Fig. 4A).
Quantification of the defects of individual HemR mutant receptors.
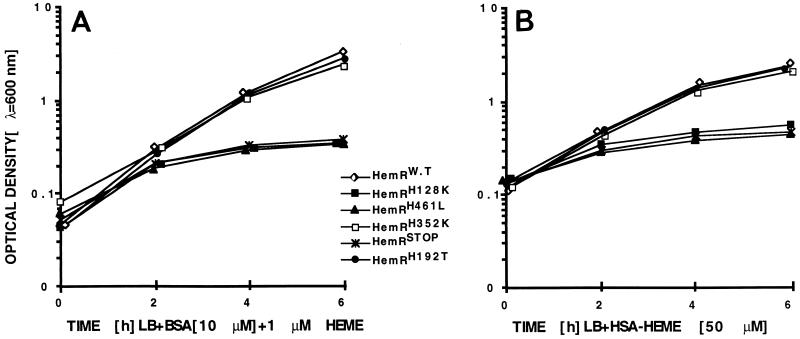
The growth defects of the mutant HemRs were quantified by expressing the mutant genes in E. coli hemA aroB (EB53) cells and measuring the kinetics of growth in liquid medium in the presence of different heme compounds. The best sources of porphyrin for the wild-type HemR-expressing bacteria were Hb and heme, allowing growth at a concentration of 1 μM. Fifty- to 75-fold higher concentrations of myoglobin were needed to obtain the same kinetics of growth (data not shown). There was a clear difference in the ability of the HemR-expressing cells to use HSA- and BSA-heme complexes; 50-fold higher concentrations of HSA-heme were needed to obtain the growth observed with BSA-heme, Hb, or heme (Fig. 6).
FIG. 6.
Ability of E. coli EB53 (hemA aroB) cells expressing HemR mutant receptors to use BSA-heme complexes (10 μM BSA and 1 μM heme) (A) and HSA-heme complexes (50 μM heme) as sources of heme. HemRW.T, wild-type HemR; HemRSTOP, pNEO1391. Two to three independent growth measurements were conducted, producing essentially identical results.
Strains expressing HemRH128K and HemRH461L could not grow in the presence of any of the heme compounds (Fig. 6 and data not shown). The phenotypes of the wild-type receptor and HemRH192T were identical, irrespective of the heme source, as were the phenotypes of receptors HemRH352A and HemRH192T,H352K (Fig. 6 and data not shown). The HemRH352K-expressing cells grew slower than the wild-type cells in the presence of 1 μM Hb, 50 μM myoglobin, and 1 μM heme; however, no difference in growth was observed with HSA-heme and BSA-heme as the sources of heme (Fig. 6).
Binding of mutant HemRs to heme- and Hb-agarose.
The abilities of HemRH461L, HemRH128K, and the wild-type receptor to bind heme and Hb were assessed by affinity purification of HemRs from bacterial OMs with heme- and Hb-agarose (9, 26). The wild-type receptor and the HemRH461L, HemRH128K, HemRH352K, HemRH461A, and HemRH128A,H461A mutant receptors were able to bind to heme- and Hb-agarose in very similar amount (Fig. 7 and data not shown). These data suggest that His128 and His461 are not the receptor residues that mediate binding to Hb and heme. Alternatively, HemR possesses several Hb and/or heme binding sites on its surface and the removal of only one site did not grossly affect the ability of the mutant HemR to bind to Hb- and heme-agarose.
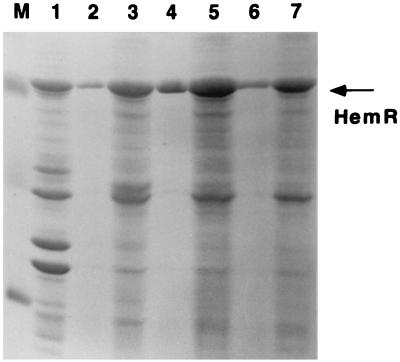
FIG. 7.
Binding of mutant HemRs and wild-type HemR (HemRWT) to heme- and Hb-agarose. Lanes: M, molecular size markers; 1, OMs of E. coli expressing HemRWT; 2, Hb-agarose-bound HemRWT; 3, heme-agarose-bound HemRWT; 4, Hb-agarose-bound HemRH128K; 5, heme-agarose-bound HemRH128K; 6, Hb-agarose-bound HemRH461L; 7, heme-agarose-bound HemRH461L. Differences in the amount of material bound to heme and Hb-agarose are due to the 30-fold-higher binding capacity of the heme- over Hb-agarose.
DISCUSSION
A very large number of microorganisms express heme and Hb receptors which allow bacteria to use heme and heme-protein complexes as the sources of iron and porphyrin (27). Our understanding of the mechanism of action of these proteins and their interactions with different heme-containing compounds is very limited. We show that the expression of the Y. enterocolitica HemR OM receptor enables E. coli to use Hb, myoglobin, heme, HSA-heme, and BSA-heme as sources of iron and porphyrin. The HemR-expressing bacteria also used catalase, haptoglobin-Hb complexes, and hemopexin but were not able to use cytochrome c. Different heme compounds were not used with equal efficiency; 50- to 75-fold-higher levels of myoglobin and HSA over those of Hb, heme, and BSA complexes were required in order to accomplish the same growth stimulation. These results are expected since human myoglobin binds heme with a higher affinity than Hb, and the affinity of human albumin for heme is 1,000 times higher than the affinity of heme for bovine albumin (5, 7).
The results of the assays using microperoxidases implicated the HemR receptor in the transport of heme-peptide conjugates across the OM. Although the transport of heme-peptides has not been demonstrated, dependence of microperoxidase-associated phenotypes on functional TonB and HemR proteins suggests an active transport process across the membrane. HemR is the first bacterial heme receptor that uses such a broad spectrum of heme-containing compounds. Indeed, the phenotypic analysis of another heme or Hb receptor, the HmbR protein of N. meningitidis, showed a very limited spectrum of usable heme sources. These phenotypic differences between the two heme and Hb receptors are in accordance with the relatively moderate level of similarity between their amino acid sequences. Conversely, a very high level of similarity (>65% identity) between HemR and its homologues found in Shigella dysenteriae, enterohemorrhagic E. coli, and Y. pestis suggests that these receptors may also allow use of a wide variety of heme-protein sources. Therefore, both amino acid comparisons and a limited phenotypic analysis indicate the existence of at least two families of heme and Hb receptors: receptors able to use a wide variety of heme sources (heme scavengers) and receptors restricted to one or two heme sources.
What is the significance of these findings for organisms that express heme and Hb receptors? It can be argued that the ability to scavenge heme from a variety of protein complexes may be advantageous to yersiniae and other members of the family Enterobacteriaceae that colonize several distinct ecological niches. Heme receptors that are less finicky may also allow microorganisms to use and/or recycle heme derivatives such as the following: heme o (2-farnesylethyl heme b) found in bacterial cytochrome complexes; heme a (8-formyl heme o) found in terminal oxidase in animal cells and bacteria; heme d (5,6-bishydroxy heme b) found in bacterial cytochrome and E. coli catalase; heme d1 which is widely present in denitrifying bacteria; and siroheme (heme b with saturated porphyrin ring), a prosthetic group of nitrite and sulfite reductases (3). Recycling of these valuable cofactors may have been the first metabolic role of heme receptors. On the other hand, highly host-adapted pathogens like Haemophilus and neisseriae, “specialized” their heme and Hb receptors to a particular heme source that is encountered in the host. These data should not be interpreted as indicating that the HemR design is superior to the HmbR design. Quite to the contrary, HmbR-expressing bacteria may be more efficient than HemR-expressing bacteria when confronted with Hb as the sole heme substrate. On the other hand, the design of HemR probably did not need any fine adjustments to fit a specific heme-protein complex because yersiniae satisfy their iron needs by producing a very powerful siderophore (16).
What is the mechanism of HemR’s function and how does HemR interact with different heme-protein complexes? The HemR receptor must be able to bind heme-protein complexes on its surface. The receptor either possesses several discrete binding sites for each heme-protein complex or has only one site that recognizes a common motif in all heme-protein complexes. After initial binding of a heme-protein complex, the receptor must release heme from the complex and transport heme into the bacterial periplasm. Both steps should involve receptor residues that specifically recognize the heme molecule. Histidines are common axial ligands of heme-iron in proteins, and it is likely that these residues play an important role in HemR function (7, 39). Indeed, site-directed mutagenesis of conserved His residues of HemR confirmed the involvement of His128 and His461 in heme utilization. The HemR His461 mutants were more severely affected in function than the HemRH128K receptor which retained some residual activity. Different phenotypes of these hemR alleles may be caused by a particular amino acid substitution. Alternatively, these results may suggest that the His128 and His461 function at different steps of the heme utilization process. However, these functionally important histidine residues are not essential for Hb binding, since HemRH128K, HemRH461L, HemRH461A, and HemRH128A,H461A mutants were still able to bind to Hb-agarose.
The His128 residue is localized in the part of HemR that is conserved among all TonB-dependent receptors (21, 31). The same region of FepA was shown to be essential for enterobactin uptake by both deletion and linker insertion analyses (2, 37). Similar analysis of homologous segments of the FhuA and BtuB receptors confirmed the importance of the amino-terminal region in receptor function (22, 23, 25). Recent crystallization studies of FhuA and FepA receptors revealed the role of the amino-terminal cork-like domain in receptor function: these domains close the receptor pore from the periplasmic side and participate in the formation of a substrate binding site (4, 10, 30). The location of His128 suggests that this residue participates in the formation of the heme binding site. Indeed, His128 aligns very well with Tyr116 of FhuA (Fig. 3B) which forms one of the apices of the amino-terminal cork domain and participates in the formation of the ferrichrome binding pocket (10, 30).
Comparison of HemR with siderophore receptors showed that the domain containing the histidine residue 461 is not well conserved in siderophore receptors but extremely well conserved among heme and Hb receptors. The whole domain is most likely specific for heme and Hb receptors. Since both the use of free and complexed heme is grossly affected in strains expressing HemRH461L and HemRH461A, His461 is probably a part of a surface-exposed, high-affinity heme binding site. This residue may participate in removal of heme from different protein complexes and in “catching” free heme molecules on the receptor surface. The ability of the HemRH461L mutant to bind heme agarose does not contradict the role of His461 in heme binding. As suggested for the ferrichrome receptor, HemR most likely possesses several weak ligand binding sites on its surface (30). These sites “concentrate” heme close to His461, improving the efficacy of the receptor. Low-affinity heme binding sites might be responsible for interactions between HemR and heme-agarose.
We postulate that two HemR histidine residues, H128 and H461, interact with the heme molecule which is reminiscent of myoglobin and Hb heme pockets where two histidines hold heme through interaction with the central iron atom (7). However, considering that the function of HemR is not to hold heme but to transport it into the cell, it seems more feasible that the heme molecule is bound to only one histidine residue at a time, thus making the dissociation of heme from the receptor more favorable (41). Conformational changes that occur upon ligand binding and after recognition of the loaded receptor by TonB (19, 24a, 29, 34, 35) would be responsible for altering the affinities of H128 and H461 in such a way as to allow transfer of heme first from H461 to H128 and then from H128 into the periplasm.
ACKNOWLEDGMENTS
Charles S. Bracken and Michael T. Baer contributed equally to this work.
We thank N. Srinivasan for excellent technical assistance. We thank E. Hansen and A. Smith for the gift of human hemopexin and R. Perry for rabbit hemopexin. We thank G. Churchward, K. Hantke, C. Moran, and W. Shafer for very helpful suggestions and interest in our work.
I.S. acknowledges financial support from NSF MCB 9728215 and NIH AI472870-01A1 grants.
REFERENCES
- 1.Aisen P, Leibman A, Zweier J. Stoichiometric and site characteristics of the binding of iron to human transferrin. J Biol Chem. 1978;253:1930–1937. [PubMed] [Google Scholar]
- 2.Armstrong S K, Francis C L, McIntosh M A. Molecular analysis of the Escherichia coli ferric enterobactin receptor FepA. J Biol Chem. 1990;265:14536–14543. [PubMed] [Google Scholar]
- 3.Beale S I. Biosynthesis of hemes. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 731–748. [Google Scholar]
- 4.Buchanan S K, Smith B S, Vankatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, Van der Helm D, Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 5.Bunn H F, Jandl J H. Exchange of heme among hemoglobins and between globin and albumin. J Biol Chem. 1968;243:465–475. [PubMed] [Google Scholar]
- 6.Cope L D, Yogev R, Muller-Eberhard U, Hansen E J. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickerson R E, Geis I. Hemoglobin: structure, function, evolution, and pathology. Menlo Park, Calif: Benjamin/Cummings Pub. Co.; 1983. Hemoglobin structure and function; pp. 19–65. [Google Scholar]
- 8.Dyer D W, West E P, Sparling P F. Effects of serum carrier proteins on the growth of pathogenic neisseriae with heme-bound iron. Infect Immun. 1987;55:2171–2175. doi: 10.1128/iai.55.9.2171-2175.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkins C, Chen C-J, Thomas C E. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect Immun. 1995;63:2194–2200. doi: 10.1128/iai.63.6.2194-2200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson A D, Hofmann E, Coulton J W, Diederichs K, Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2203. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 11.Fujimura S, Hirai K, Shibata Y, Nakayama K, Nakamura T. Comparative properties of envelope-associated arginine-gingpains and lysine-gingpain of Porphyromonas ginigivalis. FEMS Microbiol Lett. 1998;163:173–179. doi: 10.1111/j.1574-6968.1998.tb13042.x. [DOI] [PubMed] [Google Scholar]
- 12.Ghigo J M, Letoffe S, Wandersman C. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanson M S, Pelzel S E, Latimer J, Muller-Eberhard U, Hensen E J. Identification of a genetic locus of Haemophilus influenzae type b necessary for the binding and utilization of heme bound to human hemopexin. Proc Natl Acad Sci USA. 1992;89:1973–1977. doi: 10.1073/pnas.89.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 15.Harbury H A, Loach P A. Oxidation-linked proton functions in heme octa- and undecapeptides from mammalian cytochrome c. J Biol Chem. 1960;235:3640–3645. [PubMed] [Google Scholar]
- 16.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 17.Henderson D P, Payne S M. Characterization of the Vibrio cholerae outer membrane transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol. 1994;176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornung J M, Jones H A, Perry R D. The hmu locus of Yersinia pestis is essential for utilization of free hemin and heme-protein complexes as iron sources. Mol Microbiol. 1996;20:725–739. doi: 10.1111/j.1365-2958.1996.tb02512.x. [DOI] [PubMed] [Google Scholar]
- 19.Jiang X, Payne M A, Cao Z, Foster S B, Feix J B, Newton S M, Klebba P E. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science. 1997;276:1261–1264. doi: 10.1126/science.276.5316.1261. [DOI] [PubMed] [Google Scholar]
- 20.Jin H, Ren Z, Pozsgay J M, Elkins C, Whitby P W, Morton D J, Stull T L. Cloning of a DNA fragment encoding a heme-repressible hemoglobin-binding outer membrane protein from Haemophilus influenzae. Infect Immun. 1996;64:3134–3141. doi: 10.1128/iai.64.8.3134-3141.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadner R J. Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol Microbiol. 1990;4:2027–2033. doi: 10.1111/j.1365-2958.1990.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 22.Killmann H, Benz R, Braun V. Properties of the FhuA channel in the Escherichia coli outer membrane after deletion of FhuA portions within and outside the predicted gating loop. J Bacteriol. 1996;178:6913–6920. doi: 10.1128/jb.178.23.6913-6920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koebnik R, Braun V. Insertion derivatives containing segments of up to 16 amino acids identify surface- and periplasm-exposed regions of FhuA outer membrane receptor of Escherichia coli K-12. J Bacteriol. 1993;175:826–839. doi: 10.1128/jb.175.3.826-839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24a.Larsen R A, Thomas M G, Postle K. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol Microbiol. 1999;31:1809–1824. doi: 10.1046/j.1365-2958.1999.01317.x. [DOI] [PubMed] [Google Scholar]
- 25.Lathrop J T, Wei B-Y, Touchie G A, Kadner R J. Sequences of the Escherichia coli BtuB protein essential for its insertion and function in the outer membrane. J Bacteriol. 1995;177:6810–6819. doi: 10.1128/jb.177.23.6810-6819.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee B C. Isolation of hemein-binding proteins of Neisseria gonorrhoeae. J Med Microbiol. 1992;36:121–127. doi: 10.1099/00222615-36-2-121. [DOI] [PubMed] [Google Scholar]
- 27.Lee B C. Quelling the red menace: heme capture by bacteria. Mol Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- 28.Letoffe S, Ghigo J M, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Rutz J M, Klebba P E, Feix J B. A site-directed spin-labeling study of ligand-induced conformational change in the ferric enterobactin receptor, FepA. Biochemistry. 1994;33:13274–13283. doi: 10.1021/bi00249a014. [DOI] [PubMed] [Google Scholar]
- 30.Locher K P, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch J P, Moras D. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 31.Lundrigan M D, Kadner R J. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. J Biol Chem. 1986;261:10797–10801. [PubMed] [Google Scholar]
- 32.Maciver I, Latimer J L, Liem H H, Muller-Eberhard U, Hrkal Z, Hansen E J. Identification of an outer membrane protein involved in utilization of hemoglobin-haptoglobin complexes by nontypeable Haemophilus influenzae. Infect Immun. 1996;64:3703–3712. doi: 10.1128/iai.64.9.3703-3712.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matzanke B F, Berner I, Bill E, Trautwein A X, Winkelmann G. Transport and utilization of ferrioxamine-E-bound iron in Erwinia herbicola (Pantoea agglomerans) Biol Metals. 1991;4:181–185. doi: 10.1007/BF01141312. [DOI] [PubMed] [Google Scholar]
- 34.Moeck G S, Tawa P, Xiang H, Ismail A A, Turnbull J L, Coulton J W. Ligand-induced conformational change in the ferrichrome-iron receptor of Escherichia coli K-12. Mol Microbiol. 1996;22:459–471. doi: 10.1046/j.1365-2958.1996.00112.x. [DOI] [PubMed] [Google Scholar]
- 35.Moeck G S, Coulton J W, Postle K. Cell envelope signaling in Escherichia coli. Ligand binding to the ferrichrome-iron receptor FhuA promotes interaction with the energy-transducing protein TonB. J Biol Chem. 1997;272:28391–28397. doi: 10.1074/jbc.272.45.28391. [DOI] [PubMed] [Google Scholar]
- 36.Morton D J, Whitby P W, Jin H, Ren Z, Stull T L. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect Immun. 1998;67:2729–2739. doi: 10.1128/iai.67.6.2729-2739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newton S M, Igo J D, Scott D C, Klebba P E. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol Microbiol. 1999;32:1153–1165. doi: 10.1046/j.1365-2958.1999.01424.x. [DOI] [PubMed] [Google Scholar]
- 38.Nishina Y, Miyoshi S, Nagase A, Shinoda S. Significant role of an exocellular protease in utilization of heme by Vibrio vulnificus. Infect Immun. 1992;60:2128–2132. doi: 10.1128/iai.60.5.2128-2132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Halloran T V. Transition metals in control of gene expression. Science. 1993;261:715–725. doi: 10.1126/science.8342038. [DOI] [PubMed] [Google Scholar]
- 40.Perry R D, Brubaker R R. Accumulation of iron by yersiniae. J Bacteriol. 1979;137:1290–1298. doi: 10.1128/jb.137.3.1290-1298.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postle K. Active transport by customized β-barrels. Nat Struct Biol. 1999;6:3–6. doi: 10.1038/4869. [DOI] [PubMed] [Google Scholar]
- 42.Schirmer T, Cowan S W. Prediction of membrane-spanning β-strands and its application to maltoporin. Protein Sci. 1993;2:1361–1363. doi: 10.1002/pro.5560020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in Gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stojiljkovic I, Hantke K. Transport of hemin across the cytoplasmic membrane through a hemin-specific periplasmic binding protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 45.Stojiljkovic I, Hwa V, de Martin S L, Nassif X, Heffron F, So M. The Neisseria meningitidis hemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–540. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 46.Stojiljkovic I, Larson J, Hwa V, Anic S, So M. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structural conservation. J Bacteriol. 1996;178:4670–4678. doi: 10.1128/jb.178.15.4670-4678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stojiljkovic I, Srinivasan N. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in neisseriae. J Bacteriol. 1997;179:805–812. doi: 10.1128/jb.179.3.805-812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stull T L. Protein sources of heme for Haemophilus influenzae. Infect Immun. 1987;55:148–153. doi: 10.1128/iai.55.1.148-153.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:127–157. [PubMed] [Google Scholar]