Summary
Background
Pyrimidine nucleotides fuel the growth of colorectal cancer (CRC), making their associated proteins potential targets for cancer intervention. Uridine-Cytidine Kinase Like-1(UCKL1) is an enzyme involved in the pyrimidine salvage pathway. It is highly expressed in multiple cancers. But the function and underlying mechanism of UCKL1 in CRC are yet to study.
Methods
Large-scale genomic analysis was performed to search for potential CRC players related to pyrimidine metabolism. The function of UCKL1 in CRC were examined by RNA interference coupled with in vitro and in vivo assays. GSH/GSSG assay, NADP+ assay, ROS, and Lipid peroxidation assays were performed to check the function of UCKL1 in ferroptosis. Metabolomics analyses, RNA sequencing, western blotting, and rescue assays were done to reveal the underlying mechanisms of UCKL1. Xenograft mouse model was used to examine the therapeutic potential of UCKL1 as a target in combination with other ferroptosis inducers.
Findings
UCKL1 was identified to repress ferroptosis in CRC cells. It was highly expressed in CRC. It regulated CRC cells proliferation and migration. Downregulation of UCKL1 led to enhanced tumour lipid peroxidation. Intriguingly, UCKL1 reduction-mediated ferroptosis was not related to its role in catalyzing uridine monophosphate (UMP) and cytidine monophosphate (CMP) synthesis. Instead, UCKL1 stabilized Nrf2, which in turn promoted the expression of SLC7A11, a classical repressor of ferroptosis. Moreover, downregulation of UCKL1 sensitized CRC cells to GPX4 inhibitors in vitro and in vivo.
Interpretation
Our study demonstrates that UCKL1 plays a non-canonical role in repressing ferroptosis through a UCKL1-Nrf2-SLC7A11 axis in CRC cells. Combinatorial strategy in targeting ferroptosis by depletion of UCKL1 and application of GPX4 inhibitors may serve as a new effective method for CRC treatment.
Funding
This study was supported in part by fund from National Natural Science Foundation of China (Grant No. 31970674 to PY), by the Basic and Applied Basic Research Program of Guangdong Province (Grant No. 2023A1515030245 to KL), by the program of Guangdong Provincial Clinical Research Center for Digestive Diseases (2020B1111170004), and by National Key Clinical Discipline.
Keywords: Colorectal cancer (CRC), Ferroptosis, UCKL1, SLC7A11, Nrf2
Research in context.
Evidence before this study
UCKL1 is an enzyme involved in the pyrimidine salvage pathway. Previous studies have showed that UCKL1 is associated with malignant phenotype of prostate cancer, breast cancer and acute lymphoblastic leukaemia. However, the role and underling mechanism of UCKL1 in CRC is undetermined. Ferroptosis is a form of cell death induced by excessive lipid peroxidation, and ferroptosis induction is critical for tumour suppression. The relationship between UCKL1 and ferroptosis is unknown.
Added value of this study
We found that UCKL1 is amplified in CRC and correlated with poor prognosis. UCKL1 knockdown diminishes CRC cells growth through induction of ferroptosis. Interestingly, UCKL1 reduction-mediated ferroptosis is not related to its role in pyrimidine metabolism. Instead, UCKL1 stabilizes Nrf2, which in turn promotes the expression of SLC7A11. Moreover, downregulation of UCKL1 sensitizes CRC cells to GPX4 inhibitors in vitro and in vivo.
Implications of all the available evidence
Our study demonstrates that UCKL1 has a non-canonical role in ferroptosis defence in CRC. The ablation of UCKL1 inhibits CRC cells growth by inducing ferroptosis through Nrf2-SLC7A11 axis rather than pyrimidine metabolism. Targeting UCKL1 may be a new effective strategy for CRC treatment.
Introduction
Colorectal cancer (CRC) is one of the most common cancers in the world, resulting in more than 900,000 death a year.1 Many patients cannot benefit from existing standard therapies. New drug targets for CRC are urgently needed.
The crazy growth of CRC requires increased nucleotide biosynthesis to provide cell proliferation materials.2 Hence, nucleotide metabolism related proteins may be potential targets for CRC treatment. Nucleotide synthesis includes de novo synthesis and salvage synthesis. The de novo synthesis utilizes small molecules such as amino acids and ribose phosphate to synthesize nucleotides. It is the main way to synthesize nucleotides in proliferating cells.3,4 The salvage synthesis generates nucleotides rapidly by using free bases and nucleosides produced by cells after nucleic acid decomposition through simple reactions.5 The salvage synthesis is an energy and material saving pathway.6 Recently, accumulating evidence revealed a close link between pyrimidine synthesis and cancer.7, 8, 9
UCKL1, as an enzyme in pyrimidine salvage pathway, can phosphorylate uridine and cytidine into UMP and CMP respectively. It was reported that UCKL1 was associated with prostate cancer, breast cancer and leukaemia.10, 11, 12 Downregulation of UCKL1 inhibited leukaemia cell proliferation and enhanced its sensitivity to natural killer cells.12,13 However, the role of UCKL1 in CRC remains unclarified.
Ferroptosis is a form of cell death that is induced by excessive lipid peroxidation.14 It has been reported in a variety of human diseases, including cancer, kidney failure, and neurodegenerative diseases.15, 16, 17 Ferroptosis is characterized by the iron-dependent accumulation of cellular reactive oxygen species (ROS) and lipid peroxidation on cell membranes.14,18 Substantial progress has been made in understanding the role of ferroptosis in tumour biology and cancer therapy. Accumulating evidence indicates that ferroptosis induction is critical for tumour suppression. Many oncogenes and tumour suppressor genes, including p53, AMPK, mTOR and BRCA1-associated protein1 (BAP1),19, 20, 21, 22 are important regulators of ferroptosis. Ferroptosis is triggered by polyunsaturated fatty acid-containing phospholipids (PUFA-PL) synthesis and peroxidation,23,24 iron metabolism25 and mitochondrial metabolism.26 However, cancer cells have also developed anti-ferroptosis mechanisms for survival, such as SLC7A11-GSH-GPX4 system,27, 28, 29 FSP1-CoQH2 system,30,31 DHODH-CoQH2 system32 and the GCH1-BH4 system.33,34 In particular, the SLC7A11-GSH-GPX4 regulatory system plays an important role. SLC7A11, a unit of the cystine-glutamate antiporter system Xc- which transports cystine into the cytoplasm, is essential for GSH synthesis.35 In conjunction with GPX4, GSH detoxifies lipid peroxides and suppresses ferroptosis. Inactivation of SLC7A11 suppresses tumour growth.36,37 Radiotherapy and immunotherapy induce ferroptosis partly through repressing the expression of SLC7A11 too.38 In addition, inactivating SLC7A11 by ferroptosis inducers alleviate radioresistance and chemoresistance in cancer cells.39,40 Moreover, inhibition of GPX4, an enzyme essential for the clearance of lipid ROS, induces ferroptosis even when cellular GSH and cysteine levels are normal.29 However, some cancer cells are resistant to GPX4 inactivation-induced ferroptosis and the mechanism is yet to be fully investigated.41 Theoretically, application of the ferroptosis inducers or ablation of anti-ferroptosis key genes in cancer cells are both promising strategies for cancer treatment and deserved of detailed exploration.
Utilizing the CRC database, we examined the association between CRC and genes that are involved in pyrimidine pathways. We found that UCKL1 is highly expressed in CRC. Further in vitro and in vivo studies revealed that UCKL1 represses ferroptosis in CRC. Intriguingly, the anti-ferroptosis role of UCKL1 is independent of the UCKL1 metabolic role. Instead, UCKL1 stabilizes transcription factor Nrf2, which in turn promotes SLC7A11 expression, leading to the ferroptosis repression. Knockdown of UCKL1 also greatly enhanced GPX4 inhibitor induced CRC ferroptosis and tumour regression, suggesting that UCKL1 is a therapeutic target for the treatment of cancer which resists GPX4 inactivation-induced ferroptosis.
Methods
Cell lines, culture and reagents
All the cells, including 293T, DLD1, RKO, LOVO, SW480, SW620, HCT8, HCT116, HCT15, and HT29 were obtained from ATCC and confirmed free from mycoplasma. DLD1, RKO, HCT116, and HCT8 cells were cultured in RPMI 1640 (Corning, USA) supplemented with 10% (v/v) fetal bovine serum (FBS) (Sigma–Aldrich, USA) at 37 °C and 5% CO2. Other cells were maintained in complete Dulbecco’s modified Eagle’s medium (Corning, USA) supplemented with 10% FBS at 37 °C and 5% CO2.
Patients and tissue samples
Three cohorts of patients’ tissues were obtained for the study. 83 primary CRC tissues and 34 adjacent normal colon tissues for cohort 1, and 99 primary CRC tissues and 77 adjacent normal colon tissues for cohort 3 were collected from the Sixth Affiliated Hospital, Sun Yat-sen University for research under ethical approval 2022ZSLYEC-515. Cohort 2 contains 15 pairs of primary CRC tissues and adjacent normal colon tissues. It was obtained from Shanghai Outdo Biotechnology Company for research under ethical approval SHXC2021YF01. Informed consent was obtained from all patients prior to the collection of samples. The sex was not taken into account in the design of the study. The sex data was self-reported by study participants. The demographic data of all three cohorts are shown in Supplementary Table S1. The microarrays of the above samples were stained with immunohistochemistry and scanned with the Aperio Versa (Leica Biosystems). The staining intensity was quantified using Image J and the expression of UCKL1 was evaluated using the z-score. Mann–Whitney U test or Unpaired two-tailed Student’s t-test was used to compare the results.
Bioinformatics analysis
Cancer genomic analyses including mutations, copy-number, mRNA expression and protein level of the pyrimidine pathway related genes in CRC were performed using the dataset of colorectal adenocarcinoma (TCGA, Firehose Legacy) in the cBioPortal for Cancer Genomics (http://www.cbioportal.org/). The alteration frequency of UCKL1, UCK1, and UCK2 in CRC were also analysed with cBioPortal using datasets: Colorectal Adenocarcinoma (TCGA, Firehose Legacy); Colon Adenocarcinoma (CPTAC-2 Prospective, Cell 2019)42; Colorectal Adenocarcinoma (TCGA, PanCancer Atlas); Colorectal Adenocarcinoma (TCGA, Nature 2012)43 and Colorectal Adenocarcinoma (Genentech, Nature 2012).44 The expression of UCKL1, UCK1, and UCK2 in colon adenocarcinoma (TCGA) were downloaded from the University of Alabama at Birmingham Cancer data analysis Portal (http://ualcan.path.uab.edu). Datasets GSE20916 and GSE8671 were used to analyse the UCKL1 mRNA level in colon adenocarcinoma or colorectal adenoma. Kaplan–Meier disease free survival curve based on the expression level of Nrf2 or SLC7A11 in the CRC tissue from GSE106584 was downloaded from R2 Genomics Analysis and Visualization Platform.
Plasmid constructs
To generate the shRNA constructs, the following sequences were used: UCKL1 shRNA #1: CGCACACAACAACTTCAACTT.
UCKL1 shRNA #2: GCCCATTTATGACTTCACCAC.
UCK1 shRNA #1: CGGAGCTACAAGCGGACCTTT.
UCK1 shRNA #2: CGGTCACATTTGGAGTCCAGC.
UCK2 shRNA #1: GATAGCTTCTACCGTGTCCTT.
UCK2 shRNA #2: GCAGACCAATGGCTGTCTCAA.
These shRNAs were integrated into the pLKO-Tet-On vector. UCKL1, SLC7A11, and Nrf2 cDNA were cloned into PCDH or pcDNA3.1 vector. All constructs were checked by DNA sequencing.
RNA interference
Doxycycline inducible shRNA expressing lentivirus was generated following the standard protocol of Lipofectamine 2000 Transfection Reagent Kit (ThermoFisher, cat# 11668030). Briefly, 1 × 107 293T cells were co-transfected with 10 μg shRNA, 5 μg psPAX2, and 5 μg pMD2.G. Viral particles were collected from the supernatant and filtered with Millex-GP Filter Unit. Subsequently, cancer cells were infected with lentivirus in culture medium supplied with 10 μg/ml polybrene (Sigma, cat# TR-1003) for 24 h. Infected cells were selected with puromycin for 14 days to establish inducible shRNA transduced cell lines. 100 ng/ml doxycycline (Meilunbio, cat# MB1088-1) was added to the culture medium to induce shRNA expression for the following assays.
Colony formation assay
1000 cells were plated in each well of 6-well plates. After treatment under experimental conditions for 14 days, cells were fixed with methanol and stained with 1% crystal violet (Solarbio, cat# G1062). The number of colonies was counted by Image J software.
Western blotting assay
Cells or tissues were lysed using lysis buffer. The protein concentration was determined by BCA protein quantitative Kit (BestBio, cat# BB-3401-250T). Cell lysates with equal amounts of protein were separated by SDS–PAGE gel, transferred to a PVDF transfer membrane and immunoblotted with specific antibodies as indicated. Primary antibodies against UCKL1 (Proteintech, cat# 17005-1-AP, RRID: AB_2210783, 1:1000 for WB), SLC7A11 (Proteintech, cat# 26864-1-AP, RRID: AB_2880661, 1:1000 for WB), Nrf2 (Proteintech, cat# 16396-1-AP, RRID: AB_2782956, 1:1000 for WB), β-actin (Santa Cruz, cat# sc-47778, RRID: AB_626632, 1:10000 for WB), Flag (Sigma–Aldrich, cat# F1804, RRID: AB_262044, 1:2000 for WB), HA (Proteintech, cat# 51064-2-AP, RRID: AB_11042321, 1:5000 for WB) were used. Proper secondary antibodies conjugated with horseradish peroxidase (HRP) were applied accordingly. ChemiDoc imaging system (Bio-rad) was used to visualize the results.
Wound healing assay
Cancer cells were seeded into a 12-well plate that with an ibidi culture insert. The insert was removed after 24 h, and cells were washed with PBS and cultured in 2% FBS supplemented medium. Cells were photographed at 0 and 48 h. Wound closure (%) was assessed using the Image J software.
Real-time qPCR
Total RNA was extracted from cells or tissue samples using TRIzol reagent (Invitrogen, cat# 15596-026) according to the manufacturer’s instructions. RNA samples were reverse transcribed to cDNA using a cDNA synthesis kit (TransGen, cat# AE301-03). RNA sequencing was performed and analysed by GENEWIZ (Suzhou, China). Real-time qPCR analysis was carried out using an ABI QuantStudio 7 Flex with SYBR kit (TransGen, cat# AQ601-02). Three biological repeats for each sample were tested for each assay. Relative mRNA analysis was performed using the 2−ΔΔCt method. The primers used for real-time qPCR were listed in Supplementary Table S2.
Cell viability assay
For cell viability assay, cells were cultured in 96-well plates and treated with or without doxycycline, ferrostatin-1 (Sigma–Aldrich, cat# SML0583), necrostatin-1s (APExBIO, cat# A4213), Z-VAD-FMK (APExBIO, cat# A1902), N-acetyl-L-cysteine (Sigma–Aldrich, cat# 616-91-1), RSL3 (Aladdin, cat# R302648), or ML162 (Sigma–Aldrich, cat# SML2561). Subsequently, cells were exposed to medium containing 10% CCK8 for 1 h at 5% CO2 in a 37 °C incubator. The absorbance at a wavelength of 450 nm was determined by microplate reader. Three biological repeats for each sample were tested for each assay.
Lipid peroxidation and ROS assay
Cells were seeded on 6-well plates and incubated overnight. The next day, cells were treated at the indicated conditions. Fresh medium containing 5 μM BODIPY 581/591 C11 dye (Invitrogen, cat# D3861) or 10 μM DCFH-DA (Beyotime, cat# S0033M) was added to each well. BODIPY 581/591 C11 dye was used for the lipid peroxidation assay and DCFH-DA for ROS measurement. After incubation for 30 min at 37 °C, the cells were trypsinized and resuspended in PBS. ROS and lipid peroxidation levels were analysed by flow cytometry (Beckman).
Metabolomics assay
UCKL1 shRNA transduced RKO cells were seeded on 15-cm culture plates and treated with or without doxycycline. Cells were harvested at a density of about 80% confluence. Cells were washed twice with PBS and extracted with a mixture of methanol, acetonitrile and water (2:2:1, v/v/v) on ice. After rapid freezing in liquid nitrogen, samples were sent to Shanghai Applied Protein Technology for metabolomics assay. Multiple reaction monitoring (MRM) analysis of 200 metabolites in the samples was conducted using HILIC LC-MS/MS targeted method. Raw data of metabolites were extracted with MRM Analyzer to obtain the peak area of each metabolite.
GSH and GSSG assay
GSH and GSSG were measured using GSH/GSSG assay kit (Beyotime, cat# S0053) following the manufacturer’s instructions. The concentrations of GSH and GSSG were calculated from the standard curve.
NADP+/NADPH assay
NADP+/NAPDH assay kit (Beyotime, cat# S0179) was used to determine the ratio of NADP+ and NADPH. The assays were performed following the manufacturer’s instructions.
Determination of Fe2+
Iron assay kit (MesGen, cat# MIK4893) was used to measure intracellular Fe2+ levels. Assays were performed according to the manufacturer’s instructions. The relative amount of Fe2+ was calculated based on absorbance at 520 nm.
Enzymatic assays
The enzyme activity of UCKL1-wt and UCKL1-mut (aa132-aa136 were deleted) were determined by ADP-Glo™ Kinase Assay kit from Promega (Promega, cat# V6930). Cells transfected with UCKL1-wt, UCKL1-mut or empty plasmids were lysed to determine enzyme activity according to the manufacturer’s instructions.
Ubiquitination assay
Briefly, 293T cells were co-transfected with the indicated plasmids. After 48 h of transfection, cells were treated with 20 μM MG132 for 6 h and then lysed for immunoprecipitation by anti-Flag antibody. Immunoprecipitated proteins were analysed by Western blot using anti-HA antibody.
Xenograft CRC model
This study was approved by the Animal Ethical and Welfare Committee of the Sixth Affiliated Hospital, Sun Yat-sen University (IACUC-2020042801 and IACUC-2022070301). 4–6 weeks female BALB/c-nu/nu mice (about 20 g) were obtained from Guangdong GemPharmatech Co., Ltd (Guangdong, China). 5 × 106 RKO or HCT116 cells transduced with doxycycline-inducible UCKL1 shRNA were injected subcutaneously into mice to establish a xenograft model. 5 days after inoculation, mice were randomly divided into different groups. Drinking water containing 2 mg/ml doxycycline was used to induce UCKL1 knockdown. For the RSL3 treatment, 50 mg/kg RSL3 were injected intratumourally twice per week. Tumour size was measured every 3 days using a digital calliper and calculated according to the equation volume = length × width2 × 1/2. At the end of the experiment, the mice were sacrificed by CO2 suffocation. Tumours were harvested for analysis.
Immunohistochemistry
The expression of SLC7A11, UCKL1, 4-HNE, and Ki-67 in tumours were characterized by IHC. Detailed protocol is as previously described.45 Briefly, tumour sections were dewaxed in xylene, hydrated in descending concentrations of ethanol and immersed in 0.3% H2O2–methanol for 30 min. Subsequently, the samples were washed with PBS and probed with anti-UCKL1 (Proteintech, cat# 17005-1-AP, RRID: AB_2210783, 1:300), anti-SLC7A11 (Proteintech, cat# 26864-1-AP, RRID: AB_2880661, 1:300), anti-4-HNE (Abcam, cat# ab46545, RRID: AB_722490, 1:400) or anti-Ki-67 (Abcam, cat# ab15580, 1:500) antibodies at 4 °C overnight. Afterwards, the sections were incubated with biotinylated goat anti-rabbit or anti-mouse IgG at room temperature for 2 h. Immunostaining was visualized with streptavidin/peroxidase complex and diaminobenzidine, and sections were counterstained with haematoxylin. Immunohistochemistry slides were scanned with The Aperio Versa (Leica Biosystems).
Statistical analysis
All statistical analyses were performed using SPSS16.0 and GraphPad Prism 8.0 software. The data are presented as the means ± SD. Student’s t-test or Mann–Whitney U test was performed to compare means between two groups, and one-way ANOVA was used for multigroup comparisons. Paired samples were compared using a paired student’s t-test. Two-way ANOVA was used for comparison of differences between growth curves. Overall survival was evaluated by the Kaplan–Meier survival analysis and the log-rank test. Significance was defined as p < 0.05.
Role of funders
The funders were not involved in the study design, data collection, data analyses, interpretation and writing of report.
Results
UCKL1 is amplified in CRC and correlated with poor prognosis
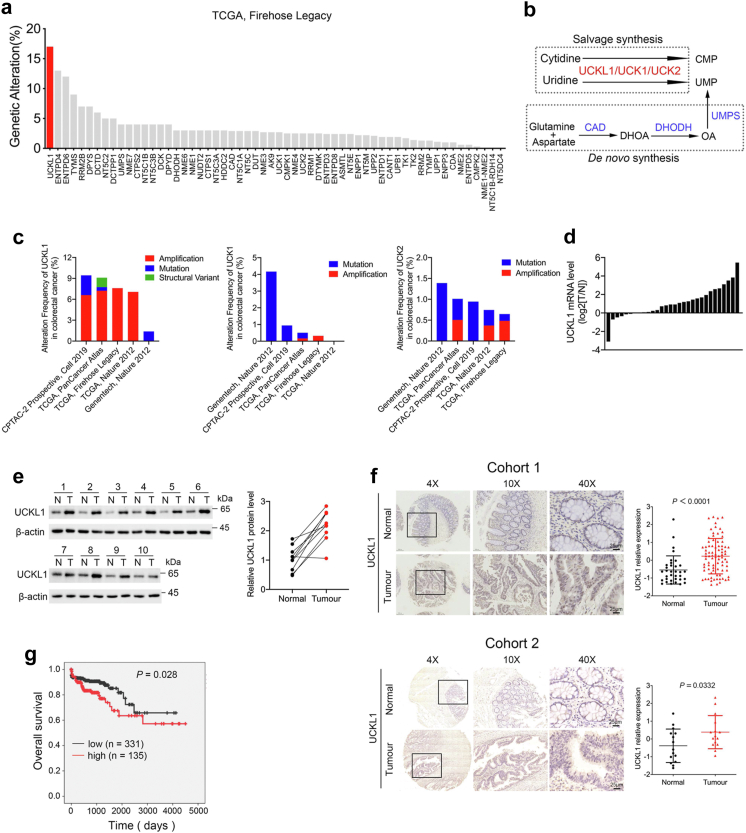
By using cBioPortal, we conducted large-scale genomic analyses in colorectal adenocarcinoma (TCGA, firehose legacy). It turned out that UCKL1 showed about 17% genetic alteration, the highest alteration frequency among pyrimidine metabolism genes in human CRC cases (Fig. 1a and Supplementary Fig. S1a). UCKL1 is a member of the Uridine-cytidine kinase (UCK) protein family, alongside UCK1 and UCK2. These proteins phosphorylate uridine and cytidine into UMP and CMP6 (Fig. 1b). Using cBioPortal, we analysed UCK1, UCK2 and UCKL1 profiles in multiple CRC datasets and confirmed that only UCKL1 is significantly amplified (Fig. 1c). Further analysis of mRNA expression from TCGA by UALCAN website revealed that UCKL1 is elevated in CRC, while the expression changes of UCK1 and UCK2 in CRC and normal tissues are relatively small (Supplementary Fig. S2a–d). In addition, UCKL1 expression level is higher in colon cancer tissues than in normal tissues in GEO cohorts GSE 20916 and GSE 8671 (Supplementary Fig. S2e and f).
Fig. 1.
UCKL1 is upregulated in CRC and associated with poor prognosis. (a) Genomic alterations of pyrimidine metabolism genes in CRC database (TCGA, Firehose Legacy) analysed by cBioPortal for Cancer Genomics. (b) Schematic map of pyrimidine synthesis pathway. (c) Genomic alterations of UCKL1, UCK1 and UCK2 in CRC database analysed by cBioPortal for Cancer Genomics. (d) Waterfall plot of the relative UCKL1 mRNA expression levels from 30 paired samples of CRC and adjacent normal tissue. N, adjacent normal tissue; T, tumour tissue. (e) UCKL1 protein level was detected from paired samples of CRC and adjacent normal tissue. N, adjacent normal tissue; T, tumour tissue. (f) Representative images of UCKL1 IHC staining in human CRC and adjacent normal tissue in cohort 1 and cohort 2. Scale bars: 25 μm. Image J was used to quantify the intensity of the staining, and z-score was calculated to evaluate the expression of UCKL1. Mann–Whitney U test and Unpaired two-tailed Student’s t-test were used to compare the results. (g) Kaplan–Meier survival curves based on UCKL1 expression in the CRC tissue from TCGA database.
To validate the database analysis, 30 paired CRC samples were examined for UCKL1 expression. Real-time qPCR assay revealed that UCKL1 mRNA levels were generally higher in CRC tissues than in adjacent normal tissues (Fig. 1d). As compared to adjacent normal tissues, the protein level of UCKL1 was upregulated in CRC tissues too (Fig. 1e). Meanwhile, IHC staining of tissue microarray assay (TMA) from three patient cohorts revealed that UCKL1 was more abundant in CRC tissues than adjacent normal tissues (Fig. 1f and Supplementary Fig. S2g). As highly expressed in CRC tissues, the protein level of UCKL1 was also increased in CRC cell lines (Supplementary Fig. S2h). Furthermore, Kaplan–Meier analysis with TCGA database showed that high UCKL1 level was correlated with poor overall survival in CRC (Fig. 1g). All these data suggest that UCKL1 is associated with CRC and may function as an oncogene.
UCKL1 is required for cell survival in CRC
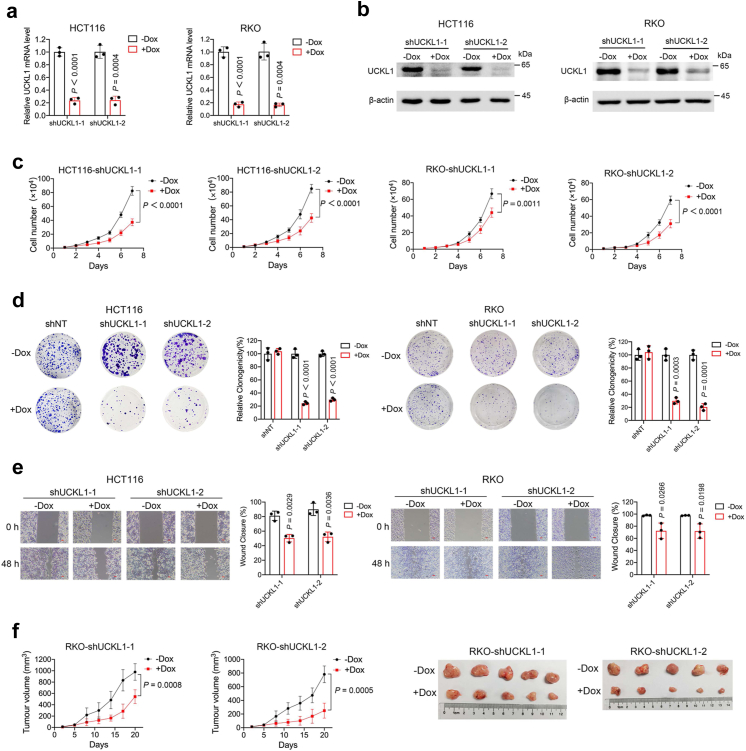
To check whether UCKL1 is an oncogene, with lentivirus-based tet-on systems, we knocked down UCKL1 expression using two short hairpin RNAs (shRNAs) that target two different loci of UCKL1. UCKL1 mRNA and protein were significantly downregulated by doxycycline induced expression of UCKL1 shRNA-1 and shRNA-2 in HCT116 cells and RKO cells (Fig. 2a and b). Meanwhile, viable cell numbers were greatly reduced (Fig. 2c). To ensure that the observation was not due to off-target effect of shRNA, we performed shRNA rescue assay by overexpressing shRNA resistant UCKL1 cDNA in UCKL1 knockdown HCT116 cells. Western blot revealed that UCKL1 shRNA depleted UCKL1 protein was efficiently rescued by expressing shRNA resistant UCKL1 cDNA (Supplementary Fig. S3a). As a result, overexpression of UCKL1 reversed the cell viability inhibition by UCKL1 knockdown in HCT116 cells (Supplementary Fig. S3b), confirming the specificity and effect of UCKL1 shRNA. Moreover, UCKL1 knockdown inhibited colony formation and cell migration (Fig. 2d and e). Hence, UCKL1 is required for the malignant behaviour of CRC cells.
Fig. 2.
UCKL1 knockdown diminishes CRC cells growth. (a) Real-time qPCR results of UCKL1 mRNA level in doxycycline-inducible UCKL1 shRNA transduced HCT116 or RKO cells treated with or without doxycycline. Unpaired two-tailed Student’s t-test was used to compare the results. (b) Western blot results of UCKL1 protein level in doxycycline-inducible UCKL1 shRNA transduced HCT116 or RKO cells treated with or without doxycycline. (c) Cell viability of doxycycline-inducible UCKL1 shRNA transduced HCT116 or RKO cells treated with or without doxycycline. The data are represented as the means ± SD. Two-way ANOVA was used to compare the results. (d) Colony formation rates of doxycycline-inducible UCKL1 shRNA transduced HCT116 or RKO cells treated with or without doxycycline. Quantification of the colony formation rates are shown in right panel. The data are represented as the means ± SD. Unpaired two-tailed Student’s t-test was used to compare the results. (e) Wound healing results of doxycycline-inducible UCKL1 shRNA transduced HCT116 or RKO cells treated with or without doxycycline. Scale bars: 50 μm. Quantitative analyses are shown in right panel. The data are represented as the means ± SD. Unpaired two-tailed Student’s t-test was used to compare the results. (f) The efficacy of UCKL1 knockdown on tumour growth was examined by subcutaneous injection of RKO cells into nude mice. UCKL1 knockdown mice were fed with water containing doxycycline (2 mg/ml), while control mice were just fed with water. Tumour growth curves are shown in the left panel. Tumour volume = length × width2 × 1/2. The data are represented as the means ± SD (n = 5). Two-way ANOVA was used for comparison of differences between growth curves. Representative images of tumours are shown in right panel.
We next asked whether other UCK family members UCK1 and UCK2 function similarly as UCKL1 in CRC cells. Using lentivirus-based RNA interference system abovementioned, we knocked down UCK1 and UCK2 expression in RKO cells (Supplementary Fig. S4a and c). However, knocking down either UCK1 or UCK2 did not reduce RKO cell viability (Supplementary Fig. S4b and d). Tumour cell colony formation capacity was not affected neither (Supplementary Fig. S4e and f). These data indicate that UCKL1, but not UCK1 and UCK2, plays a critical role in CRC cells.
To further understand UCKL1 function in vivo, we transplanted doxycycline-induced UCKL1 shRNA transduced RKO cells subcutaneously into immunodeficient mice. Xenograft tumour growth was evidently decreased by doxycycline induction (Fig. 2f). At the end of the experiment, tumour weight in the doxycycline treated group showed a significant reduction compared to the control group (Supplementary Fig. S5a). Doxycycline treatment reduced the mRNA and protein levels of UCKL1 in tumours (Supplementary Fig. S5b and c). Further immunohistochemistry assay also showed a reduction of UCKL1 and Ki-67 in doxycycline treated mouse tumour (Supplementary Fig. S5d). Together, these results indicate that UCKL1 is required for CRC growth, and that knockdown of UCKL1 leads to CRC cell death.
Downregulation of UCKL1 reduces viable CRC cells through induction of ferroptosis
Next, we set to explore what kind of cell death may be mediated by UCKL1 depletion in CRC cells. To address this, we supplied apoptosis inhibitor Z-VAD-FMK (Z-VAD), the necroptosis inhibitor necrostatin-1 (Nec-1), the ferroptosis inhibitor ferrostatin-1 (Fer-1) and the ROS scavenger N-acetyl-L-cysteine (NAC) into the culture medium of UCKL1 knockdown cells. Fer-1 and NAC partially reversed UCKL1 knockdown-induced cell viability inhibition, but Z-VAD and Nec-1 did not (Fig. 3a and Supplementary Fig. S6a). Hence, UCKL1 knockdown-induced cell death in CRC cells is likely to be ferroptosis. To substantiate this, we further examined ferroptosis related cell properties. Congruently, UCKL1 knockdown induced lipid peroxidation in HCT116 and RKO cells (Fig. 3b and Supplementary Fig. S6b). DCFH-DA staining revealed increased total ROS in UCKL1 knockdown cells (Fig. 3c and Supplementary Fig. S6c). The knockdown of UCKL1 also increased lipid peroxidation and ROS in SW620 and LOVO cells (Supplementary Fig. S7a–f). In keeping with the results, both GSSG/GSH ratio and NADP+/NADPH ratio were increased in UCKL1 knockdown cells (Fig. 3d and e and Supplementary Fig. S6d and e), which reflected the state of oxidative stress in cells. Notably, GSH showed a significant reduction as well (Fig. 3d and Supplementary Fig. S6d). The level of Fe2+, another indicator of ferroptosis, was increased in UCKL1 knockdown cells (Fig. 3f and Supplementary Fig. S6f). Transmission electron microscopy further revealed that UCKL1 knockdown cells exhibited mitochondrial abnormalities, like shrunken, enhanced membrane density and absent crista, the typical morphologic features of ferroptosis (Fig. 3g and Supplementary Fig. S6g). Conversely, overexpression of UCKL1 prevented lipid peroxidation caused by ferroptosis inducers (Fig. 3h–j). Interestingly, according to the TCGA database, UCKL1 expression level was positively correlated with the expression of GPX4 and FSP1, two master anti-ferroptosis proteins (Fig. 3k). It should be noted that neither UCK1 nor UCK2 knockdown could induce ROS and lipid peroxidation in CRC cells (Supplementary Fig. S8a–d), demonstrating a specific role of UCKL1 in ferroptosis.
Fig. 3.
UCKL1 knockdown induces ferroptosis. (a) Cell viability results of doxycycline induced UCKL1 knockdown HCT116 cells versus control cells after 2 μM Nec-1, 5 μM Z-VAD, 5 μM fer-1, 2 mM NAC, or DMSO treatment. One-way ANOVA was used to compare the results. (b) Flow cytometry analysis of C11-BODIPY staining cells to determine lipid peroxidation level in doxycycline induced UCKL1 knockdown HCT116 cells versus control cells. The lipid peroxidation level of indicated cells was shown in bar graph. Unpaired two-tailed Student’s t-test was used to compare the results. (c) Flow cytometry analysis of DCFH-DA staining cells to determine cellular total ROS level in doxycycline-induced UCKL1 knockdown HCT116 cells versus control cells. The fold change of ROS level of indicated cells was shown in bar graph. Unpaired two-tailed Student’s t-test was used to compare the results. (d) Relative GSH, GSSG, and ratios of GSSG and GSH in doxycycline-induced UCKL1 knockdown HCT116 cells versus control cells. Unpaired two-tailed Student’s t-test was used to compare the results. (e) Relative NADP+/NADPH ratios in doxycycline-induced UCKL1 knockdown HCT116 cells versus control cells. Unpaired two-tailed Student’s t-test was used to compare the results. (f) Relative Fe2+ in UCKL1 knockdown HCT116 cells versus control cells. Unpaired two-tailed Student’s t-test was used to compare the results. (g) Transmission electron microscopy images of doxycycline-induced UCKL1 knockdown HCT116 cells and control cells. Red arrows point to mitochondria. Scale bars: left, 5 μm; middle, 2 μm; right, 500 nm. (h) UCKL1 protein levels were determined by Western blot in control and UCKL1 overexpression DLD1 cells. (i) Lipid peroxidation level was determined in control and UCKL1 overexpression cells after treatment with 30 μM erastin for 48 h. One-way ANOVA was used to compare the results. (j) Lipid peroxidation level was determined in control and UCKL1 overexpression cells after treatment with 5 μM RSL3 for 48 h. (k) Correlation analysis of UCKL1 with GPX4 and FSP1 in TCGA.
UCKL1 knockdown-induced ferroptosis is independent of UMP/CMP
Next, we asked how UCKL1 is involved in ferroptosis. As UCKL1 is a key enzyme in the pyrimidine salvage pathway, we postulated that UCKL1 knockdown may cause scarcity of UMP and CMP, leading to ferroptosis in cancer cells.
To test this postulation, we performed metabolomics analyses of nucleotides in UCKL1 knockdown cells. Unexpectedly, no significant decrease of the pyrimidine metabolites was detected in UCKL1 knockdown RKO cells (Fig. 4a and b). In agreement with the metabolomics analysis results, supplement of UMP and CMP could not restore the inhibition of cell viability and colony formation caused by UCKL1 knockdown (Fig. 4c–e and Supplementary Fig. S9a–c). UMP and CMP treatment could not reverse the increased lipid peroxidation and ROS induced by UCKL1 knockdown either (Fig. 4f and g and Supplementary Fig. S9d and e). In order to confirm that UCKL1 regulates ferroptosis independent of pyrimidine metabolism, we overexpressed a UCKL1 mutant which the uridine binding motif (aa132-aa136)46,47 was deleted in DLD1 cells. UCKL1 mutant showed a reduced ability to catalyze uridine compared to wild type UCKL1 (Fig. 4h and i). However, it could still decrease erastin induced lipid peroxidation and ROS increase (Fig. 4j and k). Collectively, these observations suggested a non-canonical function of UCKL1 in repressing ferroptosis in CRC.
Fig. 4.
UCKL1 mediated ferroptosis regulation is independent of UMP/CMP. (a) The result of metabolomics in UCKL1 knockdown RKO cells. Blue dots represent reduced metabolites and red dots indicate increased metabolites. Grey dots represent substances of no significant change. UMP and CMP are presented with black dots. (b) LC-MS-based metabolomics results of indicated nucleoside in doxycycline induced UCKL1 knockdown RKO cells versus control cells. Unpaired two-tailed Student’s t-test was used to compare the results. (c) Cell viabilities of inducible UCKL1 shRNA transduced HCT116 cells with or without doxycycline and UMP treatment. The concentrations of UMP are 1, 10 or 100 μM. One-way ANOVA was used to compare the results. NS, not significant. (d) Cell viabilities of inducible UCKL1 shRNA transduced HCT116 cells with or without doxycycline and CMP treatment. The concentrations of CMP are 1, 10, or 100 μM. One-way ANOVA was used to compare the results. NS, not significant. (e) Colony formations of inducible UCKL1 shRNA transduced HCT116 cells with or without doxycycline, UMP and CMP treatment. The concentrations of UMP or CMP are 1, 10, or 100 μM. (f) Flow cytometry analysis of C11-BODIPY staining cells to resolve lipid peroxidation level in doxycycline-induced UCKL1 knockdown HCT116 cells and control cells after UMP or CMP treatment for 3 days. The lipid peroxidation level of indicated cells was shown in bar graph. One-way ANOVA was used to compare the results. NS, not significant. (g) Flow cytometry analysis of DCFH-DA staining cells to resolve ROS level in doxycycline-induced UCKL1 knockdown HCT116 cells and control cells after UMP or CMP treatment for 3 days. The fold change of ROS level of indicated cells was shown in bar graph. One-way ANOVA was used to compare the results. NS, not significant. (h) The expression of UCKL1-wt and UCKL1-mut (uridine binding domain aa132-aa136 was deleted) were determined by Western blot. (i) Luminescence of the indicated cell lysates determined by ADP-Glo™ Kinase Assay, with uridine as the substrate. One-way ANOVA was used to compare the results. (j) Lipid peroxidation level measured by flow cytometry analysis of C11-BODIPY staining at the indicated conditions. Erastin, 30 μM for 48 h. One-way ANOVA was used to compare the results. (k) ROS level determined by flow cytometry analysis of DCFH-DA staining at the indicated conditions. Erastin, 30 μM for 48 h. One-way ANOVA was used to compare the results.
UCKL1 reduction-mediated ferroptosis is SLC7A11-dependent
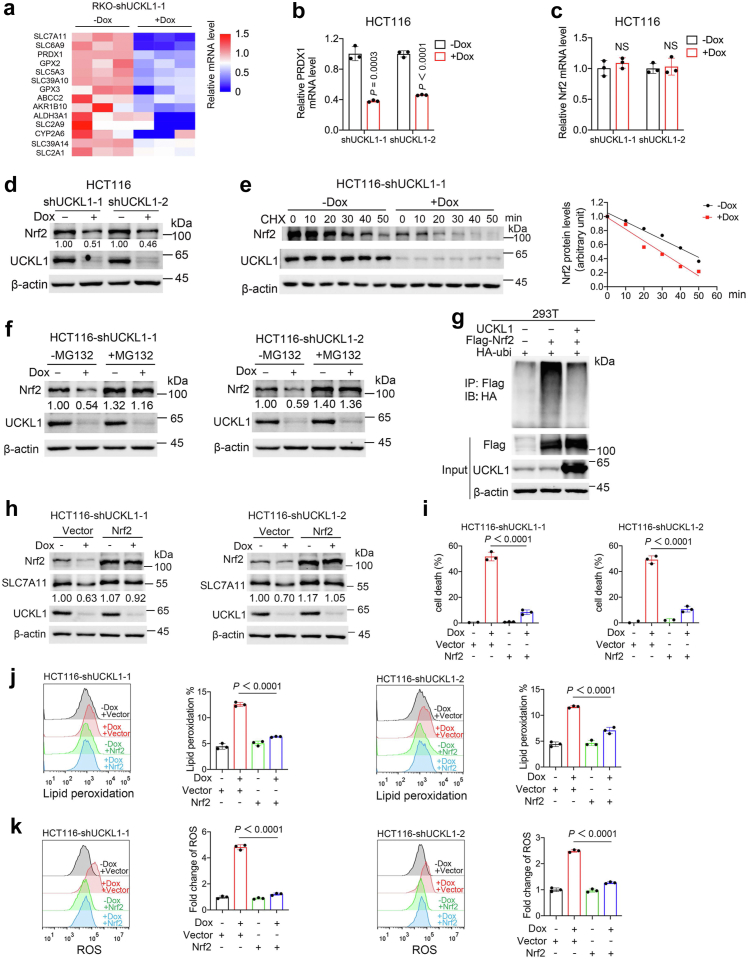
To understand how UCKL1 inhibits ferroptosis, we performed RNA sequencing assay to compare the transcriptome of the control and UCKL1 knockdown cells. SLC7A11, a key gene in ferroptosis defence, was the top gene downregulated in UCKL1 knockdown cells (Fig. 5a), suggesting that it may be a downstream effector of UCKL1. To clarify the impact of UCKL1 on SLC7A11, we checked the expression of SLC7A11 in UCKL1 knockdown CRC cells. Both SLC7A11 mRNA and protein expression were significantly downregulated upon UCKL1 knockdown (Fig. 5b and c and Supplementary Fig. S10a and b), confirming the positive correlation between UCKL1 and SLC7A11. GSH, the downstream product of SLC7A11, was reduced in UCKL1 knockdown cells too (Fig. 3d and Supplementary Fig. S6d). Considering the role of SLC7A11 in ferroptosis defence, we hypothesized that UCKL1 knockdown induces ferroptosis through SLC7A11 ablation. We tested this hypothesis by using lentivirus-mediated enforced expression of SLC7A11 in UCKL1 knockdown cells. As expected, SLC7A11 overexpression significantly alleviated the cell viability inhibition induced by UCKL1 knockdown in CRC cells (Fig. 5d and e and Supplementary Fig. S10c and d). Meanwhile, UCKL1 knockdown-induced lipid peroxidation and ROS was reversed by SLC7A11 overexpression (Fig. 5f and g and Supplementary Fig. S10e and f). Additionally, gene set enrichment analysis (GSEA) of differentially expressed genes revealed no enrichment in pyrimidine metabolism pathways, but instead showed enrichment in pathways related to lipids (Fig. 5h). This finding reinforces the connection between UCKL1 and ferroptosis which is highly associated with lipid metabolism. Furthermore, UCKL1's involvement in ferroptosis is independent of pyrimidine metabolism.
Fig. 5.
UCKL1 knockdown reduces the expression of SLC7A11. (a) Volcano plot of differentially expressed genes (DEG, identified by fold change >2 and q-value < 0.05) in doxycycline induced UCKL1 knockdown RKO cells versus control cells. Among the DEGs, SLC7A11 is the most significantly downregulated gene in UCKL1 knockdown RKO cells. (b) Real-time qPCR result of SLC7A11 mRNA expression level in inducible UCKL1 shRNA transduced HCT116 cells with or without doxycycline treatment. Unpaired two-tailed Student’s t-test was used to compare the results. (c) Western blot result of SLC7A11 protein level in doxycycline-inducible UCKL1 shRNA transduced HCT116 cells treated with or without doxycycline. (d) Western blot results of SLC7A11 protein level in doxycycline induced UCKL1 knockdown HCT116 cells versus control cells after SLC7A11 overexpression. (e) Overexpression of SLC7A11 reversed UCKL1 knockdown-induced cell death in HCT116 cells. One-way ANOVA was used to compare the results. (f) Flow cytometry analysis of C11-BODIPY staining revealed that overexpression of SLC7A11 inhibited lipid peroxidation in UCKL1 knockdown HCT116 cells. Bar graph shows lipid peroxidation level in the indicated cells. One-way ANOVA was used to compare the results. (g) Flow cytometry analysis of DCFH-DA staining revealed that overexpression of SLC7A11 rescued total ROS in UCKL1 knockdown HCT116 cells. Bar graph shows fold change of ROS level in indicated cells. One-way ANOVA was used to compare the results. (h) GSEA revealed that UCKL1 knockdown was associated with lipid storage (right) but not the pyrimidine metabolic process (left and middle).
Together, our data suggest that UCKL1 promotes ferroptosis evasion through enhancing SLC7A11 mediated lipid peroxidation scavenging.
UCKL1 promotes SLC7A11 expression through stabilizing Nrf2
We next explored how UCKL1 regulates SCL7A11. By analysing RNA sequencing data (Fig. 5a), we noticed that multiple Nrf2 targets, such as PRDX1 and GPX2, were decreased in UCKL1 knockdown RKO cells (Fig. 6a). Nrf2 is a well-known antioxidant transcriptional factor,48 we then speculated that UCKL1 regulates SLC7A11 via Nrf2. To test the speculation, we validated the RNA sequencing results. Indeed, mRNA of PRDX1 was downregulated in UCKL1 knockdown HCT116 cells and RKO cells (Fig. 6b and Supplementary Fig. S11a). However, UCKL1 knockdown did not affect Nrf2 mRNA expression (Fig. 6c and Supplementary Fig. S11b). Instead, Nrf2 protein level was obviously decreased after UCKL1 knockdown (Fig. 6d and Supplementary Fig. S11c). These observations suggested that UCKL1 might regulate Nrf2 at the protein level. Thus, we performed protein stability assay by supplying cycloheximide (CHX), the protein synthesis inhibitor, to the experimental cells. It turned out that UCKL1 knockdown increased Nrf2 turnover rate (Fig. 6e and Supplementary Fig. S11d). On the other hand, addition of MG132, a classic proteasome inhibitor, recovered UCKL1 knockdown caused Nrf2 downregulation (Fig. 6f and Supplementary Fig. S11e), suggesting that UCKL1 may be involved in the proteasome mediated Nrf2 degradation. Consistent with this, ubiquitin of Nrf2 was reduced with UCKL1 expression (Fig. 6g). According to previous studies, Kelch-like ECH-associated protein 1 (Keap1), a substrate of E3 ubiquitin ligase, binds to the ETGE and DLG motifs of Nrf2 and promotes its ubiquitination.49,50 We further confirmed that Nrf2 ubiquitination regulation by UCKL1 was abolished in Nrf2 DLG deleted mutant and ETGE deleted mutant (Supplementary Fig. S11f). This suggests that UCKL1 stabilizes Nrf2 in a Keap1-dependent manner.
Fig. 6.
UCKL1 stabilizes Nrf2 to inhibit ferroptosis in CRC cells. (a) Heatmap of the expression of Nrf2 target genes determined by RNA sequencing of control and UCKL1 knockdown RKO cells. (b) Real-time qPCR result of PRDX1 mRNA expression level in inducible UCKL1 shRNA transduced HCT116 cells with or without doxycycline treatment. Unpaired two-tailed Student’s t-test was used to compare the result. (c) Real-time qPCR result of Nrf2 mRNA expression level in inducible UCKL1 shRNA transduced HCT116 cells with or without doxycycline treatment. Unpaired two-tailed Student’s t-test was used to compare the results. NS, not significant. (d) Western blot result of Nrf2 protein level in inducible UCKL1 shRNA transduced HCT116 cells with or without doxycycline treatment. (e) Western blot analysis of the Nrf2 protein turnover rate in the control and UCKL1 knockdown HCT116 cells after treated with cycloheximide (100 μg/ml). (f) Western blot analysis of Nrf2 expression in the control and UCKL1 knockdown HCT116 cells treated with MG132 (20 μM) for 6 h. (g) Western blot analysis for HA-ubiquitin after incubation of anti-Flag-coupled beads with lysates from 293T cells transfected with the indicated constructs. (h) Western blot analysis of SLC7A11 expression after overexpression of Nrf2 in the control and UCKL1 knockdown HCT116 cells. (i) Overexpression of Nrf2 suppressed UCKL1 knockdown-induced cell death in HCT116 cells. One-way ANOVA was used to compare the results. (j) Overexpression of Nrf2 reversed UCKL1 knockdown-induced lipid peroxidation increment in HCT116 cells. The lipid peroxidation level measured by C11-BODIPY staining in the indicated cells was shown in bar graph. One-way ANOVA was used to compare the results. (k) Overexpression of Nrf2 reversed UCKL1 knockdown-induced total ROS increment in HCT116 cells. The fold change of ROS level measured by DCFH-DA staining in the indicated cells was shown in bar graph. One-way ANOVA was used to compare the results.
To further examine whether Nrf2 is a key mediator in UCKL1 regulation of SLC7A11, we overexpressed Nrf2 in UCKL1 knockdown HCT116 and RKO cells. Western blot revealed that UCKL1 knockdown caused SLC7A11 downregulation was reversed by Nrf2 overexpression (Fig. 6h and Supplementary Fig. S11g). Moreover, Nrf2 overexpression abrogated cell death induced by UCKL1 knockdown (Fig. 6i and Supplementary Fig. S11h). UCKL1 knockdown-induced lipid peroxidation and ROS were also rescued by Nrf2 expression (Fig. 6j and k and Supplementary Fig. S11i and j). Furthermore, high levels of Nrf2 and SLC7A11 are associated with poor prognosis in CRC (Supplementary Fig. S12a and b). Together, our data indicate the existence of a UCKL1-Nrf2-SLC7A11 ferroptosis defence axis in CRC.
UCKL1 knockdown enhanced the efficacy of GPX4 inhibitor in CRC suppression
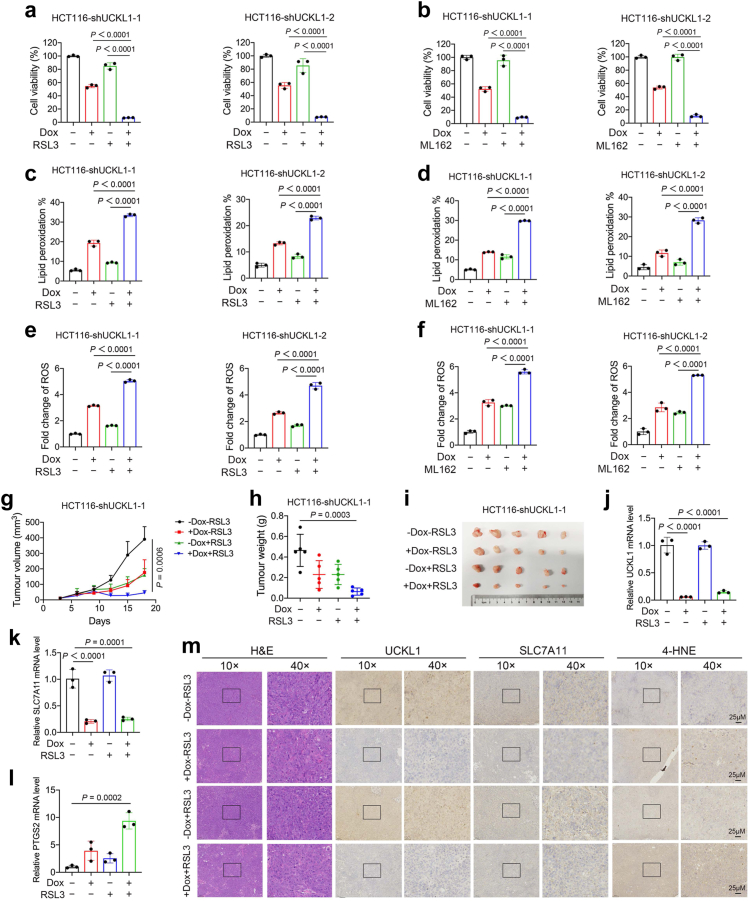
Due to UCKL1's involvement in SLC711 regulation and the crucial role of SLC7A11-GSH-GPX4 system in ferroptosis defence, knockdown of UCKL1 may enhance the antitumour efficacy of GPX4 inhibitors. To substantiate this, GPX4 inhibitor, RSL3 or ML162, was supplied at low concentration of 2 μM or 5 μM to doxycycline induced UCKL1 shRNA transduced HCT116 cells and RKO cells respectively. Without doxycycline induced UCKL1 shRNA expression, about 80∼100% CRC cells were viable upon RSL3 or ML162 treatment alone. But adding RSL3 or ML162 after doxycycline induced UCKL1 shRNA expression reduced cell viability to less than 20% (Fig. 7a and b and Supplementary Fig. S13a and b). The combinatory inhibitory effect suggests that UCKL1 knockdown sensitizes CRC cells to RSL3 or ML162. Furthermore, UCKL1 knockdown combined with RSL3 or ML162 induced more robust increase of lipid peroxidation and ROS in cancer cells than mono-treatment (Fig. 7c–f and Supplementary Fig. S13c–f), strengthening the sensitizer role of UCKL1 knockdown in ferroptosis inducer treatment in vitro.
Fig. 7.
Impeding UCKL1-SLC7A11 axis synergizes with GPX4 inhibitor in ferroptosis induction and tumour growth suppression. (a) Cell viabilities of HCT116 cells treated with UCKL1 knockdown or/and RSL3. For the combined treatment, cancer cells were pretreated with doxycycline for 2 days, and then treated with 2 μM RSL3 for 24 h. One-way ANOVA was used to compare the results. (b) Cell viabilities of HCT116 cells treated with UCKL1 knockdown or/and ML162. For the combined treatment, cancer cells were pretreated with doxycycline for 2 days, and then treated with 5 μM ML162 for 24 h. One-way ANOVA was used to compare the results. (c) Lipid peroxidation level of UCKL1 shRNA transduced HCT116 cells treated with doxycycline or/and RSL3. Cancer cells were pretreated with doxycycline for 3 days, and then treated with 5 μM RSL3 for 4 h. One-way ANOVA was used to compare the results. (d) Lipid peroxidation level of UCKL1 shRNA transduced HCT116 cells treated with doxycycline or/and ML162. Cancer cells were pretreated with doxycycline for 3 days, and then treated with 10 μM ML162 for 4 h. One-way ANOVA was used to compare the results. (e) Relative ROS level of UCKL1 shRNA transduced HCT116 cells treated with doxycycline or/and RSL3. Cancer cells were pretreated with doxycycline for 3 days, and then treated with 5 μM RSL3 for 4 h. One-way ANOVA was used to compare the results. (f) Relative ROS level of UCKL1 shRNA transduced HCT116 cells treated with doxycycline or/and ML162. Cancer cells were pretreated with doxycycline for 3 days, and then treated with 10 μM ML162 for 4 h. One-way ANOVA was used to compare the results. (g) Volume of HCT116 xenografts treated with UCKL1 knockdown or/and RSL3 (50 mg/kg) at different time points (n = 5). Drinking water containing doxycycline (2 mg/ml) was used to induce UCKL1 knockdown. RSL3 was injected intratumourally twice per week. Tumour size was measured every 3 days and calculated according to the equation volume = length × width2 × 1/2. Two-way ANOVA was used for comparison of differences between growth curves. (h) Tumour weights of HCT116 xenografts treated with doxycycline or/and RSL3 at the end point. One-way ANOVA was used to compare the result. (i) Representative images of HCT116 xenografts treated with doxycycline or/and RSL3 at the end point. (j) Representative real-time qPCR result of UCKL1 mRNA level in one of the tumour tissues of indicated mouse group. One-way ANOVA was used to compare the result. (k) Representative real-time qPCR result of SLC7A11 mRNA level in one of the tumour tissues of indicated group. One-way ANOVA was used to compare the result. (l) Representative real-time qPCR result of PTGS2 mRNA level in one of the tumour tissues of indicated group. One-way ANOVA was used to compare the result. (m) Representative images of haematoxylin and eosin staining and IHC staining of UCKL1, SLC7A11, and 4-HNE in the tumour tissues of indicated group. Scale bar: 25 μm.
Next, we tested their efficacy in vivo. Downregulation of UCKL1 or supplement of RSL3 alone was efficient in repressing tumour growth, while combined treatment showed even greater tumour repression efficacy (Fig. 7g–i and Supplementary Fig. S13g–i). Further analyses of tumour samples revealed that mRNA levels of SLC7A11 and UCKL1 were downregulated, while ferroptosis marker PTGS2 was upregulated in UCKL1 knockdown and RSL3 combined treatment samples (Fig. 7j–l). Immunohistochemistry assay showed that combined treatment led to reduction of SLC7A11 and induction of ferroptosis marker 4-HNE (Fig. 7m). Together, our data support that downregulation of UCKL1 enable CRC cells to be more sensitive to GPX4 inhibitors, contributing a potential therapeutical strategy in the future.
Discussion
The pyrimidine nucleotide metabolic pathway is closely linked to the development of cancer.51 Drugs that target pyrimidine de novo synthesis, such as 5-fluorouracil and capecitabine, are commonly used in CRC therapy.52 However, drug resistance can occur, partially due to the existence of redundant pyrimidine synthesis ways. Therefore, it is worthwhile to investigate other targets in pyrimidine metabolism for cancer treatment.6 By bioinformatics analysis of pyrimidine metabolism pathway genes, we identified UCKL1 as the most abnormal gene in CRC. UCKL1 is highly expressed in CRC and associated with poor prognosis in CRC patients.
Our subsequent in vitro and in vivo studies confirmed that UCKL1 is an oncogene for CRC. But intriguingly, the inhibition of CRC cells by UCKL1 knockdown does nothing with its pyrimidine synthesis role, as the levels of UMP and CMP in CRC cells remained unchanged after UCKL1 knockdown. As cancer cells need a stable pool of nucleotides to survive, the reduction in pyrimidine caused by UCKL1 knockdown could be compensated via feedback regulation of other pyrimidine synthesis enzymes. This often occurs in nature. For example, carbamyl phosphate synthetase II (CPSII), a key enzymes in the de novo pathway, is regulated by negative feedback from UTP.53 Chemotherapy agents can reprogram the pyrimidine de novo pathway to increase nucleotide synthesis.54 Moreover, UCK1 and UCK2 remain effective in UCKL1 knockdown cells. They have similar or even higher catalytic efficiency than UCKL1.12 Hence, it is not surprising that knockdown of UCKL1 alone did not alter the levels of UMP and CMP in CRC cells.
During our investigation into why UCKL1 knockdown reduces CRC cell viability, we discovered the ferroptosis defence role of UCKL1 in CRC. This distinguishes UCKL1 from other UCK family proteins. Our research indicates that UCKL1 promotes SLC7A11 expression through Nrf2. Subsequently, through the SLC7A11-GSH-GPX4 pathway, it protects CRC cells from ferroptosis. Nrf2 is an important transcription factor that activates several genes related to antioxidants, including SLC7A11.55 Previous research suggested that Nrf2 is primarily regulated at the protein level by Keap1.49 When Nrf2 interacts with Keap1, it is ubiquitinated and degraded by the proteasome.50 However, the interaction between Nrf2 and Keap1 can be interfered by other proteins. For instance, p21 competes with Keap1 for Nrf2 binding, leading to impaired ubiquitination.56 Additionally, iASPP can substitute Nrf2 in binding with Keap1 and stabilize Nrf2.57 In our study, we found that the regulation of Nrf2 by UCKL1 is also Keap1 dependent. If the Keap1 binding motif in Nrf2 is mutated, the effect of UCKL1 on Nrf2's protein stability is eliminated.
Last but not least, our study demonstrated that the downregulation of UCKL1 greatly enhanced the sensitivity and efficacy of GPX4 inhibitors, which may facilitate their usage in CRC treatment. Due to the increasingly recognized role of ferroptosis defence in cancer therapy resistance,38,39,41,58,59 UCKL1-based research may shed light to developing new therapeutic strategy for cancers. Additionally, since UCKL1 is expressed at high levels in cancer cells, it could also be a promising target for drug development.
Overall, our study identifies a new CRC progression related gene, UCKL1, which plays a critical role in ferroptosis repression through the UCKL1-Nrf2-SLC7A11 axis (Fig. 8). It may serve as a critical target for CRC therapy in the future.
Fig. 8.
Schematic model depicting that UCKL1 regulates ferroptosis by modulating Nrf2-SLC7A11 axis. UCKL1 stabilizes Nrf2, which in turn promotes the expression of SLC7A11. UCKL1 knockdown reduces the expression of SLC7A11 and results with ferroptosis. Targeting UCKL1 synergizes with GPX4 inhibitors thereby inhibit CRC growth via induction of ferroptosis.
Contributors
Weili Wu, Yingying Zhao, and Baifu Qin performed most of the experiments and analysed the data. Weili Wu, Kai Li, and Yingying Zhao carried out the animal experiments. Xin Jiang and Rong Hu helped design experiments and provided technical assistance. Rui Ma and Chuyue Wang conducted bioinformatics analysis. Mong-Hong Lee and Huanliang Liu provided suggestions for the project. Ping Yuan, Weili Wu, and Kai Li conceived the project, directed the research and co-wrote the manuscript. Weili Wu, Yingying Zhao, and Baifu Qin accessed and verified the data. Ping Yuan was responsible for the decision to submit the manuscript. All authors read and approved the final version of the manuscript.
Data sharing statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. RNA-seq raw data reported in this paper have been deposited in SRA with the accession number PRJNA862633.
Declaration of interests
Weili Wu, Yingying Zhao, Kai Li, and Ping Yuan have filed a patent application relating to the diagnosis, treatment, and prognosis evaluation of UCKL1 in CRC. Other authors declare no competing interests.
Acknowledgments
This study was supported in part by fund from National Natural Science Foundation of China (Grant No. 31970674 to PY), by the Basic and Applied Basic Research Program of Guangdong Province (Grant No. 2023A1515030245 to KL), by the program of Guangdong Provincial Clinical Research Center for Digestive Diseases (2020B1111170004), and by National Key Clinical Discipline.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104650.
Contributor Information
Kai Li, Email: likai39@mail.sysu.edu.cn.
Ping Yuan, Email: yuanp8@mail.sysu.edu.cn.
Appendix A. Supplementary data
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mollick T., Lain S. Modulating pyrimidine ribonucleotide levels for the treatment of cancer. Cancer Metab. 2020;8:12. doi: 10.1186/s40170-020-00218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aird K.M., Zhang R. Nucleotide metabolism, oncogene-induced senescence and cancer. Cancer Lett. 2015;356(2 Pt A):204–210. doi: 10.1016/j.canlet.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loffler M., Fairbanks L.D., Zameitat E., Marinaki A.M., Simmonds H.A. Pyrimidine pathways in health and disease. Trends Mol Med. 2005;11(9):430–437. doi: 10.1016/j.molmed.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Walter M., Herr P. Re-discovery of pyrimidine salvage as target in cancer therapy. Cells. 2022;11(4):739. doi: 10.3390/cells11040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sykes D.B., Kfoury Y.S., Mercier F.E., et al. Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell. 2016;167(1):171–186.e15. doi: 10.1016/j.cell.2016.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajzikova M., Kovarova J., Coelho A.R., et al. Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metabol. 2019;29(2):399–416.e10. doi: 10.1016/j.cmet.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Yang K., Wu Q., et al. Targeting pyrimidine synthesis accentuates molecular therapy response in glioblastoma stem cells. Sci Transl Med. 2019;11(504) doi: 10.1126/scitranslmed.aau4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng W.S., Tao H., Hu E.P., et al. Both genes and lncRNAs can be used as biomarkers of prostate cancer by using high throughput sequencing data. Eur Rev Med Pharmacol Sci. 2014;18(22):3504–3510. [PubMed] [Google Scholar]
- 11.Geiger T., Madden S.F., Gallagher W.M., Cox J., Mann M. Proteomic portrait of human breast cancer progression identifies novel prognostic markers. Cancer Res. 2012;72(9):2428–2439. doi: 10.1158/0008-5472.CAN-11-3711. [DOI] [PubMed] [Google Scholar]
- 12.Matchett E.C., Ambrose E.C., Kornbluth J. Characterization of uridine-cytidine kinase like-1 nucleoside kinase activity and its role in tumor growth. Biochem J. 2022;479(11):1149–1164. doi: 10.1042/BCJ20210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambrose E.C., Kornbluth J. Downregulation of uridine-cytidine kinase like-1 decreases proliferation and enhances tumor susceptibility to lysis by apoptotic agents and natural killer cells. Apoptosis. 2009;14(10):1227–1236. doi: 10.1007/s10495-009-0385-z. [DOI] [PubMed] [Google Scholar]
- 14.Dixon S.J., Lemberg K.M., Lamprecht M.R., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X., Kang R., Kroemer G., Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18(5):280–296. doi: 10.1038/s41571-020-00462-0. [DOI] [PubMed] [Google Scholar]
- 16.Friedmann Angeli J.P., Schneider M., Proneth B., et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichert C.O., de Freitas F.A., Sampaio-Silva J., et al. Ferroptosis mechanisms involved in neurodegenerative diseases. Int J Mol Sci. 2020;21(22):8765. doi: 10.3390/ijms21228765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiernicki B., Dubois H., Tyurina Y.Y., et al. Excessive phospholipid peroxidation distinguishes ferroptosis from other cell death modes including pyroptosis. Cell Death Dis. 2020;11(10):922. doi: 10.1038/s41419-020-03118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang L., Kon N., Li T., et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi J., Zhu J., Wu J., Thompson C.B., Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc Natl Acad Sci U S A. 2020;117(49):31189–31197. doi: 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Shi J., Liu X., et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol. 2018;20(10):1181–1192. doi: 10.1038/s41556-018-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H., Zandkarimi F., Zhang Y., et al. Energy-stress-mediated AMPK activation inhibits ferroptosis. Nat Cell Biol. 2020;22(2):225–234. doi: 10.1038/s41556-020-0461-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kagan V.E., Mao G., Qu F., et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol. 2017;13(1):81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doll S., Proneth B., Tyurina Y.Y., et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13(1):91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol Cell. 2015;59(2):298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan B. Mitochondrial regulation of ferroptosis. J Cell Biol. 2021;220(9) doi: 10.1083/jcb.202105043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishimoto T., Nagano O., Yae T., et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19(3):387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 28.Ji X., Qian J., Rahman S.M.J., et al. xCT (SLC7A11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression. Oncogene. 2018;37(36):5007–5019. doi: 10.1038/s41388-018-0307-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W.S., SriRamaratnam R., Welsch M.E., et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bersuker K., Hendricks J.M., Li Z., et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doll S., Freitas F.P., Shah R., et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575(7784):693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 32.Mao C., Liu X., Zhang Y., et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593(7860):586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraft V.A.N., Bezjian C.T., Pfeiffer S., et al. GTP cyclohydrolase 1/tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent Sci. 2020;6(1):41–53. doi: 10.1021/acscentsci.9b01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soula M., Weber R.A., Zilka O., et al. Metabolic determinants of cancer cell sensitivity to canonical ferroptosis inducers. Nat Chem Biol. 2020;16(12):1351–1360. doi: 10.1038/s41589-020-0613-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koppula P., Zhuang L., Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12(8):599–620. doi: 10.1007/s13238-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghoochani A., Hsu E.C., Aslan M., et al. Ferroptosis inducers are a novel therapeutic approach for advanced prostate cancer. Cancer Res. 2021;81(6):1583–1594. doi: 10.1158/0008-5472.CAN-20-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu K., Li K., Lv J., et al. Suppression of the SLC7A11/glutathione axis causes synthetic lethality in KRAS-mutant lung adenocarcinoma. J Clin Invest. 2020;130(4):1752–1766. doi: 10.1172/JCI124049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lang X., Green M.D., Wang W., et al. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 2019;9(12):1673–1685. doi: 10.1158/2159-8290.CD-19-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei G., Zhang Y., Koppula P., et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30(2):146–162. doi: 10.1038/s41422-019-0263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouyang S., Li H., Lou L., et al. Inhibition of STAT3-ferroptosis negative regulatory axis suppresses tumor growth and alleviates chemoresistance in gastric cancer. Redox Biol. 2022;52 doi: 10.1016/j.redox.2022.102317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viswanathan V.S., Ryan M.J., Dhruv H.D., et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547(7664):453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasaikar S., Huang C., Wang X., et al. Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell. 2019;177(4):1035–1049.e19. doi: 10.1016/j.cell.2019.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seshagiri S., Stawiski E.W., Durinck S., et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li K., Wu J.-L., Qin B., et al. ILF3 is a substrate of SPOP for regulating serine biosynthesis in colorectal cancer. Cell Res. 2019;30:163–178. doi: 10.1038/s41422-019-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomoike F., Nakagawa N., Fukui K., Yano T., Kuramitsu S., Masui R. Indispensable residue for uridine binding in the uridine-cytidine kinase family. Biochem Biophys Rep. 2017;11:93–98. doi: 10.1016/j.bbrep.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki N.N., Koizumi K., Fukushima M., Matsuda A., Inagaki F. Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase. Structure. 2004;12(5):751–764. doi: 10.1016/j.str.2004.02.038. [DOI] [PubMed] [Google Scholar]
- 48.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34(1):21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki T., Yamamoto M. Molecular basis of the Keap1-Nrf2 system. Free Radic Biol Med. 2015;88(Pt B):93–100. doi: 10.1016/j.freeradbiomed.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 50.Jaramillo M.C., Zhang D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27(20):2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma J., Zhong M., Xiong Y., et al. Emerging roles of nucleotide metabolism in cancer development: progress and prospect. Aging (Albany NY) 2021;13(9):13349–13358. doi: 10.18632/aging.202962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luengo A., Gui D.Y., Vander Heiden M.G. Targeting metabolism for cancer therapy. Cell Chem Biol. 2017;24(9):1161–1180. doi: 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin J., Mir H., Khurram M.A., et al. Allosteric regulation of CAD modulates de novo pyrimidine synthesis during the cell cycle. Nat Metab. 2023;5(2):277–293. doi: 10.1038/s42255-023-00735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown K.K., Spinelli J.B., Asara J.M., Toker A. Adaptive reprogramming of de novo pyrimidine synthesis is a metabolic vulnerability in triple-negative breast cancer. Cancer Discov. 2017;7(4):391–399. doi: 10.1158/2159-8290.CD-16-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anandhan A., Dodson M., Schmidlin C.J., Liu P., Zhang D.D. Breakdown of an ironclad defense system: the critical role of NRF2 in mediating ferroptosis. Cell Chem Biol. 2020;27(4):436–447. doi: 10.1016/j.chembiol.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen W., Sun Z., Wang X.J., et al. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34(6):663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ge W., Zhao K., Wang X., et al. iASPP is an antioxidative factor and drives cancer growth and drug resistance by competing with Nrf2 for Keap1 binding. Cancer Cell. 2017;32(5):561–573.e6. doi: 10.1016/j.ccell.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 58.Wang W., Green M., Choi J.E., et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roh J.L., Kim E.H., Jang H.J., Park J.Y., Shin D. Induction of ferroptotic cell death for overcoming cisplatin resistance of head and neck cancer. Cancer Lett. 2016;381(1):96–103. doi: 10.1016/j.canlet.2016.07.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.