Summary
Background
RNA viruses account for many human diseases and pandemic events but are often not targetable by traditional therapeutics modalities. Here, we demonstrate that adeno-associated virus (AAV) -delivered CRISPR-Cas13 directly targets and eliminates the positive-strand EV-A71 RNA virus in cells and infected mice.
Methods
We developed a Cas13gRNAtor bioinformatics pipeline to design CRISPR guide RNAs (gRNAs) that cleave conserved viral sequences across the virus phylogeny and developed an AAV-CRISPR-Cas13 therapeutics using in vitro viral plaque assay and in vivo EV-A71 lethally-infected mouse model.
Findings
We show that treatment with a pool of AAV-CRISPR-Cas13-gRNAs designed using the bioinformatics pipeline effectively blocks viral replication and reduces viral titers in cells by >99.99%. We further demonstrate that AAV-CRISPR-Cas13-gRNAs prophylactically and therapeutically inhibited viral replication in infected mouse tissues and prevented death in a lethally challenged EV-A71-infected mouse model.
Interpretation
Our results show that the bioinformatics pipeline designs efficient CRISPR-Cas13 gRNAs for direct viral RNA targeting to reduce viral loads. Additionally, this new antiviral AAV-CRISPR-Cas13 modality represents an effective direct-acting prophylactic and therapeutic agent against lethal RNA viral infections.
Funding
Agency for Science, Technology and Research (A∗STAR) Assured Research Budget, A∗STAR Central Research Fund UIBR SC18/21-1089UI, A∗STAR Industrial Alignment Fund Pre-Positioning (IAF-PP) grant H17/01/a0/012, MOE Tier 2 2017 (MOE2017-T2-1-078; MOE-T2EP30221-0005), and NUHSRO/2020/050/RO5+5/NUHS-COVID/4.
Keywords: Adeno-associated viral vectors, CRISPR-Cas13d, Bioinformatics, Enterovirus, Prophylactic, Therapeutic
Research in context.
Evidence before this study
Previous in vitro studies investigating the Cas13 family of RNA-guided endonucleases as an antiviral modality showed promising results. CasRx is one of the smallest Cas13 endonuclease enzymes known, making it an attractive candidate to be delivered using AAV vectors along with its gRNAs within the same cargo construct. Furthermore, CasRx exhibits high on-target activity and high specificity compared to other modalities, suggesting that CasRx can be translated to a safer and ideal antiviral modality. However, there is limited study in the in vivo antiviral applications of Cas13 RNA-guided endonucleases. Here, we investigated the in vivo use of AAV-CRISPR-Cas13 for multi-organ delivery and longer-lasting therapeutic expression. AAVs are attractive gene delivery vectors due to the recent successes in FDA and EMA approvals for AAV-based gene therapies. The established therapeutic applications of AAV in a wide-spanning variety of difficult-to-access tissues also made it an attractive delivery vector for nucleic acid therapeutics to treat diseases in different tissues or organs, beyond local administration. Since EV-A71 infects multiple organs and manifests pathology in the central nervous system and skeletal muscles, AAV-CRISPR-Cas13 could resolve the limitations of traditional siRNA and plasmid modalities that cannot access these key sites of EV-A71 tropism and pathology in vivo.
Added value of this study
This study demonstrates the use of a new bioinformatics pipeline, establishes AAV-CRISPR-Cas13-gRNAs design principles that confer effective antiviral function, demonstrates in vivo prophylactic and therapeutic outcomes in lethal enterovirus-infected mouse models, and shows that AAV-CRISPR-Cas13 represents an effective modality for in vivo intervention of RNA viral infection.
Implications of all the available evidence
This study bolsters the feasibility of using AAVs as an efficient delivery vector for in vivo CRISPR-Cas13 antiviral treatment. The bioinformatics pipeline, gRNAs design principles, and screening methodology can potentially be applied to develop efficient Cas13-gRNAs against other RNA viruses.
Introduction
RNA viruses account for the majority of the global viral infections that have plagued humans over the past decades.1 Currently, only a limited number of approved vaccines, drugs, or biologics can be used to prevent or treat these RNA viral infections.1 On top of that, there is a constant threat of escape mutations and antigenic drifts that could render the approved treatments ineffective.2 Current prophylactics and therapeutics mainly focus on targeting a single viral protein domain to neutralize the virus’s ability to infect or replicate within host cells. RNA viruses’ high mutation rates—stemming from error-prone viral RNA-dependent RNA polymerases–often result in the emergence of escape variants from the selective pressure posed by the drugs or biologics, as evident during the recent SARS-CoV-2 pandemic.3, 4, 5 In contrast to the backdrop of traditional small molecule drugs and antibodies, a therapeutic modality that cleaves the genome and transcripts of the RNA virus would represent a new class of therapeutics that can operate with a distinct mechanism of action, suppress the emergence of escape variants by targeting multiple conserved genomic sites, bypass the need for in-depth viral protein characterization, treat previously inaccessible tissues, and open up effective avenues against currently untreatable RNA viruses.
Enterovirus-A71 (EV-A71) are non-enveloped, positive-strand, single-stranded RNA viruses that are highly contagious and transmitted through bodily fluids. EV-A71 infection most commonly occurs in children younger than 5 years of age, with about 50%–80% of children tested seropositive for EV-A71, and is also observed in adults less frequently.6 EV-A71 is a major contributor to the disease known as Hand, Foot, and Mouth Disease (HFMD),7 and infection may occasionally result in severe neurological diseases or death.8 To date, there is only one commercially available vaccine9 and no available therapeutics against EV-A71,10,11 with clinical trials ongoing only for vaccines and not therapeutics.12,13 EV-A71 infections and HFMD reached record incidence rates in several countries, including in Southeast Asia (>200,000 HFMD cases in 2008–2018) and China (>20 million probable HFMD cases with >3700 deaths in 2008–2019), highlighting a pressing need for therapeutics against the currently incurable infections.10,14
In this study, we demonstrate the efficacy of a new class of CRISPR-Cas prophylactics and therapeutics against the EV-A71 RNA virus. To design our antiviral modality, we employed the recently discovered class 2 type VI-D CRISPR-Cas13d system derived from Ruminococcus flavefaciens XPD3002, also known as CasRx.15,16 CasRx is one of the smallest Cas13 endonuclease enzymes known, making it an attractive candidate to be delivered using AAV vectors along with its gRNAs within the same cargo construct. Furthermore, CasRx exhibits high on-target activity and high specificity without targeting the host genomic DNA and transcriptomic RNA,15 making it a safe and ideal antiviral modality. Previous in vitro studies investigating the Cas13 family of RNA-guided endonucleases as an antiviral modality showed promising results.17, 18, 19, 20, 21, 22 However, there is only one study that progressed to in vivo mouse study where nebulizer-assisted polymer-based delivery of mRNA-encoded Cas13a was used in the lung tissue,19 which resulted in a significant decrease in SARS-CoV-2 copy number of 57% in the crRNA N3.2-treated animals. Here, we investigated the in vivo use of AAV vectors that offer multi-organ delivery and longer-lasting therapeutic expression. AAVs are attractive gene delivery vectors due to the recent successes in FDA and EMA approvals for AAV-based gene therapies.23, 24, 25 The established therapeutic applications of AAV in a wide-spanning variety of difficult-to-access tissues also made it an attractive delivery vector for nucleic acid therapeutics to treat diseases in different tissues or organs, beyond local administration.26 Since EV-A71 infects multiple organs and manifests pathology in the central nervous system and skeletal muscles, AAV-CRISPR-Cas13 could resolve the limitations of traditional siRNA and plasmid modalities that cannot access these key sites of EV-A71 tropism and pathology in vivo.27 Through selecting antiviral gRNAs with a new bioinformatics pipeline, establishing AAV-CRISPR-Cas13-gRNAs design principles that confer effective antiviral function, and demonstrating in vivo prophylactic and therapeutic outcomes in infected mouse models, we showed that AAV-CRISPR-Cas13 represents a powerful new modality against previously incurable viral diseases.
Methods
Ethics statement
Animal care and housing were provided in accordance with the National Advisory Committee for Laboratory Animal Research (NACLAR) Guidelines (Guidelines on the Care and Use of Animals for Scientific Purposes). Experiments with mice were designed and approved under protocol R19-1035 by the National University of Singapore Institutional Animal Care & Use Committee (IACUC) based on NACLAR guidelines. Experiments with adeno-associated viruses with CRISPR-Cas were approved under GMAC Ref. No.: Res- 21-058.
Design and construction of AAV-vector-based CRISPR-Cas cargo plasmid
The CasRx sequence was a gift from Patrick Hsu (pXR001: EF1a-CasRX-2A-EGFP, Addgene no. 109049). Using a plasmid backbone with AAV2 ITRs, we performed EcoRI digestion to create a cut between the two ITR sequences to insert the custom-designed expression cassette. First, we derived the mammalian CMV promoter and enhancer sequence fragment from PCR amplification using pAAV-SMVP-Cas9C. Then, we assembled the CasRx with a HA tag and a rabbit polyA tail (CasRx-HA-polyA) downstream of the mammalian CMV promoter and enhancer into the AAV vector plasmid containing AAV2 ITR sequences by using Gibson Assembly reaction. Immediately downstream of the expression cassette, we assembled in the gRNA backbone driven by human U6 promoter, a gift from Patrick Hsu (pXR003: CasRx gRNA cloning backbone, Addgene no. 109053), using Gibson Assembly reaction. The expression cassette is then assembled to the EcoRI-digested plasmid with AAV2 ITRs using Gibson assembly. Individual gRNA sequences derived from the bioinformatics analysis were cloned using the BbsI digestion of the vector plasmid via Gibson Assembly of the target gRNA sequences.
Bioinformatics gRNA selection pipeline (Cas13gRNAtor)
We downloaded on 16th September 2020 and aligned 795 published EV-A71 complete genome assemblies from NCBI virus (https://www.ncbi.nlm.nih.gov/labs/virus/) using MAFFT and ran Cas13gRNAtor on these sequences. A bash script was created to align the input sequences using the MAFFT command line. We kept the length constant to the reference genome. Cas13gRNAtor utilizes, among multiple algorithms, a model and package developed by Wessel et al., 2020 (https://gitlab.com/sanjanalab/cas13) to generate additional scores for all possible gRNAs for the consensus sequence.28 Wessel employed a random forest model and a few features were selected to predict RfxCas13d on-target gRNA scores, including the crRNA folding energy, the local target ‘C’-context, and the upstream target ‘U’-context, which were determined as the most important features based on %incMSE. The random forest model utilizes a few independent software that includes RNAfold, a program to predict RNA secondary structure and minimum free energy (MFE), RNAplfold that measures RNA accessibility (Target RNA unpaired probability), and RNAhybrid that calculates RNA-RNA hybridization between the gRNA and its target site and RNA-RNA hybridization MFEs for each gRNA nucleotide. Cas13gRNAtor filters for candidate gRNA from the 3rd and 4th Quartile and removes gRNAs with T-homopolymers (>3 Ts). Conservation score at each nucleotide position was calculated as the percentage of sequences matching the consensus at that nucleotide position. A score of 1.0 indicates that the nucleotide position is fully conserved across all sequences. Shannon Entropy (SE) score calculated the propensity towards order and disorder for each nucleotide position.29 Entropy was directly proportional to the rate of disorder, i.e., a higher SE score indicated higher disorder of that particular nucleotide position. Shannon Entropy is defined as follows: SE () = −, where Pi was the probability of a given nucleotide (A, C, G, T, N; where N is an unspecified or deleted nucleotide) and n was the number of sequences used in the alignment. The summation runs over the 5 nucleotides in all the aligned sequences on every nucleotide position. The entropy range lies between 0 and log2 (5) = 2.32. A consensus sequence was generated based on the most conserved nucleotide at each position for downstream processing. For each predicted gRNAs, Cas13gRNAtor takes the entropy and conservation scores of their nucleotide positions in the consensus sequence (including in a more mutational intolerant ‘seed region’ aka ‘intolerant region’) and derives the means and standard deviations. Cas13gRNAtor scores gRNAs according to these features.
Cell lines and viruses
Human RD cells (RD-CCL-136, ATCC, Rockville, MD, USA) and mouse C2C12 (C2C12-CRL-1772, ATCC, Rockville, MD, USA; RRID:CVCL_0188) cells were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA). The cells were grown in media recommended by ATCC. The cells were tested for mycoplasma before use. AAVs were generated in-house as per previously reported.30 Briefly, AAV was packaged via a triple transfection of 293AAV cell line (AAV-100, Cell Biolabs, San Diego, CA, USA) that were plated in an HYPERFlask ‘M’ (Corning) in growth media consisting of DMEM + glutaMax + pyruvate + 10%FBS (Thermo Fisher), supplemented with 1X MEM non-essential amino acids (Gibco). Confluency at transfection was between 70% and 90%. Media was replaced with fresh pre-warmed growth media before transfection. For each HYPERFlask ‘M’, 200 μg of pHelper (Cell Biolabs), 100 μg of pRepCap [encoding capsid proteins for serotype DJ or 2], and 100 μg of pZac-CASI-GFP or pZac-CMV-CasRx-gRNAs were mixed in 5 ml of DMEM, and 2 mg of PEI “MAX” (Polysciences) (40 kDa, 1 mg/ml in H2O, pH 7.1) added for PEI: DNA mass ratio of 5:1. The mixture was incubated for 15 min and transferred drop-wise to the cell media. The day after transfection, the media was changed to DMEM + glutamax + pyruvate + 2%FBS. Cells were harvested 48–72 h after transfection by scrapping or dissociation with 1X PBS (pH7.2) + 5 mM EDTA and then pelleted at 1500 g for 12 min. Cell pellets were resuspended in 1–5 ml of lysis buffer (Tris HCl pH 7.5 + 2 mM MgCl + 150 mM NaCl), and freeze-thawed 3× between dry-ice-ethanol bath and 37 °C water bath. Cell debris was clarified via 4000 g for 5 min, and the supernatant was collected. The collected supernatant was treated with 50 U/ml of Benzonase (Sigma–Aldrich) and 1 U/ml of RNase cocktail (Invitrogen) for 30 min at 37 °C to remove unpackaged nucleic acids. After incubation, the lysate was loaded on top of a discontinuous density gradient consisting of 6 ml each of 15%, 25%, 40%, and 60% Optiprep (Sigma–Aldrich) in a 29.9 ml Optiseal polypropylene tube (Beckman–Coulter). The tubes were ultra-centrifuged at 54,000 rpm, at 18 °C, for 1.5 h, on a Type 70 Ti rotor. The 40% fraction was extracted and dialyzed with 1X PBS (pH 7.2) supplemented with 35 mM NaCl, using Amicon Ultra-15 (100 kDa MWCO) (Millipore). The titers of the purified AAV vector stocks were determined using real-time qPCR with ITR-sequence-specific primers and probe, referenced against the ATCC reference standard material 8 (ATCC). The viruses used are EV-A71 strain HFM41 (5865/sin/000009, GenBank accession no. AF316321), EV-A71 strain C4 (GenBank accession no. JQ965759.1), EV-A71 strain B5 (GenBank accession no. FJ461781.1), EV-A71 strain H (GenBank accession no. AY053402.1) and Echo7 strain Wallace (GenBank accession no. AF465516.1).
Immunofluorescence assay
10,000 human immortalized muscle cells, RD, were plated on glass slides in a 48-well plate and transduced with AAVDJ-CasRx at MOI 100K. At 3 days post-transduction, cells were fixed and permeabilized using methanol for 10 mins and blocked using 1X PBS with 5% BSA for 1 h. This is followed by primary antibody anti-HA (Cat#ab9110, abcam) incubation at 1:200 dilution for 2 h at RT. Secondary antibody staining is carried out at 1:1000 (Cat#A-21206, Thermo Fisher) for 2 h at room temperature. Slides were then washed 3X with 1X PBS and mounted onto the slide using ProLong mounting medium (Thermo). Images are taken using an Olympus microscope, exposure set at 15 ms and gain set at 6400 for brightfield and the 488 nm channel, exposure is set at 800us and gain set at 6400 for the DAPI channel.
Bio-panning assay for selection of AAV serotype
Immortalized human RD cells or mouse C2C12 cells were seeded in a 48-well plate at 10,000 cells per well in 200 μL DMEM containing 10% FBS. The cells were cultured overnight at 37 °C and allowed to adhere to the wells. A panel of AAV (1, 2, 6, 7, 8, 9, rh10, DJ, and Anc80) was used to transduce the cells at MOI of 100K or 10K in triplicates. At 72 h post-transduction, the cells were harvested, and the total GFP protein was quantitated using a GFP quantification kit (Biovision) on a multi-well plate reader (Tecan).
In vitro antiviral plaque assay with EV-A71
For screening of anti-EV-A71 activity, RD cells were seeded in 96-well plates at a density of 104 cells per well and incubated overnight in an incubator. Perform dilution of AAV2-CasRx bearing the different gRNAs for transduction individually or in pooled format at MOI 1K, 10K, 100K, and 1000K in a 100 ul volume. At 72 h post-transduction, the cells were infected with the EV-A71 virus at MOI of 1. The plate was washed twice with 1X PBS and incubated with DMEM with 2% FBS for 12 h. The supernatant from each well was harvested at 12 h post-infection and used for subsequent virus plaque assay. For virus plaque assay, RD cells were seeded in 24-well plates and incubated with 10–106 fold serially diluted supernatant samples in a 100 ul volume. Plates were washed twice with PBS and overlay media were added to each well and incubated with 15 mins rocking interval for an hour before leaving it in the incubator for 4 days. After 4 days of incubation, the overlay media was removed, crystal violet stain was added to visualize the plaques for counting, and the infectious virus titer was calculated, expressed as the average number of PFU per milliliter (PFU/ml) of the sample.
RNA-seq library preparation, sequencing, and analysis
300K RD cells were seeded in each well in a 6-well plate in triplicates. RD cells were transduced with AAV2-CasRx-EV71pool at an MOI of 1000 K or mock-infected at 8 h post-seeding. At 72 h post-transduction, the cells were infected with the EV-A71 virus at MOI of 1 for the “EV-A71-infected” and “EV-A71-infected and AAV treated” groups. Total RNA was extracted from RD cells at 12 h EV-A71 post-infection using the RNeasy Plus Mini kit from QIAGEN. Stranded mRNA libraries were prepared using the NEBNext II Ultra Directional RNA Library Prep Kit from New England Biolabs (Cat# E7760S) and sequenced on an Illumina HiSeq 4000 with 150 nt paired-end reads. ∼14 M total reads were de-multiplexed per condition per sample with Q30 above 90% and error rate of 0.02%. Sequenced reads were quality tested using FASTQC and mapped to the hg19 human genome using the 2.7.9 STAR aligner.31 Read alignment was performed using the default parameters. The genome index was constructed using the gene annotation provided with hg19 Illumina iGenomes collection and sjdbOverhang value of 100. Quantifications were performed using featureCounts v2.0.1,32 and differential gene expression was performed with DESeq2 v1.36.033 using triplicates to compute within-group dispersion and contrasts to compare between [mock vs EV-A71-infected], [EV-A71-infected vs EV-A71-infected and treated (EV-A71 gRNAs)], [EV-A71-infected and treated (EV-A71 gRNAs) vs mock] conditions (FDR < 0.1). An independent experiment was conducted to examine AAV-CasRx-3DgRNAs-only vs mock. Volcano plots were generated with Enhanced Volcano v1.18.0 in R 4.2.3, with FCcutoff = abs (0.75) and pCutoff = 0.00068083 (corresponding to adj-P = 0.1).
Flow cytometry
For titration of AAV2-GFP transduction, 10,000 cells were seeded on a 48-well plate and transduced with AAV2-GFP at MOI of 1K, 10K, 100K, and 1000K or left un-transduced as control. After 3 days, cells were harvested and suspended in flow cytometry buffer (0.5% BSA, 2 mM EDTA in 1X PBS) and passed through a 70 μm cell strainer before flow cytometry analysis by MACS Flow Cytometer (Miltenyi Biotec). To assess GFP knockdown by CasRx, 10,000 cells were seeded on a 48-well plate and transduced with AAV2-GFP at MOI of 10 K, followed by AAV2-CasRx- GFP_gRNA1, AAV2-CasRx-GFP_gRNA2, or AAV2-CasRx-GFP_gRNA1+2 at MOI 100K or 1000K or without AAV2-CasRx. After 3 days, cells were harvested and suspended in flow cytometry buffer (0.5% BSA, 2 mM EDTA in 1X PBS) and passed through a 70 μm cell strainer before flow cytometry analysis by MACS Flow Cytometer (Miltenyi Biotec). To detect CasRx-HA expression, 100,000 RD cells were seeded per well on a 12-well plate before they were transduced with AAV2-CasRx-HA at an MOI of 1000K. After 3 days, cells were harvested and rinsed once with 1X PBS before fixing with Fixation Buffer (BioLegend #420801) at room temperature for 20 min. Cells were then rinsed thrice with 1X Intracellular Staining Permeabilization Wash Buffer (BioLegend #421002) before being stained with anti-HA primary antibody (abcam ab9110, 1:50) on ice for 30 min. Cells were rinsed thrice and then stained with Alexa Fluor™ 488-conjugated secondary antibody (Invitrogen #A-21206, 1:200) on ice, in the dark, for 30 min. After 3 more rinses, cells were suspended in flow cytometry buffer (0.5% BSA, 2 mM EDTA in 1X PBS) and passed through a 70 μm cell strainer before flow cytometry analysis by MACS Flow Cytometer (Miltenyi Biotec). All flow cytometry results were analyzed using the FlowJo software.
qPCR for human innate and adaptive immune response genes profiling
In a 6-well dish, 1 million immortalized human skeletal RD cells were seeded and transduced with AAV2-CRISPR-CasRx at MOI of 100K and 1000K. RNA of the transduced cells was harvested at 72 h post-transduction using the RNeasy universal plus mini kit (Qiagen). cDNA was synthesized using Superscript III (Thermo Fisher) and the immune response gene panel was quantitated using the RT2 profiling kit (Qiagen).
Mouse infections and treatments
The EV-A71 strain HFM41 (5865/sin/000009, GenBank accession no. AF316321) was used for in vivo experiments. For toxicity analysis, BALB/c mice were injected intraperitoneally with either a dose of saline or 1 × 1012 viral genomes (vgs) of AAVDJ-GFPgRNA2 or 1 × 1011 vgs or 1 × 1012 vgs of AAVDJ-EV71gRNAs per mouse at 2 days old. For prophylaxis, BALB/c mice were injected intraperitoneally with 1 × 1011 viral genomes (vgs) or 1 × 1012 vgs of AAVDJ-EV71gRNAs per mouse at 2 days old and subsequently injected intraperitoneally with EV-A71 at a dose of 2 × 107 PFU per mouse at 5 days old. For therapeutic analysis, BALB/c mice were injected intraperitoneally with a dose of 2 × 107 PFU EV-A71 per mouse at 5 days old. After 6 h or 24 h, mice were injected intraperitoneally with a dose of 1 × 1011 viral genomes (vgs) or 1 × 1012 vgs of AAVDJ-EV71gRNAs per mouse. 1 × 1012 vgs AAVDJ-GFPgRNA2 was used as control. The survival of the mice and the clinical scores of each mouse in each treatment group was recorded daily for up to 19 days post-infection (dpi). Mice were scored in 5 categories for symptoms observed during the 19-day survival study. The scoring system includes Activity: 0 normal, 1 lethargy/abnormal posture, 2 huddled/inactive, 3 Unresponsive to stimuli, severe tremor and inability to right itself; Breathing: 0 normal, 1 rapid/shallow, 2 rapid abdominal, 3 labored, blue; Movement: 0 normal, 1 weakness, incoordination, 2 single limb dragging/paralysis, 3 multiple limb dragging/paralysis; Body weight: 0 normal, 1 loss of 5% over 24 h, 2 loss of more than 15% or up to 10% in 24 h, 3 loss of more than 10% over 24 h or 20% in total, body condition 2 or below; Dehydration: 0 normal skin tent, 1 skin tent present on dorsum, 2 Moderate skin tent, 3 Severe skin tent. A total of 6 or more points accumulated across all categories was determined as a humane endpoint, and mice with such a score were euthanized. Histological analyses were carried out on mice sacrificed at 6 days post-infection.
Histology of mouse tissues
Histological samples were fixed in 4% paraformaldehyde at 4 °C, decalcified at room temperature for 2 h, then embedded in paraffin and processed into 4 μm sections. Tissue pathology was evaluated byhematoxylin and eosin (H&E) staining and EV-A71 antigen was detected by immunohistochemistry staining (IHC) using commercially available anti-EV-A71 antibody (MAB979, Sigma Aldrich; 1:500 dilution) and image captured using the Leica Bond-Max system. For staining of CasRx, histological slides were baked at 50 °C for 10 min and de-waxed in Xylene 3x for 5 min. Slides were then rehydrated in descending grade ethanol; 100% ethanol 2x for 5 min, 95% ethanol 5 min, 80% ethanol 3 min, 70% ethanol 3 min, 50% ethanol 3 min, rinsing with water, and rinsing with 1X PBS 3x for 5 min. Antigen retrieval was conducted by 2100-Retriever steam cooker (Prestige Medical) and slides were heated for 12–15 min in 0.01 M Sodium Citrate buffer pH 6.0, cooled for 3–4 h, washed once in 1X PBS for 5 min, then washed using 1X PBS with 0.1% Triton 3x for 5 min each. Slides were blocked in 2% BSA +5% goat serum-PBS for 60 min at RT, then incubated with primary Ab 1:100 (anti-HA mouse, ab18181) in blocking buffer overnight at 4 °C. After washing with 1X PBS with 0.1% Triton 3x for 5 min, slides were incubated with secondary Ab 1:500 (Alexa Fluor 488, A21202) in washing buffer for 2 h at RT, followed by washing with 0.1% Triton-1X PBS 3x for 5 min and counterstained with DAPI. Images were captured using a confocal microscope (Leica).
Quantification of EV-A71 titre in mice tissues
EV-A71-infected mice were sacrificed on 6 days post-infection (dpi) and mice tissue, including hind limb and brain, were collected into CK14 homogenizing tubes (Bertin Corp), homogenized, and titrated by plaque-forming assay. The tissues were weighed and 1 ml and 0.5 ml of DMEM for limb tissues and brain tissues were added, respectively, into the tubes before homogenization using an orbital shaker at 6000 × g for 10 s. The process was repeated 3 times for brain tissues and 9 times for limb tissues. The homogenized tissues were centrifuged for 10 min at 8000 × g, 4 °C to pellet tissue debris. Supernatants were collected, and the viral load was titrated by viral plaque assay.34
Statistical analysis
Statistical analysis was performed using GraphPad Prism (Version 9, La Jolla, CA, USA). Differences between two groups were analyzed by student t-test (two-tailed). P-value was considered statistically significant as follow: ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001. Differences between multiple groups were analyzed by either one-way ANOVA or two-way ANOVA. P-value was considered statistically significant as follow: ∗adj-p < 0.05, ∗∗adj-p < 0.01, ∗∗∗adj-p < 0.005, ∗∗∗∗adj-p < 0.001.
Role of funding source
The funders played no part in the design, data collection, data analysis, interpretation, writing or decision to publish the results.
Results
Development of AAV-CRISPR-Cas antiviral modality for RNA virus targeting
The EV-A71 life cycle is typical of a positive-strand single-stranded RNA virus, in which the virus enters the cell, releases its RNA genome into the cytoplasm, and synthesizes the negative-sense genomic and sub-genomic viral RNAs from which new copies of the positive sense viral genomes and transcripts are synthesized.35 In this study, we proposed to use an AAV-vector-based CRISPR-Cas13d system for direct targeting and cleavage of the viral genome and mRNA to reduce viral replication and pathology (Supplementary Fig. S1a). We first built a new AAV cargo construct (Supplementary Fig. S1b) bearing the gRNAs and a CasRx that excludes the Nuclear Localization Signal (NLS) sequences so that the expressed CasRx protein has access to the viral replication complex residing outside the nucleus. To validate the expression of the CasRx protein, we transduced immortalized human muscle RD cells with AAV-CRISPR-CasRx-gRNA and confirmed the expression of the HA-tagged CasRx and its localization in the cytoplasmic region via immunofluorescence (Supplementary Fig. S1c). To test the efficacy of the new construct, we generated AAV-CRISPR-CasRx-gRNAs that target the GFP mRNA sequence and tested the viruses on GFP-expressing cells (Table 1). A titration experiment was first carried out in RD cells to determine that AAV2-GFP at MOI of 10 K can confer GFP expression in more than 98% of the cells without the GFP oversaturation observed at MOI of 100K and 1000K (Supplementary Fig. S1d). Next, we tested two different GFP-targeting gRNAs individually or in combination. The results confirmed that the AAV2-CasRx-gRNAs reduced GFP expression in a dose-dependent manner by 7.6% and 27.1% (GFPgRNA1), 7.4% and 25.2% (GFPgRNA2), and 6.5% and 21.9% (GFPgRNA1 + 2), at MOIs of 100K and 1000K (Supplementary Fig. S1e). There is no significant difference in the knockdown effect by these GFP gRNAs.
Table 1.
For GFP gRNA sequences.
| gRNA name | gRNA sequence |
|---|---|
| GFPg1 | GTGAACAGCTCCTCGCCCTTGCT |
| GFPg2 | GCTGAACTTGTGGCCGTTTAC |
Computational gRNAs design and multiplexed CRISPR-Cas targeting of RNA viruses
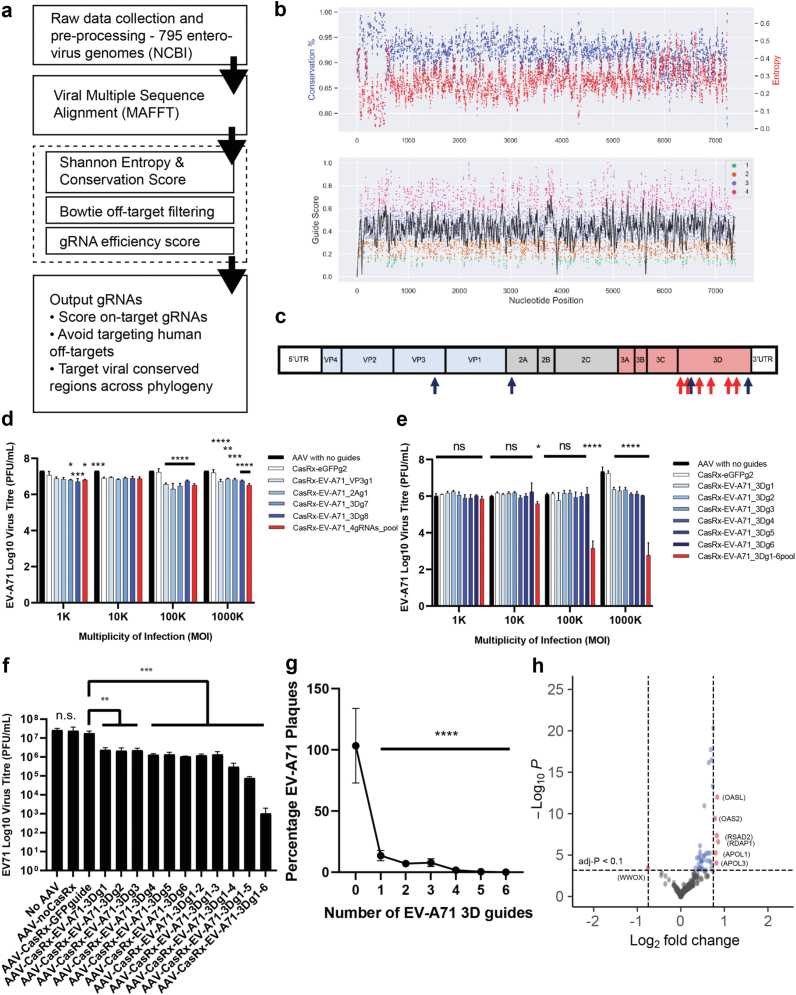
To design antiviral CRISPR-gRNAs, we developed Cas13gRNAtor, a bioinformatics tool that first generates all possible gRNA candidates for a user-provided sequence and then scores each gRNAs via multiple parameters: (i) multiple sequence alignment of target genes/genomes to defined conserved regions with entropy and conservation scores, (ii) structural information (RNA folding, hybridization energy), (iii) on-target scores for predicted efficacy,28 and (iv) off-target scores for specificity (Fig. 1a). We applied Cas13gRNAtor to curate an in silico list of candidate gRNAs that target EV-A71 within the highly conserved viral regions (to anticipate and avoid mutational escape) and are highly dissimilar to the human transcriptome (to minimize off-targeting). We filtered for the top 3rd and 4th quartiles (Q3 and Q4) of candidate gRNAs based on gRNA score (Fig. 1b), eliminated gRNAs predicted to have potential off-target sites within the human transcriptome (hg38; including non-coding RNA), and eliminated gRNAs with T-homopolymers (>3 Ts), thereby obtaining a subset of 1661 candidate gRNAs. Cas13gRNAtor further narrowed to the best 100 candidate gRNAs that target highly conserved viral regions with a mean conservation score of at least 0.9 for the target protospacer (conservation score of 1.0 represents fully conserved across all viral sequences).
Fig. 1.
CRISPR-CasRx gRNAs design strategy generates gRNAs with potent and specific antiviral activity in cells. a. Schematic of bioinformatics pipeline for gRNA prediction and scoring, as applied to enterovirus genomes. b. Visualization of scores and gRNAs against 795 enterovirus target genomic sequences. Top: Entropy Score (red) for each nucleotide of aligned viral genomes. Percentage of Conservation (blue) for each nucleotide among aligned viral genomes. Bottom: gRNAs on-target efficacy score along the target enterovirus, color-coded and divided into four quartiles. c. Designed gRNAs against the enterovirus RNA genome. Blue arrows: gRNAs with high scores. Red arrows: gRNAs with medium/low scores. d. Assessment of gRNA pools. 10K RD cells were seeded in each well of a 96-well plate and transduced with indicated AAVs. After 72 h, the cells were subjected to EV-A71 infection at MOI of 1 for 12 h, and the supernatant was then harvested for virus plaque assay (two-way ANOVA, p < 0.0001 for MOI, gRNAs, and MOIxgRNA interaction; Dunnett’s against eGFPg2). e. Assessment gRNAs against the 3D gene. 10K RD cells were seeded in each well of a 96-well plate and transduced with indicated AAVs. After 72 h, the cells were subjected to EV-A71 infection at MOI of 1 for 12 h, and the supernatant was then harvested for virus plaque assay (two-way ANOVA, p < 0.0001 for MOI, gRNAs, and MOIxgRNA interaction; Dunnett’s against eGFPg2). f. Multiplexed CasRx gRNAs on a single 3D target inhibits EV-A71 replication. RD cells were treated with AAV-CasRx bearing gRNAs at MOI of 1000K and infected with EV-A71 at MOI of 1 at 72 h later (one-way ANOVA, Dunnett’s against eGFPg2). g. Multiplexed gRNA pools inhibit EV-A71 plaque formation (all at a total MOI of 1000K) (two-way ANOVA, Dunnett’s against “0” eGFPg2). h. Specificity of CasRx-EV71_3Dguides pool in transduced human RD cells at MOI of 1000 K (n = 3 for both groups (EV-A71 gRNAs + EV-A71 infected) and (EV-A71 infected)). ∗adj-p < 0.05, ∗∗adj-p < 0.01, ∗∗∗adj-p < 0.005, ∗∗∗∗adj-p < 0.001.
Candidate gRNAs targeting protein-coding regions and their 5’ UTRs are highly conserved (>90%) across all genomes and avoid the lower conservation and higher entropy scores in more variable regions. We found that all 13 viral gene transcripts, and their respective 5’UTRs, could be targeted with Q4 gRNAs with a maximum mismatch score of 3 relative to the consensus sequence (Supplementary Fig. S2a and b). A combination of at least 3 gRNAs targets at least 99% of the EV-A71 genomes (n = 736), with 2 mismatches outside the protospacer intolerant region tolerated for each gRNA (Supplementary Fig. S3). We also analyzed another 414 Enterovirus B (EV-B) and 735 Enterovirus C (EV-C) genomes and found that a combination of 3 gRNAs targets at least 67% of the genomes with up to 3 mismatches tolerated (Supplementary Fig. S2c and d).
To efficiently deliver the CRISPR-Cas system into human skeletal RD cells that serve as the in vitro model for EV-A71 infection, we first performed an AAV serotype bio-panning experiment to determine the AAV serotype that best transduces the cells. Among 9 serotypes, we identified AAV2 as the most efficient serotype for human skeletal RD cells (Supplementary Fig. S4).
We then investigated if there were any differences in the activity of the individual gRNAs based on their target gene or gRNA scores. AAV2-CRISPR-CasRx bearing each of the individual gRNAs were used to transduce RD cells at MOI of 1K, 10K, 100K, and 1000K, followed by infection of the cells using EV-A71 at MOI of 1 at 72 h post-transduction (Fig. 1c–e). The supernatant is then harvested at 12 h post-infection for viral plaque-forming assay. Our results suggest that for individual high-scoring gRNAs, significant virus inhibition was observed at AAV-CRISPR-CasRx-gRNA at low MOIs of 1K, 10K, and 100K. For gRNAs targeting the VP3 and 3D gene, inhibition of up to 1 log reduction of virus titer (up to 90.8% of viral reduction) was observed. Low-scoring gRNAs do not exhibit significant inhibition of virus replication at AAV-CRISPR-CasRx-gRNA MOI of 1K, 10K, or 100K, but all gRNAs exhibit significant inhibition of virus replication up to 1 log reduction of virus titer at AAV-CRISPR-CasRx-gRNA MOI of 1000K. This demonstrates that Cas13gRNAtor can identify single gRNAs for efficient inhibition of viral replication. The result is also consistent with recent in vitro Cas13 antiviral studies that reported a reduction of up to 90% in virus titers or reporter activity in cell culture.17,19 To confirm that the viral inhibition is not due to any intracellular antiviral activity stimulated by the AAV delivery vector, we performed a host human innate and adaptive immune response profiling using AAV2-CasRx-GFPgRNA2 transduced RD cells at MOI of 100K and 1000K. The results showed minimal intracellular antiviral response in the AAV2-CasRx-GFPgRNA2-transduced RD cells, with only two genes (NLRP3 and SLC11A1) being significantly upregulated in a dose-dependent manner (Supplementary Fig. S5). NLRP3 and SLC11A1 are both involved in macrophage activation or recruitment but have not been shown to exhibit intracellular antiviral activity.36,37 With the minimal innate immunity induced in cells and the absence of viral inhibition from the AAV2-CasRx-GFPgRNA2 negative controls, we conclude that the antiviral function of AAV-CasRx-gRNAs is specific and caused by activity from the gRNA-targeted CRISPR-CasRx endonuclease.
To develop a more potent AAV-CRISPR-Cas antiviral, we evaluated two gRNA combinations against EV-A71, one with 4 candidate gRNAs targeting different genes (2 A, VP3, and 3D) and another with 4 candidate gRNAs targeting only one gene (3D) for comparison of antiviral efficacy (Fig. 1c, Table 2, and Supplementary Fig. S3). The multi-gRNAs multi-genes combination targeting the 2A, VP3, and 3D genes exhibits significant inhibition of viral replication titer by up to 91.1% at MOI of 100K and 1000K but is not significantly better than the single gRNAs approach (Fig. 1d). In comparison, with the multi-gRNAs single-gene combination where 4 gRNAs target the same 3D gene, we observed a significant inhibition of up to 2 logs or 98.8% of virus titer reduction (Fig. 1f). This strong antiviral effect of gRNAs targeting 3D, despite the lower aggregate gRNA scores, could reflect the function of 3D as an RNA-dependent RNA polymerase (RdRP) and an essential gene for the viral RNA synthesis in the replication cycle.38 RdRP genes are a common target for developing viral inhibitors and antiviral therapeutics via blockade of viral replication.39,40 Next, we investigated if varying the number of guides against the same gene target affects the extent of viral inhibition. Increasing the number of pooled gRNAs against 3D by up to three gRNAs did not significantly improve the inhibition, which stayed at approximately 1 log of virus titer reduction, while four gRNAs inhibited up to 2 logs of virus titer reduction (Fig. 1f and g). Further increasing the number of gRNAs targeting the same gene to five increased the inhibition up to 3 logs or 99.7% (Fig. 1f and g). The most potent cocktail consisted of 6 gRNAs, with an observed inhibition of up to 5 logs or 99.99% of virus reduction at MOI of 100K or 1000K (Fig. 1e-g). Even at a lower MOI of 10 K, significant inhibition by up to 90.1% of viral reduction is observed (Fig. 1e). Together, these data suggest that using multiple guides in a pool against the 3D gene that is critical to viral replication efficiently eliminates EV-A71 RNA viruses in vitro. Importantly, the almost complete elimination of viruses to less than 0.1% levels indicates that most infected cells were transduced with the AAV and that the CRISPR-CasRx system is functional within these cells.
Table 2.
For EV-A71 gRNAs sequences and scorings.
| gRNA name | gRNA sequence | gRNA score | Entropy score | Conservation score |
|---|---|---|---|---|
| 3Dg1 | GTTGGTCCATTGATGTTTAGTCT | 0.011814542 | 0.22192 | 0.94165 |
| 3Dg2 | TTGGAAAACAGGGCTTGTTCAAA | 0.049176595 | 0.24443 | 0.92918 |
| 3Dg3 | TAGCACGCTTCCTCCATGCTCAT | 0.56835903 | 0.17175 | 0.95543 |
| 3Dg4 | TATTTATCCATGTAGAATTTCAT | −0.06258042 | 0.17843 | 0.94914 |
| 3Dg5 | ATGATGTTGTTGATCATTGAATT | 0.080602909 | 0.14546 | 0.96782 |
| 3Dg6 | TCTGCAGGAGTCATGGTCAAACC | 0.477791799 | 0.24641 | 0.93043 |
| 3Dg7 | CAAAGTTAGGAATGGCCAACGCT | 1.303861968 | 0.30996 | 0.9162 |
| 3Dg8 | CACATACTTGGAAAACAGGGCTT | 1.074780839 | 0.22671 | 0.94192 |
| 2Ag1 | AACCACTCTGAAGTTGCCCACGT | 1.241615481 | 0.19715 | 0.94526 |
| VP3g1 | GGTGACAGATGATTGTAGCCCAA | 0.673976462 | 0.19415 | 0.95089 |
To verify that the CRISPR-CasRx system is specific, we employed differential RNA expression analysis to identify potential off-targets in the human transcriptome.15,41 No differentially expressed genes were observed in AAV-CasRx-3DgRNAs-only (MOI 1000K) or EV-A71-only treated human muscle RD cells compared to untreated cells, whereas cells treated with both EV-A71 and AAV-Cas13d-gRNAs show modest differential expression (absolute log2 fold change > 0.75; versus EV-A71-only) in 6 upregulated human genes and 1 downregulated human gene, which are predominantly involved in antiviral response (Fig. 1h).
AAV-CRISPR-CasRx-3D_gRNAs multi-gRNAs pool exhibits pan-enterovirus A71 inhibitory activity
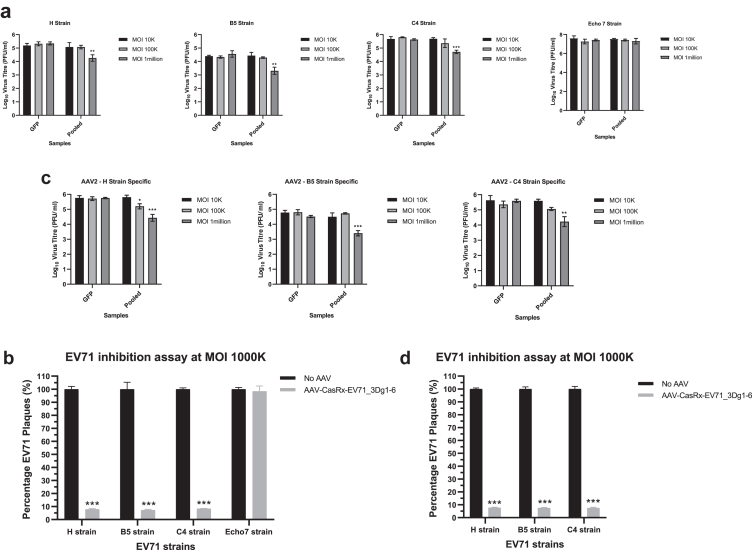
To examine if there is broad inhibitory activity across different enterovirus strains other than the lab strain HFM41, we tested the six-gRNAs pool targeting 3Dpol on three different EV-A71 strains (H, B5, and C4 strains). A picornavirus from the human enterovirus B species, Echo 7 strain, is used as a control virus. Alignments of EV71_3DgRNA1-6 against the target regions in the H, B5, and C4 strains showed non-matching (NM) bases ranging from zero to six (Supplementary Fig. S6). Results from plaque assays indicate that there is significant pan-enterovirus A71 inhibitory activity on the replication of variant strains H, B5, and C4 induced by the AAV2-CasRx-EV71_3DgRNA1-6 pool at MOI of 1000K but not at MOI of 10K or 100K (Fig. 2a), reducing viral plaque formation by 92%, 92.8%, and 91.6%, respectively, compared with cells treated using control AAV2-CasRx-GFPgRNA2 (Fig. 2b). No inhibitory activity against the control virus Echo7 was observed. When the gRNAs were re-engineered to perfectly match the sequence of target regions in their respective EV-A71 variant strains, modest improvement of inhibitory activity against the H strain was observed at the mid-dosage MOI of 100K, but not for the B5 and C4 strains (Fig. 2c). At the highest MOI of 1000K, subtle improvement in inhibitory activities were observed for the H strain (92%–92.3% mean reduction in virus plaques) and C4 strain (91.6%–92.4%) (Fig. 2d). These results suggest that the original AAV2-CasRx-EV71_3DgRNA1-6 pool exhibits significant pan-enterovirus A71 activities against other EV-A71 strains (H, B5, and C4 strains) despite the presence of sequence mismatches, and modifying the gRNAs for 100% sequence match only modestly enhances the inhibitory activities against these EV-A71 strains.
Fig. 2.
Pan-enteroviral inhibition by AAV2-CasRx-EV71_3Dguides. a. Assessment of inhibitory activity of the six-gRNAs pool targeting HFM41 strain on three other EV71 variants (H strain, B5 strain, and C4 strain). 10K RD cells were transduced with AAV2-CasRx at MOI of 10K, 100K, and 1000K bearing either GFP guide2 or EV-A71 guide(s) for 72 h and then subjected to EV-A71 infection at MOI of 1 for 12 h, after which the supernatant is harvested for virus plaque assay. b. The pool of 6 gRNAs designed against the HFM41 strain exhibits antiviral inhibition against three other EV71 variants (H strain, B5 strain, and C4 strain) at an MOI of 1000K. c. Each pool of 6 gRNAs is engineered specifically for the sequences of the H, B5, or C4 strain. d. Each re-engineered pool of 6 gRNAs exhibits antiviral inhibition against their respective H, B5, or C4 strain. In all panels, comparison between two groups (CasRx-GFPg2 and CasRx-EV71_3Dguides) was analyzed by Student t-test (two-tailed). ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001.
AAV-CRISPR-CasRx-gRNAs is prophylactic against EV-A71 infection
To ensure efficient delivery of the therapeutics into the mouse muscle tissue, which is a primary replication site for the EV-A71 virus, we performed an AAV serotype bio-panning experiment using the immortalized mouse C2C12 skeletal muscle cells, which identified AAVDJ serotype as the most efficient among the 11 serotypes assessed (Supplementary Fig. S4b). To determine if the dosages for treatment are associated with systemic toxicity, we injected 2-day-old mice with saline, AAVDJ-CRISPR-CasRx-GFPgRNA2 at 1 × 1012 total viral genomes (vgs), AAVDJ-CRISPR-CasRx-3D_gRNAs at 1 × 1011 total vgs, or AAVDJ-CRISPR-CasRx-3D_gRNAs at 1 × 1012 total vgs. Mice were monitored daily for survivability, clinical symptoms, and body weight for up to 19 days post-infection. Survival, clinical scores, and body weight of treated mice were similar to control mice without any signs of toxicity observed (Supplementary Fig. S7).
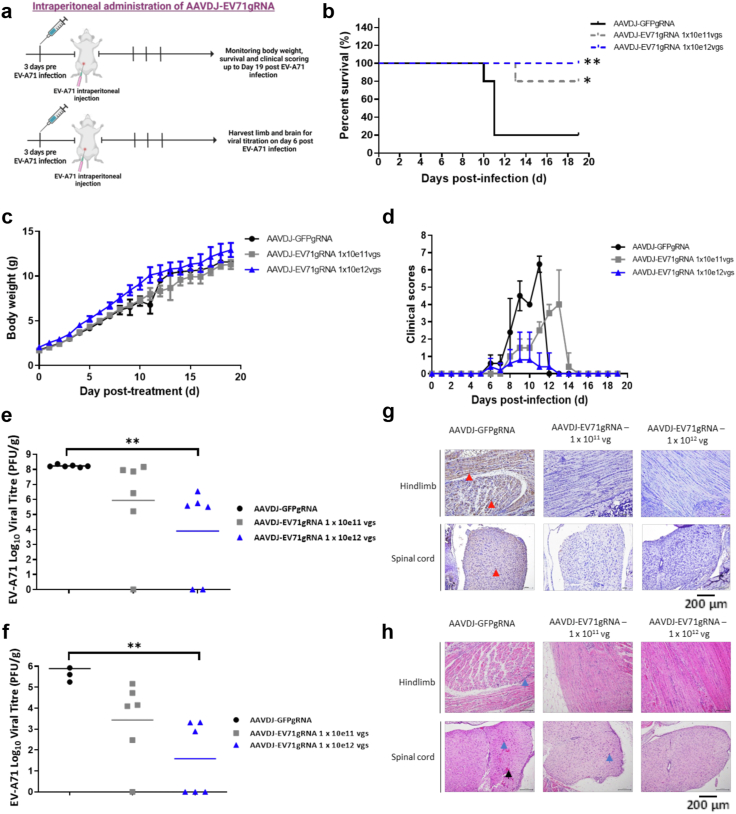
We next investigated the effectiveness of the AAV-CRISPR-CasRx antiviral modality in an established EV-A71 murine model for HFMD.42 Most of these mice succumb to the EV-A71 infection and displayed mortality. We injected 2-day-old mice with either AAVDJ-CRISPR-CasRx-GFPgRNA2 at 1 × 1012 total viral genomes (vgs) as the control group or AAVDJ-CRISPR-CasRx-3D_gRNAs at 1 × 1011 or 1 × 1012 total vgs as the treatment groups (Fig. 3a). After 3 days, the mice were injected intraperitoneally with a lethal dose of 2 × 107 PFU of EV-A71 per mouse and monitored daily for survivability, clinical symptoms, and body weight. Only 20% of the mice in the control group survived at 11 days post-infection (n = 5) (Fig. 3b). In contrast, 80% of the mice survived in the 1 × 1011 vgs AAVDJ-CRISPR-CasRx treatment group (n = 5, logrank Mantel–Cox test; ∗p = 0.0396) at up to 19 days post-infection. The higher treatment dose of 1 × 1012 vgs conferred 100% protective effect in the infected mice (n = 7, logrank Mantel–Cox test; ∗p = 0.0053) up to 19 days post lethal infection. Based on physical symptoms of body weight, activity, breathing, movement, and dehydration, we observed higher clinical scores and lower body weights of EV-A71-infected mice in the control group compared with the two treatment groups. Whereas the treatment groups showed low clinical symptoms (Fig. 3c and d), EV-A71-infected mice in the control group developed severe clinical symptoms, which include inactivity, loss of body weight, and hind limbs paralysis.
Fig. 3.
AAVDJ-CRISPR-CasRx confers prophylactic protection in a mouse model for hand, foot and mouth disease. a. BALB/c mice were injected intraperitoneally with a dose of 1 × 1011 vgs or 1 × 1012 vgs of AAVDJ-EV71gRNAs per mouse at 2 days old and subsequently injected intraperitoneally with EV-A71 at a dose of 2 × 107 PFU per mouse at 5 days old. 1 × 1012 vgs AAVDJ-GFPgRNA was used as control. b. Survival curve of treated mice over 19 days post-infection (dpi). c. The body weight of each mouse in each treatment group over 19 dpi. Comparison between two groups (CasRx-GFPg2 and CasRx-EV71_3Dguides) was analyzed by Log-rank (Mantel–Cox) test. ∗p < 0.05, ∗∗p < 0.005. d. The clinical score of mice was recorded using the mice clinical assessment scoring system (M-CASS) involving observation of five markers: activity, breathing, movement, body weight, and dehydration over 19 dpi. e. Treatment reduces virus titers in the hind limbs of mice as determined using viral plaque assay. Viral titration results were from a single experiment with multiple mice. f. Treatment reduces virus titers in the brain of mice as determined using viral plaque assay. Comparison between two groups (CasRx-GFPg2 and CasRx-EV71_3Dguides) was analyzed by Kruskal–Wallis test with Dunn's multiple comparisons post hoc test. ∗∗p < 0.005. Viral titration results were from a single experiment with multiple mice. g. Immuno-histochemistry staining for the EV-A71 VP2-specific antigen. Presence of viral antigen in the hind limbs and cervical spinal cord anterior horn cells (red arrow). h. H&E staining of the hindlimbs and spinal cord of mice. Polymorphonuclear meningitis in the spinal cord (black arrow). Necrosis and focal interstitial mononuclear cell infiltrates in the hind limbs and spinal cord (blue arrow). Magnification of H&E and IHC images are taken at 200X.
To examine the impact that the AAV-CRISPR-CasRx antiviral modality has on viral infection and pathology, muscle and brain tissues of the EV-A71-infected mice were harvested at 6 dpi and the viral titers were quantified by viral plaque assay. The results showed that while viral titers remain detectable in the hind limb and brain tissues upon treatment in a few mice, a large reduction in viral titers was observed in most treated mice. In hind limbs, viral titration showed 2 to 3-log reduction in 4 mice and viral titer was not detected in 2 of the mice in the 1 × 1012 total vgs treated mice group (p = 0.0030, Kruskal–Wallis test) compared to the control group (Fig. 3e). In brain tissues, viral titration showed a 3-log reduction in 3 mice and viral titer was not detected in 3 of the mice in the 1 × 1012 total vgs treated mice group (p = 0.0017, Kruskal–Wallis test) compared to the control group (Fig. 3f). Furthermore, no viral antigen distribution was observed in the mice treated with 1 × 1012 total vgs, minimal viral antigen was present in mice treated with 1 × 1011 total vgs, and severe viral antigen distribution was observed in control mice for both hind limbs and cervical spinal cord anterior horn neurons (Fig. 3g). Results from H&E staining performed on the limb muscle and spinal cord tissues revealed no obvious muscle necrosis in mice treated with AAVDJ-CRISPR-CasRx-3D_gRNAs at 1 × 1012 total vgs, mild necrosis in mice treated with 1 × 1011 total vgs, and severe necrosis in the control group (Fig. 3h). Through immunohistochemistry, CasRx-HA was observed in the hind limb and spinal cord tissue sections, with a higher expression in mice treated with 1 × 1012 total vgs of AAVDJ-CasRx-GFPgRNAs or AAVDJ-CRISPR-CasRx-3D_gRNAs compared to mice treated with 1 × 1011 total vgs of AAVDJ-CRISPR-CasRx-3D_gRNAs (Supplementary Fig. S8). These results indicate that AAVDJ-CRISPR-CasRx-3D_gRNAs is an effective prophylactic for preventing EV-A71 RNA virus infection and pathology in vivo.
Effective elimination of viruses in vivo in EV-A71-infected mouse model
We next evaluated the AAV-CRISPR-CasRx antiviral modality when used as a therapeutic intervention in mice already infected with EV-A71. We injected 5-day-old mice intraperitoneally with a lethal dose of 2 × 107 PFU of EV-A71 per mouse. After 6 h, we intraperitoneally injected mice with AAVDJ-CRISPR-Cas-GFPgRNA2 at 1 × 1012 vgs or AAVDJ-CRISPR-CasRx-3D_gRNAs at 1 × 1011 or 1 × 1012 total vgs and observed the mice daily for survivability, clinical symptoms, and body weight (Supplementary Fig. S9a). The results indicate that AAVDJ-CRISPR-CasRx-3D_gRNAs at 1 × 1011 vgs successfully conferred survival of 80% to the mice (n = 5, logrank Mantel–Cox test; ∗p = 0.0039) and 1 × 1012 total vgs conferred survival to 100% of the mice (n = 4, logrank Mantel–Cox test; ∗p = 0.0084) up to 19 days post lethal infection (Supplementary Fig. S9b). We observed lower clinical symptoms, better body weight, significantly reduced viral titers in hind limbs and brains, significantly reduced viral antigens in tissues, and alleviation of muscle necrosis in the treatment groups (Supplementary Fig. S9c–h).
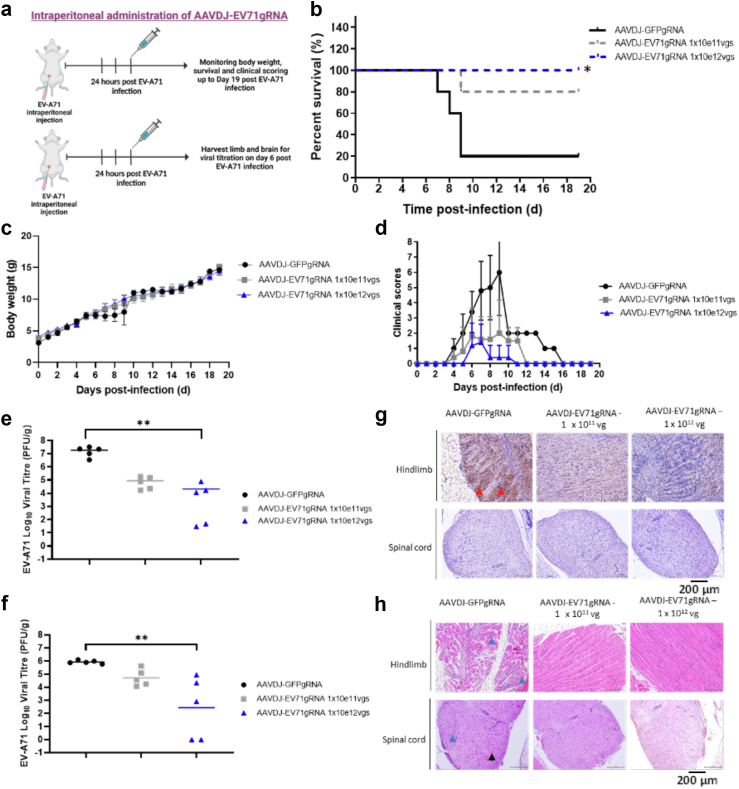
Importantly, even at an extended 24 h post-infection treatment (Fig. 4a), AAVDJ-CRISPR-CasRx-3D_gRNAs at 1 × 1011 vgs successfully conferred survival to 80% of the mice (n = 5, logrank Mantel–Cox test; ∗p = 0.053) and at 1 × 1012 total vgs conferred survival to 100% of the mice (n = 5, logrank Mantel–Cox test; ∗p = 0.015) up to 19 days post lethal infection (Fig. 4b). Treatment groups again exhibited improved clinical scores and body weights (Fig. 4c and d). Viral titers were significantly reduced by 3 logs in the hind limbs (p = 0.0018, Kruskal–Wallis test) (Fig. 4e) and 4 logs in the brain tissues (p = 0.0029, Kruskal–Wallis test) (Fig. 4f) of mice in the 1 × 1012 total vgs treatment group compared to the control group. IHC in hind limbs and cervical spinal cord anterior horn neurons showed minimal viral antigen distribution in mice treated with 1 × 1012 total vgs, modest viral antigen in mice treated with 1 × 1011 total vgs, and severe viral antigen distribution in control mice (Fig. 4g). H&E staining performed on the limb muscle and spinal cord tissues revealed that AAVDJ-CRISPR-CasRx-3D_gRNAs alleviates the severe muscle necrosis observed in the control group (Fig. 4h). Taken together, the results indicate that the AAVDJ-CRISPR-CasRx-3D_gRNAs modality is effective as a treatment against EV-A71 throughout the treatment window pre-infection and at 6 h and 24 h post-infection. Importantly, the AAVDJ-CRISPR-CasRx-3D_gRNAs treatment prevents death and eliminates up to 99.9% of the EV-A71 RNA viruses in vivo.
Fig. 4.
AAVDJ-CRISPR-CasRx prevents mortality and reduces pathology when applied 24 h after EV-A71 infection. a. BALB/c mice were injected intraperitoneally with a dose of 2 × 107 PFU EV-A71 per mouse at 5 days old. After 24 h, mice were injected intraperitoneally with a dose of 1 × 1011 viral genomes (vgs) or 1 × 1012 vgs of AAVDJ-EV71gRNAs per mouse. 1 × 1012 vgs AAVDJ-GFPgRNA was used as treatment control. b. The survival of the mice was recorded for 19 days post-infection (dpi). Comparison between two groups (CasRx-GFPg2 and CasRx-EV71_3Dguides) was analyzed by Log-rank (Mantel–Cox) test. ∗p < 0.05. c. The body weight of each mouse in each treatment group was recorded for 19 dpi. d. The clinical score of each mouse was recorded using the mice clinical assessment scoring system (M-CASS). e. Virus titers in the hind limbs of mice from different treatment groups were determined using the plaque-forming assay. Viral titration results were from a single experiment with multiple mice. f. Virus titers in the brains of mice from different treatment groups were determined using the plaque-forming assay. Comparison between two groups (CasRx-GFPg2 and CasRx-EV71_3Dguides) was analyzed by Kruskal–Wallis test with Dunn's multiple comparisons post hoc test. ∗∗p < 0.005. Viral titration results were from a single experiment with multiple mice. g. Immuno-histochemistry staining for EV-A71 specific antigen. Presence of viral antigen in the hind limbs (red arrow). h. H&E staining of the hind limbs and spinal cord of mice. Polymorphonuclear meningitis in the spinal cord (black arrow). Necrosis and focal interstitial mononuclear cell infiltrate in the hind limbs and spinal cord (blue arrow). Magnification of H&E and IHC images are taken at 200X.
Discussion
The AAV-CRISPR-Cas antiviral modality has tremendous potential as a new class of treatment against RNA viral infections. Unlike traditional small molecule drugs and biologics, CRISPR-Cas13d operates with a distinct strategy of directly cleaving the viral RNA genome and viral mRNA. Here we show that CRISPR-Cas13d eliminates an RNA virus in vivo and prevents pathology and death caused by the infection. This is important because despite EV-A71 and HFMD cases exceeding a million each year, there is currently no therapy available. Against the different classes of EV-A71 therapeutics evaluated preclinically in vivo (Supplementary Table), the AAV-CRISPR-Cas13 technology demonstrates superiority with 4 to 5-log viral inhibition in key organs, exquisite specificity, and broad pathological prevention.
Unique against other modalities that operate indirectly by disrupting viral protein function, CRISPR-Cas and RNAi function by direct nucleolytic destruction of the viral nucleic acids. Tan et al. showed that siRNA eliminates viral titers in the intestinal cells, but the authors report that the siRNA modality cannot access skeletal muscle nor the central nervous system (CNS), both of which are important reservoirs and key tissues for EV-A71 tropism and pathology.27 Our work here reports the direct elimination of EV-A71 from these key pathological tissue reservoirs. The siRNA reduced viral transcript by 40% at low dose (1 nmol) and 100% at high dose (1010 times at 10 mol) in vivo, which is also consistent with the lower efficiency of the same siRNA tested in vitro in RD cells,43 with the best result against the 3D gene achieving 77.5% reduction in viral titers and 89.3% reduction in viral plaque formation, compared to the >99.9% reduction in viral plaque formation using AAV-CRISPR-CasRx in vitro and in vivo. The CRISPR-Cas13 (specifically CasRx) approach is superior over RNAi in tissue accessibility, efficiency, and specificity,15,41,44,45 all of which contribute significantly to the safety and efficacy of the therapeutics.
We used the bioinformatics pipeline Cas13gRNAtor to design CasRx gRNAs that target conserved regions across the whole enterovirus phylogeny and showed that it identifies effective single gRNAs for viral inhibition. This pipeline focuses on gRNA design for targets with nucleotide diversity, such as viral genomes, and benefits from multiple predictive scoring algorithms, including conservation scores, entropy scores, gRNA scores,28 crRNA folding energy, local target ‘C’-context, and upstream target ‘U’-context, all in an easy-to-use format (https://github.com/chewlabSB2/Cas13gRNAtor). Cas13gRNAtor designs candidate gRNAs predicted to target conserved regions, exhibit minimal off-targets, and effect high on-target efficiency. Empirical data validates that the designed gRNAs are potent, highly specific without off-targets in the human transcriptome, and robust across different strains, which together indicate that a multi-parametric design pipeline that goes beyond sequence conservation17,21 offers a user-friendly single workflow towards designing gRNAs with multiple desired properties. The pipeline is modular, and advances in silico prediction of effective gRNAs, as exemplified by Wessels et al. for Cas13,28 could further reduce the need for empirically screening multiple gRNAs; while advances in off-target predictions, as exemplified by precedents with Cas9, could further increase specificity.
Development of new CRISPR-Cas13d antivirals could first apply Cas13gRNAtor in individual gRNA selection, followed by optimization in the number of gRNAs, to identify the best combination for antiviral activities. We used gRNA pools to target multiple regions of a crucial gene transcript for cleavage to suppress viral replication. This strategy is effective with the pooling of four gRNAs for the same gene target, and the pooling of more gRNAs enhances the efficacy of viral inhibition and reduces in vitro viral titer by more than 99.99%. A caveat is that despite gRNA pooling significantly increasing antiviral activity, our current study is limited to the few combinations used against EV-A71, and future work may uncover further mechanisms for effective combinations of gRNAs. The gRNA pool also offers potent pan-enterovirus A71 inhibitory activity, even in viral strains beyond the culture-adapted HFM41 lab strain. Despite some nucleotide variation across multiple viral strains, antiviral efficiency is only subtly improved by re-engineering the gRNAs to have a 100% match to the target sequence of each EV-A71 variant, suggesting that re-engineering is not a necessary step for a pan-enterovirus A71 targeting strategy. Despite some differences in the efficacy against the other EV-A71 strains, H, B5, and C4, this antiviral effect remains potent at a 1 log unit to 1.5 log unit reduction that translates to >90% reduction in viral replication for all the three strains of enteroviruses tested. A possible explanation for this difference could be the lower replication kinetics of these strains in vitro, which generally results in a much lower viral titre baseline following infection (Fig. 2). With a lower viral titre baseline, the effect of the antiviral reduction will be less pronounced. It is also observed that the reduction of GFP by AAV-CasRx-GFPgRNA is only 6%–28% in RD cells as these were measured by the GFP protein fluorescence level, whereas the titers from the EV-A71 plaque assay were measured in infectious units. The gRNAs against GFP targets the exogenously overexpressed GFP mRNA, resulting in downregulation of the GFP protein. The gRNAs against EV-A71 target the viral 3D polymerase mRNA/vRNA and would result in down-regulation of the viral 3D polymerase protein, which further inhibits viral production and infectious units; this 1 log reduction in viral titer is the compounded effect from RNA degradation, protein downregulation, and virus inhibition.
The suckling mice model recapitulates the clinical manifestation in pediatric human patients.34,42 We showed that death from lethal EV-A71 infection is prevented in the mice treated with AAVDJ-CasRx-EV71gRNAs. In this infected mouse model, EV-A71 clinical symptoms start from day 5 after infection, develop through day 11 after infection, and subsequently wane or progress to death (Fig. 3, Fig. 4d and Supplementary Fig. S9d and S10d). In comparison to control, mice treated with AAVDJ-CasRx-EV71gRNAs showed mild to no clinical symptoms throughout the observation period. The absence of EV-A71 VP2 protein expression in mice treated with AAVDJ-CasRx-EV71gRNA further indicates the inhibition of viral replication in the mouse hindlimb and spinal cord tissue, key sites of EV-A71 pathology that previous siRNA and plasmid modalities could not access in vivo27,45 (Supplementary Table).
The safety profile and efficacy of AAV and CRISPR-Cas is an area of active research.30,46, 47, 48 We assessed potential off-targeting by the AAV-CRISPR-Cas13-EV71_3DgRNAs using whole transcriptome DESeq, and the results are consistent with high specificity without off-targeting observed. For delivery, we used AAV vectors that are largely safe for in vivo therapeutic applications, as supported by clinical trials and FDA-approved products. The packaging limitation of AAV has been a major obstacle to the application of CRISPR-Cas technology in vivo, but recent discoveries of smaller Cas systems make it possible for all-in-one delivery of both the Cas and gRNAs within the same AAV, as demonstrated in this antiviral modality. Our study also highlights several opportunities for future investigation. First, we focused on anti-EV-A71 gRNAs; future studies could investigate if the increased potency from gRNA pooling is a general strategy effective against other targets and dissect the molecular mechanism underlying this synergy. Second, because our treatment potently eliminated EV-A71 in one dose, and because EV-A71 causes mainly acute infection similar to most other RNA viruses, re-administration of the treatment was not necessary. With other viruses that might require re-administration of the AAV-CRISPR-Cas due to reinfection or for enhanced potency, pre-existing immunity against the treatment may impose safety challenges. In such circumstances, it is possible to employ new immune-evading engineered AAV vectors49 or Cas proteins to bypass pre-existing immunity.
Our work demonstrates the potential of a new class of AAV-CRISPR-CasRx antivirals that directly target the RNA virus nucleic acid and effectively eliminate them from infected cells both in vitro and in vivo. This is an effective approach for inhibition of RNA viral replication and pathogenesis in vivo, with the potential to be further developed for clinical application in patients suffering from different types of circulating and emerging RNA viruses.
Contributors
CTK, JJHC, and CWL contributed to the conception of antiviral CRISPR-Cas application on HFMD. CTK and IBHM contributed to the conception of gRNAs scoring, selection, and screening strategy. CTK, SRV, and WLC contributed to the conception of the construction of the new Cas13d cargo plasmid for targeting RNA viruses. CTK, SRV, and DSL contributed to the production of AAVs bearing Cas13d and the different gRNAs sequences. BSC and IBHM contributed to the human transcriptome DESeq off-target experiments and analysis. CTK and BSC contributed to the flow cytometry experiments. RCHL, CTK, JJHC, and WLC contributed to the design and testing of gRNAs’ efficacy in inhibiting EV-A71 replication for in vitro screening. CTK, TY, JJHC, and WLC contributed to the design and testing of gRNAs in the efficacy of disease elimination in HFMD animal models. TY, CKM, YHW, and KG contribute to the harvesting and processing of mice tissues. CTK, TY, JJHC, and WLC contributed to the writing of the manuscript. CTK, TY, JJHC, and WLC have verified the underlying data. All the authors read and approved the final manuscript.
Data sharing statement
The data supporting the conclusions of this study are available within the article. Raw data files will be made available by inquiries to the corresponding authors (Wei Leong Chew: chewwl@gis.a-star.edu.sg and Justin Jang Hann Chu: miccjh@nus.edu.sg).
Declaration of interests
CTK, RCHL, TY, JJHC, and WLC have filed patents and provisional applications based on this work.
Acknowledgments
This work is supported by Agency for Science, Technology and Research (A∗STAR) Assured Research Budget, A∗STAR Central Research Fund UIBR SC18/21-1089UI, A∗STAR Industrial Alignment Fund Pre-Positioning (IAF-PP) grant H17/01/a0/012, MOE Tier 2 2017 (MOE2017-T2-1-078; MOE-T2EP30221-0005), and NUHSRO/2020/050/RO5+5/NUHS-COVID/4.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104682.
Contributor Information
Justin Jang Hann Chu, Email: miccjh@nus.edu.sg.
Wei Leong Chew, Email: chewwl@gis.a-star.edu.sg.
Appendix A. Supplementary data
References
- 1.Woolhouse M.E.J., Brierley L. Epidemiological characteristics of human-infective RNA viruses. Sci Data. 2018;5:1–6. doi: 10.1038/sdata.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doud M.B., Lee J.M., Bloom J.D. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-03665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belshaw R., Gardner A., Rambaut A., Pybus O.G. Pacing a small cage: mutation and RNA viruses. Trends Ecol Evol. 2008;23(4):188–193. doi: 10.1016/j.tree.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Beltran W.F., Lam E.C., St Denis K., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starr T.N., Greaney A.J., Addetia A., et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371(6531):850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ooi M.H., Wong S.C., Lewthwaite P., Cardosa M.J., Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9(11):1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 7.Jin Y., Zhang R., Wu W., Duan G. Innate immunity evasion by enteroviruses linked to epidemic hand-foot-mouth disease. Front Microbiol. 2018;9(OCT):2422. doi: 10.3389/fmicb.2018.02422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S.-M., Ho T.-S., Lin H.-C., Lei H.-Y., Wang J.-R., Liu C.-C. Reemerging of enterovirus 71 in Taiwan: the age impact on disease severity. Eur J Clin Microbiol Infect Dis. 2012;31(6):1219–1224. doi: 10.1007/s10096-011-1432-6. [DOI] [PubMed] [Google Scholar]
- 9.Hong J., Liu F., Qi H., et al. Changing epidemiology of hand, foot, and mouth disease in China, 2013-2019: a population-based study. Lancet Reg Health West Pac. 2022;20 doi: 10.1016/j.lanwpc.2021.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi E.J., Shin Y.J., Kim J.H., Kim T.G., Chang S.Y. Enterovirus 71 infection and vaccines. Clin Exp Vaccine Res. 2017;6(1):4–14. doi: 10.7774/cevr.2017.6.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan Y.W., Chu J.J.H. Protecting the most vulnerable from hand, foot, and mouth disease. Lancet Infect Dis. 2021;21(3):308–309. doi: 10.1016/S1473-3099(20)30452-7. [DOI] [PubMed] [Google Scholar]
- 12.Lin J.Y., Kung Y.A., Shih S.R. Antivirals and vaccines for enterovirus A71. J Biomed Sci. 2019;26(1):1–10. doi: 10.1186/s12929-019-0560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu F., Xu W., Xia J., et al. Efficacy, safety, and immunogenicity of an enterovirus 71 vaccine in China. N Engl J Med. 2014;370(9):818–828. doi: 10.1056/NEJMoa1304923. [DOI] [PubMed] [Google Scholar]
- 14.Teo F.M.S., Nyo M., Wong A.A., et al. Cytokine and chemokine profiling in patients with hand, foot and mouth disease in Singapore and Malaysia. Sci Rep. 2018;8(1):1–9. doi: 10.1038/s41598-018-22379-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan W.X., Chong S., Zhang H., et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70(2):327–339.e5. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbott T.R., Dhamdhere G., Liu Y., et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181(4):865–876.e12. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koujah L., Shukla D., Naqvi A.R. CRISPR-Cas based targeting of host and viral genes as an antiviral strategy. Semin Cell Dev Biol. 2019;96:53–64. doi: 10.1016/j.semcdb.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchard E.L., Vanover D., Bawage S.S., et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat Biotechnol. 2021;39(6):717–726. doi: 10.1038/s41587-021-00822-w. [DOI] [PubMed] [Google Scholar]
- 20.Li H., Wang S., Dong X., et al. CRISPR-Cas13a cleavage of dengue virus NS3 gene efficiently inhibits viral replication. Mol Ther Nucleic Acids. 2020;19:1460–1469. doi: 10.1016/j.omtn.2020.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freije C.A., Myhrvold C., Boehm C.K., et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell. 2019;76(5):826–837.e11. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashraf M.U., Salman H.M., Khalid M.F., et al. CRISPR-Cas13a mediated targeting of hepatitis C virus internal-ribosomal entry site (IRES) as an effective antiviral strategy. Biomed Pharmacother. 2021;136 doi: 10.1016/j.biopha.2021.111239. [DOI] [PubMed] [Google Scholar]
- 23.Bryant L.M., Christopher D.M., Giles A.R., et al. Lessons learned from the clinical development and market authorization of Glybera. Hum Gene Ther Clin Dev. 2013;24(2):55–64. doi: 10.1089/humc.2013.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bainbridge J.W.B., Mehat M.S., Sundaram V., et al. Long-term effect of gene therapy on Leber’s congenital amaurosis. N Engl J Med. 2015;372(20):1887–1897. doi: 10.1056/NEJMoa1414221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell S., Bennett J., Wellman J.A., et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390(10097):849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet. 2020;21(4):255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 27.Tan E.L., Tan T.M.C., Tak Kwong Chow V., Poh C.L. Inhibition of enterovirus 71 in virus-infected mice by RNA interference. Mol Ther. 2007;15(11):1931–1938. doi: 10.1038/sj.mt.6300287. [DOI] [PubMed] [Google Scholar]
- 28.Wessels H.-H., Méndez-Mancilla A., Guo X., Legut M., Daniloski Z., Sanjana N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat Biotechnol. 2020;38(6):722–727. doi: 10.1038/s41587-020-0456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon C.E. A mathematical theory of communication. Bell Syst Tech J. 1948;27(4):623–656. [Google Scholar]
- 30.Chew W.L., Tabebordbar M., Cheng J.K.W., et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13(10):868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 33.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun J., Yogarajah T., Lee R.C.H., et al. Drug repurposing of pyrimidine analogs as potent antiviral compounds against human enterovirus A71 infection with potential clinical applications. Sci Rep. 2020;10(1):8159. doi: 10.1038/s41598-020-65152-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baggen J., Thibaut H.J., Strating J.R.P.M., van Kuppeveld F.J.M. The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol. 2018;16(6):368–381. doi: 10.1038/s41579-018-0005-4. [DOI] [PubMed] [Google Scholar]
- 36.Chen I.-Y., Ichinohe T. Response of host inflammasomes to viral infection. Trends Microbiol. 2015;23(1):55–63. doi: 10.1016/j.tim.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Awomoyi A.A. The human solute carrier family 11 member 1 protein (SLC11A1): linking infections, autoimmunity and cancer? FEMS Immunol Med Microbiol. 2007;49(3):324–329. doi: 10.1111/j.1574-695X.2007.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang H., Weng L., Zhang N., et al. Biochemical characterization of enterovirus 71 3D RNA polymerase. Biochim Biophys Acta. 2011;1809(3):211–219. doi: 10.1016/j.bbagrm.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Li Y., Yu J., Qi X., Yan H. Monoclonal antibody against EV71 3D(pol) inhibits the polymerase activity of RdRp and virus replication. BMC Immunol. 2019;20(1):6. doi: 10.1186/s12865-019-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Linden L., Vives-Adrián L., Selisko B., et al. The RNA template channel of the RNA-dependent RNA polymerase as a target for development of antiviral therapy of multiple genera within a virus family. PLoS Pathog. 2015;11(3) doi: 10.1371/journal.ppat.1004733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun J., Ennis J., Turner J.D., Chu J.J.H. Single dose of an adenovirus vectored mouse interferon-α protects mice from lethal EV71 challenge. Antiviral Res. 2016;134:207–215. doi: 10.1016/j.antiviral.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sim A.C.N., Luhur A., Tan T.M.C., Chow V.T.K., Poh C.L. RNA interference against enterovirus 71 infection. Virology. 2005;341(1):72–79. doi: 10.1016/j.virol.2005.06.047. [DOI] [PubMed] [Google Scholar]
- 44.Powell J.E., Lim C.K.W., Krishnan R., et al. Targeted gene silencing in the nervous system with CRISPR-Cas13. Sci Adv. 2022;8(3) doi: 10.1126/sciadv.abk2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chin W.-X., Ang S.K., Chu J.J.H. Recent advances in therapeutic recruitment of mammalian RNAi and bacterial CRISPR-Cas DNA interference pathways as emerging antiviral strategies. Drug Discov Today. 2017;22(1):17–30. doi: 10.1016/j.drudis.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 46.Amoasii L., Hildyard J.C.W., Li H., et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362(6410):86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S., Ren S., Bai R., et al. No off-target mutations in functional genome regions of a CRISPR/Cas9-generated monkey model of muscular dystrophy. J Biol Chem. 2018;293(30):11654–11658. doi: 10.1074/jbc.AC118.004404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng H.-X., Zhai H., Shi Y., et al. Efficacy and long-term safety of CRISPR/Cas9 genome editing in the SOD1-linked mouse models of ALS. Commun Biol. 2021;4(1):396. doi: 10.1038/s42003-021-01942-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreno A.M., Palmer N., Alemán F., et al. Immune-orthogonal orthologues of AAV capsids and of Cas9 circumvent the immune response to the administration of gene therapy. Nat Biomed Eng. 2019;3(10):806–816. doi: 10.1038/s41551-019-0431-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.