Abstract
Although nitrogen (N) is known to affect mineral element homeostasis in plants, the molecular mechanisms of interactions between N and other nutrients remain largely unclear. We report here that N supply affects ion homeostasis in maize. Berberine hemisulfate staining and a propidium iodide penetration assay showed that N luxury significantly delayed Casparian strip (CS) formation in maize roots. We further demonstrated that N-mediated CS formation in maize was independent of RBOHF-activated H2O2 production. N luxury induced the expression of ZmmiR528 in whole roots and root tips. Knockdown and loss-of-function of ZmmiR528 promoted CS formation under both N-luxury and N-deficient conditions. Both ZmMIR528a and ZmMIR528b contribute to early CS formation under different N conditions. RNA-seq and real-time RT-PCR analysis demonstrated that ZmLAC3, but not ZmLAC5, responded to N treatments. Consistent with results obtained with ZmmiR528 TM transgenic maize and mir528a/b loss-of-function mutants, transgenic maize overexpressing ZmLAC3 displayed early CS formation under different N conditions. Under field conditions, K, Ca, Mn, Cu, Mg, and Zn concentrations were greater in the ear leaf of ZmLAC3-overexpressing transgenic maize than in the wild type. These results indicate that ZmmiR528 affects CS formation in maize by regulating the expression of ZmLAC3, and modification of CS formation has the potential to improve maize quality.
Key words: nitrogen, ion homeostasis, miR528, Casparian strip, maize
ZmmiR528 is important for maize lodging resistance under N-luxury conditions. This work reports that ZmmiR528 affects Casparian strip formation in maize by regulating the expression of ZmLAC3, and modification of Casparian strip formation has the potential to improve maize quality.
Introduction
Nitrogen (N) is a key environmental factor affecting crop production, nutritional quality, taste, storage, and pathogen resistance (Williams and Salt, 2009). To meet the growing demand for food production, many farmers have been applying excessive quantities of chemical N fertilizer. The excessive application of N fertilizer leads not only to environmental problems but also to increased N accumulation in the field (Ju et al., 2009; Guo et al., 2010). Excessive or deficient levels of N are known to affect how plants metabolize other nutrients. For example, N application increased the concentrations of potassium (K) and magnesium (Mg) but decreased the concentration of phosphorus (P) in the green leaves of Arabidopsis thaliana (Yan et al., 2019). Low-N stress significantly increased the concentrations of K, P, and Mg but reduced the concentrations of calcium (Ca), zinc (Zn), and manganese (Mn) in the shoots of a low-N-tolerant barley genotype (Quan et al., 2017). However, the molecular basis of interactions between N and other nutrients is unclear.
Using a multiple ion-uptake phenotyping platform, researchers recently found that plant uptake of nitrate, ammonium, K, P, and sulfate (S) was governed by shared and uncharacterized mechanisms (Griffiths et al., 2021). One possible shared mechanism could involve the formation of Casparian strips (CSs) in the endodermis, because CSs form a crucial paracellular diffusion barrier for selective nutrient uptake (Geldner, 2013). The biogenesis of CSs has been well studied in Arabidopsis, and the key genes have been identified. Initiation of CS formation involves the localization of Casparian strip membrane domain proteins (CASPs) at the CS membrane domain (Roppolo et al., 2011). The CS membrane domain recruits the respiratory burst oxidase homolog F peroxidase 64 (PER64) and the dirigent-like protein enhanced suberin 1 (ESB1) for lignin polymerization and correct deposition (Baxter et al., 2009; Hosmani et al., 2013; Lee et al., 2013).
The CS consists of a ring-like impregnation of the primary cell wall with lignin (Naseer et al., 2012). Lignin is a phenylpropanoid-derived polymer that results from oxidative polymerization of monolignol precursors in the plant cell wall (Vanholme et al., 2008). Monolignol dehydrogenation involves peroxidases and/or laccases (Vanholme et al., 2010; Zhao et al., 2013). The function of peroxidases in CS formation has been verified. However, the role of laccases in CS formation is controversial. In Arabidopsis, laccases and peroxidases co-localize in lignified secondary cell walls throughout stem development (Hoffmann et al., 2020). Pharmacological perturbation of LAC3 led to dispersed localization of CASP1 and compensatory ectopic lignification (Zhuang et al., 2020). By contrast, high-order mutants revealed an essential requirement for peroxidases but not laccases in CS lignification (Rojas-Murcia et al., 2020). However, the authors could not exclude the possibility that laccases might be required for CS formation under specific conditions and/or that upregulation of LACs could contribute to CS formation (Rojas-Murcia et al., 2020).
In Arabidopsis, miRNA397b-LACCASE2 was reported to affect root lignification under water and P deficiency (Khandal et al., 2020). miR528 is a monocot-specific miRNA that has multiple roles in rice, including antiviral responses and regulation of flowering time and pollen development (Wu et al., 2017; Yang et al., 2019; Yao et al., 2019; Zhang et al., 2019). miR528 has evolved preferences for distinct target genes in different monocots (Zhu et al., 2020). In maize, ZmmiR528 post-transcriptionally regulates the expression levels of ZmLAC3/ZmLAC5 (Sun et al., 2018). Our previous results and those of others indicate that N supply affects the mRNA abundance of ZmmiR528 in maize (Trevisan et al., 2012; Sun et al., 2018). Our results also showed that knockdown of ZmMIR528 or overexpression of ZmLAC3 increases lignin contents in soil-grown maize (Sun et al., 2018). Thus, the relationship among the miR528-LAC module, CS formation, and selective nutrient uptake in maize under N stress warrants further study.
In the present research, we show that mineral element profiles and CS formation in maize are significantly affected by N supply. We found that N-mediated CS formation in maize roots was independent of RBOHF-activated H2O2 production. We further found that, by regulating the abundance of ZmLAC3, ZmmiR528 is involved in CS formation under different N conditions.
Results
N supply affects ion homeostasis in maize
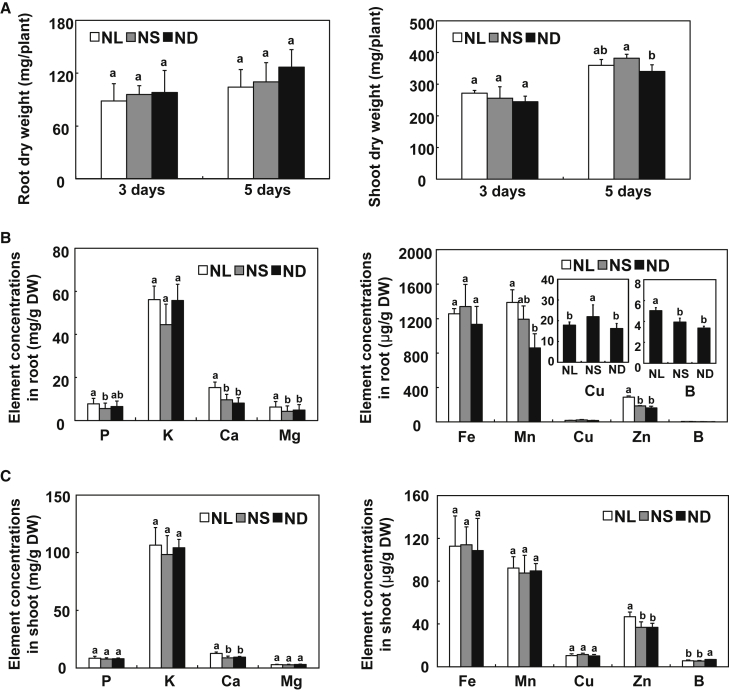
Low-N stress affects ion homeostasis in Arabidopsis thaliana and barley (Quan et al., 2017; Yan et al., 2019). To test how N affected maize ion homeostasis under different N conditions, we grew inbred line B73 under N-sufficient (NS), N-luxury (NL), or N-deficient (ND) conditions. Short-duration (3 days) N treatment had no significant effects on shoot or root dry weight of maize (Figure 1A). When N treatment was extended to 5 days, ND caused maize leaves to show a typical N-deficient phenotype, with discoloration of older leaves and accumulation of the purple flavonoid pigment anthocyanin in the stem (Supplemental Figure 1A and 1B). Although growth of maize seedlings did not differ under NS vs. NL, shoot dry weight was significantly lower under ND than under NS (Figure 1A).
Figure 1.
Effects of N supply on mineral element profiles of maize.
Inbred line B73 was grown in a hydroponic solution containing 8 mM, 4 mM, or 0.04 mM of NO3−-N for the indicated durations.
(A) Shoot and root fresh weights.
(B) Mineral element profiles of maize roots.
(C) Mineral element profiles of maize shoots. Values are means ± SD (n = 4). Means with the same letter are not significantly different at P < 0.05 according to the least signifificant difference (LSD) test. NL, NS, and ND indicate N luxury, N sufficiency, and N deficiency, respectively.
We next used inductively coupled plasma mass spectrometry to determine the effects of the three N treatments on mineral element profiles in maize shoots and roots. Except for Cu in the roots, the concentrations of mineral elements in maize shoots and roots were similar between NS and ND (Figure 1B and 1C). Relative to NS, however, NL significantly increased the concentrations of P, Ca, Mg, Zn, and B in roots and the Ca concentration in shoots (Figure 1B and 1C). These results indicated that N supply could affect ion homeostasis in maize.
N supply affects CS formation in maize
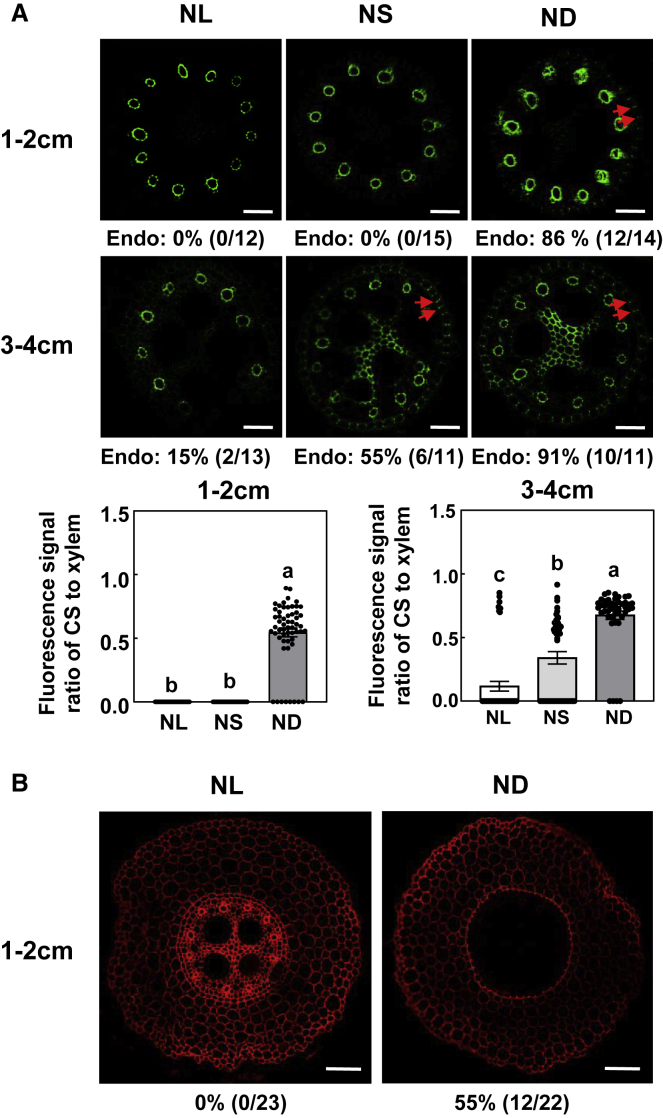
Differences in ion homeostasis caused by different N conditions could result from changes in root morphology. Although root dry weight and primary root length did not differ significantly among N treatments (Figure 1A; Supplemental Figure 2), concentrations of Ca, Mn, and Zn, which are radially transported through apoplastic or coupled transcellular pathways (White, 2001; Baxter et al., 2009; Chen et al., 2019), were affected by N supply (Figure 1B). This prompted us to investigate CS development under different N conditions because the CS is the crucial paracellular diffusion barrier. To observe CS development, we used berberine–aniline blue, which was previously found to perform equally well on a variety of plants (Brundrett et al., 1988). Because maize roots are large, we recorded the position of CS formation in terms of the distance from the root tip, instead of counting endodermal cells after the onset of root elongation as in Arabidopsis (Naseer et al., 2012; Lee et al., 2013). At the root tip (0–2 cm from the root apex) of the primary root, CSs did not form under NS or NL but formed in 86% of the observed root tips under ND (Figure 2A). In agreement with these observations, the fluorescence signal ratio of CS to xylem vessels was higher under ND than under NS or NL (Figure 2A).
Figure 2.
Casparian strip formation in inbred line B73 as affected by N supply.
Inbred line B73 was grown in a hydroponic solution containing 8 mM, 4 mM, or 0.04 mM NO3−-N for 3 days. Casparian strip formation was then examined at different distances from the apex of primary maize roots.
(A) Fluorescence images and fluorescence signal ratio of CS to xylem vessels of primary maize roots at different distances from the tip; the roots were stained with berberine hemisulfate, and Casparian strips are indicated by red arrowheads. Values are means ± SEM. Means with the same letter are not significantly different at P < 0.05 according to the LSD test.
(B) Propidium iodide (PI) penetration at 1–2 cm from the root apex under NL and ND. The maize seedlings were exposed to 100 μM PI in the dark for 6 h. Scale bars represent 50 μm in (A) and 100 μm in (B). NL, NS, ND, and Endo indicate N luxury, N sufficiency, N deficiency, and endodermis, respectively. Numbers below the photographs indicate the percentage of roots with Casparian strips.
ND also promoted the formation of exodermal CSs in the same region (Supplemental Figure 3). Although exodermal CSs were not detected at 3.0–4.0 cm from the apex of primary roots under NL or NS (Supplemental Figure 3), endodermal CSs were evident in such roots, although at a much lower rate than under ND (Figure 2A). At 5.0–6.0 cm from the apex of primary roots, the occurrence of endodermal CSs was similar under NS and ND; at this location, the occurrence was 29% higher under NS and 37% higher under ND than under NL (Supplemental Figure 3). As in other root tissues, CS development at the tips of seminal and lateral roots also increased under ND (Supplemental Figure 4). Berberine hemisulfate also stains suberin (Brundrett et al., 1988). We then used Fluorol yellow, a fluorescent suberin dye, to test whether lignin was the major contributor to the CS phenotypes. No specific signal was detected in the CS region of the root tip (Supplemental Figure 5), suggesting that lignin was the major contributor to the observed phenotypes.
The fluorescent dye propidium iodide (PI) is widely used to visualize the appearance of endodermal CSs (Naseer et al., 2012), and we therefore used PI to assess endodermal CS formation in maize. Given the similar effects of NS and NL on CS formation revealed by the use of berberine–aniline blue (Figure 2A), we limited the N treatments in the following experiments to NL and ND. PI uptake was blocked at the root tip (0–2 cm from the root apex) under ND but not under NL (Figure 2B). These results were consistent with those of roots treated with berberine–aniline blue and further demonstrated that CS formation increases in maize under N-limiting conditions.
H2O2 is not important in N-mediated CS formation in maize
Because CSs are ring-like cell wall modifications consisting of lignin polymers (Naseer et al., 2012; Geldner, 2013), we determined whether lignin deposition in root tips was affected by N conditions using Wiesner staining. Surprisingly, lignin was detected between anticlinal cells at the tips of primary roots even under NL, under which CS did not form (Supplemental Figure 6A). In the Wiesner test, lignin is stained principally through reactions with coniferaldehyde end groups (Berthet et al., 2011). Our results showed that monolignol precursors of lignin were present under both NL and ND conditions.
To explore potential regulators involved in N-mediated CS formation in maize, we compared the whole-transcriptome profiles of root tips (2 cm from the apex) subjected to NL and ND for 3 days. Three biological replicates were performed for each treatment. The six RNA libraries yielded more than 220 million raw reads, approximately 90% of which were aligned to the maize B73_RefGen_v4 genome (ftp://ftp.ensemblgenomes.org/pub/plants/release-41/fasta/zea_mays/dna/). Sequences that could not be mapped to the maize genome were discarded, and only those which mapped perfectly were analyzed further. Based on the criteria log2(fold-change ratio) ≥1 and adjusted P value ≤0.05, 10 366 differentially expressed genes (DEGs) were identified by DESeq2 software (Wang et al., 2010). Compared with the NL treatment, 5590 genes were upregulated and 4776 genes were downregulated by ND (Supplemental Figure 7). Upregulated genes involved in the phenylpropane and lignin pathways are listed in Supplemental Table 1. We then performed gene ontology (GO; http://bioinfo.cau.edu.cn/agriGO/) analysis to determine the molecular events related to the DEGs. GO analysis indicated that the 5590 ND-induced DEGs were enriched in the biological process terms phenylpropanoid biosynthetic process (GO:0009699, P = 2.8e−7), phenylpropanoid metabolic process (GO:0009698, P = 3e−9), secondary metabolic process (GO:0019748, P = 1.2e−8), lignin metabolic process (GO:0009808, P = 4.6e−6), antioxidant activity (GO:0016209, P = 3.1e−5), and response to oxidative stress (GO:0006979, P = 1.4e−5) (Supplemental Figure 7).
CSs are polymerized by oxidative coupling of monolignols resulting from the activity of specific NADPH oxidases and peroxidases. In Arabidopsis, AtRBOHF contributes to CS formation by activating NADPH oxidase-dependent localized H2O2 production at the sites of CS formation (Lee et al., 2013). The two closest homologs of AtRBOHF in maize are Zm00001d038762 and Zm00001d043543 (Supplemental Figure 8). Although CS formation obviously differed between NL and ND, RNA-seq and qPCR analysis of root tips showed that the expression levels of the two AtRBOHF homologs were similar between the two N treatments (Supplemental Figures 6B and 9A). We also assessed the expression levels of other AtRBOHF homologs in root tips. Zm00001d009349 and Zm00001d040974 were not considered owing to their low abundance (fragments per kilobase of transcript per million mapped reads <0.1). The fold changes (ND vs. NL) of these AtRBOHF homologs were ≤1.5 (Supplemental Figure 9B). Although H2O2 content was higher in maize tips under ND than under NL, staining with 3,3′-diaminobenzidine showed that H2O2 could accumulate in maize root tips under both conditions (Supplemental Figure 6C). We next determined the location of N-induced H2O2 accumulation by cerium chloride assay. With this assay, H2O2 was observed in endodermal cells at the positions of CSs (Supplemental Figure 6D). DPI pre-treatment led to almost complete absence of local H2O2 (Supplemental Figure 6D). These results indicated that the N-mediated CS formation of maize may not depend on RBOHF-activated H2O2 production.
ZmmiR528 is involved in early CS formation under different N conditions
Our previous results showed that ZmmiR528 regulates lignin biosynthesis under NL (Sun et al., 2018), and we therefore hypothesized that ZmmiR528 might contribute to CS formation under different N conditions. To test this hypothesis, we examined the expression patterns of ZmmiR528 in root tips by in situ hybridization. ZmmiR528 was detected throughout the root tip under NL (Supplemental Figure 10A). Moreover, stem-loop qRT-PCR showed that ND reduced ZmmiR528 abundance in both whole roots and root tips (Supplemental Figure 10B).
To characterize the function of ZmmiR528 in CS formation, we utilized ZmmiR528 knockdown transgenic maize (TM), which was originally generated for determining lodging resistance of maize (Sun et al., 2018). The genetic background of this transgenic maize was inbred line CZ01, not the genome-sequenced inbred line B73. We first tested the effects of N treatment on the mineral element profiles in CZ01. As was the case for B73, N supply affected ion homeostasis in CZ01 (Supplemental Figure 11). Consistent with our observations in B73, concentrations of Ca and Mn in roots and shoots of CZ01 were greater under NL than under ND (Supplemental Figure 11). By contrast, K concentrations in roots and shoots of CZ01 and B73 behaved differently under N deficiency (Supplemental Figure 11). These results suggested that the effects of N treatment on mineral element profiles were independent of maize genotype.
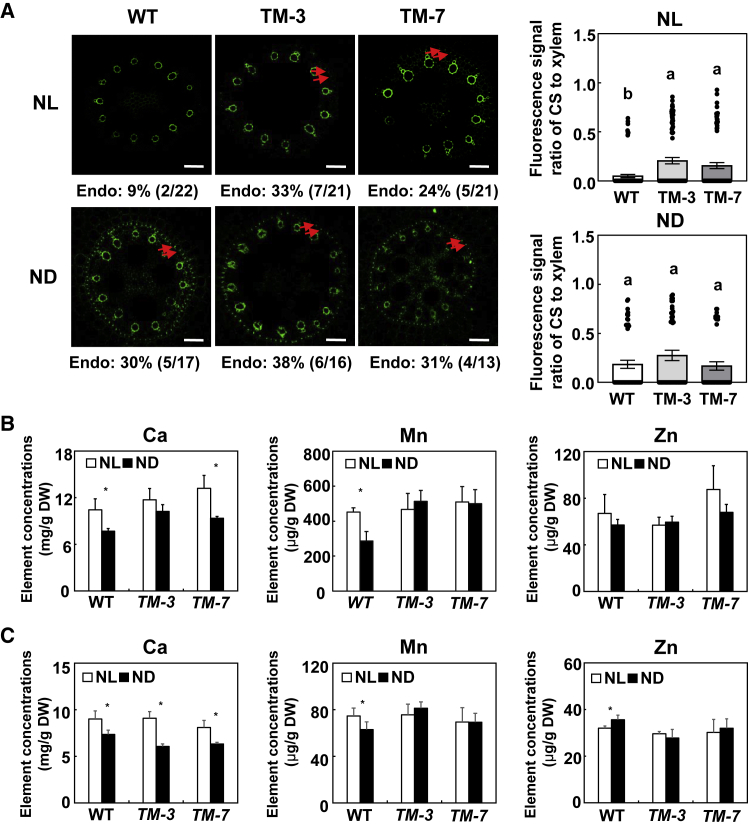
Under NL, the observed probability of endodermal CS formation and the fluorescence signal ratio of CS to xylem vessels in root tips were much higher in TM transgenic maize than in the wild type (WT), and the observed probability of endodermal CS formation in root tips of TM transgenic maize was negatively related to ZmmiR528 abundance (Figure 3A; Sun et al., 2018). By contrast, the observed probability of endodermal CS formation in root tips was similar in TM transgenic maize and the WT under ND (Figure 3A). In addition, the probability of endodermal CS formation in root tips was much lower in CZ01 than in B73 under ND conditions (Figures 2A and 3A). This difference could be due to the different tolerances of CZ01 and B73 to N stress (Supplemental Figure 12).
Figure 3.
Effects of N supply on Casparian strip formation and mineral element profiles in ZmmiR528 knockdown (TM) transgenic maize.
ZmmiR528 knockdown transgenic maize and inbred CZ01 were grown in a hydroponic solution containing 8 mM or 0.04 mM NO3−-N for 3 days.
(A) Casparian strip formation in ZmmiR528 TM transgenic maize as affected by N supply. The area 1.0–1.5 cm from the apex of roots grown under NL or ND was sampled, stained with berberine hemisulfate, and assessed for Casparian strip formation. Casparian strips are indicated by red arrowheads. Numbers below the photographs indicate the percentage of root tips with Casparian strips (% and number of roots that formed Casparian strips/total root number). Scale bars represent 50 μm. Values are means ± SEM. Means with the same letter are not significantly different at P < 0.05 according to the LSD test.
(B and C) Effects of N on Ca, Mn, and Zn concentrations in roots (B) and shoots (C) of ZmmiR528 TM transgenic maize. Values are means ± SD (n = 4). ∗P < 0.05 (Student’s t-test) indicates a significant difference from the control. NL and ND indicate N luxury and N deficiency, respectively.
We then determined the mineral element profiles in shoots and roots of hydroponically grown TM transgenic maize and WT maize under NL and ND. We focused on Ca, Mn, and Zn because of their potential apoplastic or coupled transcellular transport pathways (White, 2001; Baxter et al., 2009; Chen et al., 2019). NL increased Mn concentrations in roots and shoots of CZ01 but not of TM transgenic maize (Figure 3B and 3C). Compared with WT maize, ND increased Ca concentrations in the roots and reduced Ca concentrations in the shoots of TM transgenic maize (Figure 3B and 3C).
Both ZmMIR528a and ZmMIR528b are involved in early CS formation
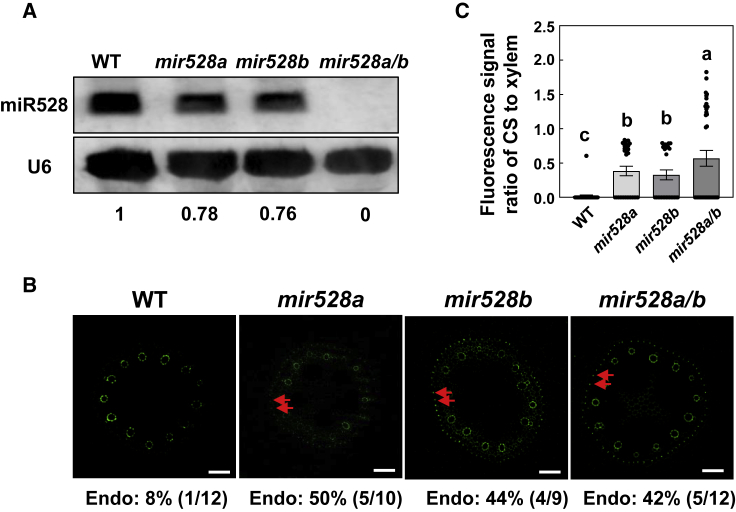
Unlike other monocots, which have only one member of the miR528 family (Wu et al., 2017; Zhu et al., 2020), maize has two members. Members of microRNA (miRNA) families usually have redundant functions (Sieber et al., 2007). However, previous research has shown that closely related miRNAs have different functions and regulatory specialization in Arabidopsis (Sieber et al., 2007; Du et al., 2017). To determine whether ZmMIR528a and ZmMIR528b have specific roles in early CS formation under different N conditions, we used the CRISPR-Cas9 system to generate ZmMIR528a and ZmMIR528b single or double loss-of-function mutants (mir528a, mir528b, and mir528a/b). Cas9-free transgenic plants were identified for phenotypic analysis. The gene sequences of these loss-of-function mutants revealed a nucleotide deletion in the mature miR528 and miR528∗ region that destroyed the miR528/miR528∗ duplexes (Supplemental Figure 13). Small RNA gel blot analysis showed that ZmmiR528 transcripts were reduced in mir528a and mir528b mutants and absent in mir528a/b loss-of-function mutants (Figure 4A). Because of the similar probability of endodermal CS formation in root tips of TM transgenic maize and WT maize under ND, we observed endodermal CS formation in mir528a, mir528b, and mir528a/b loss-of-function mutants under NL. Under NL, the observed probability of endodermal CS formation in root tips was much higher in mir528a, mir528b, and mir528a/b loss-of-function mutants than in the WT (Figure 4B). Although the observed probability of endodermal CS formation in root tips was similar among mir528a, mir528b, and mir528a/b loss-of-function mutants, the fluorescence signal ratio of CS to xylem vessels was much higher in the mir528a/b double mutant than in the mir528a and mir528b mutants (Figure 4C). These results suggested that ZmMIR528a and ZmMIR528b had both redundant and specific functions in early CS formation under NL.
Figure 4.
Casparian strip formation in mir528a, mir528b, and mir528a/b loss-of-function mutants.
(A) ZmmiR528 levels in the WT and in mir528a, mir528b, and mir528a/b loss-of-function mutants as indicated by small RNA northern blots. Numbers below each lane indicate relative expression. U6 RNA was probed as a loading control.
(B) Effects of N supply on Casparian strip formation in mir528a, mir528b, and mir528a/b loss-of-function mutants grown in a hydroponic solution containing 8 mM NO3−-N for 3 days. The area 1.0–1.5 cm from the root apex was sampled, stained with berberine hemisulfate, and assessed for Casparian strip formation. Casparian strips are indicated by red arrowheads. Numbers below the photographs indicate the percentage of root tips with Casparian strips (% and number of roots that formed Casparian strips/total root number). Scale bars represent 50 μm.
(C) Fluorescence signal ratio of Casparian strips to xylem vessels. Values are means ± SEM. Means with the same letter are not significantly different at P < 0.05 according to the LSD test.
Overexpression of ZmLAC3 promotes CS formation under different N conditions
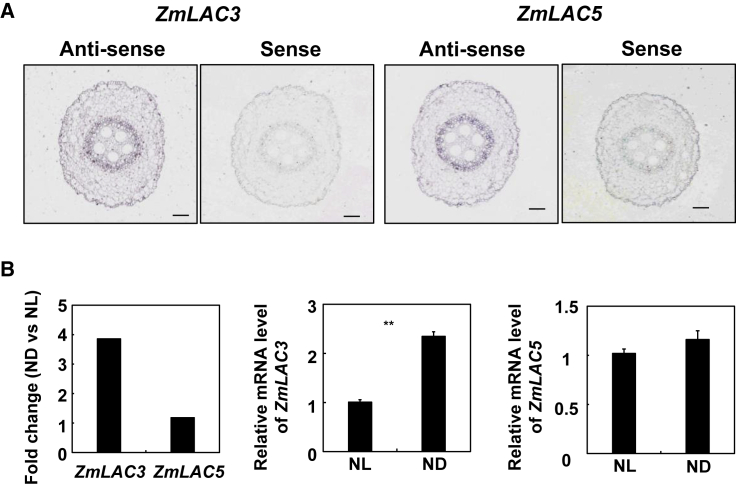
Our previous results showed that ZmmiR528 regulated lignin biosynthesis by repressing the expression levels of ZmLAC3 and ZmLAC5 (Sun et al., 2018). To determine which ZmLACs contributed to the early CS formation in ZmmiR528 knockdown transgenic maize and mir528a/b loss-of-function mutants under different N conditions, we first detected the expression patterns of ZmLAC3 and ZmLAC5 at the root tip by in situ hybridization. Anti-sense and sense probes on the images indicate the positive and negative signals, respectively. Both ZmLAC3 and ZmLAC5 were detected throughout the root tip under ND (Figure 5A), which was consistent with the location of ZmmiR528. We also analyzed the expression levels of ZmLAC3 and ZmLAC5 as indicated by RNA-seq data from the root tip. The mRNA abundance of ZmLAC3 was affected by the N treatments, but that of ZmLAC5 was not (Figure 5B). This result was verified by real-time RT-PCR (Figure 5B). We therefore hypothesized that ZmLAC3 might contribute to early CS formation in ZmmiR528 knockdown transgenic maize under different N conditions.
Figure 5.
Expression patterns of ZmLAC3 and ZmLAC5 in an area 1–2 cm from the root apex.
(A) Accumulation of ZmLAC3 and ZmLAC5 transcripts in the root tip of maize grown hydroponically under ND as determined by in situ hybridization analysis. Anti-sense and sense probes on the images indicate the positive and negative signals, respectively. Scale bars represent 100 μm.
(B) Responses of ZmLAC3 and ZmLAC5 to N treatments as determined by RNA-seq analysis and qRT-PCR. For RNA-seq analysis, the fragments per kilobase of transcript per million mapped reads value was used to determine normalized gene expression. For qRT-PCR, the expression levels were normalized to that of ZmUBQ12, and the levels of ZmLAC3 and ZmLAC5 transcripts under NL were set to 1.0. Error bars represent the SE of three independent experiments. ∗∗P < 0.01 (Student’s t-test) indicates a significant difference between NL and ND. NL and ND indicate N luxury and N deficiency, respectively.
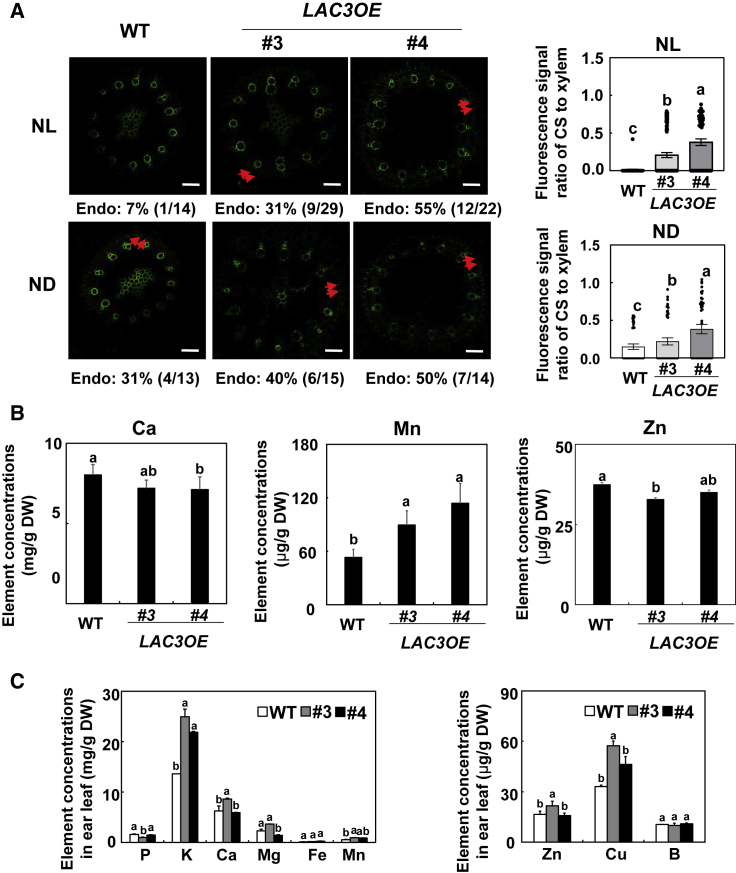
We next took advantage of the already available ZmLAC3-overexpressing transgenic maize (LAC3OE, CZ01 background) (Sun et al., 2018), and we used F4 homozygotes to determine the effect of N supply and ZmLAC3 overexpression on CS formation. The occurrence of endodermal CSs and the fluorescence signal ratio of CS to xylem vessels in root tips were much higher in ZmLAC3-overexpressing transgenic maize than in the WT under NL (Figure 6A). The occurrence of CSs in LAC3OE lines did not increase further in ND conditions (Figure 6A), hinting at its role in nitrogen dependence.
Figure 6.
Effects of ZmLAC3 overexpression on Casparian strip formation in roots, and mineral element profiles in ear leaves of field-grown maize.
(A) Casparian strip formation in ZmLAC3-overexpressing transgenic maize (LAC3OE) as affected by N supply. LAC3OE transgenic maize was grown in a hydroponic solution containing 8 mM or 0.04 mM NO3−-N for 3 days. The area 1.0–1.5 cm from the root apex under NL and ND was observed for Casparian strip formation. Casparian strips are indicated by red arrowheads. Numbers below the photographs indicate the percentage of root tips with Casparian strips. Scale bars represent 50 μm. Values are means ± SEM.
(B) Ca, Mn, and Zn concentrations in the shoots of LAC3OE transgenic maize and WT maize under NL.
(C) Mineral element profiles of LAC3OE transgenic maize grown in the field. Values are means ± SD (n = 4). Means with the same letter are not significantly different at P < 0.05 according to the LSD test. NL and ND indicate N luxury and N deficiency, respectively.
We determined the Ca, Mn, and Zn concentrations in shoots of hydroponically grown ZmLAC3-overexpressing transgenic maize and WT maize under NL. Compared with WT maize, overexpression of ZmLAC3 increased Mn concentrations and reduced Ca and Zn concentrations (Figure 6B). ZmLAC3-overexpressing transgenic maize and WT maize were then planted in the field (300 kg N ha−1, as urea) in Hainan province. Ear leaves were sampled for analysis of mineral element profiles. The concentrations of K, Mn, and Cu were significantly higher in both lines of LAC3OE transgenic maize than in the WT, and the concentrations of Ca, Mg, and Zn were significantly higher in LAC3OE #3 than in the WT (Figure 6C).
Discussion
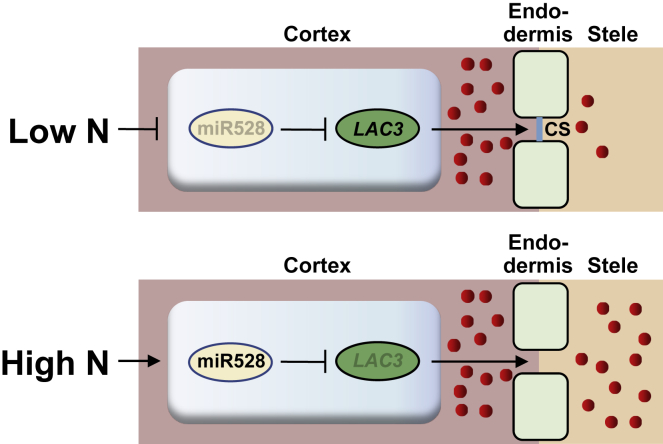
miRNAs are 20- to 24-nucleotide regulatory RNAs processed from self-complementary hairpin precursors. Although they play important roles in posttranscriptional gene regulation, their functions are less well studied in maize than in rice and Arabidopsis. Only ZmmiR156, ZmmiR164, ZmmiR165, ZmmiR166, ZmmiR172, and ZmmiR399 have been functionally identified in maize (Juarez et al., 2004; Lauter et al., 2005; Chuck et al., 2007a, 2007b; Li et al., 2012; Du et al., 2018; Kumar et al., 2021). Our previous results indicated that ZmmiR528 affects lignin biosynthesis and lodging resistance of maize under NL by negatively regulating the abundance of ZmLAC3 and ZmLAC5 mRNA. In the present research, we found that miR528 could affect mineral element profiles in maize by regulating CS formation under different N treatments (Figure 7).
Figure 7.
Proposed model of the role of miR528-LAC3 in Casparian strip formation in maize.
miR528 affects mineral element profiles in maize by regulating CS formation under different N treatments. CS indicates the Casparian strip. The red circles represent nutrients. Blunt-ended bars indicate inhibition.
Plant miRNAs are involved in almost all aspects of plant growth and development (Chen, 2012). Studies have also increasingly revealed that miRNAs regulate plant adaptive responses to stresses (Sunkar et al., 2012). However, few miRNAs have been identified that clarify the relationships between plant growth and development and stress responses. We suspect that miR528 represents a link between plant growth and development and stress responses in maize. In rice, transcriptional regulation of miR528 by OsSPL9 orchestrated antiviral responses (Yao et al., 2019), and OsmiR528 could also repress the mRNA abundance of different targets in order to affect heading under long-day conditions or during pollen intine formation (Yang et al., 2019; Zhang et al., 2019). These results showed that miR528 affected both rice development and rice responses to abiotic stress. By using ZmmiR528 knockdown transgenic maize and loss-of-function mutants (mir528a, mir528b, and mir528a/b), we found that ZmmiR528 contributed to early CS formation under different N conditions. N supply affected the mRNA abundance of ZmmiR528 in maize (Trevisan et al., 2012; Sun et al., 2018). Application of excessive N fertilizer reduces lodging resistance, and ZmmiR528 affects lodging resistance of maize by regulating lignin biosynthesis under NL (Sun et al., 2018). We therefore concluded that miR528 is an important link between N stress responses and N-mediated plant development in maize.
A CS consists of a fine band of lignin spanning the apoplastic space between adjacent endodermal cells (Hosmani et al., 2013). With CSs, endodermal cells can control passage of water and solutes into the stele and the vascular system (Geldner, 2013). The CS biogenesis pathway has been well established in Arabidopsis. In rice, OsCASP1 is required for CS formation at the endodermis (Wang et al., 2019). However, similar information is limited in maize. Even in Arabidopsis, the function of Lacs in CS formation requires further investigation. Although the lac1/3/5/7/8/9/12/13/16 nonuple mutant did not show any discernable defects in CS formation, the authors could not exclude the possibility that upregulation of LACs might contribute to CS formation (Rojas-Murcia et al., 2020). Our previous and present results showed that expression of ZmmiR528 was induced by NL but reduced by ND in both whole roots and root tips. Interestingly, ZmLAC3 responded to N treatment, but ZmLAC5 did not. Consistent with our observations that loss-of-function mutants (mir528a, mir528b, and mir528a/b) and ZmmiR528 TM transgenic maize exhibited increased CS formation under NL, overexpression of ZmLAC3 induced CS formation. By contrast, CS formation in ZmmiR528 TM transgenic maize and LAC3OE transgenic maize was similar under NL and ND. We also found that CS formation was similar in ZmLAC5-overexpressing transgenic maize and WT maize (data not shown). This appears to reflect the different physiological functions of ZmLAC5 and ZmLAC3 (Caparros-Ruiz et al., 2006; Xie et al., 2020). Taken together, our results indicate that the N-regulated miR528-LAC3 module affects CS formation in maize.
At least 13 mineral nutrients are required for normal plant growth. Imbalance of one element usually affects the contents and metabolism of other nutrients, supporting the idea that ionomic networks in plants are coordinately regulated (Huang and Salt, 2016). Nutrients enter the vascular system via the apoplastic, symplastic, and transcellular pathways (White, 2001; Baxter et al., 2009; Chen et al., 2019). The CS acts as a barrier to the apoplastic pathway and forces nutrients to reach the vascular cylinder by symplastic pathways. Here, we showed that mineral element profiles in shoots and roots of maize were affected by N supply. Transmembrane transporter activity (GO:0022857) did not rank among the top GO terms enriched in our whole-transcriptome profiles of root tips subjected to NL and ND for 3 days. This result suggested that N-induced changes in mineral element profiles could not be explained simply by altered transcript levels of transporters. The results obtained with ZmmiR528 knockdown transgenic maize and mir528a/b loss-of-function mutants demonstrated that CSs were involved in the selective uptake of mineral nutrients. Interestingly, overexpression of ZmLAC3 significantly increased the concentrations of K, Ca, Mn, Cu, Mg, and Zn in the ear leaf under field conditions. These results indicate that maize quality might be increased by modification of CS formation.
Methods
Plant materials and growth conditions
Seed surface sterilization was performed as described previously (Sun et al., 2018). The seeds were then germinated in paper rolls for 8 days. After their endosperms were removed, the maize seedlings were transferred to 3-L pots; they were grown hydroponically in modified half-strength Hoagland’s nutrient solution for 2 days and then full-strength Hoagland’s nutrient solution for another 2 days. At day 5 and thereafter, NS, NL, and ND were simulated using 4 mM NO3− (original NO3− concentration in full-strength Hoagland’s nutrient solution), 8 mM NO3−, or 0.04 mM NO3−, respectively. The plants were grown in a growth chamber with 14 h light/10 h dark and a 28/22°C day/night temperature regime. Samples were collected at the indicated times after initiation of N treatment.
For the field experiment, maize was planted in rows at the Yacheng Agriculture Experimental Station of the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (18°23′57″N, 109°11′54″E). There were 55 cm between rows and 25 cm between plants in rows.
Constructs and generation of transgenic maize
CRISPR-Cas9-mediated generation of ZmMIR528a and ZmMIR528b single or double loss-of function mutants (mir528a, mir528b, and mir528a/b) was performed as described by Wang et al. (2020). In brief, we designed two guide RNAs to target sequences located at nucleotides 4 and 16 of mature miR528 and nucleotides 10 and 18 of miR528∗ to gene-edit both ZmMIR528a and ZmMIR528b. The fragment was cloned into the pBUE411 vector using the BsaI restriction site by a T4 ligase reaction. The primers are listed in Supplemental Table 2.
The plasmid was electroporated into Agrobacterium tumefaciens EHA105 and transformed into immature embryos of the maize inbred line CZ01 at Weimi Biotechnology (Changzhou, China). Transformants were selected with bialaphos. Cas9-free transgenic maize was identified for phenotypic analysis.
RNA analysis
Total RNA extraction and purification, first-strand cDNA synthesis, qRT-PCR, and stem-loop qRT-PCR were performed as described previously (Sun et al., 2018). Each analysis was based on three replications, and the comparative Ct method was used. The specific primers are listed in Supplemental Table 2.
RNA-seq analysis
Total RNA was extracted from root tips (2 cm from the apex) that had been subjected to NL or ND for 3 days. Three biological replicates were performed for each treatment. RNA libraries were constructed and raw data analyzed as described previously (Wang et al., 2020). Clean reads were obtained after exclusion of low-quality reads, 5′ and 3′ adaptor contaminants, and rRNA sequences obtained from GenBank. Reads were then aligned to the maize B73_RefGen_v4 genome (ftp://ftp.ensemblgenomes.org/pub/plants/release-41/fasta/zea_mays/dna/) using HISAT2 v2.1.0 with default parameters (Kim et al., 2015). Only perfectly matching sequences were considered for further analysis. The count information was used to calculate normalized gene expression levels as fragments per kilobase of transcript per million mapped reads (Pertea et al. 2015). Genes were considered to be differentially expressed if the log2(fold-change ratio) was ≥1 and the adjusted P value was <0.05 according to the DESeq2 method (Wang et al., 2010). GO enrichment was performed using AgriGO v2.0 with default parameters (Tian et al., 2017).
Histochemical analysis of Casparian strip and lignin
Roots subjected to NS, NL, and ND for 3 days were sampled and embedded in 3% agarose. A Leica VT1000 S vibratome was used to obtain 50-μm cross sections. To observe CSs, the sections were stained with 0.1% (w/v) berberine hemisulfate for 1 h and with 0.5% (w/v) aniline blue for 30 min (Brundrett et al., 1988; Vishal et al., 2019). Images were obtained with a Zeiss LSM 700 confocal microscope with excitation at 450–490 nm and detection at 520 nm.
Wiesner staining was used to observe lignin as described previously (Berthet et al., 2011), and images were collected with a Leica DM6000 M microscope.
Propidium iodide staining
The PI penetration assay was performed as described by Naseer et al. (2012) with some modifications. In brief, maize seedlings were exposed to 100 μM PI in the dark for 6 h, and the area 1–2 cm from the root apex was sampled. A Leica VT1000 S vibratome was used to obtain 50-μm cross sections. Images were obtained with a Zeiss LSM 700 confocal microscope with excitation at 488 nm and detection at 500–550 nm.
Histochemical detection of H2O2
For in situ detection of H2O2, maize roots that had been subjected to NL or ND for 3 days were detached and stained with 3,3′-diaminobenzidine solution for 8 h in the dark at 37°C (Giraud et al., 2009; Ng et al., 2013). The samples were then destained by boiling in a mixture of ethanol:acetic acid:glycerol (3:1:1, v/v/v). Images were collected with a Leica M165 FC microscope.
Transmission electron microscopy for cerium-chloride-based H2O2 detection
Roots subjected to NL or ND for 3 days were sampled for cerium-chloride-based H2O2 detection using previously described procedures (Lee et al., 2013). Images were collected with an HT7700 transmission electron microscope (Hitachi High Technologies, Tokyo, Japan) operating at 80 kV.
In situ hybridization
Tissue fixation and RNA in situ hybridization for ZmmiR528 and ZmLAC5 were performed as described by Sun et al. (2018). For ZmLAC3, a specific 315-bp fragment was amplified and cloned into the pGEM-T-easy vector (Promega). Sense and anti-sense RNA probes were generated with the DIG RNA Labeling Kit (Roche). The procedures for in situ hybridization of ZmLAC3 were described previously (Zhang et al., 2007). Images were collected with a Leica DM6000 M microscope. The specific primers are listed in Supplemental Table 2.
Elemental assay
Maize roots and shoots subjected to NL, NS, or ND for 5 days were collected. All the samples were dried at 65°C for 72 h and milled to a fine powder. The dried samples were digested with HNO3 at ≥240°C. The elemental concentrations were determined by inductively coupled plasma mass spectrometry (Agilent 7700x, Agilent Technologies, USA).
Funding
This work was supported by the National Key Research and Development Program of China (2021YFF1000500), the National Natural Science Foundation of China (grant number 31861143004), and the Agricultural Science and Technology Innovation Program of CAAS to WXL.
Author contributions
W.-X.L. designed the research; Y.G., Y.W., H.C., Q.D., Z.W., X.G., and Q.S. performed the research; W.-X.L., Y.G., and Y.W. analyzed the data. W.-X.L. wrote the article.
Acknowledgments
The authors declare no conflict of interest.
Published: January 21, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Accession numbers
Transcriptomic data generated from the research have been deposited in BIG sub (https://ngdc.cncb.ac.cn) with the accession number CRA009546.
Supplemental information
References
- Baxter I., Hosmani P.S., Rus A., Lahner B., Borevitz J.O., Muthukumar B., Mickelbart M.V., Schreiber L., Franke R.B., Salt D.E. Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet. 2009;5:e1000492. doi: 10.1371/journal.pgen.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cézard L., Le Bris P., Borrega N., Hervé J., Blondet E., Balzergue S., et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011;23:1124–1137. doi: 10.1105/tpc.110.082792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett M.C., Enstone D.E., Peterson C.A. A berberine-aniline blue fluorescent staining procedure for suberin, lignin, and callose in plant tissue. Protoplasma. 1988;146:133–142. [Google Scholar]
- Caparros-Ruiz D., Fornale S., Civardi L., Puigdomenech P., Rigau J. Isolation and characterization of a family of laccases in maize. Plant Sci. 2006;171:217–225. [Google Scholar]
- Chen A., Husted S., Salt D.E., Schjoerring J.K., Persson D.P. The intensity of manganese deficiency strongly affects root endodermal suberization and ion homeostasis. Plant Physiol. 2019;181:729–742. doi: 10.1104/pp.19.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Small RNAs in development - insights from plants. Curr. Opin. Genet. Dev. 2012;22:361–367. doi: 10.1016/j.gde.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Cigan A.M., Saeteurn K., Hake S. The heterochronic maize mutant corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- Chuck G., Meeley R., Irish E., Sakai H., Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 2007;39:1517–1521. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- Du Q., Zhao M., Gao W., Sun S., Li W.X. microRNA/microRNA∗ complementarity is important for the regulation pattern of NFYA5 by miR169 under dehydration shock in Arabidopsis. Plant J. 2017;91:22–33. doi: 10.1111/tpj.13540. [DOI] [PubMed] [Google Scholar]
- Du Q., Wang K., Zou C., Xu C., Li W.X. The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol. 2018;177:1743–1753. doi: 10.1104/pp.18.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N. The endodermis. Annu. Rev. Plant Biol. 2013;64:531–558. doi: 10.1146/annurev-arplant-050312-120050. [DOI] [PubMed] [Google Scholar]
- Griffiths M., Roy S., Guo H., Seethepalli A., Huhman D., Ge Y., Sharp R.E., Fritschi F.B., York L.M. A multiple ion-uptake phenotyping platform reveals shared mechanisms affecting nutrient uptake by roots. Plant Physiol. 2021;185:781–795. doi: 10.1093/plphys/kiaa080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Van Aken O., Ho L.H.M., Whelan J. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 2009;150:1286–1296. doi: 10.1104/pp.109.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.H., Liu X.J., Zhang Y., Shen J.L., Han W.X., Zhang W.F., Christie P., Goulding K.W.T., Vitousek P.M., Zhang F.S. Significant acidification in major Chinese croplands. Science. 2010;327:1008–1010. doi: 10.1126/science.1182570. [DOI] [PubMed] [Google Scholar]
- Hoffmann N., Benske A., Betz H., Schuetz M., Samuels A.L. Laccases and peroxidases co-localize in lignified secondary cell walls throughout stem development. Plant Physiol. 2020;184:806–822. doi: 10.1104/pp.20.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmani P.S., Kamiya T., Danku J., Naseer S., Geldner N., Guerinot M.L., Salt D.E. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl. Acad. Sci. USA. 2013;110:14498–14503. doi: 10.1073/pnas.1308412110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.Y., Salt D.E. Plant ionomics: from elemental profiling to environmental adaptation. Mol. Plant. 2016;9:787–797. doi: 10.1016/j.molp.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Ju X.T., Xing G.X., Chen X.P., Zhang S.L., Zhang L.J., Liu X.J., Cui Z.L., Yin B., Christie P., Zhu Z.L., Zhang F.S. Reducing environmental risk by improving N management in intensive Chinese agricultural systems. Proc. Natl. Acad. Sci. USA. 2009;106:3041–3046. doi: 10.1073/pnas.0813417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C.P. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- Khandal H., Singh A.P., Chattopadhyay D. The microRNA397b -LACCASE2 module regulates root lignification under water and phosphate deficiency. Plant Physiol. 2020;182:1387–1403. doi: 10.1104/pp.19.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam V., Singh A., Yadav S., Singh S., Kumar P., Sarkar Das S., Sarkar A.K. Conserved LBL1-ta-siRNA and miR165/166-RLD1/2 modules regulate root development in maize. Development. 2021;148:dev190033. doi: 10.1242/dev.190033. [DOI] [PubMed] [Google Scholar]
- Lauter N., Kampani A., Carlson S., Goebel M., Moose S.P. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA. 2005;102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Rubio M.C., Alassimone J., Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153:402–412. doi: 10.1016/j.cell.2013.02.045. [DOI] [PubMed] [Google Scholar]
- Li J., Guo G., Guo W., Guo G., Tong D., Ni Z., Sun Q., Yao Y. miRNA164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.) BMC Plant Biol. 2012;12:220. doi: 10.1186/1471-2229-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer S., Lee Y., Lapierre C., Franke R., Nawrath C., Geldner N. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl. Acad. Sci. USA. 2012;109:10101–10106. doi: 10.1073/pnas.1205726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Ivanova A., Duncan O., Law S.R., Van Aken O., De Clercq I., Wang Y., Carrie C., Xu L., Kmiec B., et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell. 2013;25:3450–3471. doi: 10.1105/tpc.113.113985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan X., Zeng J., Han Z., Zhang G. Ionomic and physiological responses to low nitrogen stress in Tibetan wild and cultivated barley. Plant Physiol. Biochem. 2017;111:257–265. doi: 10.1016/j.plaphy.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Rojas-Murcia N., Hématy K., Lee Y., Emonet A., Ursache R., Fujita S., De Bellis D., Geldner N. High-order mutants reveal an essential requirement for peroxidases but not laccases in Casparian strip lignification. Proc. Natl. Acad. Sci. USA. 2020;117:29166–29177. doi: 10.1073/pnas.2012728117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roppolo D., De Rybel B., Dénervaud Tendon V., Pfister A., Alassimone J., Vermeer J.E.M., Yamazaki M., Stierhof Y.D., Beeckman T., Geldner N. A novel protein family mediates Casparian strip formation in the endodermis. Nature. 2011;473:380–383. doi: 10.1038/nature10070. [DOI] [PubMed] [Google Scholar]
- Sieber P., Wellmer F., Gheyselinck J., Riechmann J.L., Meyerowitz E.M. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development. 2007;134:1051–1060. doi: 10.1242/dev.02817. [DOI] [PubMed] [Google Scholar]
- Sun Q., Liu X., Yang J., Liu W., Du Q., Wang H., Fu C., Li W.X. MicroRNA528 affects lodging resistance of maize by regulating lignin biosynthesis under nitrogen-luxury conditions. Mol. Plant. 2018;11:806–814. doi: 10.1016/j.molp.2018.03.013. [DOI] [PubMed] [Google Scholar]
- Sunkar R., Li Y.F., Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., Xu W., Su Z. agriGO v2.0: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2017;45:W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisan S., Nonis A., Begheldo M., Manoli A., Palme K., Caporale G., Ruperti B., Quaggiotti S. Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant Cell Environ. 2012;35:1137–1155. doi: 10.1111/j.1365-3040.2011.02478.x. [DOI] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 2010;153:895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Morreel K., Ralph J., Boerjan W. Lignin engineering. Curr. Opin. Plant Biol. 2008;11:278–285. doi: 10.1016/j.pbi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Vishal B., Krishnamurthy P., Ramamoorthy R., Kumar P.P. OsTPS8 controls yield-related traits and confers salt stress tolerance in rice by enhancing suberin deposition. New Phytol. 2019;221:1369–1386. doi: 10.1111/nph.15464. [DOI] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu W., Wang H., Du Q., Fu Z., Li W.X., Tang J. ZmEHD1 is required for kernel development and vegetative growth through regulating auxin homeostasis. Plant Physiol. 2020;182:1467–1480. doi: 10.1104/pp.19.01336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yamaji N., Huang S., Zhang X., Shi M., Fu S., Yang G., Ma J.F., Xia J. OsCASP1 is required for Casparian strip formation at endodermal cells of rice roots for selective uptake of mineral elements. Plant Cell. 2019;31:2636–2648. doi: 10.1105/tpc.19.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P.J. The pathways of calcium movement to the xylem. J. Exp. Bot. 2001;52:891–899. doi: 10.1093/jexbot/52.358.891. [DOI] [PubMed] [Google Scholar]
- Williams L., Salt D.E. The plant ionome coming into focus. Curr. Opin. Plant Biol. 2009;12:247–249. doi: 10.1016/j.pbi.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Yang R., Yang Z., Yao S., Zhao S., Wang Y., Li P., Song X., Jin L., Zhou T., et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants. 2017;3:16203. doi: 10.1038/nplants.2016.203. [DOI] [PubMed] [Google Scholar]
- Xie T., Liu Z., Wang G. Structural basis for monolignol oxidation by a maize laccase. Nat. Plants. 2020;6:231–237. doi: 10.1038/s41477-020-0595-5. [DOI] [PubMed] [Google Scholar]
- Yan Z., Hou X., Han W., Ma S., Shen H., Guo Y., Fang J. Effects of nitrogen and phosphorus supply on stoichiometry of six elements in leaves of Arabidopsis thaliana. Ann. Bot. 2019;123:441–450. doi: 10.1093/aob/mcy169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Li P., Mei H., Wang D., Sun J., Yang C., Hao L., Cao S., Chu C., Hu S., et al. Fine-tuning of MiR528 accumulation modulates flowering time in rice. Mol. Plant. 2019;12:1103–1113. doi: 10.1016/j.molp.2019.04.009. [DOI] [PubMed] [Google Scholar]
- Yao S., Yang Z., Yang R., Huang Y., Guo G., Kong X., Lan Y., Zhou T., Wang H., Wang W., et al. Transcriptional regulation of miR528 by OsSPL9 orchestrates antiviral response in rice. Mol. Plant. 2019;12:1114–1122. doi: 10.1016/j.molp.2019.04.010. [DOI] [PubMed] [Google Scholar]
- Zhang Y.C., He R.R., Lian J.P., Zhou Y.F., Zhang F., Li Q.F., Yu Y., Feng Y.Z., Yang Y.W., Lei M.Q., et al. OsmiR528 regulates rice-pollen intine formation by targeting an uclacyanin to influence flavonoid metabolism. Proc. Natl. Acad. Sci. USA. 2019;117:727–732. doi: 10.1073/pnas.1810968117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Madi S., Borsuk L., Nettleton D., Elshire R.J., Buckner B., Janick-Buckner D., Beck J., Timmermans M., Schnable P.S., Scanlon M.J. Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet. 2007;3:e101. doi: 10.1371/journal.pgen.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., Shao H., Wang X., Wang Z.Y., Dixon R.A. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25:3976–3987. doi: 10.1105/tpc.113.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Chen C., Zeng J., Yun Z., Liu Y., Qu H., Jiang Y., Duan X., Xia R. MicroRNA528, a hub regulator modulating ROS homeostasis via targeting of a diverse set of genes encoding copper-containing proteins in monocots. New Phytol. 2020;225:385–399. doi: 10.1111/nph.16130. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Zuo D., Tao Y., Cai H., Li L. Laccase3-based extracellular domain provides possible positional information for directing Casparian strip formation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2020;117:15400–15402. doi: 10.1073/pnas.2005429117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.