Abstract
There are no effective therapeutic targets or strategies that simultaneously inhibit tumour growth and promote cardiac function recovery. Here, we analyzed targets for cancer treatments and cardiac repair, with demethylation emerging as a common factor in these candidate lists. As DNA methyltransferase 1 (DNMT1) majorly responds to methylation, a natural compound library is screened, identifying dioscin as a novel agent targeted at DNMT1, widely used for heart diseases. Dioscin was found to reduce DNMT activities and inhibits growth in breast cancer cells. Combined with analyses of RNA-seq and MeDIP-seq, the promoters of antioxidant genes were demethylated after dioscin, recruiting NRF2 and elevating their expression. In Nrf2 knockout mice, the cardiac protection role of dioscin was blocked by Nrf2-loss. Furthermore, in tumour-bearing mice with hypertrophy, dioscin was observed to inhibit tumour growth and alleviate cardiac injury simultaneously. This study is the first to identify dioscin as a novel demethylation agent with dual functions of anti-cancer and cardio-protection.
Keywords: DNMT inhibitors, Demethylation, NRF2, Hypertrophy, Cancer treatment
Highlights
-
•
Dioscin abolishes methylation in cancer cells and cardiomyocytes.
-
•
Dioscin demethylates the modified promoters of antioxidant genes transcriptionally regulatedD by NRF2 in hypertrophy.
-
•
Dioscin provides a novel strategy for the joint treatment of cancer and cardiovascular diseases for the first time.
1. Introduction
Cardiovascular disease (CVD) is currently the primary cause of morbidity and mortality among survivors of cancer treatment [[1], [2], [3]]. These survivors suffer from adverse cardiovascular events, possibly due to cancer treatment or an exacerbation of underlying CVD. A recent comprehensive review of breast cancer survivors across the United States highlighted a significantly increased risk of death from CVD, presenting with various clinical manifestations, including a reduction in left ventricular ejection fraction (LVEF) [4]. This risk surpasses their likelihood of death from initial cancer or recurrent disease. To alleviate cardiac dysfunction in cancer patients with underlying CVD, it is imperative to identify optimal therapeutic targets that can concurrently inhibit tumour growth and promote cardiac function recovery.
DNA methylation, a universal epigenetic feature, is a reversible process that alters gene expression profiles by modifying their promoters. Due to differences in DNA methylation patterns between normal and malignant breast tissues, it is considered for DNA methylation to serve as a novel diagnostic strategy for breast cancer potentially [[5], [6], [7]]. Additionally, aberrant methylation landmark is an obvious risk factor for breast cancer, reflecting increased methylation levels within promoters and declined expression of tumour suppressor associated with an elevated risk of breast cancer. Promoter methylation of tumour suppressors interferon regulatory factor-4 (Irf4) [8]and breast cancer gene 1 (Brca1) was observed in the breast cancer tissues, resulting in depression of IRF4 and BRCA1 [9]. Conversely, demethylation correlates with reduced tumorigenicity, often related to re-expression of tumour suppressors [10].
Cardiac hypertrophy is associated with transcriptional reprogramming due to alterations in mechanisms that regulate gene expression [11]. In both clinics and the experimental hypertrophic models, the aberrant methylation profile was observed both in heart tissues and peripheral blood [12]. Furthermore, the potential involvement of DNA methylation in chronic hypertension-induced cardiac injury as treatment with the demethylating agent 5Aza (5-Azacytidine) was shown to attenuate cardiac hypertrophy and fibrosis in the spontaneous hypertensive rat (SHR) model [13]. This indicated that the epigenome, specifically DNA methylation, undergoes reprogramming in heart failure, potentially driving a pathological shift in cardiac gene expression. The properties of methylation that potentiate its detrimental effects on tumour growth and cardiac dysfunction suggest potential targets for anti-tumour treatment in patients with cardiovascular disease. This highlights the importance of understanding the role of DNA methylation in oncology and cardiology and the potential for therapeutic interventions that target this epigenetic modification.
DNA methylation involves the addition of a methyl group to the cytosine pyrimidine ring in CpG dinucleotides, a process facilitated by DNA methyltransferases (DNMTs), including DNMT1, DNMT3a, and DNMT3b identified in humans [14,15]. DNMT1 primarily recognizes and methylates CpG dinucleotides during DNA replication [16], while DNMT3a and DNMT3b [17] predominantly function as de novo methyltransferases, establishing DNA methylation. Currently, the demethylating agent 5-azacytidine-2′-deoxycytidine (decitabine) is approved by the FDA for the treatment of haematological neoplasms, indicating that clinical benefits of targeting the epigenetic machinery. However, several side-effects occurred at high doses of 5-azacytidine (5Aza) and decitabine, such as nausea and weight loss, suggesting a need for other DNMT1 antagonists with low side effects. What's more, 5Aza can integrate into RNA to inhibit mRNA translation [18] and reduce the overall protein profile. Given these limitations of these demethylation agents, there is an urgent need to discover novel drug candidates that can effectively target at DNA methylation with fewer side effect to inhibit tumour growth and ameliorate cardiac dysfunction simultaneously.
In this current study, we screened a natural product library and identified a novel DNMT1 antagonist, dioscin, which exhibits excellent physiochemical properties and biological activities. We confirmed the inhibitory effect of dioscin on demethylation both in vitro and in vivo. Furthermore, we identified dioscin that inhibits cancer growth and recovers cardiac function, offering a new molecular compound for consideration in the development of tumour therapies for patients with a CVD background.
2. Material and methods
An expanded section of methods is available in the Supplementary material online.
Further data and materials that support the findings of this study are available from the authors upon reasonable request.
3. Results
3.1. Dioscin causes demethylation in breast cancer cells
To explore potential simultaneous therapeutics with cancer treatment and cardiac function recovery, we first screened the therapeutic targets used in clinical cancer treatments and trials. Then combined with the drug candidates that possess effective prevention/treatment of heart failure (HF), identifying DNA methylation increased both in carcinoma and HF heart tissues. As references, demethylated agents 5Aza and RG108 inhibit both tumour growth and the progress of heart failure, accompanied by the manifestation of nausea and weight loss.
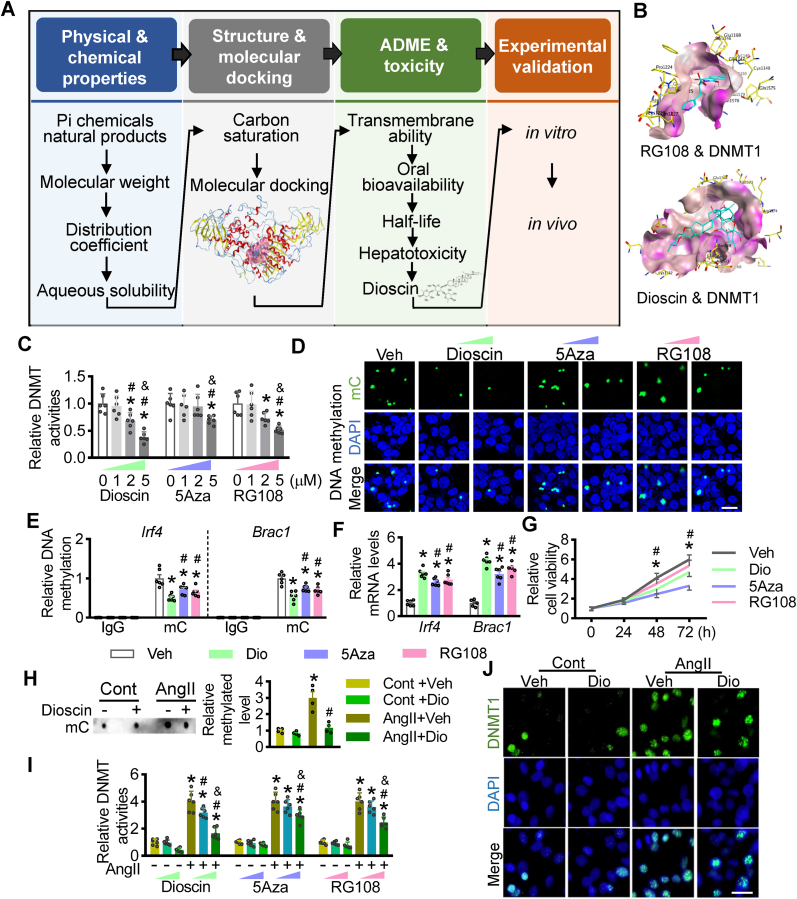
Due to this, we have to explore the fewer side-effect compounds targeted at methylation. Then, we proposed a computer-aided analysis strategy to screen for novel DNMT1 antagonists within the Pi compounds of a natural product library containing 418 compounds. The screening strategy involves four steps with physical and chemical properties, molecular docking, toxicity, and experimental validation (Fig. 1A). Initially, 152 compounds were selected after considering molecular weight, distribution coefficient, and aqueous solubility. Substantially, Molecular Operating Environment was used to dock these candidates and score their potential complementarity with putative binding sites of DNMT1, compared with the positive controls of RG108 and 5Aza. After docking analysis, 27 compounds demonstrated higher molecular docking interaction energy than RG108. Considering the oral bioavailability and half-life, only one compound, dioscin, was selected.
Fig. 1.
Screening and validation of DNMT1 antagonists. (A) Schematic diagram is displaying a screening strategy to explore DNMT1 inhibitors. There are 4 steps: 1) physical and chemical properties; 2) molecular docking; 3) biological and toxic properties; 4) experimental validation. (B) Binding modes of RG108 and dioscin with human DNMT1 were predicted by AutoDock, respectively. RG108 and dioscin were shown in blue. (C and D) Breast cancer cells MDA-MB-231 were incubated with different doses of dioscin, 5Aza and RG108 at the concentration of 0, 3 and 5 μM. (C) DNMT activities were determined with these cellular lysates (n = 6 per group; *P < 0.05 vs. 0 μM, #P < 0.05 vs. 1 μM, & P < 0.05 vs. 2 μM). (D) The cells were stained with antibody against methylcytosine (mC) and DAPI. (E-G) MDA-MB-231 cells were cultured with dioscin, 5Aza and RG108, respectively. (E) ChIP-qPCR assay was performed with these lysates. The antibody against mC was used, and then the specific primers for the promoters of Irf4 and Brac4 were used to qualify the methylated levels of these two gene promoters. IgG was as the negative control for ChIP-qPCR assay. (F) Quantitative PCR (qPCR) assays were performed, showing the mRNA levels of Irf4 and Brac4 with different treatment in MDA-MB-231 cells. (G) The cell viabilities of MDA-MB-231 with dioscin, 5Aza and RG108 treatment were measured using CCk8 kits, respectively (n = 6 per group; *P < 0.05 vs. Veh, #P < 0.05 vs. Dio). (H and J) Dioscin was incubated in the neonatal mouse cardiomyocytes (NMCMs) cultured with AngII. (H) Dot bot assay was performed to monitor the levels of total mC in NMCMs (n = 4 per group; *P < 0.05 vs. Cont, #P < 0.05 vs. Veh). (I) AngII-pretreated NMCMs were incubated with different doses (0, 3 and 5 μM) of dioscin, 5Aza and RG108. qPCR assays were performed to monitor the mRNA levels of Dnmt1, Dnmt3a and Dnmt3b in NMCMs (n = 6 per group; *P < 0.05 vs. Cont, #P < 0.05 vs. 0 μM, & P < 0.05 vs. 3 μM). (J) Representative images of fluorescence staining were presented for DNMT1. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In detail, 5Aza, a cytosine-like chemical, incorporates to DNA, irreversibly binds to DNMT1 at Asn1578, and traps the enzymes on DNA. RG108 and dioscin could be bound to DNMT1 at the same site of Asn1578, some additional hydrogen bonds with DNMT1 at Phe1145 and Glu1168 were formed with dioscin (Fig. 1B, Supplementary materials online, Fig. S1). These results show that the screened DNMT1 antagonist, dioscin, can bind tightly with DNMT1.
To test the effect of dioscin on demethylation in cancer, we first evaluated the DNMT activities in two breast cancer cell lines, MDA-MB-231 and BT474, in response to dioscin treatment. In MDA-MB-231 cells, both DNMT activities and Dnmt1 mRNA levels decreased dose-dependent with dioscin treatment (Fig. 1C, Supplementary materials online, Fig. S2). High doses of dioscin suppressed the mRNA levels of Dnmt3a and Dnmt3b, indicating that DNMT1 is the most sensitive member of the DNMTs family in response to dioscin (Supplementary materials online, Fig. S2). The methylated levels were decreased with the incubation of dioscin (Fig. 1D). In detail, the declined methylation at the regions of Irf4 and Brac1 promoters was caused by dioscin treatment (Fig. 1E), leading to the reduction of Irf4 and Brac1 mRNA levels (Fig. 1F) and cell proliferation (Fig. 1G).
What's more, we verified the inhibitory effect of dioscin on DNMT1 activities in BT474 cells. Consistent with the results in MDA-MB-231 cells, dioscin caused the reduction of DNMT activities and the reduction of Dnmt1 mRNA levels in BT474 cells (Supplementary materials online, Figs. S3A and S3B), resulting in decreased proliferation abilities (Supplementary materials online, Fig. S3C). And dioscin erased the methylated modification located at the promoters of Irf4 and Brac1 and consequently increasing their mRNA levels (Supplementary materials online, Figs. S3D and S3E). These data suggested that dioscin demethylates gene promoters and causes the reduction of their expression via DNMT1 inhibition in breast cancer cells.
3.2. Dioscin ameliorates cardiac hypertrophy
Furthermore, we evaluated the efficiency of dioscin in inhibiting DNMT activities in cardiomyocytes. Considering that the enriched methylated profile was observed both in the hypertrophic cardiac tissues and AngII-pretreated cardiomyocytes, we first detected the methylated levels in neonatal mouse cardiomyocytes (NMCMs) incubated with dioscin in response to AngII. Dot blot assay indicated that dioscin repressed the elevated methylated levels in AngII-pretreated NMCMs (Fig. 1H). DNMT activities were increased in NMCMs after AngII treatment, as previously reported, and dioscin prevented this AngII-induced increase of DNMT activities (Fig. 1I). Additionally, dioscin decreased Dnmt1 mRNA levels in a dose-dependent way in NMCMs, while the reduction of Dnmt3a and Dnmt3b mRNA levels were reduced at the high dose of dioscin (Supplementary materials online, Fig. S4). DNMT1 staining further confirmed that dioscin reduced DNMT1 expression in NMCMs (Fig. 1J). These results suggested that, in vitro, dioscin acts as a DNMT1 antagonist to prevent AngII-induced increase of methylation in NMCMs.
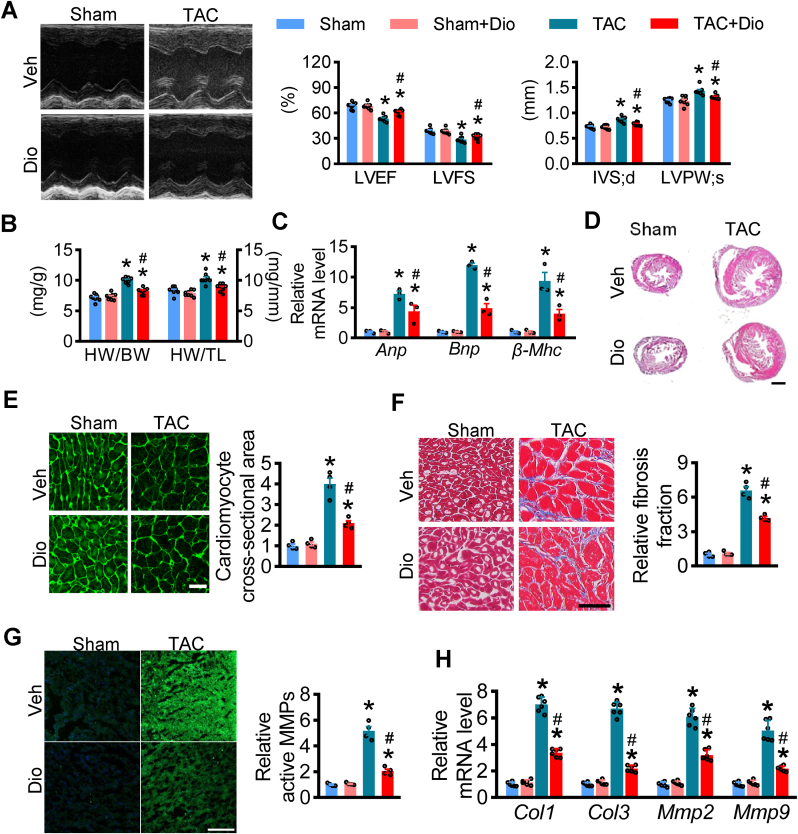
To explore whether, in vivo, dioscin ameliorates cardiac function in hypertrophy or not, a hypertrophic model with transverse aorta constriction (TAC) surgery was used and administrated dioscin for four weeks. Echocardiography showed that both left ventricle ejection fraction (LVEF) and left ventricle fractional shortening (LVFS) decreased in hypertrophic mice, while the interventricular septal thickness at end-diastole (IVS; d) and left ventricular posterior wall dimension at end-systole (LVPW; s) increased after TAC surgery, as previously described (Fig. 2A). Dioscin alleviated the reduction of LVEF and LVFS and the increased thickness of IVS and LVPW in hypertrophic mice (Fig. 2A). Additionally, dioscin suppressed the increase in both ratios of heart weight to body weight (HW/BW) and heart weight to tibia length (HW/TL) (Fig. 2B). Hypertrophic biomarkers were measured in heart tissues, showing that dioscin repressed the increase of natriuretic peptide A (Anp), natriuretic peptide B (Bnp) and beta-myosin heavy chain (β-Mhc) at mRNA levels in hypertrophic hearts (Fig. 2C). TAC-induced enlargement of heart size was prevented after dioscin administration (Fig. 2D). Meanwhile, wheat germ agglutinin (WGA) staining showed that the sizes of cardiomyocytes increased in hypertrophic hearts compared to those from the sham mice, and this increase was inhibited with dioscin (Fig. 2E). These data suggested that dioscin ameliorated cardiac hypertrophy in vivo.
Fig. 2.
Dioscin alleviates cardiac dysfunction in TAC-induced hypertrophy. C57BL/6 mice were subjected to sham or TAC operations. Mice were treated with dioscin or vehicle for 4 weeks from day 1 after operation (n = 8 per group). (A) Representative M-mode echocardiography is presented with left ventricular ejection fraction (LVEF, %), left ventricular fractional shortening (LVFS, %), interventricular septal thickness at diastole (IVS; d, mm), left ventricular end systolic posterior wall dimension (LVPW; s, mm). (B) The ratios for heart weight/body weight (HW/BW, mg/g) and heart weight/tibia length (HW/TL, mg/mm) post sham or TAC surgery were shown. (C) Gene expression levels of heart failure markers Anp, Bnp and β-Mhc were determined by qPCR assays using heart tissues. (D-G) Representative images of cardiac histological examination, including hematoxylin and eosin (HE) staining (D), white germ agglutinin (WGA) staining (E), Masson staining (scale bar = 50 μm) (F), and active matrix metalloproteinases (MMPs) staining (G). (H)The expression of Col1, Col3, Mmp2, and Mmp9 at mRNA levels in hearts were presented. N = 6 per group; *P < 0.05 vs. Sham, #P < 0.05 vs. TAC.
In the process of hypertrophy, cardiac fibrosis occurs with enlarged cardiomyocytes. We performed Masson staining on these heart tissues and found that the cardiac fibrosis was suppressed in hypertrophy mice after disocin administration (Fig. 2F). Additionally, dioscin could modulate the increase of active matrix metalloproteinases (MMPs), which occurred as interstitial collagens (Fig. 2G). The expression of fibrotic markers, including MMP2, MMP9, COL1 (Collagen1), and COL3 (Collagen3), increased in TAC hearts both at the protein and mRNA levels as previously reported, while dioscin repressed their increases in expression (Fig. 2H, Supplementary materials online, Fig. S5).
Given that dioscin alleviated cardiac dysfunction in hypertrophic mice, we verified the cardiac-protective role of dioscin in vitro. Dioscin inhibited the AngII-induced increase of hypertrophic markers Anp, Bnp, and β-Mhc in NMCMs (Supplementary materials online, Fig. S6A). AngII increased the sizes of NMCMs, while the enlargement of cellular sizes was repressed by dioscin (Supplementary materials online, Fig. S6B). As previously reported, AngII caused an increase in the mRNA levels of Mmp2, Mmp9, Col1, and Col3 and the enhanced MMP activation in NMCMs. Dioscin suppressed such increases (Supplementary materials online, Figs. S6C and S6D). Our data suggested that dioscin prevented AngII-induced hypertrophy and fibrosis in vitro, consistent with the in vivo results.
3.3. Dioscin causes demethylation in hypertrophic hearts via targeting at DNMT1
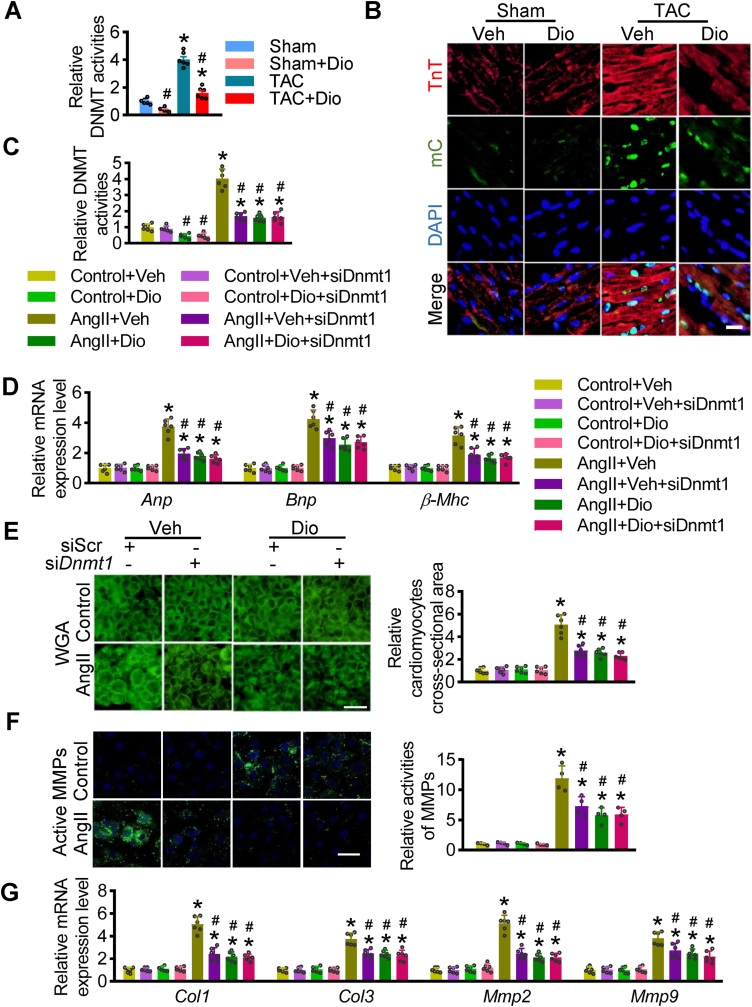
Dioscin plays a cardioprotective role in response to hypertrophy. As revealed in our earlier results, we further measured the efficiency of DNMT inhibition in hypertrophic hearts after dioscin treatment. Dioscin repressed the elevated DNMT activities in hypertrophic hearts (Fig. 3A). Immunostaining showed TAC-elevated methylation was repressed after dioscin administration (Fig. 3B). In detail, dioscin significantly reduced the Dnmt1 mRNA levels (Supplementary materials online, Fig. S7A). The reduction of DNMT1 protein levels was observed in hypertrophic hearts after dioscin administration (Supplementary materials online, Fig. S7B). Furthermore, we analyzed the binding abilities of dioscin with DNMTs, identifying the highest affinity of DNMT1 with dioscin among DNMTs (Supplementary materials online, Fig. S7C).
Fig. 3.
Dioscin mediates demethylation in hypertrophic hearts. C57BL/6 mice were subjected to sham or TAC operations. Mice were treated with dioscin or vehicle for 4 weeks from day 1 after operation. (A) DNMT activities were determined in heart tissues from each group (n = 6 per group; *P < 0.05 vs. Sham, #P < 0.05 vs. TAC). (B) Representative images of fluorescence staining for mC staining (green) were presented, and TnT was as cardiomyocytic marker (red). (C-G) AngII-incubated NMCMs were transfected with siRNA against Dnmt1 and then treated with dioscin. (C) DNMT activities were determined in different treated cells. (D) The mRNA levels of Anp, Bnp and β-Mhc were determined by qPCR assay in different groups. WGA staining (E) and active MMPs staining (F) were presented (n = 4 per group). (G) Real-time PCR analysis was performed to detect the mRNA levels of Mmp2, Mmp9, Col1, and Col3 genes in NMCMs, n = 6 per group. *P < 0.05 vs. Control, #P < 0.05 vs. AngII + Veh. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In confirm the target of dioscin is DNMT1, we transfected siRNA against Dnmt1 into AngII-incubated NMCMs with/without dioscin treatment (Fig. 3B). Once Dnmt1 was knockdown, dioscin did not affect DNMT1 expression (Fig. 3B, Supplementary materials online, Figs. S8A and S8B). Although the DNMT activities declined after the Dnmt1 knockdown, this reduction did not worsen with dioscin (Fig. 3C). In response to Dnmt1 knockdown, the elevated hypertrophic markers, including Anp, Bnp, and β-Mhc mRNAs, and the enlarged cellular sizes were reduced in AngII-incubated NMCMs, while dioscin did not exacerbate these increases (Fig. 3D and E). The active MMPs levels and the expression of Col1, Col3, Mmp2, and Mmp9 mRNA were detected, revealing that dioscin did not further repress Dnmt1-knockdown-caused reduction of fibrosis in AngII-incubated MNCMs (Fig. 3F and G). These data showed that dioscin loses its cardio-protective role in AngII-cultured NMCMs after Dnmt1 knockdown, indicating dioscin can mainly protect cardiomyocytes in hypertrophic conditions via DNMT1.
3.4. Dioscin mediates antioxidant stress response via demethylation/NRF2 axis
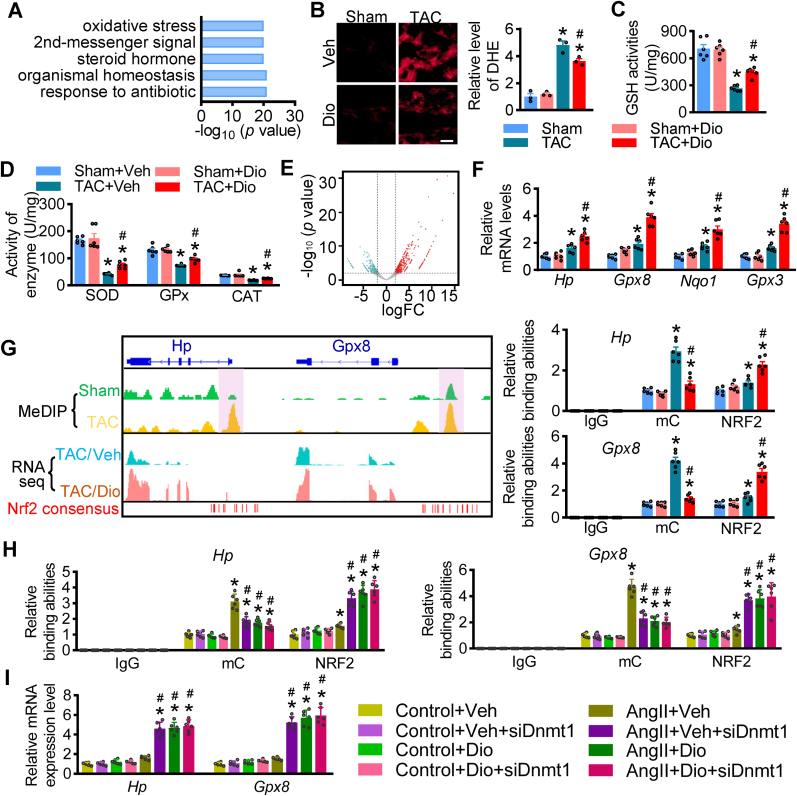
To further elucidate the underlying molecular mechanism by which dioscin protects cardiac function in hypertrophy, we performed RNA sequencing analysis using heart tissues from hypertrophic mice with/without dioscin administration. The gene ontology (GO) analysis showed that oxidative stress was regulated by dioscin in hypertrophic hearts (Fig. 4A). Dihydroethidium (DHE) staining was performed on heart tissue sections, confirming that reactive oxygen species (ROS) were significantly repressed by dioscin administration after TAC surgery (Fig. 4B). Furthermore, the enzymes eliminating ROS, including glutathione (GSH), superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GPx), were up-regulated with dioscin (Fig. 4C and D). Finally, RNAseq analysis showed up-regulated antioxidant genes, including Hp, Gpx8, Nqo1, and Gpx3 (Fig. 4E), with their elevated expression confirmed by qPCR assay (Fig. 4F).
Fig. 4.
Dioscin alleviates oxidative stress via NRF2 transcriptional ability in hypertrophic hearts. (A) The gene ontology (GO) analysis of RNA-sequencing data using heart tissues from hypertrophic mice treated with dioscin. (B) Representative images of DHE staining in heart sections and its quantification (scale bar = 50 μm), n = 3 per group. (C) The measurement of GSH activity was performed using heart tissues from each group. (D) The enzymes activities of SOD, GPx and CAT were measured using heart tissues from mice. (E) The expression profile is presented in volcano map after RNAseq analysis. Compared with hearts from TAC mice, the up- (red) and down-regulated (blue) genes were indicated in hearts from TAC mice with dioscin administration. (F) qPCR assay is performed to detect the mRNA levels of some antioxidant genes up-regulated, including Hp, Gpx8, Nqo1 and Gpx3, after dioscin administration. (G) Left panel: Genome browser tracks displays DNA methylation levels, gene expression levels, differentially methylation regions (shaded vertical bars) and predicted NRF2-binding sites of Hp and Gpx8; right panel: ChIP-qPCR analysis is performed to detect the methylated levels and NRF2 binding abilities among the promoters of Hp and Gpx8 in heart tissues from mice using mC and NRF2 antibodies, respectively. IgG is considered as negative control (n = 6 per group, *P < 0.05 vs. Sham, #P < 0.05 vs. TAC). (H and I) AngII-incubated NMCMs were transfected with siRNA against Dnmt1 and then treated with dioscin. (H) ChIP-qPCR analysis was performed to detect the methylated levels and NRF2 binding abilities among the promoters of Hp and Gpx8 in NMCMs, respectively. (I) The mRNA expression of Hp and Gpx8 levels in AngII-cultured NMCMs after siDnmt1 transfection (n = 6 per group; *P < 0.05 vs. Control, #P < 0.05 vs. AngII + Veh, & P < 0.05 vs. AngII + Dio). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To investigate the mechanism by which dioscin up-regulates antioxidant gene expression, we analyzed the methylated profile using the MeDIP sequencing with heart tissues from sham and TAC mice. The promoters of these antioxidant genes, Hp, Gpx8, Nqo1, and Gpx3 were methylated in hypertrophic hearts (Fig. 4G, Supplementary materials online, Figs. S9A and S9B), while chromatin immunoprecipitation-qPCR assay (ChIP-qPCR) indicated that dioscin demethylated the promoters of these genes (Fig. 4G, Supplementary materials online, Figs. S9A and S9B). Substantially, these regions with methylated change along these anti-antioxidant genes were analyzed using transfactors/enhancers prediction tool JASPER, identifying multiple NRF2-binding consensus sites located at/around the methylated-changed regions Fig. 4G, Supplementary materials online, Figs. S9A and S9B). We confirmed that the demethylation compound dioscin causes more NRF2 boundary to the demethylated regions located at the promoters of antioxidant genes Hp, Gpx8, Nqo1, and Gpx3 by ChIP-qPCR assays (Fig. 4G, Supplementary materials online, Figs. S9A and S9B).
We subsequently confirmed that dioscin alleviates oxidant stress via the DNMT1-mediated demethylation/NRF2 axis in vitro. Dioscin was found to inhibit the accumulation of AngII-induced ROS product, while, once Dnmt1 was knockdown, dioscin did not repress ROS levels (Supplementary materials online, Fig. S10). In AngII-treated NMCMs, dioscin demethylated the promoters of Hp, Gpx8, Nqo1, and Gpx3 and recruited more NRF2 binding to these demethylated regions, leading to the elevated mRNA levels of Hp, Gpx8, Nqo1, and Gpx3 (Fig. 4H and I, Supplementary materials online, Figs. S11A and S11B). Once Dnmt1 was knockdown, the effects of dioscin were missing, as reflected by the unchanged levels in the expression of Hp, Gpx8, Nqo1, and Gpx3 (Fig. 4H and I, Supplementary materials online, Figs. S11A and S11B).
3.5. Nrf2 deficiency abolishes the effect of dioscin-mediated demethylation in hypertrophic hearts
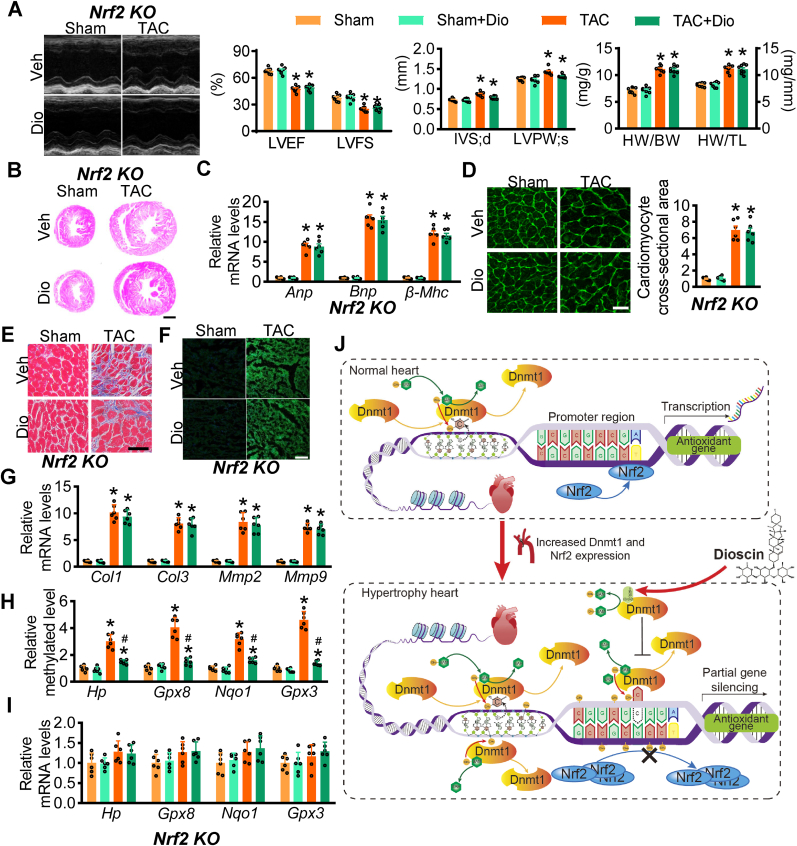
To confirm the protective role of dioscin via DNMT1/NRF2 axis in cardiac hypertrophy, Nrf2 knockout mice were subjected to TAC surgery and then treated with dioscin. Four weeks after TAC surgery, echocardiography was performed, showing the reduction of LVEF and LVFS after TAC surgery in Nrf2-deficient mice, and Nrf2 deficiency did not cause significant changes of LVEF and LVFS among administration of dioscin and vehicle after TAC surgery (Fig. 5A). Dioscin exhibited no effect on IVS or LVPW in Nrf2 deficient TAC mice (Fig. 5A). The same results were observed for HW/BW and HW/TL (Fig. 5A). These data indicated that the recovery of cardiac function by dioscin administration was abolished after Nrf2 deficiency.
Fig. 5.
Nrf2 deficiency abolishes the protective role of dioscin in hypertrophic mice.Nrf2 KO mice were subjected to sham or TAC operations, and substantially treated with dioscin or vehicle for 4 weeks after operation (n = 8 per group). (A) Representative M-mode echocardiography, the values for LVEF, LVFS, IVS and LVPW and the ratios for HW/BW and HW/TL were showed in hypertrophic Nrf2 KO mice. (B) Hematoxylin and eosin (HE) staining is shown (scale bar, 1 mm). (C) The mRNA levels of heart failure markers Anp, Bnp and β-Mhc in hearts were determined by qPCR assay. (D) Representative images of WGA staining (scale bar, 100 μm) were presented. Masson staining (scale bar, 50 μm) (E) and the active-matrix metalloproteinases (MMPs) (F) were showed. (G) The mRNA levels of Col1, Col3, Mmp2, and Mmp9 were detected in hearts. (H) The methylation at the promoters of Hp, Gpx8, Nqo1 and Gpx3 in heart tissues from Nrf2 deficient mice were detected with ChIP-qPCR assay. (I) qPCR assays were performed to monitor the mRNA levels of Hp, Gpx8, Nqo1 and Gpx3 in hearts from Nrf2 deficient mice. (J) The outline for dioscin in hypertrophic hearts was indicated. N = 6 per group; *P < 0.05 vs. Sham, #P < 0.05 vs. TAC.
HE staining suggested that dioscin did not affect the sizes of hearts in hypertrophic Nrf2-deficient mice (Fig. 5B). The expression levels of hypertrophic markers Anp, Bnp, and b-Mhc mRNAs were unchanged between dioscin and vehicle administration in the Nrf2 deficient condition (Fig. 5C). This result was consistent with WGA analysis (Fig. 5D).
Masson staining results suggested that Nrf2-deficiency abolished the anti-fibrotic role of dioscin in hypertrophic hearts (Fig. 5E). Meanwhile, active MMPs staining demonstrated no evident improvement for dioscin in eliminating active MMPs (Fig. 5F, Supplementary materials online, Figs. S12A and S12B). Dioscin did not change the increase of Mmps and Cols expression at mRNA levels after TAC surgery in Nrf2-deficient mice (Fig. 5G). In summary, Nrf2 deficiency eliminates the cardio-protective role of dioscin in cardiac hypertrophy.
Although Nrf2 deficiency abolishes the protective role of dioscin in hypertrophic mice, it is intriguing to know the DNMT activities in Nrf2 KO hypertrophic mice. Even with Nrf2 deficiency, dioscin declined the DNMT activities in hearts, accompanied by the reduction of Dnmt1 mRNA levels (Supplementary materials online, Figs. S12C and S12D). And Nrf2 loss did not affect dioscin-induced demethylation around the promoters of Hp, Gpx8, Nqo1, and Gpx3 (Fig. 5H). Conversely, due to Nrf2 deficiency, the mRNA levels of Hp, Gpx8, Nqo1, and Gpx3 did not increase after dioscin administration (Fig. 5H). These results suggested that dioscin plays an antioxidant role via two independent ways of DNMT1/demethylation axis and NRF2 transcriptional ability (Fig. 5J).
3.6. Dioscin alleviates cardiac hypertrophy in tumour-bearing mice
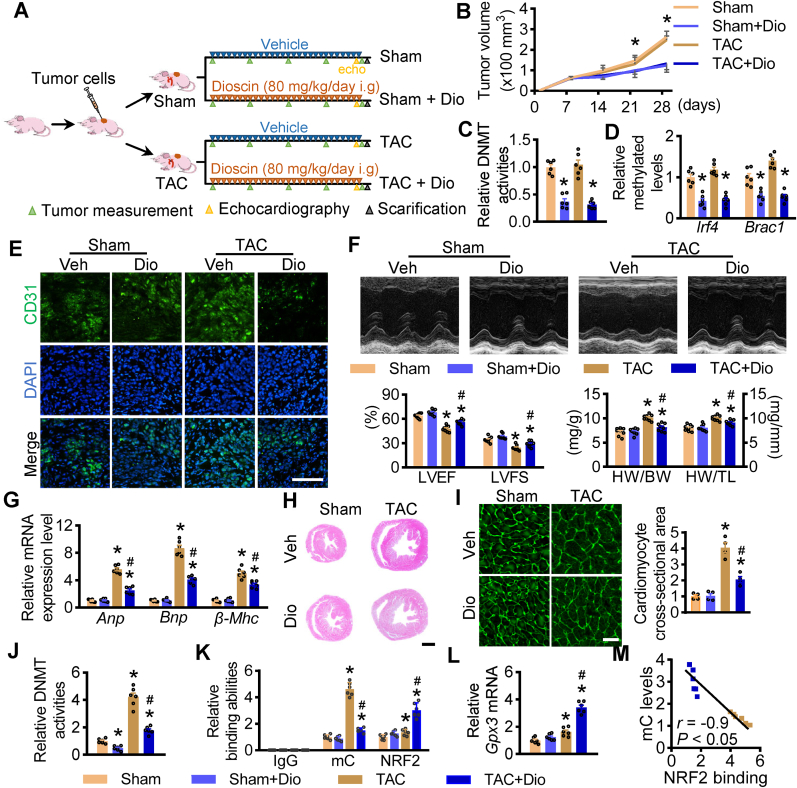
To confirm the dual effect of dioscin on tumour treatment and cardiac function recovery, BALB/c nude mice were transplanted with breast cancer cells MDA-MB-231 and then suffered with TAC surgery. After aortic constriction, these mice were randomly administrated with either vehicle or dioscin (Fig. 6A). Dioscin administration significantly delayed tumour growth compared to the vehicle (Fig. 6B). As a novel demethylated compound, dioscin dramatically repressed DNMT activities and Dnmt1 mRNA levels in tumour tissues (Fig. 6C, Supplementary materials online, Fig. S13). Dioscin demethylated the promoters of Irf4 and Brac1, resulting in the reduction of these gene expressions (Fig. 6D, Supplementary materials online, Fig. S14). Additionally, dioscin repressed angiogenesis in tumour tissues from tumour-bearing mice (Fig. 6E). These results indicated that dioscin prevented tumour growth in hypertrophic mice.
Fig. 6.
Effect of dioscin in tumour-bearing mice of hypertrophy. (A) The schematic outline of dioscin treatment for tumour-bearing mice of hypertrophy. Nude mice were implanted with MDAMB- 231 cells. One week later, these mice were randomly subgrouped and subjected with transverse aorta constriction. Then these tumour-bearing mice were administrated with dioscin for 3 weeks (n = 8 per group). (B) The volume for tumor were measured each week (*P < 0.05 vs. Sham). (C) DNMT activities were determined in tumor tissues. (D) The methylation levels of Irf4 and Brac4 promoters were detected in tumor tissues. (E) The representative images for CD31 staining were presented in tumor tissues. (F) Representative M-mode echocardiography, the values for LVEF and LVFS and the ratios for HW/BW and HW/TL were showed in hypertrophic tumour-bearing mice with/without dioscin administration. (G) The mRNA levels of heart failure markers Anp, Bnp and β-Mhc in hearts were determined by qPCR assay. (H) Hematoxylin and eosin (HE) staining is shown (scale bar = 1 mm). (I) Representative images of WGA staining were presented (scale bar = 100 μm). (J) DNMT activities were measured in heart tissues from tumour-bearing mic. (K) ChIP-qPCR assay was performed to detect the methylation and NRF2 binding abilities at the promoters of Gpx3 (n = 6 per group). (L) The mRNA levels of Gpx3 were up-regulated in heart tissues from tumour-bearing hypertrophy after dioscin administration. (M) Correlation between mC level and NRF2 binding ability at the promoter of Gpx3 in heart tissues. N = 6 per group; *P < 0.05 vs. Sham, #P < 0.05 vs. TAC.
Here, we confirmed the cardioprotective role of dioscin in nude mice suffered with tumour. Dioscin ameliorated the declines in LVEF and LVFS, reduced both IVS and LVPW in tumour-bearing hypertrophic mice, and also suppressed the increase in both ratios of HW/BW and HW/TL (Fig. 6F). The hypertrophic biomarkers, including Anp, Bnp and β-Mhc at mRNA levels, were inhibited after dioscin administration in hypertrophic mice (Fig. 6G). Both sizes of the hearts and cardiomyocytes were increased, while dioscin repressed this elevation (Fig. 6H and I). And dioscin alleviated cardiac fibrosis accompanied by the reduction of Col1, Col3, Mmp2, and Mmp9 mRNAs (Supplementary materials online, Figs. S15A and S15B).
Importantly, it was observed that dioscin prevents TAC-caused elevated DNMT activities and Dnmt1 mRNA levels in heart tissues of tumour-bearing mice (Fig. 6J, Supplementary materials online, Fig. S16). In detail, hypertrophy enhanced methylated levels at the promotion of these anti-oxidative genes Hp, Gpx8, Nqo1, and Gpx3. Dioscin inhibited the occurrence of DNA methylation, promoting NRF2 binding to the demethylated regions, leading to the induction of their expression (Fig. 6K, L, Supplementary materials online, Figs. S17A, S17B, S17C). Furthermore, the methylated level at the promoter of Gpx3 was negatively correlated with NRF2 binding abilities (Fig. 6M). These suggested that dioscin has a cardioprotective effect on hypertrophy in tumour-bearing mice.
4. Discussion
In this study, we sought to identify therapeutic targets that could effectively combat carcinoma while offering cardioprotection. Compared with the anticancer targets and underlying mechanisms through which heart failure occurs and develops, we identified the potential of demethylation to serve dual functions in anti-tumour treatment and cardioprotection. Subsequently, we identified dioscin as a novel demethylation compound with advantages of physical and chemical properties through unbiased natural product library screening. In breast cancer cells, dioscin inhibits tumour growth by recovery of tumour suppressor gene expression via the demethylation of their promoters. Meanwhile, in cardiomyocytes, dioscin erases DNMT1-mediated methylation located at the promoters of antioxidant genes and recruits more NRF2 boundary to the demethylated regions accompanying their elevated expression, thus leading to rescue cardiac function. These findings identify a promising therapeutic compound for tumour treatment, most importantly, provide a novel therapeutic strategy for anticancer treatment-promoted heart failure in cancer patients with heart injury. This dual-action approach could potentially revolutionize treatment protocols for cancer and heart disease patients.
DNA methylation-induced silencing of tumour suppressors is a common feature in cancer [[19], [20], [21]], and reversal of promoter DNA hypermethylation has proven effective in anti-tumour treatment. Apart from decitabine and 5Aza, which are the only demethylating drugs approved by the FDA for the treatment of myelodysplastic syndrome (MDS) [22,23], other DNMT inhibitors, such as guadecitabine, hydralazine, and MG98, have also been investigated as anticancer agents [24]. Among these demethylation agents [25], decitabine is widely used in clinics [26,27], but its anti-tumour effect varies in solid tumours. In solid cancers, decitabine has been primarily studied at high doses, which have been shown to rapidly induce DNA damage and substantial cytotoxicity [28,29], thereby limiting the clinical development of decitabine for solid tumours. This mechanism differs from the mechanism observed at lower doses. Despite the toxicity, the anti-cancer activity of decitabine was observed, with response rates ranging from 6% in female reproductive cancer to 50% in breast cancer. This demonstrated that DNMT activities are more sensitiveto the novel demethylated compound dioscin than decitabine, indicating that dioscin at low doses could demethylate promoters of tumour suppressor genes and inhibit tumour growth from being potential for solid tumour treatment.
Consistent with the finding of a hypertrophic heart, the promotion of hypertrophy and failing heart due to methylation induced by anti-tumour treatment is indeed significant. Nonspecific DNMT inhibitors, including 5Aza and decitabine, have beneficial effects in TAC-induced [30]hypertrophy and cardiac fibrosis. Furthermore, the deficiency of Dnmt3a and Dnmt3b in cardiomyocytes has no abnormalities, under both basal and stressed conditions, of maintenance with chromatin organization and expression profile despite changes in DNA methylation. In this way, specific DNMT1 inhibitor would be a more effective demethylation agent for cardioprotection during or after anti-tumour treatment. Herein, we identified a novel demethylation agent, dioscin, after screening with DNMT1 structure from the natural compounds.
Additionally, the non-nucleosidic DNMT inhibitor [31] RG108 does not need to be incorporated into the DNA for its action [32], showing faster interaction, particularly in virtually non-dividing cells like cardiomyocytes. In contrast, the action of nucleosidic DNMT inhibitors is more biased towards mitotic cells as it relies on DNA synthesis. Moreover, nucleosidic DNMT inhibitors exhibit typical cytotoxic side effects of anti-neoplastic compounds by predominantly affecting all rapidly dividing cells of the body [33,34]. The common side effects of these nucleoside-like analogues include mutagenic risk and genomic instability. In this study, dioscin is more like RG108 as non-nucleosidic DNMT1 inhibitor and is binds to DNMT1 more tightly. This suggests that dioscin might primarily protect against the detrimental consequences of methylation-mediated cardiac injury, offering a potentially safer and more effective therapeutic approach.
There are multiple ways for demethylation via DNMT1-mediated regulation. A prior study has shown that the demethylation agent decitabine induces proteosomal-dependent DNMT1 degradation by APC/CCdh1 in HeLa cervical cancer cells and HCT116 colon cancer cells [35,36]. Here we identified that DNMT1 was degraded in response to low doses of dioscin in breast cancer cells MDA-MB-231 and BT474 and in hypertrophic cardiomyocytes and hearts. Considering the correlation between demethylation and the therapeutic effect of demethylation, our findings suggested that dioscin abolishes methylation in cancer cells and cardiomyocytes in two steps: by decaying the DNMT1 expression and inhibiting DNMT activities, thus leading to the demethylation profile in tumour tissues and injury hearts via these two different ways.
While numerous studies have proposed that oxidative stress [37,38] impairs the cardiovascular system and leads to cardiovascular diseases [39], it is disappointing that these therapeutic strategies targeted only at oxidative stress have failed in preclinical and clinical trials. Although the endogenous NRF2 protein levels are significantly elevated in hypertrophic hearts, the expression of downstream antioxidant genes Hp, Gpx8, Nqo1, and Gpx3 regulated by NRF2 is slightly increased. Specifically, hypertrophy-induced methylation blocks NRF2 from getting close to the promoters of antioxidant genes, leading to declines in their expression. In this way, it is possible that merely overexpression of NRF2 may not have a significant cardioprotective effect on hypertrophic hearts, which could explain the failure of unique antioxidant therapeutic strategies due to DNA epigenetics. Here, it is evident that demethylation is more effective in antioxidant response. Dioscin demethylates the modified promoters of antioxidant genes transcriptionally regulated by NRF2 in hypertrophy, abolishes the blockage of methylation, and recruits more NRF2 boundary to the promoters, thus presenting the antioxidant potency and a cardioprotective role.
Although this study underscores the critical role of demethylation in inhibiting tumour growth and providing cardioprotection, it is crucial to develop therapeutic strategies synergistically with demethylation in multiple diseases, such as cancers and cardiovascular diseases. Currently, demethylation is served as an amplifier to enhance response and promote therapeutic effects. In most clinical trials involving demethylation agents, these DNMT inhibitors are combined synergistically with other compounds [40] to amplify the anti-cancer effect of others. Therefore, further studies on dioscin would be needed to identify other compounds combined with dioscin administration for the dual purpose of inhibiting tumour growth and providing cardioprotection. Consequently, additional preclinical and clinical studies are required to validate all these findings.
5. Conclusion
In summary, our study has discovered a novel demethylation compound, dioscin, with dual functions of cancer treatment and cardiac function recovery. Dioscin demethylates tumour-suppressors and increases their expression in the tumour. Additionally, dioscin erases hypertrophy-induced methylation at the promoters of antioxidant genes, promotes NRF2 binding to these demethylated sites and elevates their expression, alleviating the cardiac injury. Hence, we have found the potential therapeutic target for cancer treatment, without cardiotoxicity, even with a cardioprotective role. Our findings not only show that dioscin is a promising compound for clinical therapy of cardiac dysfunction and even heart failure, but also provide a novel therapeutic strategy for the joint treatment of cancer and cardiovascular diseases for the first time in this area.
Funding
This study was supported by the grants from the National Natural Science Foundation of China (82073928, 81970414, 82270272), CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-5–011), Leading Technology Foundation Research Project of Jiangsu Province (BK20192005), Sanming Project of Medicine in Shenzhen (SZSM201801060).
Declaration of competing interest
None.
Acknowledgements
Q. L. and G. W. designed the study. X. L. performed ChIP-qPCR experiments. Y. S. and J. L. performed analysis of MeDIP-seq and RNAseq data. Z. Z. contributed to analysis and interpretation of data. D. S and C.H. performed the animal experiments and staining. X. L. and D. L. contributed to data analysis. X. L., Z. Z., G. W. and Q. L. wrote the paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102785.
Contributor Information
Qiulun Lu, Email: qllu@cpu.edu.cn.
Guangji Wang, Email: guangjiwang@hotmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Jemal A., Ward E., Hao Y., Thun M. Trends in the leading causes of death in the United States, 1970-2002. JAMA. 2005;294:1255–1259. doi: 10.1001/jama.294.10.1255. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N., Ries L.A.G., Mariotto A.B., Reichman M.E., Ruhl J., Cronin K.A. Improved estimates of cancer-specific survival rates from population-based data. J. Natl. Cancer Inst. 2010;102:1584–1598. doi: 10.1093/jnci/djq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodai B.I., Tuso P. Breast cancer survivorship: a comprehensive review of long-term medical issues and lifestyle recommendations. Perm. J. 2015;19:48–79. doi: 10.7812/TPP/14-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zamorano J.L., Lancellotti P., Rodriguez Muñoz D., Aboyans V., Asteggiano R., Galderisi M., Habib G., Lenihan D.J., Lip G.Y.H., Lyon A.R., Lopez Fernandez T., Mohty D., Piepoli M.F., Tamargo J., Torbicki A., Suter T.M., Zamorano J.L., Aboyans V., Achenbach S., Agewall S., Badimon L., Barón-Esquivias G., Baumgartner H., Bax J.J., Bueno H., Carerj S., Dean V., Erol Ç., Fitzsimons D., Gaemperli O., Kirchhof P., Kolh P., Lancellotti P., Lip G.Y.H., Nihoyannopoulos P., Piepoli M.F., Ponikowski P., Roffi M., Torbicki A., Vaz Carneiro A., Windecker S. Authors/task force members, ESC committee for practice guidelines (CPG), document reviewers, 2016 ESC position paper on cancer treatments and cardiovascular toxicity developed under the auspices of the ESC committee for practice guidelines: the task force for cancer treatments and cardiovascular toxicity of the European society of cardiology (ESC) Eur. J. Heart Fail. 2017;19:9–42. doi: 10.1002/ejhf.654. [DOI] [PubMed] [Google Scholar]
- 5.van Veldhoven K., Polidoro S., Baglietto L., Severi G., Sacerdote C., Panico S., Mattiello A., Palli D., Masala G., Krogh V., Agnoli C., Tumino R., Frasca G., Flower K., Curry E., Orr N., Tomczyk K., Jones M.E., Ashworth A., Swerdlow A., Chadeau-Hyam M., Lund E., Garcia-Closas M., Sandanger T.M., Flanagan J.M., Vineis P. Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis. Clin. Epigenet. 2015;7:67. doi: 10.1186/s13148-015-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Severi G., Southey M.C., English D.R., Jung C., Lonie A., McLean C., Tsimiklis H., Hopper J.L., Giles G.G., Baglietto L. Epigenome-wide methylation in DNA from peripheral blood as a marker of risk for breast cancer. Breast Cancer Res. Treat. 2014;148:665–673. doi: 10.1007/s10549-014-3209-y. [DOI] [PubMed] [Google Scholar]
- 7.Delgado-Cruzata L., Wu H.-C., Liao Y., Santella R.M., Terry M.B. Differences in DNA methylation by extent of breast cancer family history in unaffected women. Epigenetics. 2014;9:243–248. doi: 10.4161/epi.26880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vernier M., McGuirk S., Dufour C.R., Wan L., Audet-Walsh E., St-Pierre J., Giguère V. Inhibition of DNMT1 and ERRα crosstalk suppresses breast cancer via derepression of IRF4. Oncogene. 2020;39:6406–6420. doi: 10.1038/s41388-020-01438-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice J.C., Ozcelik H., Maxeiner P., Andrulis I., Futscher B.W. Methylation of the BRCA1 promoter is associated with decreased BRCA1 mRNA levels in clinical breast cancer specimens. Carcinogenesis. 2000;21:1761–1765. doi: 10.1093/carcin/21.9.1761. [DOI] [PubMed] [Google Scholar]
- 10.Phan N.L.-C., Trinh N.V., Pham P.V. Low concentrations of 5-aza-2’-deoxycytidine induce breast cancer stem cell differentiation by triggering tumor suppressor gene expression. OncoTargets Ther. 2016;9:49–59. doi: 10.2147/OTT.S96291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura M., Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018;15:387–407. doi: 10.1038/s41569-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 12.Papait R., Serio S., Pagiatakis C., Rusconi F., Carullo P., Mazzola M., Salvarani N., Miragoli M., Condorelli G. Histone methyltransferase G9a is required for cardiomyocyte homeostasis and hypertrophy. Circulation. 2017;136:1233–1246. doi: 10.1161/CIRCULATIONAHA.117.028561. [DOI] [PubMed] [Google Scholar]
- 13.Watson C.J., Horgan S., Neary R., Glezeva N., Tea I., Corrigan N., McDonald K., Ledwidge M., Baugh J. Epigenetic therapy for the treatment of hypertension-induced cardiac hypertrophy and fibrosis. J. Cardiovasc. Pharmacol. Therapeut. 2016;21:127–137. doi: 10.1177/1074248415591698. [DOI] [PubMed] [Google Scholar]
- 14.Mattei A.L., Bailly N., Meissner A. DNA methylation: a historical perspective. Trends Genet. 2022;38:676–707. doi: 10.1016/j.tig.2022.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Jeltsch A. Molecular enzymology of mammalian DNA methyltransferases. Curr. Top. Microbiol. Immunol. 2006;301:203–225. doi: 10.1007/3-540-31390-7_7. [DOI] [PubMed] [Google Scholar]
- 16.Li E., Bestor T.H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 17.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 18.Aimiuwu J., Wang H., Chen P., Xie Z., Wang J., Liu S., Klisovic R., Mims A., Blum W., Marcucci G., Chan K.K. RNA-dependent inhibition of ribonucleotide reductase is a major pathway for 5-azacytidine activity in acute myeloid leukemia. Blood. 2012;119:5229–5238. doi: 10.1182/blood-2011-11-382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T., Wu H.-J., Liang Y., Liang X.-J., Huang H.-C., Zhao Y.-Z., Liao Q.-C., Chen Y.-Q., Leng A.-M., Yuan W.-J., Zhang G.-Y., Peng J., Chen Y.-H. Tumor-specific expression of shVEGF and suicide gene as a novel strategy for esophageal cancer therapy. World J. Gastroenterol. 2016;22:5342–5352. doi: 10.3748/wjg.v22.i23.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H., Li H., Wang L., Li Y., Yang C. A 5-gene DNA methylation signature is a promising prognostic biomarker for early-stage cervical cancer. J. Obstet. Gynaecol. 2022;42:327–332. doi: 10.1080/01443615.2021.1907563. [DOI] [PubMed] [Google Scholar]
- 21.Jiang L., Chen T., Xiong L., Xu J.-H., Gong A.-Y., Dai B., Wu G., Zhu K., Lu E., Mathy N.W., Chen X.-M. Knockdown of m6A methyltransferase METTL3 in gastric cancer cells results in suppression of cell proliferation. Oncol. Lett. 2020;20:2191–2198. doi: 10.3892/ol.2020.11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Issa J.-P.J., Kantarjian H.M., Kirkpatrick P. Azacitidine, Nat Rev Drug Discov. 2005;4:275–276. doi: 10.1038/nrd1698. [DOI] [PubMed] [Google Scholar]
- 23.Gore S.D., Jones C., Kirkpatrick P. Decitabine, Nat Rev Drug Discov. 2006;5:891–892. doi: 10.1038/nrd2180. [DOI] [PubMed] [Google Scholar]
- 24.Linnekamp J.F., Butter R., Spijker R., Medema J.P., van Laarhoven H.W.M. Clinical and biological effects of demethylating agents on solid tumours - a systematic review. Cancer Treat Rev. 2017;54:10–23. doi: 10.1016/j.ctrv.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Pappalardi M.B., Keenan K., Cockerill M., Kellner W.A., Stowell A., Sherk C., Wong K., Pathuri S., Briand J., Steidel M., Chapman P., Groy A., Wiseman A.K., McHugh C.F., Campobasso N., Graves A.P., Fairweather E., Werner T., Raoof A., Butlin R.J., Rueda L., Horton J.R., Fosbenner D.T., Zhang C., Handler J.L., Muliaditan M., Mebrahtu M., Jaworski J.-P., McNulty D.E., Burt C., Eberl H.C., Taylor A.N., Ho T., Merrihew S., Foley S.W., Rutkowska A., Li M., Romeril S.P., Goldberg K., Zhang X., Kershaw C.S., Bantscheff M., Jurewicz A.J., Minthorn E., Grandi P., Patel M., Benowitz A.B., Mohammad H.P., Gilmartin A.G., Prinjha R.K., Ogilvie D., Carpenter C., Heerding D., Baylin S.B., Jones P.A., Cheng X., King B.W., Luengo J.I., Jordan A.M., Waddell I., Kruger R.G., McCabe M.T. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat. Can. (Que.) 2021;2:1002–1017. [PMC free article] [PubMed] [Google Scholar]
- 26.Sidaway P. Haematological cancer: TP53 mutations sensitize to decitabine. Nat. Rev. Clin. Oncol. 2017;14:72. doi: 10.1038/nrclinonc.2016.215. [DOI] [PubMed] [Google Scholar]
- 27.DiNardo C.D., Pratz K.W., Letai A., Jonas B.A., Wei A.H., Thirman M., Arellano M., Frattini M.G., Kantarjian H., Popovic R., Chyla B., Xu T., Dunbar M., Agarwal S.K., Humerickhouse R., Mabry M., Potluri J., Konopleva M., Pollyea D.A. Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: a non-randomised, open-label, phase 1b study. Lancet Oncol. 2018;19:216–228. doi: 10.1016/S1470-2045(18)30010-X. [DOI] [PubMed] [Google Scholar]
- 28.Christman J.K. 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 29.Jüttermann R., Li E., Jaenisch R. Toxicity of 5-aza-2’-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao D., Dasgupta C., Chen M., Zhang K., Buchholz J., Xu Z., Zhang L. Inhibition of DNA methylation reverses norepinephrine-induced cardiac hypertrophy in rats. Cardiovasc. Res. 2014;101:373–382. doi: 10.1093/cvr/cvt264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stenzig J., Schneeberger Y., Löser A., Peters B.S., Schaefer A., Zhao R.-R., Ng S.L., Höppner G., Geertz B., Hirt M.N., Tan W., Wong E., Reichenspurner H., Foo R.S.-Y., Eschenhagen T. Pharmacological inhibition of DNA methylation attenuates pressure overload-induced cardiac hypertrophy in rats. J. Mol. Cell. Cardiol. 2018;120:53–63. doi: 10.1016/j.yjmcc.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Stresemann C., Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 33.Stresemann C., Brueckner B., Musch T., Stopper H., Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 34.Murakami T., Li X., Gong J., Bhatia U., Traganos F., Darzynkiewicz Z. Induction of apoptosis by 5-azacytidine: drug concentration-dependent differences in cell cycle specificity. Cancer Res. 1995;55:3093–3098. [PubMed] [Google Scholar]
- 35.Patel K., Dickson J., Din S., Macleod K., Jodrell D., Ramsahoye B. Targeting of 5-aza-2’-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38:4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Q., Ma Z., Ding Y., Bedarida T., Chen L., Xie Z., Song P., Zou M.-H. Circulating miR-103a-3p contributes to angiotensin II-induced renal inflammation and fibrosis via a SNRK/NF-κB/p65 regulatory axis. Nat. Commun. 2019;10:2145. doi: 10.1038/s41467-019-10116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J., Wang Y., Bao C., Liu T., Li S., Huang J., Wan Y., Li J. Curcumin-loaded PEG-PDLLA nanoparticles for attenuating palmitate-induced oxidative stress and cardiomyocyte apoptosis through AMPK pathway. Int. J. Mol. Med. 2019;44:672–682. doi: 10.3892/ijmm.2019.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S., Fan B. Myricetin protects cardiomyocytes from LPS-induced injury. Herz. 2018;43:265–274. doi: 10.1007/s00059-017-4556-3. [DOI] [PubMed] [Google Scholar]
- 39.Chen L., Tian Q., Shi Z., Qiu Y., Lu Q., Liu C. Melatonin alleviates cardiac function in sepsis-caused myocarditis via maintenance of mitochondrial function. Front. Nutr. 2021;8 doi: 10.3389/fnut.2021.754235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gravina G.L., Festuccia C., Marampon F., Popov V.M., Pestell R.G., Zani B.M., Tombolini V. Biological rationale for the use of DNA methyltransferase inhibitors as new strategy for modulation of tumor response to chemotherapy and radiation. Mol. Cancer. 2010;9:305. doi: 10.1186/1476-4598-9-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.