Abstract
The nucleoids in Mycoplasma capricolum cells were visualized by phase-combined fluorescence microscopy of DAPI (4′,6-diamidino-2-phenylindole)-stained cells. Most growing cells in a rich medium had one or two nucleoids in a cell, and no anucleate cells were found. The nucleoids were positioned in the center in mononucleoid cells and at one-quarter and three-quarters of the cell length in binucleoid cells. These formations may have the purpose of ensuring delivery of replicated DNA to daughter cells. Internucleoid distances in binucleoid cells correlated with the cell lengths, and the relationship of DNA content to cell length showed that cell length depended on DNA content in binucleoid cells but not in mononucleoid cells. These observations suggest that cell elongation takes place in combination with nucleoid movement. Lipid synthesis was inhibited by transfer of cells to a medium lacking supplementation for lipid synthesis. The transferred cells immediately stopped dividing and elongated while regular spaces were maintained between the nucleoids for 1 h. After 1 h, the cells changed their shapes from rod-like to round, but the proportion of multinucleoid cells increased. Inhibition of protein synthesis by chloramphenicol induced nucleoid condensation and abnormal positioning, although partitioning was not inhibited. These results suggest that nucleoid partitioning does not require lipid or protein synthesis, while regular positioning requires both. When DNA replication was inhibited, the cells formed branches, and the nucleoids were positioned at the branching points. A model for the reproduction process of M. capricolum, including nucleoid migration and cell division, is discussed.
Mycoplasmas are wall-less parasitic bacteria possessing extremely small genomes (9). In mycoplasmas, chromosome replication has been reported to start at a fixed site, followed by bidirectional progression (21–23, 28). In mycoplasmas, as in walled bacteria (36), the dnaA gene is expressed and is believed to have important roles in the initiation of replication (21–23, 35). The outline of DNA replication in mycoplasmas is presumably similar to that of walled bacteria. However, the coupling of chromosome replication and cell division in mycoplasmas is unclear. This area of investigation is difficult to explore because mycoplasma cells have high plasticity owing to the lack of a peptidoglycan layer.
In a previous paper, we demonstrated that the basic cell shape of Mycoplasma capricolum is a simple rod and that the bacterium divides by binary fission (34). DNA replication is believed to use the whole cell division interval, because the time required for DNA replication corresponded to the doubling time of living cells and most DNA molecules were in intermediate forms (34). Next, to elucidate the coupling of cell division and DNA replication we have examined the spatial behavior of the chromosome.
In Escherichia coli, a cell maintains the DNA molecule in a condensed form, the nucleoid, at its center. After the completion of DNA replication, the nucleoid partitions, moves, and positions itself in every quarter of the cell length to ensure the delivery of the chromosome into progeny cells (8, 15). In cells deficient in division, nucleoids are separated and positioned at regular intervals, suggesting the existence of markers for nucleoid positioning (33). Nucleoid positioning is also observed for Bacillus subtilis (16), which is phylogenetically related to mycoplasmas (38). However, these concepts cannot be applied simply to mycoplasmas, because they lack a peptidoglycan layer, which plays important roles in nucleoid partitioning and cell division in walled bacteria (6). Unique patterns of nucleoid partitioning and positioning are expected for mycoplasmas.
In this study, we observed the nucleoids of DAPI (4′,6-diamidino-2-phenylindole)-stained M. capricolum cells by phase-combined fluorescence microscopy (14) and examined cellular mechanisms for nucleoid migration in mycoplasmas.
MATERIALS AND METHODS
Cultivation.
M. capricolum ATCC 27343 was grown at 37°C. Modified Edward medium (30) was used with some modifications as previously described (34); i.e., 5% horse serum was replaced by 2 mg of bovine serum albumin/ml, 20 μg of cholesterol/ml, 10 μg of palmitic acid/ml, and 12 μg of oleic acid/ml (31). For supplementation with nucleosides, 40 μg (each) of adenosine, guanosine, and uridine/ml and 20 μg of thymidine/ml were added to the medium. Frozen cultures were inoculated into the medium and grown overnight to reach an optimal density at 600 nm (OD600) of around 0.05. The cultures were diluted as the OD600 became 10−4 and were used for assay after several hours.
Microscopic observation.
M. capricolum cells in culture were fixed with 4.2% glutaraldehyde for 60 min at room temperature. The fixed cells were collected by centrifugation at 10,000 × g at 4°C for 3 min, washed, and resuspended in phosphate-buffered saline. For phase-combined fluorescence microscopy (14), fixed cells were mixed with an equal volume of 20-μg/ml DAPI solution and incubated for 10 min at room temperature. Microscopic images were photographed with Fuji Super G 400 film. The cell length and the positions of fluorescent nucleoids were measured on printed images as shown in Fig. 1F. Fluorescence density was used to calculate DNA content, as previously described (34). For transmission electron microscopy, cells were fixed in the cultures with the addition of 4.2% glutaraldehyde for 60 min at room temperature and directly placed on a 180A grid supported by a collodion membrane. Alternatively, the fixed cells were collected by centrifugation at 10,000 × g at 4°C for 3 min, suspended in phosphate-buffered saline, and then placed on a grid. The cells on grids were allowed to dry, negatively stained with 2% ammonium molybdate for 1.5 min, and observed with a transmission electron microscope (5, 34).
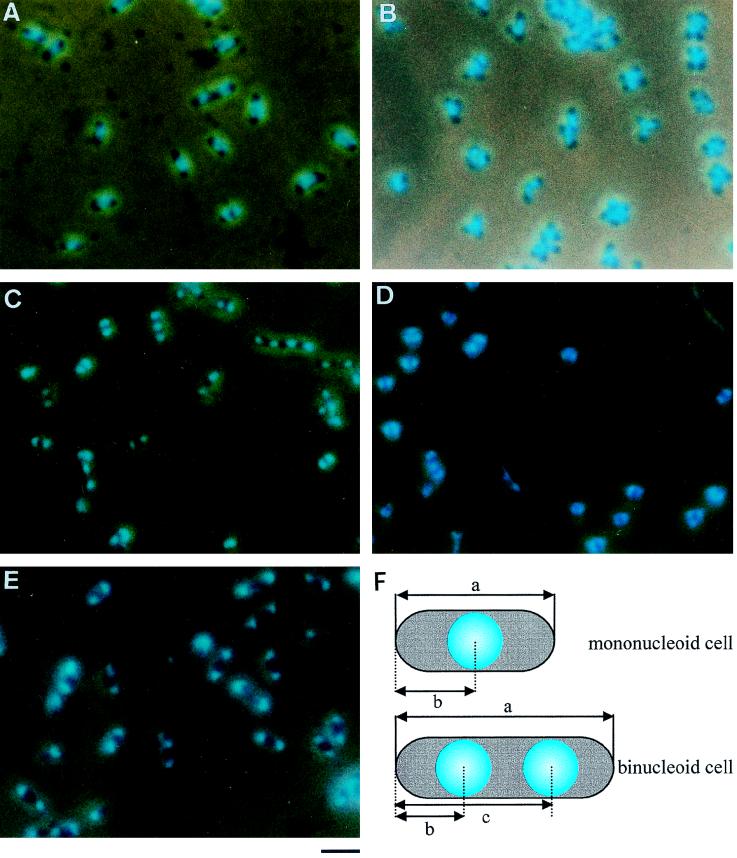
FIG. 1.
Images of M. capricolum cells observed by phase-combined fluorescence microscopy. Cells were fixed, stained with DAPI, and photographed. (A) Culture was grown to an OD600 of 0.05 in the standard medium. (B) Culture was grown to an OD600 of 0.4 without nucleoside supplementation. (C and D) Cells grown to an OD600 of 0.05 in the standard medium were transferred to a medium without supplementation for lipid synthesis and incubated for 1 and 2 h, respectively. (E) Cells grown to an OD600 of 0.05 in the standard medium were incubated with chloramphenicol at 37°C for 1.5 h. Bar, 2 μm. (F) Schematic illustration of M. capricolum cells. a, cell length; b and c, nucleoid positions. Internucleoid distance was calculated by subtraction of b from c.
Measurement of phospholipid synthesis.
H332PO4 was added to a final concentration of 925 kBq/ml (2.7 nM). Radiolabeled cells were collected and treated with a solvent consisting of methanol-chloroform (1:2) at room temperature for 6 h (1). The extract was washed twice with a buffer consisting of 10 mM Tris-HCl (pH 7.6) and 1 mM EDTA, mixed with scintillation cocktail, and measured with a scintillation counter.
RESULTS
Nucleoid observation.
In a previous paper, we showed that nucleoside supplementation is necessary for proper progression of DNA replication (34). Therefore, we used modified Edward medium supplemented with nucleoside as the standard medium of this study. To study the position of nucleoids, M. capricolum cells growing in the standard medium were stained with DAPI and observed by phase-combined fluorescence microscopy (Fig. 1A). Fluorescent nucleoids were condensed and occupied a part of the cell interior. Cells containing one, two, and three or more separate nucleoids accounted for 32.8, 67.8, and 0.4% of total cells, respectively. We observed more than 1,000 cells but did not find anucleate cells marked with weak fluorescence, which is known to be emitted from RNA molecules (14). These results suggest that the replicated chromosomes have the purpose of ensuring segregation into nascent progeny cells in the standard medium.
Inhibition of DNA replication by nucleoside starvation has been reported to induce branching (34). We induced branching by culturing the mycoplasma cells to an OD600 of 0.4 in medium without nucleoside supplementation and observed cells by phase-combined fluorescence microscopy (Fig. 1B). In the branched cells, the nucleoids were located at branching points. No anucleate cells were found in this culture.
Position of nucleoids in cells growing in the standard medium.
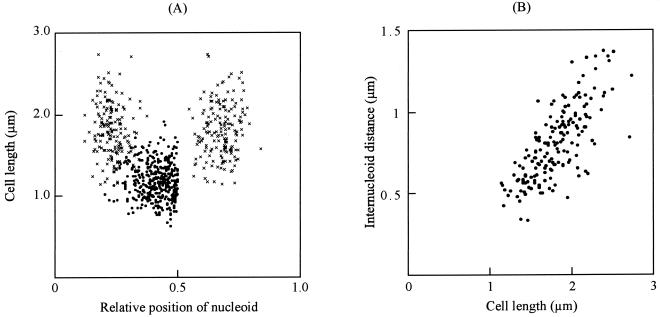
We determined the intracellular positions of nucleoids in cells growing in the standard medium (Fig. 2A). The average length of mononucleoid cells was 1.18 μm, and the nucleoids were positioned around the center of cell length. The average length of binucleoid cells was 1.80 μm, and the nucleoids in these cells were positioned at 0.23 and 0.68 of cell length, respectively. These results suggest that replicated chromosomes are segregated from the center to each quarter of the cell prior to cell division. We examined internucleoid distances in binucleoid cells (Fig. 1B). They correlated with cell length, suggesting that nucleoid movement occurs in combination with cell elongation.
FIG. 2.
Nucleoid positions of cells growing in the standard medium. (A) Positions of nucleoids are presented relative to position 0. The nucleoid position in a mononucleoid cell is indicated by a closed circle (n = 376). The nucleoid positions in a binucleoid cell are indicated by multiplication signs (n = 169). The average lengths of mono- and binucleoid cells are 1.18 and 1.80 μm with SDs of 0.22 and 0.32 μm, respectively. Nucleoids are positioned at the center with an SD of 0.11 and at 0.23 and 0.68 with SDs of 0.05 and 0.06 of cell length, respectively, in mono- and binucleoid cells. (B) Internucleoid distances in binucleoid cells are plotted against cell lengths (n = 169).
Timing of cell elongation.
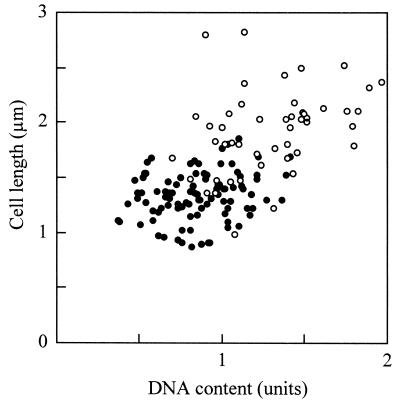
Previously, we reported that most DNA molecules are in intermediate forms in the standard medium (34). Assuming that the intermediates are always undergoing reaction, it is possible to order the individual cell images by titrating the DNA content. Therefore, we estimated the DNA content in a cell from the fluorescence of DAPI-stained images and plotted the cell length against the DNA content (Fig. 3). The lengths of binucleoid cells were apparently greater than those of mononucleoid cells and correlated with the DNA content as expected. However, the cell lengths were not related to the DNA content in mononucleoid cells, suggesting that the cell elongation does not take place until nucleoid partitioning occurs.
FIG. 3.
Relationship of DNA content and cell length in rod cells. Cells containing one and two nucleoids are indicated by closed and open circles, respectively (n = 150). The average DNA content in each cell is normalized to 1 U.
Nucleoid migration without lipid synthesis.
To determine if growth of the cell membrane promotes nucleoid migration, we analyzed cells with inhibited lipid synthesis, because they must take up fatty acid and sterol, by using serum albumin as a carrier for lipid synthesis (30, 32). We therefore transferred the mycoplasma cells to a medium not supplemented for lipid synthesis and monitored the content of phosphate from the solvent extract of culture (Fig. 4A). Radiolabeled phosphate was added to cells in the standard medium at an OD600 of 0.01. At an OD600 of 0.05, half of the culture was transferred to the nonsupplemented medium and the other half was transferred to the supplemented medium. At 1 h after transfer, the phospholipid content in the nonsupplemented culture increased to 1.57 times the original content, and at 2 h, the content decreased to 1.38 times the original content, while phospholipid content in the supplemented culture continued to increase. These results show that removing the supplement inhibits lipid synthesis.
FIG. 4.
The effect of removing supplementation for lipid synthesis on phospholipid synthesis and cell division. (A) Phospholipid synthesis. Culture was supplemented with labeled phosphate at −3 h and transferred at time zero to a new medium supplemented (solid circles) or not supplemented (open circles) for lipid synthesis. The value at time zero was normalized to 1 U. (B) CFU are shown for the cultures with and without supplementation for lipid synthesis by solid and open circles, respectively.
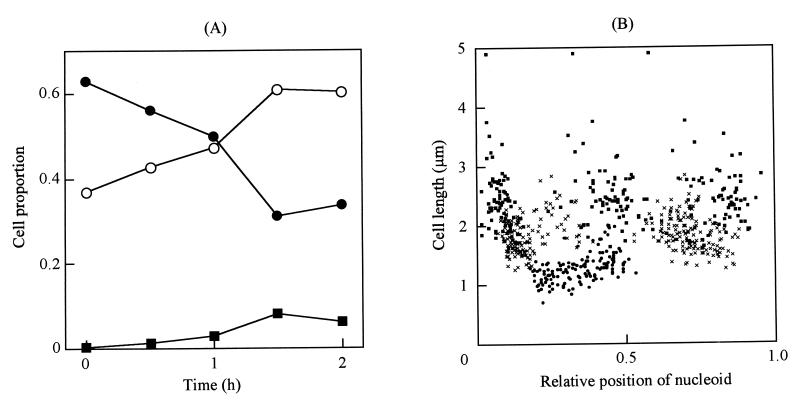
CFU did not increase in nonsupplemented culture, indicating that cell division was inhibited immediately after transfer (Fig. 4B). We microscopically examined the transferred cells (Fig. 1C and D) and found that the proportion of binucleoid cells increased from 32.8% at time zero to 43.9 and 54.4% at 1 and 2 h, respectively (Fig. 5). Cells containing three or more nucleoids increased in number, reaching 14.1 and 30.8% of the population at 1 and 2 h, respectively. In the control culture transferred into the standard medium, the nucleoid number did not change. The nucleoid positions were examined in the cells at 1 h after transfer to the nonsupplemented medium (Fig. 6). The nucleoids in mononucleoid cells were at the center with a standard deviation (SD) of 0.11 of relative cell length, and those in binucleoid cells were positioned at 0.22 and 0.68 of cell length, respectively. Cells with three or more nucleoids also had nucleoid positioning at regular intervals. In the control cells transferred into the standard medium, the nucleoid positioning was not significantly different from that observed in Fig. 2A (data not shown). At 2 h after the transfer into the nonsupplemented medium, the cell length in binucleoid cells and cells with three or more nucleoids decreased (Fig. 5C), i.e., the average lengths at 1 h after the transfer were 1.77 and 2.49 μm for binucleoid and multinucleoid cells, respectively, but those at 2 h were 1.19 and 1.31 μm for binucleoid and multinucleoid cells, respectively. At 2 h, most cells were round (Fig. 1D), while the proportion of multinucleoid cells increased from 1 to 2 h (Fig. 5C). At 2 h, nucleoid spacing was irregular in most cells (Fig. 1D). The lengths of cells transferred into the standard medium did not change significantly over time (Fig. 5D). These results suggest that inhibition of lipid synthesis stops cell elongation, division, nucleoid movement, and nucleoid positioning but that nucleoid partitioning still occurs.
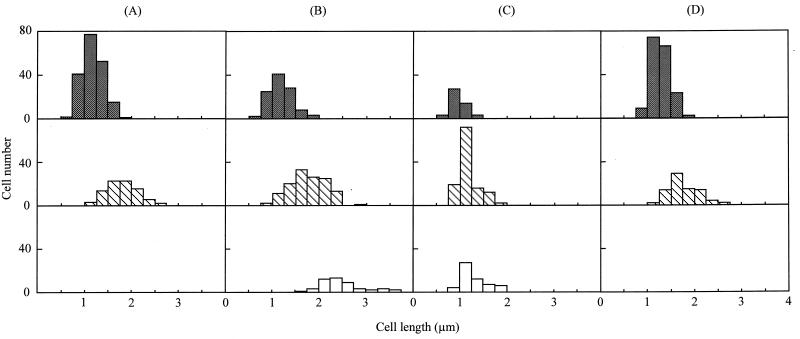
FIG. 5.
Distribution of cell length with inhibition of lipid synthesis. The distribution of cell lengths in cells with one, two, and three or more nucleoids is shown by shaded, hatched, and open bars, respectively. The cells at 0, 1, and 2 h after the transfer into nonsupplemented medium are shown in columns A (n = 273), B (n = 286), and C (n = 234), respectively. The cells at 2 h after transfer into the standard medium are indicated in column D (n = 174). The average lengths of cells in columns A, B, C, and D are 1.37, 1.63, 1.17, and 1.43 μm with SDs of 0.39, 0.57, 0.25, and 0.32 μm, respectively.
FIG. 6.
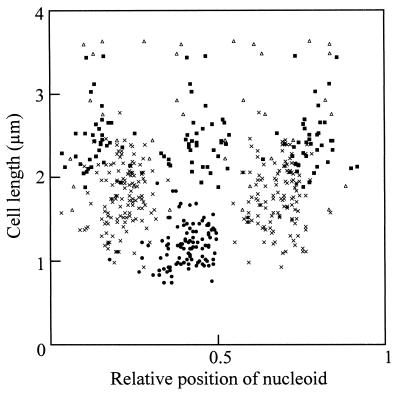
Nucleoid positions in cells incubated for 1 h without supplementation for lipid synthesis. The positions of nucleoids are indicated relative to position 0. The positions in cells containing one, two, three, and four nucleoids are indicated by closed circles, multiplication signs, solid squares, and open triangles, respectively (n = 107, 132, 40, and, 10, respectively).
Effect of inhibition of protein synthesis on nucleoid migration.
We examined the requirement for protein synthesis for nucleoid migration by adding 100 μg of chloramphenicol/ml to the standard culture. The nucleoids were condensed into more compact structures (Fig. 1E). The proportions of binucleoid and trinucleoid cells increased from 32.8 and 0.4% of total cells at time zero to 60.8 and 8.1% at 1.5 h, respectively (Fig. 7A). The intracellular positions of nucleoids at 1.5 h were significantly disturbed and preferentially associated with cell poles (Fig. 7B). No changes were observed in both the cell type proportion and the nucleoid spacing in the control culture at 1.5 h (data not shown).
FIG. 7.
Nucleoid numbers and positions in cells incubated with chloramphenicol. (A) Nucleoid numbers in a cell. The proportions of cells containing one, two, and three or more nucleoids are indicated by solid circles, open circles, and solid squares, respectively. More than 300 cells were examined at each time. (B) Nucleoid positions in cells incubated with chloramphenicol for 1.5 h. The positions of nucleoids are indicated relative to position 0. The positions in cells containing one, two, and three nucleoids are indicated by solid circles, multiplication signs, and solid squares, respectively (n = 143, 148, and 73, respectively).
Electron microscopy and effect of centrifugation on fixed cells.
In order to examine the effects of centrifugation on the morphology of fixed cells, we compared electron microscopic images before and after the centrifugation. We examined cultures in the standard medium (Fig. 8A) for inhibition of lipid synthesis (Fig. 8B and C), inhibition of protein synthesis (Fig. 8D), and depletion of nucleotides (data not shown). No differences in cell appearance were found between images before centrifugation and those after centrifugation (data not shown). The average cell length in the standard culture was 0.941 μm after centrifugation, not significantly different from the value before centrifugation, 0.910 μm. These results indicate that the change in cellular morphology was not caused by fusion or fragmentation due to the centrifugation.
FIG. 8.
Cell images observed by electron microscopy. Cells were fixed in the cultures, directly placed on copper grids, negatively stained, and observed. (A) Cells were grown to an OD600 of 0.05 in the standard medium. (B and C) Cells grown in the standard medium were transferred to a medium without supplementation for lipid synthesis and incubated for 1 and 2 h, respectively. (D) Cells grown in the standard medium were incubated with chloramphenicol at 37°C for 1.5 h. Bar, 0.5 μm.
DISCUSSION
In mycoplasmas, the spatial behavior of chromosomal DNA has been unknown. We have previously observed chromosomal DNA by fluorescence microscopy of DAPI-stained M. capricolum cells; however, no information about intracellular position was obtainable because the entire cytoplasmic space was fluorescent (34). In this study, we observed the nucleoids by phase-combined fluorescence microscopy and found that the nucleoids were condensed and occupied only a part of the cytoplasmic space (Fig. 1). Presumably, visual light reduced the weak fluorescence emitted from RNA molecules and intracellular halation (14). In cells growing in the standard medium, the nucleoids were positioned at the centers of mononucleoid cells and at each quarter in binucleoid cells, and no anucleate cells were found (Fig. 1A and 2A). These results show that the chromosomes migrate synchronously with cell division to ensure their delivery to progeny cells. It is remarkable that the mechanism for nucleoid migration is so well organized in mycoplasma cells, which do not have cell walls and have little genome information (3, 10, 12, 24, 29). The analyses of timing of the nucleoid separation show that the nucleoid movement occurred in combination with the cell elongation (Fig. 2B and 3). What kind of force propels and localizes the nucleoids? We tried characterizing each step of nucleoid migration, i.e., partitioning, movement, and positioning.
Generally, mycoplasmas cannot synthesize lipid without supplementation with fatty acid, sterol, and serum albumin (30, 32) (Fig. 4A). We utilized this dependency to examine the role of lipid synthesis in nucleoid migration. The cells ceased division immediately after deprivation of lipid synthesis supplements, and the proportion of multinucleoid cells increased (Fig. 4A and 5). This indicates that nucleoid partitioning occurs independently of cell division. At 1 h after deprivation, the spacing of nucleoids was normal (Fig. 6). This observation rules out the possibility that the position of the nucleoid is determined by the balance of tension from cell poles as is the case for the metaphase plate of eukaryotic cells (26), because it is unlikely that the mechanisms for proper force generation are produced following cell elongation caused by the reduction of lipid synthesis.
Until 1 h after deprivation, lipid synthesis stopped entirely (Fig. 4A). Subsequently, the length of multinucleoid cells decreased, while the proportion of partitioned nucleoids continued to increase (Fig. 5). This observation shows that the nucleoid partitions independently of lipid synthesis and cell elongation, although they are required for movement and positioning of nucleoids. Garnier et al. reported that the cytoplasmic membrane elongates around the center and propels the chromosome in Spiroplasma citri (11), which is phylogenetically closely related to M. capricolum (38). However, our results do not support this possibility in M. capricolum.
Nucleoid partitioning did not require de novo protein synthesis in M. capricolum (Fig. 7). This result is different from that obtained with E. coli (15). Woldringh et al. proposed that the bacterial chromosomes are anchored to the cytoplasmic membrane or peptidoglycan layer by nascent mRNA molecules on which nascent proteins are being translated (39). In this model, the nucleoids are propelled by motive force generated by transcription and translation. The results obtained here rule out this hypothesis for M. capricolum.
In E. coli, it has been suggested that nucleoid positioning is related to a solid structure, the periseptal annuli, which divide the periplasmic space into two components (13). This is believed to be so because periseptal annuli are duplicated in newborn cells, move toward each quarter of the cell length, and are immobile throughout the cell division cycle (7), and the nucleoid position corresponds to those of the periseptal annuli (15). Mycoplasma cells probably lack a solid structure like the periseptal annulus because they have no peptidoglycan layer. Therefore, the structure required for nucleoid migration is probably more fragile and temporal in mycoplasmas. Our results indicating that the nucleoid positions were disturbed by the inhibition of protein synthesis (Fig. 1E and 7) may reflect the character of this structure.
Cytoskeleton-like structures have been identified in some mycoplasmas (17, 18, 20, 27, 37). In Mycoplasma pneumoniae, actin-like filaments have been reported to form a network complex (20). In Mycoplasma gallisepticum, a eukaryotic tubulin-like filament was found to form a submembrane structure (18). In both mycoplasmas, subcellular structures for cytoadherence supported by the filamentous structures duplicate prior to cell division, synchronizing with DNA replication (2, 25). It is possible that the filamentous structures are involved in nucleoid migration (19). This hypothesis is consistent with our observations that cell elongation was coupled with nucleoid movement and that the positioning was disturbed with protein synthesis inhibition.
In this study, we observed various cell morphologies of M. capricolum (Fig. 1), including branching, filamentation, and rounding, which were induced by stresses, as summarized in Fig. 9. We released the cells from the stresses and found that all types of cells were viable. They returned to the rod type and started reproduction within 3 h after the release (data not shown). Buxton and Fraser (4) suggested that Mycoplasma mycoides, which is closely phylogenetically related to M. capricolum (38), reproduces via these characteristic types of cells, namely, it develops from an elementary rounded body into a branched and filamentous form, and then new rounded elementary bodies develop within these filaments and are released when mature. Our previous and current observations rule out the presence of such characteristic cells in the reproduction process under rich conditions (34). However, it is still possible that the characteristic types of cells are involved in the reproduction process as stressed forms in nature, because stresses similar to those examined here can occur in the tissues of hosts.
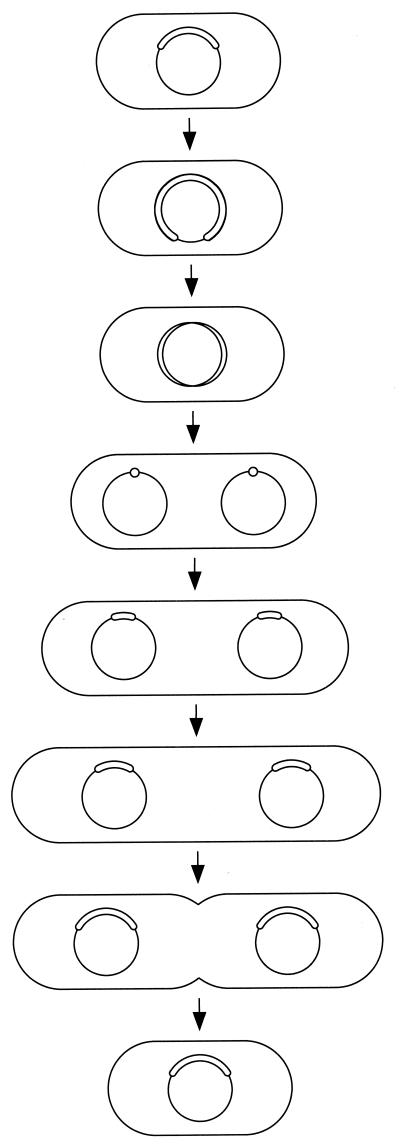
FIG. 9.
Schematic illustration of cell type conversion by stresses.
We have previously proposed a reproduction model for the cell division cycle of M. capricolum (34). To that, we add the information about nucleoid migration obtained here (Fig. 10). DNA replication uses almost the whole cell division interval. After the completion of DNA replication, replicated chromosomes migrate to each quarter of a cell, and then cell elongation occurs in combination with movement of nucleoids, until the double cell length is achieved. The nucleoid partitions independently of de novo synthesis of lipid or protein, but nucleoid movement and positioning depend on both syntheses.
FIG. 10.
Model for nucleoid migration and cell division of M. capricolum cells. The state of chromosomal DNA replication is indicated by circles in the cells.
REFERENCES
- 1.Acher D B. Modification of the membrane composition of Mycoplasma mycoides subsp. capri by the growth medium. J Gen Microbiol. 1975;88:329–338. doi: 10.1099/00221287-88-2-329. [DOI] [PubMed] [Google Scholar]
- 2.Biberfeld G, Biberfeld P. Ultrastructural features of Mycoplasma pneumoniae. J Bacteriol. 1970;102:855–861. doi: 10.1128/jb.102.3.855-861.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bork P, Ouzounis C, Casari G, Schneider R, Sander C, Dolan M, Gilbert W, Gillevet P M. Exploring the Mycoplasma capricolum genome: a minimal cell reveals its physiology. Mol Microbiol. 1995;16:955–967. doi: 10.1111/j.1365-2958.1995.tb02321.x. [DOI] [PubMed] [Google Scholar]
- 4.Buxton A, Fraser G. Animal microbiology. I. Oxford, United Kingdom: Blackwell Scientific Publications Ltd.; 1977. Mycoplasma, Acholeplasma and L-form of bacteria; pp. 267–283. [Google Scholar]
- 5.Cole R M. Transmission electron microscopy: basic techniques. In: Razin S, Tully J S, editors. Methods in mycoplasmology. New York, N.Y: Academic Press; 1983. pp. 43–50. [Google Scholar]
- 6.Cooper S. Segregation of cell structure. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella; cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1652–1661. [Google Scholar]
- 7.de Boer P A J, Cook W R, Rothfield L I. Bacterial cell division. Annu Rev Genet. 1990;24:249–274. doi: 10.1146/annurev.ge.24.120190.001341. [DOI] [PubMed] [Google Scholar]
- 8.Donachie W D, Begg K J. Chromosome partition in Escherichia coli requires postreplication protein synthesis. J Bacteriol. 1989;171:5405–5409. doi: 10.1128/jb.171.10.5405-5409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dybvig K, Voelker L L. Molecular biology of mycoplasmas. Annu Rev Microbiol. 1996;50:25–57. doi: 10.1146/annurev.micro.50.1.25. [DOI] [PubMed] [Google Scholar]
- 10.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman R D, Weidman J F, Small K V, Sandusky M, Fuhrmann J, Nguyen D, Utterback R, Saudek D M, Phillips C A, Merrick J M, Tomb J-F, Dougherty B A, Bott K F, Hu P-C, Lucier T S, Peterson S N, Smith H O, Hutchison III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 11.Garnier M, Clerc M, Bove J M. Growth and division of Spiroplasma citri: elongation of elementary helices. J Bacteriol. 1984;158:23–28. doi: 10.1128/jb.158.1.23-28.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiraga S. Chromosome partition in Escherichia coli. Curr Opin Genet Dev. 1993;3:789–801. doi: 10.1016/s0959-437x(05)80100-5. [DOI] [PubMed] [Google Scholar]
- 14.Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffe A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiraga S, Ogura T, Niki H, Ichinose C, Mori H. Positioning of replicated chromosomes in Escherichia coli. J Bacteriol. 1990;172:31–39. doi: 10.1128/jb.172.1.31-39.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ireton K, Gunther N W, IV, Grossman A D. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kessel M, Peleg I, Muhlrad A, Kahane I. Cytoplasmic helical structure associated with Acholeplasma laidlawii. J Bacteriol. 1981;147:653–659. doi: 10.1128/jb.147.2.653-659.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korolev E V, Nikonov A V, Brudnaya M S, Snigirevskaya E S, Sabinin G V, Komissarchik Y Y, Ivanov P I, Borchsenius S N. Tubular structures of Mycoplasma gallisepticum and their possible participation in cell motility. Microbiology. 1994;140:671–681. doi: 10.1099/00221287-140-3-671. [DOI] [PubMed] [Google Scholar]
- 19.Krause D C. Mycoplasma pneumoniae cytadherence: organization and assembly of the attachment organelle. Trends Microbiol. 1998;6:15–18. doi: 10.1016/S0966-842X(97)01168-2. [DOI] [PubMed] [Google Scholar]
- 20.Meng K E, Pfister R M. Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J Bacteriol. 1980;144:390–399. doi: 10.1128/jb.144.1.390-399.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyata M, Fukumura T. Asymmetrical progression of replication forks just after initiation on Mycoplasma capricolum chromosome revealed by two-dimensional gel electrophoresis. Gene. 1997;193:39–47. doi: 10.1016/s0378-1119(97)00075-9. [DOI] [PubMed] [Google Scholar]
- 22.Miyata M, Sano K-I, Okada R, Fukumura T. Mapping of replication initiation site in Mycoplasma capricolum genome by two-dimensional gel-electrophoretic analysis. Nucleic Acids Res. 1993;21:4816–4823. doi: 10.1093/nar/21.20.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyata M, Wang L, Fukumura T. Localizing the replication origin region on the physical map of the Mycoplasma capricolum genome. J Bacteriol. 1993;175:655–660. doi: 10.1128/jb.175.3.655-660.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyata M, Wang L, Fukumura T. Physical mapping of the Mycoplasma capricolum genome. FEMS Microbiol Lett. 1991;79:329–334. doi: 10.1016/0378-1097(91)90107-l. [DOI] [PubMed] [Google Scholar]
- 25.Morowitz H J, Maniloff J. Analysis of the life cycle of Mycoplasma gallisepticum. J Bacteriol. 1966;91:1638–1644. doi: 10.1128/jb.91.4.1638-1644.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray A, Hunt T. The cell cycle: an introduction. W. H. New York, N.Y: Freeman and Company; 1993. [Google Scholar]
- 27.Peterson J E, Rodwell A W, Rodwell E S. Occurrence and ultrastructure of a variant (rho) form of Mycoplasma. J Bacteriol. 1973;115:411–425. doi: 10.1128/jb.115.1.411-425.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pyle L E, Finch L R. Preparation and FIGE separation of infrequent restriction fragments from Mycoplasma mycoides DNA. Nucleic Acids Res. 1988;16:2263–2268. doi: 10.1093/nar/16.5.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Razin S. Molecular properties of mollicutes: a synopsis. In: Razin S, Tully J G, editors. Molecular and diagnostic procedures in mycoplasmology. 1st ed. Vol. 1. San Diego, Calif: Academic Press, Inc.; 1995. pp. 1–25. [Google Scholar]
- 30.Razin S, Rottem S. Techniques for the manipulation of mycoplasma membranes. In: Maddy A H, editor. Biochemical analysis of membranes. London, United Kingdom: Chapman & Hall; 1976. pp. 3–25. [Google Scholar]
- 31.Rodwell A W, Peterson J E, Rodwell E S. Macromolecular synthesis and growth of mycoplasmas. Ciba Found. Symp. 1972. pp. 123–139. [Google Scholar]
- 32.Rottem S, Shinar D, Bittman R. Symmetrical distribution and rapid transbilayer movement of cholesterol in Mycoplasma gallisepticum membrane. Biochim Biophys Acta. 1981;649:572–580. doi: 10.1016/0005-2736(81)90161-9. [DOI] [PubMed] [Google Scholar]
- 33.Schmid M B, von Freiesleben U. Nucleoid segregation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1662–1671. [Google Scholar]
- 34.Seto S, Miyata M. Cell reproduction and morphological changes in Mycoplasma capricolum. J Bacteriol. 1998;180:256–264. doi: 10.1128/jb.180.2.256-264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seto S, Murata S, Miyata M. Characterization of dnaA gene expression in Mycoplasma capricolum. FEMS Microbiol Lett. 1997;150:239–247. doi: 10.1016/s0378-1097(97)00121-3. [DOI] [PubMed] [Google Scholar]
- 36.Skarstad K, Boye E. The initiator protein DnaA: evolution, properties and function. Biochim Biophys Acta. 1994;1217:111–130. doi: 10.1016/0167-4781(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 37.Townsend R, Acher D B, Plaskitt K A. Purification and preliminary characterization of spiroplasma fibrils. J Bacteriol. 1980;142:694–700. doi: 10.1128/jb.142.2.694-700.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisburg W G, Tully J G, Rose D L, Petzel J P, Oyaizu H, Yang D, Mandelco L, Sechrest J, Lawrence T G, Van Etten J, Maniloff J, Woese C R. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989;171:6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woldringh C L, Jensen P R, Westerhoff H V. Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces? FEMS Microbiol Lett. 1995;131:235–242. doi: 10.1111/j.1574-6968.1995.tb07782.x. [DOI] [PubMed] [Google Scholar]