Abstract
Flowering is the primary stage of the plant developmental transition and is tightly regulated by environmental factors such as light and temperature. However, the mechanisms by which temperature signals are integrated into the photoperiodic flowering pathway are still poorly understood. Here, we demonstrate that HOS15, which is known as a GI transcriptional repressor in the photoperiodic flowering pathway, controls flowering time in response to low ambient temperature. At 16°C, the hos15 mutant exhibits an early flowering phenotype, and HOS15 acts upstream of photoperiodic flowering genes (GI, CO, and FT). GI protein abundance is increased in the hos15 mutant and is insensitive to the proteasome inhibitor MG132. Furthermore, the hos15 mutant has a defect in low ambient temperature–mediated GI degradation, and HOS15 interacts with COP1, an E3 ubiquitin ligase for GI degradation. Phenotypic analyses of the hos15 cop1 double mutant revealed that repression of flowering by HOS15 is dependent on COP1 at 16°C. However, the HOS15–COP1 interaction was attenuated at 16°C, and GI protein abundance was additively increased in the hos15 cop1 double mutant, indicating that HOS15 acts independently of COP1 in GI turnover at low ambient temperature. This study proposes that HOS15 controls GI abundance through multiple modes as an E3 ubiquitin ligase and transcriptional repressor to coordinate appropriate flowering time in response to ambient environmental conditions such as temperature and day length.
Key words: Arabidopsis, flowering response, low ambient temperature, HOS15, GIGANTEA
Flowering time is tightly regulated by multiple pathways. This study shows that HOS15 accelerates low ambient temperature–mediated GI degradation. HOS15 integrates photoperiodic and thermosensory flowering pathways by controlling GI abundance as an E3 ubiquitin ligase and transcriptional repressor to coordinate appropriate flowering time.
Introduction
Earth dynamics induce diurnal and seasonal fluctuations in light and temperature across the planet. Plants sense these environmental input signals, and the accumulated diurnal information in plant cells drives various developmental outputs, including flowering (Song, 2016). Flowering is a major developmental transition from the vegetative to reproductive state that determines crop production, including that of vegetables and grains (Jung and Müller, 2009). Flowering time is delicately determined by various pathways, including the photoperiodic (day length), thermosensory (temperature), hormonal, autonomous (genetic makeup), and vernalization (prolonged exposure to cold) pathways. The genetic pathway regulates expression of floral integrator genes such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), which activate downstream floral identity genes (such as APETALA1 and LEAFY) to induce flowering (Kim et al., 2009; Andrés and Coupland, 2012; Song et al., 2015).
The photoperiodic pathway is responsible for predicting upcoming environmental changes and precisely aligning flowering time under favorable conditions (Andrés and Coupland, 2012). Long-day (LD) conditions accelerate flowering by promoting the floral activator FT. The diurnal cycles of CONSTANS (CO), a transcriptional activator of FT, are controlled by the complex of CYCLING DOF FACTOR (CDF), FLAVIN-BINDING KELCH REPEAT F-BOX 1 (FKF1), and GIGANTEA (GI) proteins (Imaizumi, 2005; Sawa et al., 2007; Fornara et al., 2009). CDF proteins are transcriptional repressors of CO during the morning (Fornara et al., 2009) and are targeted by the E3 ubiquitin ligase FKF1 (Imaizumi, 2005) and a unique plant-specific protein, GI (Kim et al., 2007). Under LD conditions, FKF1 and GI proteins form a complex in a blue light-dependent manner (Sawa et al., 2007) and then degrade CDF proteins to activate CO transcription in the afternoon (Fornara et al., 2009; Sawa et al., 2007). CO protein activates SOC1 via FT to promote flowering (Yoo et al., 2005; Nakamichi et al., 2007).
GI, a nucleocytoplasmic protein, is involved in various biological processes, including the circadian clock, flowering time, starch accumulation, chlorophyll accumulation, hypocotyl elongation, and a wide range of responses to stresses such as drought, cold, salt, and oxidative stress (Mishra and Panigrahi, 2015). GI is known to function as a mediator of red light reception (photoreceptor) and the circadian rhythm (Fowler et al., 1999; Huq et al., 2000), as well as a floral promoter (Koornneef et al., 1991). GI transcripts oscillate under the control of the circadian clock, peaking at 8–10 h after sunrise (Fowler et al., 1999; Park et al., 1999). CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1), a core clock component in the morning loop, reduces GI expression in the morning by directly associating with the CCA1-binding site of the GI promoter (Lu et al., 2012). GI transcripts accumulate in the afternoon when CCA1 is repressed by another core clock component, TIMING OF CAB EXPRESSION 1 (Gendron et al., 2012; Huang et al., 2012; Lu et al., 2012; Pokhilko et al., 2012). EARLY FLOWERING 3 (ELF3), a GI upstream interactor, positively regulates GI transcription because the rhythm of GI transcripts is disrupted in elf3 mutants under continuous light (LL) conditions (Fowler et al., 1999). Recently, we showed that GI is regulated transcriptionally through an epigenetic mechanism during the regulation of photoperiodic flowering (Park et al., 2019). The evening complex (EC), which is composed of ELF3, ELF4, and LUX ARRHYTHMO (LUX), binds to the LUX binding site (LBS) on the GI promoter with the histone deacetylase complex consisting of HISTONE DEACETYLASE 9 (HDA9) and HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 15 (HOS15). The repressor complex forms a de-acetylated heterochromatin structure, leading to decreased GI expression in the evening and concomitant photoperiodic flowering (Nusinow et al., 2011; Park et al., 2019). By contrast, MUT9P-LIKE KINASE 4 (MLK4), which phosphorylates histone H2A, associates with CCA1 and regulates the accumulation of H2A.Z, a variant form of H2A, and the acetylation of histone 4 at the GI promoter, thereby inducing GI expression (Su et al., 2017). Recently, ELF3 was shown to directly interact with the SWI2/SNF2-RELATED 1 (SWR1) complex to control the deposition of H2A.Z at EC target genes and repress their expression (Tong et al., 2020).
The diel cyclic patterns of GI protein abundance observed in a constitutive overexpressor of GI suggest that post-translational modifications affect the stability and lifetime of GI protein (David et al., 2006). GI proteins that accumulate in blue light are degraded by the 26S proteasome in the dark (David et al., 2006); this process is mediated by ELF3 and CONSTITUTIVE PHOTOMORPHOGENENIC 1 (COP1), which act as a RING domain E3 ligase and a negative photomorphogenesis regulator, respectively (Yu et al., 2008; Lau and Deng, 2012). In addition, COP1 is stabilized at low ambient temperature and triggers proteasomal degradation of GI, resulting in delayed flowering at low ambient temperature, independent of the CO pathway (Jang et al., 2015). Thus, the amount of GI protein is fine-tuned to respond to various environmental changes for proper growth and development.
The major differences between natural and laboratory conditions for plants are the ratio of red/far-red light and the diel temperature fluctuation. In particular, the diel temperature fluctuation under natural conditions is sufficient to mimic natural flowering regulation by reducing FT expression in the afternoon (Song et al., 2018). Therefore, low ambient temperature–mediated repression of flowering is important for synchronizing flowering time in nature. Here, we report the implications of HOS15-mediated GI degradation in the low ambient temperature–mediated flowering response. HOS15 can interact with GI and promote its degradation through the 26S proteasome. Furthermore, HOS15 has a COP1-dependent role in flowering at low ambient temperature. However, the protein interaction between HOS15 and COP1 is attenuated at low ambient temperature, and HOS15 plays a role in low ambient temperature–mediated GI turnover independently of COP1. Taken together, our results suggest that HOS15 plays a role in flowering repression by promoting GI degradation in response to low ambient temperature.
Results
HOS15 is required for flowering repression in response to low ambient temperature
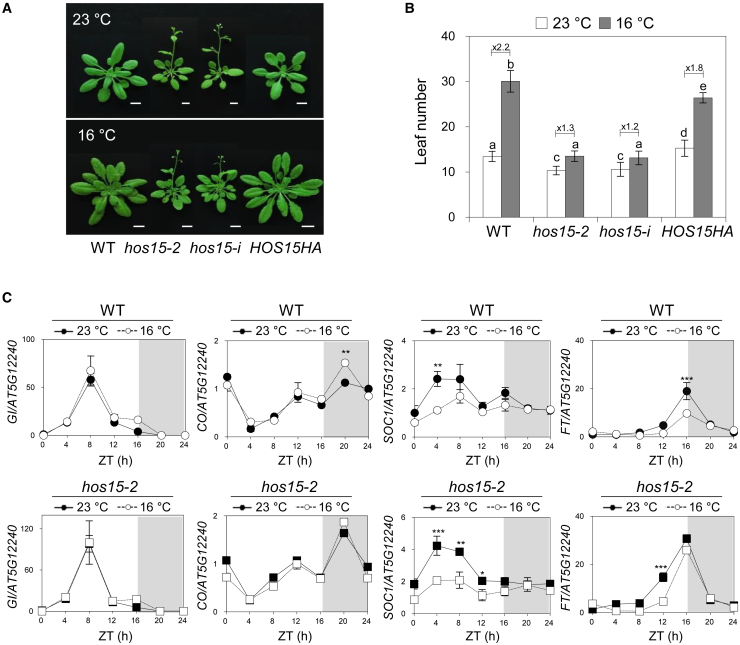
Flowering is tightly controlled by the integration of external factors such as temperature and day length with internal factors such as the circadian clock and phytohormones (Shim et al., 2017). HOS15 plays an essential role in the regulation of both photoperiodic flowering and the cold tolerance response through chromatin remodeling (Zhu et al., 2008a, 2008b; Park et al., 2019). This suggests that HOS15 may be involved in the integration of flowering and temperature response. To test this possibility, the flowering times of a T-DNA insertion line (hos15-2) and an RNAi line (hos15-i) of HOS15 were measured under normal (23°C) and low ambient temperature (16°C) conditions (Figure 1A). The flowering times of wild-type (WT) and HOS15-complemented transgenic plants (proHOS15:HOS15-HA hos15-2, HOS15HA) were delayed at 16°C, with flowering time ratios (16°C/23°C) of approximately 2.2 and 1.8, respectively. On the other hand, hos15-2 and hos15-i were insensitive to low ambient temperature–mediated repression of flowering compared with the WT, with a flowering time ratio of approximately 1.2–1.3 (Figure 1B), indicating that HOS15 is involved in low ambient temperature–mediated repression of flowering. To gain insight into the role of HOS15 in low ambient temperature–mediated flowering, we examined the transcript and protein levels of HOS15 at 23°C and 16°C. Protein and transcript levels of HOS15 did not change at 16°C compared with 23°C during the day (Supplemental Figures 1A and 2B), indicating that HOS15 acts as an upstream regulator in thermosensory flowering.
Figure 1.
HOS15-impaired mutants are insensitive to low ambient temperature–mediated repression of flowering.
(A) Effect of low ambient temperature on flowering of wild-type Col-0 (WT), hos15-2, hos15-i, and proHOS15:HOS15-HA hos15-2 (HOS15HA). Plants were grown under LDs (16L/8D) at 23°C or 16°C. The photographs were taken when hos15-2 and hos15-i mutants bolted. Scale bars correspond to 1 cm.
(B) The numbers of rosette and cauline leaves were counted at bolting to measure flowering time. Data are presented as means ± SD of three independent replicates, each including at least 15 plants. Different letters denote statistically significant differences (ANOVA, Bonferroni post-hoc test, P < 0.05).
(C) Transcript levels of GI, CO, SOC1, and FT at low ambient temperatures. Twelve-day-old plants at 23°C or 16°C in LDs were harvested every 4 h, and the transcript levels of GI, CO, SOC1, and FT were measured by quantitative real-time PCR. Three independent replicates were averaged, and error bars represent the SD of the mean. Asterisks indicate statistically significant differences between 23°C and 16°C in WT and hos15-2 (Student’s t-test; ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05).
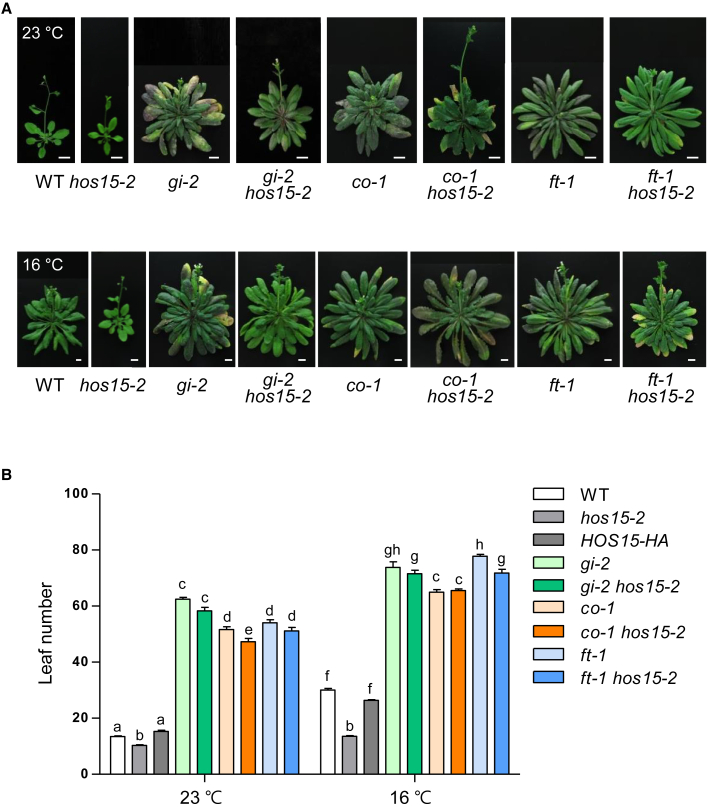
Figure 2.
HOS15 is upstream of the photoperiodic flowering-related genes at low ambient temperature.
(A) Effect of low ambient temperature on flowering of double mutants of hos15-2 crossed with mutants of flowering-related genes. Plants were grown under LDs (16L/8D) at 23°C or 16°C. The photographs were taken when plants bolted. Scale bars correspond to 1 cm.
(B) The numbers of rosette and cauline leaves were counted at bolting to measure flowering time. Data are presented as means ± SD of three independent replicates, each including at least 15 plants. Different letters denote statistically significant differences (ANOVA, Bonferroni post-hoc test; P < 0.05).
HOS15 has been shown to function as a transcriptional repressor of GI, which activates photoperiodic flowering (Park et al., 2019). Therefore, the regulation of GI transcription by HOS15 may underlie the functions of HOS15 in thermosensory flowering. However, the transcript levels of GI in WT and hos15-2 were not altered at 16°C compared with 23°C, suggesting that transcriptional repression of GI by HOS15 is not a major factor in the thermosensory flowering response (Figure 1C). In addition, the expression of CO, which is known to be independent of ambient temperature changes (Lee et al., 2012a; Jang et al., 2015), was not significantly changed at 16°C during the daytime in WT and hos15-2 (Figure 1C). Low ambient temperature signals are associated with floral integrators such as SOC1 and FT (Blazquez et al., 2003). Consistent with the phenotype results, SOC1 and FT expression levels were reduced at low ambient temperature in both WT and hos15-2, and their transcripts in hos15-2 at 16°C were maintained as much as those in the WT at 23°C, indicating that HOS15 acts upstream of SOC1 and FT to prevent the initiation of flowering under low ambient temperature (Figure 1C).
HOS15 is an upstream regulator of photoperiodic flowering genes at low ambient temperature
Photoperiod and temperature are major external signals that control flowering time. Photoperiodic and thermosensory flowering pathways communicate with each other to promote flowering at high ambient temperature and delay flowering at low ambient temperature (Fernandez et al., 2016; Song, 2016). To examine the genetic relationship between the HOS15-mediated thermosensory pathway and the photoperiodic pathway in the control of flowering time, we generated double mutants by crossing hos15-2 with the photoperiodic flowering mutants gi-2, co-1, and ft-1 (Figure 2A). Analysis of flowering time in these double mutants at 23°C and 16°C showed that the photoperiodic flowering mutants were completely or mostly epistatic to hos15-2, indicating that HOS15 acts upstream of the photoperiodic flowering genes at 23°C and 16°C (Figure 2B). Thus, HOS15 is involved in low ambient temperature–mediated repression of flowering through GI–CO–FT photoperiodic flowering pathways. Although GI transcript levels were not altered in WT and hos15-2 at 16°C compared with 23°C (Figure 1C), the flowering time of the gi-2 hos15-2 double mutant was comparable to that of gi-2 under low ambient temperature (Figure 2B). This suggests the possibility that HOS15 regulates GI in a post-translational manner to control flowering time.
HOS15 participates in 26S proteasome–mediated GI degradation
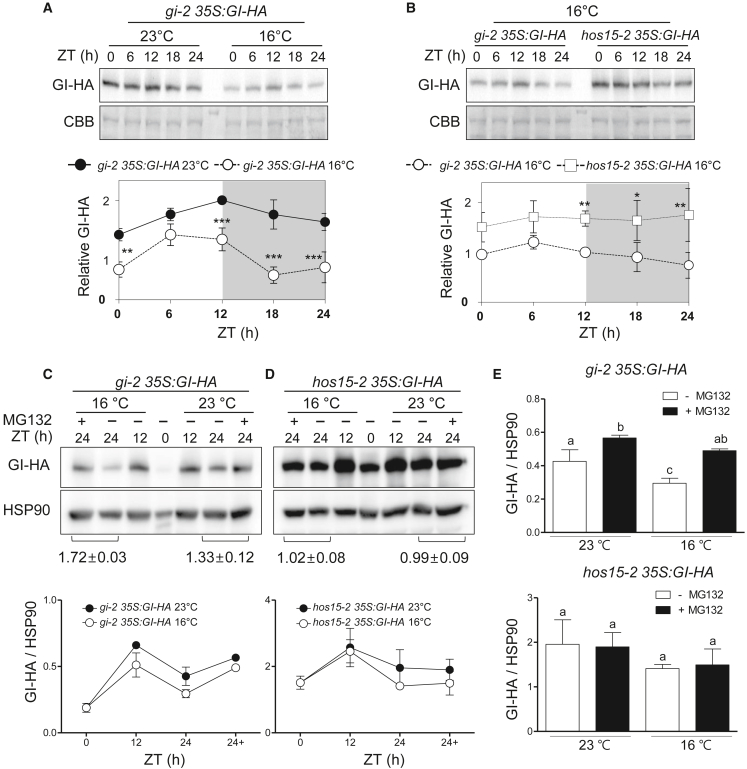
To understand how HOS15 regulates GI to control flowering in response to low ambient temperature, we examined the effect of HOS15 on GI protein abundance at 16°C (Figure 3A and 3B). Previous research has shown that GI protein oscillation is stronger under short-day conditions than under long-day conditions (David et al., 2006). To observe clear changes in GI abundance at low ambient temperature, we investigated GI protein levels under a 12-h light/12-h dark (12L/12D) cycle. The hos15-2 mutants showed an early flowering phenotype at 16°C under 12L/12D conditions (Supplemental Figure 3), consistent with the flowering phenotype under long-day conditions (Figure 1). The abundance of GI decreased at 16°C compared with 23°C in the WT (Figure 3A); however, the abundance of GI was higher in hos15-2 than in the WT at 16°C (Figure 3B), suggesting that HOS15 contributes to the reduction in GI abundance at low ambient temperature. Previous research has shown that GI turnover is accelerated by the COP1-mediated 26S proteasome at low ambient temperature (Jang et al., 2015). HOS15 also functions as a substrate receptor of the 26S proteasome in the forms of a CULLIN 4 (CUL4)-based E3 ubiquitin ligase complex and an SKP1/CUL1/F-box (SCF) E3 ubiquitin ligase complex (Park et al., 2018a; Shen et al., 2020). Therefore, to further test whether lower abundance of GI at low ambient temperature is mediated by the HOS15-based 26S proteasome, we compared GI protein levels in hos15-2 treated with the proteasome inhibitor MG132 at 16°C and 23°C. In WT plants, approximately 72% and 33% of the GI proteins were additionally stabilized by the proteasome inhibitor treatment at 16°C and 23°C, respectively, indicating preferential degradation of GI at low temperature by the ubiquitin-proteasome degradation pathway (Figure 3C and 3E). In the hos15-2 mutant (hos15-2 35S:GI-HA), GI abundance was insensitive to MG132 treatment at both 23°C and 16°C: the ratios of GI protein levels with and without MG132 treatment for 24 h were 0.99 ± 0.09 and 1.02 ± 0.08 at 23°C and 16°C, respectively (Figure 3D and 3E). These results indicate that HOS15 is involved in 26S proteasome–mediated GI degradation at low and normal ambient temperature.
Figure 3.
26S proteasome-mediated GI degradation is impaired in hos15-2 under low ambient temperature.
(A) GI protein abundance at 23°C and 16°C in the WT.
(B) GI protein levels in the hos15-2 mutant under low ambient temperatures. gi-2 35S:GI-HA and hos15-2 35S:GI-HA plants were grown for 10 days under 12L/12D conditions at 23°C and then transferred to 16°C for an additional 2 days and harvested every 4 h. The GI-HA protein levels were normalized to a Coomassie brilliant blue (CBB)-stained protein. (A and B) Data are presented as means ± SD of three independent replicates. Asterisks indicate statistically significant differences (Student’s t-test; ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05).
(C and D) GI protein abundance in gi-2 35S:GI-HA(C) and hos15-2 35S:GI-HA(D) plants at 23°C and 16°C with or without MG132. Ten-day-old plants (35S:GI-HA and hos15-2 35S:GI-HA) grown under 12L/12D at 23°C were transferred to 16°C at ZT 0 with or without MG132 (50 μM) treatment and harvested at the indicated time points. The GI-HA protein levels were normalized to HSP90. Data are presented as means ± SD of three independent replicates. Numbers below the blots are a ratio of relative GI-HA levels with and without MG132 treatment at ZT24.
(E) The relative GI-HA protein levels with and without MG132 treatment at ZT24, shown in (C) and (D), were statistically analyzed. Different letters denote statistically significant differences (ANOVA, Bonferroni post-hoc test; P < 0.05).
HOS15-mediated GI degradation is dependent on low ambient temperature, not darkness
GI proteins are accumulated during the day and degraded through dark- and low ambient temperature–induced proteolysis by the 26S proteasome (David et al., 2006; Jang et al., 2015). To understand whether HOS15-mediated GI degradation is a response to dark or low ambient temperature, hos15-2 and cop1-4 mutants were grown at 23°C under 12L/12D conditions and treated with the protein synthesis inhibitor cycloheximide at ZT12 (23°C dark). They were then transferred to different conditions to distinguish the responses to dark and low ambient temperature in GI degradation: 16°C (16°C dark), subjective night at 23°C (23°C light), and subjective night at 16°C (16°C light) (Figure 4A). GI degradation occurred rapidly under 23°C dark, 16°C light, and 16°C dark conditions compared with the 23°C light condition in the gi-2 35S:HA-GI complementation line (Figure 4A). GI protein levels under each condition were compared as a ratio relative to GI protein levels under the 23°C light condition (Figure 4B). In hos15-2 mutants, GI protein degradation was induced by darkness but was insensitive to 16°C, indicating that HOS15-mediated GI turnover responds to low ambient temperature but not to darkness (Figure 4B). Consistent with previous studies in which COP1-mediated GI turnover was accelerated by both darkness and low ambient temperature (Yu et al., 2008; Jang et al., 2015), GI protein levels in cop1-4 mutants were insensitive to both dark and 16°C conditions (Figure 4B). Taken together, our results suggest that HOS15 and COP1 facilitate GI degradation at low ambient temperature. However, HOS15 does not contribute to dark-induced GI degradation, unlike COP1.
Figure 4.
hos15-2 has a defect in GI degradation induced by low ambient temperature but not by darkness.
(A) GI protein levels in gi-2 35S:GI-HA, hos15-2 35S:GI-HA, and cop1-4 35S:GI-HA in response to low ambient temperature, darkness, and a combination of conditions after cycloheximide treatment. Plants were grown for 10 days under 12L/12D conditions at 23°C and then transferred to 16°C, darkness, and a combination of conditions at ZT12 with cycloheximide treatment. A schematic diagram shows the experimental process.
(B) Quantification of GI amount under each condition relative to the GI amount under continuous light at 23°C. The levels of GI-HA protein under each condition were normalized to that of HSP90 and compared with the levels under the 23°C light condition. Data are presented as means ± SD of three independent replicates. Different letters denote statistically significant differences among gi-2 35S:GI-HA, hos15-2 35S:GI-HA, and cop1-4 35S:GI-HA under the same experimental conditions at the same time point (two-way ANOVA, Bonferroni post-hoc test; P < 0.05).
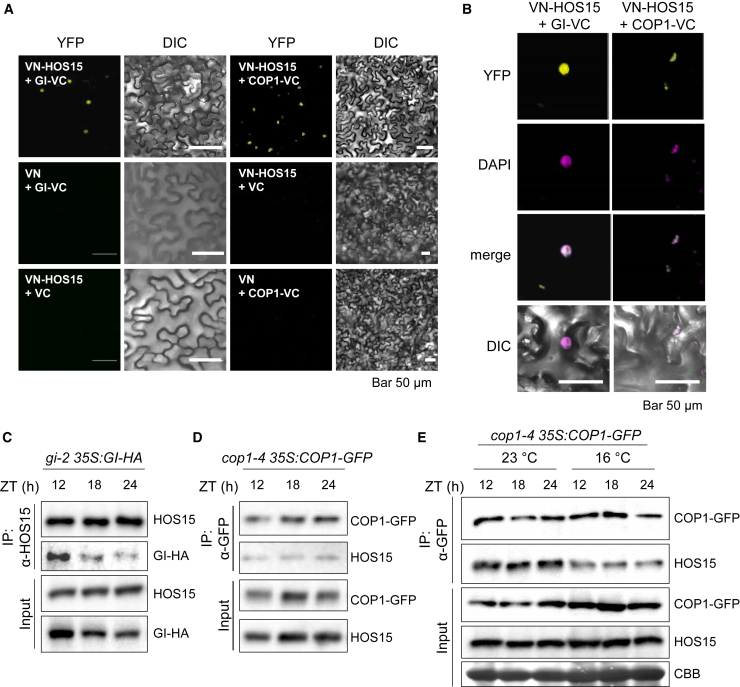
HOS15 forms a complex with GI and COP1 in vivo
Both HOS15 and COP1 function as substrate receptors of the CUL4-based E3 ubiquitin ligase complex (Chen et al., 2010; Zhu et al., 2015; Park et al., 2018a). Because the CUL4-based E3 ubiquitin ligase makes a tetramer formation (Mohamed et al., 2021), we hypothesized that HOS15 and COP1 might function together as substrate receptors to degrade GI proteins at low ambient temperature. To gain insight into the molecular mechanisms underlying the functional relationship between HOS15 and COP1 in GI degradation, we tested the interactions of HOS15 with GI and COP1 in vivo using bimolecular fluorescence complementation (BiFC) (Figure 5A). Although GI and COP1 are located in the nucleus and cytosol (Stacey et al., 1999; Kim et al., 2013; Pacín et al., 2013), HOS15 interacted with GI and COP1 in the nucleus (Figure 5B). To further confirm HOS15 complex formation with GI and COP1 in vivo, we performed a co-immunoprecipitation (Co-IP) assay using transgenic plants overexpressing GI-HA and COP1-GFP (gi-2 35S:GI-HA and cop1-4 35S:COP1-GFP) at ZT12, 18, and 24 (Figure 5C and 5D). The interaction between GI and HOS15 was highest at ZT12 and decreased over time (Figure 5C) because GI protein levels are reduced through dark-induced degradation. By contrast, HOS15 interacted with COP1 at steady-state levels (Figure 5D). We next investigated the interaction between HOS15 and COP1 in response to low ambient temperature. Unexpectedly, the interaction between HOS15 and COP1 was weaker at 16°C than at 23°C (Figure 5E), suggesting that HOS15 and COP1 may have distinct roles in low ambient temperature responses. HOS15 acts as a substrate receptor for a CULLIN 4 (CUL4)-based E3 ubiquitin ligase complex (CUL4HOS15) and an SCF E3 ubiquitin ligase complex (SCFHOS15) for the proteolysis of target proteins (Park et al., 2018a; Shen et al., 2020). Therefore, one possibility is that HOS15 forms a COP1-independent E3 ubiquitin ligase complex at low ambient temperature. Sequence-based prediction revealed that HOS15 contains an F-box-like motif and a LisH domain at the N-terminal region and a WD40 repeat domain at the C-terminal region (Park et al., 2018a; Shen et al., 2020). COP1 is composed of RING, coiled-coil, and WD40-repeat domains, and it directly binds to the F-box and KELCH domain of FKF1 through the RING domain (Lee et al., 2017). Therefore, HOS15 may directly bind to the RING domain of COP1 through the F-box-like motif. To identify the interacting domain of HOS15 and COP1, full-length and truncated COP1 proteins were transiently expressed in tobacco with full-length HOS15 and analyzed by Co-IP (Supplemental Figure 4). All GFP-tagged protein bands shifted up by about 15 kDa after immunoprecipitation with GFP antibody. This could be explained by high-affinity molecules such as the light chain of GFP antibody (25 kDa) that strongly bind to GFP under SDS–PAGE conditions. Thus, the N-terminal region of HOS15 containing a LisH domain and an F-box-like motif interacted with the WD40 domain of COP1 (Supplemental Figure 4).
Figure 5.
HOS15 forms a complex with COP1 in vivo.
(A) BiFC assay for interaction between HOS15 and GI or COP1. The Venus fluorescent protein N terminus (VN) alone or fused to HOS15 (VN-HOS15) and the Venus fluorescent protein C terminus (VC) alone or fused to GI (GI-VC) and COP1 (VC-COP1) were transiently expressed in tobacco leaves, and the fluorescence was observed by confocal microscopy with an excitation wavelength of 515 nm for YFP. Scale bar corresponds to 50 μm.
(B) BiFC signal analysis with DAPI staining.
(C and D) Co-immunoprecipitation assay for HOS15 and GI interaction (C) and COP1 and HOS15 interaction (D). Transgenic plants overexpressing GI-HA(C) or COP1-GFP(D) were grown in 12D/12 L at 23°C for 12 days and harvested at the indicated ZT time points. Total protein was extracted and immunoprecipitated with α-HOS15 and α-GFP, respectively.
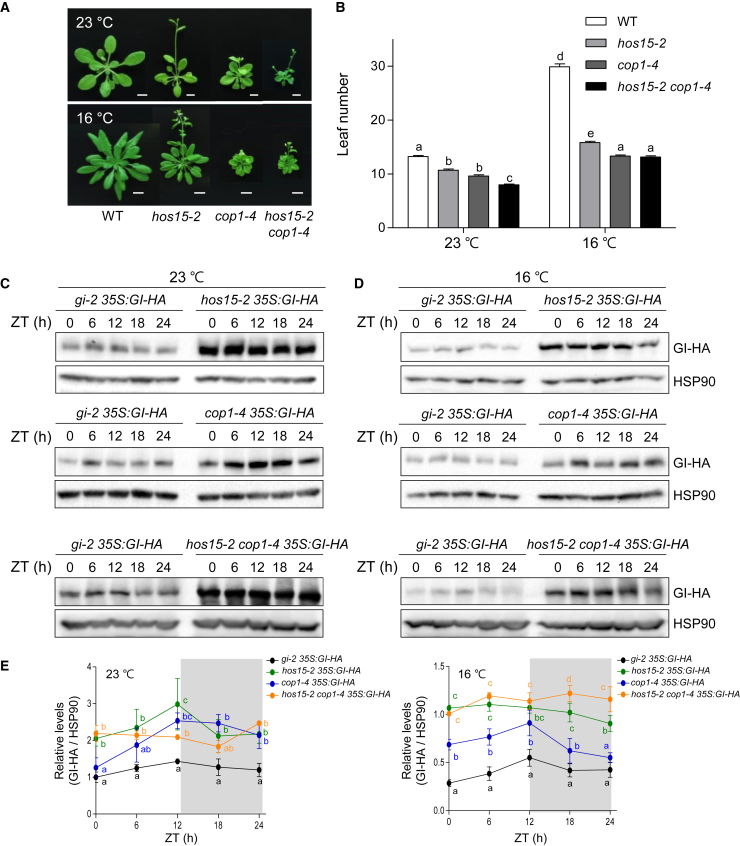
(E) COP1 and HOS15 interaction at low ambient temperature. Ten-day-old 35S:COP1-GFP overexpressing plants were transferred to 16°C for an additional 2 days and harvested at the indicated ZT time points. Total protein extracts were immunoprecipitated with α-GFP.
HOS15 repression of flowering depends on COP1 at low ambient temperature, but HOS15 works independently of COP1 in GI turnover
To confirm the genetic interaction between HOS15 and COP1 in low ambient temperature–mediated flowering, we measured the flowering times of hos15-2, cop1-4, and the hos15-2 cop1-4 double mutant under LD conditions at 23°C and 16°C. The cop1-4 mutants flowered earlier than hos15-2 at 23°C and 16°C. The hos15-2 cop1-4 double mutants exhibited an additive phenotype at 23°C and a phenotype similar to cop1-4 at 16°C (Figure 6A and 6B), indicating that HOS15 regulation of flowering time is independent of COP1 at 23°C but dependent on COP1 at 16°C. To gain insight into the relationship between HOS15 and COP1 in GI turnover, we compared GI protein levels in hos15-2, cop1-4, and hos15-2 cop1-4 mutants at 23°C and 16°C. HOS15 and COP1 did not affect each other’s transcript and protein levels in response to temperature (Supplemental Figures 1B, 2A, and 2B). At 23°C, GI-HA protein levels were higher in hos15-2 cop1-4 mutants than in GI-HA control plants and similar to those of hos15-2 and cop1-4 (Figure 6C and 6E). At 16°C, GI protein levels were higher in hos15-2 plants than in cop1-4. Furthermore, GI protein levels in the hos15-2 cop1-4 double mutant exhibited an additive increase at 16°C compared with hos15-2 and cop1-4 at ZT18 and ZT24 (Figure 6D and 6E), indicating that HOS15-mediated GI turnover is independent of COP1 at low ambient temperature. To further investigate the relationship between HOS15 and COP1 in GI degradation, we measured GI protein levels in the hos15-2 cop1-4 double mutant treated with cycloheximide under continuous light conditions at 23°C or 16°C (Supplemental Figure 5). GI degradation patterns of the hos15-2 cop1-4 double mutant at 23°C and 16°C were similar to those of hos15-2, and there were no statistically significant differences in GI degradation between the hos15-2 cop1-4 double mutant and each single mutant (Supplemental Figure 6). Taken together, these results show that HOS15 plays a COP1-dependent role in low ambient temperature–mediated flowering but participates in GI turnover independently of COP1 (Figure 7). Our results indicate that the HOS15-based 26S proteasome plays a role in the control of thermosensory flowering by regulating the protein abundance of GI, a photoperiodic floral activator.
Figure 6.
HOS15 is genetically dependent on COP1 in low ambient temperature–mediated flowering but independent of COP1 in low ambient temperature-induced GI turnover.
(A) Flowering phenotypes of WT, hos15-2, cop1-4, and hos15-2 cop1-4 mutants grown under LDs (16L/8D) at 23°C or 16°C. The photographs were taken when hos15-2 bolted. Scale bars correspond to 1 cm.
(B) The numbers of rosette and cauline leaves were counted at bolting to measure flowering time. Data are presented as means ± SD of three independent replicates, each including at least 15 plants. Different letters denote statistically significant differences (ANOVA, Bonferroni post-hoc test; P < 0.05).
(C and D) GI protein levels in hos15-2, cop1-4, and hos15-2 cop1-4 plants at 23°C (C) or 16°C (D). Plants were grown for 10 days under 12L/12D conditions at 23°C and then transferred to 16°C for an additional 2 days.
(E) Quantification of GI protein levels in hos15-2 35S:GI-HA, cop1-4 35S:GI-HA, and hos15-2 cop1-4 35S:GI-HA. The levels of GI-HA protein were normalized to HSP90 and statistically analyzed. Data are presented as means ± SD of three independent replicates. Different letters denote statistically significant differences (ANOVA, Bonferroni post-hoc test; P < 0.05).
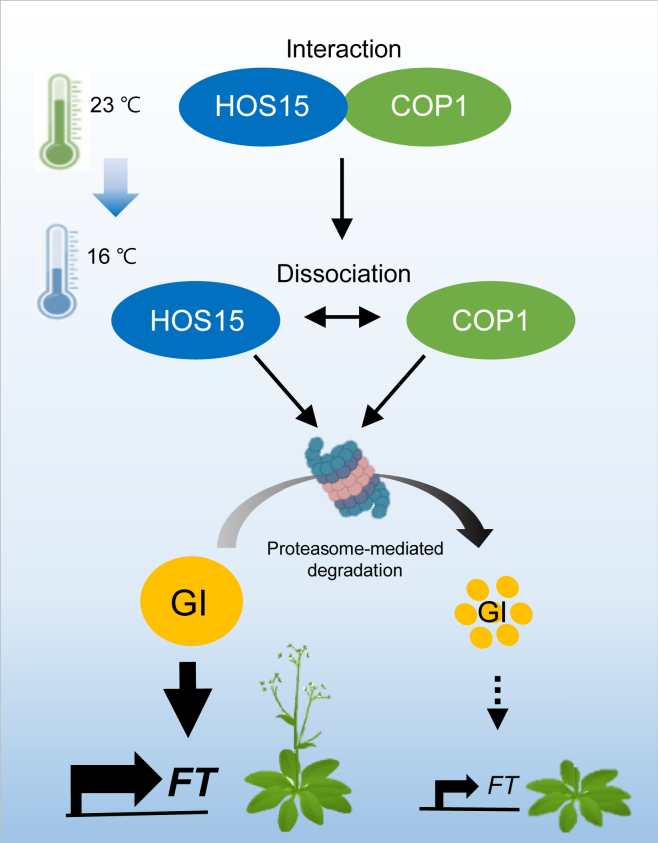
Figure 7.
Model of post-translational regulation of GI abundance by HOS15 and COP1.
HOS15 represses flowering by promoting low ambient temperature–mediated degradation of GI protein, a floral activator. Low ambient temperature (16°C) promotes the dissociation of HOS15 and COP1, allowing HOS15 and COP1 to play independent roles in low ambient temperature-induced GI turnover. Therefore, FT expression is reduced and flowering is delayed under low ambient temperature.
Discussion
Flowering is regulated by the integration of complex regulatory responses to photoperiod, temperature, hormones, and metabolites (Andrés and Coupland, 2012). In particular, the red/far-red light ratio and daily temperature oscillation are key environmental parameters needed to mimic natural long-day flowering regulation under laboratory growth conditions. Daily temperature fluctuation is required to reduce afternoon FT expression levels and mimic the natural pattern (Song et al., 2018), and low ambient temperature-induced daily flowering repression is important for synchronizing flowering time. In WT plants, GI protein levels were lower at 16°C (Figure 3A), but transcript levels of GI were unchanged (Figure 1C). Therefore, low ambient temperature–mediated GI degradation may be a key mechanism of daily flowering repression.
Previously, we showed that HOS15 forms a complex with the EC and HDA9 to repress transcriptional activation of the GI gene (Park et al., 2019). Plants defective in the HOS15-mediated histone deacetylase complex exhibit various developmental phenotypes, including early flowering, small plant size, and cold responses (Mizuno et al., 2014; Jang et al., 2015; Park et al., 2018a, 2019). For example, hos15-2, hda9-1, pwr, elf3-1, and pcl1-1/lux mutants flower earlier than the WT (Park et al., 2019). The transcript levels of the cold-responsive genes CBFs and DREB are upregulated in hos15-2, lux, and elf3 mutants upon cold exposure (Keily et al., 2013; Ezer et al., 2017; Park et al., 2018a). Mutants of the EC components ELF3 and LUX, and mutants involving target genes such as PSEUDO-RESPONSE REGULATOR 7 (PRR7), PSEUDO-RESPONSE REGULATOR 9 (PRR9), PHYTOCHROME INTERACTING FACTOR 4 (PIF4), and PIF5 (prr7, prr9, pif4, and pif5) are insensitive to ambient temperature (Mizuno et al., 2014; Ezer et al., 2017). Based on these results, we hypothesized that HOS15 might be involved in thermosensory flowering. Our data showed that the hos15-2 mutant was insensitive to low ambient temperature–mediated repression of flowering and that HOS15 acted upstream of the photoperiodic flowering genes at low ambient temperature (Figures 1 and 2).
HOS15 acts as a substrate receptor of the CUL4-based E3 ubiquitin ligase complex as well as the SCF E3 ubiquitin ligase complex (Park et al., 2018a; Shen et al., 2020). The CUL4 system is assembled with its linker protein Damage-specific DNA binding protein 1 (DDB1) and a WD40-containing substrate receptor (Biedermann and Hellmann, 2011; Iovine et al., 2011). DDB1 directly binds to the substrate receptor, which is known as DDB1-BINDING WD40 (DWD) protein. HOS15 has a DWD motif and interacts with DDB1 for protein degradation by associating with the CUL4-based E3 ligase complex (Lee et al., 2008; Park et al., 2018a). COP1 and SPAs (SUPPRESSOR OF PHYA-105) were identified as DDB1-CUL4-associated factors (DCAFs) because mutations in their WDxR motif impair protein binding to DDB1 (Chen et al., 2010; Fonseca and Rubio, 2019). Structural studies using X-ray crystallography have shown that the WDxR motif of Arabidopsis COP1 is buried and cannot be accessed without a break involving the β propeller domain, unlike the human COP1 protein (Uljon et al., 2016). In addition, the WD domain of COP1 is not sufficient for COP1/SPA activity (Kerner et al., 2021). This may be supported by other domains such as the helix-loop-helix domain for interactions with DDB1 (Fischer et al., 2011; Uljon et al., 2016). For example, COP1 could recruit another WD40 protein into the complex, such as SPA1, which adds complex stability and serves as a substrate receptor. (Bosu and Kipreos, 2008; Hua and Vierstra, 2011; Errington et al., 2012; Yasukawa et al., 2020; Kerner et al., 2021). The Arabidopsis COP1/SPA1 complex forms via its coiled-coil domains in a light-dependent manner and cooperates to suppress photomorphogenesis (Hoecker and Quail, 2001; Saijo et al., 2003; Zhu et al., 2008a, 2008b). Unlike the COP1/SPA1 complex, HOS15 bound to the WD40 domain of COP1 (Figure 6A).
The interaction between HOS15 and COP1 was attenuated at 16°C compared with 23°C (Figure 5E), suggesting that HOS15 and COP1 work together at 23°C but that their functions separate at 16°C. GI protein levels were lower in cop1-4 than in the WT and hos15-2 under continuous light at 23°C, but not at 16°C (Supplemental Figure 6). This phenomenon was recovered in the hos15-2 cop1-4 double mutant, suggesting that COP1 contributes to GI protein stabilization in a HOS15-dependent manner under light conditions at 23°C. Therefore, it is possible that COP1 can work together with HOS15 in GI stabilization at 23°C under light conditions and dissociate at 16°C for an independent function. In addition, the functions of COP1 are regulated by its subcellular partitioning between the nucleus and cytosol (Stacey et al., 1999). The subcellular behavior of COP1 proteins may therefore be closely related to the HOS15-COP1 interaction. For example, warm temperature (28°C) triggers the nuclear accumulation of COP1 for the regulation of thermomorphogenesis (Park et al., 2017; Blanco-Touriñán et al., 2020). Thus, the subcellular localization of COP1 is closely related to its function in response to temperature. To investigate the dynamic behaviors of COP1 in response to low ambient temperature, we observed the subcellular localization of COP1-GFP at 23°C and 16°C. COP1-GFP formed condensates in the nucleus and cytosol at 23°C, but the cytosolic condensates of COP1 disappeared in response to low ambient temperature (Supplemental Figure 7). Protein solubility affects condensate formation (Riback and Brangwynne, 2020). Therefore, COP1 protein may be more soluble at low temperature, allowing COP1-containing condensates to disperse and COP1 to form other protein complexes. Recently, the formation of biomolecular condensates was characterized as a plant thermosensory mechanism (Hahm et al., 2020; Jung et al., 2020). Therefore, it is tempting to speculate that condensate formation may affect the interaction between COP1 and HOS15 for temperature responses.
GI is degraded by COP1 in response to both dark and low ambient temperature (Yu et al., 2008; Jang et al., 2015; Figure 4). However, HOS15-mediated GI turnover responds to low ambient temperature but not to darkness (Figure 4). These findings suggest that HOS15-mediated proteolysis is involved in regulating flowering in response to temperature, but not photoperiod. In Arabidopsis, FLOWERING LOCUS M (FLM) is a crucial factor in the regulation of thermosensory flowering. It is alternatively spliced in response to temperature as two predominant splice variants: FLM-β (floral repressor) and FLM-δ (floral activator) (Lutz et al., 2015). The relative abundance of FLM-β and FLM-δ changes in response to ambient temperature, and FLM-β and FLM-δ compete with each other for protein interaction with SHORT VEGETATIVE PHASE (SVP), a floral repressor (Posé et al., 2013). The transcript expression of FLM-δ was less sensitive to temperature changes than that of FLM-β in 13 ambient temperature–sensitive and –insensitive wild accessions (Lee et al., 2013). Thus, an additional mechanism for downregulation of FLM-δ protein levels may be required for rapid activation of flowering repression mediated by the SVP–FLM-β complex. Therefore, FLM-δ may be a candidate for a direct target of HOS15-mediated degradation in response to low ambient temperature.
GI protein levels were decreased in cop1-4 compared with the WT and hos15-2 under continuous light conditions at 23°C (Figure 4 and Supplemental Figure 6A). There are several potential explanations for this phenomenon. One possibility is that COP1 stabilizes GI under light conditions at 23°C. For example, the COP1/SPA1 complex stabilizes PIF3 by preventing its phosphorylation and degradation, which are controlled by BIN2 (Ling et al., 2017). Thus, COP1 may stabilize GI by interfering with an interaction between GI and factors that cause its destabilization or degradation. The decreased protein levels of GI in cop1-4 were recovered in the hos15-2 cop1-4 double mutant (Supplemental Figure 6), indicating that COP1 stabilizes GI in an HOS15-dependent manner at 23°C. Taken together, these results suggest that COP1 may have a role in the inhibition of HOS15-mediated GI destabilization at 23°C.
How temperature is sensed is a very intriguing question in plant biology, especially how ambient temperature is merged into photoperiodic flowering time or plant morphogenesis pathways from the perspective of plant growth and development. The first aspect of this question is how plants sense ambient temperature: a few temperature sensors have been characterized, including photosensors, phytochrome B and phototropin, the circadian EC component ELF3, and an RNA switch (Hayes et al., 2021). Even though COP1 is involved in thermosensory flowering (Yu et al., 2008; Jang et al., 2015) and hypocotyl thermomorphogenesis (Park et al., 2017; Nieto et al., 2020), there is also evidence that ELF3, an interacting partner of COP1, functions as a thermosensory regulator (Zhang et al., 2021). In particular, HOS15 interacts with ELF3 and its EC components, LUX and ELF4, and the HOS15–EC–HDA complex appears to repress transcription of GI in the evening (Park et al., 2019). This suggests that COP1 and HOS15 are involved in degradation of GI protein, and ELF3 might provide complex stability and ambient temperature sensing in the HOS15–COP1–ELF3 complex. Epigenetic mechanisms have also been implicated in the transcriptional memory of environmental stresses such as heat (Bäurle and Trindade, 2020). HOS15, a component of the histone deacetylase complex in the HOS15–COP1–ELF3 complex, might therefore contribute to another epigenetic regulatory layer of thermomorphogenesis.
The list of putative HOS15-interacting proteins included components of proteasomal degradation, mRNA processing, nuclear organization (genomic integrity), chromatin remodeling, the cell cycle (proliferation), DNA repair, and floral development. These interactors can be identified by affinity chromatography followed by mass spectrometry to isolate putative targets of HOS15 and study the molecular function of HOS15 (Park et al., 2018b; Mayer et al., 2019). In particular, the HOS15-mediated chromatin remodeling complex is associated with a complex composed of ATROPOS (ATO) or arginine/serine-rich splicing factor 35 (RSP35), which regulates pre-spliceosome formation and mRNA splicing (Reddy, 2004; Tanny, 2014; Zhou et al., 2014; Chen et al., 2015; Park et al., 2018b). Recently, plant COP1 was shown to participate in microRNA biogenesis because levels of multiple miRNAs are reduced in light-grown cop1 mutants with lower protein levels of HYPONASTIC LEAVES1 (HYL1), a miRNA-processing factor (Cho et al., 2014; Achkar et al., 2018). Thus, HOS15 and COP1 may be closely related to various modes of action for the regulation of plant development.
Here, we showed that the WD40 DCAF HOS15 could form a complex with another WD40 DCAF, COP1, and collaboratively regulate the protein abundance of GI, a key regulator of photoperiodic flowering. Thus, HOS15 determines flowering time via multiple modes, not only as a transcriptional repressor in the photoperiodic pathway but also as an E3 ubiquitin ligase in the thermosensory pathway.
Methods
Plant materials and growth conditions
Mutants and transgenic plants used in this study are included in Supplemental Table 1. All of the mutants used in this study were in the Arabidopsis thaliana Columbia (Col) background. The ft-1 (Col) allele, which is originally from the Landsberg erecta ecotype, was introgressed seven times into the Col background (Yoo et al., 2005). Plants were grown in a growth room (16 h light/8 h dark [16L/8D] cycles, 100 μmol photons m−2 s−1 of white cool fluorescent light) at 23°C or 16°C to measure flowering time or to check the expression of floral regulators by quantitative real-time PCR. The total number of rosette and cauline leaves were counted at bolting (about 3 cm) as an indicator of flowering time. The 10-day-old seedlings grown at 23°C under 12L/12D were transferred to 16°C for 2 days to check the abundance of proteins by western blotting or to test the interaction between proteins by Co-IP as indicated in the figure legends. For MG132 treatment, 10-day-old plants (35S:GI-HA, hos15-2 35S:GI-HA, and cop1-4 35S:GI-HA) grown at 23°C under 12L/12D cycles were transferred to 16°C for 2 days. Plants were vacuum-infiltrated in liquid medium containing MG132 (50 μM) or DMSO at ZT0 at greater than −0.1 MPa until the liquid was at a good rolling boil for 3 min; the vacuum was released gently and the plants removed at the indicated time points. To observe protein turnover, 10-day-old plants (35S:GI-HA, hos15-2 35S:GI-HA, and cop1-4 35S:GI-HA) grown at 23°C under 12L/12D cycles were vacuum-infiltrated in liquid medium containing cycloheximide (100 μM) or DMSO at ZT12 and transferred to various conditions.
Quantitative real-time PCR analysis
Total RNA was isolated from 12-day-old seedlings using a Plant RNeasy kit (QIAGEN, Hilden, Germany), and first-strand cDNA was synthesized using a cDNA synthesis kit (RevertAid First Strand cDNA synthesis kit, Thermo Scientific) according to the manufacturer’s instructions. To measure transcript levels of genes, quantitative PCR was performed using the QuantiMix SYBR kit (PKT, Philekorea Technology, Seoul, Korea) and a CFX384 Real-time PCR Detection System and CFX Manager software (Bio-Rad, Hercules, CA). The program was 50°C for 10 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s. The primers used for quantitative real-time PCR analysis are listed in Supplemental Table 2.
Immunoblotting and Co-IP assay
Total proteins from the Arabidopsis plants indicated in the figure legends were extracted in extraction buffer (100 mM Tris–HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, 3 mM DTT, and protease inhibitors) and incubated with α-HA cross-linked to protein A Agarose (Invitrogen), α-GFP cross-linked to protein A Agarose (Invitrogen), or protein A agarose fused to α-HOS15 (Lee et al., 2012b; Park et al., 2018a) or α-COP1 (Lee et al., 2017) at 4°C for 2 h. After SDS–PAGE electrophoresis, the proteins were transferred onto a polyvinylidene fluoride membrane (Immobilon-P, Millipore). Immunoblotting was performed using α-HA (Roche), α-GFP (Abcam), α-HOS15, and α-COP1 antibodies (1:1000 dilution). The antigen protein was detected by chemiluminescence using an ECL detection reagent (Thermo Fisher Scientific). Experimental data were statistically analyzed with GraphPad Prism (GraphPad Software, San Diego, CA).
Plasmid construction
Fragmented HOS15 (aa 1–243 and 244–613) and COP1 (aa 1–104, 121–213, 371–675, and 121–675) were amplified from full-length cDNA of HOS15 and COP1 by PCR. These PCR products were cloned into the pPCR8/GW/TOPO vector (Invitrogen). HOS15 fragment constructs were subcloned into a pEarleyGate 201 binary vector, and COP1 fragment constructs were subcloned into a pMDC43 binary vector using LR clonase II (Invitrogen).
BiFC assay and imaging
HOS15 was fused in-frame to Venus aa 1–173, the N-terminal fragment of the Venus fluorescent protein. GI and COP1 were fused in-frame to Venus aa 156–239, the C-terminal fragment of Venus (Supplemental Table 3) (Gehl et al., 2009). For co-expression, Agrobacterium tumefaciens (GV 3101) cells harboring the construct for each protein were infiltrated into the leaves of 3-week-old Nicotiana benthamiana plants and grown for 2 days (Sparkes et al., 2006). Fluorescence was detected via confocal laser-scanning microscopy (Olympus FV3000, Olympus, Shinjuku, Tokyo, Japan) under an excitation wavelength of 515 nm for YFP.
Funding
This research was supported by National Research Foundation of Korea (NRF) grants funded by the Korean Government (MSIT-2022R1A5A1031361 and MSIT-2020R1A2C3014814 to W.-Y.K.) and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2021R1I1A1A01059532 to G.A. and NRF-2019R1I1A1A01041422 to H.J.P.).
Author contributions
G.A., H.J.P., S.Y.J., and W.-Y.K. designed the research. G.A., H.J.P., S.Y.J., G.-I.S., M.G.J., and J.-Y.C. conducted the experiments. G.A., H.J.P., S.Y.J., J.-Y.C., J.K., M.G.K., D.-J.Y., and W.-Y.K. analyzed data or wrote the paper.
Acknowledgments
We thank the Plant Biological Rhythm Research Center (PBRRC) and BK21 four program for financial assistance. No conflict of interest declared.
Published: March 2, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Achkar N.P., Cho S.K., Poulsen C., Arce A.L., Re D.A., Giudicatti A.J., Karayekov E., Ryu M.Y., Choi S.W., Harholt J., et al. A quick HYL1-dependent reactivation of MicroRNA production is required for a proper developmental response after extended periods of light deprivation. Dev. Cell. 2018;46:236–247.e6. doi: 10.1016/j.devcel.2018.06.014. [DOI] [PubMed] [Google Scholar]
- Andrés F., Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Bäurle I., Trindade I. Chromatin regulation of somatic abiotic stress memory. J. Exp. Bot. 2020;71:5269–5279. doi: 10.1093/jxb/eraa098. [DOI] [PubMed] [Google Scholar]
- Biedermann S., Hellmann H. WD40 and CUL4-based E3 ligases: lubricating all aspects of life. Trends Plant Sci. 2011;16:38–46. doi: 10.1016/j.tplants.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Blanco-Touriñán N., Legris M., Minguet E.G., Costigliolo-Rojas C., Nohales M.A., Iniesto E., García-Leόn M., Pacín M., Heucken N., Blomeier T., et al. COP1 destabilizes DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2020;117:13792–13799. doi: 10.1073/pnas.1907969117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez M.A., Ahn J.H., Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 2003;33:168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- Bosu D.R., Kipreos E.T. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Huang X., Gusmaroli G., Terzaghi W., Lau O.S., Yanagawa Y., Zhang Y., Li J., Lee J.-H., Zhu D., et al. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22:108–123. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Cui P., Xiong L. The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2015;43:8283–8298. doi: 10.1093/nar/gkv751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.K., Ben Chaabane S., Shah P., Poulsen C.P., Yang S.W. COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat. Commun. 2014;5:5867. doi: 10.1038/ncomms6867. [DOI] [PubMed] [Google Scholar]
- David K.M., Armbruster U., Tama N., Putterill J. Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett. 2006;580:1193–1197. doi: 10.1016/j.febslet.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Errington W.J., Khan M.Q., Bueler S.A., Rubinstein J.L., Chakrabartty A., Privé G.G. Adaptor protein self-assembly drives the control of a cullin-RING ubiquitin ligase. Structure. 2012;20:1141–1153. doi: 10.1016/j.str.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Ezer D., Jung J.H., Lan H., Biswas S., Gregoire L., Box M.S., Charoensawan V., Cortijo S., Lai X., Stöckle D., et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants. 2017;3:17087. doi: 10.1038/nplants.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández V., Takahashi Y., Le Gourrierec J., Coupland G. Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J. 2016;86:426–440. doi: 10.1111/tpj.13183. [DOI] [PubMed] [Google Scholar]
- Fischer E.S., Scrima A., Böhm K., Matsumoto S., Lingaraju G.M., Faty M., Yasuda T., Cavadini S., Wakasugi M., Hanaoka F., et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- Fonseca S., Rubio V. Arabidopsis CRL4 complexes: surveying chromatin states and gene expression. Front. Plant Sci. 2019;10:1095. doi: 10.3389/fpls.2019.01095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K.C.S., Gissot L., Sauerbrunn N., Rühl M., Jarillo J.A., Coupland G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Morris B., Coupland G., Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C., Waadt R., Kudla J., Mendel R.-R., Hänsch R. New GATEWAY vectors for high throughput analyses of protein–protein interactions by bimolecular fluorescence complementation. Mol. Plant. 2009;2:1051–1058. doi: 10.1093/mp/ssp040. [DOI] [PubMed] [Google Scholar]
- Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA. 2012;109:3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm J., Kim K., Qiu Y., Chen M. Increasing ambient temperature progressively disassembles Arabidopsis phytochrome B from individual photobodies with distinct thermostabilities. Nat. Commun. 2020;11:1660. doi: 10.1038/s41467-020-15526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S., Schachtschabel J., Mishkind M., Munnik T., Arisz S.A. Hot topic: thermosensing in plants. Plant Cell Environ. 2021;44:2018–2033. doi: 10.1111/pce.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U., Quail P.H. The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 2001;276:38173–38178. doi: 10.1074/jbc.M103140200. [DOI] [PubMed] [Google Scholar]
- Hua Z., Vierstra R.D. The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- Huq E., Tepperman J.M., Quail P.H. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2000;97:9789–9794. doi: 10.1073/pnas.170283997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. FKF1 F-Box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Iovine B., Iannella M.L., Bevilacqua M.A. Damage-specific DNA binding protein 1 (DDB1): a protein with a wide range of functions. Int. J. Biochem. Cell Biol. 2011;43:1664–1667. doi: 10.1016/j.biocel.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Jang K., Lee H.G., Jung S.-J., Paek N.-C., Seo P.J. The E3 ubiquitin ligase COP1 regulates thermosensory flowering by triggering GI degradation in Arabidopsis. Sci. Rep. 2015;5:12071. doi: 10.1038/srep12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Müller A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009;14:563–573. doi: 10.1016/j.tplants.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Barbosa A.D., Hutin S., Kumita J.R., Gao M., Derwort D., Silva C.S., Lai X., Pierre E., Geng F., et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature. 2020;585:256–260. doi: 10.1038/s41586-020-2644-7. [DOI] [PubMed] [Google Scholar]
- Keily J., MacGregor D.R., Smith R.W., Millar A.J., Halliday K.J., Penfield S. Model selection reveals control of cold signalling by evening-phased components of the plant circadian clock. Plant J. 2013;76:247–257. doi: 10.1111/tpj.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner K., Nagano S., Lübbe A., Hoecker U. Functional comparison of the WD-repeat domains of SPA1 and COP1 in suppression of photomorphogenesis. Plant Cell Environ. 2021;44:3273–3282. doi: 10.1111/pce.14148. [DOI] [PubMed] [Google Scholar]
- Kim D.-H., Doyle M.R., Sung S., Amasino R.M. Vernalization: winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- Kim W.-Y., Fujiwara S., Suh S.-S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H.G., Somers D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- Kim Y., Han S., Yeom M., Kim H., Lim J., Cha J.-Y., Kim W.-Y., Somers D.E., Putterill J., Nam H.G., et al. Balanced nucleocytosolic partitioning defines a spatial network to coordinate circadian physiology in plants. Dev. Cell. 2013;26:73–85. doi: 10.1016/j.devcel.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C.J., van der Veen J.H. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Lee B.-D., Kim M.R., Kang M.-Y., Cha J.-Y., Han S.-H., Nawkar G.M., Sakuraba Y., Lee S.Y., Imaizumi T., McClung C.R., et al. The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nat. Commun. 2017;8:2259. doi: 10.1038/s41467-017-02476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Kim J.J., Kim S.H., Cho H.J., Kim J., Ahn J.H. The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1802–1814. doi: 10.1093/pcp/pcs123. [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Terzaghi W., Gusmaroli G., Charron J.-B.F., Yoon H.-J., Chen H., He Y.J., Xiong Y., Deng X.W. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell. 2008;20:152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Park H.C., Bae S., Hong J., Choi J., Hong K., Jhun H., Kim K., Kim E., Jo S., et al. Monoclonal antibodies against recombinant AtHOS15. Hybridoma. 2012;31:118–124. doi: 10.1089/hyb.2011.0096. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Ryu H.S., Chung K.S., et al. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science. 2013;342:628–632. doi: 10.1126/science.1241097. [DOI] [PubMed] [Google Scholar]
- Ling J.J., Li J., Zhu D., Deng X.W. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. USA. 2017;114:3539–3544. doi: 10.1073/pnas.1700850114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Webb C.J., Knowles S.M., Kim S.H.J., Wang Z., Tobin E.M. CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012;158:1079–1088. doi: 10.1104/pp.111.189670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz U., Posé D., Pfeifer M., et al. Modulation of ambient temperature-dependent flowering in Arabidopsis thaliana by natural variation of FLOWERING LOCUS M. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K.S., Chen X., Sanders D., Chen J., Jiang J., Nguyen P., Scalf M., Smith L.M., Zhong X. HDA9-PWR-HOS15 is a core histone deacetylase complex regulating transcription and development. Plant Physiol. 2019;180:342–355. doi: 10.1104/pp.18.01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Panigrahi K.C. GIGANTEA-an emerging story. Front. Plant Sci. 2015;6:8. doi: 10.3389/fpls.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Nomoto Y., Oka H., Kitayama M., Takeuchi A., Tsubouchi M., Yamashino T. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:958–976. doi: 10.1093/pcp/pcu030. [DOI] [PubMed] [Google Scholar]
- Mohamed W.I., Schenk A.D., Kempf G., Cavadini S., Basters A., Potenza A., Abdul Rahman W., Rabl J., Reichermeier K., Thomä N.H. The CRL4DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state. EMBO J. 2021;40:e108008. doi: 10.15252/embj.2021108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kita M., Niinuma K., Ito S., Yamashino T., Mizoguchi T., Mizuno T. Arabidopsis clock-associated Pseudo-Response Regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007;48:822–832. doi: 10.1093/pcp/pcm056. [DOI] [PubMed] [Google Scholar]
- Nieto C., Luengo L.M., Prat S. Regulation of COP1 function by Brassinosteroid signaling. Front. Plant Sci. 2020;11:1151. doi: 10.3389/fpls.2020.01151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacín M., Legris M., Casal J.J. COP1 re-accumulates in the nucleus under shade. Plant J. 2013;75:631–641. doi: 10.1111/tpj.12226. [DOI] [PubMed] [Google Scholar]
- Park D.H., Somers D.E., Kim Y.S., Choy Y.H., Lim H.K., Soh M.S., Kim H.J., Kay S.A., Nam H.G. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- Park H.J., Baek D., Cha J.-Y., Liao X., Kang S.-H., McClung C.R., Lee S.Y., Yun D.-J., Kim W.-Y. HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell. 2019;31:37–51. doi: 10.1105/tpc.18.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lim C.J., Shen M., Park H.J., Cha J.Y., Iniesto E., Rubio V., Mengiste T., Zhu J.K., Bressan R.A., et al. Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. USA. 2018;115:E5400–E5409. doi: 10.1073/pnas.1721241115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lim C.J., Khan I.U., Jan M., Khan H.A., Park H.J., Guo Y., Yun D.-J. Identification and molecular characterization of HOS15-interacting proteins in Arabidopsis thaliana. J. Plant Biol. 2018;61:336–345. [Google Scholar]
- Park Y.J., Lee H.J., Ha J.H., Kim J.Y., Park C.M. COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol. 2017;215:269–280. doi: 10.1111/nph.14581. [DOI] [PubMed] [Google Scholar]
- Pokhilko A., Fernández A.P., Edwards K.D., Southern M.M., Halliday K.J., Millar A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012;8:574. doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D., Verhage L., Ott F., Yant L., Mathieu J., Angenent G.C., Immink R.G.H., Schmid M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature. 2013;503:414–417. doi: 10.1038/nature12633. [DOI] [PubMed] [Google Scholar]
- Reddy A.S.N. Plant serine/arginine-rich proteins and their role in pre-mRNA splicing. Trends Plant Sci. 2004;9:541–547. doi: 10.1016/j.tplants.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Riback J.A., Brangwynne C.P. Can phase separation buffer cellular noise? Science. 2020;367:364–365. doi: 10.1126/science.aba0446. [DOI] [PubMed] [Google Scholar]
- Saijo Y., Sullivan J.A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003;17:2642–2647. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Lim C.J., Park J., Kim J.E., Baek D., Nam J., Lee S.Y., Pardo J.M., Kim W.Y., Mackey D., et al. HOS15 is a transcriptional corepressor of NPR1-mediated gene activation of plant immunity. Proc. Natl. Acad. Sci. USA. 2020;117:30805–30815. doi: 10.1073/pnas.2016049117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J.S., Kubota A., Imaizumi T. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 2017;173:5–15. doi: 10.1104/pp.16.01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Shim J.S., Kinmonth-Schultz H.A., Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015;66:441–464. doi: 10.1146/annurev-arplant-043014-115555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H. The effect of fluctuations in photoperiod and ambient temperature on the timing of flowering: time to move on natural environmental conditions. Mol. Cell. 2016;39:715–721. doi: 10.14348/molcells.2016.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Kubota A., Kwon M.S., Covington M.F., Lee N., Taagen E.R., Laboy Cintrón D., Hwang D.Y., Akiyama R., Hodge S.K., et al. Molecular basis of flowering under natural long-day conditions in Arabidopsis. Nat. Plants. 2018;4:824–835. doi: 10.1038/s41477-018-0253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- Stacey M.G., Hicks S.N., von Arnim A.G. Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell. 1999;11:349–364. doi: 10.1105/tpc.11.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Wang S., Zhang F., Zheng H., Liu Y., Huang T., Ding Y. Phosphorylation of histone H2A at serine 95: a plant-specific mark involved in flowering time regulation and H2A.Z deposition. Plant Cell. 2017;29:2197–2213. doi: 10.1105/tpc.17.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J.C. Chromatin modification by the RNA Polymerase II elongation complex. Transcription. 2014;5:e988093. doi: 10.4161/21541264.2014.988093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong M., Lee K., Ezer D., Cortijo S., Jung J., Charoensawan V., Box M.S., Jaeger K.E., Takahashi N., Mas P., et al. The evening complex establishes repressive chromatin domains via H2A.Z deposition. Plant Physiol. 2020;182:612–625. doi: 10.1104/pp.19.00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uljon S., Xu X., Durzynska I., Stein S., Adelmant G., Marto J.A., Pear W.S., Blacklow S.C. Structural basis for substrate selectivity of the E3 ligase COP1. Structure. 2016;24:687–696. doi: 10.1016/j.str.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T., Tsutsui A., Tomomori-Sato C., Sato S., Saraf A., Washburn M.P., Florens L., Terada T., Shimizu K., Conaway R.C., et al. NRBP1-Containing CRL2/CRL4A regulates amyloid β production by targeting BRI2 and BRI3 for degradation. Cell Rep. 2020;30:3478–3491.e6. doi: 10.1016/j.celrep.2020.02.059. [DOI] [PubMed] [Google Scholar]
- Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005;139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.W., Rubio V., Lee N.Y., Bai S., Lee S.Y., Kim S.S., Liu L., Zhang Y., Irigoyen M.L., Sullivan J.A., et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell. 2008;32:617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.L., Shao Y.J., Ding L., Wang M.J., Davis S.J., Liu J.X. XBAT31 regulates thermoresponsive hypocotyl growth through mediating degradation of the thermosensor ELF3 in Arabidopsis. Sci. Adv. 2021;7:eabf4427. doi: 10.1126/sciadv.abf4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-L., Luo G., Wise J.A., Lou H. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res. 2014;42:701–713. doi: 10.1093/nar/gkt875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Maier A., Lee J.H., Laubinger S., Saijo Y., Wang H., Qu L.J., Hoecker U., Deng X.W. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell. 2008;20:2307–2323. doi: 10.1105/tpc.107.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Jeong J.C., Zhu Y., Sokolchik I., Miyazaki S., Zhu J.-K., Hasegawa P.M., Bohnert H.J., Shi H., Yun D.-J., et al. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. USA. 2008;105:4945–4950. doi: 10.1073/pnas.0801029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Bu Q., Xu X., Paik I., Huang X., Hoecker U., Deng X.W., Huq E. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 2015;6:7245. doi: 10.1038/ncomms8245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.