Abstract
Wheat (Triticum aestivum, BBAADD) is an allohexaploid species that originated from two polyploidization events. The progenitors of the A and D subgenomes have been identified as Triticum urartu and Aegilops tauschii, respectively. Current research suggests that Aegilops speltoides is the closest but not the direct ancestor of the B subgenome. However, whether Ae. speltoides has contributed genomically to the wheat B subgenome and which chromosome regions are conserved between Ae. speltoides and the B subgenome remain unclear. Here, we assembled a high-quality reference genome for Ae. speltoides, resequenced 53 accessions from seven species (Aegilops bicornis, Aegilops longissima, Aegilops searsii, Aegilops sharonensis, Ae. speltoides, Aegilops mutica [syn. Amblyopyrum muticum], and Triticum dicoccoides) and revealed their genomic contributions to the wheat B subgenome. Our results showed that centromeric regions were particularly conserved between Aegilops and Triticum and revealed 0.17 Gb of conserved blocks between Ae. speltoides and the B subgenome. We classified five groups of conserved and non-conserved genes between Aegilops and Triticum, revealing their biological characteristics, differentiation in gene expression patterns, and collinear relationships between Ae. speltoides and the wheat B subgenome. We also identified gene families that expanded in Ae. speltoides during its evolution and 789 genes specific to Ae. speltoides. These genes can serve as genetic resources for improvement of adaptability to biotic and abiotic stress. The newly constructed reference genome and large-scale resequencing data for Sitopsis species will provide a valuable genomic resource for wheat genetic improvement and genomic studies.
Key words: Aegilops, Sitopsis, polyploid wheat, B subgenome, conserved blocks
Aegilops speltoides is the closest species to the wheat B subgenome. A high-quality reference genome for Ae. speltoides and 53 resequenced Aegilops and Triticum accessions revealed blocks/genes conserved between Aegilops and the wheat B subgenome, as well as expanded and unique genes in Ae. speltoides. These valuable genomic resources can facilitate wheat genetic improvement.
Introduction
Bread wheat (Triticum aestivum) is the most important staple crop cultivated worldwide and contributes approximately 20% of the calories in the human diet (Shewry and Hey, 2015). T. aestivum is an allohexaploid (BBAADD) that originated from two rounds of recent polyploidization events. A natural interspecific hybridization occurred less than 0.8 million years ago between the two wild diploid species that contributed the A and B subgenomes. A further hybridization with Aegilops tauschii contributed the D subgenome (Tanno and Willcox, 2006; Kilian et al., 2007; Feldman and Levy, 2012). These polyploidizations have improved grain yields and nutrient content, thus enhancing the adaptability of bread wheat in diverse environments. The progenitor of the wheat A subgenome is generally believed to be Triticum urartu, and the D subgenome reportedly derives from goat grass, Ae. tauschii (Dvorak et al., 1993; Salamini et al., 2002; Dubcovsky and Dvorak, 2007; Peng et al., 2011). Although great efforts have been made to determine the origin of the B subgenome, the exact donor species remains unknown. Based on similarities in spikelet morphology and karyotype structure between polyploid wheat and Sitopsis species, the B subgenome is thought to be closely related to the S genomes of the five Sitopsis species (Aegilops bicornis, Aegilops longissima, Aegilops searsii, Aegilops sharonensis, Aegilops speltoides) (Mori et al., 1995; Daud and Gustafson, 1996; Maestra and Naranjo, 1998; Haider, 2013). Molecular phylogenetic evidence suggests that Ae. speltoides could be the progenitor of the B subgenome (Liu et al., 2003; El Baidouri et al., 2017; Zhang et al., 2018).
Recent genomic analyses, however, have cast doubt on whether Ae. speltoides is the direct ancestor of the wheat B subgenome. Analysis of several Sitopsis genome assemblies suggested that the donor of the wheat B subgenome may have been an extinct species that was closely related to Ae. speltoides (Avni et al., 2022; Li et al., 2022). In addition, at the tetraploid level, previous studies have also rejected the polyphyletic origin of the wheat B subgenome (El Baidouri et al., 2017). Nucleotide polymorphisms analyzed with transcriptomic data revealed that the rate of genome evolution has varied during allopolyploid plant evolution (Miki et al., 2019). The genome sizes of different Ae. speltoides accessions (5.13 Gb in Avni et al., 2022; 4.11 Gb in Li et al., 2022) in the two studies varied by more than 20%, suggesting the presence of high genomic diversity across Sitopsis genomes. Owing to the lack of large-scale resequencing data for Sitopsis species in previous studies, it is still unclear which chromosome regions are more conserved between Aegilops and Triticum and how Ae. speltoides may have contributed genetic elements during wheat improvement.
During long-term domestication and breeding, the genetic basis of wheat has continually narrowed (Bellon, 1996; Heal et al., 2004; Zhang et al., 2018). Wheat wild relatives have long been considered potential sources of allelic diversity for defense against biotic and abiotic stresses and improvements in other agronomically important traits (Pour-Aboughadareh et al., 2021). Distant hybridization is an efficient way to introduce novel genes for wheat improvement (Whitford et al., 2013). Wide crosses have been made successfully between wheat and plants from diverse Triticeae genera, including (but not limited to) Aegilops, Agropyron, Dasypyrum, Elymus, Hordeum, Leymus, Psathyrostachys, Secale, and Thinopyrum, and superior agronomic traits have been transferred to wheat (Rasheed et al., 2018). Aegilops is the closest genus to Triticum, and Aegilops species are considered the ancestors of both the D and B wheat subgenomes (Wang et al., 2013). Ae. tauschii is the donor of the D subgenome and can be readily crossed with tetraploid and hexaploid wheat (Luo et al., 2017; Zhao et al., 2017). Moreover, Aegilops species in the Sitopsis section contain many useful traits for disease resistance and abiotic stress tolerance (Anikster et al., 2005; Olivera et al., 2007; Huang et al., 2018; Kishii, 2019). For example, several powdery mildew resistance genes of Sitopsis have been successfully transferred to wheat through hybridization, including Pm12 (Jia et al., 1996), Pm32 (Hsam et al., 2003), and Pm53 (Petersen et al., 2015) from Ae. speltoides, Pm57 from Ae. searsii (Liu et al., 2017), and Pm13 (Cenci et al., 1999) and Pm66 (Li et al., 2020) from Ae. longissima. In addition, Ae. sharonensis provided the stripe rust resistance gene Yr38 (Marais et al., 2006). However, use of superior genetic resources from Sitopsis species depends on distant hybridization or genetic engineering technology, which remains a challenge for wheat genetic improvement. Identifying Sitopsis species-specific genes and functionally diverse genes from the B subgenome would help wheat breeders to evaluate the potential applications of S genome genes and increase detection efficiency in breeding programs.
In this study, we aimed to discover conserved chromosome regions between Sitopsis genomes and the wheat B subgenome, to evaluate the genomic contribution of Ae. speltoides to the B subgenome, and to characterize unique genetic resources in Aegilops for wheat improvement. To this end, we generated a high-quality reference genome of Ae. speltoides and resequenced 53 accessions from seven species: Ae. bicornis, Ae. longissima, Ae. mutica, Ae. searsii, Ae. sharonensis, Ae. speltoides, and Triticum dicoccoides. We revealed that relatively high conservation at the centromere region was common among the Aegilops and Triticum genera, and we discovered 0.17 Gb of conserved regions between Ae. speltoides and the wheat B subgenome. In addition, we detected the genomic contribution of Sitopsis species to the wheat B subgenome by identifying Ae. speltoides-specific genes that have significant sequence diversity and exhibit different expression patterns in Sitopsis species and the B subgenome as potential genetic resources for wheat breeding.
Results
Genome sequencing of Sitopsis species
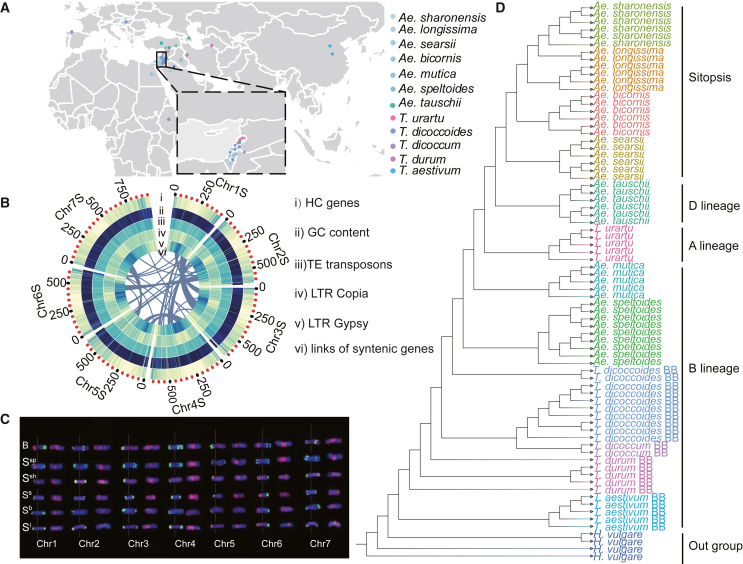
To characterize genomic resources in Aegilops, we de novo assembled the genome of Ae. speltoides using Oxford Nanopore long-read sequencing technology, and we resequenced 53 accessions of germplasm resources collected from the Fertile Crescent. Forty-seven accessions were examined by whole-exome sequencing (average sequencing depth 50× to 184×), and the remaining six accessions were analyzed by whole-genome sequencing (approximately 30×). In addition, we obtained resequencing data for six species from a public database to broaden our analysis (Figure 1A; Supplemental Figure 1; Supplemental Spreadsheet 1). We obtained a total of 429.24 Gb of Nanopore single-cell reads and 646.70 Gb of Illumina clean reads and assembled the reads into 7059 contigs with an N50 of 1.71 Mb, which formed the basis of a total assembly of 4.68 Gb (Supplemental Table 1). The 3D proximity information obtained from Hi-C sequencing data was used to correct instances of mis-joining. Consequently, 96.74% of the assembled sequences were effectively anchored, structured, and oriented into seven pseudochromosomes (Supplemental Figure 2). The final assembled Ae. speltoides genome was 4.88 Gb in length with a scaffold N50 of 640.45 Mb (Supplemental Table 1). The average chromosome length was 674.06 Mb across the whole genome (Supplemental Table 2). The nucleotide composition of the Ae. speltoides genome was 26.68% A, 26.69% T, 23.32% C, and 23.31% G (Supplemental Table 3). BUSCO (Benchmarking Universal Single-Copy Orthologs) analysis revealed that 1440 (94.1%) conserved orthologous genes could be identified in the Ae. speltoides genome.
Figure 1.
Chromosome-level assembly and annotation of Ae. speltoides.
(A) Geographic distribution of the 12 species used in this research: Ae. bicornis, Ae. longissima, Ae. mutica, Ae. searsii, Ae. sharonensis, Ae. speltoides, Ae. tauschii, T. aestivum, T. dicoccoides, T. dicoccum, T. durum, and T. urartu.
(B) Circos plot of genomic features of the assembled Ae. speltoides genome: (i) density of HC genes, (ii) distribution of GC content, (iii) TE density along each chromosome, (iv) density of Copia TEs, (v) density of Gypsy elements, and (vi) links between syntenic genes.
(C) FISH results with the probes (left) Oligo-pSc119.2 (green) + Oligo-pTa71 (red) and the probe (right) Oligo-(GAA)7 (red) for the chromosomes of Ae. speltoides (Ssp), Ae. searsii (Ss), Ae. bicornis (Sb), Ae. longissima (Sl), Ae. sharonensis (Ssh), and the wheat B subgenome (B). The numbers 1–7 represent homoeologous chromosome groups 1–7, respectively.
(D) Phylogenetic tree constructed with data from 13 species using the neighbor-joining method; barley was used as the outgroup species.
We selected 17 different tissues of Ae. speltoides at different developmental stages (Supplemental Spreadsheet 1) and sequenced their whole-genome transcriptome to annotate the protein-coding genes. A total of 46 348 high-confidence protein-coding genes were annotated, with an average length of 3159 bp (Figure 1B; Supplemental Tables 4 and 5). Approximately 0.15% of the genome assembly was annotated as noncoding RNAs, which comprised 55 853 microRNAs (miRNAs), 2234 tRNAs, 480 rRNAs, and 1909 small nuclear RNAs (snRNAs) (Supplemental Table 6). Combined homology-based and de novo predictions were used to identify repetitive sequences. A total of 3.91 Gb, representing 83.56% of the Ae. speltoides genome assembly (without Ns), was annotated as repetitive sequences, which comprised 4.08 million elements from 4689 families (Supplemental Table 7). Among these repetitive elements, long terminal repeats (LTRs), including Gypsy and Copia elements, were among the most abundant, accounting for 44.82% and 22.70%, respectively. These results indicated that LTR-type repetitive sequences may have contributed to genome expansion in Ae. speltoides.
Figure 5.
Sitopsis species provide resources for wheat improvement.
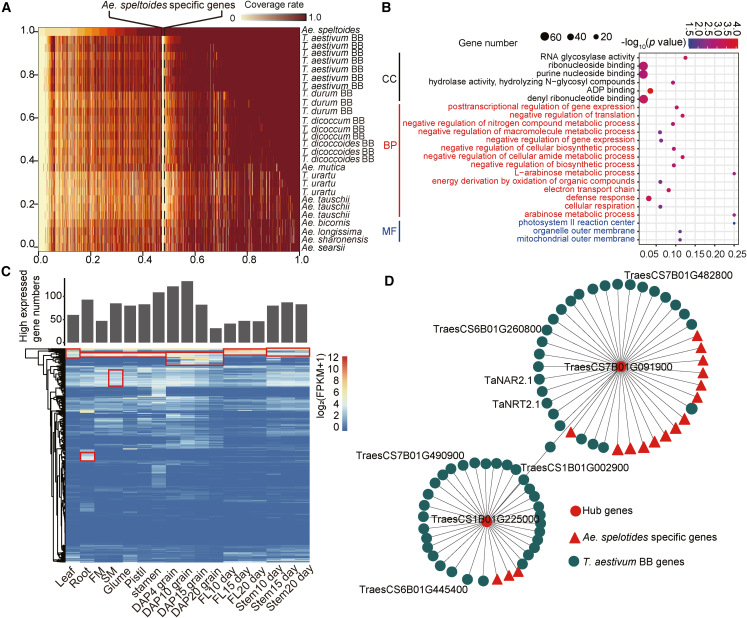
(A) Identification of Ae. speltoides-specific genes. The right column shows the different species, and the horizontal axis represents the gene coverage rate. The region between the black dotted lines indicates the 789 Ae. speltoides-specific genes, and the remaining regions represent shared genes.
(B) GO enrichment analysis of the 789 Ae. speltoides-specific genes. The horizontal axis indicates the enrichment factor values, and the vertical axis represents the GO terms. The circle size represents the gene numbers, and the circle colors indicate the significance of the regulatory pathway, which is indicated by −log10(P value).
(C) Expression levels of the 789 Ae. speltoides-specific genes. The histogram above the heatmap shows the numbers of highly expressed genes, which are marked with red rectangles in the heatmap.
(D) Co-expression networks of wheat TraesCS1B01G225000 and TraesCS7B01G091900; each node represents a gene. Red circles represent hub genes, red triangles represent Ae. speltoides-specific genes, and darkcyan circles represent T. aestivum BB genes.
We also made a detailed comparison between our genome and the other two reported genomes (Avni et al., 2022; Li et al., 2022), including aspects of genome assembly, repetitive sequences, and protein-coding genes (Supplemental Table 8). Total genome size was largest in Avni et al. (2022) (5.13 Gb), whereas our genome size was 4.88 Gb and that of Li et al. (2022) was 4.11 Gb. The average chromosome lengths were 672.85 Mb (current study), 574.28 Mb (Avni et al., 2022), and 587.14 Mb (Li et al., 2022). Avni et al. (2022) had the largest chromosome N50 (3.11 Mb) compared with the current study (1.71 Mb) and Li et al. (2022) (1.78 Mb). There were no obvious differences in repetitive sequence content among the three genomes (3.91 Gb in this study, 4.04 Gb in Avni et al., 2022, and 3.54 Gb in Li et al., 2022). The numbers of high-confidence genes were 46 348 (current study), 36 928 (Avni et al., 2022), and 37 607 (Li et al., 2022). We next performed a collinear gene analysis among the three reference genomes (Supplemental Figure 3); the numbers of collinear genes between genome pairs were 29 168 (current study vs. Li et al., 2022), 27 942 (current study vs. Avni et al., 2022), and 27 914 (Li et al., 2022 vs. Avni et al., 2022). The three genomes were thus highly consistent in most respects, and they differed mainly in total genome size, single chromosome length, and numbers of high-confidence genes. Ae. speltoides is a cross-pollinating species, and the three genomes originated from different geographic environments. The Ae. speltoides used in this study was from Iraq, whereas the others were from Israel, and intraspecific differences among the three genomes may have caused the observed differences. We also discovered that the ChrUn of Avni et al. (2022) was 1.11 Gb and was significantly larger than the other chromosomes, suggesting that different assembly pipelines may also have led to differences.
We used fluorescence in situ hybridization (FISH) to examine chromosome morphological structures between the wheat B subgenome and the five Sitopsis species (Ae. speltoides, Ae. bicornis, Ae. longissima, Ae. searsii, and Ae. sharonensis). Ae. speltoides and wheat displayed a similar FISH pattern of Oligo-pTa71, which was located on the short arms of groups 1S and 6S. Oligo-pTa71 was on groups 5S and 6S in other Aegilops species. The hybridization patterns of Oligo-pSc119.2 of Ae. speltoides were also similar to those of the wheat B subgenome, whereas the Oligo-pSc119.2 signals of the other four Sitopsis species were observed only on both ends of chromosomes. The hybridization patterns of Oligo-(GAA)7 in the five Aegilops species were similar to that of the wheat B subgenome and were distributed throughout the arms, particularly around the pericentromeric regions (Tang et al., 2014) (Figure 1C).
Phylogenetic relationships among Sitopsis species and the wheat B subgenome
We analyzed 13 species to further reveal the phylogenetic relationships between the Aegilops and Triticum genera. First, we constructed a genetic variation map of the 13 species by aligning the resequencing data to the IWGSC RefSeq v1.0 B subgenome. The average mapping percentage of Ae. speltoides accessions was 85.6%, which was higher than that of other Sitopsis species and Ae. mutica (∼80%) but lower than that of hexaploid and tetraploid wheat accessions (>90%) (Supplemental Figure 4A). We obtained 5 626 275 to 27 593 890 high-quality single-nucleotide polymorphism (SNP) markers from whole-exome sequencing samples (Supplemental Figure 4B), and a total of 1 333 497 SNPs were ultimately used to build a phylogenetic tree after quality control. Barley (Hordeum vulgare) was selected as the outgroup species. B-lineage genomes (T. aestivum BB, Triticum durum BB, Triticum dicoccum BB, T. dicoccoides BB, Ae. speltoides, and Ae. mutica) were closest to barley, followed by the A-lineage genome of T. urartu and D-lineage genomes of Ae. tauschii as well as four Sitopsis species (Ae. searsii, Ae. bicornis, Ae. longissima, and Ae. sharonensis) (Figure 1D). These findings are consistent with those of recent studies and showed that Ae. speltoides is most closely related to the wheat B subgenome (Glemin et al., 2019; Miki et al., 2019; Li et al., 2022). In addition, we discovered that Ae. speltoides and Ae. mutica were separate from the rest of the Sitopsis species, which may have implications for their taxonomic positions in the Sitopsis section.
We next calculated the genomic nucleotide diversity (π) and population differentiation (FST) of seven species. Among these species, nucleotide diversity was lowest in the B subgenome of bread wheat (0.37 × 10−3), followed by the genome of Ae. speltoides (1.16 × 10−3). Ae. searsii harbored the highest genomic nucleotide diversity of 1.40 × 10−3 (Supplemental Figure 4C). FST was used to measure population differentiation and genetic distance with reference to the B subgenome of bread wheat. Ae. speltoides had the lowest genomic differentiation relative to the B subgenome with an FST value of 0.234, followed by Ae. mutica at 0.257. The Ae. searsii genome was most distantly related to the Triticum B subgenome, with and FST of 0.262 (Supplemental Figure 4C). These results revealed that the genome of Ae. speltoides is closer to the wheat B subgenome compared with other Aegilops species.
Centromeric regions are conserved between the Ae. speltoides genome and the T. aestivum B subgenome
A conserved block analysis was used to identify conserved regions between Aegilops and Triticum (Brinton et al., 2020). Using MUMmer, we performed whole-chromosome pairwise alignments among the 13 (sub)genomes and identified conserved blocks relative to the B subgenome of T. aestivum (Figure 2A; Supplemental Figure 5A and 5B); we assumed that the scattered blocks derived from a common ancestor. Because non-syntenic retrotransposons displayed a median length of 9584 bp in wheat (Wicker et al., 2018), alignment blocks with a total length shorter than 10 kb were excluded from subsequent analysis. In total, 109 808 and 104 013 conserved blocks were identified in Ae. tauschii vs. T. aestivum DD and in T. urartu vs. T. aestivum AA, whereas approximately a tenth as many were identified in Ae. speltoides vs. T. aestivum B (11 472). The total lengths of the conserved blocks in Ae. tauschii vs. T. aestivum DD (2.92 Gb) and T. urartu vs. T. aestivum AA (2.13 Gb) were significantly higher than that in Ae. speltoides vs. T. aestivum BB (0.17 Gb) (Figure 2B). Further analysis showed that the conserved blocks between Ae. speltoides and the wheat B subgenome were mainly found in centromeric regions (Figure 2A). The sequence identity of conserved blocks showed the following pattern: Ae. tauschii vs. T. aestivum DD (99.11%) > T. urartu vs. T. aestivum AA (97.36%) >> Ae. speltoides vs. T. aestivum BB (95.28%) (Supplemental Figure 6; Supplemental Spreadsheet 2).
Figure 2.
Conserved blocks between the Triticum B genome and Aegilops species are enriched in centromeric regions.
(A) Chromosomal distribution of conserved genomic blocks with the T. aestivum B subgenome as the background.
(B) Boxplots of conserved genomic block lengths with the T. aestivum B, A, or D subgenome as the background.
(C) Evolutionary patterns of orthologous gene pairs estimated using a Ks distribution density plot.
(D) Ks distribution in the IWGSC RefSeq v1.0 B subgenome. The figure represents the Ks distribution of an average of 50 genes. The red dotted lines indicate chromosome boundaries.
(E) Boxplots of the variation frequency in the gene-coding regions of 12 species. The red dotted line represents the average variation frequency of Ae. speltoides. The top figure shows SNP variation frequency in centromeric regions, and the bottom figure shows SNP variation frequency in non-centromeric regions.
(F) SNP variation frequency of different TEs indicated by SNP density values, which ranged from 0 to 0.08. Red represents higher variation frequency, and blue represents lower frequency.
To compare the divergence times of Sitopsis species, we calculated the synonymous substitution rate (Ks) values of single-copy genes using the genomes of T. dicoccoides and T. durum as the foreground branches and a definite (T. urartu for the A genome and Ae. tauschii for the D genome) or undetermined (Ae. speltoides for the B subgenome) background to reveal their divergence times (Figure 2C). The distribution of Ks values of all gene pairs for the different comparisons peaked at approximately 0.001 for T. urartu vs. T. aestivum AA, 0.013 for Ae. speltoides vs. T. aestivum BB, and 0.001 for Ae. tauschii vs. T. aestivum DD (Supplemental Figure 7). The average Ks values displayed the following pattern: Ae. speltoides vs. T. aestivum BB (0.0770) >> Ae. tauschii vs. T. aestivum DD (0.0241) ≈ T. urartu vs. T. aestivum AA (0.0226). These results indicated that Ae. speltoides is highly divergent from T. aestivum BB and may not be the direct ancestor of the wheat B subgenome.
Sequence similarity can reflect the degree of evolutionary relatedness between species. Therefore, a BLAST search was performed to determine the optimal target genes in different subgenomes for the T. aestivum A, B, and D subgenomes (Supplemental Figure 8A). The optimal pairs were classified into seven categories based on sequence identity. We found a similar sliding-to-sliding block pattern of sequence identity for the T. dicoccoides A subgenome, the T. dicoccoides B subgenome, and Ae. tauschii, suggesting that these subgenomes experienced the process of hexaploid wheat at a similar time (Supplemental Figure 8B). Remarkably, compared with Ae. tauschii vs. T. aestivum D subgenome with a 99.5–100 identity region of 49.02%, the proportion between T. urartu and T. aestivum A was 26.56% (99.5–100 identity region). These findings are consistent with the fact that common wheat has experienced two rounds of allopolyploidization, with T. urartu involved in the first polyploidization event and Ae. tauschii in the second. Compared with T. urartu vs. T. aestivum A subgenome and Ae. tauschii vs. T. aestivum D subgenome, a significant reduction was observed for Ae. speltoides vs. T. aestivum B subgenome (99.5–100 identity region, 7.07%) (Supplemental Spreadsheet 3). The discrepancy in highly conserved sequence composition between the different comparisons again indicated that Ae. speltoides may not be the direct ancestor of the B subgenome of polyploid wheat.
We next evaluated which chromosome parts were conserved between the Sitopsis and B genomes using the Ks method. The results showed that all the species shared most of the conserved regions around the centromere region rather than the distal regions (Figure 2D), consistent with the distribution of conserved blocks (Figure 2A). We also compared the SNP variation frequency in centromeric and non-centromeric regions between Aegilops and Triticum, including nine accessions of Ae. speltoides, five Ae. mutica, six Ae. longissima, six Ae. sharonensis, six Ae. searsii, six Ae. bicornis, ten T. dicoccoides, one Ae. tauschii, one T. urartu, two T. dicoccum, two T. durum, and two T. aestivum. SNP variation frequency was lower in centromeric regions than in non-centromeric regions, and the log2(SNP variation frequency) was lower in Ae. speltoides accessions than in other non-B lineage species. The log2(SNP variation frequency) of Ae. speltoides in the centromeric region was −7.3, compared with −6.3 in the non-centromeric region (Figure 2E).
We also compared the SNP variation frequency of different transposable elements (TEs) between Aegilops and Triticum. Hierarchical clustering of TEs showed that Ae. speltoides was in the cluster closest to the B lineage genomes (Figure 2F). In addition, DTC (CACTA), DHH (Harbinger), DXX (Unclassified class 2), RLC (Copia), and RLG (Gypsy) had lower variation, similar to findings for the B-lineage genomes, whereas DTT (Mariner), DTX (Unclassified with TIRs), and RIX (LINE) showed higher variation frequencies (Figure 2F). We analyzed the degree of enrichment of TEs in the conserved blocks. The DNA transposon DHH and non-LTR retrotransposon SIX (SINE) were among the most enriched TEs, whereas DTA was less likely to be enriched in conserved blocks (Supplemental Figure 9).
Expansion of gene families in Ae. speltoides
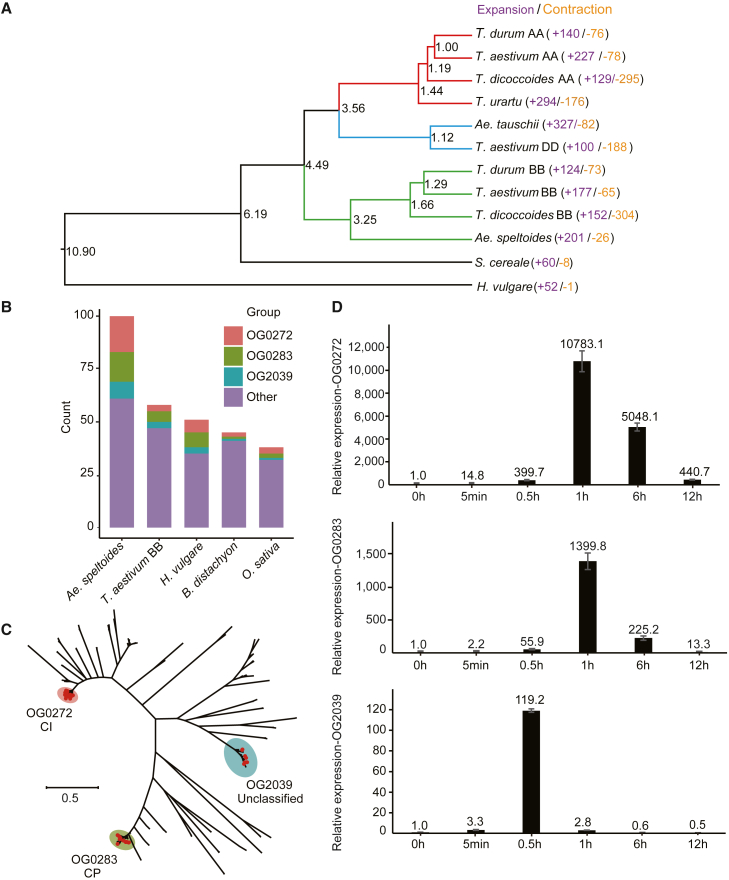
Orthologous clustering analysis revealed a total of 35 617 gene families, 10 454 (29.35%) of which were shared by all these subgenomes (Supplemental Figure 10A). A total of 4923 of the 10 454 shared gene families with one copy in each subgenome were designated as single-copy orthologous genes and subsequently used to construct a species phylogenetic tree (Figure 3A). Clustering results revealed that the A, B, and D subgenomes were grouped into three separate clades, consistent with the results of previous studies (Zhou et al., 2020). The species with a Triticum A subgenome and Aegilops D subgenome exhibited a shorter genetic distance than species with the B subgenome. Within the phylogenetic tree, Ae. speltoides was clustered together with the B subgenomes. The divergence time between Ae. speltoides and the B subgenomes of polyploid wheat was earlier than that between Ae. tauschii vs. other D subgenomes and between T. urartu vs. other A subgenomes. These results indicated that the divergence times between the tetraploid wheat B subgenome vs. Ae. speltoides and the tetraploid wheat A subgenome vs. T. urartu were not consistent.
Figure 3.
Phylogenetic relationships of genes from Aegilops/Triticum species.
(A) Comparative evolutionary relationships of Ae. speltoides and other Aegilops and Triticum genomes. The number of gene families that were expanded (purple) or contracted (orange) in each species was estimated using CAFÉ.
(B) Statistical analysis of heat shock protein 20 (HSP20) genes in five representative species. OG, orthologous groups.
(C) Neighbor-joining phylogeny of HSP20 genes in Ae. speltoides. CI and CP represent HSP20 subfamilies.
(D) Relative expression level of three HSP20 genes under heat stress treatments of different durations measured by RT-qPCR.
Analysis of the expansion and contraction of the 35 617 gene families as indicated by the phylogenetic tree revealed 201 expanded gene families composed of 2092 genes in Ae. speltoides. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses revealed that the expanded genes were highly enriched in various biological processes (Supplemental Figure 10B and 10C). Notable among the expanded genes was the heat shock protein 20 (HSP20) family. This is a major heat shock protein family whose members respond to high temperature stress in plants (Waters et al., 1996; Sung et al., 2003). HSP20s are the most abundantly produced proteins in many higher plants under heat stress and are encoded by a polygenic family (Charng et al., 2006). In previous studies, Ae. speltoides has been characterized as highly thermotolerant, potentially making it an important genetic resource for wheat improvement (Pradhan et al., 2012; Awlachew et al., 2016). The number of HSP20 proteins in Ae. speltoides (100) was approximately twice that in the T. aestivum B subgenome (58) and the genomes of H. vulgare (51), Brachypodium distachyon (45), and Oryza sativa (38) (Figure 3B). Further analysis showed that orthologous groups (OGs) OG0272, OG0283, and OG2039 contributed greatly to the expansion of the HSP20 gene family. With reference to the classification scheme for Arabidopsis thaliana and O. sativa, OG0272, OG0283, and OG2039 were assigned to the CI (Cytosol I), CP (Chloroplast), and unclassified subfamilies (Figure 3C). HSP20 genes in the same subfamily tended to have similar gene structures and conserved domain distributions, whereas these patterns were highly variable among subfamilies, which supported the phylogenetic relationships (Supplemental Figure 11A). In addition, gene expression data showed that OG0283, OG0272, and OG2039 had differentiated gene expression patterns (Supplemental Figure 11B), suggesting that the expanded HSP20 genes might be involved in the regulation of plant growth and development. To determine whether the expression level of HSP20 genes responded to heat stress, three genes from OG0283, OG0272, and OG2039 were selected for RT-qPCR. Under heat stress, the relative expression of these three genes increased significantly compared with the control group (0 h). The highest expression levels of OG0283 and OG0272 were found after 1 h of heat stress, whereas OG2039 expression was highest after 0.5 h of heat stress. These results clearly demonstrated that expression levels of HSP20 genes were upregulated under high temperatures (Figure 3D), indicating that they might function in heat responses.
Identification of conserved and non-conserved genes between Aegilops and Triticum
To effectively use the genetic resources of Aegilops, we analyzed the conserved and non-conserved gene groups between Aegilops and the Triticum B subgenome. We classified the genes based on their SNP variation frequency compared with those of the Triticum B subgenome using the k-means algorithm (k = 7). In general, the B subgenome genes can be divided into five categories (Figure 4A; Supplemental Spreadsheet 4): (1) conserved gene groups with low variation frequency in Aegilops and Triticum species, which were named the B lineage:Sitopsis conserved gene groups (B:S conserved) and included G1 (12 140 genes) and G6 (11 461 genes); (2) gene groups with significant SNP variation between Aegilops and Triticum, which were named B lineage:Sitopsis non-conserved gene groups (B:S non-conserved); these included G2 (1470 genes) and G3 (1247 genes), which we merged into one group; (3) genes that were uniquely similar between Ae. speltoides and B lineage species (G4, 5732 genes), which were named B lineage:Sitopsissp conserved genes (B:Ssp conserved); (4) B lineage:Sitopsissp non-conserved genes (G5, 1404 genes), which were uniquely similar between the B lineage genome and non-Ae. speltoides Sitopsis section species (B:Ssp non-conserved); and (5) B lineage conserved genes (G7:2066 genes), which were particularly conserved in the wheat B lineage species (B conserved). The B:S conserved and B conserved genes were mainly distributed around the centromere regions compared with genes in other groups (Figure 4B), consistent with the distribution of conserved blocks (Figure 2A).
Figure 4.
Identifying conserved and non-conserved genes between Aegilops and Triticum.
(A) Gene classification between Aegilops and Triticum species on the basis of SNP variation frequency. Red and blue represent high and low SNP variation frequency between Aegilops and Triticum, respectively. The horizontal axis shows groups of genes defined by SNP variation frequency, including seven groups (G1–7) and five types of genes. G1 and G6 had similar patterns of SNP variation frequency, as did G2 and G3. The vertical axis shows the different species.
(B) Distribution of five types of genes in the wheat B subgenome: (1) B lineage:Sitopsis (B:S) conserved genes, (2) B:S non-conserved genes, (3) B:Ssp conserved genes, (4) B:Ssp non-conserved genes, and (5) B lineage conserved genes.
(C) Heatmap of gene expression levels of the non-conserved and conserved gene groups. The vertical axis represents different RNA-seq tissues, and the horizontal axis shows the different types of genes: B:S conserved genes, B:S non-conserved genes, B:Ssp conserved genes, B:Ssp non-conserved genes, and B conserved genes.
(D) Distribution proportion of the five types of genes in conserved blocks.
(E) Distribution of collinear genes in the conserved and non-conserved gene groups.
(F) Distribution of differentially expressed genes and non-differentially expressed genes in the conserved and non-conserved gene groups.
We performed a GO analysis of the conserved and non-conserved genes between Aegilops and Triticum to explore the biological processes in which they participate. The B:S conserved gene group was mainly involved in regulation of basic molecular mechanisms; however, the B:S non-conserved gene group was mainly involved in tissue growth and development, material transport, nitrogen metabolism, and response to biotic and abiotic stresses (Supplemental Figure 12A; Supplemental Spreadsheet 5). We then obtained the expression patterns from the co-expression network analysis (Supplemental Figure 12B) and examined the expression patterns of conserved and non-conserved gene groups in seven wheat tissues (Figure 4C). The B:S conserved genes were highly expressed in all tissues. The B:S non-conserved and B:Ssp non-conserved genes were highly expressed in grain tissues (days after pollination [DAP]10 grain, DAP15 grain, and DAP20 grain), but showed lower expression in the leaf, floral meristem (FM), DAP4 grain, and root, which may indicate that B:S non-conserved and B:Ssp non-conserved genes are strongly associated with grain development. In addition, the B:Ssp conserved gene group and B conserved genes had similar expression patterns, but the B conserved genes generally showed lower expression in all tissues compared with the other four groups (Figure 4C).
We next calculated the distribution of genes from conserved blocks (Ae. speltoides vs. wheat B subgenome) in the conserved and non-conserved groups; 1091 genes (86.9%) in the conserved blocks were in the B:S conserved group, 83 genes (6.6%) were in the B:Ssp conserved group, and the remaining genes were in the other three gene groups (Figure 4D). We also calculated the distribution of collinear gene pairs in the five gene groups and found that most of the collinear genes were in the B:S conserved gene groups (19 988) and B:Ssp conserved gene group (3942). By contrast, non-collinear genes were mainly in the non-conserved gene groups (Figure 4E). In addition, we calculated the number of genes that were differentially expressed in the five groups. There were more non-differentially expressed genes than differentially expressed genes in the B:S conserved gene groups, but there were no significant differences in the other groups (Figure 4F).
Identification of agronomic gene resources from the Sitopsis group
Sitopsis species represent a secondary gene pool and germplasm resources for wheat genetic improvement. To further identify genetic resources for wheat improvement, we mapped 33 reported agronomic genes cloned in wheat by forward and homology-based strategies (Gaurav et al., 2022) (Supplemental Spreadsheet 4) to the conserved and non-conserved gene groups. We found that 16 reported genes were in the B:S conserved gene groups (G1 and G6), including growth and development-related genes such as Rht-B1, TB-B1, Vrn3-7B, WFZP-2B, and GNI1-B1 (Supplemental Spreadsheet 4). However, the disease resistance genes were mainly found in the other four groups. For example, eight disease resistance genes were found in the B:S non-conserved groups, including the powdery mildew resistance genes Pm60, Pm5e, and Pm1A; stripe rust resistance genes Yr5a and Yr36; leaf rust resistance genes Lr10 and Lr14a; and septoria nodorum blotch-related gene Snn1. Four disease-related genes (Pm17, Pm24, Yr7, and Lr1) were in the B:Ssp conserved gene group. Three disease-related genes (Lr21, Lr13, and Tsn1) and two disease-related genes (Pm12 and Sr13) were found in the B conserved gene group and B:Ssp non-conserved gene group, respectively (Supplemental Spreadsheet 4).
We identified 789 Ae. speltoides-specific genes, including 45 disease resistance genes (Figure 5A; Supplemental Spreadsheet 6). GO enrichment analysis revealed that these 789 genes were mainly involved in the photosynthetic reaction (GO: 0009539) and organonitrogen compound metabolic process (GO: 1901564) (Figure 5B; Supplemental Spreadsheet 5). We calculated the expression levels of these genes in 17 tissues of Ae. speltoides and discovered that they showed significant tissue specificity; a 90th percentile threshold was used to identify highly expressed genes (Figure 5C). Sixty genes were highly expressed in the leaf, 93 genes in the root, 47 genes in the FM, 85 genes in the spikelet, 80 genes in the glume, 83 genes in the pistil, 109 genes in the stamen, 122 genes in DAP4 grain, 133 genes in DAP10 grain, 82 genes in DAP15 grain, 31 genes in DAP20 grain, 41 genes in the flag leaf at DAP10, 46 genes in the flag leaf at DAP15, 47 genes in the flag leaf at DAP20, 80 genes in the stem at DAP10, 87 genes in the stem at DAP15, and 83 genes in the stem at DAP20 (Figure 5C).
Nitrogen is an essential element for wheat growth and development, and nitrogen utilization significantly affects wheat yield (Cormier et al., 2016). The Ae. speltoides-specific genes and B:S conserved/non-conserved genes were enriched in GO terms associated with the organonitrogen compound metabolic process and nitrogen compound transport. A previous study found that Ae. speltoides was tolerant of low-nitrogen environments (Gorny and Garczyński, 2008), and we therefore speculated that these pathways may be associated with adaptation to low-nitrogen conditions. In addition, previous studies have reported that OsTCP19 (Liu et al., 2021) and OsNRT1.1B (Zhang et al., 2019) play important roles in the regulation of nitrogen metabolism in rice. BLAST searches showed that the homologs of OsTCP19 and OsNRT1.1B in wheat are TraesCS7B01G091900 and TraesCS1B01G225000, respectively. Co-expression network analysis showed that 111 Ae. speltoides-specific genes out of 15 940 genes were co-expressed with OsTCP19 (Figure 5D; Supplemental Spreadsheet 7). In addition, 8553 genes were associated with TraesCS1B01G225000 (Supplemental Spreadsheet 7), including 58 Ae. speltoides-specific genes. We therefore concluded that nitrogen-related genes identified by weighted gene co-expression network analysis (WGCNA) may be related to adaptation to low-nitrogen conditions, and these genes could be used to improve wheat low-nitrogen tolerance through distant hybridization or genetic engineering.
Discussion
The Aegilops genus is considered the diploid progenitor of the wheat B and D subgenomes (Zhang et al., 2018). Based on morphological similarity and molecular phylogenetic relationships, Ae. speltoides has been considered the possible donor of the wheat B subgenome (Petersen et al., 2006). In this study, we assembled a high-quality Ae. speltoides genome and resequenced 53 accessions across seven species. The total genome size of Ae. speltoides is approximately 4.88 Gb, similar to the size of the wheat B subgenome (Appels et al., 2018). Our Ae. speltoides genome assembly is larger than that reported by Li et al. (2022) (4.11 Gb) but smaller than that reported by Avni et al. (2022) (5.13 Gb), suggesting that there may be substantial variation in genome size among different Ae. speltoides accessions. On the basis of similarity in identical gene numbers, gene identity, conserved block length and counts, and SNP density, we suggest that the Ae. speltoides genome is the closest to the B subgenome, but it may not be the direct donor of the B subgenome, consistent with previous studies (Avni et al., 2022; Li et al., 2022). Few conserved blocks were identified in the euchromatic regions of other Sitopsis species, indicating that the conserved blocks were inherited from the most recent common ancestor of Aegilops and Triticum rather than arising from genetic introgressions. Therefore, these results also reject the hypothesis of the polyphyletic origin of the wheat B subgenome at the hexaploid level. The above analysis also showed that there were 0.17 Gb of conserved blocks between Ae. speltoides and the wheat B subgenome and that the centromere regions were more conserved than other regions (Figure 2A and 2D).
Aegilops species have been shown to contain many beneficial traits, especially for resistance to different diseases and tolerance of high-temperature and low-nutrient environments (Pradhan et al., 2012; Awlachew et al., 2016; Kishii, 2019); they thus represent important genetic resources that could be exploited for wheat improvement. Distant hybridization is commonly used to enhance the genetic diversity of modern wheat (Mujeeb-Kazi et al., 2013; Alvarez and Guzman, 2018; Cui et al., 2018). In this study, we first identified five gene groups that were conserved or non-conserved between Aegilops and Triticum by analyzing population resequencing data and revealed their biological characteristics. By mapping the reported genes to conserved and non-conserved gene groups, we revealed that most of the disease resistance genes were found in the B:S non-conserved, B:Ssp conserved, and B:Ssp non-conserved groups (Supplemental Spreadsheet 4). By contrast, growth- and development-associated genes were mainly found in the B:S conserved gene group (Supplemental Spreadsheet 4). It is likely that gene segments with high differentiation or variation between Sitopsis and the B subgenome could confer disease resistance (Kishii, 2019). We demonstrate that the strategy of identifying conserved and non-conserved gene groups between Sitopsis and B-lineage species could be an effective method for discovering potentially important agronomic gene resources. We found that the HSP20 gene family was expanded in Ae. speltoides, indicating that Ae. speltoides could provide a genetic resource for tolerance to high-temperature stress. We also identified Ae. speltoides-specific genes and found that several of these genes were involved in nitrogen compound metabolic processes, responses to stress, and plant growth and development pathways. We therefore suggested that these nitrogen-related genes could be potential resources for improving tolerance to low-nitrogen conditions.
The high-quality genome assembly of Ae. speltoides and large-scale resequencing of Sitopsis species not only contribute to understanding the genomic contribution of Aegilops to Triticum but also help to reveal conserved chromosome regions between Aegilops and the wheat B subgenome. The newly identified conserved and non-conserved genes and Ae. speltoides-specific genes can be important genetic resources for wheat improvement.
Methods
Plant materials and data collection
We selected 47 accessions from seven species worldwide to perform exome capture sequencing. These included eight Ae. speltoides, six Ae. bicornis, six Ae. searsii, six Ae. longissima, six Ae. sharonensis, five Ae. mutica, and 10 T. dicoccoides accessions. We also selected six accessions to perform whole-genome resequencing, including one each of Ae. speltoides, Ae. bicornis, Ae. searsii, Ae. longissima, Ae. sharonensis, and Ae. mutica. To further detect the genetic variation in B-lineage species, we downloaded whole-genome resequencing data for six species (Ae. tauschii, H. vulgare, T. aestivum, T. dicoccoides, T. dicoccum, and T. urartu) from a public database (Supplemental Spreadsheet 1) (Jayakodi et al., 2020; Zhou et al., 2020).
Sequencing and assembly of the Ae. speltoides genome
Fresh leaves of Ae. speltoides ssp. speltoides (Y2032) were collected and used to extract genomic DNA with the Qiagen Genomic DNA Extraction Kit according to standard operating procedures. A NanoDrop One UV-VIS spectrophotometer (Thermo Fisher Scientific, USA) was used to detect DNA purity, and a Qubit 3.0 fluorometer (Invitrogen, USA) was used for accurate DNA quantification. After a sample passed the quality inspection, large DNA fragments were collected using the BluePippin system (Sage Science, USA). After DNA damage repair and end repair using polishing enzymes, the A nucleotide was added to the 3′ end. The library was processed using a ligation sequencing kit (SQK-LSK109, Oxford Nanopore) according to the manufacturer’s protocol, and the sample was quantified using the Qubit 3.0 fluorometer. Sequencing of gDNA long reads was performed on a MinION Nanopore sequencer (Oxford Nanopore Technologies, UK). After data quality control, 30.46 million Nanopore reads (∼429.24 Gb of data) were obtained.
For Hi-C sequencing, nuclear DNA from fresh leaves of Ae. speltoides was cross-linked, extracted, and digested with a restriction enzyme. The sticky ends of the digested fragments were biotinylated, diluted, and randomly ligated. Sequencing was performed on an Illumina HiSeq 4000 platform, and fastp (version 0.12.6) (https://github.com/OpenGene/fastp) was used for quality control. A total of 2.91 Gb of clean paired-end reads were generated. NextDenovo (https://github.com/Nextomics/NextDenovo) was used with default parameters to correct the Nanopore reads. After initial assembly, three rounds of iterative polishing were performed using NextPolish with default parameters.
To further anchor the contigs to chromosomes, the corrected assembly was then subjected to redundancy elimination using Hi-C technology. The sequence contigs of the genome assembly were clustered and oriented into pseudochromosomes using LACHESIS (https://github.com/shendurelab/LACHESIS) with the following parameters: cluster min re sites = 100, cluster max link density = 2.5, cluster noninformative ratio = 1.4, order min n res in trunk = 60, and order min n res in shreds = 60. The Hi-C contact heatmap was used to evaluate the orientation of the contigs on the pseudochromosomes. The completeness and accuracy of the assembled genome were evaluated with BUSCO version 3.0.2 (Embryophyta dataset version odb9) to identify the presence of single-copy genes in the assembled genome.
RNA sequencing and analysis
Ae. speltoides plants were grown in a growth chamber under conditions of 16 h light, 20°C/8 h dark, 18°C. The following fresh tissues were harvested: leaves and roots at the three-leaf stage; stamens, pistils, and glumes before flowering; grains at 4, 10, 15, and 20 DAP; flag leaf and stems under spikes at 10, 15, and 20 DAP; and young spikes with initiating spikelet meristems (SMs) and initial FMs (Supplemental Spreadsheet 1). Harvested tissues were immediately frozen in liquid nitrogen, and the TRIzol method was used to extract RNA. RNA sequencing (RNA-seq) libraries were constructed following the Illumina standard pipeline, and 150-bp paired-end sequencing was performed on each sample using the Illumina NovaSeq platform. RNA-seq data were also generated from seven hexaploid wheat Chinese Spring tissues at the same developmental stages as Ae. speltoides, including leaf, root, FM, and grain tissues at 4, 10, 15, and 20 DAP (Supplemental Spreadsheet 1).
RNA-seq data were first processed for quality control using Trimmomatic (version 3) (http://www.usadellab.org/cms/?page=trimmomatic) with the following parameters: ILLUMINACLIP:1.adapter.list:2:30:10 LEADING:10 TRAILING:10 SLIDINGWINDOW:1:10 MINLEN:50. Then, FastQC (version 0.11.5) (https://bio.tools/FastQC) was used to evaluate read quality. Principal-component analysis (PCA) (Supplemental Figure 13A) and correlation analysis (Supplemental Figure 13B) of different samples confirmed that the RNA-seq data were of high quality and could be used for subsequent analysis. Pertea’s experimental procedure was used to perform RNA-seq analysis (Pertea et al., 2016). HISAT2 (version 2.5.3a) (http://daehwankimlab.github.io/hisat2/) was used to map the clean paired-end data to the reference genome, StringTie (version 1.3.3b) (https://ccb.jhu.edu/software/stringtie/) was used to assemble transcripts with default parameters, and the Ballgown R package (version 2.26) (Frazee et al., 2015) was used to estimate gene expression levels as fragments per kilobase of exon model per million mapped fragments (FPKM).
Repetitive sequence annotation
A combination of homology-based and de novo strategies was used to identify and annotate repetitive sequences in the Ae. speltoides genome. For the homology-based approach, the Repbase TE library (version 15.02) and the TE protein database (http://www.girinst.org/repbase) were used to query the whole genome with RepeatMasker (version 4.0.5) and RepeatProteinMask (http://www.repeatmasker.org/), respectively. For the de novo approach, LTR_FINDER (version 1.0.7) (https://github.com/xzhub/LTR_Finder), Piler (version 1.06) (http://www.drive5.com/piler/), RepeatScout (version 1.0.5) (http://www.repeatmasker.org/), and RepeatModeler (version 1.0.11) (http://www.repeatmasker.org/RepeatModeler/) were used to construct a de novo repetitive sequence library. RepeatMasker was then used to search against the library. Tandem repeat sequences were identified using Tandem Repeats Finder (TRF) (version 4.09) (http://tandem.bu.edu/trf/trf.html).
Gene annotations
Protein-coding genes were annotated using a combination of three independent approaches: homology-based, ab initio-based, and transcriptome-based prediction. The sequences of homologous proteins from four related plant genomes (Ae. tauschii, T. dicoccoides, T. durum, and T. aestivum) were downloaded from the EnsemblPlants database. genBlastA (https://anaconda.org/bioconda/genblasta) was used to compare protein sequences encoded in the related genomes with those of Ae. speltoides. GeneWise (version 2.42.0) (http://www.ebi.ac.uk/Tools/psa/genewise/) was used to annotate the gene structures of corresponding genomic regions by taking their orthologous genes as queries. For transcriptome-based predictions, RNA-seq data were mapped onto the reference genome using TopHat (version 2.0.8) (https://ccb.jhu.edu/software/tophat/index.shtml), and gene models were assembled with Cufflinks (version 2.1.1) (http://cole-trapnell-lab.github.io/cufflinks/). The RNA-seq data were also assembled using Trinity (version 2.4) (https://bio.tools/trinity), and the assembled sequences were directly mapped to the Ae. speltoides genome and reconstructed with PASA (version 2.1.0) (https://github.com/PASApipeline/PASApipeline). This gene set was denoted the PASA Trinity set (PASA-T-set) and used to train ab initio gene prediction programs. Five ab initio programs were used to predict coding regions in the repeat-masked genome: Augustus (version 3.0.2) (https://bioinf.uni-greifswald.de/augustus/), GeneScan v1.0 (http://hollywood.mit.edu/GENSCAN.html), GlimmerHMM (version 3.0.2) (https://ccb.jhu.edu/software/glimmerhmm/), GeneID (version 1.4) (https://genome.crg.es/software/geneid/), and SNAP (version 4.0) (https://bio.tools/snap). The gene models generated by all methods were integrated into a non-redundant set of gene structures using EVidenceModeler (EVM) (version 1.1.1) (https://github.com/EVidenceModeler/EVidenceModelerr).
Predicted genes were functionally annotated using two integrated protein sequence databases, Swiss-Prot and Non-redundant (nr), by BLASTP with an e-value of 1e−5. Domains of the protein-coding genes were identified by searching against the Pfam (version 27.0) (https://pfam.xfam.org/) and InterPro (version 5.16) (https://www.ebi.ac.uk/interpro/) databases using HMMER (version 3.1) (http://hmmer.org/) and InterProScan, respectively. GO terms were assigned to the genes on the basis of the Pfam and InterPro entries. KEGG pathways in which the genes participated were annotated using BLAST searches against the KEGG dataset with an e-value cutoff of 1e−5.
tRNAs were predicted using tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/). rRNAs were identified by performing a BLAST search against rRNA sequences with an e-value of 1e−10. Infernal (version 1.1.1) (http://eddylab.org/infernal/) was used to identify miRNAs and snRNAs by searching against the Rfam database (release 9.1) (https://rfam.org).
Whole-genome resequencing
A total of six accessions of the six species were selected for whole-genome resequencing (Supplemental Spreadsheet 1). Seeds of these accessions were sown in a growth chamber under conditions of 16 h light, 20°C/8 h dark, 18°C. A total of 4 μg of genomic DNA from each accession at the three-leaf stage was fragmented into 300-bp fragments. The fragmented DNA was size-selected for an average insert size of 300 bp using NucleoMag technology (Macherey-Nagel, Duren, Germany) according to the manufacturer’s instructions. Barcode libraries were constructed using the VAHTS Universal DNA Library Prep Kit. DNA was end-repaired with an end-repair enzyme, a deoxyadenosine was added to the 3′ ends of the fragments, and the final libraries were sequenced on the Illumina NovaSeq platform.
Whole-exome resequencing
We selected 47 accessions of seven species (Supplemental Spreadsheet 1) for whole-exome resequencing. A total of 2 μg of genomic DNA from each accession at the three-leaf stage was fragmented into 250-bp fragments, and the total fragmented DNA was subjected to size selection with magnetic beads according to the manufacturer’s instructions. Pre-libraries were constructed using the VAHTS Universal DNA Library Prep Kit for Illumina NovaSeq. Selected DNA was end-repaired with an end-repair enzyme, and a deoxyadenosine was added to the 3′ ends of the fragments. VAHTS DNA barcodes and VAHTS indexing adapters were ligated to the sample libraries. Pre-capture amplification was performed using the VAHTS protocol with 5 PCR cycles. Equal amounts of products from six libraries were pooled to obtain a total of 2 μg of DNA for hybridization. Hybridization of sample libraries was performed at 47°C for ∼72 h using the SeqCap EZ library (Roche/NimbleGen, Madison, WI) and custom-designed exome capture probes (Tcuni Technologies, Chengdu, China). The hybridized sequences were enriched using capture beads from the SeqCap EZ pure capture bead kit, washed, and amplified by ligation-mediated PCR. The quality of the captured libraries was assessed using the Agilent 4200 TapeStation. qPCR was used to quantify the libraries, which were then sequenced on the Illumina NovaSeq platform to obtain 150-bp paired-end reads.
FISH analysis
Seeds of Aegilops accessions were germinated in an incubator at 22°C–23°C. When the roots of seedlings reached 2–3 cm, they were used for chromosome preparation of mitotic metaphase by the air-drop method as described by Lang et al. (2019). The sequential nondenaturing FISH (ND-FISH) protocol using synthesized probes was described by Fu et al. (2015). The sequences of Oligo probes Oligo-pSc119.2, Oligo-pTa71, and Oligo-(GAA)7 were described by Tang et al. (2014) (Supplemental Table 9). The Oligo probes were synthesized by Shanghai Invitrogen Biotechnology (Shanghai, China). The synthetic Oligo probe sequences were either 5′-end-labeled with 6-carboxyfluorescein (6-FAM) of Oligo-pSc119.2 for green signals or 6-carboxytetramethylrhodamine (Tamra) of Oligo-pTa71 and Oligo-(GAA)7 for red signals. Image collection of the ND-FISH signals was performed with a fluorescence microscope (BX53, Olympus) equipped with a DP-80 charge-coupled device (CCD) camera.
Variant calling and coverage analysis
All sequenced reads from the materials were mapped to the IWGSC v1.0 B subgenome to obtain SNPs. Because the materials included diploid, tetraploid, and hexaploid accessions, we first split the IWGSC RefSeq v1.0 sequence into three subgenomes according to their genome components, which were named BB, AABB, and AABBDD, respectively. BWA (version 0.7.17) (http://bio-bwa.sourceforge.net/) was used to build an index of the three-type reference genome. Clean data from the diploid, tetraploid, and hexaploid accessions were then mapped to the BB, AABB, and AABBDD reference genomes with default parameters. The alignment results were saved as SAM files, and SAMtools (version 1.9) (http://www.htslib.org/) was used to calculate the average sequencing depth of each BAM file. We used GATK (version 4.1.8) (McKenna et al., 2010) to perform MarkDuplicates on the BAM files. The HaplotypeCaller function of GATK was used to detect variants in each sample, and the raw gVCF file was filtered using VCFtools (version 0.1.16) (http://vcftools.sourceforge.net/) with the parameters --minDP 3 -max-missing 1. Finally, high-quality variant information was obtained for subsequent analysis.
To explore the SNP variation frequency and coverage of the gene-coding region, we first extracted the sequence of the gene-coding region from the IWGSC RefSeq v1.0 annotation file. We selected homozygous variation sites (0/0, 0|0) and non-homozygous variation sites (1/1, 1|1) to calculate the SNP variation frequency. We calculated the coverage using the following process: (1) homozygous variation frequency + non-homozygous variation frequency; (2) gene variation frequency was calculated as non-homozygous frequency/(homozygous frequency + non-homozygous frequency), and, if the coverage was 0 in the coding region, the gene variation frequency was set to NA. To further reveal which genes were similar to the IWGSC RefSeq v1.0 B subgenome according to the gene variation frequency, we used the k-means clustering method of the R package Pheatmap (version 1.0.12) (https://cran.r-project.org/web/packages/pheatmap/) to cluster genes with similar variation patterns and the R package factoextra (https://github.com/kassambara/factoextra) to determine the most appropriate k value for the k-means analysis. The distribution of genes of different groups throughout the chromosomes was visualized using the R package RIdeogram (https://cran.r-project.org/web/packages/RIdeogram/).
Calculation of nucleotide diversity (π) and fixation index (FST)
To explore the detailed population genetic diversity changes between the Sitopsis species and the wheat B subgenome, we selected six Ae. bicornis, six Ae. longissima, five Ae. mutica, six Ae. sharonensis, six Ae. searsii, nine Ae. speltoides, and 15 T. aestivum BB genomes for calculation of π and FST values. We calculated π and FST using VCFtools software with a 10-kb window size. A mean FST value in a window less than 0 was set to 0 when FST was calculated.
Phylogenetic tree construction and divergence time estimation
To reveal the phylogenetic relationships between the genera Aegilops and Triticum, we constructed a phylogenetic tree of 13 species with barley as the outgroup using 1 085 451 SNPs common to the 13 species. VCF2Dis (version 1.44) (https://github.com/BGI-shenzhen/VCF2Dis) was used to calculate the p-distance matrix of the species, Fastme (version 2.1.6.1) (http://www.atgc-montpellier.fr/fastme/) was used to calculate the phylogenetic relationships based on the neighbor-joining method, and Interactive Tree of Life (iTOL) (https://itol.embl.de/) was used to visualize the tree.
We constructed another phylogenetic tree using 4923 shared single-copy gene families. Based on the gene-family alignment data, an ultrametric time-scaled phylogenetic tree was constructed using the penalized likelihood method implemented in r8s (version 1.71) (http://loco.biosci.arizona.edu/r8s/). The time calibrations were obtained based on the divergence times of H. vulgare vs. Ae. tauschii (10.90 MYA) and Ae. tauschii vs. T. urartu (3.56 MYA) from fossil records available at TimeTree (http://timetree.org).
Identification of conserved chromosome regions between Ae. speltoides and the wheat B subgenome
To reveal the conserved gene regions between Ae. speltoides and the wheat B subgenome, 10 species were selected and used to calculate Ks changes in gene-coding regions (Ae. sharonensis, Ae. longissima, Ae. bicornis, Ae. searsii, Ae. tauschii, T. urartu, Ae. mutica, Ae. speltoides, T. dicoccoides BB, and T. aestivum BB). First, SnpEff (version 4.3t) (http://pcingola.github.io/SnpEff/) was used to annotate the gVCF file of each accession. Then missense and synonymous variant sites of the annotated VCF file were filtered, and a custom Python script (https://github.com/MerrimanLab/selectionTools/blob/master/extrascripts/kaks.py) was used to calculate the Ks value of each accession at the whole-genome level. We then extracted the Ks values of the protein-coding genes of the IWGSC RefSeq v1.0 B subgenome, and filtered genes with abnormal Ks values (e.g., NA or higher than 0.1). A sliding window method of 50 genes was used to show the distribution of Ks values with the Pheatmap R package.
Gene-family cluster, expansion, and contraction analysis
The genome sequences of eight species (T. urartu, Ae. speltoides, Ae. tauschii, S. cereale, H. spontaneum, H. vulgare, T. dicoccoides, and T. aestivum) were used for orthologous gene clustering. First, the nucleotide and amino acid sequences were downloaded from MBKBASE (http://www.mbkbase.org/Tu) (Peng et al., 2020) and the Ensembl Plants database (http://plants.ensembl.org). The tetraploid and hexaploid wheat genomes were manually split into diploid subgenomes, i.e., A, B, and/or D. Only the longest transcripts were reserved for subsequent analysis. OGs across these species were inferred using OrthoFinder (version 2.3.1) (https://github.com/davidemms/OrthoFinder) with default settings. Computational Analysis of Gene Family Evolution (CAFÉ) (version 4.0.1) (De Bie et al., 2006) was used to determine gene-family expansion and contraction patterns, and a P value of 0.05 was used to indicate a possible expansion or contraction event.
Species tree, expansion patterns, and expression profiles of the HSP20 gene family
The HMM profile of the HSP20 domain (PF00011) was downloaded from the Pfam database (http://pfam.sanger.ac.uk/). HMMER (version 3.1b2) was used to build the HMM profile and search against all proteins in the genome with an e-value cutoff of 0.001. Multiple sequence alignments were constructed using ClustalW (version 2.1) (http://www.clustal.org/) with default parameters. A neighbor-joining tree was generated using MEGA X with the following criteria: 1000 bootstrap replicates, 95% partial deletion, and the Poisson correction model. The Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/) was used to infer the gene structure of the HSP20 genes. Conserved domains were identified using the SMART online tool (https://smart.embl.de/).
RT-qPCR of the HSP20 gene family
Ae. speltoides seeds were placed in an incubator for normal processing under growth conditions of 16 h light, 20°C/8 h dark, 18°C. When plants had grown to the trilobal stage, they were placed in a heat treatment incubator under the following conditions: 16 h light, 40°C/8 h dark, 40°C. Six seedlings were separately sampled at 0 h, 5 min, 0.5 h, 1 h, 6 h, and 12 h after application of heat stress. We selected three genes from OG0272, OG0283, and OG2039 for RT-qPCR, which was performed as described previously (Yang et al., 2021). The primer sequences used in this research are listed in Supplemental Table 10.
Construction of conserved blocks in Triticum and Aegilops
Chromosome-level NUCmer (NUCleotide MUMer) alignments were performed to construct conserved blocks. For all 13 (sub)genomes, the complete chromosome set was divided into individual chromosomes, and pairwise chromosome alignments were performed with the T. aestivum A, B, and D subgenomes. Alignments were performed using the NUCmer tool embedded in MUMmer (version 4.0) (https://mummer4.github.io). The data were filtered with the option “-l 10 000” (minimum length of alignment, 10 000 bp). The chromosome distribution of the conserved blocks was displayed using the RIdeogram R package.
Gene identity, genomic sequence identity, and Ka/Ks calculations
A BLASTP search with an e-value cutoff of 1e−5 and max_target_seq of 1 was performed, taking the A, B, and D subgenomes as the reference sequence. The distribution intervals were divided into 0–80, 80–90, 90–95, 95–98, 98–99.5, and 99.5–100 according to the percentage sequence identity. Nonsynonymous (Ka) and Ks substitution rates were calculated using the Codeml program embedded in PAML (version 4.9e) (http://abacus.gene.ucl.ac.uk/software/paml.html).
Identification of collinear gene pairs between Ae. speltoides and the hexaploid wheat B subgenome
To identify collinear genes between Ae. speltoides and the wheat B subgenome, the Python version of MCscan (https://github.com/tanghaibao/mcscan) was used to perform collinear gene pair analysis using the parameter --minspan = 30 to detect collinear gene regions.
GO enrichment analysis
The GO enrichment analysis was performed using agriGO online software (https://systemsbiology.cpolar.cn/agriGOv2/). The GO analysis was based on Fisher’s exact test with a false discovery rate (FDR) correction for multiple testing. We identified significantly enriched GO terms based on a P value of less than 0.05.
Identification of specific genes of Ae. speltoides
To identify Ae. speltoides-specific genes, we used whole-genome resequencing data from one Ae. bicornis, one Ae. longissima, one Ae. mutica, one Ae. searsii, one Ae. sharonensis, one Ae. speltoides, three Ae. tauschii, three T. urartu, three T. dicoccoides, three T. dicoccum, three T. durum, and seven T. aestivum. Clean data from these materials were mapped to the Ae. speltoides reference genome assembled in the current study: HISAT2 software was used to build an index of the Ae. speltoides reference genome, the clean reads were mapped to the reference genome with default parameters, and the mapping information was stored in a BAM file. We removed unmapped reads and reads with MAPQ values less than 20 using SAMtools (version 1.9) with the parameters -bF 4 -q 20. We calculated the coverage of each BAM file with high-confidence genes using BEDtools (version 2.29.2) (https://bedtools.readthedocs.io/en/latest/). The coverage of Ae. speltoides was taken as the standard with which to identify Ae. speltoides-specific genes; that is, if the gene coverage of other materials was lower than one-fifth that of the Ae. speltoides gene, this gene was considered to be an Ae. speltoides-specific gene.
Calculation of TE variation frequency
To measure changes in SNP variation frequency in noncoding regions, we calculated the variation frequency changes in TEs. TEs can be classified into three types: (1) DNA transposons, including CACTA (DTC), Mutator (DTM), Unclassified with TIRs (DTX), Harbinger (DTH), Mariner (DTT), Unclassified class 2 (DXX), hAT (DTA), and Helitrons (DHH); (2) LTR retrotransposons, including Gypsy (RLG), Copia (RLC), and Unclassified LTR-RT (RLX); and (3) non-LTR retrotransposons, including LINE (RIX) and SINE (SIX). The method for calculating SNP variation frequency in TEs was similar to that used for gene variation. First, we calculated the average values of each accession, and then we normalized the average values by log2 transformation. The Pheatmap R package was used to perform hierarchical clustering. We also extracted the TEs from the conserved block regions to determine the extent of TE enrichment using the mean variation frequency. Pheatmap was used to perform the hierarchical clustering and visualize the results.
Construction of the nitrogen co-expression network
In this study, we identified several nitrogen metabolic and transport pathways that indicated that Sitopsis species can provide nitrogen-related genetic resources; we therefore aimed to discover nitrogen-related genes by constructing a nitrogen co-expression network using the WGCNA R package (version 1.70–3) (Langfelder and Horvath, 2008). We used seven common RNA-seq tissues between Ae. speltoides and wheat to perform the co-expression network analysis. Two previously mentioned genes, OsTCP19 (TraesCS7B01G091900) and OsNRT1.1B (TraesCS1B01G225000), were used to build the co-expression network, and weight values calculated by the WGCNA method were used to measure the degree of association between pairs of genes; higher-weighted values indicate a higher degree of association between two genes. Cytoscape (version 3.7.2) (https://cytoscape.org/) was used to visualize the nitrogen-related co-expression network.
Data availability
The Ae. speltoides genome assembly was deposited in NCBI GenBank under BioProject accession number PRJNA802349. The raw sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under the BioProject accession numbers PRJNA893435 (Nanopore raw data), PRJNA802349 (WGS), and PRJNA802730 (Hi-C). The RNA-seq data, whole-exome resequencing data, and gVCF files were saved in the GEO database with accession number GSE197593, and the whole-genome sequencing data were saved in the SRA database with accession number PRJNA806672. We also deposited the reference genome of Aegilops speltoides and annotation file to ScienceDB (https://www.scidb.cn/en/s/VFrU7z). The whole-genome resequencing data, whole-exome resequencing data, and RNA-seq data were saved in the China National Genomics Data Center with the accession number CRA009743.
Funding
This research was supported by the National Natural Science Foundation of China (grant no. 31991213), the Talent Program and Agricultural Science and the Technology Innovation Program of CAAS, the China Postdoctoral Science Foundation (grant no. 2022M713430), and the Central Public-interest Scientific Institution Basal Research Fund (grant no. S2022ZD02). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
X.Y.K., J.Z.J., and X.L. conceived the research. X.Y.K., J.Z.J., and Z.F.L. designed and supervised the project. Y.X.Y., L.C.C., Z.F.L., G.R.L., Z.J.Y., G.Y.Z., C.Z.K., D.P.L., Y.Y.C., Z.C.X., Z.X.C., L.C.Z., and C.X. analyzed the data and performed the experiments. Y.X.Y., L.C.C., Z.F.L., J.Z.J., and X.Y.K. wrote and revised the manuscript. All authors discussed the results and approved the manuscript.
Acknowledgments
We thank Dr. Cheng Liu from the Crop Research Institute, Shandong Academy of Agricultural Sciences, for providing some accessions of Sitopsis species, we thank Prof. Weining Song from Northwest A&F University for offering several accessions of T. dicoccoides and T. durum, and we also thank PhD students Hao Cheng, Zhan Li, and Zhe Yang of our team for their help with the heat stress experiment. No conflict of interest is declared.
Published: February 28, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Zefu Lu, Email: luzefu@caas.cn.
Xu Liu, Email: liuxu03@caas.cn.
Jizeng Jia, Email: jiajizeng@caas.cn.
Xiuying Kong, Email: kongxiuying@caas.cn.
Supplemental information
References
- Alvarez J.B., Guzman C. Interspecific and intergeneric hybridization as a source of variation for wheat grain quality improvement. Theor. Appl. Genet. 2018;131:225–251. doi: 10.1007/s00122-017-3042-x. [DOI] [PubMed] [Google Scholar]
- Anikster Y., Manisterski J., Long D.L., Leonard K.J. Resistance to leaf rust, stripe rust, and stem rust in Aegilops spp. in Israel. Plant Dis. 2005;89:303–308. doi: 10.1094/PD-89-0303. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium IWGSC, Appels R., Eversole K., Feuillet C., Keller B., Rogers J., Stein N., Pozniak C.J., Choulet F., Distelfeld A., et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361:eaar7191. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- Avni R., Lux T., Minz-Dub A., Millet E., Sela H., Distelfeld A., Deek J., Yu G., Steuernagel B., Pozniak C., et al. Genome sequences of three Aegilops species of the section Sitopsis reveal phylogenetic relationships and provide resources for wheat improvement. Plant J. 2022;110:179–192. doi: 10.1111/tpj.15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awlachew Z.T., Singh R., Kaur S., Bains N.S., Chhuneja P. Transfer and mapping of the heat tolerance component traits of Aegilops speltoides in tetraploid wheat Triticum durum. Mol. Breeding. 2016;36:78. doi: 10.1007/s11032-016-0499-2. [DOI] [Google Scholar]
- Bellon M.R. The dynamics of crop infraspecific diversity: a conceptual framework at the farmer level. Econ. Bot. 1996;50:26–39. doi: 10.1007/Bf02862110. [DOI] [Google Scholar]
- Brinton J., Ramirez-Gonzalez R.H., Simmonds J., Wingen L., Orford S., Griffiths S., 10 Wheat Genome Project. Haberer G., Spannagl M., Walkowiak S., et al. A haplotype-led approach to increase the precision of wheat breeding. Commun. Biol. 2020;3:712. doi: 10.1038/s42003-020-01413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci A., D'Ovidio R., Tanzarella O.A., Ceoloni C., Porceddu E. Identification of molecular markers linked to Pm13, an Aegilops longissima gene conferring resistance to powdery mildew in wheat. Theor. Appl. Genet. 1999;98:448–454. doi: 10.1007/s001220051090. [DOI] [Google Scholar]
- Charng Y.Y., Liu H.C., Liu N.Y., Hsu F.C., Ko S.S. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 2006;140:1297–1305. doi: 10.1104/pp.105.074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier F., Foulkes J., Hirel B., Gouache D., Moënne-Loccoz Y., Le Gouis J. Breeding for increased nitrogen-use efficiency: a review for wheat (T. aestivum L.) Plant Breed. 2016;135:255–278. doi: 10.1111/pbr.12371. [DOI] [Google Scholar]
- Cui L., Ren Y., Murray T.D., Yan W., Guo Q., Niu Y., Sun Y., Li H. Development of perennial wheat through hybridization between wheat and wheatgrasses: a review. Engineering. 2018;4:507–513. doi: 10.1016/j.eng.2018.07.003. [DOI] [Google Scholar]
- Daud H.M., Gustafson J.P. Molecular evidence for Triticum speltoides as a B-genome progenitor of wheat (Triticum aestivum) Genome. 1996;39:543–548. doi: 10.1139/g96-069. [DOI] [PubMed] [Google Scholar]
- De Bie T., Cristianini N., Demuth J.P., Hahn M.W. café: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J., Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorák J., Terlizzi P., Zhang H.B., Resta P. The evolution of polyploid wheats: identification of the A genome donor species. Genome. 1993;36:21–31. doi: 10.1139/g93-004. [DOI] [PubMed] [Google Scholar]
- El Baidouri M., Murat F., Veyssiere M., Molinier M., Flores R., Burlot L., Alaux M., Quesneville H., Pont C., Salse J. Reconciling the evolutionary origin of bread wheat (Triticum aestivum) New Phytol. 2017;213:1477–1486. doi: 10.1111/nph.14113. [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A.A. Genome evolution due to allopolyploidization in wheat. Genetics. 2012;192:763–774. doi: 10.1534/genetics.112.146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazee A.C., Pertea G., Jaffe A.E., Langmead B., Salzberg S.L., Leek J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015;33:243–246. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Chen L., Wang Y., Li M., Yang Z., Qiu L., Yan B., Ren Z., Tang Z. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 2015;5:10552. doi: 10.1038/srep10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaurav K., Arora S., Silva P., Sánchez-Martín J., Horsnell R., Gao L., Brar G.S., Widrig V., John Raupp W., Singh N., et al. Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol. 2022;40:422–431. doi: 10.1038/s41587-021-01058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S., Scornavacca C., Dainat J., Burgarella C., Viader V., Ardisson M., Sarah G., Santoni S., David J., Ranwez V. Pervasive hybridizations in the history of wheat relatives. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górny A.G., Garczyński S. Nitrogen and phosphorus efficiency in wild and cultivated species of wheat. J. Plant Nutr. 2008;31:263–279. doi: 10.1080/01904160701853878. [DOI] [Google Scholar]
- Haider N. The origin of the B-genome of bread wheat (Triticum aestivum L.) Russ. J. Genet. 2013;49:263–274. doi: 10.1134/S1022795413030071. [DOI] [PubMed] [Google Scholar]
- Heal G., Walker B., Levin S., Arrow K., Dasgupta P., Daily G., Ehrlich P., Maler K.G., Kautsky N., Lubchenco J., et al. Genetic diversity and interdependent crop choices in agriculture. Resour. Energy Econ. 2004;26:175–184. doi: 10.1016/j.reseneeco.2003.11.006. [DOI] [Google Scholar]
- Hsam S.L.K., Lapochkina I.F., Zeller F.J. Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 8. Gene Pm32 in a wheat-Aegilops speltoides translocation line. Euphytica. 2003;133:367–370. doi: 10.1023/A:1025738513638. [DOI] [Google Scholar]
- Huang S., Steffenson B.J., Sela H., Stinebaugh K. Resistance of Aegilops longissima to the rusts of wheat. Plant Dis. 2018;102:1124–1135. doi: 10.1094/PDIS-06-17-0880-RE. [DOI] [PubMed] [Google Scholar]
- Jayakodi M., Padmarasu S., Haberer G., Bonthala V.S., Gundlach H., Monat C., Lux T., Kamal N., Lang D., Himmelbach A., et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature. 2020;588:284–289. doi: 10.1038/s41586-020-2947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J., Devos K.M., Chao S., Miller T.E., Reader S.M., Gale M.D. RFLP-based maps of the homoeologous group-6 chromosomes of wheat and their application in the tagging of Pm12, a powdery mildew resistance gene transferred from Aegilops speltoides to wheat. Theor. Appl. Genet. 1996;92:559–565. doi: 10.1007/BF00224558. [DOI] [PubMed] [Google Scholar]
- Kilian B., Ozkan H., Deusch O., Effgen S., Brandolini A., Kohl J., Martin W., Salamini F. Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol. Biol. Evol. 2007;24:217–227. doi: 10.1093/molbev/msl151. [DOI] [PubMed] [Google Scholar]
- Kishii M. An update of recent use of Aegilops species in wheat breeding. Front. Plant Sci. 2019;10:585. doi: 10.3389/fpls.2019.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T., Li G., Wang H., Yu Z., Chen Q., Yang E., Fu S., Tang Z., Yang Z. Physical location of tandem repeats in the wheat genome and application for chromosome identification. Planta. 2019;249:663–675. doi: 10.1007/s00425-018-3033-4. [DOI] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Dong Z., Ma C., Xia Q., Tian X., Sehgal S., Koo D.H., Friebe B., Ma P., Liu W. A spontaneous wheat-Aegilops longissima translocation carrying Pm66 confers resistance to powdery mildew. Theor. Appl. Genet. 2020;133:1149–1159. doi: 10.1007/s00122-020-03538-8. [DOI] [PubMed] [Google Scholar]
- Li L.F., Zhang Z.B., Wang Z.H., Li N., Sha Y., Wang X.F., Ding N., Li Y., Zhao J., Wu Y., et al. Genome sequences of five Sitopsis species of Aegilops and the origin of polyploid wheat B subgenome. Mol. Plant. 2022;15:488–503. doi: 10.1016/j.molp.2021.12.019. [DOI] [PubMed] [Google Scholar]
- Liu B., Segal G., Rong J.K., Feldman M. A chromosome-specific sequence common to the B genome of polyploid wheat and Aegilops searsii. Plant Systemat. Evol. 2003;241:55–66. doi: 10.1007/s00606-003-0015-0. [DOI] [Google Scholar]
- Liu W., Koo D.H., Xia Q., Li C., Bai F., Song Y., Friebe B., Gill B.S. Homoeologous recombination-based transfer and molecular cytogenetic mapping of powdery mildew-resistant gene Pm57 from Aegilops searsii into wheat. Theor. Appl. Genet. 2017;130:841–848. doi: 10.1007/s00122-017-2855-y. [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang H., Jiang Z., Wang W., Xu R., Wang Q., Zhang Z., Li A., Liang Y., Ou S., et al. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature. 2021;590:600–605. doi: 10.1038/s41586-020-03091-w. [DOI] [PubMed] [Google Scholar]
- Luo M.C., Gu Y.Q., Puiu D., Wang H., Twardziok S.O., Deal K.R., Huo N., Zhu T., Wang L., Wang Y., et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature. 2017;551:498–502. doi: 10.1038/nature24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestra B., Naranjo T. Homoeologous relationships of Aegilops speltoides chromosomes to bread wheat. Theor. Appl. Genet. 1998;97:181–186. doi: 10.1007/s001220050883. [DOI] [Google Scholar]
- Marais G.F., McCallum B., Marais A.S. Leaf rust and stripe rust resistance genes derived from Aegilops sharonensis. Euphytica. 2006;149:373–380. doi: 10.1007/s10681-006-9092-9. [DOI] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y., Yoshida K., Mizuno N., Nasuda S., Sato K., Takumi S. Origin of wheat B-genome chromosomes inferred from RNA sequencing analysis of leaf transcripts from section Sitopsis species of Aegilops. DNA Res. 2019;26:171–182. doi: 10.1093/dnares/dsy047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N., Liu Y.G., Tsunewaki K. Wheat phylogeny determined by RFLP analysis of nuclear DNA.2. wild tetraploid wheats. Theor. Appl. Genet. 1995;90:129–134. doi: 10.1007/Bf00221006. [DOI] [PubMed] [Google Scholar]
- Mujeeb-Kazi A., Kazi A.G., Dundas I., Rasheed A., Ogbonnaya F., Kishii M., Bonnett D., Wang R.R.C., Xu S., Chen P., et al. Genetic diversity for wheat improvement as a conduit to food security. Adv. Agron. 2013;122:179–257. doi: 10.1016/B978-0-12-417187-9.00004-8. [DOI] [Google Scholar]
- Olivera P.D., Kolmer J.A., Anikster Y., Steffenson B.J. Resistance of sharon goatgrass (Aegilops sharonensis) to fungal diseases of wheat. Plant Dis. 2007;91:942–950. doi: 10.1094/PDIS-91-8-0942. [DOI] [PubMed] [Google Scholar]
- Peng H., Wang K., Chen Z., Cao Y., Gao Q., Li Y., Li X., Lu H., Du H., Lu M., et al. MBKbase for rice: an integrated omics knowledgebase for molecular breeding in rice. Nucleic Acids Res. 2020;48:D1085–D1092. doi: 10.1093/nar/gkz921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J.H., Sun D., Nevo E. Domestication evolution, genetics and genomics in wheat. Mol. Breeding. 2011;28:281–301. doi: 10.1007/s11032-011-9608-4. [DOI] [Google Scholar]
- Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G., Seberg O., Yde M., Berthelsen K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum) Mol. Phylogenet. Evol. 2006;39:70–82. doi: 10.1016/j.ympev.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Petersen S., Lyerly J.H., Worthington M.L., Parks W.R., Cowger C., Marshall D.S., Brown-Guedira G., Murphy J.P. Mapping of powdery mildew resistance gene Pm53 introgressed from Aegilops speltoides into soft red winter wheat. Theor. Appl. Genet. 2015;128:303–312. doi: 10.1007/s00122-014-2430-8. [DOI] [PubMed] [Google Scholar]
- Pour-Aboughadareh A., Kianersi F., Poczai P., Moradkhani H. Potential of wild relatives of wheat: ideal genetic resources for future breeding programs. Agronomy. 2021;11:1656. doi: 10.3390/agronomy11081656. [DOI] [Google Scholar]
- Pradhan G.P., Prasad P.V.V., Fritz A.K., Kirkham M.B., Gill B.S. High temperature tolerance in Aegilops species and its potential transfer to wheat. Crop Sci. 2012;52:292–304. doi: 10.2135/cropsci2011.04.0186. [DOI] [Google Scholar]
- Rasheed A., Mujeeb-Kazi A., Ogbonnaya F.C., He Z., Rajaram S. Wheat genetic resources in the post-genomics era: promise and challenges. Ann. Bot. 2018;121:603–616. doi: 10.1093/aob/mcx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamini F., Ozkan H., Brandolini A., Schäfer-Pregl R., Martin W. Genetics and geography of wild cereal domestication in the Near East. Nat. Rev. Genet. 2002;3:429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- Shewry P.R., Hey S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015;4:178–202. doi: 10.1002/fes3.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung D.Y., Kaplan F., Lee K.J., Guy C.L. Acquired tolerance to temperature extremes. Trends Plant Sci. 2003;8:179–187. doi: 10.1016/S1360-1385(03)00047-5. [DOI] [PubMed] [Google Scholar]
- Tang Z., Yang Z., Fu S. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 2014;55:313–318. doi: 10.1007/s13353-014-0215-z. [DOI] [PubMed] [Google Scholar]
- Tanno K.I., Willcox G. How fast was wild wheat domesticated? Science. 2006;311:1886. doi: 10.1126/science.1124635. [DOI] [PubMed] [Google Scholar]
- Wang J., Luo M.C., Chen Z., You F.M., Wei Y., Zheng Y., Dvorak J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013;198:925–937. doi: 10.1111/nph.12164. [DOI] [PubMed] [Google Scholar]
- Waters E.R., Lee G.J., Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J. Exp. Bot. 1996;47:325–338. doi: 10.1093/jxb/47.3.325. [DOI] [Google Scholar]
- Whitford R., Fleury D., Reif J.C., Garcia M., Okada T., Korzun V., Langridge P. Hybrid breeding in wheat: technologies to improve hybrid wheat seed production. J. Exp. Bot. 2013;64:5411–5428. doi: 10.1093/jxb/ert333. [DOI] [PubMed] [Google Scholar]
- Wicker T., Gundlach H., Spannagl M., Uauy C., Borrill P., Ramírez-González R.H., De Oliveira R., International Wheat Genome Sequencing Consortium. Mayer K.F.X., Paux E., Choulet F. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 2018;19:103. doi: 10.1186/s13059-018-1479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhang X., Wu L., Zhang L., Liu G., Xia C., Liu X., Kong X. Transcriptome profiling of developing leaf and shoot apices to reveal the molecular mechanism and co-expression genes responsible for the wheat heading date. BMC Genom. 2021;22:468. doi: 10.1186/s12864-021-07797-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu Y.X., Zhang N., Hu B., Jin T., Xu H., Qin Y., Yan P., Zhang X., Guo X., et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019;37:676–684. doi: 10.1038/s41587-019-0104-4. [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhang M., Zhu X., Cao Y., Sun Q., Ma G., Chao S., Yan C., Xu S.S., Cai X. Molecular cytogenetic and genomic analyses reveal new insights into the origin of the wheat B genome. Theor. Appl. Genet. 2018;131:365–375. doi: 10.1007/s00122-017-3007-0. [DOI] [PubMed] [Google Scholar]
- Zhao G., Zou C., Li K., Wang K., Li T., Gao L., Zhang X., Wang H., Yang Z., Liu X., et al. The Aegilops tauschii genome reveals multiple impacts of transposons. Nat. Plants. 2017;3:946–955. doi: 10.1038/s41477-017-0067-8. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhao X., Li Y., Xu J., Bi A., Kang L., Xu D., Chen H., Wang Y., Wang Y.G., et al. Triticum population sequencing provides insights into wheat adaptation. Nat. Genet. 2020;52:1412–1422. doi: 10.1038/s41588-020-00722-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Ae. speltoides genome assembly was deposited in NCBI GenBank under BioProject accession number PRJNA802349. The raw sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under the BioProject accession numbers PRJNA893435 (Nanopore raw data), PRJNA802349 (WGS), and PRJNA802730 (Hi-C). The RNA-seq data, whole-exome resequencing data, and gVCF files were saved in the GEO database with accession number GSE197593, and the whole-genome sequencing data were saved in the SRA database with accession number PRJNA806672. We also deposited the reference genome of Aegilops speltoides and annotation file to ScienceDB (https://www.scidb.cn/en/s/VFrU7z). The whole-genome resequencing data, whole-exome resequencing data, and RNA-seq data were saved in the China National Genomics Data Center with the accession number CRA009743.