Abstract
Heterosis refers to the superior performance of a hybrid compared with its parental lines. Although several genetic and molecular models have been proposed to explain heterosis, it remains unclear how hybrid cells integrate complementary gene expression or activity to drive heterotic growth. In this work, we show that accumulation of growth-promoting and energy metabolism proteins, enhanced energy metabolism activities, and increased protein lysine acetylation were associated with superior growth of the panicle meristem in the elite hybrid rice Shanyou 63 relative to its parental varieties. Metabolism of nuclear/cytosolic acetyl-coenzyme A was also enhanced in the hybrid, which paralleled increases in histone H3 acetylation to selectively target the expression of growth-promoting and metabolic genes. Lysine acetylation of cellular proteins, including TARGET OF RAPAMYCIN complex 1, ribosomal proteins, and energy metabolism enzymes, was also augmented and/or remodeled to modulate their activities. The data indicate that an enhanced network of energy-producing metabolic activity and growth-promoting histone acetylation/gene expression in the hybrid could contribute to its superior growth rate and may constitute a model to explain heterosis.
Key words: heterosis, TORC1, r-protein, lysine acetylation, energy metabolism, hybrid rice
This study revealed an enhanced regulatory network in elite hybrid rice that involves energy metabolism, lysine acetylation of histones and cellular proteins, growth-promoting signaling pathways, and epigenetic activation of genes related to cell division and energy metabolism. This reinforcing feedback loop likely contributes to hybrid vigor.
Introduction
Heterosis refers to the superior performance of an F1 hybrid compared with its parental lines in terms of growth, yield, and adaptation to the environment. Although heterosis is widely utilized in agriculture and several quantitative genetics models have been proposed to explain heterosis, the molecular basis underlying this phenomenon remains elusive (Chen, 2013; Goff and Zhang, 2013; Ouyang et al., 2022). It has been suggested that functional complementation between dominant superior loci in hybrids triggers heterosis (Hua et al., 2003; Huang et al., 2016) and that allelic-specific expression drives expression complementation in hybrids and is associated with heterosis (Baldauf et al., 2018; Shao et al., 2019). Heterotic quantitative trait loci mapping and genome-wide association studies in rice have identified numerous superior alleles that contribute to yield-related heterosis (Huang et al., 2015, 2016). Single genes responsible for heterosis have also been discovered, including rice Ghd7 for yield heterosis (Xue et al., 2008), tomato SINGLE FLOWER TRUSS for fruit yield heterosis (Krieger et al., 2010), and Arabidopsis CCA1, a central circadian oscillator gene, for bacterial defense heterosis (Yang et al., 2021). Several other studies have suggested that both transcriptional and epigenetic variations (including DNA methylation, small RNAs, and histone modifications) in the hybrid may play a role in heterosis (Groszmann et al., 2011; Kawanabe et al., 2016; Yang et al., 2016; Ma et al., 2021).
Although quantitative genetic models such as dominance, overdominance, and epistasis have been proposed to explain heterosis, studies of genomic architecture in Arabidopsis have revealed only a limited contribution of genome-wide heterozygosity of hybrids to heterosis (Yang et al., 2017). No locus shows an obvious overdominance effect in hybrids, whereas the accumulation of superior alleles of genes involved in metabolic and cellular processes improves the development and growth of hybrids (Yang et al., 2017). Other studies have identified regulatory genes of circadian rhythm and stress responses and enhanced salicylic acid biosynthesis as contributing to growth heterosis in Arabidopsis (Yang et al., 2021). Recent work comparing time-series transcriptomes of an Arabidopsis hybrid and its parents indicated that expression complementation between fundamental biological pathways such as cell division and photosynthesis contributes to biomass heterosis (Liu et al., 2021). However, whether transcript levels reflect protein accumulation and function in heterosis remains to be determined. How complemented fundamental biological pathways are coordinated to drive heterosis growth is also unclear. In maize, the temporal regulation of metabolites and proteins of photosynthetic and photorespiratory pathways was shown to contribute to heterosis (Li et al., 2020). Nonetheless, it remains to be determined how cells integrate metabolic and protein variations for heterotic growth.
The elite hybrid rice Shanyou 63 (SY63) generated from a cross between Zhenshan 97 (ZS97; indica I) and Minghui 63 (MH63; indica II) exhibits superiority for yield and a large number of agronomic traits and has been widely cultivated in China (Zhang et al., 2016; Xie and Zhang, 2018). This particular hybrid rice system has also been used as a model for genetic and molecular studies of heterosis (Yu et al., 1997; Hua et al., 2003; Zhou et al., 2012; Xie et al., 2015; Shao et al., 2019). The availability of high-quality reference genome sequences of the ZS97 and MH63 parental lines and their detailed comparative annotation, as well as relevant transcriptomes and epigenomes, provide a unique opportunity to test theoretical models and pathways of heterosis (Zhang et al., 2016; Shao et al., 2019; Ma et al., 2021).
In this work, we performed an integrative analysis of proteomes, metabolomes, transcriptomes, epigenomes, and protein lysine acetylomes, together with biochemical tests of SY63 and its parental lines. The results revealed an inter-dependent network of enhanced energy-producing metabolism and increased histone acetylation and expression of growth-promoting and metabolism genes in the hybrid, suggesting that inter-connected growth-promoting cellular processes are implicated in the superior growth rate of the hybrid.
Results
Comparative proteomics of young panicles from elite hybrid rice SY63 and its parental lines
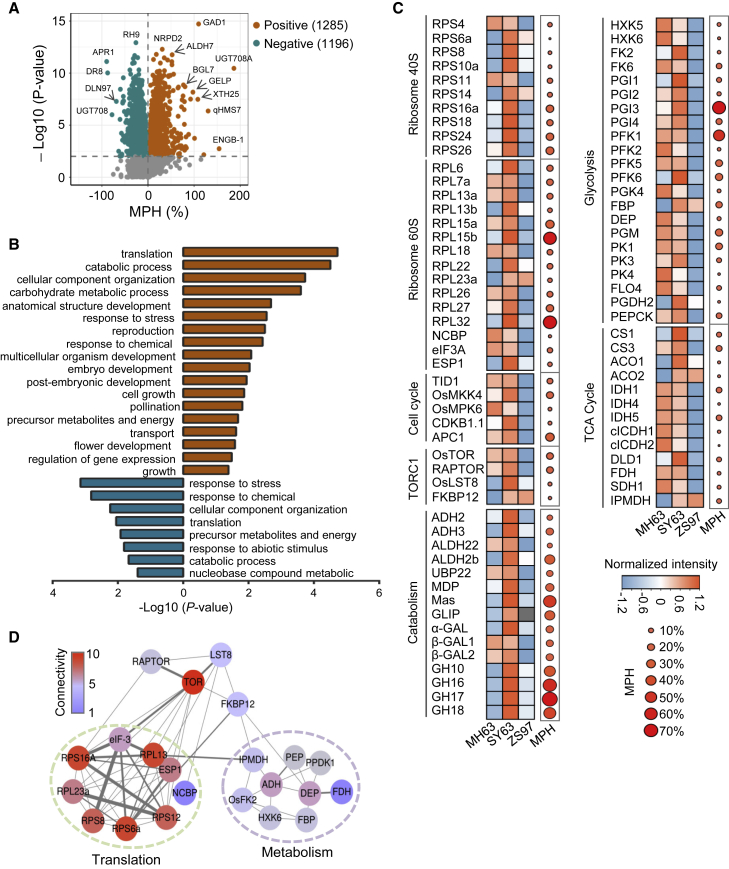
Panicle size is the major heterotic yield trait of the hybrid rice SY63 (Xing and Zhang, 2010). To investigate whether heterotic panicle growth is related to accumulation of specific proteins, we performed quantitative proteomic analysis of panicle (inflorescence) meristems (at the 2-mm length stage) of the hybrid and the parental lines using label-free liquid chromatography tandem mass spectrometry (LC–MS/MS) (Supplemental Figure 1A). Three biological replicates were tested for each genotype (Pearson’s r = 0.99–1) (Supplemental Figure 1B). A total of 6952 proteins and 43 713 peptide fragments were identified from the samples, and 5725 of the proteins could be accurately quantified (Supplemental Figure 1C; Supplemental Table 1; Supplemental Dataset 1). Principal-component analysis (PCA) revealed that the SY63 proteome differed from that of the parental lines and was closer to that of MH63 than that of ZS97 (Supplemental Figure 2A). The numbers of total proteins identified in the three genotypes were comparable, and 4773 of the proteins were detected in all genotypes (Supplemental Figure 2B). There were 999 differentially accumulated proteins (DAPs; >1.5-fold, P < 0.05) between the parental lines, 640 DAPs between SY63 and ZS97, and 175 DAPs between SY63 and MH63 (Supplemental Figure 2C).
Using the formula for mid-parent value (MPV) heterosis (MPH) (MPH = [(F1 − MPV)/MPV]×100%) and a P < 0.01 cutoff (Student’s t-test) (Li et al., 2020), we identified 1285 proteins with positive MPH and 1196 proteins with negative MPH in the hybrid (Figure 1A; Supplemental Dataset 2). The negative-MPH proteins were mainly enriched in stress-response functions (Figure 1B), represented by abscisic stress ripening proteins 1 and 5, heat shock proteins, ascorbate peroxidases, peroxidases, superoxide dismutases, Xa21 binding protein 21, Xa21 binding protein 25, and pathogenesis-related protein 10 (Supplemental Figure 3B). These data were consistent with previous findings that stress-response genes are downregulated in Arabidopsis hybrids relative to their parents (Yang et al., 2017).
Figure 1.
Identification of proteins with mid-parent heterosis (MPH) in the hybrid SY63.
(A) Volcano plot showing the positive- and negative-MPH proteins identified in the hybrid SY63. MPH = [(F1 − MPV)/MPV]×100%, MPV = (P1 + P2)/2, P < 0.01, two-tailed t-test.
(B) Biological process GO terms enriched in positive-MPH (brown bars) and negative-MPH (blue bars) proteins in SY63.
(C) Heatmap showing the accumulation of ribosomal proteins, translational elongation factors, TOR signaling components, and key enzymes involved in glycolysis, TCA, and catabolic pathways in panicle meristems of MH63, ZS97, and SY63.
(D) The protein–protein interaction network of TORC1 includes ribosomal proteins and enzymes involved in energy metabolism.
The positive-MPH proteins were enriched in translation, catabolism, carbohydrate metabolism, and reproductive development functions (Figure 1B). Among the positive-MPH proteins involved in development (Supplemental Figure 3A), many were previously reported to function in the control of inflorescence development and heading date, such as GF14c, GF14f, MADS50, ABERRANT PANICLE ORGANIZATION 1 and 2, Heading date 6, VERNALIZATION INSENSITIVE 3-LIKE 2, and Fertilization Independent Endosperm 2 (Purwestri et al., 2009; Ikeda-Kawakatsu et al., 2012; Wang et al., 2013; Li et al., 2014; Huang et al., 2021); gametophyte development, such as API5-INTERACTING PROTEIN2, DWARF AND RUNTISH SPIKELET1 and 2, Cannot Reach the Roof 1, and Rice G protein beta subunit 1 (Li et al., 2011a; Song et al., 2016; Pu et al., 2017); and panicle architecture, such as PLASTOCHRON1 (PLA1), PLA2, and PLA3 and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 14 and 17 (Kawakatsu et al., 2009; Jiao et al., 2010; Wang et al., 2021a). In addition, proteins involved in regulating grain size and starch synthesis, such as GRAIN LENGTH 3.1, GRAIN SIZE5, SLENDER GRAIN, and Starch synthase IIb and IIc, also showed positive MPH in the young panicle of the hybrid (Supplemental Figure 3A) (Li et al., 2011b; Qi et al., 2012; Feng et al., 2016). Increases in the levels of these proteins are potentially related to heterotic growth of the hybrid panicle.
Plant hormones such as cytokinin and auxin play important roles in rice panicle development (Kurakawa et al., 2007; Wang et al., 2018). The protein levels of cytokinin biosynthetic phosphoribohydrolases LOGL1 and LOGL7 were increased in the hybrid panicle, whereas those of LOGL4 and the catabolic cytokinin oxidase CKX11 were decreased (Supplemental Figure 3A). The levels of proteins involved in auxin polar transport such as PIN1b and PIN1d were decreased in the hybrid, and those of several auxin, gibberellin, and jasmonic acid signaling proteins were increased (Supplemental Figure 3A).
Accumulation of growth-promoting and energy metabolism proteins in the hybrid panicle
Among the positive-MPH proteins were many translational proteins, including ribosomal proteins (r-proteins), elongation factor eIF3A, eukaryotic peptide chain release factor 1, and novel cap-binding protein (Figure 1C). Proteins involved in cell proliferation and the cell cycle, such as TWISTED DWARF1, mitogen-activated protein kinase kinase 4 (OsMKK4), mitogen-activated protein kinase 6 (OsMPK6), cyclin-dependent kinase B1-1, and anaphase-promoting complex 1, also showed positive MPH in the hybrid (Figure 1C). Interestingly, protein levels of key TOR complex 1 (TORC1) subunits, including TOR, PAPTOR, and LST8, were also increased in the hybrid (Figure 1C) (Xiong and Sheen, 2014). Prediction of TORC1 protein–protein interactions in rice using the STRING database (v.11.5, https://cn.string-db.org) revealed several translational proteins such as eS6z (RPS6a), eS8y (RPS8), eS12z (RPS12), eL13z (RPL13), uS9z (RPS16a), and uL23z (RPL23a) (Scarpin et al., 2022a); novel cap-binding protein; and metabolic enzymes such as 3-isopropylmalate dehydrogenase and dehydratase-enolase-phosphatase (Figure 1D). Levels of these proteins also showed a positive MPH (Figure 1C).
Metabolic enzymes with positive MPH were mainly related to energy-producing pathways such as glycolysis (hexokinase-5 and -6, glucose-6-phosphate isomerase 1/2/3/4, phosphofructokinase 1 [PFK1] and PFK2/5/6, and pyruvate kinase 1 [PK1] and PK3/4); the tricarboxylic acid (TCA) cycle (citrate synthase 1 [CS1] and CS2, aconitase 1 [ACO1] and ACO2, isocitrate dehydrogenase 1 [IDH1] and IDH4/5, fumarate dehydrogenase, and succinate dehydrogenase 1); and other carbohydrate catabolic processes (alcohol dehydrogenases 2/3, acetaldehyde dehydrogenases 2b/22, and glycosyl hydrolase 10 [GH10] and GH16/17/18) (Figure 1C). These data indicated that growth-promoting protein levels and energy-producing metabolism were enhanced in the hybrid.
Metabolomics revealed higher cellular energy levels in the hybrid
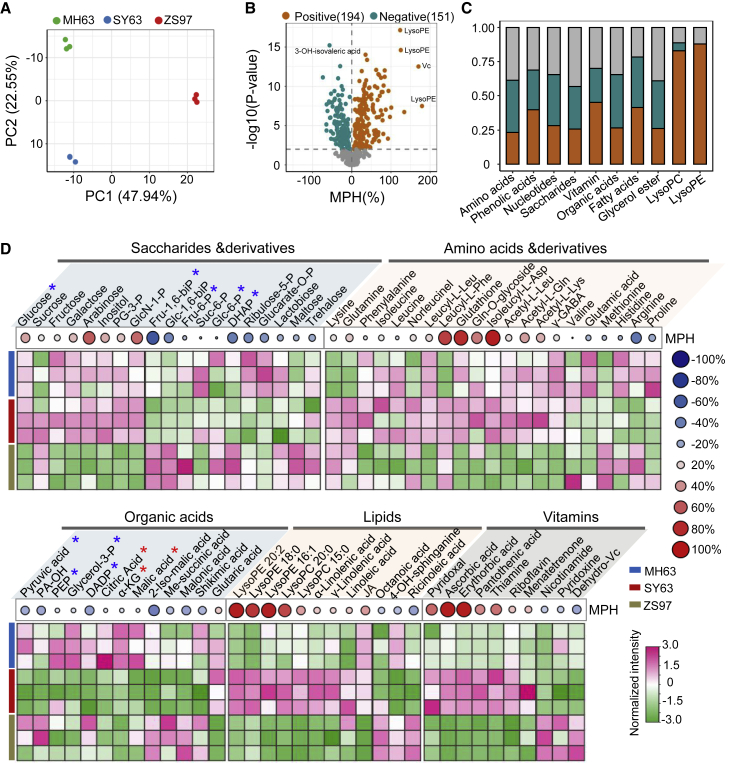
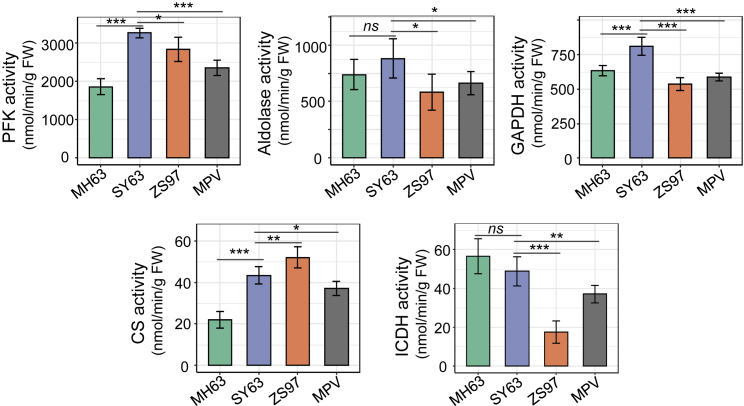
Cellular energy metabolites are the main upstream signals for activation of the TOR pathway that, in turn, modulates cellular metabolism and activates growth-related gene expression and/or protein synthesis (Xiong et al., 2013; Dobrenel et al., 2016a). To determine whether the accumulation of growth-promoting and energy metabolism proteins in the hybrid was associated with the production of energy metabolites, we analyzed the panicle meristem metabolomes of SY63 and its parental lines by ultra-performance LC–MS/MS (UPLC–MS/MS). Three biological replicates were performed for each genotype (Figure 2A; Supplemental Figure 4A and 4B). PCA indicated that the SY63 metabolome was distinct from that of its parents but closer to that of MH63 (Figure 2A). A total of 581 non-redundant metabolites from different categories were identified in the panicles (Supplemental Figure 4C; Supplemental Dataset 3). In the hybrid, 194 metabolites with a positive MPH and 151 with a negative MPH were identified in the different categories (Figure 2B; Supplemental Dataset 4). Among the saccharides, sugars (especially glucose, arabinose, and galactose) showed clear positive MPH, whereas phosphorylated or activated sugar molecules (glucose-6-P, fructose-6-P, fructose-1,6-BP, glucose-1,6-BP), glycerol-3-P, and glyceraldehyde-3-phosphate, which are substrates or intermediate products of glycolysis, displayed negative MPH (Figure 2D). Interestingly, nearly all lysophosphatidylcholine and lysophosphatidylethanolamine molecules showed positive MPH in the hybrid (Figure 2C and 2D). Lysophosphatidylcholines and lysophosphatidylethanolamines have been shown to promote plant growth (Jasieniecka-Gazarkiewicz et al., 2017), and their accumulation may be related to heterotic growth of the hybrid. By contrast, organic acids (pyruvate, succinate, α-ketoglutaric acid, malate), which are substrate and intermediate products of the TCA cycle, displayed negative MPH (Figure 2D). These data suggested that glycolysis and TCA activities were higher in the hybrid, consistent with the increased protein abundance of rate-limiting enzymes of glycolysis (PFK, PK) and the TCA cycle (CS and IDH/cICDH) (Figure 1C). Tests of glycolytic enzyme activities (PFK, aldolase, glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) and TCA enzyme activities (CS and ICDH) confirmed the hypothesis (Figure 3). Together, these data indicated a higher energy-producing metabolism in the hybrid. In addition, levels of amino acids (in particular, derivatives of leucine, isoleucine, and glutamine) and vitamins (such as ascorbic acid and erythorbic acid) were clearly increased in the hybrid (Figure 2D; Supplemental Dataset 4).
Figure 2.
Comparison of panicle meristem primary metabolomes between SY63 and its parental lines.
(A) PCA of the panicle meristem metabolomes of MH63, ZS97, and SY63. Three biological replicates per genotype are shown.
(B) Volcano plot showing the numbers of metabolites with positive and negative MPH identified in SY63.
(C) Percentage of the different categories of metabolites with positive and negative MPH.
(D) Heatmap showing differential accumulation of saccharides, amino acids and their derivatives, organic acids, lipids, and vitamins in panicle meristems of MH63, ZS97, and SY63. Percentages of positive (red) and negative (blue) MPH are indicated.
Figure 3.
Activities of glycolytic (PFK, aldolase, and GAPDH) and TCA (CS and ICDH) enzymes in panicles of MH63, ZS97, and SY63.
Proteins extracted from about 0.1 g rice panicle meristem tissues were tested for enzyme activities. Six biological replicates were used. Bars indicate the mean ± SD (n = 6). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ns, P > 0.05 (two-tailed t-test).
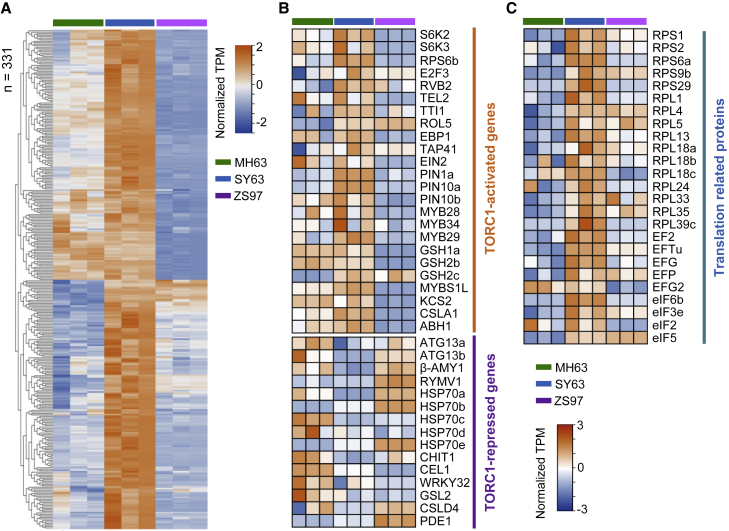
To determine whether the higher energy-producing metabolism detected in the hybrid was associated with a cellular energy–promoted gene expression program, we analyzed transcriptomic data from hybrid and parent panicles (Ma et al., 2021). Among the upregulated genes in the hybrid, 331 were related to energy functions, carbohydrate and lipid metabolism, and catabolic processes (Figure 4A). In addition, marker genes activated by a high energy state (or TORC1) were upregulated in the hybrid, whereas marker genes repressed by a high energy state (or TORC1) were downregulated (Figure 4B) (Xiong and Sheen, 2014; Brunkard et al., 2020; Fu et al., 2020). Twenty-five translational protein genes were also upregulated in the hybrid (Figure 4C). We validated the expression of 10 of the genes by quantitative real-time PCR (Supplemental Figure 5A). Finally, higher rRNA levels were detected in the hybrid (Supplemental Figure 5B), consistent with its higher r-protein levels.
Figure 4.
TORC1-regulated gene expression program is enhanced in the SY63 panicle meristem.
(A) Heatmap showing that the genes involved in energy metabolism or catabolic processes were upregulated in the young panicle of SY63. A total of 331 genes are shown, which were enriched in the GO terms generation of precursor metabolites and energy, catabolic process, carbohydrate metabolic process, and lipid metabolic process.
(B) Heatmap showing expression levels of marker genes activated or repressed by a high cellular energy state or TORC1 in panicles of MH63, SY63, and ZS97.
(C) Heatmap showing the transcript levels of 25 ribosomal protein genes in the panicle meristem of SY63 compared with its parental lines.
Histone H3 acetylation was increased and redistributed in the hybrid
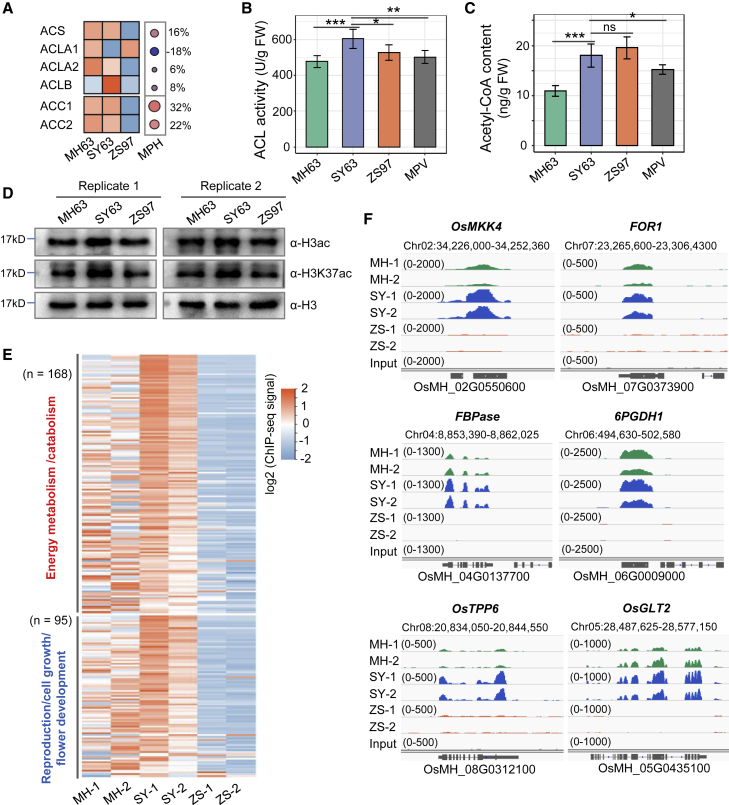
In yeast and mammalian cells, high cellular glucose (energy) levels fuel the production of cytoplasmic/nuclear acetyl-coenzyme A (CoA), which is produced mainly by ATP-citrate lyase (ACL). Accumulation of ACL-sourced acetyl-CoA stimulates histone acetylation of growth-promoting genes under nutrition-sufficient conditions (Wellen et al., 2009; Shen et al., 2015; Shi and Tu, 2015; Hsieh et al., 2022). Plant ACL has two subunits, ACLA and ACLB. In rice, ACLA is encoded by three genes and ACLB by one gene. The ACLB subunit showed a clear positive MPH, whereas the A subunit proteins showed both positive and negative MPH (Figure 5A). ACL activity was higher in the hybrid than in either parental line (Figure 5B), and acetyl-CoA synthase, which uses acetate to generate acetyl-CoA, also showed a positive MPH in the hybrid (Figure 5A). Protein levels of acetyl-CoA carboxylase, which consumes acetyl-CoA, also showed a positive MPH (Figure 5A), indicating a highly active acetyl-CoA metabolism in the hybrid. Nonetheless, the total acetyl-CoA level in the hybrid was higher than the MPV (Figure 5C).
Figure 5.
Acetyl-CoA content and histone H3 acetylation level were increased in the hybrid panicle meristem.
(A) Protein abundance of cytosolic acetyl-CoA biosynthetic and metabolic enzymes in the three genotypes. ACS, acetyl-CoA synthase; ACL, ATP-citrate lyase; ACC, acetyl-CoA carboxylase.
(B) ATP-citrate lyase activities in the three genotypes.
(C) Acetyl-CoA contents in the panicle meristems of the three genotypes. Bars in (B) and (C) indicate means ± SD (n = 6). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ns, P > 0.05 (two-tailed t-test).
(D) Immunoblots of acetylation levels of histone H3 and H3K37 in panicle meristems of MH63, SY63, and ZS97. Anti-H3 antibody was used as a loading control.
(E) Heatmap showing the H3K9 acetylation levels of energy metabolism and growth-promoting genes in the three genotypes.
(F) Integrative Genomics Viewer screenshot showing the H3K9 acetylation levels of six genes related to cell division (mitogen-activated protein kinase 4 and FOR) and metabolic processes (6PGDH1, OsGLT2, FBPase, and OsTPP6) in SY63 and the parental lines.
To test whether the higher acetyl-CoA metabolism stimulated histone acetylation in the hybrid, we performed immunoblot analysis and detected some increases in H3 acetylation in SY63 compared with its parents (Figure 5D), which were confirmed by MS (see below). We next performed chromatin immunoprecipitation sequencing (ChIP-seq) to analyze the distribution of H3K9 acetylation in the hybrid and parental genomes (Supplemental Figure 6A; Supplemental Table 2). We identified 20 216 H3K9ac-marked genes in SY63, 19 440 in MH63, and 17 481 in ZS97 replicates (Supplemental Figure 6B). Metaplots indicated that the overall genic H3K9 acetylation levels were similar between the hybrid and its parents (Supplemental Figure 6D). However, 1826 genes gained H3K9ac and a similar number of genes lost this mark in the hybrid compared with either parent (Supplemental Figure 6C; Supplemental Dataset 5). Changes in H3K9ac modification between the hybrid and its parents were positively correlated with changes in gene expression (Supplemental Figure 6E). We found that 168 of the 331 upregulated energy metabolism genes in SY63 also gained H3K9ac in the hybrid (Figure 5E; Supplemental Dataset 6). In addition, 95 growth-promoting genes also displayed higher H3K9ac in the hybrid than in the parents (Figure 5E and 5F; Supplemental Dataset 6). These data indicated a redistribution of the histone acetylation mark in favor of energy metabolism and growth-promoting genes in the hybrid.
The overall protein acetylation level was increased in the hybrid panicle
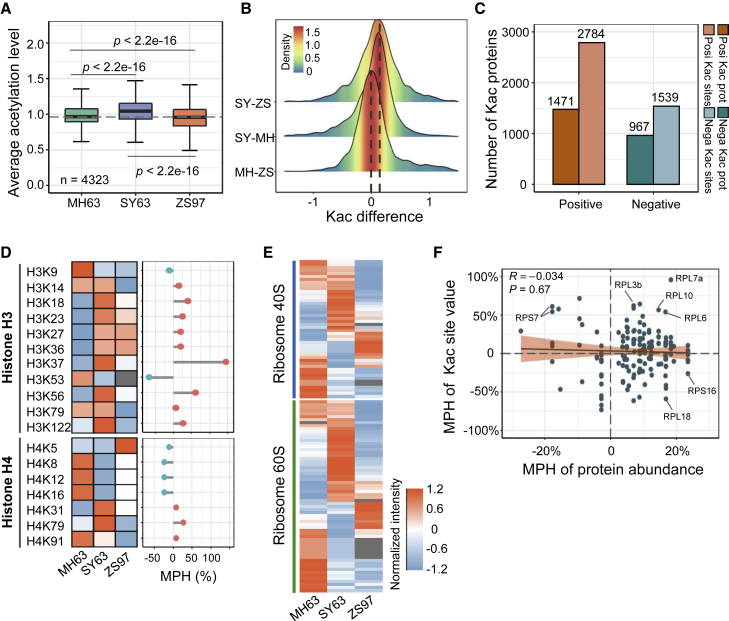
To further test whether the higher energy metabolism also stimulated cellular protein lysine acetylation, we performed quantitative acetyl-proteomic analysis using label-free LC–MS/MS (Supplemental Figure 1A). The three biological replicates of each genotype were highly correlated (Supplemental Figure 7A). PCA indicated that the SY63 acetylome was closer to that of MH63 than that of ZS97 (Supplemental Figure 7B). In the panicle, we detected 10 852 acetylated lysine (Kac) residues in 3994 proteins, and 7948 Kac sites in 3222 proteins were quantifiable (Supplemental Figure 7C; Supplemental Table 1). Normalization of the acetylome data with the proteome revealed 6069 Kac sites in 2290 proteins (Supplemental Table 1; Supplemental Dataset 7). These numbers were 2–5 times higher than those of previously identified Kac sites and acetylated proteins in rice seedlings or other tissues/organs (Supplemental Figure 7D). The identified Kac proteins accounted for approximately 60% of the total proteins (Supplemental Figure 7E), and more than half of the acetylated proteins had ≥2 Kac sites (Supplemental Figure 7C).
Gene Ontology (GO) enrichment showed that the Kac proteins were mainly involved in translation; carbohydrate, amino acid, and energy metabolism; and transcription and protein processing (Supplemental Figure 8A). Subcellular localization prediction showed that more than 60% of the Kac proteins were located in the cytoplasm and plastid, 20% in the nucleus, and 7% in the mitochondrion (Supplemental Figure 8B). Large numbers of Kac sites were detected in ARGONAUTE4a/b, elongation factor1-α and -2, sucrose synthase 1/2, and ACOs (ACO1/2) (Supplemental Figure 8C), and 148 transcription factors from 40 different families were found to be acetylated (Supplemental Figure 9). Many of the acetylated transcription factors had previously been reported to regulate plant development and responses to environmental cues. These data provide a comprehensive resource for the rice protein acetylome.
Boxplots revealed that the average level of overall protein Kac was significantly higher in SY63 than in MH63 and ZS97 (Figure 6A). Density-plot analysis also indicated a higher overall Kac level in SY63 than the parents (Figure 6B). There were 1471 proteins (2784 Kac sites) with positive MPH of Kac and 967 proteins (1539 Kac sites) with negative MPH (Figure 6C), indicating a remodeling of the protein acetylome in the hybrid. For histones, we found that acetylation of several histone lysine residues, especially H3K37 and H3K56, was enhanced in the hybrid (Figure 6D), consistent with the immunoblot data (Figure 5D). For proteins involved in translation, the most enriched GO term of the protein acetylome (Supplemental Figure 8A), we found that the Kac of r-proteins displayed opposite profiles in the hybrid and the parents: many Kac sites gained acetylation in the hybrid, whereas others lost this modification, indicating that there was extensive reconfiguration of r-protein Kac in the hybrid (Figure 6E). Several r-proteins showed net loss or gain of overall Kac relative to the MPV (Supplemental Dataset 8). Previous results have shown that r-protein Kac may affect translational efficiency or stability in rice (Xu et al., 2021). The abundance of many r-proteins was increased in the hybrid (Figure 1C). However, there was no general correlation between protein abundance and changes in Kac level, except for uL3y (RPL3b), eL6z (RPL6), eL8z (RPL7a), and uL16y (RPL10), which showed clear increases in both protein abundance and Kac levels, and uS9z (RPS16) and eL20z (RPL18), which showed increased protein abundance but decreased Kac levels (Figure 6F). Possibly, Kac may have different effects on the stability and/or function of different r-proteins. We speculate that reconfiguration of r-protein Kac, together with higher r-protein abundance, may alter translational activity in the hybrid. Recent results indicated that acetyl-CoA promotes mouse TORC1 activity through acetylation of the mTORC regulator Raptor (Son et al., 2019). We found that the TORC1 complex proteins were all acetylated and that higher Kac levels were detected in the hybrid at most Kac sites of these proteins (Supplemental Table 3).
Figure 6.
Comparison of panicle meristem protein acetylomes of MH63, ZS97, and SY63.
(A) Boxplot showing the differences in average lysine acetylation levels of the acetylated proteins identified in panicles of MH63, ZS97, and SY63. P values were calculated by the Wilcoxon rank-sum test.
(B) Density plot showing differences in total protein acetylation levels between the hybrid and its parents.
(C) Numbers of acetylated proteins and lysine acetylation (Kac) sites with positive and negative MPH identified in SY63 panicles.
(D) Heatmap showing differences in Kac levels of histones H3 and H4 in the three genotypes. Lollipop chart (right) shows the MPH levels of the indicated Kac sites in SY63.
(E) Heatmap showing the lysine acetylation levels of ribosome proteins in MH63, ZS97, and SY63.
(F) Scatterplot showing the relationship between ribosomal protein abundance and Kac levels.
Lysine acetylation has been shown to alter activities of TCA cycle and glycolysis enzymes (Zhang et al., 2017; Xu et al., 2021; Balparda et al., 2022). In the hybrid, several enzymes showed higher Kac, and others displayed lower Kac. For instance, lysine acetylation of PFK2/6/8, PK2/5/6, CS3, IDH2/4, acetyl-CoA synthase, and ACLA1/2 was increased in the hybrid (Supplemental Figure 10A–10C), whereas that of aldolase1/2, GAPDH1/2/5/6, cICDH2, and MDH2/3/5 was decreased (Supplemental Figure 10D and 10E). These enzymes all showed activities higher than the MPV in the hybrid (Figures 1C, 3, and 5B), suggesting that variation in Kac in the hybrid may be involved in modulating their activities. Together, these results suggest that Kac changes driven by high energy metabolism in the hybrid may feedback to regulate metabolic, translational, and TORC1 activities.
Discussion
Several molecular models have been proposed to explain heterosis (Chen, 2013; Goff and Zhang, 2013; Shao et al., 2019; Li et al., 2020; Liu et al., 2021). The present data suggest that an enhanced network of energy-producing metabolic activity and growth-promoting histone acetylation/gene expression is a mechanism that promotes superior growth of the hybrid.
The enhanced levels of translational (ribosome and elongation) and cell-cycle proteins, higher energy metabolism activities, and stimulated program of growth-promoting gene expression detected in SY63 (Figures 1C, 2, 3, 4, and 5; Supplemental Figure 5) suggest that TOR signaling may be enhanced in the hybrid. One of the key downstream activities of TOR is the control of the protein synthesis machinery. This is achieved, in part, via coordinated regulation of mRNAs that contain a terminal oligopyrimidine (CCUUUCU) tract (TOP) at their 5′ terminal end (Meyuhas and Kahan, 2015; Scarpin et al., 2022b). TOP motifs are found mainly in mRNAs that encode r-proteins and translation factors (Fonseca et al., 2014; Iadevaia et al., 2014). TOP-like motifs are abundant in the 5′ UTRs of r-protein mRNAs in plants (Dobrenel et al., 2016b; Scarpin et al., 2020, 2022b). TORC1 activates translation of TOP-containing mRNA, regulating ribosome activity of protein synthesis and thus cell growth (Meyuhas and Kahan, 2015). We speculate that TOR signaling may be enhanced to stimulate translation of TOP-containing mRNAs that contribute to cell growth in the hybrid, although it may also upregulate the expression of growth-promoting genes, including some translational protein genes (Figure 4C).
In addition to translational proteins, a number of developmental regulators and metabolic enzymes also showed increased abundance in the hybrid (Figure 1C; Supplemental Figure 3A). The increased protein levels of energy metabolism enzymes were consistent with the upregulation of energy metabolism genes and metabolic activities in the hybrid (Figures 3 and 4A). This is in line with previous results showing that regulation of the metabolome and proteome in the photosynthetic and photorespiratory pathways contributes to heterosis in maize (Li et al., 2020). The enhanced energy metabolism in hybrids is likely achieved through the activation of the energy-sufficient gene expression program (Figure 4A and 4B).
Enhanced cytosolic acetyl-CoA biosynthesis activities were associated with increases in histone H3 acetylation in the hybrid (Figures 5D, 6A, and 6B). In yeast, several regulatory and metabolic protein genes show dynamic histone acetylation precisely in phase with acetyl-CoA oscillations or in response to glucose levels in the medium (Cai et al., 2011). Here, the increase in H3 lysine acetylation was consistent with the glucose accumulation observed in the hybrid, corroborating results showing that histone acetylation can be triggered by high glucose uptake in yeast and mammalian cells and that increased H3 acetylation mainly targets growth-related genes (Wellen et al., 2009; Cai et al., 2011; Shi and Tu, 2015). Histone H3 lysine acetylation is tightly associated with gene activation. Although the overall level of H3K9 acetylation was not clearly altered, this mark appeared to be redistributed to genes related to metabolism and cell division in the hybrid genome (Figure 5E and 5F). Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are involved in histone acetylation homeostasis. The transcript levels of HAT and HDAC genes appeared unchanged in the hybrid relative to the parental lines (Supplemental Figure 11). However, the abundance of HAT proteins, especially GCN5, was clearly increased in the hybrid, whereas the abundance of HDACs (HDA711 and HDT701) was reduced (Supplemental Figure 11). GCN5 is a major H3 acetyltransferase and a transcriptional co-activator in plants (Benhamed et al., 2006; Zhou et al., 2017); its accumulation may contribute to increased H3 acetylation and higher expression of genes related to energy metabolism and growth in the hybrid.
The increase in overall lysine acetylation levels also attests to a higher energy metabolism in the hybrid cells. Translational and energy metabolism proteins are major targets of lysine acetylation of non-histone cellular proteins (Park et al., 2016; Zhang et al., 2017; Xu et al., 2021). Both gain and loss of lysine acetylation observed for the translational and metabolic pathways in the hybrid (Figure 6E; Supplemental Figure 10) indicate a remodeling of the protein acetylome. The effect of lysine acetylation on target protein function remains largely unknown. Lysine acetylation may affect r-protein stability and/or translational efficiency, although its effects may differ among specific r-proteins (Xu et al., 2021). We speculate that the differential lysine acetylation may target distinct r-proteins and selectively affect the efficiency of translation. Previous results have indicated that lysine acetylation may inhibit some enzymes of glycolysis (Finkemeier et al., 2011; Park et al., 2016; Zhang et al., 2017; Bontemps-Gallo et al., 2018). Similarly, acetylation of an active-site lysine residue inhibits the activity of acetyl-CoA synthases (Starai et al., 2002), indicating that lysine acetylation also acts as a feedback response to high energy metabolism and acetyl-CoA levels in the cell. The enhanced activities of glycolysis and acetyl-CoA metabolism detected in SY63 suggest that the remodeling of lysine acetylation of relevant enzymes helps to sustain high energy and acetyl-CoA metabolism in the hybrid.
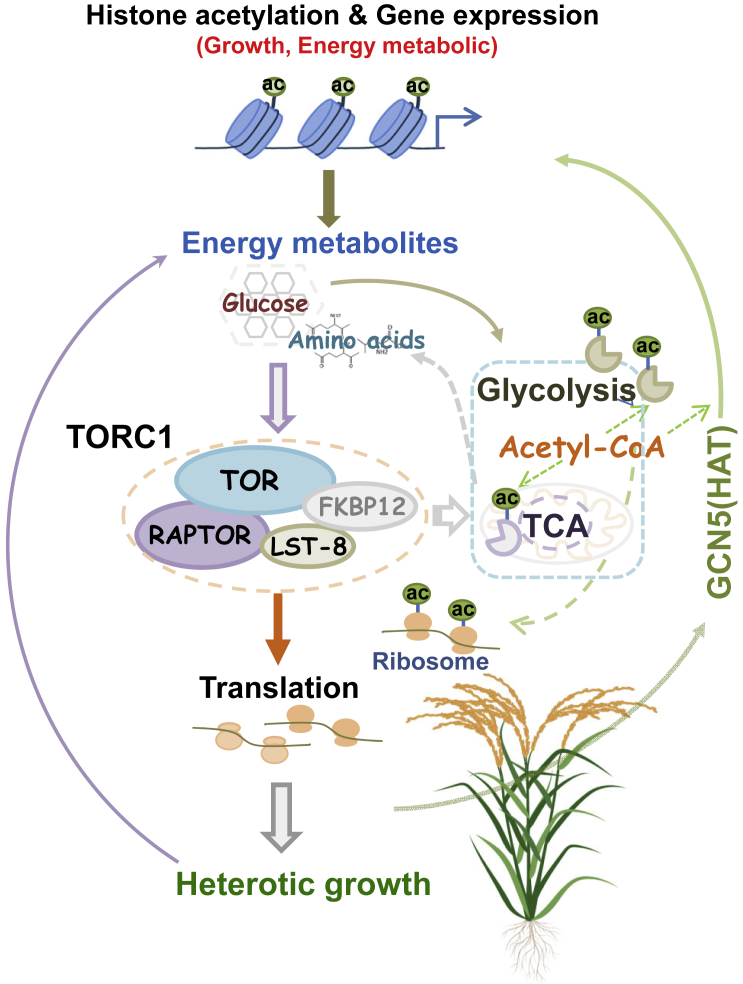
Based on these data, we propose that TORC1 may be activated by glucose accumulation in the hybrid to stimulate energy metabolism, protein translation, and anabolic or biosynthetic processes and cell growth rate, which in turn stimulate TORC1 activity. Higher energy/acetyl-CoA metabolism leads to increases in histone acetylation. Enhanced histone H3 acetylation that may involve the HAT GCN5 epigenetically activates the expression of genes related to metabolism and cell division, contributing to heterotic growth. Increases and remodeling of lysine acetylation of cellular proteins might help to sustain the positive feedback loop between TORC1 signaling, energy metabolism, and cell growth-promoting activity (translation and cell division) (Figure 7). Our model proposes that enhanced activity within this loop due to genetic and/or enzymatic complementation or interaction of the parental genomes in the hybrid will trigger and/or sustain heterotic growth.
Figure 7.
Proposed model of the positive feedback loop among TOR signaling, energy metabolism, protein lysine acetylation, and histone modification/gene expression to promote superior growth of hybrid rice.
In the hybrid, accumulation of glucose or any component or activity within the loop may lead to higher TORC1 signaling, thereby enhancing energy metabolism, protein translation, and anabolic or biosynthetic processes that, in turn, reinforce TORC1 activity. Higher energy/acetyl-CoA metabolism leads to increases in histone acetylation that involve the histone acetyltransferase GCN5 to epigenetically activate the expression of metabolism and cell division genes, helping to sustain TORC1 signaling. Increases and remodeling of lysine acetylation of cellular proteins are also likely to contribute to sustaining the positive feedback loop among TORC1 signaling, energy metabolism, and cell growth-promoting activity (translation and cell division).
Methods
Plant materials and sample collection
Rice (Oryza sativa spp. indica/xian) varieties ZS97 and MH63 and their hybrid SY63 (with ZS97 as the female parent and MH63 as the male parent) were used in this study. Seeds were soaked in water for 72 h at 32°C in darkness and then sown in a field for germination. After 3 weeks, the rice seedlings were transplanted to a paddy field under normal agricultural rice growth conditions in Wuhan, China. Young panicles (2 mm in length, at the early stage of floral differentiation) were collected, immediately placed in liquid nitrogen, and stored at −80°C. Three biological replicates of each variety were harvested for each type of analysis, with 15 plants per replicate.
Label-free proteomics and acetyl-proteomics
Protein extraction and trypsin digestion
About 0.3 g frozen panicles were ground in liquid nitrogen to a fine powder and then transferred to a tube with 2 mL lysis buffer (1% Triton X-100, 10 mM DTT, 2 mM EDTA, 1% Protease Inhibitor Cocktail [Roche], 3 μM TSA, and 50 mM NAM) and sonicated three times on ice using a high-intensity ultrasonic processor (Scientz, Ningbo, China). An equal volume of Tris-saturated phenol (pH 8) was added and centrifuged at 5000 × g for 10 min at 4°C. The supernatant was transferred to a tube and precipitated with ammonium sulfate–saturated methanol at −20°C for 6 h. Protein precipitates were washed with cold acetone three times and suspended in 8 M urea. The protein concentration was determined with a BCA kit (Beyotime Biotechnology). Prior to digestion, cysteines were reduced with 5 mM DTT and alkylated with 11 mM idoacetamide in darkness. The protein solution was then diluted with 100 mM TEAB to a urea concentration less than 2 M. Finally, trypsin was added to a trypsin/protein ratio of 1:50 for the first digestion overnight and a trypsin/protein ratio of 1:100 for the second digestion for 4 h.
Kac peptide enrichment
To enrich Kac peptides, the digested peptides were incubated with 20 μl anti-acetyl-lysine beads (PTM-104, PTM Bio, Hangzhou, China) in NETN buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, 0.5% NP-40 [pH 8]) at 4°C overnight. After washing with NETN buffer and distilled H2O, the bound peptides were eluted from the beads with 0.1% trifluoroacetic acid. For LC–MS/MS analysis, the acetylated peptides were vacuum dried and desalted with C18 ZipTips (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions.
LC–MS/MS analysis
The digested peptides (for the proteome) or enriched acetyl-peptides (for the acetylome) were dissolved in solvent A (0.1% formic acid in water) and loaded onto a reversed-phase analytical column (15 cm, 75 μm inner diameter). Solvent B was composed of 0.1% formic acid and 98% acetonitrile. The liquid phase gradient was 6%–23% B, 0–38 min; 23%–35% B, 38–52 min; 35%–80% B, 52–56 min; and 80% B, holding for 4 min. The operations were performed at a constant flow rate of 400 nl/min in an EASY-nLC 1200 UPLC system (Thermo Fisher Scientific, Waltham, MA, USA). The peptides were subjected to a nano electrospray ionization source followed by MS/MS on a Q Exactive Plus instrument (Thermo Fisher Scientific) coupled online to the UPLC. The electrospray voltage was applied at 2 kV. The m/z scan range was 350–1800 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 70 000. Peptides were then selected for MS/MS using an NCE setting of 28, and the fragments were detected in the Orbitrap at a resolution of 17 500. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans with 15-s dynamic exclusion was used. Automatic gain control was set to 5E4. Fixed first mass was set to 100 m/z.
Protein database search
The raw MS/MS data were extracted and searched using MaxQuant (v.1.6.6.0). MS/MS spectra were searched against the Oryza sativa subsp. indica MH63RS2 protein database (http://rice.hzau.edu.cn/rice_rs2/) and concatenated with the reverse decoy database to remove contamination. Trypsin/P was specified as the cleavage enzyme, and up to two miscleavages were allowed. The minimum peptide length was set to seven amino acid residues and five modifications per peptide. The mass tolerance for precursor ions was set to 40 ppm in “First” search and 5 ppm in “Main” search, and that for fragment ions was set to 0.04 Da. Carbamidomethylation of cysteine residues was specified as a fixed modification, and oxidation on methionines, acetylation on lysines, and N-terminal acetylation on proteins were defined as variable modifications. The false discovery rate threshold for proteins, peptides, and modified sites was specified as less than 0.01, and the minimum score for modified peptides was set to >40.
Proteomics and acetyl-proteomics data analysis
The label-free quantification intensity (I) of each protein in different samples was transformed by centralization and used to obtain the relative quantitative value (R). The formula was as follows: Rij = Iij/Mean(Ij), where i indicates the sample and j indicates the protein.
GO enrichment of proteins was performed using the GO&KEGG toolkit of TBtools software (v.1.086). The GO background file was generated from the MH63RS2 genome annotation (http://rice.hzau.edu.cn/rice_rs2/). Subcellular localizations of proteins were predicted using the online tool WoLF PSORT (https://wolfpsort.hgc.jp/). The protein–protein interaction network was predicted using the STRING database (v.11.5, https://cn.string-db.org/). PCA was performed using the factoextra R package (https://rpkgs.datanovia.com/factoextra/). The bar plots and volcano plots were produced with the ggplot2 package in RStudio (v.4.1.2, https://posit.co/download/rstudio-desktop/). Heatmaps were generated using the Graphics toolkit of TBtools software (v.1.086) (Chen et al., 2020).
Metabolomics
Metabolite extraction
For metabolite extraction, samples of young rice panicles were freeze dried in a vacuum freeze dryer (Scientz-100F) and ground into fine powder using a mixer mill (MM400, Retsch) with a zirconia bead for 1.5 min at 30 Hz. An aliquot (100 mg) of powder was extracted with 1.2 mL 70% aqueous methanol (methanol:H2O, 70:30, v/v) overnight at 4°C. The mixture was centrifuged at 12 000 × g for 10 min at 4°C. The supernatant was collected and filtered through a 0.22-μm filter membrane (SCAA-104, ANPEL Shanghai, China) for UPLC–MS/MS analysis by a method described previously (Chen et al., 2013).
UPLC–MS/MS
The metabolite extracts were analyzed with a UPLC–electrospray ionization–MS/MS system (UPLC, Shimadzu Nexera X2; MS, Applied Biosystems QTRAP 4500) following the manufacturer’s instructions (Metware Biotechnology). Three biological replicates were performed for each genotype. The electrospray ionization source operation parameters were as follows: ion source, turbo spray; source temperature, 550°C; ion spray voltage, 5500/−4500 V (positive ion/negative ion mode); ion source gas I, gas II, and curtain gas set to 50, 60, and 25 psi, respectively. The collision-activated dissociation was high. Instrument tuning and mass calibration were performed with 10 and 100 μmol/l polypropylene glycol solutions in triple-quadrupole and LIT modes, respectively. The exported MS/MS files were processed with Analyst software (v.1.6.3, AB Sciex) and searched against the local metabolomics database (Metware Biotechnology). Metabolite quantification was performed by the multiple reaction monitoring method on the triple-quadrupole mass spectrometer.
Determination of acetyl-CoA contents
Acetyl-CoA contents were measured using an acetyl-CoA ELISA kit (D751001, BBI, Sangon Biotech, Shanghai, China) following the manufacturer’s instructions. Six biological replicates were performed for each genotype. Frozen young rice panicles were ground to a fine powder in liquid nitrogen; 100 mg of powder was transferred to a tube with 900 μl PBS buffer (0.01 M [pH 7.4], MonPro), vortexed for 1 min, and sonicated three times on ice using an ultrasonic processor (Scientz, Ningbo, China). After centrifugation for 10 min (5000 × g, 4°C), the supernatant was transferred to a tube. An acetyl-CoA standard solution (20 ng/ml) was serially diluted to 20, 10, 5, 2.5, 1.25, 0.63, 0.31, and 0 ng/ml. The 100 μl standard dilution or a sample extract was added to each reaction well of an ELISA plate and incubated for 90 min at 37°C. We then discarded the liquid and dried the wells in air for 2 min, added 100 μl biotin-conjugated antibody solution to each well, and incubated the plate for 60 min at 37°C. The liquid in each reaction well was discarded, and each well was washed four times with 350 μl washing solution. HRP-conjugated streptavidin solutions (100 μl) were added to each well and incubated for 30 min at 37°C, then discarded. Each well was washed four times. Finally, 90 μl substrate reagents were added to each well and incubated for 15 min at 37°C in darkness. After addition of 50 μl stop solution, the absorbance of each well at 450 nm was immediately measured using a multimode microplate reader (Spark, TECAN, Männedorf, Switzerland). The standard curve was constructed in Microsoft Excel with the standard concentrations on the x axis and the OD values on the y axis.
Enzyme activity measurement
Activities of the glycolytic enzymes ATP-dependent 6-PFK (EC 2.7.1.11, catalog number PFK-1-Y), fructose-bisphosphate aldolase (FBA; EC4.1.2.13, FBA-1-G), and GAPDH (EC1.2.1.12, GAPDH-1-Y) and TCA enzymes CS (EC 2.3.3.1, CS-1-Y) and ICDH (EC 1.1.1.41, ICDHM-1-Y) were determined using commercial kits (Keming Biotechnology) following the manufacturer’s instructions. In brief, 0.1 g young rice panicle was homogenized in 1 ml enzyme extraction buffer and then centrifuged at 4°C for 10 min; the supernatant was transferred to a microplate well and mixed with enzyme reaction buffers. The absorbance was measured at a specific wavelength (PFK, FBA, GAPDH, and ICDH at 340 nm and CS at 412 nm) using a multimode microplate reader (Spark, TECAN). Six biological replicates were performed for each genotype. Enzyme activity was calculated from the absorbance value and the sample fresh weight using the formula provided in the enzyme kit instructions.
Activity of ACL was measured using a commercial kit (Cat.# BC4225, Solarbio Life Sciences) following the manufacturer’s protocol. In brief, 0.1 g rice young panicle was ground in liquid nitrogen to a fine powder and homogenized in 1 ml extraction buffer, then centrifuged at 4°C 8000 × g for 10 min; the supernatant was transferred 10 μl to a microplate well and mixed with reaction buffers (volumes of reagents I, II, III, IV, and V were 161, 4, 20, 4, and 1 μl, respectively), and we measured the absorbance at 340 nm and calculated the enzyme activity using the formula
| ACL activity (U/g fresh weigh) = [ΔA∗Vt∗109/(ε∗d)]/ (W∗Vs∗Vst)/T, |
where Vt is the total volume of the reaction (2∗10−4 l); ε is the NADH molar extinction coefficient (6.22∗103 l/mol/cm); d is the optical distance of cuvette (1 cm); W is the sample weight; Vs is the volume of extract for a reaction (10 μl); and Vst is the volume of the extraction buffer.
Quantitative real-time PCR
Total RNA were extracted from the young panicles using TRIzol reagent (Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol. Two μg total RNA was used to synthesize first-strand cDNA with a reverse transcription kit (Invitrogen). Quantitative real-time PCR was conducted using SYBR Premix ExTaq (TaKaRa) in a real-time system. The transcript level of Actin was used as the internal control. Three biological replicates were performed for each quantitative real-time PCR analysis. Primer sequences are listed in Supplemental Table 4.
Histone extraction and western blot
Total histones were extracted from the frozen panicle samples using the EpiQuik Total Histone Extraction kit (Epigentek, Cat.# OP-0006) following the protocol. Concentration of the eluted protein was measured by the Bradford method according to the manufacturer’s instructions (Bio-Rad protein assay, Hercules, CA, USA). For immunoblotting, the histone samples were diluted with SDS loading buffer, and 20 μg proteins of each sample were separated by 16.5% SDS-PAGE and electro-blotted onto PVDF blotting membrane (Amersham Hybond, Cytiva). The blot was probed with anti-acetyl-histone H3 antibody (Millipore, Cat.# 06-599, 1:1000 dilution), anti-acetyl-histone H3K37 antibody (Active Motif, Cat.# 61 587, 1:1000 dilution), or anti-histone H3 antibody (Sigma, Cat.# SAB5701101, 1:1000 dilution).
ChIP-seq and data analysis
Chromatin immunoprecipitation experiments were performed as described previously (Wang et al., 2021b). In brief, about 2 g rice seedling leaves were crosslinked with 1% (v/v) formaldehyde (30 min) and used for chromatin extraction. Chromatin was fragmented to approximately 200 bp by sonication using a Bioruptor Plus System (Diagenode). The chromatin fragments were incubated overnight with antibody-conjugated beads (anti-acetyl-H3K9, Millipore, catalog number 07-352). After washing, immunoprecipitated chromatin was de-crosslinked, and DNA was purified; non-precipitated chromatin was used as input.
DNA isolated by ChIP was used to construct sequencing libraries following the protocol provided with the Illumina TruSeq ChIP Sample Prep Set A and sequenced on the Illumina HiSeq 2500 platform. Cutadapt software (v.3.5) was used to filter out low-quality reads. Clean reads were aligned to the rice genome (MH63RS2, http://rice.hzau.edu.cn/rice_rs2/) using Bowtie 2 (v.2.2.8), allowing up to two mismatches. SAMtools (v.0.1.14) was used to remove potential PCR duplicates. MACS2 (v.2.2.7.1) software was used to call histone modification peaks with default parameters (bandwidth, 300 bp; model fold, 5, 50; P value, 1E−5) (Zhang et al., 2008), and the input was used as a control. Histone modification peaks were annotated with HOMER (v.4.11) software (Heinz et al., 2010). Heatmaps were generated with TBtools (v.1.086) (Chen et al., 2020) and deepTools2.0 software (Ramirez et al., 2016).
Funding
This work was supported by grants from the National Natural Science Foundation of China (31730049); the National Key Research and Development Program of China (2016YFD0100802); the Huazhong Agricultural University Scientific & Technological Selfinnovation Foundation (2016RC003); and the Fundamental Research Funds for the Central Universities (2662015PY228).
Author contributions
X. Ma performed most of the experimental work and data analysis; Q.J., S.L., Z.C., and X. Ming assisted X. Ma in collection of plant tissue samples and participated in experimental work; Y.Z. and D.-X.Z. designed and supervised the work; D.-X.Z. wrote the paper with input from X. Ma.
Acknowledgments
We would like to thank Dr. Zhongyi Cheng for proteomic and acetylomic analysis and Mr. Yuan Liu and Drs. Xianghua Li, Qinghua Zhang, and Jinghua Xiao for assistance. We are also thankful for the support of the BaiChuan Fellowship of the College of Life Science and Technology, Huazhong Agricultural University. No conflict of interest is declared.
Published: February 11, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Accession numbers
The MS source data produced by this study are deposited into the PRIDE/ProteomeXchange consortium repository (BioProject: PXD035495 and PXD035543). The ChIP-seq data are deposited at the NCBI Sequence Read Archive (BioProject: PRJNA860563) and the Genome Sequence Archive (GSA: CRA009670) of the China National Genomics Data Center (BioProject: PRJCA014647).
Supplemental information
References
- Baldauf J.A., Marcon C., Lithio A., Vedder L., Altrogge L., Piepho H.P., Schoof H., Nettleton D., Hochholdinger F. Single-parent expression is a general mechanism driving extensive complementation of non-syntenic genes in maize hybrids. Curr. Biol. 2018;28:431–437.e4. doi: 10.1016/j.cub.2017.12.027. [DOI] [PubMed] [Google Scholar]
- Balparda M., Elsässer M., Badia M.B., Giese J., Bovdilova A., Hüdig M., Reinmuth L., Eirich J., Schwarzländer M., Finkemeier I., et al. Acetylation of conserved lysines fine-tunes mitochondrial malate dehydrogenase activity in land plants. Plant J. 2022;109:92–111. doi: 10.1111/tpj.15556. [DOI] [PubMed] [Google Scholar]
- Benhamed M., Bertrand C., Servet C., Zhou D.X. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell. 2006;18:2893–2903. doi: 10.1105/tpc.106.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontemps-Gallo S., Gaviard C., Richards C.L., Kentache T., Raffel S.J., Lawrence K.A., Schindler J.C., Lovelace J., Dulebohn D.P., Cluss R.G., et al. Global profiling of lysine acetylation in Borrelia burgdorferi B31 reveals its role in central metabolism. Front. Microbiol. 2018;9:2036. doi: 10.3389/fmicb.2018.02036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkard J.O., Xu M., Scarpin M.R., Chatterjee S., Shemyakina E.A., Goodman H.M., Zambryski P. TOR dynamically regulates plant cell-cell transport. Proc. Natl. Acad. Sci. USA. 2020;117:5049–5058. doi: 10.1073/pnas.1919196117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Sutter B.M., Li B., Tu B.P. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Chen W., Gong L., Guo Z., Wang W., Zhang H., Liu X., Yu S., Xiong L., Luo J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant. 2013;6:1769–1780. doi: 10.1093/mp/sst080. [DOI] [PubMed] [Google Scholar]
- Chen Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013;14:471–482. doi: 10.1038/nrg3503. [DOI] [PubMed] [Google Scholar]
- Dobrenel T., Caldana C., Hanson J., Robaglia C., Vincentz M., Veit B., Meyer C. TOR signaling and nutrient sensing. Annu. Rev. Plant Biol. 2016;67:261–285. doi: 10.1146/annurev-arplant-043014-114648. [DOI] [PubMed] [Google Scholar]
- Dobrenel T., Mancera-Martínez E., Forzani C., Azzopardi M., Davanture M., Moreau M., Schepetilnikov M., Chicher J., Langella O., Zivy M., et al. The Arabidopsis TOR kinase specifically regulates the expression of nuclear genes coding for plastidic ribosomal proteins and the phosphorylation of the cytosolic ribosomal protein S6. Front. Plant Sci. 2016;7:1611. doi: 10.3389/fpls.2016.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Wu C., Wang C., Roh J., Zhang L., Chen J., Zhang S., Zhang H., Yang C., Hu J., et al. SLG controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. J. Exp. Bot. 2016;67:4241–4253. doi: 10.1093/jxb/erw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I., Laxa M., Miguet L., Howden A.J.M., Sweetlove L.J. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 2011;155:1779–1790. doi: 10.1104/pp.110.171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca B.D., Smith E.M., Yelle N., Alain T., Bushell M., Pause A. The ever-evolving role of mTOR in translation. Semin. Cell Dev. Biol. 2014;36:102–112. doi: 10.1016/j.semcdb.2014.09.014. [DOI] [PubMed] [Google Scholar]
- Fu L., Wang P., Xiong Y. Target of rapamycin signaling in plant stress responses. Plant Physiol. 2020;182:1613–1623. doi: 10.1104/pp.19.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., Zhang Q. Heterosis in elite hybrid rice: speculation on the genetic and biochemical mechanisms. Curr. Opin. Plant Biol. 2013;16:221–227. doi: 10.1016/j.pbi.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Groszmann M., Greaves I.K., Albertyn Z.I., Scofield G.N., Peacock W.J., Dennis E.S. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc. Natl. Acad. Sci. USA. 2011;108:2617–2622. doi: 10.1073/pnas.1019217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh W.C., Sutter B.M., Ruess H., Barnes S.D., Malladi V.S., Tu B.P. Glucose starvation induces a switch in the histone acetylome for activation of gluconeogenic and fat metabolism genes. Mol. Cell. 2022;82:60–74.e5. doi: 10.1016/j.molcel.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J., Xing Y., Wu W., Xu C., Sun X., Yu S., Zhang Q. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA. 2003;100:2574–2579. doi: 10.1073/pnas.0437907100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Hua K., Xu R., Zeng D., Wang R., Dong G., Zhang G., Lu X., Fang N., Wang D., et al. The LARGE2-APO1/APO2 regulatory module controls panicle size and grain number in rice. Plant Cell. 2021;33:1212–1228. doi: 10.1093/plcell/koab041. [DOI] [PubMed] [Google Scholar]
- Huang X., Yang S., Gong J., Zhao Q., Feng Q., Zhan Q., Zhao Y., Li W., Cheng B., Xia J., et al. Genomic architecture of heterosis for yield traits in rice. Nature. 2016;537:629–633. doi: 10.1038/nature19760. [DOI] [PubMed] [Google Scholar]
- Huang X., Yang S., Gong J., Zhao Y., Feng Q., Gong H., Li W., Zhan Q., Cheng B., Xia J., et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 2015;6:6258. doi: 10.1038/ncomms7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia V., Liu R., Proud C.G. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin. Cell Dev. Biol. 2014;36:113–120. doi: 10.1016/j.semcdb.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K., Maekawa M., Izawa T., Itoh J.I., Nagato Y. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 2012;69:168–180. doi: 10.1111/j.1365-313X.2011.04781.x. [DOI] [PubMed] [Google Scholar]
- Jasieniecka-Gazarkiewicz K., Lager I., Carlsson A.S., Gutbrod K., Peisker H., Dörmann P., Stymne S., Banaś A. Acyl-CoA:Lysophosphatidylethanolamine acyltransferase activity regulates growth of Arabidopsis. Plant Physiol. 2017;174:986–998. doi: 10.1104/pp.17.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T., Taramino G., Itoh J.I., Allen J., Sato Y., Hong S.K., Yule R., Nagasawa N., Kojima M., Kusaba M., et al. PLASTOCHRON3/GOLIATH encodes a glutamate carboxypeptidase required for proper development in rice. Plant J. 2009;58:1028–1040. doi: 10.1111/j.1365-313X.2009.03841.x. [DOI] [PubMed] [Google Scholar]
- Kawanabe T., Ishikura S., Miyaji N., Sasaki T., Wu L.M., Itabashi E., Takada S., Shimizu M., Takasaki-Yasuda T., Osabe K., et al. Role of DNA methylation in hybrid vigor in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2016;113:E6704–E6711. doi: 10.1073/pnas.1613372113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger U., Lippman Z.B., Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010;42:459–463. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Li S., Zhou B., Peng X., Kuang Q., Huang X., Yao J., Du B., Sun M.X. OsFIE2 plays an essential role in the regulation of rice vegetative and reproductive development. New Phytol. 2014;201:66–79. doi: 10.1111/nph.12472. [DOI] [PubMed] [Google Scholar]
- Li X., Gao X., Wei Y., Deng L., Ouyang Y., Chen G., Li X., Zhang Q., Wu C. Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5'-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell. 2011;23:1416–1434. doi: 10.1105/tpc.110.082636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., Shao D., Xu C., Li X., Xiao J., et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011;43:1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- Li Z., Zhu A., Song Q., Chen H.Y., Harmon F.G., Chen Z.J. Temporal regulation of the metabolome and proteome in photosynthetic and photorespiratory pathways contributes to maize heterosis. Plant Cell. 2020;32:3706–3722. doi: 10.1105/tpc.20.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., He G., Deng X.W. Biological pathway expression complementation contributes to biomass heterosis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2023278118. e2023278118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Xing F., Jia Q., Zhang Q., Hu T., Wu B., Shao L., Zhao Y., Zhang Q., Zhou D.X. Parental variation in CHG methylation is associated with allelic-specific expression in elite hybrid rice. Plant Physiol. 2021;186:1025–1041. doi: 10.1093/plphys/kiab088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O., Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta. 2015;1849:801–811. doi: 10.1016/j.bbagrm.2014.08.015. [DOI] [PubMed] [Google Scholar]
- Ouyang Y., Li X., Zhang Q. Understanding the genetic and molecular constitutions of heterosis for developing hybrid rice. J Genet Genomics. 2022;49:385–393. doi: 10.1016/j.jgg.2022.02.022. [DOI] [PubMed] [Google Scholar]
- Park S.H., Ozden O., Liu G., Song H.Y., Zhu Y., Yan Y., Zou X., Kang H.J., Jiang H., Principe D.R., et al. SIRT2-Mediated deacetylation and tetramerization of pyruvate kinase directs glycolysis and tumor growth. Cancer Res. 2016;76:3802–3812. doi: 10.1158/0008-5472.CAN-15-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu C.X., Han Y.F., Zhu S., Song F.Y., Zhao Y., Wang C.Y., Zhang Y.C., Yang Q., Wang J., Bu S.L., et al. The rice receptor-like kinases DWARF and RUNTISH SPIKELET1 and 2 repress cell death and affect sugar utilization during reproductive development. Plant Cell. 2017;29:70–89. doi: 10.1105/tpc.16.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwestri Y.A., Ogaki Y., Tamaki S., Tsuji H., Shimamoto K. The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 2009;50:429–438. doi: 10.1093/pcp/pcp012. [DOI] [PubMed] [Google Scholar]
- Qi P., Lin Y.S., Song X.J., Shen J.B., Huang W., Shan J.X., Zhu M.Z., Jiang L., Gao J.P., Lin H.X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012;22:1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpin M.R., Busche M., Martinez R.E., Harper L.C., Reiser L., Szakonyi D., Merchante C., Lan T., Xiong W., Mo B., et al. An updated nomenclature for plant ribosomal protein genes. Plant Cell. 2022:koac333. doi: 10.1093/plcell/koac333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpin M.R., Leiboff S., Brunkard J.O. Parallel global profiling of plant TOR dynamics reveals a conserved role for LARP1 in translation. Elife. 2020;9:e58795. doi: 10.7554/eLife.58795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpin M.R., Simmons C.H., Brunkard J.O. Translating across kingdoms: target of rapamycin promotes protein synthesis through conserved and divergent pathways in plants. J. Exp. Bot. 2022;73:7016–7025. doi: 10.1093/jxb/erac267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L., Xing F., Xu C., Zhang Q., Che J., Wang X., Song J., Li X., Xiao J., Chen L.L., et al. Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc. Natl. Acad. Sci. USA. 2019;116:5653–5658. doi: 10.1073/pnas.1820513116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Wei W., Zhou D.X. Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 2015;20:614–621. doi: 10.1016/j.tplants.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Shi L., Tu B.P. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr. Opin. Cell Biol. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S.M., Park S.J., Lee H., Siddiqi F., Lee J.E., Menzies F.M., Rubinsztein D.C. Leucine signals to mTORC1 via its metabolite acetyl-coenzyme A. Cell Metabol. 2019;29:192–201.e7. doi: 10.1016/j.cmet.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Wang R., Zhang L., Wang Y., Yao S. CRR1 encoding callose synthase functions in ovary expansion by affecting vascular cell patterning in rice. Plant J. 2016;88:620–632. doi: 10.1111/tpj.13287. [DOI] [PubMed] [Google Scholar]
- Starai V.J., Celic I., Cole R.N., Boeke J.D., Escalante-Semerena J.C. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- Wang J., Hu J., Qian Q., Xue H.W. LC2 and OsVIL2 promote rice flowering by photoperoid-induced epigenetic silencing of OsLF. Mol. Plant. 2013;6:514–527. doi: 10.1093/mp/sss096. [DOI] [PubMed] [Google Scholar]
- Wang L., Ming L., Liao K., Xia C., Sun S., Chang Y., Wang H., Fu D., Xu C., Wang Z., et al. Bract suppression regulated by the miR156/529-SPLs-NL1-PLA1 module is required for the transition from vegetative to reproductive branching in rice. Mol. Plant. 2021;14:1168–1184. doi: 10.1016/j.molp.2021.04.013. [DOI] [PubMed] [Google Scholar]
- Wang W., Lu Y., Li J., Zhang X., Hu F., Zhao Y., Zhou D.X. SnRK1 stimulates the histone H3K27me3 demethylase JMJ705 to regulate a transcriptional switch to control energy homeostasis. Plant Cell. 2021;33:3721–3742. doi: 10.1093/plcell/koab224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang T., Wang R., Zhao Y. Recent advances in auxin research in rice and their implications for crop improvement. J. Exp. Bot. 2018;69:255–263. doi: 10.1093/jxb/erx228. [DOI] [PubMed] [Google Scholar]
- Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., Thompson C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F.M., Zhang J.F. Shanyou 63: an elite mega rice hybrid in China. Rice. 2018;11:17. doi: 10.1186/s12284-018-0210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Wang G., Yuan M., Yao W., Lyu K., Zhao H., Yang M., Li P., Zhang X., Yuan J., et al. Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc. Natl. Acad. Sci. USA. 2015;112:E5411–E5419. doi: 10.1073/pnas.1515919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Zhang Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010;61:421–442. doi: 10.1146/annurev-arplant-042809-112209. [DOI] [PubMed] [Google Scholar]
- Xiong Y., McCormack M., Li L., Hall Q., Xiang C., Sheen J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496:181–186. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Sheen J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014;164:499–512. doi: 10.1104/pp.113.229948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Liu Q., Chen Z., Yue Y., Liu Y., Zhao Y., Zhou D.X. Histone deacetylases control lysine acetylation of ribosomal proteins in rice. Nucleic Acids Res. 2021;49:4613–4628. doi: 10.1093/nar/gkab244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., Zhang Q. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yang L., Liu P., Wang X., Jia A., Ren D., Tang Y., Tang Y., Deng X.W., He G. A central circadian oscillator confers defense heterosis in hybrids without growth vigor costs. Nat. Commun. 2021;12:2317. doi: 10.1038/s41467-021-22268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Wang X., Huang H., Ren D., Su Y., Zhu P., Zhu D., Fan L., Chen L., He G., Deng X.W. Natural variation of H3K27me3 modification in two Arabidopsis accessions and their hybrid. J. Integr. Plant Biol. 2016;58:466–474. doi: 10.1111/jipb.12443. [DOI] [PubMed] [Google Scholar]
- Yang M., Wang X., Ren D., Huang H., Xu M., He G., Deng X.W. Genomic architecture of biomass heterosis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2017;114:8101–8106. doi: 10.1073/pnas.1705423114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.B., Li J.X., Xu C.G., Tan Y.F., Gao Y.J., Li X.H., Zhang Q., Saghai Maroof M.A. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA. 1997;94:9226–9231. doi: 10.1073/pnas.94.17.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhao Y., Zhou D.X. Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res. 2017;45:12241–12255. doi: 10.1093/nar/gkx825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Chen L.L., Xing F., Kudrna D.A., Yao W., Copetti D., Mu T., Li W., Song J.M., Xie W., et al. Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc. Natl. Acad. Sci. USA. 2016;113:E5163–E5171. doi: 10.1073/pnas.1611012113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Chen Y., Yao W., Zhang C., Xie W., Hua J., Xing Y., Xiao J., Zhang Q. Genetic composition of yield heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA. 2012;109:15847–15852. doi: 10.1073/pnas.1214141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Jiang W., Long F., Cheng S., Yang W., Zhao Y., Zhou D.-X. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell. 2017;29:1088–1104. doi: 10.1105/tpc.16.00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.