Abstract
Identifying sources of phytopathogen inoculum and determining their contributions to disease outbreaks are essential for predicting disease development and establishing control strategies. Puccinia striiformis f. sp. tritici (Pst), the causal agent of wheat stripe rust, is an airborne fungal pathogen with rapid virulence variation that threatens wheat production through its long-distance migration. Because of wide variation in geographic features, climatic conditions, and wheat production systems, Pst sources and related dispersal routes in China are largely unclear. In the present study, we performed genomic analyses of 154 Pst isolates from all major wheat-growing regions in China to determine Pst population structure and diversity. Through trajectory tracking, historical migration studies, genetic introgression analyses, and field surveys, we investigated Pst sources and their contributions to wheat stripe rust epidemics. We identified Longnan, the Himalayan region, and the Guizhou Plateau, which contain the highest population genetic diversities, as the Pst sources in China. Pst from Longnan disseminates mainly to eastern Liupan Mountain, the Sichuan Basin, and eastern Qinghai; that from the Himalayan region spreads mainly to the Sichuan Basin and eastern Qinghai; and that from the Guizhou Plateau migrates mainly to the Sichuan Basin and the Central Plain. These findings improve our current understanding of wheat stripe rust epidemics in China and emphasize the need for managing stripe rust on a national scale.
Key words: Puccinia striiformis f. sp. tritici, stripe rust, disease epidemics, population genetics, genome sequencing
On the basis of genomic analyses, trajectory tracking, and field investigations, this study proposes that Longnan, the Himalayan region, and the Guizhou Plateau are the sources of the wheat stripe rust pathogen in China. Migration pathways originating from these sources and their effects on disease epidemics across the country were analyzed.
Introduction
Plant disease epidemics have changed the course of history in human civilization. With changes in climate, cropping systems, and human activities, the increasing risk of plant disease outbreaks continues to threaten food security (Fisher et al., 2012; Ristaino et al., 2021). Long-distance airborne dispersal is an important route by which numerous pathogens, especially fungal and fungal-like pathogens, invade and establish in new territories, rapidly causing large-scale epidemics. Spores of these airborne pathogens can be transported passively by the wind for up to thousands of kilometers to spread diseases (Bowden et al., 1971; Dyer et al., 1993; Brown and Hovmøller, 2002; Wellings, 2007). Investigating pathogen sources and dispersal routes is essential for designing disease monitoring strategies, predicting disease development on a large scale, and developing control measures at an appropriate scale, including the use of resistant cultivars, fungicides, and quarantine measures (Nagarajan and Singh, 1990; Goss, 2015).
Wheat stripe (yellow) rust, caused by Puccinia striiformis f. sp. tritici (Pst), is a devastating airborne disease that threatens wheat production in more than 60 countries and is responsible for significant crop losses each year (Chen, 2005; Hovmøller et al., 2011). Major stripe rust epidemics have been reported in China (Chen et al., 2007), the USA (Chen, 2005), the UK (Johnson and Taylor, 1972), New Zealand (Beresford, 1982), Australia (Wellings, 2007), Pakistan (Duveiller et al., 2007), India (Prashar et al., 2007), and many other countries (Wellings, 2011; Chen, 2020), resulting in grain losses of up to tens of millions of tons annually. Through analysis of Pst isolates from different continents, the Himalayan region was putatively identified as the center of Pst origin because of its high Pst genotypic diversity, high possibility for sexual reproduction, and independent differentiated populations (Ali et al., 2014). The Mediterranean to Asian regions were proposed as the broader center of stripe rust origin because of their high Pst genotypic diversity and long history of cereal cultivation (Stubbs, 1985; Sharma-Poudyal et al., 2020). In China, epidemics of stripe rust occur almost every year, causing average yield losses of 1 million tonnes annually (Chen et al., 2007). The most severe epidemics occurred in 1950, 1964, 1990, 2002, 2017, and 2020 and resulted in a cumulative yield loss of nearly 12.0 million tonnes (Li and Zeng, 2002; Wan et al., 2004; Liu et al., 2022). Located in the putative center of Pst origin, China has higher genotypic diversity and allelic richness in Pst populations than most other regions worldwide (Ali et al., 2014; Sharma-Poudyal et al., 2020), causing rapid variations in Pst virulence. Wheat stripe rust exhibits unique epidemic characteristics in China because of its complicated topographies, diverse climatic conditions, and different cropping systems (Zeng and Luo, 2006); sources and related migration routes of Pst thus remain unclear on a national scale.
Among all Pst epidemic regions in China, Longnan, a mountainous region in southern Gansu, is considered to be the center of variation in Pst virulence from which new and aggressive isolates emerge (Wan et al., 2007; Chen et al., 2009). Population genetic analyses have indicated that the genotypic and genetic diversity of the Pst population are higher in Longnan than in adjacent regions (Liang et al., 2013, 2016; Wan et al., 2015; Hu et al., 2017; Wang et al., 2022). According to earlier field surveys, stripe rust epidemic levels in Longnan are directly related to those in the Sichuan Basin and Huang-Huai-Hai wheat-growing regions in most years (Li and Zeng, 2002). Emerging aggressive Pst races in Longnan can spread to main wheat-growing regions, causing the breakdown of resistant cultivars.
However, exceptions to this scenario were reported in the wheat-growing seasons of 2000–2001 and 2016–2017 (Wan et al., 2002; Huang et al., 2018). In 2001, stripe rust occurred intensively in the Sichuan Basin and southwestern China but only lightly in Longnan (Wan et al., 2002). Stripe rust was severe in the Huang-Huai-Hai region in 2017 but occurred at a low level in Longnan in autumn 2016, the lowest since 2000 (Huang et al., 2018). Because these observations challenge our previous understanding, further studies are needed to determine the presence of Pst sources in China and delineate their dispersal routes at a finer scale. Recently, Yunnan in southwestern China was proposed as a probable center of Pst inoculum because of an ancestral haplotype detected there (Li et al., 2021a). However, more evidence is needed to support this conclusion and clarify the inoculum center(s) of Pst in China.
Rapid development of genomic and transcriptomic sequencing technologies, bioinformatics, and aerodynamics modeling has enabled the identification of Pst sources and dispersal routes through integrated studies. Using a wealth of variants obtained by high-throughput sequencing, genetic diversity and migration patterns in Pst populations have been investigated in the UK and Australia (Hubbard et al., 2015; Bueno-Sancho et al., 2017; Ding et al., 2021). In the present work, we studied Pst sources and their contributions in China using population genetics analysis, trajectory cluster calculations, and field surveys. We identified Longnan, the Himalayan region, and the Guizhou Plateau as three inoculum centers of Pst in China, from which Pst spreads to the main wheat-growing regions. We specifically analyzed the dispersal routes derived from these sources and assessed the contributions of each source to nationwide Pst epidemics.
Results
Sampling, sequencing, and variant detection
Samples were collected from wheat stripe rust epidemic areas in 11 provinces and autonomous regions across China. According to geographic and topographic features, nine geographic populations were identified: Longnan (G1), Himalayan regions (G2), Guizhou Plateau (G3), eastern Liupan Mountain (G4), Sichuan Basin (G5), Central Plain (G6), eastern Qinghai (G7), Yunnan Plateau (G8), and Yili Valley (G9) (Table 1, Supplemental Table 1 and Figure 1A). After multiplication of urediniospores in the greenhouse, Pst DNA was extracted from individual samples and sequenced. After filtering, a total of 893.54 Gb clean reads were obtained from 154 isolates (Table 1 and Supplemental Table 2). The quality and completeness of selected Pst genomes (see methods) were evaluated to identify an appropriate reference genome (Supplemental Tables 3 and 4). On the basis of scaffold number, N50 scaffold length, and percentage of complete benchmarking universal single-copy orthologs (BUSCOs), DK09_11 was selected as the reference genome. More than 1.83 million variants were generated from alignments with DK09_11, using GATK for variant calling. The mapping rates ranged from 58.36% to 95.65% for individual isolates, with an average of 88.95% (Supplemental Table 5). After further quality filtering, approximately 840 000 high-quality single nucleotide polymorphisms (SNPs) were obtained. These SNPs were primarily located in intergenic, downstream, and upstream regions, and deleterious and high impacts on gene functions were caused mainly by missense mutations, gain and loss of start and stop codons, and splice donor and acceptor variants (Supplemental Table 6).
Table 1.
Summary of Pst samples and genome sequencing.
| Geographic population | Ecological region | Sample size | Clean bases (bp) | Q30 (%) | Mapping rate (%) | Average coverage depth (×) |
|---|---|---|---|---|---|---|
| G1 | Longnan | 7 | 5 650 801 629 | 88.62 | 80.23 | 45.48 |
| G2 | Himalayan region | 33 | 5 805 693 836 | 90.73 | 89.95 | 56.82 |
| G3 | Guizhou Plateau | 18 | 5 747 079 400 | 91.44 | 90.85 | 54.68 |
| G4 | Eastern Liupan Mountain | 12 | 6 380 583 750 | 89.42 | 81.19 | 54.73 |
| G5 | Sichuan Basin | 23 | 6 003 921 013 | 91.34 | 91.58 | 61.38 |
| G6 | Central Plain | 12 | 5 536 370 450 | 90.18 | 81.91 | 48.83 |
| G7 | Eastern Qinghai | 25 | 5 563 350 732 | 90.86 | 92.59 | 58.16 |
| G8 | Yunnan Plateau | 18 | 5 576 530 183 | 91.58 | 90.83 | 54.63 |
| G9 | Yili Valley | 6 | 6 398 406 850 | 91.40 | 86.71 | 53.81 |
| Total/average | 154 | 893 536 323 600 | 90.81 | 88.95 | 55.79 |
Figure 1.
Geographic distribution and population genetic structure of 154 Pst isolates.
(A) Geographic distribution and geographic populations of 154 Pst isolates collected from 11 provinces in China. Different colors represent different sampling sites.
(B) Maximum likelihood phylogenetic tree of 154 Pst isolates from China and sixteen alien isolates using SNPs pruned by linkage disequilibrium. Asterisks on the branches indicate greater than or equal to 60% bootstrap support. Samples collected from different provinces are represented with circles in various colors. The blue and red dots represent cluster 1 and cluster 2, respectively.
(C) Ancestry coefficient analysis of the Pst accessions. The accessions were grouped into two clusters with K = 2.
Figure 4.
Optimal models of historical demographic events between Pst populations.
Numbers along with the arrows indicate the migration ratio. The number of generations represents the divergence time between different populations.
(A) Divergence and migration of Pst populations between Longnan and adjacent regions.
(B) Divergence and migration of Pst populations between the Himalayan region and the Sichuan Basin and eastern Qinghai.
(C) Divergence and migration of Pst populations between Guizhou Plateau and the Sichuan Basin and Central Plain.
Phylogeny and population structure of Pst
To explore the population genetic structure of Pst in China and its relationship with Pst in other countries, we constructed a phylogenetic tree using 55 107 linkage disequilibrium–pruned SNPs (Figure 1B). With P. striiformis f. sp. hordei (Psh) as the outgroup, we found that Pst isolates from India and the UK belong to two independent lineages. The lineage from India shows a close genetic relationship with isolates in China, indicating that possible incursions may have happened between these two regions. In China, two lineages (cluster 1 and cluster 2) were identified (Figure 1B) from a phylogeny based on the maximum likelihood method. This finding was partially suggested by principal component analysis (PCA): the first two PCs, explaining 27.42% and 12.50% of the total variance, indicated possible separation of cluster 1 and cluster 2 (Supplemental Figure 1). The two-cluster population structure was also supported by admixture analysis (Figure 1C and Supplemental Figure 2). Pst isolates in cluster 1 showed a close genealogical relationship to the divergence of Pst and Psh, suggesting that cluster 1 probably represents an ancestral lineage in China compared with cluster 2. To further investigate the relationships among isolates within each cluster, the two lineages from the phylogeny were analyzed separately. Cluster 1 was partitioned into three sub-groups containing isolates from Longnan (cluster 1.1), the Himalayan region (cluster 1.2), and the Guizhou Plateau (cluster 1.3) as indicated by bootstrap values; these subgroups were also supported by PCA and admixture analysis (Supplemental Figures 3 and 4), suggesting that Pst populations in Longnan, the Himalayan region, and the Guizhou Plateau could separate from one another and further develop into subgroups independently. Likewise, cluster 2 was partitioned into four subgroups using an unrooted phylogenetic tree; these subgroups were also supported by admixture analysis and partially suggested by PCA (Supplemental Figure 4). Samples in cluster 2.1 comprised isolates from multiple locations, and those in cluster 2.2 were mainly from the Sichuan Basin and the Yunnan Plateau in southwest China. Clusters 2.3 and 2.4 comprised isolates mostly from winter-spring wheat areas in southwest and northwest China (Supplemental Figures 4 and 5).
Figure 5.
Major migration routes of Pst from sources in China.
Migration routes were inferred from population genetics analysis, air trajectories, and disease occurrence. The red arrows represent migration routes supported by all analyses. The dark blue arrows show migration routes supported by population genetics and air trajectories. The green arrows indicate migration routes supported by disease surveys and air trajectories. The solid lines indicate direct migration from the Pst sources, whereas the dashed lines indicate indirect migration. The width of each arrow shows the probability of the migration route.
Genetic diversity and heterozygosity of the Pst population
Genetic diversity of each Pst geographic population was calculated using the nucleotide diversity parameters π and θω. Among all nine populations, the Guizhou Plateau (G3) had the highest diversity indices (π = 0.0037, θω = 0.0024), followed by Longnan (G1) (π = 0.0034, θω = 0.0023), and the Himalayan region (G2) (π = 0.0034, θω = 0.0022); these populations differed significantly from populations in other regions (P < 0.05). The lowest diversity parameters were detected in the Yili valley (G9) (π = 0.0008, θω = 0.0006) and Yunnan Plateau (G8) (π = 0.0011, θω = 0.0008) (Figure 2A). Heterozygosity identification showed that the observed heterozygosity was significantly higher (Padjusted < 0.05) than the expected heterozygosity in all geographic populations except for eastern Liupan Mountain (G4) and the Yili Valley (G9) (Figure 2B). A neutrality test showed that the highest value of Tajima’s D was observed in Longnan (2.34), followed by the Himalayan region (2.11) and the Guizhou Plateau (2.03); values in other populations were less than 2 and were significantly lower than those in G1 to G3 (P < 0.05) (Figure 2C). Similar calculations were performed to determine the genetic diversity and heterozygosity between cluster 1 and cluster 2. High values of genetic diversity (π = 0.0045, θω = 0.0025) and Tajima’s D (2.80) were observed in cluster 1 and were significantly greater than those in cluster 2 (π = 0.0045, θω = 0.0025, Tajima’s D = 1.69) (Supplemental Figure 6).
Figure 2.
Genetic diversity and heterozygosity of Pst populations in China.
(A) Boxplot of the nucleotide diversity of Pst populations. Genetic diversity was assessed with π and θω. Letters above boxes indicate significant differences determined with Kruskal-Wallis tests (P < 0.05). The median of each population is marked in the box.
(B) Histogram of the observed and expected heterozygosity of each cluster. A permutation test was performed to examine the significant variation between observed and expected heterozygosity. P values were adjusted using the Benjamini–Hochberg method. ∗Padjusted < 0.05. Error bars represent the standard errors of observed and expected heterozygosity for samples in each population.
(C) Neutrality test of Pst populations. The width of the violin plot represents the density of Tajima’s D values. The diamond symbols represent the median Tajima’s D values of populations. Letters indicate significant differences determined with the Kruskal-Wallis test (P < 0.05).
Correlation between population genetic and geographic distance
Because early studies showed that wind dispersal is important for long-distance Pst transport, we assumed that airflow would contribute to the spread of Pst in China. To examine this hypothesis, we performed a correlation analysis between population genetic distance and geographic distance using the Mantel test (Supplemental Tables 7 and 8). The lack of a significant correlation indicated that airflow was likely to be the major driving force for wide dispersal of Pst in China (Supplemental Figure 7).
Trajectory simulation and traceability analysis of Pst migration
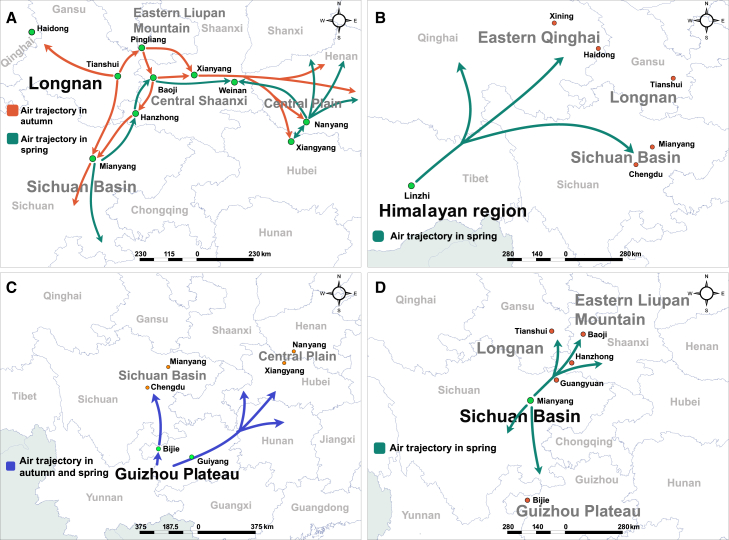
To identify the Pst source populations and their contributions to other populations, we performed air trajectory tracking analyses as an initial test. On the basis of climate conditions for oversummering and overwintering, cropping systems, and genetic diversity of Pst, Longnan, the Himalayan region, and the Guizhou Plateau were selected to perform Pst spread simulations. The annual cycle of stripe rust on winter wheat comprises four stages: oversummering, seedling infection in autumn, overwintering, and spring epidemic. Pst migrations in autumn and spring are therefore the key periods that account for disease epidemics. At 1000–3000 m above ground level (AGL), Pst from Longnan dispersed mainly to eastern Liupan Mountain with an average trajectory frequency (ATF) of 23%, and then to the Sichuan Basin (12%) and eastern Qinghai (8%) (Supplemental Figure 8), which were regarded as the major potential Pst epidemic regions threatened by inoculum from Longnan in autumn with regard to the distribution of wheat planting (Figure 3A). Further trajectory analysis in autumn indicated that Pst inocula in eastern Liupan Mountain were responsible for autumn seedling infection in southwestern Shaanxi (29%), central Shaanxi (17%), the Sichuan Basin (15%), and the Central Plain (5%) (Figure 3A and Supplemental Figure 8).
Figure 3.
Trajectory analysis of Pst migration routes in essential regions in China
The gray and orange arrows show the air trajectory in spring and autumn, respectively. The blue arrow indicates the air trajectory in both spring and autumn.
(A) The simulation period was October 1 to November 30 in Tianshui (Longnan) and Pingliang (eastern Liupan Mountain), January 1 to April 10 in Mianyang (Sichuan Basin), and February 20 to April 10 in Nanyang and Xiangyang (Central Plain).
(B) The air trajectory from Linzhi in the Himalayan region in spring. The simulation period was April 1 to May 31.
(C) Aerial transport from the Guizhou Plateau. The blue arrows show the air trajectory in spring and autumn. The simulation periods in the Guizhou Plateau were October 1 to November 30 and February 20 to April 10.
(D) The air trajectory from the Sichuan Basin in spring for the simulation period January 1 to April 10.
Because outbreaks of stripe rust in the Himalayan region occur in late spring, meteorological data from April to May was selected to perform trajectory simulations. At 3000–5000 m AGL, inocula from the Himalayan region were mainly responsible for disease incidence in the Sichuan Basin with an ATF greater than 16%. In addition to the Sichuan Basin, another possible dispersal route was to southern Qinghai and eastern Qinghai with a frequency of 3%–5% (Figure 3B and Supplemental Figure 9). The average trajectory distribution of the Guizhou Plateau mainly covered areas in the Sichuan Basin in autumn and spring with an ATF of 23%. Pst dispersal from the Guizhou Plateau to the Central Plain was also possible, with 5% ATF in autumn and 12% in spring (Figure 3C and Supplemental Figure 10). To examine whether Pst inocula in the Guizhou Plateau could have migrated from the adjacent Yunnan Plateau, a parallel simulation was performed for the Yunnan Plateau. Trajectory analysis confirmed Pst inocula from Yunnan dispersed to the Sichuan Basin in autumn with an ATF of greater than 20%, whereas ATF values for most areas in the Guizhou Plateau were less than 3% (Supplemental Figure 11). Pst trajectories from Longnan, the Himalayan region, and the Guizhou Plateau intersected in the Sichuan Basin, and Pst inocula in this region mainly circulated inside the basin with a possibility of greater than 50% and dispersed to eastern Liupan Mountain (7%), Longnan (5%), and northwest of the Guizhou Plateau (3%) in spring at 100–1000 m AGL (Figure 3D and Supplemental Figure 12).
To confirm and trace sources of Pst from overwintering regions in the Sichuan Basin and Central Plain, backward trajectory analyses were performed in each representative location. Backward trajectory aggregation suggested that Pst inocula in the Sichuan Basin were most likely to have come from Longnan and eastern Liupan Mountain, and those in the Central Plain were likely to have come from eastern Liupan Mountain and central Shaanxi (Supplemental Figure 13).
Historical demographic events between Pst populations
On the basis of the initial air trajectory tests, we further explored historical population demographic events between Longnan, the Himalayan region, the Guizhou Plateau, and other geographic populations using coalescent simulations and site frequency spectra. From 21 tested scenarios, the 7 best models were selected on the basis of agreement between estimated and observed maximum likelihoods (Figure 4A–4C and Supplemental Table 9). Pst populations in Longan were found to be the source of eastern Liupan Mountain populations by one-way migrations with a ratio of 0.047, and the divergence time was estimated to be 500–600 generations ago (Figure 4A). Historical demographic events between populations in Longnan and the Sichuan Basin indicated that their divergence occurred more than 40 000 generations ago, and migrations were detected in both directions with comparable migration ratios (Figure 4A). The Longnan population was also found to have diverged from that of eastern Qinghai more than 5000 generations ago. Although bidirectional migrations were identified between Longnan and eastern Qinghai, an order of magnitude higher migration ratio indicated that the dispersal mainly occurred from Longnan (Figure 4A).
Similar paired analyses of Pst populations were performed for the Himalayan region and the Guizhou Plateau with adjacent regions where airflow could reach. The divergence between the Himalayan region and Sichuan Basin populations was estimated to have occurred within the past 100 generations. Two-way migrations were found between these regions, with a migration ratio of 0.0048–0.0057 (Figure 4B). The divergence time between the Himalayan region and eastern Qinghai Pst populations was estimated at approximately 1000 generations ago. An asymmetric migration pattern mainly from the Himalayan region to eastern Qinghai was detected on the basis of the migration ratio (Figure 4B). Coalescent-based analysis showed that the Guizhou Plateau population diverged from the Sichuan Basin population 1000–2000 generations ago and from the Central Plain population more than 20 000 generations ago (Figure 4C). Bidirectional migrations were found within the diverged populations; migration ratios from the Guizhou Plateau were higher, indicating that more migration events derived from this region (Figure 4C).
Genetic exchanges among Pst populations
To further examine the findings from air trajectory simulations and historical demographic analyses, we investigated genetic introgressions and historical relationships among populations using the ABBA-BABA test and TreeMix. Thirty-four taxon topologies were acquired from phylogenetic analysis with D statistics (Supplemental Table 10). Significant genetic introgressions between P2 and P3 (see methods) were identified in the 24 topologies (Padjusted < 0.01). Among all the significant topologies, genetic exchanges between Longnan and eastern Liupan Mountain, Longnan and the Sichuan Basin, the Himalayan region and the Sichuan Basin, the Guizhou Plateau and the Central Plain, and the Guizhou Plateau and the Sichuan Basin were confirmed (Supplemental Table 10). The results of TreeMix further supported the recent migrations from Longnan toward eastern Liupan Mountain and from the Guizhou Plateau toward the Central Plain (Supplemental Figure 14).
Sequence of disease occurrence based on field investigations
To investigate whether the Pst migration pattern was consistent with the sequence of wheat stripe rust occurrence, we performed field surveys of disease onset in different regions during epidemic years. Because wheat stripe rust epidemics caused yield losses of 4.28 and 2.49 million tonnes in 2017 and 2020, respectively (Liu et al., 2022), the onset of Pst occurrence in the 2016–2017 and 2019–2020 cropping seasons was surveyed. Generally, the onset of disease occurrence was earlier in the 2019–2020 crop season than in 2016–2017 in most regions, but the temporal sequence of starting dates in different regions was essentially the same (Supplemental Tables 11 and 12). Stripe rust on wheat seedlings was first found in Longnan and eastern Liupan Mountain in October to early November, followed by the Sichuan Basin, Central Plain, and Yunnan Plateau from mid-November to December. The onset of stripe rust in spring epidemic regions, including Anhui, Jiangsu, Shanxi, Shandong, and Hebei, occurred in March to April the following spring. Although field surveillance might be insufficiently accurate, the basic dispersal routes could be inferred from the temporal sequence of disease occurrence in different regions.
Sources of Pst and their contributions to disease epidemics in China
By integrating population genetic diversity and structure analyses, air trajectories, coalescent-based analysis, temporal sequence of disease occurrence, and cropping systems in different regions, we deduced that Longnan, the Himalayan region, and the Guizhou Plateau were the most important Pst sources in China, and we proposed the main dispersal routes of Pst derived from these three sources (Figure 5). We concluded that Longnan is the most important source of Pst in China. Pst from this region is directly associated with disease occurrence in wheat-growing regions in eastern Liupan Mountain, the Sichuan Basin, and eastern Qinghai and may also affect central Shaanxi and the Central Plain. In autumn, the major dispersal routes from Longnan are to eastern Liupan Mountain, the Sichuan Basin, and eastern Qinghai. Once in eastern Liupan Mountain, Pst can proliferate and disperse in the same period (autumn), mainly to central Shaanxi, the Sichuan Basin, and the Central Plain, where urediniospore multiplication may occur during the winter. In the following spring, Pst from the overwintering regions can spread both northward and eastward to spring epidemic regions, including Shanxi, Hebei, Anhui, Jiangsu, and Shandong.
The Himalayan region is another Pst source, and its inoculum mainly contributes to disease epidemics in the Sichuan Basin and eastern Qinghai. Because stripe rust occurs in the Himalayan region mostly from April to June, Pst urediniospore inoculum from this region is likely to affect the disease mainly in the northern Sichuan Basin, where wheat is harvested in early June. Pst inoculum from the Himalayan region may also occasionally contribute to stripe rust in eastern Qinghai, where winter and spring wheat is harvested from July to October. The Guizhou Plateau, located in southwestern China, is another central Pst source, mainly contributing to disease in the Sichuan Basin and Central Plain. After oversummering, Pst inoculum in the Guizhou Plateau may be dispersed to the Sichuan Basin in the autumn, where it can multiply over winter. In addition, exchanges of Pst between the Guizhou Plateau and the Central Plain can take place in the autumn and spring in epidemic years. As the Sichuan Basin is a recipient of urediniospores from all three major sources, urediniospores produced in this area are mainly responsible for epidemics inside the basin, although urediniospores can also spread from the basin to Longnan and the Guizhou Plateau (Figure 5).
Discussion
As a distinct epidemiological region of stripe rust, China shows greater variation in Pst than other regions (Ali et al., 2014; Sharma-Poudyal et al., 2020). However, its specific inoculum centers and their related dispersal routes have not been clear. Recently, much intensive research has been performed to investigate Pst dispersal patterns in China, although it has focused on studying relationships among Pst populations in a few regions using a handful of molecular markers (Liang et al., 2013, 2016, 2021; Wan et al., 2015; Wang et al., 2020, 2022; Huang et al., 2021, 2022; Zhan et al., 2022). Analyses based on a limited number of markers and sampling sites may not provide sufficient resolution to dissect the genetic diversity and structure of Pst populations (Hubbard et al., 2015; Bueno-Sancho et al., 2017). In the present study, we dissected the population genetic structure of Pst in China, as well as other alien isolates. Using air trajectories as the initial tests, we performed coalescent-based analysis to propose Longnan, the Guizhou Plateau, and the Himalayan region as three major Pst sources in China, and we determined their population divergence from adjacent regions. Combining genetic introgression, migration event detection, and field disease monitoring, we proposed dispersal routes of Pst derived from each source.
Longnan has long been regarded as the Pst inoculum source responsible for stripe rust epidemics throughout China because of its high diversity of Pst pathotypes and genotypes (Wan et al., 2004; Chen et al., 2009; Hu et al., 2017; Liang et al., 2021; Wang et al., 2022). We confirmed that this region is a major Pst source on the basis of the population structure, high genetic diversity, and heterozygosity of Pst populations from major wheat-growing areas. Earlier studies proposed that Pst from Longnan could migrate to the Sichuan Basin (Liang et al., 2016) and eastern Liupan Mountain (Liang et al., 2013; Wang et al., 2022) based on gene flows. In the current study, we systematically evaluated the relationships between Longnan and adjacent regions by population genetic analyses and identified the directions of Pst migration using air trajectories and coalescent-based analysis. Pst originating in Longnan could migrate directly to eastern Liupan Mountain, the Sichuan Basin, and the eastern Qinghai regions, and eastern Liupan Mountain was the main Pst recipient (Figure 5), a result that was supported by genetic introgression and migration detection. Using genetic relationships among regions adjacent to Longnan from our earlier study (Wang et al., 2022), we found that Pst from Longnan could also contribute to disease epidemics in the Central Plain, located in the vast Huang-Huai-Hai wheat-growing region, via the bridge zones of eastern Liupan Mountain and central Shaanxi. Although a previous study inferred migration between Gansu and Sichuan Basin populations (Liang et al., 2016), distant divergence times and a low migration ratio between these regions indicate that such migration occurred infrequently. Wan et al. (2015) detected genetic exchanges between eastern Qinghai and Longnan by examining the frequency of shared genotypes. We found that eastern Qinghai, whose population diverged from that of Longnan approximately 5000 generations ago, was another Pst recipient of Longnan, with a relatively low migration ratio. Asymmetric migration patterns indicated that migrations derived mainly from Longnan.
Tibet in the Himalayan region was found to be a highly differentiated epidemic region because of Pst race and genotype composition in recent decades (Hu et al., 2012, 2017). Awais et al. (2022) proposed that Tibet was an independent epidemic zone that shared the fewest genotypes with other regions. These studies agreed that Pst in the Himalayan region was separate from and rarely exchanged with that of other areas in China. Our investigations confirmed that the Himalayan region was another independent Pst inoculum center with high genetic diversity, but we found that it contributed to disease epidemics in other wheat-growing areas, especially the Sichuan Basin and eastern Qinghai. It is interesting to note that the Pst populations in the Himalayan region diverged from those in the Sichuan Basin within the most recent 100 generations. A high migration ratio indicated that frequent migrations have occurred between these two regions. Although bidirectional migration events were detected, the air trajectory simulation indicated that these migrations originated mostly from the Himalayan region. In addition to the Sichuan Basin, migrations also appear to have occurred from the Himalayan region to eastern Qinghai, although with a lower frequency.
Studies on the relationships of Pst populations in southwestern China with those of adjacent areas have lagged behind studies of other epidemic regions. Wang et al. (2010) discovered long-distance spore transport among Guizhou, Sichuan, and Yunnan in southwestern China by air trajectory analysis with meteorological data. Recently, Jiang et al. (2022) explored the genetic relationships of Pst in southwestern and northwestern China. Zhan et al. (2022) and Huang et al. (2022) identified Pst migration routes from the Yunnan-Guizhou Plateau in southwestern China to the Central Plain by population genetic analyses. Awais et al. (2022) confirmed migrations between Pst populations in the southwest and the east in China and speculated that Yunnan in the Yunnan-Guizhou Plateau could play an important role in the spread of Pst inocula. Despite the similar topological features of the Yunnan and Guizhou Plateaus, Pst in these regions cannot be considered a single population, as our results show them to be distinct in genetic diversity, phylogenetic lineage, and trajectory distribution. We found that Pst populations from the Guizhou Plateau had the greatest genetic diversity (π), as well as the highest Watterson’s θω. As one of the proposed inoculum sources in China, the Guizhou Plateau mainly provided Pst inoculum to the Sichuan Basin and Central China, where Pst cannot oversummer. Based on divergence time and migration ratio with the Guizhou Plateau, the Sichuan Basin was determined to be the region where more migration events had occurred. Despite the fact that these populations diverged more than 20 000 generations ago, detection of a historical migration event from the Guizhou Plateau to the Central Plain indicated the importance of this migration route.
All three Pst sources are located in mountainous areas of China, and some isolates have rarely spread from such areas to other regions, resulting in preservation of ancient Pst races. A striking example is that CYR27 and CYR28, the predominant Pst races in the 1990s, were not detected in any regions except for Longnan in a nationwide race identification in 2010–2011. Similarly, CYR21 was detected only in Longnan, the Himalayan region, and Shaanxi (adjacent to Longnan) in race tests (Liu et al., 2012). High diversity of Pst populations in its inoculum centers can be explained by the coexistence of ancient and currently prevalent races. The mountainous terrain also provides suitable conditions for growth of barberry plants. Since these alternate hosts of Pst were found in 2010 (Jin et al., 2010), many Berberis spp. susceptible to Pst have been reported in Longnan (Zhao et al., 2013), the Himalayan region (Wang et al., 2016), and the Guizhou Plateau (Li et al., 2021b), together with natural Pst infection on alternate host plants. The high probability of sexual recombination in these regions may have contributed to their high Pst diversity, making them sources of emerging races.
The Huang-Huai-Hai plain is the major wheat-producing region in China, producing approximately 70% of the national wheat grain (Wang et al., 2009), and inocula that overwinter in the Central Plain directly affect spring rust occurrence in this vast region (Li and Zeng, 2002). Our field surveys showed that stripe rust is always found first on lower and older wheat seedlings in the Central Plain of the Huang-Huai-Hai region in spring, suggesting that the epidemic is largely caused by the local overwintering Pst inoculum. Trajectory tracking and field surveys in the current study indicated that Longnan is the primary source of Pst inoculum and is responsible for overwintering inoculum in the Central Plain via eastern Gansu and central Shaanxi. Previous field observations found that disease incidence in Longnan and central Shaanxi is related to that in the Central Plain in most years. However, during the severe stripe rust epidemic of 2017, rust in the Central Plain was more closely related to epidemics in southwestern China than in Longnan. More recently, studies proposed the profound effect of Pst inoculum from the southwest on the Central Plain (Awais et al., 2022; Huang et al., 2022; Zhan et al., 2022). However, we found that disease epidemics in the Central Plain mainly result from local overwintering inoculum, which may have arrived from the Guizhou Plateau the previous autumn. Because trajectory analysis indicated that the frequency of Pst dispersal from the Guizhou Plateau to the Central Plain is approximately 5% in the autumn, inoculum from the Guizhou Plateau can contribute to disease epidemics in the Huang-Huai-Hai region, but this depends largely on inoculum dose and climate conditions. Although the Himalayan region is another center of Pst inoculum, inoculum from this region is unlikely to cause rust in the Huang-Huai-Hai wheat-producing region directly; instead, it mainly contributes to rust development in the Sichuan Basin. Although the Huang-Huai-Hai region may be affected by Pst inocula from the Himalayan region by successive migrations across the Sichuan Basin, further evaluations are needed to assess this possibility.
Pst urediniospores can be dispersed for up to thousands of kilometers by airflow (Brown and Hovmøller, 2002; Hovmøller et al., 2002; Kim and Beresford, 2008), resulting in recurrent exchanges of spores among different regions. In contrast to patterns observed in humans, domestic animals, and plants, continual interchanges and coexistence of multiple races of Pst inocula in some regions may decouple phylogenetic clusters from geographic regions. A lack of correlation between genetic similarity and geographic distance indicated that constant wind dispersal may be a major factor driving exchanges of Pst nationwide. Similar results were reported for a study of wheat powdery mildew in individual geographic populations, including those in China (Sotiropoulos et al., 2022). In the current study, observed heterozygosity was significantly higher than expected in most geographic populations. Given that the wind plays an important role in Pst dispersal, we speculated that frequent genetic exchanges caused by airflow could be the reason for isolate-breaking effects, resulting in deviations from Hardy-Weinberg equilibrium.
Evaluation of sources and migration patterns of wheat stripe rust cannot solely rely on shared genotypes, population structure, or fixation index without considering pathogen migratory direction, air trajectory, and cropping systems. We performed integrative investigations to identify Pst inoculum sources and possible key dispersal routes, which are valuable for implementing effective Pst monitoring and management strategies. Reducing Pst oversummering inoculum at its source(s) and performing effective rust management in areas along important dispersal routes should be an efficient strategy for prevention of severe rust in main wheat production regions. Selection of resistant cultivars and application of fungicides when needed can be used to reduce Pst inoculum in oversummering regions. These approaches should also be considered in other epidemic regions in case rust control in key overwintering regions is not as effective as desired. Pst isolates analyzed in the present study were collected in 2015 instead of multiple years. In future investigations, the spatial and temporal distribution of samples will be considered.
Methods
Sample collection, purification, and multiplication
A total of 154 Pst-infected wheat leaves were sampled in 11 provinces throughout China in 2015, and only one leaf bearing uredinia was sampled from a single infection spot in a single field. Collected samples were stored in a desiccator at 4°C for no longer than 1 week before being used to inoculate wheat seedlings for inoculum multiplication. To obtain fresh urediniospores for inoculation, the infected leaf was placed on a water-soaked paper towel in a dew chamber for 16 h in the dark at 10°C. Urediniospores from a single fresh uredium were used to inoculate the seedlings of Mingxian 169, a wheat cultivar that is highly susceptible to all Pst races identified to date in China. Incubation of inoculated seedlings and subsequent collection of urediniospores from infected seedlings followed previously described procedures (Wan and Chen, 2014; Li et al., 2019a). Multiplication of urediniospores was repeated until there was a sufficient quantity for DNA extraction.
DNA extraction and whole-genome sequencing
Genomic DNA was extracted directly from harvested urediniospores using the cetyltrimethyl ammonium bromide method with some modifications (Chen et al., 1993). Quality and quantity of genomic DNA were examined with 0.8% agarose gel electrophoresis and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Whole-genome sequencing was performed on the Illumina NovaSeq 6000 platform by Novogene Co., Ltd. (Beijing, China). A 150-bp, paired-end DNA library for each Pst isolate was constructed using the NEB Next Ultra DNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA). All raw sequence reads were deposited in the National Center for Biotechnology Information under BioProject PRJNA809046. The read-depth coverage of each isolate was assessed using SAMtools v1.9 (Li et al., 2009). Raw read quality was evaluated with FastQC v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and low-quality reads were removed using Trimmomatic v0.39 (Bolger et al., 2014).
Detection and annotation of variants
Genomic variants were identified using a previously reported framework (DePristo et al., 2011). To select a reference genome, Pst genomes 11-281 (Li et al., 2019b), 93-210 (Xia et al., 2018), 104E137A (Schwessinger et al., 2018), DK09_11 (Schwessinger et al., 2020), and PST-130 (Vasquez-Gross et al., 2020) sequenced by third-generation sequencing technology were evaluated by statistical assessment with BUSCO v3.0.2 (https://gitlab.com/ezlab/busco). Error-corrected reads of 154 Pst isolates were aligned to the selected reference genome using BWA-MEM v0.7.17 with default settings (Li and Durbin, 2009). The obtained files in SAM format were converted to BAM format using SAMtools. The BAM files were cleaned, sorted, and validated with Picard tools v2.23.9 (https://broadinstitute.github.io/picard/). Variant calling was performed with Genome Analysis Toolkit (GATK) v4.1.9.0 HaplotypeCaller and ApplyBQSR (https://gatk.broadinstitute.org/) using the indexed filtered BAM files as input. Two rounds of calling were performed to identify SNPs and indels throughout the genome as previously reported (Li et al., 2020). Hard filtering was used to tease apart substandard variants and select high-quality SNPs and indels using GATK VariantFiltration. Parameters were set to “QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0 || SOR>3” according to the recommendation of the GATK Best Practice Pipeline, with modifications (https://github.com/gatk-workflows/). Filtered variants of each isolate were merged into a single genome variant call format (gVCF) file using GATK CombineGVCFs. Joint genotyping was performed on the combined file using GATK GenotypeGVCFs. The output file was filtered using VCFtools v0.1.13 (https://vcftools.github.io) with the parameters “--maf 0.05 --mac 3 --minQ 200 --max-missing 0.3”. To retain high-quality variants whose read-depth coverage deviated less than 30% from the average in the 154 Pst isolates, the parameters “--min-meanDP 38 --max-meanDP 71” were set using VCFtools. Variants were further annotated with SnpEff v4.3t (Cingolani et al., 2012).
Phylogenetic and population structure analyses
To ensure that adjacent SNPs were not linked, the squared correlation (r2) was calculated to estimate the linkage disequilibrium between each pair of SNPs in a window of 50 SNPs with 1 SNP shifted forward across the genome using PLINK v1.9 (Stamatakis, 2014). SNPs with r2 greater than 0.2 were removed. Remaining unlinked SNPs were used to construct maximum likelihood phylogenetic trees with 1000 replications using RAxML v8.2.12 (Alexander et al., 2009), and a rapid bootstrap analysis was performed with the GTRGAMMAX model. SNPs of Pst isolates from India (Race 31, Race K, and Race Yr9) (Kiran et al., 2017), the UK (87/66, 03/07, 01/34, 98/83, 87/66, 87/27, 08/501, 11/140, WYR 88.45SS, WYR88.44SS3, WYR 88.55SS, and WYR88.5SS1) (Hubbard et al., 2015), and the United States (93-210) (Xia et al., 2018) detected using the same approach were included in the phylogenetic analysis. Psh isolate 93TX-2 (Xia et al., 2018) was used as an outgroup in the phylogenetic analysis. Phylogenetic trees were visualized and annotated using iTOL v6 (https://itol.embl.de/). A bootstrap value of at least 60 indicated that the node was well supported. Population genetic structure was examined using ADMIXTURE v1.3.0 with K values ranging from 2 to 10 for 500 replications (Watterson, 1975). PCA was performed based on the SNP data using PLINK v1.9 (Purcell et al., 2007), and the first two PCs were plotted using the ggplot2 package in R v4.1.2.
Genetic diversity and heterozygosity identification
The genetic diversity of Pst isolates in each cluster identified on the basis of phylogeny and population genetic structure was evaluated using nucleotide diversity parameters (π and θω). π was computed in 5000-bp sliding windows using VCFtools v0.1.13 (https://vcftools.github.io). θω was calculated using the formulas and , where K is the proportion of segregating sites in the window and n is the total number of haploids (Watterson, 1975). The significance level of differences in π and θω between different groups was assessed with a nonparametric Kruskal–Wallis test. To determine the genetic variation in the population, observed and expected heterozygosity were estimated with the formulas and using PLINK v1.9, where N is the total number of alleles, and and are the observed and expected number of homologous alleles, respectively (Chen et al., 2016). A permutation test was performed to examine significant variation between observed and expected heterozygosity. A neutrality test with Tajima’s D was used to confirm the heterozygosity levels among different populations using VCFtools v0.1.13 with a 5000-bp sliding window. Significance levels derived from Tajima’s D among different populations were determined with the Kruskal–Wallis test.
Correlation between genetic and geographic distance
A Mantel test was performed to examine the correlation between genetic distance and geographic distance of Pst populations. The population genetic distance was calculated by Pst paired geographic population fixation statistics (FST) using VCFtools v0.1.13. The dist () command in the geosphere v1.5-14 R package (Karney, 2013) was used to calculate the geographic distance. The matrix of FST/(1 – FST) (Rousset, 1997) and the matrix of geographic population were used to run the mantel () command in the vegan v2.6-4 R package (Oksanen, 2007) with 9999 permutations. The kde2d () command from the MASS v7.3-53 R package (https://cran.r-project.org/web/packages/MASS/index.html) was used to calculate the two-dimensional kernel density estimate for all correlated points in the genetic/geographic matrix.
Trajectory simulation and traceability analysis of Pst migration
On the basis of earlier epidemiological studies of Pst in China, representative locations were selected and used to perform trajectory simulation of Pst with Hybrid Single-Particle Lagrangian Integrated Trajectory version 4 (HYSPLIT-4) (Draxler and Hess, 1998). Weekly meteorological data from 2010 to 2019 were obtained from the National Oceanic and Atmospheric Administration (NOAA) website (ftp://arlftp.arlhq.noaa.gov/pub/archives/gdas1). The data were converted to 1° latitude-longitude (360 × 181) grids using NOAA’s Air Resource Laboratory archiving program. Altitudes and simulation time periods were selected based on different geographic locations to mimic biological events of Pst dispersal (Supplemental Tables 13 and 14). Using the meteorological data as input for the Trajectory model in HYSPLIT-4, trajectory frequencies were calculated for Pst dispersal from each location at 100–1000 m, 1000–3000 m, and 3000–5000 m AGL, depending on the geographic characteristics of the locations. Because Pst spores can survive in the air for up to 120 h (Wang et al., 2010), each simulation was run for 120 h. The ATF spanning the entire 10 years for each location was calculated. The range of these highly frequent trajectories was visualized using the ggplot2, tmap, and sp Gallery packages in R v4.1.2. To trace the potential source of Pst dispersal in the Sichuan Basin, Henan, and Hubei from October 1 to November 30 (Supplemental Table 13), MeteoInfoMap and TrajStat software were used for backward trajectory simulation analysis (http://meteothink.org/index.html) using the meteorological data from the NOAA. The angle distance clustering method was used to compute backward trajectory clusters in the altitude from 100 m to 3000 m AGL (http://meteothink.org/docs/trajstat/cluster_cal.html).
Inference of population demographic history
We used fastsimcoal2 to estimate population demographic history by simulating historical events (Excoffier et al., 2021). The VCF files we combined in each geographic population were used to calculate the site frequency spectrum (SFS) using the easySFS.py script (https://github.com/isaacovercast/easySFS). We used the folded observed SFS to perform 1 000 000 coalescent simulations (-n) using 40 optimization cycles (-L) during likelihood maximization (-M) using fastsimcoal2 v2.7.0.9. The mutation rate was set to 2.0 × 10−8 per base pair per year according to previous Pst studies (Hovmøller and Justesen, 2007; Xia et al., 2018). A total of 21 scenarios were tested with different models. The optimal scenarios were selected according to the best-supported likelihoods.
Determination of genetic exchanges among Pst populations
Genetic introgression analyses were performed using D statistics (also known as the ABBA-BABA test) to identify evidence of genetic introgression between populations using Dsuite software (Malinsky et al., 2021). The D and f4-ratio can be presented as a given four-taxon topology ((P1, P2), P3, O). In the ABBA pattern, P1 harbors an ancestral allele “A” derived from outgroup O, whereas P2 and P3 share a different allele “B” at a SNP site. In the BABA pattern, P1 and P3 share the allele “B,” whereas P2 retains the allele “A” derived from outgroup O. Under the null hypothesis, which assumes that no genetic introgression occurs, equal frequencies should be observed in ABBA and BABA patterns. A significant deviation indicates that introgression occurred between P3 and P1 or P2. The f4-ratio shows the mixing proportions of admixture events (Patterson et al., 2012). Migration events among different populations in the maximum likelihood phylogeny were estimated using TreeMix with high-quality SNPs filtered by linkage disequilibrium (Pickrell and Pritchard, 2012). The number of migration events (m) from 1 to 7 between any two populations was tested with 20 replications, and the optimal number of events was determined using the OptM package in R v4.1.2 (Fitak, 2021).
Field surveillance surveys of stripe rust occurrence
The occurrence of wheat stripe rust in the main epidemic regions in China was investigated by the co-authors at the National Agro-Tech Extension and Service Center, universities, and regional academic institutes. The two most recent epidemics in 2017 and 2020 were selected to estimate the temporal sequence of disease occurrence among the selected regions. Disease onset in the representative locations was estimated on the basis of field monitoring. Pst dispersal routes were inferred from the earliest dates of stripe rust observation and the dates of sowing and harvesting.
Funding
This work was supported by the National Key Research and Development Program of China to X.H. and Y.L. (2021YFD1401000), the National Natural Science Foundation of China (31471731 and 31772102) to X.H., and the Postdoctoral Science Foundation of China to Y.L. (2021M690130).
Author contributions
X.H. and Z.K. designed and managed the research project; Y.L. assisted with the experimental design; X.H. and Y.L. performed data analysis and wrote the manuscript; J.D. conducted the population genetic analysis; T.Z. performed air trajectory analysis; Y.L. and S.Z. performed the coalescent-based analyses; J.Z. and C.W. participated in inoculating and harvesting spores; B.W., Q.Y., M.L., C.L., Y.P., S.Q., H.G., W.H., X.F., Y.B., and Z.Q. participated in disease surveys and sampling; G.Z. and F.T. assisted with spore production; B.W., C.H., and W.L. conducted disease surveys; H.S., J.Z., X.X., and X.C. offered suggestions for the study and revised the manuscript. All the authors read and approved the manuscript.
Acknowledgments
We acknowledge the high-performance computing clusters at the State Key Laboratory of Crop Stress Biology for Arid Areas for providing computing resources. No conflict of interest is declared.
Published: February 21, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Zhensheng Kang, Email: kangzs@nwsuaf.edu.cn.
Xiaoping Hu, Email: xphu@nwsuaf.edu.cn.
Accession numbers
Raw sequenced reads from 154 samples have been deposited at the National Center for Biotechnology Information in BioProject database PRJNA809046 containing SRA accessions SRR18091433 to SRR18091586 and are publicly available at the China National Genomics Data Center database under BioProject PRJCA014799. Code analyzed in this study has been described in the Methods and is available on GitHub at https://github.com/yuxiang-li/code_Pst_population.
Supplemental information
References
- Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Gladieux P., Leconte M., Gautier A., Justesen A.F., Hovmøller M.S., Enjalbert J., de Vallavieille-Pope C. Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f. sp. tritici. PLoS Pathog. 2014;10:e1003903. doi: 10.1371/journal.ppat.1003903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awais M., Ali S., Ju M., Liu W., Zhang G., Zhang Z., Li Z., Ma X., Wang L., Du Z., et al. Countrywide inter-epidemic region migration pattern suggests the role of southwestern population in wheat stripe rust epidemics in China. Environ. Microbiol. 2022;24:4684–4701. doi: 10.1111/1462-2920.16096. [DOI] [PubMed] [Google Scholar]
- Beresford R.M. Stripe rust (Puccinia striiformis), a new disease of wheat in New Zealand. Cereal Rusts Bulletin. 1982;10:35–41. [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J., Gregory P.H., Johnson C.G. Possible wind transport of coffee leaf rust across the Atlantic Ocean. Nature. 1971;229:500–501. doi: 10.1038/229500b0. [DOI] [PubMed] [Google Scholar]
- Brown J.K.M., Hovmøller M.S. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- Bueno-Sancho V., Persoons A., Hubbard A., Cabrera-Quio L.E., Lewis C.M., Corredor-Moreno P., Bunting D.C.E., Ali S., Chng S., Hodson D.P., et al. Pathogenomic analysis of wheat yellow rust lineages detects seasonal variation and host specificity. Genome Biol. Evol. 2017;9:3282–3296. doi: 10.1093/gbe/evx241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Cosgrove E.J., Bowman R., Fitzpatrick J.W., Clark A.G. Genomic consequences of population decline in the endangered Florida scrub-Jay. Curr. Biol. 2016;26:2974–2979. doi: 10.1016/j.cub.2016.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Xu S., Wu L. Epidemiology and sustainable management of wheat stripe rust caused by Puccinia striiformis West. in China: a historical retrospect and prospect. (In Chinese with English abstr.) Sci. Agric. Sin. 2007;40:177–183. [Google Scholar]
- Chen W.Q., Wu L.R., Liu T.G., Xu S.C., Jin S.L., Peng Y.L., Wang B.T. Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis. 2009;93:1093–1101. doi: 10.1094/PDIS-93-11-1093. [DOI] [PubMed] [Google Scholar]
- Chen X., Line R.F., Leung H. Relationship between virulence variation and DNA polymorphism in Puccinia striiformis. Phytopathology. 1993;83:1489–1497. [Google Scholar]
- Chen X.M. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. J. Indian Dent. Assoc. 2005;27:314–337. [Google Scholar]
- Chen X. Pathogens which threaten food security: Puccinia striiformis, the wheat stripe rust pathogen. Food Secur. 2020;12:239–251. [Google Scholar]
- Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., Del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Cuddy W.S., Wellings C.R., Zhang P., Thach T., Hovmøller M.S., Qutob D., Brar G.S., Kutcher H.R., Park R.F. Incursions of divergent genotypes, evolution of virulence and host jumps shape a continental clonal population of the stripe rust pathogen Puccinia striiformis. Mol. Ecol. 2021;30:6566–6584. doi: 10.1111/mec.16182. [DOI] [PubMed] [Google Scholar]
- Draxler R.R., Hess G.D. An overview of the HYSPLIT_4 modeling system of trajectories, dispersion, and deposition. Aust. Meteorol. Mag. 1998;47:295–308. [Google Scholar]
- Duveiller E., Singh R.P., Nicol J.M. The challenges of maintaining wheat productivity: pests, diseases, and potential epidemics. Euphytica. 2007;157:417–430. [Google Scholar]
- Dyer A., Matusralt M., Drenth A., Cohen A., Splelman L. Historical and recent migrations of Phytophthora infestans: chronology, pathways, and implications. Plant Dis. 1993;77:653–661. [Google Scholar]
- Excofffier L., Marchi N., Marques D.A., Matthey-Doret R., Gouy A., Sousa V.C. fastsimcoal2: demographic inference under complex evolutionary scenarios. Bioinformatics. 2021;37:4882–4885. doi: 10.1093/bioinformatics/btab468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M.C., Henk D.A., Briggs C.J., Brownstein J.S., Madoff L.C., McCraw S.L., Gurr S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitak R.R. OptM: estimating the optimal number of migration edges on population trees using Treemix. Biol. Methods Protoc. 2021;6:bpab017. doi: 10.1093/biomethods/bpab017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss E.M. Genome-enabled analysis of plant-pathogen migration. Annu. Rev. Phytopathol. 2015;53:121–135. doi: 10.1146/annurev-phyto-080614-115936. [DOI] [PubMed] [Google Scholar]
- Hovmøller M.S., Justesen A.F. Rates of evolution of avirulence phenotypes and DNA markers in a northwest European population of Puccinia striiformis f. sp. tritici. Mol. Ecol. 2007;16:4637–4647. doi: 10.1111/j.1365-294X.2007.03513.x. [DOI] [PubMed] [Google Scholar]
- Hovmøller M.S., Justesen A.F., Brown J.K.M. Clonality and long-distance migration of Puccinia striiformis f. sp. tritici in north-west Europe. Plant Pathol. 2002;51:24–32. [Google Scholar]
- Hovmøller M.S., Sørensen C.K., Walter S., Justesen A.F. Diversity of Puccinia striiformis on cereals and grasses. Annu. Rev. Phytopathol. 2011;49:197–217. doi: 10.1146/annurev-phyto-072910-095230. [DOI] [PubMed] [Google Scholar]
- Hu X., Li J., Wang Y., Wang B., Li Q., Kang Z., Yang M., Peng Y., Liu T., Chen W., et al. Race composition of Puccinia striiformis f. sp. tritici in Tibet, China. Plant Dis. 2012;96:1615–1620. doi: 10.1094/PDIS-10-11-0884-RE. [DOI] [PubMed] [Google Scholar]
- Hu X., Ma L., Liu T., Wang C., Peng Y., Pu Q., Xu X. Population genetic analysis of Puccinia striiformis f. sp. tritici suggests two distinct populations in Tibet and the other regions of China. Plant Dis. 2017;101:288–296. doi: 10.1094/PDIS-02-16-0190-RE. [DOI] [PubMed] [Google Scholar]
- Huang C., Jiang Y.Y., Li P.L., Peng H., Cui Y., Yang J., Xie F.Z. Epidemics analysis of wheat stripe rust in China in 2017. J. Plant Prot. 2018;44:162–166. [Google Scholar]
- Huang L., Yang H., Xia C., Li H., Wang J., Wang A., Zhang M., Kang X., Gao L., Zhou Y., et al. Long-distance transport of Puccinia striiformis f. sp. tritici by upper airflow on the Yunnan–Guizhou Plateau disrupts the balance of agricultural ecology in central China. Plant Dis. 2022;106:2940–2947. doi: 10.1094/PDIS-01-22-0038-RE. [DOI] [PubMed] [Google Scholar]
- Huang M., Liu T., Cao S., Yuen J., Zhan J., Jia Q., Gao L., Liu B., Chen W., Berlin A. Analyses of wheat yellow rust populations reveal sexual recombination and seasonal migration pattern of Puccinia striiformis f. sp. tritici in Gangu, Northwestern China. Phytopathology. 2021;111:2268–2277. doi: 10.1094/PHYTO-12-20-0558-R. [DOI] [PubMed] [Google Scholar]
- Hubbard A., Lewis C.M., Yoshida K., Ramirez-Gonzalez R.H., de Vallavieille-Pope C., Thomas J., Kamoun S., Bayles R., Uauy C., Saunders D.G.O. Field pathogenomics reveals the emergence of a diverse wheat yellow rust population. Genome Biol. 2015;16:1–15. doi: 10.1186/s13059-015-0590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B., Wang C., Guo C., Lv X., Gong W., Chang J., He H., Feng J., Chen X., Ma Z. Genetic relationships of Puccinia striiformis f. sp. tritici in southwestern and northwestern China. Microbiol. Spectr. 2022;10:e0153022. doi: 10.1128/spectrum.01530-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Szabo L.J., Carson M. Century-old mystery of Puccinia striiformis life history solved with the identification of Berberis as an alternate host. Phytopathology. 2010;100:432–435. doi: 10.1094/PHYTO-100-5-0432. [DOI] [PubMed] [Google Scholar]
- Johnson R., Taylor A.J. Isolates of Puccinia striiformis collected in England from the wheat varieties Maris Beacon and Joss Cambier. Nature. 1972;238:105–106. [Google Scholar]
- Karney C.F.F. Algorithms for geodesics. J. Geodes. 2013;87:43–55. [Google Scholar]
- Kim K.S., Beresford R.M. Use of a spectrum model and satellite cloud data in the simulation of wheat stripe rust (Puccinia striiformis) dispersal across the Tasman Sea in 1980. Agric. For. Meteorol. 2008;148:1374–1382. [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhang Y., Chen W., Duan X., Liu T., Jia Q., Cao S., Xu Z. Evidence for Yunnan as the major origin center of the dominant wheat fungal pathogen Puccinia striiformis f. sp . tritici. Australas. Plant Pathol. 2021;50:241–252. [Google Scholar]
- Li S.n., Chen W., Ma X.y., Tian X.x., Liu Y., Huang L.l., Kang Z.s., Zhao J. Identification of eight Berberis species from the Yunnan-Guizhou plateau as aecial hosts for Puccinia striiformis f. sp. tritici, the wheat stripe rust pathogen. J. Integr. Agric. 2021;20:1563–1569. [Google Scholar]
- Li Y., Wang M., See D.R., Chen X. Ethyl-methanesulfonate mutagenesis generated diverse isolates of Puccinia striiformis f. sp. tritici, the wheat stripe rust pathogen. World J. Microbiol. Biotechnol. 2019;35:28. doi: 10.1007/s11274-019-2600-6. [DOI] [PubMed] [Google Scholar]
- Li Y., Xia C., Wang M., Yin C., Chen X. Genome sequence resource of a Puccinia striiformis isolate infecting wheatgrass. Phytopathology. 2019;109:1509–1512. doi: 10.1094/PHYTO-02-19-0054-A. [DOI] [PubMed] [Google Scholar]
- Li Y., Xia C., Wang M., Yin C., Chen X. Whole-genome sequencing of Puccinia striiformis f. sp. tritici mutant isolates identifies avirulence gene candidates. BMC Genom. 2020;21:247–322. doi: 10.1186/s12864-020-6677-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.Q., Zeng S.M. China Agriculture Press; Beijing, China: 2002. Wheat Rust in China. [Google Scholar]
- Liang J., Liu X., Li Y., Wan Q., Ma Z., Luo Y. Population genetic structure and the migration of Puccinia striiformis f. sp. tritici between the Gansu and Sichuan Basin populations of China. Phytopathology. 2016;106:192–201. doi: 10.1094/PHYTO-03-15-0081-R. [DOI] [PubMed] [Google Scholar]
- Liang J., Liu X., Tsui C.K.M., Ma Z., Luo Y. Genetic structure and asymmetric migration of wheat stripe rust pathogen in western epidemic areas of China. Phytopathology. 2021;111:1252–1260. doi: 10.1094/PHYTO-06-20-0236-R. [DOI] [PubMed] [Google Scholar]
- Liang J., Wan Q., Luo Y., Ma Z. Population genetic structures of Puccinia striiformis in Ningxia and Gansu provinces of China. Plant Dis. 2013;97:501–509. doi: 10.1094/PDIS-01-12-0072-RE. [DOI] [PubMed] [Google Scholar]
- Liu T.G., Wang B.T., Jia Q.Z., Zhang Z.Y., Li Q., Cao S.Q., Peng Y.L., Jin S.L., Li M.J., Liu B. Physiologic specialization of Puccinia striiformis f. sp. tritici in China during 2010-2011. J. Triticeae Crop. 2012;32:574–578. [Google Scholar]
- Liu W.C., Zhao Z.H., Wang B.T., Li Y., Wang X.J., Kang Z.S. Analysis on the contribution rate of plant protection to the control of wheat stripe rust in China. China Plant Prot. 2022;42:5–9+53. [Google Scholar]
- Kiran K., Rawal H.C., Dubey H., Jaswal R., Bhardwaj S.C., Prasad P., Pal D., Devanna B.N., Sharma T.R. Dissection of genomic features and variations of three pathotypes of Puccinia striiformis through whole genome sequencing. Sci. Rep. 2017;7:42419. doi: 10.1038/srep42419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M., Matschiner M., Svardal H. Dsuite-Fast D-statistics and related admixture evidence from VCF files. Mol. Ecol. Resour. 2021;21:584–595. doi: 10.1111/1755-0998.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan S., Singh D.V. Long-distance dispersion of rust pathogens. Annu. Rev. Phytopathol. 1990;28:139–153. doi: 10.1146/annurev.py.28.090190.001035. [DOI] [PubMed] [Google Scholar]
- Oksanen J. Vegan: community ecology package. R package version 1.8-5. 2007. http://www.cran.r-project.org
- Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J., Pritchard J. Inference of population splits and mixtures from genome-wide allele frequency data. Nat. Prec. 2012:1. doi: 10.1038/npre.2012.6956.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashar M., Bhardwaj S.C., Jain S.K., Datta D. Pathotypic evolution in Puccinia striiformis in India during 1995–2004. Aust. J. Agric. Res. 2007;58:602–604. [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristaino J.B., Anderson P.K., Bebber D.P., Brauman K.A., Cunniffe N.J., Fedoroff N.V., Finegold C., Garrett K.A., Gilligan C.A., Jones C.M., et al. The persistent threat of emerging plant disease pandemics to global food security. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2022239118. e2022239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Chen Y.J., Tien R., Vogt J.K., Sperschneider J., Nagar R., McMullan M., Sicheritz-Ponten T., Sørensen C.K., Hovmøller M.S., et al. Distinct life histories impact dikaryotic genome evolution in the rust fungus Puccinia striiformis causing stripe rust in wheat. Genome Biol. Evol. 2020;12:597–617. doi: 10.1093/gbe/evaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Sperschneider J., Cuddy W.S., Garnica D.P., Miller M.E., Taylor J.M., Dodds P.N., Figueroa M., Park R.F., Rathjen J.P. A near-complete haplotype-phased genome of the dikaryotic wheat stripe rust fungus Puccinia striiformis f. sp. tritici reveals high interhaplotype diversity. mBio. 2018;9 doi: 10.1128/mBio.02275-17. e02275022755-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma-Poudyal D., Bai Q., Wan A., Wang M., See D., Chen X. Molecular characterization of international collections of the wheat stripe rust pathogen Puccinia striiformis f. sp. tritici reveals high diversity and intercontinental migration. Phytopathology. 2020;110:933–942. doi: 10.1094/PHYTO-09-19-0355-R. [DOI] [PubMed] [Google Scholar]
- Sotiropoulos A.G., Arango-Isaza E., Ban T., Barbieri C., Bourras S., Cowger C., Czembor P.C., Ben-David R., Dinoor A., Ellwood S.R., et al. Global genomic analyses of wheat powdery mildew reveal association of pathogen spread with historical human migration and trade. Nat. Commun. 2022;13:4315–4414. doi: 10.1038/s41467-022-31975-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs, R. (1985). Stripe rust: the cereal rusts II: diseases, distribution, epidemiology, and control. (Eds.): A.P. Roelfs and W.R. Bushnell. Academic Press, Inc. New York.
- Vasquez-Gross H., Kaur S., Epstein L., Dubcovsky J. A haplotype-phased genome of wheat stripe rust pathogen Puccinia striiformis f. sp. tritici, race PST-130 from the Western USA. PLoS One. 2020;15:e0238611. doi: 10.1371/journal.pone.0238611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan A.M., Chen X.M., He Z.H. Wheat stripe rust in China. Aust. J. Agric. Res. 2007;58:605–619. [Google Scholar]
- Wan A., Chen X. Virulence characterization of Puccinia striiformis f. sp. tritici using a new set of Yr single-gene line differentials in the United States in 2010. Plant Dis. 2014;98:1534–1542. doi: 10.1094/PDIS-01-14-0071-RE. [DOI] [PubMed] [Google Scholar]
- Wan A.M., Wu L.R., Jin S.L., Jia Q.Z., Yao G., Yang J.X., Wang B.T., Li G.B., Yuan Z.Y. Occurrence of wheat stripe rust and monitoring of physiological races of Puccinia striiformis f. sp. tritici in China in 2000 ∼ 2001. J. Plant Prot. 2002;28:5–8. [Google Scholar]
- Wan A., Zhao Z., Chen X., He Z., Jin S., Jia Q., Yao G., Yang J., Wang B., Li G., et al. Wheat stripe rust epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002. Plant Dis. 2004;88:896–904. doi: 10.1094/PDIS.2004.88.8.896. [DOI] [PubMed] [Google Scholar]
- Wan Q., Liang J., Luo Y., Ma Z. Population genetic structure of Puccinia striiformis in northwestern China. Plant Dis. 2015;99:1764–1774. doi: 10.1094/PDIS-02-15-0144-RE. [DOI] [PubMed] [Google Scholar]
- Wang C., Li L., Jiang B., Zhang K., Chu B., Luo Y., Ma Z. Genetic diversity and population structure of Puccinia striiformis f. sp. tritici reveal its migration from central to eastern China. Crop Protect. 2020;128:104974. [Google Scholar]
- Wang C., Li Y., Wang B., Hu X. Genetic analysis reveals relationships among populations of Puccinia striiformis f. sp. tritici from the Longnan, Longdong and central Shaanxi regions of China. Phytopathology. 2022;112:278–289. doi: 10.1094/PHYTO-07-20-0312-R. [DOI] [PubMed] [Google Scholar]
- Wang E., Chen C., Yu Q. 18th World IMACS/MODSIM Congress. Cairns; Australia: 2009. Modeling the response of wheat and maize productivity to climate variability and irrigation in the North China Plain; pp. 2742–2748. [Google Scholar]
- Wang H., Yang X.B., Ma Z. Long-distance spore transport of wheat stripe rust pathogen from Sichuan, Yunnan, and Guizhou in southwestern China. Plant Dis. 2010;94:873–880. doi: 10.1094/PDIS-94-7-0873. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhao J., Chen X., Peng Y., Ji J., Zhao S., Lv Y., Huang L., Kang Z. Virulence variations of Puccinia striiformis f. sp. tritici isolates collected from Berberis spp. in China. Plant Dis. 2016;100:131–138. doi: 10.1094/PDIS-12-14-1296-RE. [DOI] [PubMed] [Google Scholar]
- Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wellings C.R. Puccinia striiformis in Australia: a review of the incursion, evolution, and adaptation of stripe rust in the period 1979–2006. Aust. J. Agric. Res. 2007;58:567–575. [Google Scholar]
- Wellings C.R. Global status of stripe rust: a review of historical and current threats. Euphytica. 2011;179:129–141. [Google Scholar]
- Xia C., Wang M., Yin C., Cornejo O.E., Hulbert S.H., Chen X. Genomic insights into host adaptation between the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici) and the barley stripe rust pathogen (Puccinia striiformis f. sp. hordei) BMC Genom. 2018;19:664. doi: 10.1186/s12864-018-5041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S.M., Luo Y. Long-distance spread and interregional epidemics of wheat stripe rust in China. Plant Dis. 2006;90:980–988. doi: 10.1094/PD-90-0980. [DOI] [PubMed] [Google Scholar]
- Zhan G., Ji F., Chen X., Wang J., Zhang D., Zhao J., Zeng Q., Yang L., Huang L., Kang Z. Populations of Puccinia striiformis f. sp. tritici in winter spore production regions were spread from southwestern oversummering areas in China. Plant Dis. 2022;106:2856–2865. doi: 10.1094/pdis-09-21-2070-re. [DOI] [PubMed] [Google Scholar]
- Zhao J., Wang L., Wang Z., Chen X., Zhang H., Yao J., Zhan G., Chen W., Huang L., Kang Z. Identification of eighteen Berberis species as alternate hosts of Puccinia striiformis f. sp. tritici and virulence variation in the pathogen isolates from natural infection of barberry plants in China. Phytopathology. 2013;103:927–934. doi: 10.1094/PHYTO-09-12-0249-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.