Abstract
Gene duplication is assumed to be the major force driving the evolution of metabolite biosynthesis in plants. Freed from functional burdens, duplicated genes can mutate toward novelties until fixed due to selective fitness. However, the extent to which this mechanism has driven the diversification of metabolite biosynthesis remains to be tested. Here we performed comparative genomics analysis and functional characterization to evaluate the impact of gene duplication on the evolution of triterpenoid biosynthesis using Panax species as models. We found that whole-genome duplications (WGDs) occurred independently in Araliaceae and Apiaceae lineages. Comparative genomics revealed the evolutionary trajectories of triterpenoid biosynthesis in plants, which was mainly promoted by WGDs and tandem duplication. Lanosterol synthase (LAS) was likely derived from a tandem duplicate of cycloartenol synthase that predated the emergence of Nymphaeales. Under episodic diversifying selection, the LAS gene duplicates produced by γ whole-genome triplication have given rise to triterpene biosynthesis in core eudicots through neofunctionalization. Moreover, functional characterization revealed that oxidosqualene cyclases (OSCs) responsible for synthesizing dammarane-type triterpenes in Panax species were also capable of producing ocotillol-type triterpenes. Genomic and biochemical evidence suggested that Panax genes encoding the above OSCs originated from the specialization of one OSC gene duplicate produced from a recent WGD shared by Araliaceae (Pg-β). Our results reveal the crucial role of gene duplication in diversification of triterpenoid biosynthesis in plants and provide insight into the origin of ocotillol-type triterpenes in Panax species.
Key words: gene duplication, Panax genomes, whole-genome duplications, triterpenoid biosynthesis, ocotillol-type triterpenes, specialization
This study reports a chromosome-level genome of Panax vietnamensis var. fuscidiscus and updates the genome of Panax notoginseng. Using comparative genomics, it provides evidence that the evolution of triterpenoid biosynthesis in plants has mainly been promoted by repeated whole-genome duplications (WGDs) and tandem duplications. Functional characterization suggests that genes responsible for synthesizing dammarane-type triterpenes in Panax species are also capable of producing ocotillol-type triterpenes.
Introduction
Plants have evolved to synthesize a diverse array of metabolites that play essential roles in various biological processes. The adaptivity derived from these metabolites has driven the evolution of plants and even their interactors. For decades, biologists have been intrigued by the evolutionary mechanism underlying the diversification of metabolite biosynthesis in the plant kingdom. Gene duplication is proposed to be the major force driving the evolution of metabolite biosynthesis: relaxed from functional constraints, one duplicate can accumulate mutations. In most cases, such mutations will result in gene loss, but some may be fixed owing to selective advantages conferred by their altered function, whether neofunctionalization, subfunctionalization, or specialization (Ober, 2005). These novelties in function or expression pattern would gradually reshape the biosynthetic pathway for metabolites. In land plants, pervasive whole-genome duplications (WGDs) or polyploidizations serve as the primary sources of gene duplicates. These frequent WGDs are thought to have a key causal role in species diversification, phenotypic evolution, and chemical diversification in both gymnosperm and angiosperm lineages (Landis et al., 2018; Lichman et al., 2020; Stull et al., 2021). The causal linkage between WGDs and diversification of metabolite biosynthesis, although supported on a theoretical basis, remains to be rigorously tested.
Triterpenoids are one of the most diverse metabolites present in plants. Their biosynthesis is catalyzed by enzymes known as oxidosqualene cyclases (OSCs), which can cyclize the precursors 2,3-oxidosqualene and 2,3; 22,23-dioxidosqualene. Two different types of substrate conformation exist during the cyclization process: the chair-boat-chair (CBC) conformation and the chair-chair-chair (CCC) conformation (Thimmappa et al., 2014). Sterols, including cycloartenol and lanosterol, are produced via CBC folding, whereas triterpenes are produced via CCC folding. Based on the catalytic products, plant OSCs can be broadly classified into cycloartenol synthase (CAS), lanosterol synthase (LAS), lupeol synthase (LUS), β-amyrin synthase, and other multifunctional triterpene synthases (bAS and other mTTSs). Sterols function as important membrane components and also as plant hormones that regulate growth and development (Schaller, 2003). The “nonessential” triterpenes are considered to have more specialized functions in plant defense and microbiome interactions (Delis et al., 2011; Khakimov et al., 2015; Miettinen et al., 2018; Huang et al., 2019; Li et al., 2021b; Busta et al., 2021). Genomic screening of the Viridiplantae phylogeny revealed that angiosperms are a hotspot of OSC diversification. Both divergent and convergent evolutionary processes are thought to have influenced the evolution of OSCs, and it is generally accepted that expansion of OSCs has been driven mainly by tandem duplications and that the triterpene synthases of eudicots likely originated from LAS rather than CAS (Pichersky and Lewinsohn, 2011; Xue et al., 2012; Zhou et al., 2016; Cárdenas et al., 2019; Dong et al., 2021). However, the impact of WGDs on the diversification of OSCs and the corresponding evolutionary trajectory remain unresolved.
The genus Panax L. (Araliaceae), which contains seven well-defined species and one species complex, is one of the most medicinally important plant genera. The pharmaceutical activities of Panax species have been attributed mainly to ginsenosides (glycosylated triterpenoids) (Leung and Wong, 2010; Fan et al., 2020). Biochemical approaches have revealed a wide variety of triterpenoids in Panax species, including the dammarane, α/β-amyrin, and ocotillol types (Hou et al., 2021). To date, OSC genes responsible for synthesis of dammarane-type triterpenes have been reported for several Panax species (Tansakul et al., 2006; Wang et al., 2014), but the biosynthetic pathway of ocotillol-type triterpenes remains unclear. As one Panax species with high medicinal value, Panax vietnamensis var. fuscidiscus is widely cultivated in Yunnan, China. The high content of ocotillol-type saponins in P. vietnamensis var. fuscidiscus make it a suitable model for exploring the mechanism of ocotillol-type triterpene biosynthesis (Zhang et al., 2015). Panax species have experienced several rounds of WGD in their evolutionary history, but whether extra WGDs have occurred in the common ancestor of all Apiales species after the γ whole-genome triplication (WGT) remains a topic of controversy (Kim et al., 2018a; Li et al., 2021a; Yang et al., 2021a, 2021b; Song et al., 2021). Regardless of disputes about WGD history, genomic and phytochemical evidence indicates that the evolution of triterpenoid biosynthesis in Panax species is likely to have been affected by WGDs (Li et al., 2021a). The diversity of triterpenoids and the presence of WGDs in Panax species make this genus a suitable model for examining the effects of WGDs on the evolution and diversification of OSCs.
Here we report a high-quality chromosome-level assembly for P. vietnamensis var. fuscidiscus, together with an improved assembly for Panax notoginseng. We found that WGDs have occurred independently in Araliaceae and Apiaceae species rather than being shared by Apiales. Comparative genomics revealed that the diversification of triterpenoid biosynthesis was promoted mainly by WGDs and tandem duplications. Notably, the dammarenediol-II synthases (DDSs) in Panax species were functionally characterized as mTTSs. These Panax DDS genes originated from the specialization of one OSC gene duplicate produced by the Pg-β WGD. Our findings systematically reveal how gene duplication drives the diversification of triterpenoid biosynthesis in plants and reveal the origin of ocotillol-type triterpenes in Panax species.
Results
Panax genome sequencing, assembly, and annotation
PacBio long reads were used to build a de novo assembly for P. vietnamensis var. fuscidiscus (Supplemental Figure 1A). This preliminary assembly was polished with Illumina short reads and scaffolded using Hi-C technology. The final chromosome-level assembly of P. vietnamensis var. fuscidiscus spans 1.73 Gb, with a scaffold N50 of 144.08 Mb (Supplemental Table 1). The largest 12 scaffolds, which correspond to the karyotype of P. vietnamensis var. fuscidiscus (2n = 2x = 24), covered 91.04% of the assembly (1.57 Gb) (Supplemental Figures 1B, 2A). The size of the pseudochromosomes is close to the flow cytometry (1.61 Gb) and k-mer-based estimates (1.43 Gb) (Supplemental Figure 3A; Supplemental Tables 2, 3). To evaluate the quality of the P. vietnamensis var. fuscidiscus genome, 229.47 Gb of the Illumina short reads (132.64×) were mapped to the assembly. The mapping rate of properly paired reads and genome coverage rate were 94.47% and 97.40%, respectively (Supplemental Table 4). We annotated 36 454 protein-coding genes in the P. vietnamensis var. fuscidiscus genome, with an average gene length of 6166.47 bp (Supplemental Table 5). A total of 33 570 (92.09%) predicted genes could be functionally annotated (Supplemental Table 6). Benchmarking Universal Single-Copy Orthologs (BUSCO) completeness of the assembly and annotated genes were 95.3% and 92.6% (Supplemental Figure 3B; Supplemental Table 7). The reported genome assembly size for P. vietnamensis Ha et Grushv. (2n = 2x = 24) is 3.00 Gb (Tien et al., 2021), which is 1.73-fold larger than the genome size of P. vietnamensis var. fuscidiscus. Thus, P. vietnamensis var. fuscidiscus is likely to be an independent species rather than a variety of P. vietnamensis Ha et Grushv. If not, P. vietnamensis var. fuscidiscus is still worth studying for its different genome size compared with P. vietnamensis Ha et Grushv. and well-developed biosynthetic modules based on P. vietnamensis var. fuscidiscus.
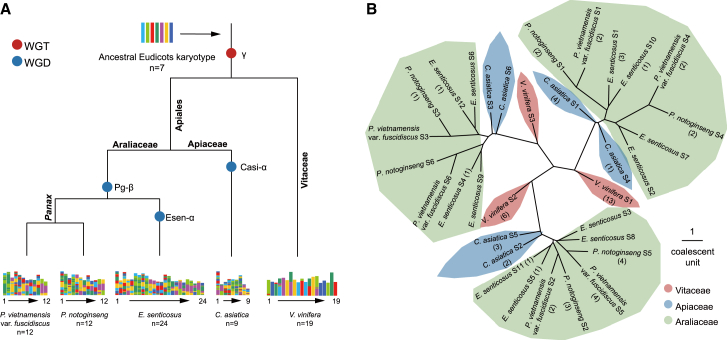
Figure 2.
Inference of polyploidization and speciation history in Apiales.
(A) The inferred phylogeny of Apiales species with placement of polyploidizations. Karyotypes were painted in seven colors, corresponding to the seven ancestral eudicot chromosomes.
(B) Synteny-based coalescent species tree showing independent WGDs in Apiaceae and Araliaceae. Genomes were classified into collinear subsets based on polyploidization history (denoted with S). Branch lengths are shown in coalescent units. Because the ASTRAL tree leaves the branch length of terminal branches empty, the lengths of terminal branches were all set to one. Numbers in parentheses represent the number of putative OSC genes.
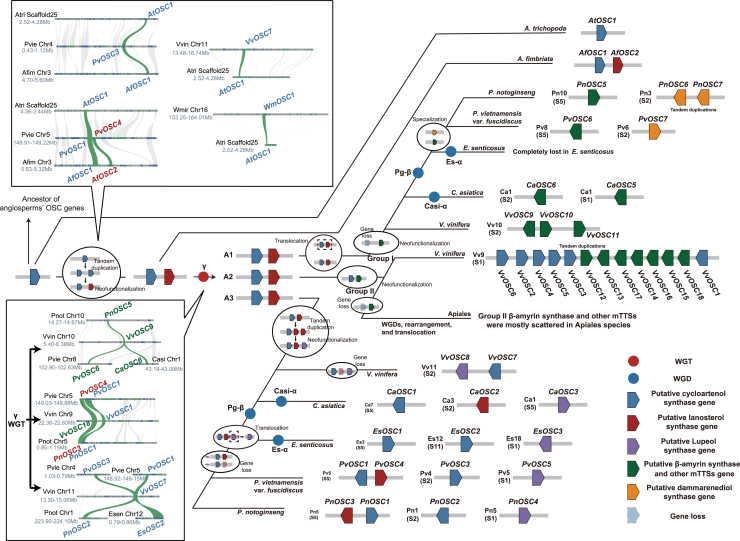
Figure 3.
The inferred evolutionary trajectory of OSC genes in plants.
Polyploidizations are shown with blue and red circles. Pentagons with a solid border represent OSC genes. Gene loss is shown by pentagons with a dashed border. Interspecific micro-syntenic relationships of putative OSC genes are shown in boxes. Direct collinear relationships between putative OSC genes are highlighted in green (Vvin, V. vinifera; Pvie, P. vietnamensis var. fuscidiscus; Pnot, P. notoginseng; Atri, A. trichopoda; Afim, A. fimbriata; Wmir, W. mirabilis). Inferred gene duplication, neofunctionalization, translocation, and loss are highlighted in circles.
We also provide an improved chromosome-level assembly of P. notoginseng (2n = 2x = 24) created using a previous contig-level assembly (Supplemental Table 8). More sequences (94.29%) were anchored to the 12 pseudochromosomes compared with the previous assembly (86.87%) (Supplemental Figure 2B). We annotated 36 747 protein-coding genes in the updated P. notoginseng genome, 92.79% of which were functionally annotated (Supplemental Tables 5 and 9). BUSCO analysis suggested 97.5% and 93.3% completeness of the updated P. notoginseng assembly and annotated genes (Supplemental Figure 3B; Supplemental Table 7).
Species-specific LTR expansion produced genome size variation in Panax
Repetitive elements constitute 86.79% and 88.18% of the P. vietnamensis var. fuscidiscus and P. notoginseng assemblies. LTR-RTs are the most abundant type of transposable elements (TEs) in both Panax species, accounting for 78.94% and 80.66% of the P. vietnamensis var. fuscidiscus and P. notoginseng assemblies. Among the LTR-RTs in P. vietnamensis var. fuscidiscus, Gypsy elements (54.52% of the genome) are far more abundant than Copia elements (5.67%). A similar phenomenon was observed in the P. notoginseng genome, with Gypsy and Copia elements accounting for 55.49% and 4.55% of the genome. DNA transposons are the second most abundant type of TE and constitute 2.90% and 3.13% of the P. vietnamensis var. fuscidiscus and P. notoginseng genomes (Supplemental Tables 10 and 11). Based on intact LTR-RTs (21 251 in P. vietnamensis var. fuscidiscus and 24 899 in P. notoginseng), we estimated the insertion times for LTR-RTs. P. vietnamensis var. fuscidiscus was found to have experienced a more recent burst of LTRs compared with P. notoginseng (Supplemental Figure 4). Clade-level classification of TEs revealed that the numbers of several Gypsy-clade elements (mainly Tekay, Ogre, and Athila) are much higher in P. notoginseng than in P. vietnamensis var. fuscidiscus (Supplemental Tables 12 and 13). These results indicate that P. notoginseng experienced a more intense expansion of LTR insertions compared with P. vietnamensis var. fuscidiscus after their divergence; the difference in genome sizes between the two Panax species (∼680 Mb) can be attributed mainly to the more intense expansion of LTRs in P. notoginseng (LTR size difference, ∼609 Mb).
Phylogenomics and evolution of P. vietnamensis var. fuscidiscus
To study the evolutionary history of Panax species, we first performed gene family analysis using 12 eudicots: Vitis vinifera, Coffea canephora, Codonopsis pilosula, Lactuca sativa, Lonicera japonica, Centella asiatica, Daucus carota, Apium graveolens, Eleutherococcus senticosus, Panax ginseng, P. notoginseng, and P. vietnamensis var. fuscidiscus. A total of 30 074 ortholog groups, harboring 93.1% of all the studied genes, were identified for the 12 species, and 168 groups are presented as single-copy orthogroups (Supplemental Figure 5A; Supplemental Table 14). Investigation of gene families in P. vietnamensis var. fuscidiscus and five other Apiales species suggested that P. vietnamensis var. fuscidiscus and P. notoginseng contain 436 and 673 unique gene families, respectively (Supplemental Figure 5B).
Single-copy orthogroups were used to construct maximum likelihood (ML) phylogenetic trees. The species trees inferred by the concatenation method and coalescence-based phylogenetic analysis are identical and well supported (Supplemental Figure 6A and 6B). P. vietnamensis var. fuscidiscus is placed as a sister lineage to P. notoginseng rather than P. ginseng, consistent with a Panax phylogeny based on chloroplast genomes and ribosomal DNA (Ji et al., 2019). Divergence times were estimated using MCMCTree with time calibrations. The estimated divergence between Araliaceae and Apiaceae occurred ∼56.3–68.3 million years ago (Mya). In the Panax genus, the speciation of P. ginseng occurred first (∼8.3–10.4 Mya), followed by the divergence of P. vietnamensis var. fuscidiscus and P. notoginseng (7.3–9.3 Mya) (Figure 1A). We also noted the early divergence of C. asiatica in the family Apiaceae, which occurred approximately 48.3–59.1 Mya, validating the basal group position of C. asiatica in Apiaceae (Li et al., 2020).
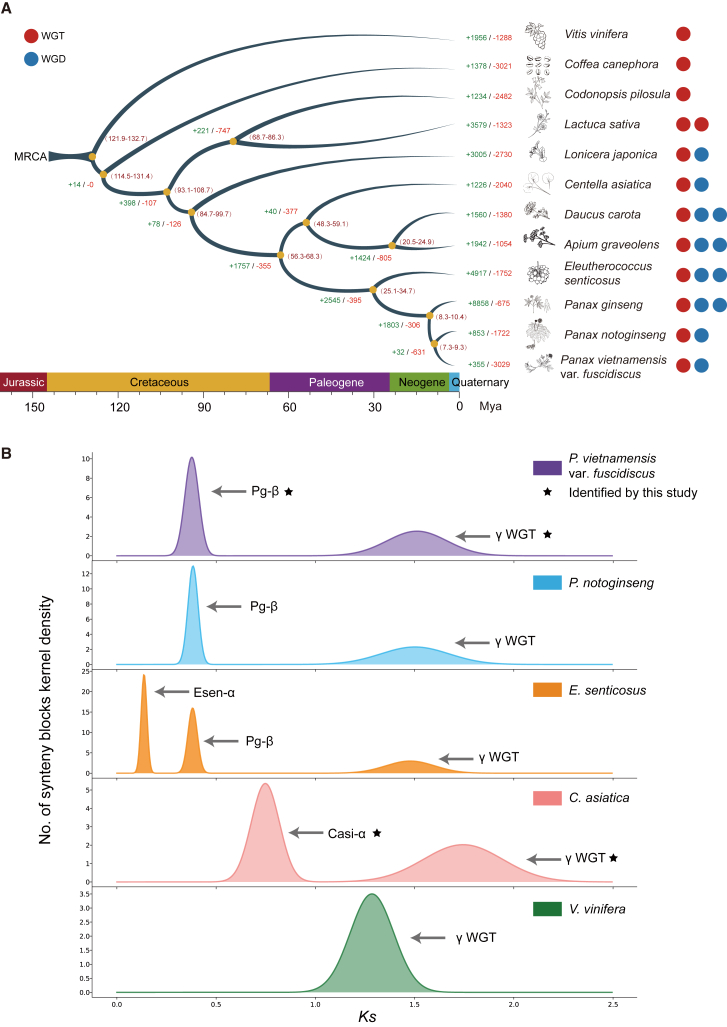
Figure 1.
Evolutionary analysis of P. vietnamensis var. fuscidiscus.
(A) Species tree for 12 eudicots including P. vietnamensis var. fuscidiscus. Numbers in parentheses indicate estimated divergence times in Mya with 95% confidence intervals. Expansion and contraction of gene families are indicated with plus and minus signs. Whole-genome duplications (WGDs) and whole-genome triplications (WGTs) in each species are shown in blue/red circles. MRCA, most recent common ancestor.
(B)Ks distribution of intraspecific collinear blocks. Ks peaks of polyploidizations are labeled for each species. Esen, E. senticosus; Casi, C. asiatica.
Finally, we estimated the expansion and contraction of gene families during the phylogenetic history of the 12 species using the resolved species tree. In P. vietnamensis var. fuscidiscus, 355 gene families had undergone expansion, whereas 3029 gene families had undergone contraction (P < 0.05) (Figure 1A). Expanded gene families in P. vietnamensis var. fuscidiscus showed functional enrichment in sesquiterpenoid and triterpenoid biosynthesis (P < 0.05) (Supplemental Figure 7A; 7B, Supplemental Tables 15 and 16).
Polyploidization history in Apiales
Polyploidizations in Apiales were systematically characterized to study their impact on the evolution of triterpenoid biosynthesis. The V. vinifera genome was used as an outgroup because only one polyploidization event (γ WGT) occurred during its evolution. We inferred WGDs and speciation events by examining the synonymous substitutions per synonymous site (Ks) of collinear gene pairs and intra/interspecific syntenic relationships. Two clear peaks were observed in the Ks distribution of intraspecific collinear gene pairs for P. vietnamensis var. fuscidiscus, suggesting an extra round of WGD after the γ WGT (Figure 1B). Interspecific synteny comparison between genomes of P. vietnamensis var. fuscidiscus and V. vinifera revealed that for each genomic region in V. vinifera, there are up to six syntenic matches in P. vietnamensis var. fuscidiscus, validating the extra round of WGD in the latter species (Supplemental Figure 8). In addition to recent peaks attributed to speciation, extra peaks were detected in the Ks distribution of collinear gene pairs between P. vietnamensis var. fuscidiscus and the other two Araliaceae species (P. notoginseng and E. senticosus) (Supplemental Figure 9). The ancient peaks indicate the shared γ WGT, and the relatively young peaks may represent the Pg-β WGD, which is presumed to be shared by Araliaceae species. The Ks peak values for the Pg-β WGD and the γ WGT are nearly identical in the three Araliaceae species, suggesting little variation in evolutionary rates among Araliaceae. Synteny comparisons of the updated P. notoginseng assembly with that of E. senticosus showed exactly 1:2 ratios for the best-matched regions in the largest 12 pseudochromosomes, validating the high quality of the updated P. notoginseng assembly compared with an older version (Supplemental Figure 10A and 10B). C. asiatica experienced two WGDs according to our analysis (Figure 1B). The Ks distribution of interspecific collinear gene pairs between C. asiatica and P. vietnamensis var. fuscidiscus showed two peaks, which correspond to speciation (∼0.53) and the shared γ WGT (∼1.63) (Supplemental Table 17). The absence of additional peaks suggested that the younger WGDs in Apiaceae and Araliaceae may have occurred independently after their speciation (Figure 2A).
To examine Apiales evolution with greater resolution, we performed synteny-based phylogenetic analysis. Five species (V. vinifera, C. asiatica, E. senticosus, P. notoginseng, and P. vietnamensis var. fuscidiscus) that exhibit a well-preserved ancestral eudicot karyotype (AEK) were selected for the analysis. Using the AEK and the V. vinifera genome as references, collinear regions were partitioned into different copies for each species with consideration of WGDs (Supplemental Figures 11–15; Supplemental Table 18). Based on 2255 collinear gene pairs (23 821 genes), ASTRAL produced a phylogenetic tree for the five species with a normalized quartet score of 0.8146. The topology of the synteny-based species tree provides solid evidence that the Pg-β WGD occurred independently in Araliaceae and was shared by Araliaceae species (Figure 2B and Supplemental Figure 16). Interestingly, the collinear subsets for C. asiatica in all three lineages produced by the γ WGT did not form sister groups but split successively instead. This suggested that the relatively recent WGD in C. asiatica may have been induced by an ancient hybridization.
Evolution of OSCs was mainly promoted by WGDs and tandem duplications
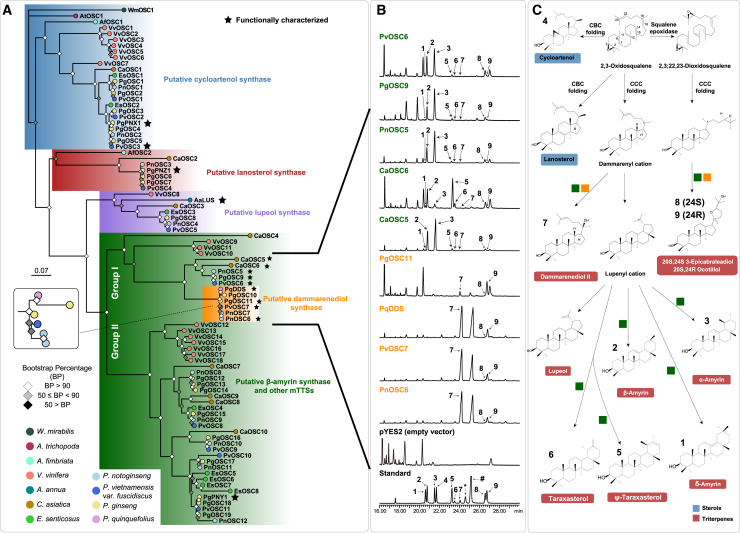
Previous studies have suggested that OSCs for sterol biosynthesis in Eukarya have a bacterial origin and that plant OSCs have likely undergone divergent evolution, with triterpene biosynthesis derived from sterol biosynthesis (Xue et al., 2012; Santana-Molina et al., 2020). Thus CAS likely served as the foundation of OSC evolution. Here, we performed phylogenetic and comparative genomics analyses to clarify the evolution of OSCs in plants. Nine species (Amborella trichopoda, Aristolochia fimbriata, V. vinifera, C. asiatica, E. senticosus, P. ginseng, P. vietnamensis var. fuscidiscus, P. notoginseng, and Panax quinquefolius) were included in the analysis, including six Apiales species selected for their diversity in triterpenoid biosynthesis and well-characterized phylogenetic history. We included A. trichopoda (ANA-grade) and A. fimbriata (Magnoliids) in the analysis for their absence of WGD since the emergence of flowering plants (Amborella Genome Project, 2013; Qin et al., 2021). First, we performed genome-wide identification of OSCs based on conserved protein domains. For P. quinquefolius, which lacks a reference assembly, one DDS was used (Supplemental Table 19). In contrast to the abundant OSC genes in eudicots, we identified only one putative OSC in A. trichopoda and two putative OSCs in A. fimbriata, implying an important role for WGDs in the expansion of OSCs. An ML phylogenetic tree was built for the identified putative OSCs using codon alignments (Figure 4A). On the basis of conserved motifs (Supplemental Figure 17) and phylogenetic relationships with functionally characterized OSCs, the OSCs were classified into putative functional groups (CAS, LAS, LUS, and bAS and other mTTSs) (Chen et al., 2021a). Functionally characterized DDSs from Panax species were nested within bAS and other mTTSs, indicating their close phylogenetic relationship (Tansakul et al., 2006; Wang et al., 2014). We also noticed that members of bAS and other mTTSs were recovered in two lineages (group I and group II) with high support, suggesting their distinct origins.
Figure 4.
Phylogenetic analysis and functional characterization of OSCs.
(A) ML phylogenetic tree of OSCs based on codon alignments. Bootstraps are shown as colored squares at each node, and species are shown as colored circles at each terminal branch.
(B) Functional characterization of nine OSCs using heterologous expression. The asterisk (∗) and hash (#) in the total ion chromatograms (TICs) represent the epoxydammaranes mono-trimethylsilyl ether and dammarenediol-II mono-trimethylsilyl ether, respectively. 1, δ-amyrin; 2, β-amyrin; 3, α-amyrin; 4, cycloartenol; 5, ψ-taraxasterol; 6, taraxasterol; 7, dammarenediol-II; 8, 20S,24S-3-epicabraleadiol; 9, 20S,24R-ocotillol.
(C) Schematic for triterpenoid biosynthesis with sterols highlighted in blue and triterpenes highlighted in red. Compound numbers correspond to the numbers in TICs from (B). Colored squares indicate functions of enzymes in (B).
The distribution pattern of OSCs on the synteny-based species tree suggested that the expansion of OSCs was affected by WGDs (Figure 2B). In addition, the scarcity of OSCs on the lineage leading to V. vinifera S3 indicated that gene loss or translocation events had occurred before speciation. We next performed inter/intraspecific synteny analysis to investigate the evolutionary trajectory of OSCs with differentiation of paralogous and orthologous syntenic regions produced by polyploidizations and speciation. Intraspecific synteny comparisons revealed OSCs produced from recent WGDs (Casi-α and Pg-β WGD) in Apiales species (Supplemental Figure 18). In P. vietnamensis var. fuscidiscus, no direct syntenic relationship was found for PvOSC7 (DDS gene) and PvOSC6 (bAS and other mTTSs), yet both genes were located on highly syntenic chromosomal regions produced by the Pg-β WGD. The same phenomena were also observed in P. notoginseng between PnOSC5 (bAS and other mTTSs) and the tandemly-duplicated PnOSC6/PnOSC7 (DDS genes) (Supplemental Figure 18). Thus, we speculate that DDS in Panax species likely originated from neofunctionalization of a group I bAS and other mTTS copy produced from the Pg-β WGD. We observed only one syntenic relationship between VvOSC12 (from chromosome [chr] 9) and VvOSC9 (from chr 10) in V. vinifera. Considering the fact that OSCs in grape are found on only three chromosomes (chr 9, 10, 11) and that chr 9, chr 11, and a part of chr 4 were produced by the γ WGT, grape OSCs from chr 9, 10, and 11 are likely to share the same origin. After the γ WGT, a chromosomal region harboring OSCs in chr 4 was translocated to chr 10. This assumption was verified by interspecific synteny comparisons between V. vinifera and Apiales species, in which the grape CASs VvOSC7 (from chr 11) and VvOSC1 (from chr 9) showed syntenic relationships with the same P. vietnamensis var. fuscidiscus CAS, PvOSC1 (Figure 3). We also compared OSC syntenic relationships of P. vietnamensis var. fuscidiscus and V. vinifera with species with non-duplicated genomes (A. trichopoda and A. fimbriata). The CASs produced by the Pg-β WGD (PvOSC3 and PvOSC1) and an LAS (PvOSC4) from P. vietnamensis var. fuscidiscus showed clear syntenic relationships with OSC genes from A. trichopoda (AtOSC1) and A. fimbriata (AfOSC1 and AfOSC2). A syntenic relationship was also found between grape VvOSC7 and A. trichopoda AtOSC1 (Figure 3). Such conservation was even detected between A. trichopoda and the gymnosperm Welwitschia mirabilis (Figure 3), demonstrating that CAS genes are spatially conserved in higher plants.
With the above information, we deduced the evolutionary trajectory of OSCs (Figure 3). OSCs were conserved for sterol biosynthesis during the early stages of plant evolution, as only CASs were found in the genomes of lower plants (Xue et al., 2012). Following the emergence of angiosperms, one CAS duplicate (possibly produced by tandem duplication) may have diversified to give rise to LAS through neofunctionalization. The absence of LAS in Amborella suggests that the duplication probably occurred after Amborella speciation. The chromosomal region harboring CAS and LAS was triplicated into three copies by the γ WGT (A1–3). Several changes were inferred for the triplicated copies. Before the speciation of grape and Apiales, A1 experienced translocation followed by functional diversification of LAS to group I bAS and other mTTSs. The newly formed group I bAS and other mTTSs was duplicated by the Pg-β WGD, with one copy then neofunctionalizing to DDS in the ancestor of extant Panax species. For A2, neofunctionalization of LAS gave rise to group II bAS and other mTTSs. In the lineage leading to Apiales, CAS was lost, and the group II bAS and other mTTSs was likely affected by reshuffling, resulting in a non-syntenic distribution pattern. By contrast, CAS and group II bAS and other mTTSs were retained and proliferated through tandem duplications in grape. LUS may have been produced by neofunctionalization of a tandemly duplicated copy of LAS in A3 before the speciation of grape and Apiales. In the Apiales lineage, the LUS probably experienced translocation, as no syntenic relationships were found between LUS and other OSCs.
Functional characterization revealed the origin of ocotillol-type triterpenes in Panax
To verify the proposed Pg-β origin of DDSs in Panax species, we performed functional analysis to determine the catalytic activities of each tested OSC. Nine OSC genes (five from group I bAS and other mTTS clades and four from the DDS clade) were selected for the analysis (Figure 4B). The OSC genes were heterologously expressed in mutant yeast strain GIL77, which was engineered to accumulate the precursor oxidosqualene (Morita et al., 1997). The products were identified through GC–MS and NMR (Supplemental Figures 19–26; Supplemental Tables 20 and 21). Nine products were identified for every OSC from group I bAS and other mTTSs. For PvOSC6, PgOSC9, PnOSC5, and CaOSC5, α-amyrin, β-amyrin, ψ-taraxasterol, and 3-epicabraleadiol were identified as the main products, with trace amounts of δ-amyrin, taraxasterol, dammarenediol-II, ocotillol, and an unidentified product. The product profile of CaOSC6 was slightly different, with an increased proportion of ψ-taraxasterol and a decrease in α-amyrin content (Figures 4B, 4C, and 27; Supplemental Table 22). In a recent study, CaOSC5 was functionally characterized as a multifunctional OSC producing δ-amyrin, α-amyrin, β-amyrin, ψ-taraxasterol, taraxasterol, and an unidentified product in a ratio of 1:67:26:4:1:1 (Kim et al., 2018b). The previously reported catalytic activities of CaOSC5 are highly consistent with our results, except for the weak ability to produce dammarane-type triterpenes (dammarenediol-II, ocotillol, and 3-epicabraleadiol) identified in our study. Surprisingly, ocotillol and 3-epicabraleadiol were also detected in addition to dammarenediol-II as catalytic products of DDSs from Panax species (PgOSC11, PqDDS, PvOSC7, and PnOSC6) (Figure 4B and Supplemental Figure 27; Supplemental Table 22). This multifunctional nature of DDSs in Panax species has not previously been reported. We also noted that PgOSC11 produces mainly ocotillol, whereas the other DDSs predominantly produce dammarenediol-II. These findings validate our assumption that DDS in Panax species originated from a duplicate of a group I bAS and other mTTSs produced from a WGD. After the Pg-β WGD, one copy of group I bAS and other mTTSs retained its original function (similar to homologs in C. asiatica), whereas the other copy experienced neofunctionalization. Judging by the catalytic products, this neofunctionalization should be viewed as a specialization process: from a generalist ancestor to a more specialized state.
Selective forces underlying evolution of OSCs
According to their deduced evolutionary trajectory, OSC genes have experienced several rounds of independent neofunctionalization and specialization events. It is expected that the diversifications occurred under various selection pressures. To characterize the selective forces driving the evolution of OSC genes, we performed various branch-specific tests.
Branch-site unrestricted statistical test for episodic diversification (BUSTED) analysis found evidence (likelihood ratio test [LRT], P < 0.05) of gene-wide episodic diversifying selection on at least one site on at least one branch in the phylogeny (Supplemental Figure 28). Adaptive branch-site random effects likelihood (aBSREL) and mixed effects model of evolution (MEME) were then used to determine the exact lineages and sites that were under positive selection. With a priori knowledge that CAS genes serve as a blueprint for functional diversification of OSCs, CAS lineages were labeled as background in the branch-site analysis. Analysis with aBSREL found evidence of episodic diversifying selection on 20 out of 159 branches in the tested phylogeny (LRT, P ≤ 0.05), with only four in the CAS and LAS clades and the rest distributed in lineages leading to LUS, bAS and other mTTSs, and DDS (Supplemental Figure 29). The fact that almost all of the CAS genes were under negative or neutral selection could be explained by the importance of their cycloartenol product, which is the precursor for almost all plant sterols and plays an essential role in plant developmental processes (Gas-Pascual et al., 2014). Notably, episodic diversifying selection was detected on internal branches leading to LUS and group I/II bAS and other mTTSs (nodes 3–5), in which the major neofunctionalization of OSCs occurred (Supplemental Figure 29). This indicates that the diversification of sterol biosynthesis toward triterpene biosynthesis in core eudicots was driven by episodic positive selection, possibly due to the better adaptability conferred by the triterpene products. Notably, the specialization of DDS from group I bAS and other mTTSs in Panax species was predicted to be under neutral or negative selection. This could be explained by trade-offs during specialization: for an enzyme in the generalist state, specialization toward certain functions requires a decrease in the rest. In most cases, reduced promiscuity was shaped by negative selection (Tokuriki et al., 2012; Noda-Garcia and Tawfik, 2020; Tawfik and Gruic-Sovulj, 2020).
MEME found evidence of episodic diversifying selection at 84 sites (P < 0.05) (Supplemental Figure 30A). Most of these sites were located near the N/C terminus and the putative active center (Supplemental Figure 30B). Residues from several function-related motifs were found to be under episodic positive selection. For the motif M(W/L)C(Y/H)CR, which has been proposed to stabilize tetracyclic or pentacyclic intermediates, the second site (W/L) was identified as being under episodic positive selection (Kushiro et al., 1999; Ito et al., 2016). Motif Y410, which has been proposed to play an important role in ceiling formation of the active center or in D-ring formation, was also under diversifying selection. The site is conserved as Y in CBC-folding OSCs and F in CCC-folding OSCs (Ma et al., 2016; Chen et al., 2021a). These results provide insights into the evolution of OSCs at the molecular level.
Discussion
The discovered evolutionary trajectories for triterpenoid biosynthesis demonstrate the prominent role of gene duplication in creating a diverse array of triterpenoids in plants. WGDs and tandem duplications are the main forces driving the diversification of OSCs. An ancient tandem duplication of CAS during the early evolution of angiosperms may have given rise to LAS. This event possibly predates the emergence of Nymphaeales species, given the presence of LAS orthologs in Nymphaea colorata (Wang et al., 2022). The expansion and diversification of OSCs in core eudicots were attributed mainly to the γ WGT, with subsequent neofunctionalization occurring in the LAS triplicates. Specifically, triterpene synthases in core eudicots originated from independent neofunctionalization of LAS copies. This finding supports the hypothesis that eudicot triterpene biosynthesis derives from LAS rather than CAS (Kolesnikova et al., 2006; Xue et al., 2012). Indeed, experimental evidence suggested that LAS can supplement the biosynthesis of phytosterols in plants. However, the methyl jasmonate/bacteria-induced regulatory mechanism and tissue-specific expression pattern of LAS suggest its similarity to triterpene synthases (Zimmermann et al., 2004; Kolesnikova et al. 2006). The altered regulation and expression pattern of LAS may represent the initial step toward triterpene biosynthesis. Major angiosperm lineages such as monocots also exhibit great potential for the synthesis of various triterpenes (Inagaki et al., 2011); thus, the revealed γ WGT origin of triterpene biosynthesis in core eudicots suggests convergent evolution in OSC diversification in addition to the prevalent divergent evolution. We also revealed that group I and group II bAS and other mTTSs in eudicots originated from different LAS copies, thus explaining their distant phylogenetic relationship. A recent study of the evolutionary path of OSC genes based on phylogenetic trees inferred several major duplication events for OSC genes, including one ancient duplication event generating the CAS and LAS lineages and another three separate duplication events generating triterpene synthase (LUS and bAS) (Wang et al., 2022). Our results show that the former ancient duplication event was likely caused by tandem duplication of the ancestral OSC gene, whereas the latter three duplication events actually resulted from a single duplication event, the γ WGT. This finding also demonstrates the limitations of using only phylogenetic trees when inferring the evolutionary paths of genes.
Dammarane-type and ocotillol-type triterpenes are abundant mainly in Panax species. Several Panax DDS genes have been functionally characterized as producing dammarenediol-II, but the genes responsible for synthesizing ocotillol-type triterpenes remain unclear. Our analysis revealed the origin of the DDS gene family in Panax species and its multifunctional nature. Future studies using site-directed mutagenesis and crystal structure analysis may provide insight into the reaction mechanism that underlies the shift in product profile of these OSCs.
In principle, reshaping of metabolite biosynthesis after gene duplication is strongly affected by selection. Our results suggest that most of the WGD-derived OSC gene copies were lost during evolution, possibly owing to accumulation of negative mutations under relaxed selection pressure. However, some OSC duplicates acquired altered functions through mutations, which were then fixed by fitting ecological opportunities. This scenario was illustrated by the effect of episodic diversifying selection on neofunctionalization of LAS copies toward triterpene synthases in core eudicots. Such functional innovations of duplicates are not always driven by positive selection. In ancestral lineages of Panax species, one group I bAS and mTTS copy (which mainly produced amyrin) that derived from the Pg-β WGD experienced functional specialization under negative/neutral selection, eventually giving rise to DDS. The absence of positive selection might be explained by the trade-off in catalytic activities between amyrin-type and dammarane-type triterpenes (Noda-Garcia and Tawfik, 2020). Accordingly, it should be noted that the absence of positive selection during the creation of novelties does not necessarily indicate a lack of improvement in adaptivity.
In summary, the revealed origin and evolutionary history of triterpenoid biosynthesis in angiosperms provide insight into how gene duplication can drive the diversification of metabolite biosynthesis. In plants, triterpenoids are often further modified by tailoring enzymes (e.g., cytochrome P450s, glycosyltransferases, and acyltransferases) (Thimmappa et al., 2014). Studies have suggested that genes for OSCs and tailoring enzymes are likely functionally co-opted by gene duplication, resulting in diversification in gene regulation and expression patterns for triterpenoid biosynthesis (Li et al., 2021b; Su et al., 2021). Future studies on the interactions between these tailoring enzymes and OSCs and their origins will deepen our understanding of the evolution of metabolite biosynthesis.
Methods
Genome sequencing and assembly
Plant samples of P. vietnamensis var. fuscidiscus were collected from individuals cultivated in Jinping County, Yunnan, China. Fresh leaves, stems, and roots were stored in liquid nitrogen and sent to Novogene for sequencing (Beijing, China). High-molecular-weight genomic DNA was extracted from leaves using the cetyltrimethylammonium bromide (CTAB) method and purified with a QIAGEN Genomic Kit (Qiagen, USA). For long-read sequencing, 20-kb SMRTbell libraries were generated and sequenced on the PacBio Sequel platform. This produced ∼67.7× PacBio long reads. We also generated ∼132.6× Illumina short reads. Four libraries with an insert size of 300 bp were prepared and sequenced on the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA). High-throughput chromosome conformation capture (Hi-C) libraries were prepared and sequenced on the Illumina HiSeq 4000 platform. In brief, chromatin was cross-linked with formaldehyde and digested with the restriction enzyme DpnII before sequencing. For the purpose of gene prediction, total RNA was isolated from leaves, stems, and roots using the RNAprep Pure Plant Kit (TIANGEN). RNA libraries with an insert size of 300 bp were generated and sequenced on the Illumina HiSeq 2000 platform (Supplemental Table 23).
The genome size of P. vietnamensis var. fuscidiscus was estimated using flow cytometry (BD FACSCalibur) and GenomeScope (v2.0) with kmer frequencies counted from 132.6× Illumina reads using Jellyfish (v2.2.10) (Marçais and Kingsford, 2011; Ranallo-Benavidez et al., 2020). The PacBio reads were assembled using NextDenovo (v2.4.0) (https://github.com/Nextomics/NextDenovo), followed by two rounds of polishing with NextPolish (v1.3.1) (Hu et al., 2020). After removing allelic contigs using Purge Haplotigs (v1.1.1) (Roach et al., 2018), we performed scaffolding using Juicer (v1.6.2) (Durand et al., 2016) and the three-dimensional (3D) de novo assembly (3D-DNA) pipeline (Dudchenko et al., 2017). Mis-joins were manually corrected on the basis of Hi-C contact signals. For transcriptome assembly, raw reads were trimmed with fastp (v0.20.1) (Chen et al., 2018) and assembled using Trinity (v2.11.0) (Grabherr et al., 2011).
The quality of the genome assemblies was evaluated using BUSCO (v5.1.2) (Manni et al., 2021) with dataset eudicots_odb10. We also mapped Illumina reads to the genome using BWA-MEM (v0.7.12) (Li and Durbin, 2009a) and calculated the mapping statistics using SAMtools (v1.9) (Li et al., 2009b).
Genome annotation
We used LTR_FINDER_parallel (v1.1) (Ou and Jiang, 2019) and LTRharvest (v1.0) (Ellinghaus et al., 2008) to predict long terminal repeat retrotransposons (LTR-RTs). The identified LTR-RT candidates were passed to LTR_retriever (v2.8) (Ou and Jiang, 2018b) to filter out the false positives and generate a genome LTR assembly index (LAI) (Ou et al., 2018a). Only intact LTRs were retained for insertion time estimation. The equation T = K/2μ was used for time estimation, where K is the LTR divergence rate and μ is the neutral mutation rate (1.3 × 10−8 mutations per site per year). We also used RepeatModeler (v2.0) (Flynn et al., 2020) to detect novel repeat sequences. Repetitive elements generated by LTR_retriever and RepeatModeler were fed to RepeatMasker (v4.0.9) (http://www.repeatmasker.org) for de novo prediction. For evidence-based methods, repetitive elements were predicted using RepeatMasker and RepeatProteinMask (v4.0.9) (http://www.repeatmasker.org) with Repbase (v24.06) (Bao et al., 2015) as the reference. The tandem repeats were annotated using Tandem Repeat Finder (v4.09) (Benson, 1999). The predicted LTR-RTs were further classified by TEsorter (v1.2.5) (Zhang et al., 2022) with REXdb Viridiplantae (v2.2) (Neumann et al., 2019).
Gene structures were predicted using a combination of ab initio-, homology-, and transcript-based methods. GenScan (v1.0) (Aggarwal and Ramaswamy, 2002), GlimmerHMM (v3.0.3) (Majoros et al., 2004), geneid (v1.4.4) (Alioto et al., 2018), Augustus (v3.2.2) (Stanke et al., 2008), and SNAP (v1.0) (Korf, 2004) were used for ab initio prediction of protein-coding genes. For the homology-based method, proteomes of A. thaliana, V. vinifera, E. senticosus, D. carota, and P. ginseng were searched against the genomes using TBLASTN (v2.2.29+) (Altschul et al., 1990) with 1e−5 as the cutoff e-value. Gene models were predicted by GenomeThreader (v1.7.3) (Gremme et al., 2005) using the above hits. For the transcript-based method, Program to Assemble Spliced Alignments (PASA) (v2.4.1) (Haas et al., 2003) was used for gene prediction by comparing Trinity transcripts with genomes. Finally, all gene models were integrated using EVidenceModeler (v1.1.1) (Haas et al., 2008) and updated with PASA. Functional annotation was performed with eggNOG-mapper (v2.1.7) (Cantalapiedra et al., 2021) by searching the eggNOG database (v5.0.2) (Huerta-Cepas et al., 2019) (Viridiplantae-33090) using DIAMOND (v2.0.14) (Buchfink et al., 2015).
Phylogenomics and evolutionary analysis
Orthogroups were identified using OrthoFinder (v2.5.4) (van Dongen, 2000; Emms and Kelly, 2019) based on protein sequences of 12 species (Supplemental Table 24). The Venn diagram was visualized using Evenn (Chen et al., 2021b).
Species trees were inferred based on single-copy orthogroups. Protein sequences from each single-copy orthogroup of 12 species were extracted and aligned using MAFFT (v7.475) (Katoh and Standley, 2013). Then, the protein alignments were converted to codon alignments using PAL2NAL (v14) (Suyama et al., 2006). Poorly aligned regions from codon alignments were trimmed using trimAl (v2.rev0) (Capella-Gutiérrez et al., 2009). For the concatenation-based method, an ML phylogenetic tree was built based on concatenated codon alignments using IQ-TREE (v2.0.3) (Nguyen et al., 2015) with the best-fit substitution model determined using ModelFinder (Kalyaanamoorthy et al., 2017). Branch supports were estimated using 1000 replicates with ultrafast bootstrap approximation (UFBoot2) (Hoang et al., 2018). For the coalescent-based method, a species tree was estimated using ASTRAL (v5.7.7) (Zhang et al., 2018) based on ML trees produced from IQ-TREE. We estimated species divergence times using MCMCTree from the PAML package (v4.9j) (Yang, 2007) with molecular clock and nucleotide substitution set as correlated rates and JC69 model. The MCMC process was run for 100 000 iterations with a burn-in of 50 000 and a sampling frequency of five. The tree was calibrated with the following constraints: divergence time of D. carota and A. graveolens (∼22–37 Mya), divergence time of Araliaceae and Apiaceae (∼45–70 Mya), and divergence time of V. vinifera and the other studied species (∼111–131 Mya) (Kumar et al., 2017). Phylogenetic trees were visualized using FigTree (v1.4.4) (http://tree.bio.ed.ac.uk/software/figtree/).
Changes in gene family size during species evolution were estimated using CAFE (v5) (Mendes et al., 2020). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of gene families were performed using clusterProfiler (v4.2.2) (Wu et al., 2021) and TBtools (v1.098685) (Chen et al., 2020), respectively, with P values adjusted by the Benjamini and Hochberg method.
WGD and speciation analysis
The WGDI toolkit (v0.5.1) (Sun et al., 2022) was used to detect WGD and speciation events. First, BLASTP (v2.2.29+) was used to search for homologs with a 1e−5 cutoff e-value. Collinear genes were identified by WGDI on the basis of the identified homologs using the parameter -icl. Ks values of collinear gene pairs were then calculated using the YN00 program from the PAML package with the Nei-Gojobori method (Nei and Gojobori, 1986). The median Ks values of inter/intraspecific collinear blocks were fitted using Gaussian kernel density estimation with the parameter -pf. The intraspecific syntenic relationships within P. vietnamensis var. fuscidiscus, together with the GC content, TE density, and gene density, were visualized using Circos (v0.69-9) (Krzywinski et al., 2009).
Synteny-based phylogenetic analysis was used to infer the WGD and speciation history. On the basis of the similarity and completeness of inter/intraspecific syntenic blocks, syntenic regions were assigned to WGD-related putative sets for V. vinifera, C. asiatica, E. senticosus, P. notoginseng, and P. vietnamensis var. fuscidiscus with parameters -bi and -a. Collinear genes from the characterized sets were extracted and used to construct ML phylogenetic trees separately using IQ-TREE. Collinear gene pairs encompassing genes from all studied species were retained for ASTRAL analysis.
Inferring evolutionary trajectories of OSC genes
Putative OSCs were identified using HMMER (v3.1b2) (Eddy, 1998) by searching with the squalene-hopene cyclase N-terminal domain (PF13249) and C-terminal domain (PF13243) from Pfam (v35.0) (Mistry et al., 2021) with the parameter -cut_tc. Sequences that contained both domains were retained for analysis. For phylogenetic analysis, protein sequences of the putative OSCs were aligned using MAFFT. The protein alignments were converted to codon alignments by PAL2NAL, followed by trimming with trimAl. IQ-TREE was used to construct an ML phylogenetic tree for the putative OSCs. The tree and motifs were visualized using the R packages ggtree (v2.4.1) (Yu et al., 2018) and ggmsa (v1.0.0) (http://yulab-smu.top/ggmsa/). To assist with classification of putative OSCs, sequences of functionally characterized OSCs were downloaded from NCBI and included in the analysis. One P. vietnamensis var. fuscidiscus OSC (PvOSC3) was also functionally characterized (Supplemental Figure 31). For synteny-based analysis, the syntenic relationships among putative OSC genes were identified with WGDI. We used JCVI utility libraries (v1.1.23) (Tang et al., 2008) to visualize the micro-synteny of OSCs.
Functional characterization of OSCs
Nucleotide coding sequences of putative OSC genes were synthesized and ligated into the yeast expression vector pYES2 (Invitrogen) under the control of the GAL1 promotor by GeneCreate (Wuhan, China). Vectors carrying putative OSC genes were transformed into DH5α competent cells. The resulting plasmid DNAs were transformed into the mutant yeast strain GIL77 by the lithium acetate/single-stranded carrier DNA/PEG method (Gietz and Schiestl, 2007). Yeast strains transformed with the empty vector were used as controls. Yeast strains were incubated in synthetic medium containing ergosterol (20 μg ml−1), hemin chloride (13 μg ml−1), and Tween 80 (5 μg ml−1) for 3 days followed by 48-h Gal induction and another 24-h incubation. Cells were harvested and refluxed in 20% KOH/50% EtOH for 10 min and extracted with petroleum ether three times. The organic phase was concentrated in vacuo. Gas chromatography–mass spectrometry (GC–MS) analysis was performed using an Agilent 7890A and Agilent 6540 Accurate-Mass Q-TOF (Santa Clara, USA). NMR analysis was performed on a Bruker AV 600 MHz spectrometer (Billerica, USA) (see supporting information Methods S1).
Selection analysis
The codon alignments and ML phylogenetic tree for putative OSCs from the previous step were used for selection analysis. We used HYPHY (v2.5.32) (http://hyphy.org/) to perform the BUSTED (Murrell et al., 2015), aBSREL (Smith et al., 2015), and MEME (Murrell et al., 2012) analyses. The 3D protein structure of P. ginseng CAS was downloaded from UniProt (UniProt Consortium, 2021) with identifier AF-O82139-F1 (predicted by AlphaFold [Jumper et al., 2021]). PyMOL was used for visualization of protein structures (The PyMOL Molecular Graphics System, Version 2.5, Schrödinger, LLC.).
Funding
This work was supported by Digitalization of biological resources (202002AA100007), the Guangxi Innovation-Driven Development Project (GuiKe AA18242040), the General Project for Basic Research in Yunnan (grant no. 202201AT070266), and the National Natural Science Foundation of China (81860680).
Author contributions
S.Y. and Y.D. designed the research. Z.Y., X.L., L.Y., W.S., and G.Z. collected the data. Z.Y., X.L., L.Y., and S.P. performed the data and experimental analyses. Z.Y., W.C., S.Y., and Y.D. wrote the manuscript with contributions from all authors.
Acknowledgments
We thank Pengchuan Sun from SiChuan University and Yue Wang for technical support. No conflict of interest is declared.
Published: March 16, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Shengchao Yang, Email: shengchaoyang@163.com.
Yang Dong, Email: dongyang@dongyang-lab.org.
Accession numbers
The raw sequencing data, assembly, and annotation files of P. vietnamensis var. fuscidiscus and P. notoginseng have been deposited at CNGBdb (https://db.cngb.org/) under project accession numbers CNP0002878 and CNP0003588.
Supplemental information
References
- Aggarwal G., Ramaswamy R. Ab initio gene identification: prokaryote genome annotation with GeneScan and GLIMMER. J. Biosci. 2002;27:7–14. doi: 10.1007/BF02703679. [DOI] [PubMed] [Google Scholar]
- Alioto T., Blanco E., Parra G., Guigó R. Using geneid to identify genes. Curr. Protoc. Bioinformatics. 2018;64:e56. doi: 10.1002/cpbi.56. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- Bao W., Kojima K.K., Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Busta L., Schmitz E., Kosma D.K., Schnable J.C., Cahoon E.B. A co-opted steroid synthesis gene, maintained in sorghum but not maize, is associated with a divergence in leaf wax chemistry. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2022982118. e2022982118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalapiedra C.P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. EggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021;38:5825–5829. doi: 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas P.D., Almeida A., Bak S. Evolution of structural diversity of triterpenoids. Front. Plant Sci. 2019;10:1523. doi: 10.3389/fpls.2019.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Chen K., Zhang M., Ye M., Qiao X. Site-directed mutagenesis and substrate compatibility to reveal the structure-function relationships of plant oxidosqualene cyclases. Nat. Prod. Rep. 2021;38:2261–2275. doi: 10.1039/d1np00015b. [DOI] [PubMed] [Google Scholar]
- Chen T., Zhang H., Liu Y., Liu Y.X., Huang L. EVenn: easy to create repeatable and editable Venn diagrams and Venn networks online. J. Genet. Genomics. 2021;48:863–866. doi: 10.1016/j.jgg.2021.07.007. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delis C., Krokida A., Georgiou S., Peña-Rodríguez L.M., Kavroulakis N., Ioannou E., Roussis V., Osbourn A.E., Papadopoulou K.K. Role of lupeol synthase in Lotus japonicus nodule formation. New Phytol. 2011;189:335–346. doi: 10.1111/j.1469-8137.2010.03463.x. [DOI] [PubMed] [Google Scholar]
- Dong L., Almeida A., Pollier J., Khakimov B., Bassard J.E., Miettinen K., Stærk D., Mehran R., Olsen C.E., Motawia M.S., et al. An independent evolutionary origin for insect deterrent cucurbitacins in Iberis amara. Mol. Biol. Evol. 2021;38:4659–4673. doi: 10.1093/molbev/msab213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O., Batra S.S., Omer A.D., Nyquist S.K., Hoeger M., Durand N.C., Shamim M.S., Machol I., Lander E.S., Aiden A.P., Aiden E.L. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand N.C., Shamim M.S., Machol I., Rao S.S.P., Huntley M.H., Lander E.S., Aiden E.L. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D., Kurtz S., Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W., Huang Y., Zheng H., Li S., Li Z., Yuan L., Cheng X., He C., Sun J. Ginsenosides for the treatment of metabolic syndrome and cardiovascular diseases: pharmacology and mechanisms. Biomed. Pharmacother. 2020;132:110915. doi: 10.1016/j.biopha.2020.110915. [DOI] [PubMed] [Google Scholar]
- Flynn J.M., Hubley R., Goubert C., Rosen J., Clark A.G., Feschotte C., Smit A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gas-Pascual E., Berna A., Bach T.J., Schaller H. Plant oxidosqualene metabolism: cycloartenol synthase-dependent sterol biosynthesis in Nicotiana benthamiana. PLoS One. 2014;9:e109156. doi: 10.1371/journal.pone.0109156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremme G., Brendel V., Sparks M.E., Kurtz S. Engineering a software tool for gene structure prediction in higher organisms. Inf. Softw. Technol. 2005;47:965–978. doi: 10.1016/j.infsof.2005.09.005. [DOI] [Google Scholar]
- Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K., Jr., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., White O., Buell C.R., Wortman J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou M., Wang R., Zhao S., Wang Z. Ginsenosides in Panax genus and their biosynthesis. Acta Pharm. Sin. B. 2021;11:1813–1834. doi: 10.1016/j.apsb.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Fan J., Sun Z., Liu S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2020;36:2253–2255. doi: 10.1093/bioinformatics/btz891. [DOI] [PubMed] [Google Scholar]
- Huang A.C., Jiang T., Liu Y.X., Bai Y.C., Reed J., Qu B., Goossens A., Nützmann H.W., Bai Y., Osbourn A. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science. 2019;364:eaau6389. doi: 10.1126/science.aau6389. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J., et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y.S., Etherington G., Geisler K., Field B., Dokarry M., Ikeda K., Mutsukado Y., Dicks J., Osbourn A. Investigation of the potential for triterpene synthesis in rice through genome mining and metabolic engineering. New Phytol. 2011;191:432–448. doi: 10.1111/j.1469-8137.2011.03712.x. [DOI] [PubMed] [Google Scholar]
- Ito R., Nakada C., Hoshino T. β-Amyrin synthase from Euphorbia tirucalli L. functional analyses of the highly conserved aromatic residues Phe413, Tyr259 and Trp257 disclose the importance of the appropriate steric bulk, and cation-π and CH-π interactions for the efficient catalytic action of the polyolefin cyclization cascade. Org. Biomol. Chem. 2016;15:177–188. doi: 10.1039/c6ob02539k. [DOI] [PubMed] [Google Scholar]
- Ji Y., Liu C., Yang Z., Yang L., He Z., Wang H., Yang J., Yi T. Testing and using complete plastomes and ribosomal DNA sequences as the next generation DNA barcodes in Panax (Araliaceae) Mol. Ecol. Resour. 2019;19:1333–1345. doi: 10.1111/1755-0998.13050. [DOI] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakimov B., Kuzina V., Erthmann P.Ø., Fukushima E.O., Augustin J.M., Olsen C.E., Scholtalbers J., Volpin H., Andersen S.B., Hauser T.P., et al. Identification and genome organization of saponin pathway genes from a wild crucifer, and their use for transient production of saponins in Nicotiana benthamiana. Plant J. 2015;84:478–490. doi: 10.1111/tpj.13012. [DOI] [PubMed] [Google Scholar]
- Kim N.H., Jayakodi M., Lee S.C., Choi B.S., Jang W., Lee J., Kim H.H., Waminal N.E., Lakshmanan M., van Nguyen B., et al. Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol. J. 2018;16:1904–1917. doi: 10.1111/pbi.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O.T., Um Y., Jin M.L., Kim J.U., Hegebarth D., Busta L., Racovita R.C., Jetter R. A novel multifunctional C-23 Oxidase, CYP714E19, is involved in asiaticoside biosynthesis. Plant Cell Physiol. 2018;59:1200–1213. doi: 10.1093/pcp/pcy055. [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova M.D., Xiong Q., Lodeiro S., Hua L., Matsuda S.P.T. Lanosterol biosynthesis in plants. Arch. Biochem. Biophys. 2006;447:87–95. doi: 10.1016/j.abb.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Kushiro T., Shibuya M., Ebizuka Y. Chimeric triterpene synthase. a possible model for multifunctional triterpene synthase. J. Am. Chem. Soc. 1999;121:1208–1216. doi: 10.1021/ja983012h. [DOI] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis J.B., Soltis D.E., Li Z., Marx H.E., Barker M.S., Tank D.C., Soltis P.S. Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 2018;105:348–363. doi: 10.1002/ajb2.1060. [DOI] [PubMed] [Google Scholar]
- Leung K.W., Wong A.S.T. Pharmacology of ginsenosides: a literature review. Chin. Med. 2010;5:20. doi: 10.1186/1749-8546-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Xie X., Li F., Tian E., Shu Y., Chao Z. The complete chloroplast genome sequence of Centella asiatica (Linnaeus) Urban. Mitochondrial DNA. B Resour. 2020;5:2149–2150. doi: 10.1080/23802359.2020.1768922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.R., Ding N., Lu T., Zhao J., Wang Z.H., Jiang P., Liu S.T., Wang X.F., Liu B., Li L.F. Evolutionary contribution of duplicated genes to genome evolution in the ginseng species complex. Genome Biol. Evol. 2021;13:evab051. doi: 10.1093/gbe/evab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Leveau A., Zhao Q., Feng Q., Lu H., Miao J., Xue Z., Martin A.C., Wegel E., Wang J., et al. Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals. Nat. Commun. 2021;12:2563. doi: 10.1038/s41467-021-22920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichman B.R., Godden G.T., Buell C.R. Gene and genome duplications in the evolution of chemodiversity: perspectives from studies of Lamiaceae. Curr. Opin. Plant Biol. 2020;55:74–83. doi: 10.1016/j.pbi.2020.03.005. [DOI] [PubMed] [Google Scholar]
- Ma Y., Zhou Y., Ovchinnikov S., Greisen P., Jr., Huang S., Shang Y. New insights into substrate folding preference of plant OSCs. Sci. Bull. 2016;61:1407–1412. doi: 10.1007/s11434-016-1103-1. [DOI] [Google Scholar]
- Majoros W.H., Pertea M., Salzberg S.L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- Manni M., Berkeley M.R., Seppey M., Simão F.A., Zdobnov E.M. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021;38:4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes F.K., Vanderpool D., Fulton B., Hahn M.W. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics. 2020;36:5516–5518. doi: 10.1093/bioinformatics/btaa1022. [DOI] [PubMed] [Google Scholar]
- Miettinen K., Iñigo S., Kreft L., Pollier J., De Bo C., Botzki A., Coppens F., Bak S., Goossens A. The TriForC database: a comprehensive up-to-date resource of plant triterpene biosynthesis. Nucleic Acids Res. 2018;46:D586–D594. doi: 10.1093/nar/gkx925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Shibuya M., Lee M.S., Sankawa U., Ebizuka Y. Molecular cloning of pea cDNA encoding cycloartenol synthase and its functional expression in yeast. Biol. Pharm. Bull. 1997;20:770–775. doi: 10.1248/bpb.20.770. [DOI] [PubMed] [Google Scholar]
- Murrell B., Wertheim J.O., Moola S., Weighill T., Scheffler K., Kosakovsky Pond S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B., Weaver S., Smith M.D., Wertheim J.O., Murrell S., Aylward A., Eren K., Pollner T., Martin D.P., Smith D.M., et al. Gene-wide identification of episodic selection. Mol. Biol. Evol. 2015;32:1365–1371. doi: 10.1093/molbev/msv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Neumann P., Novák P., Hoštáková N., Macas J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA. 2019;10:1. doi: 10.1186/s13100-018-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda-Garcia L., Tawfik D.S. Enzyme evolution in natural products biosynthesis: target- or diversity-oriented? Curr. Opin. Chem. Biol. 2020;59:147–154. doi: 10.1016/j.cbpa.2020.05.011. [DOI] [PubMed] [Google Scholar]
- Ober D. Seeing double: gene duplication and diversification in plant secondary metabolism. Trends Plant Sci. 2005;10:444–449. doi: 10.1016/j.tplants.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Ou S., Chen J., Jiang N. Assessing genome assembly quality using the LTR Assembly Index (LAI) Nucleic Acids Res. 2018;46:e126. doi: 10.1093/nar/gky730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S., Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S., Jiang N. LTR_FINDER_parallel: parallelization of LTR_FINDER enabling rapid identification of long terminal repeat retrotransposons. Mob. DNA. 2019;10:48. doi: 10.1186/s13100-019-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E., Lewinsohn E. Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 2011;62:549–566. doi: 10.1146/annurev-arplant-042110-103814. [DOI] [PubMed] [Google Scholar]
- Qin L., Hu Y., Wang J., Wang X., Zhao R., Shan H., Li K., Xu P., Wu H., Yan X., et al. Insights into angiosperm evolution, floral development and chemical biosynthesis from the Aristolochia fimbriata genome. Nat. Plants. 2021;7:1239–1253. doi: 10.1038/s41477-021-00990-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez T.R., Jaron K.S., Schatz M.C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 2020;11:1432. doi: 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach M.J., Schmidt S.A., Borneman A.R. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinf. 2018;19:460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana-Molina C., Rivas-Marin E., Rojas A.M., Devos D.P. Origin and evolution of polycyclic triterpene synthesis. Mol. Biol. Evol. 2020;37:1925–1941. doi: 10.1093/molbev/msaa054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller H. The role of sterols in plant growth and development. Prog. Lipid Res. 2003;42:163–175. doi: 10.1016/s0163-7827(02)00047-4. [DOI] [PubMed] [Google Scholar]
- Smith M.D., Wertheim J.O., Weaver S., Murrell B., Scheffler K., Kosakovsky Pond S.L. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 2015;32:1342–1353. doi: 10.1093/molbev/msv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Sun P., Yuan J., Gong K., Li N., Meng F., Zhang Z., Li X., Hu J., Wang J., et al. The celery genome sequence reveals sequential paleo-polyploidizations, karyotype evolution and resistance gene reduction in Apiales. Plant Biotechnol. J. 2021;19:731–744. doi: 10.1111/pbi.13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Diekhans M., Baertsch R., Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Stull G.W., Qu X.J., Parins-Fukuchi C., Yang Y.Y., Yang J.B., Yang Z.Y., Hu Y., Ma H., Soltis P.S., Soltis D.E., et al. Gene duplications and phylogenomic conflict underlie major pulses of phenotypic evolution in gymnosperms. Nat. Plants. 2021;7:1015–1025. doi: 10.1038/s41477-021-00964-4. [DOI] [PubMed] [Google Scholar]
- Su W., Jing Y., Lin S., Yue Z., Yang X., Xu J., Wu J., Zhang Z., Xia R., Zhu J., et al. Polyploidy underlies co-option and diversification of biosynthetic triterpene pathways in the apple tribe. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2101767118. e2101767118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1.Sun P., Jiao B., Yang Y., et al. WGDI: a user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotype. Mol. Plant. 2022;15:1841–1851. doi: 10.1016/j.molp.2022.10.018. [DOI] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Bowers J.E., Wang X., Ming R., Alam M., Paterson A.H. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Tansakul P., Shibuya M., Kushiro T., Ebizuka Y. Dammarenediol-II synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng. FEBS Lett. 2006;580:5143–5149. doi: 10.1016/j.febslet.2006.08.044. [DOI] [PubMed] [Google Scholar]
- Tawfik D.S., Gruic-Sovulj I. How evolution shapes enzyme selectivity - lessons from aminoacyl-tRNA synthetases and other amino acid utilizing enzymes. FEBS J. 2020;287:1284–1305. doi: 10.1111/febs.15199. [DOI] [PubMed] [Google Scholar]
- Thimmappa R., Geisler K., Louveau T., O'Maille P., Osbourn A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 2014;65:225–257. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- Tien N.Q.D., Ma X., Man L.Q., et al. De novo whole-genome assembly and discovery of genes involved in triterpenoid saponin biosynthesis of Vietnamese ginseng (Panax vietnamensis Ha et Grushv.) Physiol. Mol. Biol. Plants. 2021;27:2215–2229. doi: 10.1007/s12298-021-01076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N., Jackson C.J., Afriat-Jurnou L., Wyganowski K.T., Tang R., Tawfik D.S. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat. Commun. 2012;3:1257. doi: 10.1038/ncomms2246. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen S. University of Utrecht; 2000. Graph clustering by flow simulation. PhD thesis. [Google Scholar]
- Wang J., Guo Y., Yin X., Wang X., Qi X., Xue Z. Diverse triterpene skeletons are derived from the expansion and divergent evolution of 2,3-oxidosqualene cyclases in plants. Crit. Rev. Biochem. Mol. Biol. 2022;57:113–132. doi: 10.1080/10409238.2021.1979458. [DOI] [PubMed] [Google Scholar]
- Wang L., Zhao S.J., Cao H.J., et al. The isolation and characterization of dammarenediol synthase gene from Panax quinquefolius and its heterologous co-expression with cytochrome P450 gene PqD12H in yeast. Funct. Integr. Genomics. 2014;14:545–557. doi: 10.1007/s10142-014-0384-1. [DOI] [PubMed] [Google Scholar]
- Wu T., Liu W., Huang S., Chen J., He F., Wang H., Zheng X., Li Z., Zhang H., Zha Z., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Mater. Today. Bio. 2021;12:100141. doi: 10.1080/10409238.2021.1979458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Duan L., Liu D., Guo J., Ge S., Dicks J., ÓMáille P., Osbourn A., Qi X. Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 2012;193:1022–1038. doi: 10.1111/j.1469-8137.2011.03997.x. [DOI] [PubMed] [Google Scholar]
- Yang Z. Paml 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z., Liu G., Zhang G., Yan J., Dong Y., Lu Y., Fan W., Hao B., Lin Y., Li Y., et al. The chromosome-scale high-quality genome assembly of Panax notoginseng provides insight into dencichine biosynthesis. Plant Biotechnol. J. 2021;19:869–871. doi: 10.1111/pbi.13558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Chen S., Wang S., Hu Y., Zhang G., Dong Y., Yang S., Miao J., Chen W., Sheng J. Chromosomal-scale genome assembly of Eleutherococcus senticosus provides insights into chromosome evolution in Araliaceae. Mol. Ecol. Resour. 2021;21:2204–2220. doi: 10.1111/1755-0998.13403. [DOI] [PubMed] [Google Scholar]
- Yu G., Lam T.T.Y., Zhu H., Guan Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol. Biol. Evol. 2018;35:3041–3043. doi: 10.1093/molbev/msy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.H., Ma C.H., Zhang J.J., Chen J.W., Tang Q.Y., He M.H., Xu X.Z., Jiang N.H., Yang S.C. Transcriptome analysis of Panax vietnamensis var. fuscidiscus discovers putative ocotillol-type ginsenosides biosynthesis genes and genetic markers. BMC Genom. 2015;16:159. doi: 10.1186/s12864-015-1332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Rabiee M., Sayyari E., Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018;19:153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.G., Li G.Y., Wang X.L., Dainat J., Wang Z.X., Ou S., Ma Y. TEsorter: an accurate and fast method to classify LTR-retrotransposons in plant genomes. Hortic. Res. 2022;9:uhac017. doi: 10.1093/hr/uhac017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Ma Y., Zeng J., Duan L., Xue X., Wang H., Lin T., Liu Z., Zeng K., Zhong Y., et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat. Plants. 2016;2:16183. doi: 10.1038/nplants.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.