Abstract
Higd1a is a conserved gene in evolution which is widely expressed in many tissues in mammals. Accumulating evidence has revealed multiple functions of Higd1a, as a mitochondrial inner membrane protein, in the regulation of metabolic homeostasis. It plays an important role in anti-apoptosis and promotes cellular survival in several cell types under hypoxic condition. And the survival of porcine Sertoli cells facilitated by Higd1a helps to support reproduction. In some cases, Higd1a can serve as a sign of metabolic stress. Over the past several years, a considerable amount of studies about how tumor fate is determined and how cancerous proliferation is regulated by Higd1a have been performed. In this review, we summarize the physiological functions of Higd1a in metabolic homeostasis and its pathophysiological roles in distinct diseases including cancer, nonalcoholic fatty liver disease (NAFLD), type II diabetes and mitochondrial diseases. The prospect of Higd1a with potential to preserve mammal health is also discussed. This review might pave the way for Higd1a-based research and application in clinical practice.
Keywords: Apoptosis, Cancer, Diabetes, Mitochondrial homeostasis, NAFLD
An overview of Higd1a and its other family members
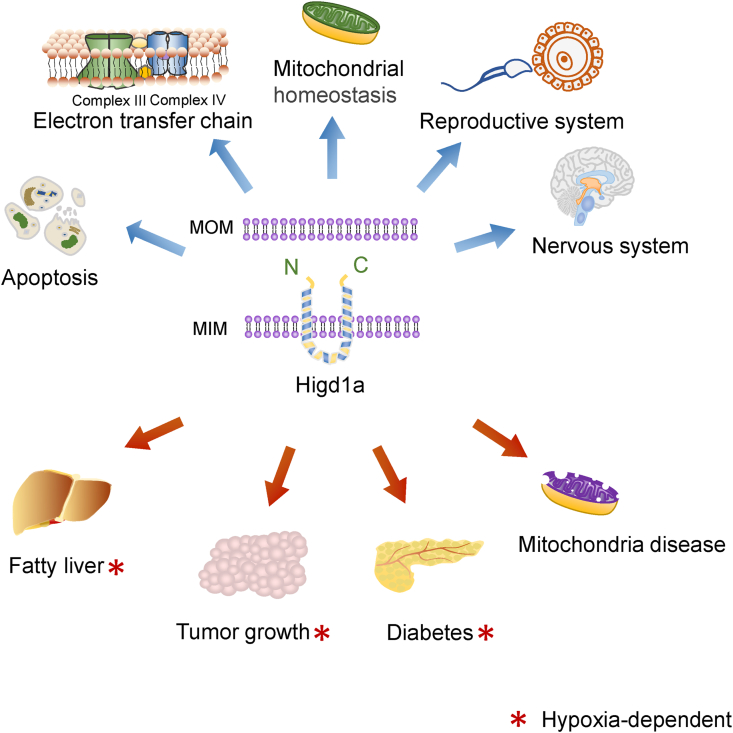
Hypoxia induced gene domain family-1a (Higd1a) is a 10.4 kDa protein attached to mitochondrial inner membrane, also known as HIMP1a, HIG1 and Rcf1, whose expression could be induced under the conditions of hypoglycemia and hypoxia.1 It has two transmembrane domains with a loop-like structure inside of mitochondrial inner membrane (MIM) and both its N-terminal and C-terminal are presented outside of MIM (Fig. 1). Higd1a, expressed in eukaryotes ranging from fungi to human, is a member of hypoxia inducible gene domain (HIGD) family that is conserved in eukaryotes.2, 3, 4 Initially, it was discovered in cultured human cervical epithelial cells, hypoxic neuron-enriched primary cultures, and mouse embryo fibroblasts treated with nickel.5,6 Then, it was found to be ubiquitously expressed in many tissues, such as colon, heart, brain, kidney and liver, with high expression levels in cerebellum, frontal lobe and cortex in mice.5
Figure 1.
Higd1a, as a mitochondrial inner protein, plays important roles in various physiological and pathophysiological processes. Higd1a has two transmembrane domains with a loop-like structure inside of mitochondrial inner membrane and both its N-terminal and C-terminal are presented outside of mitochondrial inner membrane. Higd1a is involved in multiple physiological processes, such as inhibiting apoptosis of the cells, facilitating electron transport chain (ETC) assembly, maintaining mitochondrial homeostasis, regulating the function of reproduction system and the development of nervous system. In pathophysiological processes, Higd1a exhibits diverse roles in regulating tumor growth, displays protective roles against hepatic lipotoxicity, diabetes and mitochondrial diseases. It is known that the roles of Higd1a in the processes of hepatic lipotoxicity, tumor growth and diabetes are hypoxia-dependent, which have been labeled by “∗”. However, whether the functions of Higd1a in other processes, which are not labeled with “∗”, are hypoxia-dependent or not needs further investigation. MOM: mitochondria outer membrane; MIM: mitochondria inner membrane.
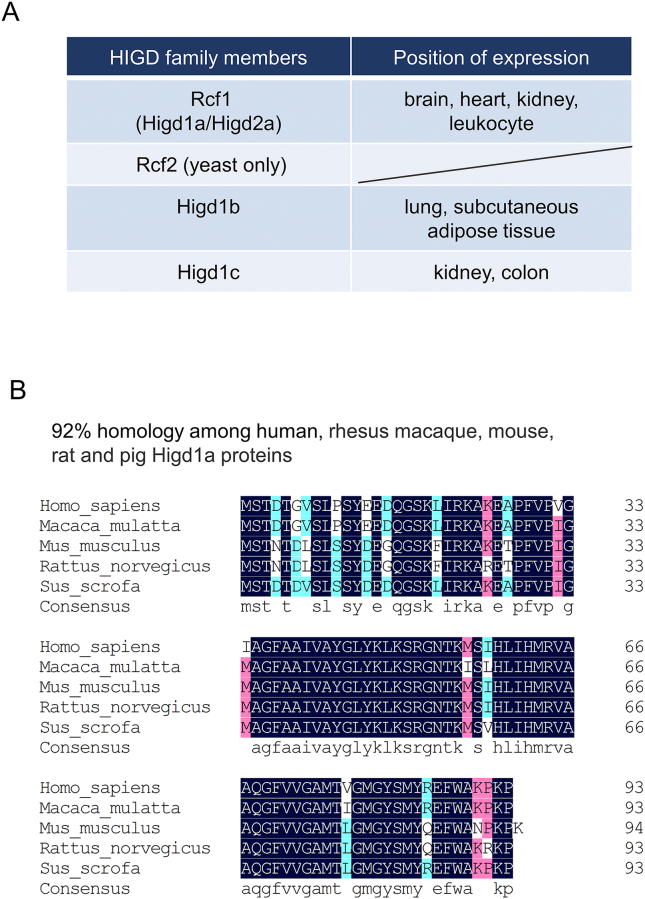
Members of HIGD family regulate multiple critical biological processes. Besides Higd1a, there are several homologous proteins like Higd1b, Higd1c, Higd1d, Higd2a in HIDG family with tissue specific expression profiles (Fig. 2A). Higd1b is regarded as a substitutional formation of Higd1a on account of its N-terminal 17 amino acids that are different from those of Higd1a.6,7 Higd1a and Higd2a are primarily expressed in brain, heart, kidney and leukocyte, while Higd1b is highly expressed in lung and subcutaneous adipose tissue and Higd1c is expressed in kidney and colon. In the yeast, Higd proteins are recognized as respiratory supercomplex factors (Rcfs), mediating the organization of the components of respiratory chain. The yeast Rcf1 is also found in mammals but Rcf2 merely exists in yeast. Higd1a and Higd2a are the mammalian homologs of Rcf1. Rcf1 an Rcf2 cooperatively stabilize the oligomerization of a subclass in respiratory chain supercomplex.8, 9, 10, 11
Figure 2.
Tissue expression profiles of hypoxia inducible gene domain (HIGD) family genes, and sequence alignment of Higd1a proteins among species. (A) The tissue expression profiles of HIGD family genes in mammals, including Higd1a, Higd1b, Higd1c, Higd1d and Higd2a. Rcf1 and Rcf2 are the names for HIGD genes in yeast. The yeast Rcf1 is also found in mammals, but Rcf2 merely exists in yeast. Higd1a and Higd2a are the mammalian homologs of Rcf1. (B) Comparison of Higd1a protein sequences of human, rhesus macaque, mouse, rat and pig. Among the 5 sequences, 3 identical residues are represented in blue; 4 identical residues are represented in red; 5 identical residues are represented in dark blue.
The homology among human, rhesus macaque, mouse, rat and pig Higd1a proteins is as high as 92% by comparison of protein sequence (Fig. 2B). The high homology of Higd1a among species suggests a critical role of Higd1a in biological processes. Indeed, accumulating evidence indicates that Higd1a is involved in multiple physiological and pathophysiological processes (Fig. 1), which will be reviewed and discussed here.
Functional role of Higd1a
Inhibiting apoptosis and promoting survival under hypoxia
Hypoxia is an important cellular stress threatening cellular survival, resulting in severe cellular destruction and even apoptosis.12 It has been broadly recognized that apoptosis is an evolutionarily conserved form of programmed cell death with little ability to induce inflammatory responses. Apoptosis is mediated by the effect of a kind of serine proteases known as caspases, which is activated by the death signals.13, 14, 15 Higd1a, which is induced by hypoxia-inducible factor-1 α (HIF1α), is proved to exert its anti-apoptotic effect mainly through restraining cytochrome C (Cc) release and inhibiting the activation of caspases. Cc could act as a pro-apoptotic protein because its release into the cytosol from the mitochondria is the initiative step of apoptotic process.16
Hyun-Jung An et al disclosed the effect of Higd1a on Cc release and caspases activation. Higd1a increased cell survival by inhibiting Cc release and reducing the activities of caspases. RAWMock, RAWHigd1a (RAW264.7 macrophages stably transfected with Higd1a) and RAWHigd−1a−ΔNT (RAW264.7 macrophages stably transfected with Higd1a with the deletion of its N-terminal 26 amino acids) cells were cultured under hypoxia for 4 days. Then they performed an immunoblot analysis after fractionating the cells into mitochondrial and cytosolic fractions. Four days of hypoxia led to the release of Cc from the mitochondria of the RAWMock and RAWHigd−1a−ΔNT cells, but not from those of the RAWHigd1a cells. Moreover, stably transfected cells were cultured in hypoxia for 6 days to examine the activation of caspases. The RAWHigd1a cells contained less caspase 3 and caspase 9 activity than the RAWMock cells.7 The N-terminal 26 amino acids region of Higd1a turned out to be essential for the survival effect of Higd1a. Furthermore, Higd1a not only helps to inhibit Cc release, but also directly bind with Cc to protect cells from death. The result of co-immunoprecipitation assay illustrated the interaction between Cc and Higd1a. And confocal analysis displayed that Higd1a-mCherry appeared in the same position as Cc-GFP in Heltog cells (a HeLa cell line constitutively expressing green fluorescent protein (GFP)-tagged Cc),8 which indicates the co-localization of Higd1a and Cc. When Higd1a cooperated with Y97pCMF, a phosphomimetic Cc mutant, higher efficiency of electron transport chain (ETC) flow was evidenced than with wild type Cc, which indicates that phosphorylation of Cc could promote the collaborative effect of Higd1a and Cc on facilitating proper cellular response to hypoxic stress.4
Apart from Cc, Higd1a interacts with cytochrome c oxidase (CcO) to protect cells from apoptosis as well. The ETC complex IV, or CcO is the terminal complex of eukaryotic mitochondrial respiratory chain in mitochondria. This complex couples the reduction of molecular oxygen (O2) to water (H2O) and migration of protons from the internal mitochondrial matrix to the inter membrane space (IMS). Adenosine triphosphate (ATP), as the chemical energy required for many biological processes, is produced during this process.17,18 Recently, Higd1a is reported to directly interact with CcO after screening hypoxia-inducible genes in cardiomyocytes. Higd1a binds to CcO, leading to its structural alternation at heme A (the active center of CcO), thereby promoting CcO activity and subsequent mitochondrial oxidative phosphorylation and ATP synthesis. In this case, downregulating endogenous Higd1a caused increased cellular apoptosis under hypoxia owing to decreased oxygen consumption and ATP synthesis as tested by mitochondria-targeted ATP biosensor. On the contrary, exogenous expression of Higd1a was able to reverse the above effect.19
Moreover, despite of the fact that Higd1a is highly expressed in pancreatic α cells but not β cells, it still regulates the survival of β cells upon ectopic expression. Higd1a is defined as an anti-apoptosis factor for the apoptosis rate of MIN6 and βTC3 cells with ectopic expression of Higd1a was significantly lower than the control cells under hypoxic or low glucose situation.4 Further research applying the methods of trypan blue staining, immunoblot, intraperitoneal glucose tolerance test (IPGTT), and glucose-stimulated insulin secretion (GSIS) showed that β-cell lines transfected with Higd1a cDNAs, which expressed relative lower level of Higd1a rather than high level of Higd1a, strengthened the tolerance of the cells to low oxygen/low glucose insults, and promoted insulin biosynthesis, as well as insulin secretion.20 Therefore, Higd1a might be beneficial for insulin production and secretion and protect against diabetes by promoting β cells survival and preserving the mass of pancreatic β cells.21 Taken together, Higd1a preserves cells from apoptosis mainly via influencing some important components involved in mitochondrial homeostasis and such effects of Higd1a remain to be further explored and investigated whether the anti-apoptosis effect of Higd1a is ubiquitous.
Facilitating electron transport chain (ETC) assembly
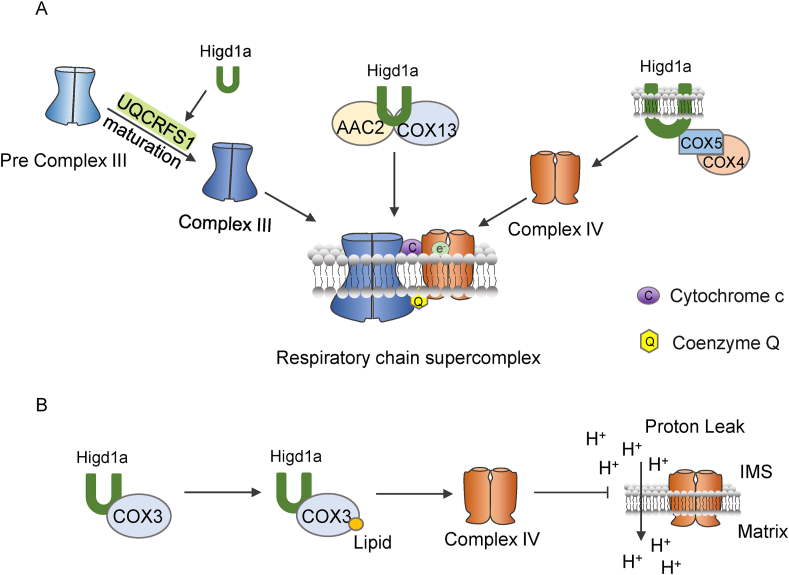
To prohibit the production of reactive oxygen species (ROS), the components of ETC are installed into massive conduction machines as a supercomplexes (SCs) to accelerate electronic flow.22 Higd1a can participate in the assembly of complex III, complex IV and the SCs. The SCs are composed of both complex III (cytochrome bc1 complex) and complex IV (cytochrome c oxidase complex) and perhaps complex II in Saccharomyces cerevisiae. In mammals, SCs usually also contain complex I. Higd1a is involved in the assembling of them by interacting with AAC2 and cytochrome C oxidase 13 (COX13). AAC2 is the yeast homologous protein of human adenine nucleotide translocase 1 (ANT1) and was demonstrated to participate in assembling the ETC complexes.23,24 And the loss of AAC2 caused a synergistic survival defect in both the Higd1a deficient and cardiolipin synthase deficient mutants. The mutant with double deficiency of Rcf1 (Higd1a homolog in yeast) and COX13, on the other hand, demonstrated a more substantial loss of the III2/IV2 complex than the mutant with single deficiency of either gene. Other studies reported a similar role of Higd1a in human embryonic kidney 293T (HEK293T) cells grown under standard normoxic conditions. Higd1a was found to interact with complex IV subunits COX4 and COX5 to further participate in SCs assembly as tested by BN-PAGE analysis and immunoblot. In addition, Higd1a plays a significant role in the final stage of maturation in complex III2 to which Higd1a boosts the fusion of ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 (UQCRFS1) into final assembling stage of complex III as the incorporation of the Rieske Fe–S protein UQCRFS1 to the complex III2 structure was attenuated in the mitochondria of cells with Higd1a deficiency.25,26 The roles of Higd1a described above are illustrated in Figure 3A.
Figure 3.
Higd1a is involved in electron transport chain (ETC) assembly. (A) Higd1a boosts the fusion of ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 (UQCRFS1) into final assembling stage of complex III, thereby facilitating the final stage of maturation in complex III2. Higd1a is involved in the assembling of respiratory chain supercomplex by interacting with AAC2 and COX13. Higd1a could interact with complex IV subunits COX4 and COX5 to further participate in respiratory chain supercomplex assembly. (B) Higd1a is able to stimulate activity of complex IV through mediating the formation of electron-transfer bridge between complex III and complex IV by a tightly bound cytochrome c. Higd1a interplays with complex IV via Cox3, which prevents complex IV from inactivation through impeding lipid loss of COX3 protein in order to block proton leak. IMS: mitochondrial intermembrane space.
In the yeast, Higd1a interplays with complex IV via COX3 (Fig. 3B).27 It prevents complex IV from inactivation through impeding lipid loss of COX3 protein in order to block proton leak because the leaking extent showed a positive correlation with the degree of inactive CIV in MIM in the yeast strains lacking Higd1a (Fig. 3B).28 Besides, through mediating the formation of electron-transfer bridge between complex III and complex IV by a tightly bound cytochrome c, Higd1a is able to stimulate activity of complex IV.29 Moreover, quantitative mass spectrometry analysis modified the cytochrome c oxidase assembly model from a sequential pathway to a module-based process, and it has been verified that Higd1a is engaged in the beginning stage of modular assembly. Higd1a combines with the CcO subunit 1 (MT-Co1), which is inserted in the MIM, to regulate activation of CcO.19,30
In conclusion, Higd1a is involved in the assembling processes of complex III, complex IV and supercomplex to improve the efficiency of ETC (Fig. 3), highlighting the essential role of Higd1a in the function of mitochondrial respiratory chain.
Maintaining dynamic equilibrium of mitochondria
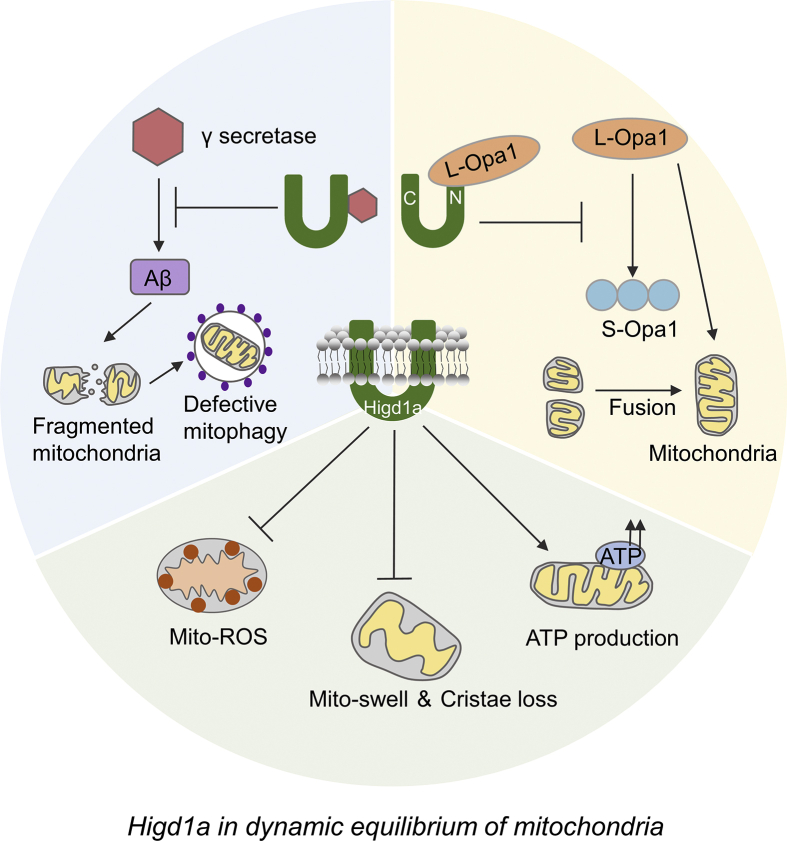
Regular mitochondrial function is preserved by dynamic fusion and fission process.31 Higd1a contributes to the fusion of mitochondrial inner membrane by means of interacting with optic atrophy 1 (OPa1) on its N-terminal tail and inhibiting the cleavage of the long Opa1 isoform (L-Opa1) to short Opa1 (S-Opa1) isoform to keep mitochondrial homeostasis.32,33 The L-Opa1 anchors on MIM and is generated by cleavage of the mitochondrial targeting sequence of Opa1 precursor protein. The S-Opa1 is produced by further proteolyzing the N terminus of L-Opa1. The reciprocal co-IP experiments proved that Higd1a mutant without the N-terminal 26 amino acids totally lost the ability to interact with OPa1. And L OPa1 isoform was found cracked at once in HEK-293T cells under hypoxia, but such event could be dulled by Higd1a. Moreover, dysfunction such as loss of mtDNA, disorganization of cristae, and growth retardation of the cells occurs in the absence of Higd1a.34
Furthermore, the fact that 53–93 aa of the transmembrane domain of Higd1a is necessary for its connection with γ secretase complex was sought out, which illustrates the critical role of Higd1a in supporting mitochondrial function. Higd1a is verified as a negative regulator of γ secretase complex activation via high-throughput screening. γ-Secretase complex is a multi-subunit enzyme composed of catalytic presenilin (PSEN), presenilin enhancer-2 (PEN-2), anterior pharynx defective-1 (APH-1), and Nicastrin (NCT). Its activity drives the cleavage procedure of multiple type-I transmembrane substrates. One of the cleavage products is amyloid-β (Aβ), which induces excessive fragmentation of mitochondria and causes defective mitophagy, thereby impairing the dynamic equilibrium of mitochondria and promoting neurodegeneration.35, 36, 37 Some studies uncovered that low oxygen induces the activation of the γ secretase complex in neuronal cells. Nonetheless, stable expression of Higd1a attenuated this effect to a great extent, with a 50.8% reduction examined in SK-N-SH cells, the human neuroblastoma cells. Consistent with less activation of γ secretase complex, Higd1a also brings about less mitochondrial damage as indicated by reduced ROS production and mitochondrial swelling, restored mitochondrial cristae and increased ATP production.38
All of these results suggest that, under hypoxia, Higd1a plays an important role in blunting γ secretase complex activation and alleviating mitochondrial disorder.38 To sum up, Higd1a, through interplaying with Opa1 and γ secretase complex, keeps mitochondrial viability and integrity to maintain the dynamic balance of mitochondria (Fig. 4), which is pivotal to mitochondrial function.
Figure 4.
Higd1a regulates the dynamic equilibrium of mitochondria. By interacting with OPa1, Higd1a postpones the cleavage of the long Opa1 (L-Opa1) isoform to the short Opa1 (S-Opa1) isoform, which facilitates the fusion of mitochondria. The L-Opa1 anchors on MIM and is generated by cleavage of the mitochondrial targeting sequence of Opa1 precursor protein. The S-Opa1 is produced by further proteolyzing the N terminus of L-Opa1. Silencing of Higd1a leads to impaired mitochondrial fusion, loss of mtDNA, disorganization of cristae, and growth retardation of the cells. Higd1a functions as a negative regulator of γ secretase complex activation to decrease the production of amyloid-β (Aβ), which induces excessive fragmentation of mitochondria and causes defective mitophagy. Consistent with less activation of γ secretase complex, Higd1a attenuates mitochondrial damage as indicated by reduced reactive oxygen species (ROS) production, less severe mitochondrial swelling and increased ATP production, which indicates improved dynamic equilibrium of mitochondria.
Higd1a as potential marker of metabolic stress
Usually, metabolic stress in vivo ultimately causes detrimental clinical illness such as cancer and heart disease.39,40 Apoptosis-inducing factor (AIF) is a flavoprotein originally identified as a soluble 57-kDa protein, and it responds to metabolic stress through subcellular re-localization. Cleaved AIF triggered by metabolic stress is released from mitochondria to cytosol and then moves to nucleus in a caspase-independent manner.41 In the analysis of immunofluorescence confocal laser scanning microscopy, co-staining for Higd1a-GFP and AIF showed the presence of both factors within the nucleus after exposure to etoposide, an apoptotic stimuli inducing DNA damage. Cells treated by etoposide demonstrated a greater extent of nuclear Higd1a accumulation, which was further confirmed by experiment of immunoblot analyses. Additionally, Higd1a interacted and co-localized with AIF in nucleus in a BAX and BAK dependent manner. In mouse embryonic fibroblasts (MEFs) with BAX/BAK double knockout, etoposide-induced localization of Higd1a and AIF within nucleus was attenuated. Moreover, in some disease models, such as myocardial infarction, hypoxic-ischemic encephalopathy (HIE), and different types of cancer, co-location of Higd1a and AIF in the nucleus was also observed.5 These studies demonstrate the correlation between nuclear Higd1a and metabolic disease under certain harmful stimuli. Consequently, Higd1a could serve as a warning signal of metabolic stress, which merits further investigation.
A critical role of Higd1a in reproduction
Higd1a also exerts beneficial effects on procreative system. Sertoli (ST) cells, an important type of supportive cells of spermatogenesis, facilitate male germ cell proliferation and differentiation. Thus, Sertoli cells are critical for male mammals in aspect of sperm fertility.42 Guo et al demonstrated that Higd1a plays a significant role in the development of testicular tissue and interacts with miR-375 to regulate proliferation of pig Sertoli cells. miR-375 is an important factor in functional reproductive system of mammals. It inhibits estradiol (E2) synthesis by means of controlling corticotropin releasing hormone (CRH) pathway.43 Moreover, Higd1a has been proclaimed acting as the target gene of miR-375 and the special targeting of Higd1a mRNA by miR-375 was verified by luciferase assays.44 Another related research demonstrated that ST cells derived from Junmu No. 1 white pigs overexpressing miR-375 showed a significantly higher apoptotic rate than those cells overexpressing Higd1a. In addition, sertoli cells transfected with miR-375 mimic was able to expedite apoptosis in high-glucose or low-glucose condition yet Higd1a curbed such effect.45 As mentioned before, Higd1a is the target gene of miR-375, suggesting that Higd1a can induce a reduction in apoptosis and miR-375 can promote apoptosis at least partially by downregulating the expression level of Higd1a.45 Collectively, these results demonstrate a close connection of miR-375 and Higd1a with the survival of ST cells. Therefore, Higd1a emerges as a functional regulator in reproduction system.
Potential role of Higd1a in nervous system
Higd1a is shown to play a significant role in nervous system. The expression of Higd1a in central nervous system (CNS) exhibits great plasticity as the level of its mRNA shows a gradually increasing trend in the spinal cord in 15 days after parturition in rats.46 Higd1a expression level is significantly increased in the parietal cortex in both male and female rats at day 8 after parturition and reaches the peak at day 15 in either parietal cortex or diencephalon. In the meantime, it exhibits a tendency to be higher at day 5 after parturition in the spinal cord. These results indicate that the significant expression level change of Higd1a is distinct in different ages and in different parts of CNS.46 Other features of Higd1a expression in nervous system include spatial specificity and high expression in amygdala and in many nuclei of the brainstem. Also, diverse expression patterns of Higd1a protein in the nervous system of male Wistar rats were discovered, shifting from neurons at first day of postnatal life to glial cells at the ninetieth day of postnatal life.47 In short, Higd1a is expressed in different kinds of cells in central nervous system with temporal change, strongly hinting its significance in physiological processes of neural proliferation, cell fate specification and neuronal migration and its potential role in cellular preservation when programmed cell death occurs during CNS maturation.
Role of Higd1a in diseases
Higd1a in cancers
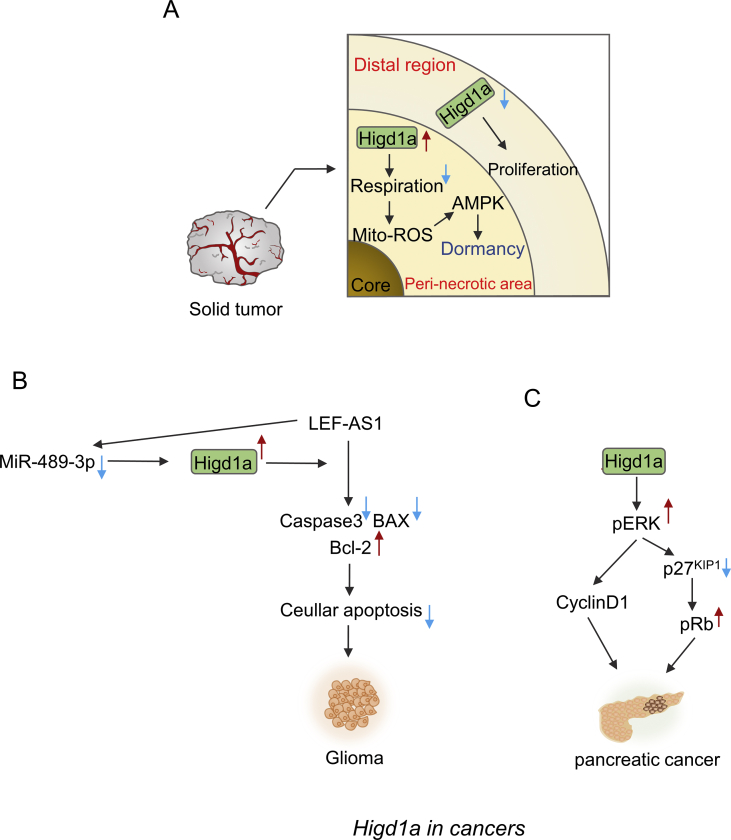
Higd1a possesses an important role in tumorigenesis, with both tumor-promoting and tumor-suppressing function. Higd1a was supposed to be the target gene of HIF-1α. However, recent research demonstrated that it could be induced in an HIF-1α independent manner in certain region of solid tumor. The core zone of the solid tumor is basically necrotic and the area around the core zone called the peri-necrotic region is usually exposed to serious hypoxia and poor vascularity, resulting in nutrient deficiency all the time (Fig. 5A). In the peri-necrotic area around the necrotic core, the activity of DNA methyltransferase 1 (DNMT1), which is in charge of DNA methylation in cells of mammals, is inhibited due to serious glucose deprivation and ischemia. Therefore, the expression of Higd1a is enhanced since the human Higd1a gene is found in the location of many tumor suppressor genes that are repressed by DNA methylation.3,48 In these settings, Higd1a reduced mitochondrial respiration during glucose deprivation with the subsequent total ROS reduction relieving cell death. This may be because that Higd1a-triggered mitochondrial superoxide (O2−) summons activation of AMPK pathway for adaptive response. A considerable leap of AMPK activity in HIF-1α-deficient MEFs expressing Higd1a during glucose starvation was observed but MitoQ, acting as mitochondria-specific antioxidant, dulled such effect. Activation of AMPK pathway improves pentose phosphate shunt-mediated NADPH production, attenuates oxidative stress49 and finally brings about dormancy of tumor cells. Thus, Higd1a hijacked the tumors to deter further development of cancers and kept them in dormancy (Fig. 5A).
Figure 5.
Diverse roles of Higd1a in cancers. (A) In the peri-necrotic area of some solid tumors, where serious hypoxia and poor vascularity occur, Higd1a expression is elevated, which is owing to decreased expression of DNMT1 that leads to the decreased DNA methylation on Higd1a promoter. In this area, Higd1a inhibits mitochondrial respiration of the tumor cells, which will elevate mitochondrial superoxide (O2−) to activate AMPK pathway for adaptive response. Activation of AMPK pathway will attenuate oxidative stress and finally bring about dormancy of tumor cells. In this way, Higd1a hijacks the tumors to deter further development of cancers and keeps them in dormancy. In contrast, in the distal region to necrosis with mild to moderate hypoxia, Higd1a expression is inhibited as a result of up-regulation of DNMT1. In this area, Higd1a functions as suppressor for the growth of tumor cells. Cancer cell shows a rapid growth when Higd1a expression level is repressed in this area. (B, C) However, in some other cases, Higd1a has the ability to expedite the proliferation of tumor cells. (B) In glioma, miR-489-3p can bind to the mRNA of Higd1a to inhibit Higd1a expression, while LEF1-AS1 can bind to miR-489-3p to block the inhibitory effect of miR-489-3p on Higd1a expression. Higd1a could facilitate LEF1-AS1-mediated inhibition of apoptosis as indicated by the decrease of cleaved caspase 3 and Bax expression, along with the increase of Bcl-2 expression, which will promote the growth of glioma. (C) In pancreatic cancer cells, Higd1a will increase ERK activity, thereby decreasing the expression of p27KIP1 and increasing the expression of cyclin D1. Moreover, decreased p27KIP1 boosts phosphorylation of Rb protein to repress the function of Rb protein. The effects above will ultimately promote the proliferation of pancreatic cancer cells.
However, in addition to the dormant effect of Higd1a in tumorous cell, it has been proved that tumor proliferation might be controlled as long as Higd1a is expressed in the distal region to necrosis with mild to moderate hypoxia (Fig. 5A). In general, expression of HIF-1α is increased in the tumor region of less severe nutritious deprivation, but together with up-regulation of DNMT1, Higd1a expression was inhibited as a result of elevated DNA methylation. Cancer cell shows a rapid growth if Higd1a expression level is repressed.3 In this case, Higd1a functions as suppressor for the growth of tumor cells. HIF-1α-deficient transformed mouse embryonic fibroblasts (MEFs) were generated and xenografts were established in order to construct the state that Higd1a is induced in HIF-1α independent manner. It turns out that tumor mass formed from the HIF-1α-deficient MEFs with the overexpression of Higd1a was obviously less than their HIF-1α-deficient counterparts without Higd1a over-expression. Therefore, it could be speculated that Higd1a undermines the proliferation of tumor cells and keeps them in dormancy in core zone of the tumors.3,50
However, some studies showed that, not only in glioma but also in pancreatic cancer, Higd1a has the ability to expedite the proliferation of tumor cells (Fig. 5B, C). The fact that long non-coding (lncRNA) lymphoid enhancer-binding factor 1 antisense RNA 1 (LEF1-AS1) causes carcinogenesis in multiple kinds of cancers is well recognized.51,52 With the evidence from flow cytometry analysis and TUNEL assay, Higd1a hindered apoptosis of glioma cells caused by the silence of LEF1-AS1. LEF-AS1 can suppress the function of MiR-489-3p, which targets Higd1a mRNA to impede its translation into Higd1a protein, thus enhancing the expression of Higd1a and eventually promoting glioma cells survival (Fig. 5B). Consistently, depletion of LEF1-AS1 promoted apoptosis as indicated by the increase of cleaved caspase 3 and Bax expression, along with the decrease of Bcl-2 expression (Fig. 5B). In this case, aiming to curb glioma proliferating, miR-489-3p-mediated downregulation of Higd1a might restrict the developing process of glioma.53 In line with antiapoptotic consequence of Higd1a in glioma, An and Ryu also noticed that knockdown of Higd1a results in growth retardation of cancer cells without cell death in the pancreas. Upon the deficiency of Higd1a induced by short hairpin RNA (shRNA) in both pancreatic cancer cells PANC-1 and ASPC-1, phosphorylated level of extracellular regulated protein kinases (pERK) cuts back, which increases the expression of p27KIP1 and inhibits the expression of G0/G1 check point gene cyclin D1 (Fig. 5C). Then, when applying 10 μM U0126 (the chemical inhibitor of MEK) to AsPC-1 cells to suppress ERK activation, p27KIP1 level was observed to increase, suggesting that cell cycle arrest is mediated by the downregulation of the ERK activity and the subsequent induction of p27KIP1 in pancreatic cancer cells. Moreover, increased p27KIP1 dents phosphorylation of Rb protein to enhance the function of Rb protein (Fig. 5C). These data suggest an important role of Higd1a in promoting the proliferation of pancreatic cancer cells through blocking the p27KIP1/Rb pathway.53 In addition, Higd1a is highly relevant to cell senescence as its depletion increases the number of flattened and enlarged cells, which are proposed to be the hallmark of senescence. Meanwhile, cells infected with lentiviruses expressing Higd-1a shRNA seemed to induce SA-β-gal activity, which is considered as a specific marker of senescence.54 Hence, it is likely that depletion of Higd1a blunts tumor growth via blocking the cell cycle in phase of G0/G1 and promoting senescence phenotype through the pERK/p27kip1/pRB axis.1
To sum up, Higd1a seems to play distinct roles in tumor growth. In some cases, it acts as a negative regulator in tumorous proliferation since the bulk of tumorous mass of some solid tumors is tremendously less in the presence of Higd1a. When exposed to an extremely tough environment, the process of tumor dormancy is initiated by Higd1a. Nonetheless, in some other cases, Higd1a might play an opposite role. As a result, the relationship between Higd1a and tumor growth fails to be straightforward or black and white. Whether it acts as a mediator in other pathways in tumor growth or whether there is an expression level-dependent effect of Higd1a on tumor biology remains to be explored.
Protective role of Higd1a against lipotoxicity in the liver
It has been predicted that incidence of worldwide population suffering from nonalcoholic fatty liver disease (NAFLD) is about 25% and NAFLD is already the fastest growing cause of hepatocellular carcinoma (HCC) in many developed countries.55, 56, 57 NAFLD is a recapitulative term including a continuum of liver abnormalities ranging from nonalcoholic fatty liver (NAFL) to nonalcoholic steatohepatitis (NASH), which has a variable course and can lead to cirrhosis and liver cancer.58,59,60 In the process of NAFLD, the liver's capacity to handle the primary metabolic energy substrates, including carbohydrates and fatty acids, is overwhelmed, leading to the accumulation of toxic lipid species in hepatocytes.61 Higd1a is shown to effectively improve lipotoxicity of hepatocytes and prevent those cells from apoptosis. The 0.4 mM oleic acid and 0.2 mM palmitate were added to the growth media of HepG2 and LO2 cells for 72 h to establish the lipotoxicity model of hepatocytes. The role of Higd1a in regulating apoptosis rate and mitochondrial transmembrane potential (MMP) in the above cells was examined. It was discovered that overexpression of Higd1a brings about a lower risk of apoptosis, indicating the protective effect of Higd1a against the stress induced by high-fat exposure. On the contrary, knockdown of Higd1a led to an opposite result. Further experiment manifests that Higd1a is induced by PGC-1α and HIF-1α in a synergistic effect. PGC-1α and HIF1α expression is elevated by ROS, which is induced upon high-fat exposure, to increase Higd1a expression in order to preserve HepG2 and LO2 cells from lipotoxic attack. Collectively, Higd1a, induced by PGC-1α and HIF-1α under the oxidative stress condition, can help to protect hepatocytes from lipotoxicity and cell death.2 These data suggest a potential protective role of Higd1a against NAFLD, which merits further investigation.
Protective role of Higd1a in type II diabetes
Type 2 Diabetes Mellitus (T2DM) is a common but complicated metabolic dysfunction triggered by both genetic and environmental factors.62, 63, 64, 65 The main symptoms of it is islet failure resulting from mass and function loss of pancreatic β cells together with more glucagon secretion, which triggers insulin resistance.66,67 Experiments in vivo and ex vivo have uncovered that the damage of β cell can be partly rescued via ectopic expression of Higd1a. Wang and coworkers discovered that Higd1a expressing in MIN β cells triggers a protective procedure against mitochondrial stress and exerts a positive effect on β cells survival and protects β cells from apoptosis under low glucose or low O2 condition. Higd1a is identified to be expressed in pancreatic α cells rather than β cells according to double immunostaining of pancreas sections. However, there was a higher percentage of viability in the models of MIN6 and βTC3 cells transfected with Higd1a than control cells under a 5% hypoxic environment. Furthermore, when facing either high (25 mM) or low glucose (2.5 mM) levels, βTC3 cells with ectopic expression of Higd1a had markedly lower rate of apoptosis than that of control cells, strongly suggesting the protective role of Higd1a in the survival of pancreatic β cells.4 Later study applying a DNA fragment (364 bp) specifically from the MIP-Higd1a expression cassette, which allows ectopic Higd1a overexpression merely within pancreatic β cells by the control of mouse insulin 1 gene promoter (MIP) in rodents to steadily enhance Higd1a expression at three distinct levels in pancreatic β cells, revealed a more concrete result. Relatively lower extent of Higd1a overexpression (least level of overexpression) effectively facilitated the tolerance of the mice to low oxygen and low glucose threats, and promoted basal insulin secretion and basal (pro)insulin biosynthesis.20 In short, mitochondrial stress could be alleviated through appropriate overexpression of Higd1a, which protects pancreatic β cells from apoptosis under low/high glucose or hypoxia conditions and boosts inulin synthesis and secretion, which are beneficial effects in rivalry to T2DM.
Role of Higd1a in mitochondrial diseases
Mitochondrial diseases are defined as inherited disorders due to dysregulated oxidative phosphorylation originating from impaired function of electronic transport chain (ETC). It has been discovered that mutations in either mitochondrial function-related nuclear genes or mitochondrial genes had negative effects on oxidative phosphorylation (OXPHOS), bringing about a group of lethal dysfunctions.68 The whole process of oxidative phosphorylation occurs in mitochondria. The normal function of ETC is required for OXPHOS, exerting the redox reactions of cellular respiration and generating the proton motive force for the synthesis of ATP.37 Mitochondrial dysfunction could cause detrimental consequences because mitochondria is the power source of the body.69,70
Higd1a has been shown to have beneficial roles either in vivo or in vitro in maintaining functional respiration chain as suggested by various kinds of models of mitochondrial disease. It has been verified that the activity of respiratory chain complex IV would be depressed under hypoxia, and results in dysfunction of respiratory chain, such as decline of proton pumping and mitochondrial ATP production. All of these are the main characteristics of mitochondrial diseases.71 Zebrafish hypoxia model is applied to explore the role of Higd1a in mitochondrial disease. In a zebrafish model, which contain the mitochondria-targeted FRET-based ATP sensor to evaluate ATP synthesis in time in heart, Higd1a was expressed exogenously via tol2 transposon system. As an autonomous transposable element, tol2 itself can encode a transposase, and so far, it is the only transposon with autonomous transposable activity found in vertebrates. In this way, the exogenous gene can replace the transposase coding sequence to construct a Tol2 transposable vector, and be transformed into the host cell with the transposase cDNA vector plasmid, and the exogenous gene will be integrated into the genome without causing any rearrangement and modification of target sites in a “cut-and-paste” manner. Via fluorescent, it is likely to observe embryos and detect heart motion at the same time after inducing Higd1a by a tol2 vector with transposes in oocytes from the casper zebrafish lacking pigmentation. The result is that heart beat better and mitochondrial ATP production was higher in those zebrafish with exogenous expression of Higd1a compared with the control group under low oxygen, suggesting an ameliorative effect of Higd1a in mitochondrial diseases in vivo.72
The role of ectopic expression of Higd1a was also assessed in four distinct models of mitochondrial dysfunction, NDUFS4 depleted HEK293T cell line, NSUN3 knockout cells, and A3243G mutation-induced mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS), respectively. NDUFS4 is a subunit of complex I of the ETC and its loss leads to mitochondrial dysfunction characterized by progressive focal neurodegenerative lesions in specific regions of the brain.73 NSUN3 is the 5-methylcytosine (5mC) methyltransferase, which is encoded by NOP2/Sun RNA methyltransferase 3 (NSUN3). Mutations of NSUN3 had multisystem mitochondrial disease associated with a combined oxidative phosphorylation (OXPHOS) deficiency.74 And MELAS is a rare mitochondrial disease with various manifestations induced by multisystem disorder,23,75 which was due to damaged respiratory chain components. In those mitochondrial disease models, dysfunction of mitochondria was improved by exogenous expression of Higd1a, in which CcO activity was increased, and OCR and ATP level was elevated after exogenous Higd1a introduction.72 What is more, the correlation between mitochondrial dysfunction and neurodegeneration has been evidenced.76,77 Higd1a, as previously mentioned, interplays with γ secretase to reduce excessive fragmentation of mitochondria to protect against Alzheimer's Disease. Those findings imply that Higd1a-based therapeutic strategies to treat mitochondrial diseases merits further investigation.
Conclusions
Higd1a was originally found in cervical epithelial cells from human as a protein of mitochondrial inner membrane. It plays important roles in multiple biological processes and diseases. Higd1a can communicate with most parts of respiratory chain (mainly with Complex III and IV) and participate in supercomplex assembling to boost ETC function. Through interacting with Opa1 to improve fusion of mitochondrial inner membrane and with γ secretase complex to rescue mitochondrial dysfunction, Higd1a contributes to maintaining the balance of mitochondrial function. Consistent with these data, mitochondrial disease is improved by Higd1a to a certain degree. Based on these results, we reasoned that Higd1a may participate in mediating the functions of skeletal muscle tissue, brown adipose tissue and myocardial tissue since mitochondria are enriched in those tissue. Because skeletal muscle and heart are two important organs involving in exercise,78, 79, 80 Higd1a could play a part in exercise-mediated improvement of health, which needs further investigation.
In terms of cancer, Higd1a is able to activate AMPK pathway to cut down total ROS and push tumorous cells into dormant status in serious oxygen and glucose deprivation. In less severe condition, however, Higd1a could inhibit the growth of some tumors. In other cases, Higd1a may function to promote tumor growth. Moreover, Higd1a is regarded as a positive regulator to raise ST cell survival rate and to attenuate the lipotoxicity of hepatocytes in high fat condition. Thus, distinct roles of Higd1a are proved in the regulation of tumor growth and in response to various harmful stimulation. And exploration of novel Higd1a-based therapeutic strategies and underlying mechanisms are needed for better application of this target. Besides, identifying the functions of the other HIGD family members would provide more detailed understanding of the roles of this class of hypoxia-induced proteins in various kinds of physiological and pathophysiological processes.
Author contributions
The search and collection of literatures was performed by J.-Y.Z., M.C., W.-J.M., H.-Y.L., and L.G. The first draft of the manuscript was written by J.-Y.Z. and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Conflict of interests
Authors declare no conflict of interests.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31871435 and 32070751 to L.G.).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Abbreviations
- 5mC
5-methylcytosine
- AIF
apoptosis-inducing factor
- ANT1
adenine nucleotide translocase 1
- APH-1
anterior pharynx defective-1
- ATP
adenosine triphosphate
- Aβ
amyloid-β
- Cc
cytochrome C
- CcO
cytochrome c oxidase
- CNS
central nervous system
- COX13
cytochrome C oxidase 13
- CRH
corticotropin releasing hormone
- DNMT1
DNA methyltransferase1
- ETC
electron transport chain
- E2
estradiol
- GFP
green fluorescent protein
- GSIS
glucose-stimulated insulin secretion
- HCC
hepatocellular carcinoma
- HEK293T
human embryonic kidney 293T
- Higd1a
hypoxia induced gene domain family-1a
- HIGD
hypoxia inducible gene domain
- HIF1α
hypoxia-inducible factor-1 α
- HIE
hypoxic-ischemic encephalopathy
- IMS
inter membrane space
- IPGTT
intraperitoneal glucose tolerance test
- lncRNA
long non-coding RNA
- LEF1-AS1
lymphoid enhancer-binding factor 1 antisense RNA 1
- MEFs
mouse embryonic fibroblasts
- MELAS
mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes
- MIM
mitochondrial inner membrane
- MIP
mouse insulin 1 gene promoter
- MMP
mitochondrial transmembrane potential
- MOM
mitochondria outer membrane
- MT-Co1
CcO subunit 1
- NCT
Nicastrin
- NAFLD
nonalcoholic fatty liver disease
- NAFL
nonalcoholic fatty liver
- NASH
nonalcoholic steatohepatitis
- OPa1
optic atrophy 1
- OXPHOS
oxidative phosphorylation
- O2
oxygen
- O2−
superoxide
- PEN-2
presenilin enhancer-2
- PSEN
presenilin
- Rcfs
respiratory supercomplex factors
- ROS
reactive oxygen species
- shRNA
short hairpin RNA
- SCs
supercomplexes
- ST
Sertoli
- T2DM
type 2 diabetes mellitus
- UQCRFS1
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
References
- 1.An H.J., Ryu M., Jeong H.J., et al. Higd-1a regulates the proliferation of pancreatic cancer cells through a pERK/p27 KIP1/pRB pathway. Cancer Lett. 2019;461:78–89. doi: 10.1016/j.canlet.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Li T., Xian W.J., Gao Y., et al. Higd1a protects cells from lipotoxicity under high-fat exposure. Oxid Med Cell Longev. 2019;2019:6051262. doi: 10.1155/2019/6051262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ameri K., Jahangiri A., Rajah A.M., et al. HIGD1A regulates oxygen consumption, ROS production, and AMPK activity during glucose deprivation to modulate cell survival and tumor growth. Cell Rep. 2015;10(6):891–899. doi: 10.1016/j.celrep.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J., Cao Y., Chen Y., Chen Y., Gardner P., Steiner D.F. Pancreatic beta cells lack a low glucose and O2-inducible mitochondrial protein that augments cell survival. Proc Natl Acad Sci U S A. 2006;103(28):10636–10641. doi: 10.1073/pnas.0604194103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ameri K., Rajah A.M., Nguyen V., et al. Nuclear localization of the mitochondrial factor HIGD1A during metabolic stress. PLoS One. 2013;8(4):e62758. doi: 10.1371/journal.pone.0062758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denko N., Schindler C., Koong A., Laderoute K., Green C., Giaccia A. Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin Cancer Res. 2000;6(2):480–487. [PubMed] [Google Scholar]
- 7.An H.J., Shin H., Jo S.G., et al. The survival effect of mitochondrial Higd-1a is associated with suppression of cytochrome C release and prevention of caspase activation. Biochim Biophys Acta. 2011;1813(12):2088–2098. doi: 10.1016/j.bbamcr.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 8.Guerra-Castellano A., Díaz-Quintana A., Pérez-Mejías G., et al. Oxidative stress is tightly regulated by cytochrome c phosphorylation and respirasome factors in mitochondria. Proc Natl Acad Sci U S A. 2018;115(31):7955–7960. doi: 10.1073/pnas.1806833115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strogolova V., Furness A., Robb-McGrath M., Garlich J., Stuart R.A. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32(8):1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vukotic M., Oeljeklaus S., Wiese S., et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15(3):336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Shoubridge E.A. Supersizing the mitochondrial respiratory chain. Cell Metab. 2012;15(3):271–272. doi: 10.1016/j.cmet.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Guo Y., Tan J., Miao Y., Sun Z., Zhang Q. Effects of microvesicles on cell apoptosis under hypoxia. Oxid Med Cell Longev. 2019;2019:5972152. doi: 10.1155/2019/5972152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X., Ferrell J.E., Jr. Apoptosis propagates through the cytoplasm as trigger waves. Science. 2018;361(6402):607–612. doi: 10.1126/science.aah4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen B.D., Sørensen C.S. The caspase-activated DNase: apoptosis and beyond. FEBS J. 2017;284(8):1160–1170. doi: 10.1111/febs.13970. [DOI] [PubMed] [Google Scholar]
- 15.Xu X., Lai Y., Hua Z.C. Apoptosis and apoptotic body: disease message and therapeutic target potentials. Biosci Rep. 2019;39(1) doi: 10.1042/BSR20180992. BSR20180992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santucci R., Sinibaldi F., Cozza P., Polticelli F., Fiorucci L. Cytochrome c: an extreme multifunctional protein with a key role in cell fate. Int J Biol Macromol. 2019;136:1237–1246. doi: 10.1016/j.ijbiomac.2019.06.180. [DOI] [PubMed] [Google Scholar]
- 17.Brischigliaro M., Zeviani M. Cytochrome c oxidase deficiency. Biochim Biophys Acta Bioenerg. 2021;1862(1):148335. doi: 10.1016/j.bbabio.2020.148335. [DOI] [PubMed] [Google Scholar]
- 18.Watson S.A., McStay G.P. Functions of cytochrome c oxidase assembly factors. Int J Mol Sci. 2020;21(19):7254. doi: 10.3390/ijms21197254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi T., Asano Y., Shintani Y., et al. Higd1a is a positive regulator of cytochrome c oxidase. Proc Natl Acad Sci U S A. 2015;112(5):1553–1558. doi: 10.1073/pnas.1419767112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., Degenstein L., Cao Y., Stein J., Osei K., Wang J. beta-Cells with relative low HIMP1 overexpression levels in a transgenic mouse line enhance basal insulin production and hypoxia/hypoglycemia tolerance. PLoS One. 2012;7(3):e34126. doi: 10.1371/journal.pone.0034126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheuner D., Kaufman R.J. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev. 2008;29(3):317–333. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genova M.L., Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochim Biophys Acta. 2014;1837(4):427–443. doi: 10.1016/j.bbabio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Dallabona C., Baruffini E., Goffrini P., Lodi T. Dominance of yeast aac2 R96H and aac2 R252G mutations, equivalent to pathological mutations in ant1, is due to gain of function. Biochem Biophys Res Commun. 2017;493(2):909–913. doi: 10.1016/j.bbrc.2017.09.122. [DOI] [PubMed] [Google Scholar]
- 24.Dienhart M.K., Stuart R.A. The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol Biol Cell. 2008;19(9):3934–3943. doi: 10.1091/mbc.E08-04-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y.C., Taylor E.B., Dephoure N., et al. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15(3):348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timón-Gómez A., Garlich J., Stuart R.A., Ugalde C., Barrientos A. Distinct roles of mitochondrial HIGD1A and HIGD2A in respiratory complex and supercomplex biogenesis. Cell Rep. 2020;31(5):107607. doi: 10.1016/j.celrep.2020.107607. [DOI] [PubMed] [Google Scholar]
- 27.Garlich J., Strecker V., Wittig I., Stuart R.A. Mutational analysis of the QRRQ motif in the yeast Hig1 type 2 protein Rcf1 reveals a regulatory role for the cytochrome c oxidase complex. J Biol Chem. 2017;292(13):5216–5226. doi: 10.1074/jbc.M116.758045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoang N.H., Strogolova V., Mosley J.J., Stuart R.A., Hosler J. Hypoxia-inducible gene domain 1 proteins in yeast mitochondria protect against proton leak through complex IV. J Biol Chem. 2019;294(46):17669–17677. doi: 10.1074/jbc.RA119.010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rydström Lundin C., von Ballmoos C., Ott M., Ädelroth P., Brzezinski P. Regulatory role of the respiratory supercomplex factors in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2016;113(31):E4476–E4485. doi: 10.1073/pnas.1601196113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vidoni S., Harbour M.E., Guerrero-Castillo S., et al. MR-1S interacts with PET100 and PET117 in module-based assembly of human cytochrome c oxidase. Cell Rep. 2017;18(7):1727–1738. doi: 10.1016/j.celrep.2017.01.044. [DOI] [PubMed] [Google Scholar]
- 31.Ren L., Chen X., Chen X., Li J., Cheng B., Xia J. Mitochondrial dynamics: fission and fusion in fate determination of mesenchymal stem cells. Front Cell Dev Biol. 2020;8:580070. doi: 10.3389/fcell.2020.580070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishihara N., Fujita Y., Oka T., Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25(13):2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olichon A., Emorine L.J., Descoins E., et al. The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 2002;523(1–3):171–176. doi: 10.1016/s0014-5793(02)02985-x. [DOI] [PubMed] [Google Scholar]
- 34.An H.J., Cho G., Lee J.O., Paik S.G., Kim Y.S., Lee H. Higd-1a interacts with Opa1 and is required for the morphological and functional integrity of mitochondria. Proc Natl Acad Sci U S A. 2013;110(32):13014–13019. doi: 10.1073/pnas.1307170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia W. gamma-Secretase and its modulators: twenty years and beyond. Neurosci Lett. 2019;701:162–169. doi: 10.1016/j.neulet.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Escamilla-Ayala A.A., Sannerud R., Mondin M., et al. Super-resolution microscopy reveals majorly mono- and dimeric presenilin1/γ-secretase at the cell surface. eLife. 2020;9 doi: 10.7554/eLife.56679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pradeepkiran J.A., Reddy P.H. Defective mitophagy in Alzheimer's disease. Ageing Res Rev. 2020;64:101191. doi: 10.1016/j.arr.2020.101191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi H., Nakagami H., Takeichi M., et al. HIG1, a novel regulator of mitochondrial γ-secretase, maintains normal mitochondrial function. FASEB J. 2012;26(6):2306–2317. doi: 10.1096/fj.11-196063. [DOI] [PubMed] [Google Scholar]
- 39.Peoples J.N., Saraf A., Ghazal N., Pham T.T., Kwong J.Q. Mitochondrial dysfunction and oxidative stress in heart disease. Exp Mol Med. 2019;51(12):1–13. doi: 10.1038/s12276-019-0355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munir R., Lisec J., Swinnen J.V., Zaidi N. Lipid metabolism in cancer cells under metabolic stress. Br J Cancer. 2019;120(12):1090–1098. doi: 10.1038/s41416-019-0451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo S.M., Martinez P.A., Marques E.F., et al. Oxidation of apoptosis-inducing factor (AIF) to disulfide-linked conjugates. Arch Biochem Biophys. 2020;692:108515. doi: 10.1016/j.abb.2020.108515. [DOI] [PubMed] [Google Scholar]
- 42.Parekh P.A., Garcia T.X., Hofmann M.C. Regulation of GDNF expression in Sertoli cells. Reproduction. 2019;157(3):R95–R107. doi: 10.1530/REP-18-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu C., Li M., Wang Y., et al. miR-375 mediates CRH signaling pathway in inhibiting E2 synthesis in porcine ovary. Reproduction. 2017;153(1):63–73. doi: 10.1530/REP-16-0323. [DOI] [PubMed] [Google Scholar]
- 44.Guo J., Liu X., Yang Y., et al. miR-375 down-regulation of the rearranged L-myc fusion and hypoxia-induced gene domain protein 1A genes and effects on Sertoli cell proliferation. Asian Australas J Anim Sci. 2018;31(8):1103–1109. doi: 10.5713/ajas.17.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo J., Yang C., Zhang S., et al. MiR-375 induces ROS and apoptosis in ST cells by targeting the HIGD1A gene. Gene. 2019;685:136–142. doi: 10.1016/j.gene.2018.10.086. [DOI] [PubMed] [Google Scholar]
- 46.López L., Zuluaga M.J., Lagos P., Agrati D., Bedó G. The expression of hypoxia-induced gene 1 (Higd1a) in the central nervous system of male and female rats differs according to age. J Mol Neurosci. 2018;66(3):462–473. doi: 10.1007/s12031-018-1195-y. [DOI] [PubMed] [Google Scholar]
- 47.Bedó G., Lagos P., Agrati D. Temporal distribution of Hig-1 (hypoxia-induced gene 1) mRNA and protein in rat spinal cord: changes during postnatal life. J Mol Neurosci. 2012;47(3):666–673. doi: 10.1007/s12031-012-9713-9. [DOI] [PubMed] [Google Scholar]
- 48.Mishima Y., Brueckner L., Takahashi S., et al. Enhanced processivity of Dnmt1 by monoubiquitinated histone H3. Gene Cell. 2020;25(1):22–32. doi: 10.1111/gtc.12732. [DOI] [PubMed] [Google Scholar]
- 49.Wu S.B., Wei Y.H. AMPK-mediated increase of glycolysis as an adaptive response to oxidative stress in human cells: implication of the cell survival in mitochondrial diseases. Biochim Biophys Acta. 2012;1822(2):233–247. doi: 10.1016/j.bbadis.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 50.Ameri K., Maltepe E. HIGD1A-mediated dormancy and tumor survival. Mol Cell Oncol. 2015;2(4):e1030537. doi: 10.1080/23723556.2015.1030537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Congrains-Castillo A., Niemann F.S., Santos Duarte A.S., Olalla-Saad S.T. LEF1-AS1, long non-coding RNA, inhibits proliferation in myeloid malignancy. J Cell Mol Med. 2019;23(4):3021–3025. doi: 10.1111/jcmm.14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu D.C., Song L.L., Liang Q., Hao L., Zhang Z.G., Han C.H. Long noncoding RNA LEF1-AS1 silencing suppresses the initiation and development of prostate cancer by acting as a molecular sponge of miR-330-5p via LEF1 repression. J Cell Physiol. 2019;234(8):12727–12744. doi: 10.1002/jcp.27893. [DOI] [PubMed] [Google Scholar]
- 53.Cheng Z., Wang G., Zhu W., Luo C., Guo Z. LEF1-AS1 accelerates tumorigenesis in glioma by sponging miR-489-3p to enhance HIGD1A. Cell Death Dis. 2020;11(8):690. doi: 10.1038/s41419-020-02823-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noren Hooten N., Evans M.K. Techniques to induce and quantify cellular senescence. J Vis Exp. 2017;(123):55533. doi: 10.3791/55533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawano Y., Cohen D.E. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol. 2013;48(4):434–441. doi: 10.1007/s00535-013-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo L., Guo Y.Y., Li B.Y., et al. Enhanced acetylation of ATP-citrate lyase promotes the progression of nonalcoholic fatty liver disease. J Biol Chem. 2019;294(31):11805–11816. doi: 10.1074/jbc.RA119.008708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo L., Zhang P., Chen Z., et al. Hepatic neuregulin 4 signaling defines an endocrine checkpoint for steatosis-to-NASH progression. J Clin Investig. 2017;127(12):4449–4461. doi: 10.1172/JCI96324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang D.Q., El-Serag H.B., Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2021;18(4):223–238. doi: 10.1038/s41575-020-00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24(7):908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo L., Zhou S.R., Wei X.B., et al. Acetylation of mitochondrial trifunctional protein α-subunit enhances its stability to promote fatty acid oxidation and is decreased in nonalcoholic fatty liver disease. Mol Cell Biol. 2016;36(20):2553–2567. doi: 10.1128/MCB.00227-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng W.Q., Xiao G., Li B.Y., Guo Y.Y., Guo L., Tang Q.Q. L-theanine activates the browning of white adipose tissue through the AMPK/α-ketoglutarate/Prdm16 axis and ameliorates diet-induced obesity in mice. Diabetes. 2021;70(7):1458–1472. doi: 10.2337/db20-1210. [DOI] [PubMed] [Google Scholar]
- 62.Mu W.J., Zhu J.Y., Chen M., Guo L. Exercise-mediated browning of white adipose tissue: its significance, mechanism and effectiveness. Int J Mol Sci. 2021;22(21):11512. doi: 10.3390/ijms222111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Javeed N., Matveyenko A.V. Circadian etiology of type 2 diabetes mellitus. Physiology. 2018;33(2):138–150. doi: 10.1152/physiol.00003.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo Y.Y., Li B.Y., Peng W.Q., Guo L., Tang Q.Q. Taurine-mediated browning of white adipose tissue is involved in its anti-obesity effect in mice. J Biol Chem. 2019;294(41):15014–15024. doi: 10.1074/jbc.RA119.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandez-Vizarra E., Zeviani M. Mitochondrial disorders of the OXPHOS system. FEBS Lett. 2021;595(8):1062–1106. doi: 10.1002/1873-3468.13995. [DOI] [PubMed] [Google Scholar]
- 66.Indrieri A., Carrella S., Romano A., et al. miR-181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol Med. 2019;11(5):e8734. doi: 10.15252/emmm.201708734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435(2):297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nagao T., Shintani Y., Hayashi T., et al. Higd1a improves respiratory function in the models of mitochondrial disorder. FASEB J. 2020;34(1):1859–1871. doi: 10.1096/fj.201800389R. [DOI] [PubMed] [Google Scholar]
- 69.Johnson S.C., Kayser E.B., Bornstein R., et al. Regional metabolic signatures in the Ndufs4(KO) mouse brain implicate defective glutamate/α-ketoglutarate metabolism in mitochondrial disease. Mol Genet Metab. 2020;130(2):118–132. doi: 10.1016/j.ymgme.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paramasivam A., Meena A.K., Venkatapathi C., Pitceathly R.D.S., Thangaraj K. Novel biallelic NSUN3 variants cause early-onset mitochondrial encephalomyopathy and seizures. J Mol Neurosci. 2020;70(12):1962–1965. doi: 10.1007/s12031-020-01595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhatia K.D., Krishnan P., Kortman H., Klostranec J., Krings T. Acute cortical lesions in MELAS syndrome: anatomic distribution, symmetry, and evolution. AJNR Am J Neuroradiol. 2020;41(1):167–173. doi: 10.3174/ajnr.A6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moreira P.I., Zhu X., Wang X., et al. Mitochondria: a therapeutic target in neurodegeneration. Biochim Biophys Acta. 2010;1802(1):212–220. doi: 10.1016/j.bbadis.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Calvo-Rodriguez M., Bacskai B.J. Mitochondria and calcium in Alzheimer's disease: from cell signaling to neuronal cell death. Trends Neurosci. 2021;44(2):136–151. doi: 10.1016/j.tins.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 74.Luan X., Tian X., Zhang H., et al. Exercise as a prescription for patients with various diseases. J Sport Health Sci. 2019;8(5):422–441. doi: 10.1016/j.jshs.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang R., Tian H., Guo D., Tian Q., Yao T., Kong X. Impacts of exercise intervention on various diseases in rats. J Sport Health Sci. 2020;9(3):211–227. doi: 10.1016/j.jshs.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo S., Huang Y., Zhang Y., Huang H., Hong S., Liu T. Impacts of exercise interventions on different diseases and organ functions in mice. J Sport Health Sci. 2020;9(1):53–73. doi: 10.1016/j.jshs.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen M., Zhu J.Y., Mu W.J., Guo L. Cysteine dioxygenase type 1 (CDO1): its functional role in physiological and pathophysiological processes. Gene Dis. 2023;10(3):877–890. doi: 10.1016/j.gendis.2021.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo L., Guo Y.Y., Li B.Y., Peng W.Q., Tang Q.Q. Histone demethylase KDM5A is transactivated by the transcription factor C/EBPbeta and promotes preadipocyte differentiation by inhibiting Wnt/beta-catenin signaling. J Biol Chem. 2019;294(24):9642–9654. doi: 10.1074/jbc.RA119.008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Y., Peng W.Q., Guo Y.Y., Liu Y., Tang Q.Q., Guo L. Krüppel-like factor 10 (KLF10) is transactivated by the transcription factor C/EBPβ and involved in early 3T3-L1 preadipocyte differentiation. J Biol Chem. 2018;293:14012–14021. doi: 10.1074/jbc.RA118.004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li B.Y., Guo Y.Y., Xiao G., Guo L., Tang Q.Q. SERPINA3C ameliorates adipose tissue inflammation through the CathepsinG/Integrin/AKT pathway. Mol Metabol. 2022;61:101500. doi: 10.1016/j.molmet.2022.101500. [DOI] [PMC free article] [PubMed] [Google Scholar]