Abstract
Studies on carcinogenesis continue to provide new information about different disease-related processes. Among others, much research has focused on the involvement of cancer stem cells (CSCs) in tumor initiation and progression. Studying the similarities and differences between CSCs and physiological stem cells (SCs) allows for a better understanding of cancer biology. Recently, it was shown that stem cell identity is partially governed by the Krϋppel-associated box domain zinc finger proteins (KRAB-ZFPs), the biggest family of transcription regulators. Several KRAB-ZFP factors exert a known effect in tumor cells, acting as tumor suppressor genes (TSGs) or oncogenes, yet their role in CSCs is still poorly characterized. Here, we review recent studies regarding the influence of KRAB-ZFPs and their cofactor protein TRIM28 on CSCs phenotype, stemness features, migration and invasion potential, metastasis, and expression of parental markers.
Keywords: Cancer, Cancer stem cell, Epigenetic, KRAB-ZFP, Transcription factors
Introduction
Genetic changes and epigenetic processes in cancer cells may initiate and influence each other.1 Mutations within the genome, as well as multiple epigenetic changes, frequently result in augmented tumor heterogeneity. Indeed, in most cases, cancer contains various cell subpopulations, including cancer stem cells (CSCs). An increasing amount of research indicates that CSCs may be responsible for metastasis, resistance to therapies, and subsequently, worse patient prognosis.2,3
The function of many proteins is frequently perturbed in cancer cells,4,5 including Krϋppel-associated box domain zinc finger proteins (KRAB-ZFPs). KRAB-ZFPs belong to a large family of epigenetic repressors. Some of these factors play an essential role in stem cell biology and differentiation processes.6, 7, 8 Moreover, the pan-cancer transcriptomic analysis revealed that many KRAB-ZFPs are commonly downregulated or overexpressed in multiple tumor types.9,10 A growing amount of data indicate that some of them may function as oncogenes, while others as tumor suppressor genes (TSGs).11 Nevertheless, the specific role of the majority of KRAB-ZFP family members remains unexplored, and consequently, little is known about their functioning in CSCs. Combining the knowledge about individual factors and their functioning in physiological SCs like embryonic stem cells (ESCs), adult stem cells (ASCs) or hematopoietic stem cells (HSCs) and cancer cells may shed some light on CSCs biology. This issue needs to be better studied in order to reveal the potential role of KRAB-ZFPs in the maintenance of pluripotency and cancer aggressiveness. Physiological SCs and CSCs share many similarities; hence it is possible that KRAB-ZFPs will have a common effect on both cell types. Currently, many studies are focused on the improvement of anti-cancer therapies. As mentioned above, CSCs may be responsible for cancer recurrence, metastasis, and poor patient outcome, so it is crucial to characterize them in more detail. Below, we will focus on these observations to summarize the data indicating that KRAB-ZFPs may have an important role in CSCs.
Cancer formation via interconnected genetic and epigenetic aberrancies
The knowledge regarding cancer origin, biology, and treatment options is still expanding, yet it remains the second leading cause of death.12 During oncogenesis accumulating genetic and epigenetic aberration lead to the acquisition of hallmark cancer features, including constant signaling of proliferation, avoidance of growth suppressors, replicative immortality, resistance to cell death, induction of tumor angiogenesis, invasiveness and metastasis, presence of mutations in the genome and its instability, inflammation that promotes tumor formation, avoidance of immune responses, and reprogrammed cellular metabolism.13,14 All of these features influence the course of carcinogenesis, drug susceptibility, and patient life expectancy.
A wide array of driver mutations were identified to date across multiple tumor types through classical methods or next-generation sequencing.15, 16, 17 Mutations may affect the DNA sequence of a single gene or non-coding RNA (ncRNA), as well as a chromosome structure and copy number. These mutations may be inherited or stimulated by a variety of external and internal factors, e.g., smoking, viral infections, inflammation, irradiation, etc. As mentioned above, apart from the genetic and genomic alterations, cancer formation may be driven by disturbed epigenetic signaling.4 The epigenomic profile of a given cell drives its transcriptional signature. Thus, the cells with the same genotype but different epigenomes are able to perform distinct transcriptional programs, and as such – exhibit divergent functions.18,19 In certain cases, epigenetic changes may affect cancer cell phenotype more profoundly than genetic mutations, as may be observed in several childhood malignancies, including retinoblastoma20 or medulloblastoma.21,22 Nevertheless, in the majority of cancers, genetic mutations and epigenetic aberrations (so-called epimutations) may coincide and influence each other.
One of the best-characterized epigenetic events is cytosine methylation within CpG dinucleotides. Cancer cells frequently harbor a specific pattern of DNA methylation: genome-wide hypomethylation accompanied by local CpG hypermethylation.4,23 Hypermethylation in cancer frequently occurs in the promoters of TSGs and DNA repair genes and may lead to a decline in their expression.24 In contrast, global DNA hypomethylation may disturb chromatin structure and epigenetic regulation. Moreover, it may contribute to the augmented genomic instability and activation of oncogene transcription.4,25 Besides changes within DNA methylation signature, cancer cells harbor numerous epigenetic alterations related to histone post-translational modifications (PTMs), ncRNA expression profile, and 3D chromatin organization. As could be expected, many chromatin modifiers exhibit aberrant functioning in cancer due to mutations or changes in their expression level.5,26,27 In many cancer types, the chromatin factors responsible for the maintenance of heterochromatin are overexpressed, while those involved in chromatin relaxation are downregulated or mutated.4,5 Therefore, PTMs may become largely re-distributed in tumors compared to their normal counterparts,28,29 as well as between cancer cells at various differentiation states, e.g., metastatic vs. non-metastatic cells.30 These global alterations rewire the transcriptional program that supports the maintenance or switch between distinct cancerous phenotypes.30, 31, 32 For instance, H3K9me2 is a repressive mark that accumulates during cell differentiation within transcriptionally silenced sites. These regions are significantly lost in cancer cells, which likely contribute to enhanced phenotypic plasticity observed as well in ESCs.33 Also, H3K9me3 levels may drop during carcinogenic transformation as demonstrated in the case of oncogene promoters, pericentromeric heterochromatin, and transposable elements (TEs).29,34, 35, 36 Such an event may lead to genomic instability via inappropriate chromosome segregation,35 as well as TE-mediated insertional mutagenesis predisposing to recombination.36 Interestingly, the upregulation of H3K9 methyltransferases: SET domain bifurcated 1/2 (SETDB1/2) and subsequent increase in H3K9me3 deposition are frequently linked to the acquired drug resistance in many tumor types.32,37,38 A study by Al Emran and colleagues focused on histone modifications in induced drug tolerance in cancer cells (IDTCs) .37 By examining melanoma cell lines with IDTCs, it was shown that SETDB1 levels were higher in cells with drug resistance, thereby increasing the presence of H3K9me3, possibly affecting gene repression in IDTCs.
Epigenetic alterations occurring in cancer cells may increase cellular plasticity in a growing tumor.4 The available data indicate that tumor mass may be highly heterogeneous, and the composition of distinct cell populations may vary during the course of the disease.39, 40, 41, 42 Such changes in heterogeneity within the tumor may be related to the presence of cells with stem cell-like features known as CSCs.
Stem cells and cancer stem cells
CSCs make up a small part of the tumor mass and may be identified at various cancer stages.3,43 They are slowly growing, self-renewing cells capable of withstanding harsh conditions, such as hypoxia, nutrient deprivation, or anti-cancer drug administration. Furthermore, CSCs may undergo epithelial-to-mesenchymal transition (EMT) that facilitates mobility and metastasis.44, 45, 46 Such plasticity of CSCs may be ascribed to epigenetic events that enable the fast acquisition of traits supporting adaptation to changes within the cancer environment.
Embryonic SCs (ESCs) have the ability to differentiate into virtually any cell type.47 Also, CSCs produce various classes of neoplastic cells.3,48,49 Nevertheless, the origin of CSCs is difficult to trace. Hypothetically, they may appear as a consequence of perturbed balance between self-renewal and differentiation of physiological stem cells. Alternatively, they may arise upon the induction of de novo self-renewal capacity within somatic cells. Due to their unique features, CSCs may initiate tumor growth and maintain cell division.47,50 However, not every cell has such a founder cell potential.43
After the first discovery of CSCs in AML, they were identified in many types of tumors, including breast,51 lung,52 and brain cancers.53 A subset of cells with stemness properties may be isolated from the tumor mass based on the presence of specific surface markers. In breast cancer, they are detected by CD44+/CD24−/low phenotype.51 Isolated breast cancer cells with CD44+/CD24− profile are able to form mammospheres in vitro and xenografts in mice model in vivo. Additionally, these cells express the pluripotency-related gene OCT4.54 Of note, in many cases, the identification of CSCs within the tumor of particular origin is based on the gene expression signature that is also characteristic to normal stem cells.
Indeed, physiological SCs and CSCs share many common characteristics, although several essential differences can be noted (Table 1). They demonstrate high telomerase activity, resistance to DNA damage, and the ability to self-renew and differentiate into more specialized cell lineages.47,55 However, in CSC, the differentiation process may be disturbed, and the emerging cells may be incorrect.47 Instead of engaging proper differentiation signaling pathways, CSCs utilize some of the pathways active in normal stem cells. An example is the JAK/STAT pathway, where STAT3 protein is up-regulated in a population of stem-like breast cancer cells in comparison to non-stem-like breast cancer cells.56 STAT3 is known for its positive effect on maintaining the undifferentiated state in mouse ESCs (mESCs)57 and for promoting hMSCs (human mesenchymal stem cells) migration to breast cancer cells.58 This may suggest that upregulation of STAT3 in CSCs is necessary for its stem-like nature.
Table 1.
Comparison of physiological SCs, CSCs and regular cancer cells. All types of cells possess cell-specific characteristics but also share some common features.
| Features | Physiological stem cells | Cancer stem cells | Regular cancer cells |
|---|---|---|---|
| Self-renewal ability | Enables tissues to be built from progenitor cells59 | Enables production of cancer cells that form a tumor mass59 | No self-renewal capability |
| Telomerase | Active | Active | Active in about 85% of all tumor types60 |
| Pluripotency markers | OCT4, NANOG, SOX2,61 SALL4,62 STAT3,63 c-Myc,64 Klf465 | OCT4, NANOG, SOX2,66 SALL4,67 STAT3,68 c-Myc,64 Klf469 | SALL4,70 STAT3,71 c-Myc72 |
| Susceptibility to chemotherapy | Resistant | Resistant | Sensitive according to tumor type |
| Markers | CD29high/+,73,74 CD34+,75,76 CD44+,74 CD49fhigh/+,73, 74, 75, 76, 77 CD90+,74,76 CD117+,76 CD133+,73 CD184+,76 CD200+,77 Sca1+74,77 | Annexin II+ (AII+),78 ALDHhigh,78 CD34low/high,79 CD44+/CD24−/low,51 CD49f+,78,82, 81, 80 CD133+,82,83 CK-17,78 p63+78 | CCR10high,84 CD9,85 CD63,86 CD82,87 CD151,88 CD182low,84 CD184high/+,84,89 CD197high84 |
| Differentiation | Differentiate to almost every type of cell; Possibility to change into CSC |
Change into a cancer cell | Already differentiated cells |
| Function of cells in relation to their place of origin | Replacing old cells in the body; Creating different germ layers | Forming metastasis | Forming tumor mass |
| Signaling pathways | Signal pathways functioning properly | Altered in comparison to the regular functioning of pathways JAK-STAT, Hedgehog pathway – overexpression of involved genes WNT – activation of the pathway PI3K – repressed by miR-12690 |
Altered pathway genes according to tumor type; The most commonly altered pathways – RTK/RAS/MAP-Kinase (46% of examined cancer samples), cell cycle (45%), PI3K (33%), TP53 (29%), NOTCH (23%), WNT (15%), MYC (11%), HIPPO (10%), TGF-Beta (7%) and NRF2 (1%)91 |
As mentioned above, CSCs may be recognized by the biomarkers that are also characteristic for physiological stem cells. For example, cell surface markers, such as CD133, may be found in both CSCs and regular SCs like HSCs or ASCs.47,92,93 CD133 detected on CSCs in glioblastoma, pancreatic adenocarcinoma, hepatocellular carcinoma (HCC), and brain tumor,50,94, 95, 96 is also a marker of HSCs and neural stem cells (NSCs).50,97 Research conducted by Ma and colleagues showed that CD133+ HCC cells also had enhanced expression of various stemness factors (e.g., β-catenin, OCT3/4, iBMI, SMO, and NOTCH1).95 In glioblastoma, CD133 positive cancer cells exhibit resistance to chemotherapy.98 Also, ALDH1 shows high expression in physiological SCs, and in CSCs in various tissues. Augmented ALDH1 levels in breast, bladder, and prostate cancers correlate with poor prognosis.99, 100, 101 In thyroid neoplasm, ALDHHigh CSCs show resistance to chemotherapy due to the activation of Hedgehog pathway.102 Cells with high ALDH levels are also characterized by increased expression of stemness genes, i.e., OCT4, NANOG, and SOX2.103
It is well established that CSCs are responsible for tumor migration and metastasis.3 Thus, they were divided into two subtypes responsible for different functions: primary CSCs (pCSCs), which form the tumor and exist in its environment, and migrating CSCs (mCSCs), which are responsible for metastasis.50 Studies conducted on breast CD44+/CD24− CSCs showed that OCT4 and NANOG have a significant impact on metastasis. Silencing both factors simultaneously reduced the expression of genes that positively affect EMT. The opposite effect was noted in the case of overexpression – the levels of EMT-related genes were increased. Also, the same relationship occurred in in vitro culture of CSCs in terms of cell migration.104 Similar results were observed in lung adenocarcinoma cell line A549.105 A study by Xu and colleagues on colorectal cancer also showed that higher NANOG-expression correlated with more frequent postoperative liver metastases.106 Furthermore, Lu and colleagues showed that Nanog induced migratory, invasive, and metastatic potential of mammary cancer cells in a mouse model.107 They observed metastases to the lungs, liver, and kidneys. Notably, the cells with high Nanog expression were characterized by an increased expression of Pdgfrα, which positively affects carcinogenesis and metastasis.107 Nevertheless, stem cell markers may also show a negative impact on metastasis. For example, the study of Shen and colleagues in in vitro and in vivo mouse models showed that the overexpression of Oct4 reduced the metastatic potential of breast cancer cells. This effect was ascribed to the findings that Oct4 reduces the expression of Rnd1, which is responsible for cell adhesion.108 Such opposing observations may be related to the differences in utilized research models, and particularly to their genomic and epigenomic make-up. Clearly, further studies are needed to improve our understanding of SC markers' involvement in cancer biology.
KRAB-ZFPs
The aforementioned OCT4 and NANOG are well-known factors regulating pluripotency-related features. The knowledge about other proteins affecting physiological and cancer stem cells is yet still expanding. One example is TRIM28 (tripartite motif protein 28), also known as KRAB-associated protein 1 (KAP1). It interacts and co-operates with KRAB-ZFP factors in mediating epigenetic repression. TRIM28 and several KRAB-ZFP factors were shown to participate in the maintenance of pluripotency in human embryonic stem cells (hESCs).7 As mentioned earlier, the expression of many KRAB-ZFPs is deregulated in multiple tumor types,9,10 and accumulating evidence suggests that some of the KRAB-ZFP factors play a role in oncogenesis.11 Based on the connection between KRAB-ZFP, stemness, and cancer, we will review the available literature that points towards the potential involvement of TRIM28 and KRAB-ZFPs in CSCs phenotype. KRAB-ZFPs belong to the relatively young family of transcriptional repressors containing a KRAB domain and a variable number of zinc finger motifs. They emerged around 420 million years ago in a common ancestor of coelacanths lungfish and tetrapods.109 The human genome comprises approximately 381 KRAB-ZFP genes, which may form over 800 transcripts.110 KRAB-ZFPs play an essential role in the transcriptional regulation of various signaling pathways, thus affecting many biological processes, including apoptosis, cell cycle, development, and stem cell differentiation.7,11,111,112 The KRAB domain may be composed of two parts: A-box, which is necessary for the repressive function of the protein, and B-box, which enhances the repressive effect.6,113 KRAB-ZFPs may also contain additional domains, such as SRE-ZBP, CTfin51, AW-1, and Number18 cDNA domain (SCAN) or Domain of Unknown Function 3669 (DUF3669).6
The canonical function of KRAB-ZFPs is epigenetic suppression through the deposition of repressive chromatin marks. In addition, KRAB-ZFPs may also facilitate DNA methylation within the affected locus. However, this effect seems to occur only in the cells with stem cell phenotype.7,8 KRAB-ZFPs bind DNA via zinc finger motifs, whereas the KRAB domain recruits its corepressor – TRIM28. The KRAB domain interacts with the Ring finger/B box/Coiled-Coil (RBCC) domain at the N-terminus of TRIM28.111,112,114,115 The C-terminal domain and bromodomain recruit Nucleosome Remodelling Deacetylase (NuRD) complex and histone lysine methyltransferase SET Domain Bifurcated 1 (SETDB1), thus mediating histone deacetylation and H3K9me3 deposition, respectively. The central part of TRIM28 binds to Heterochromatin Protein 1 (HP1), which recognizes H3K9me3 and promotes regional heterochromatization.112,116,117 The repressive functions conferred by KRAB-ZFPs may affect gene promoters and 3′ ends, imprinted control regions, and various classes of TEs.109
An increasing number of published reports indicate that KRAB-ZFP may play an important role in cancer, affecting cancer cell phenotype, as well as disease prognosis and patient survival (reviewed in11). The data from various tumor tissues and experimental models show that KRAB-ZFPs may act both as TSGs (e.g., ZNF23, ZNF382, ZNF471, ZNF545)118, 119, 120, 121 or oncogenes (e.g., ZNF300, ZKSCAN3).122,123 Their imbalanced expression results in the aberrant stimulation of various cancer-related signaling pathways (e.g., p53, NFkB, and Wnt/β-catenin).11 An interesting example of oncogenic transcriptional repression over several regions was demonstrated in the case of ZNF304 in colorectal cancer.124 ZNF304, whose expression rises upon mutant KRAS signaling, was shown to mediate the coordinated silencing of several loci, thus contributing to the CpG island methylator phenotype. In the study by Serra and colleagues, ZNF304 attached to the INK4-ARF locus that encodes p14ARF, p15INK4B, and p16INK4A TSGs, and recruited other epigenetic corepressors, including TRIM28, SETDB1, and DNMT1.124 Of note, the published evidence suggests that KRAB-ZFPs may act not only as gene co-suppressors but also as co-activators depending on the protein partners interacting with a given KRAB-ZFP factor.11,125, 126, 127
KRAB-ZFP/TRIM28 influence on stem cells and cancer stem cells
TRIM28
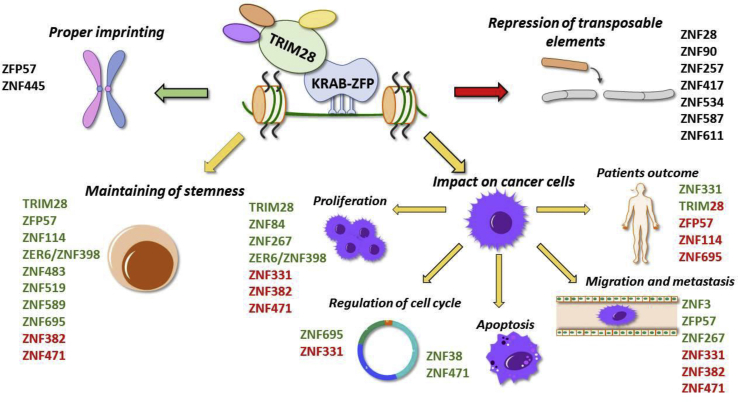
KRAB-ZFP cofactor protein, TRIM28, influences both stem cells and cancer stem cells (Fig. 1).114,128 Its essential role in the functioning of SCs was demonstrated in the research carried out by Hu and colleagues in the mESCs.129 Lack of Trim28 triggered ES differentiation into the primitive ectoderm lineage. Interestingly, Trim28-targeted transcription regulators of the cell cycle, cell death, and cancer, supporting the idea that self-renewal-specific pathways may also be active in certain cancers.129 In another study, Seki and colleagues demonstrated that induced overexpression of Trim28 sustained the pluripotency of mESCs. In contrast, Trim28 silencing resulted in a decreased growth, change in ESCs morphology (cells became stretched and less tightly compacted), and reduced expression of pluripotency markers, namely SSEA1 and Nanog.128 It is worth mentioning that Trim28 is indispensable for early development and mouse embryos with Trim28 knockout are lethal.130,131 Therefore, all changes observed in ESCs upon Trim28 knock-down should be analyzed carefully to confirm that they are not a side effect in dying cells.
Figure 1.
TRIM28 and KRAB-ZFPs are involved in different biological processes in physiological and cancer stem cells.
In hESCs, TRIM28 silencing triggered differentiation manifested with morphological changes as well as the reduction of expression of pluripotency markers: NANOG, OCT3/4, and TRA-1-60.7 Similar to physiological SCs, TRIM28 affects stem cell features in the cancer setting. The study conducted by Czerwińska and colleagues114 revealed increased expression of TRIM28 in 14 tumor types from The Cancer Genome Atlas (TCGA) project, e.g., lung, breast, gastric, and pancreatic cancer. Moreover, a high TRIM28 level was associated with the patient's outcome. For example, in gastric cancer, high TRIM28 expression correlated with lower survival rate.114 Conversely, in the early stage of lung cancer, high level of TRIM28 expression correlated with better overall patient survival.132 Knock-down of TRIM28 in vitro had no major effect on proliferation and stemness in breast cancer cell lines. However, in in vivo experiments, a lower TRIM28 level was shown to be related to weaker self-renewal and tumor formation properties. Xenografts obtained from the TRIM28KD MDA-MB-231 breast cancer cell line (that contains an enriched CD44+/CD24−/low subpopulation) exhibited reduced tumor formation. This effect was not observed in the TRIM28KD MCF7 cell line with observed low content CD44+/CD24−/low cells. Molecular profiling revealed that TRIM28 silencing was associated with lower expression of stem cell markers, including OCT3/4, SOX2, and NANOG, and with the metabolic switch from oxphos to glycolysis.114 Altogether, these data indicate that TRIM28 participates in the stemness maintenance in both physiological and cancer stem cells.
KRAB-ZFPs
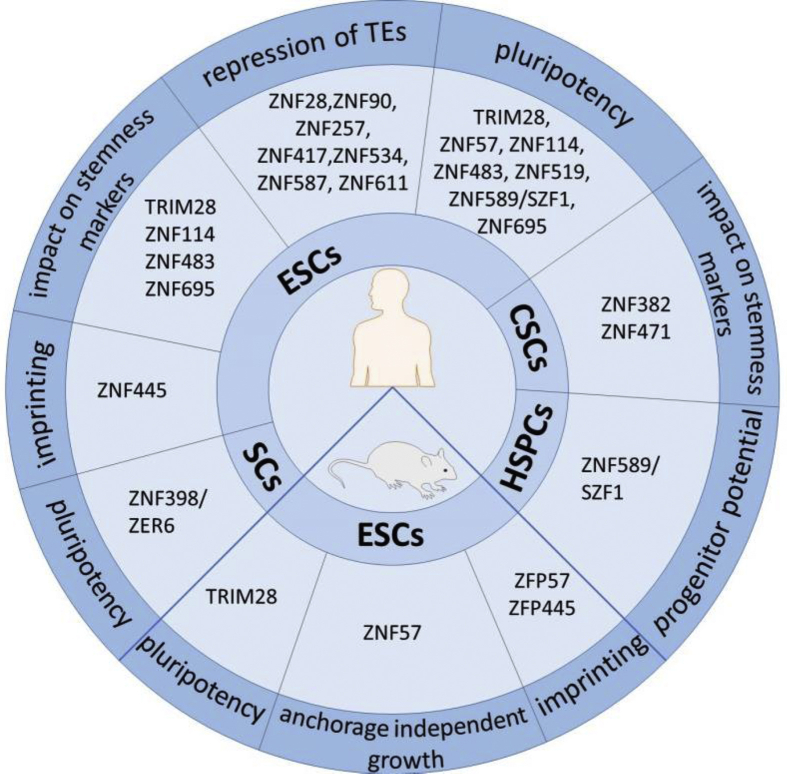
As mentioned above, KRAB-ZFP factors take part in the functioning of differentiated and undifferentiated cells (Fig. 2).7 In our previous research, we used transcriptomic data from home-derived dermal fibroblasts and iPSCs, as well as the Progenitor Cell Biology Consortium (PCBC), focused on the iPS induction and differentiation to various cell lineages.133 In both datasets, we identified 38 KRAB-ZFPs up- and down-regulated in pluripotent stem cells compared to their more differentiated counterparts. We confirmed that at least 10 of them are up-regulated in iPSCs compared to fibroblasts. Furthermore, their level increased during somatic cell reprogramming to iPSC and vice versa – their level decreased with iPSCs differentiation into somatic cells. To determine the functional effect of certain KRAB-ZFPs on pluripotency, ZFP57, ZNF114, ZNF483, ZNF519, ZNF589, and ZNF695, as well as TRIM28, were knocked down in hESCs. The silencing caused changes in cell morphology – cell elongation, dispersion, and loss of colony boundaries, which suggested cell differentiation. Besides, silencing TRIM28, ZNF483 and ZNF695 was followed by a decrease in NANOG and OCT3/4 levels. The data showed as well that KRAB-ZFPs may target the promoters of genes implicated in developmental biology, cell cycle, and extracellular matrix organization. Moreover, during reprogramming of dermal fibroblasts to iPSCs, KRAB/TRIM28 axis mediated promoter silencing via deposition of H3K9me3 and DNA methylation.7
Figure 2.
KRAB-ZFP/TRIM28 protein complex is involved in activation (green) and repression (red) of several biological processes.
Another function important for stemness that occurs during embryonic development is the ability of KRAB-ZFP factors to restrict various classes of transposable elements. For example, ZNF90, ZNF257, and ZNF534 repress human endogenous retrovirus K (HERVK), while ZNF28 and ZNF611 silence SINE-VNTR-Alu (SVA) during embryonic genome activation (EGA). These factors target recent TEs to rewire transcriptional programs in naive hESCs.134 Other KRAB-ZFPs associated with ESCs and TEs are ZNF417 and ZNF587. These factors repress HERVK and SVA during EGA in ESCs, which later differentiate into neurons.135 TEs take part in the regulation of pluripotency – a lot of binding sites intended for pluripotency factors are found within endogenous retroviruses (ERVs).6 Almost one-fifth of the Oct4–Sox2 binding sites are located in mouse endogenous retrovirus K (ERVK) and may therefore be affected by KRAB-ZFP.136 Moreover, ZFP57 and ZNF445/ZFP445 are involved in the control of gene imprinting.137 In mESCs, both factors are responsible for the proper imprinting pattern within imprinting control regions (ICRs). However, in hESCs, ZNF445 has greater importance in the regulation of imprinting.137
Transcriptomic profiling of different cell populations within the tumor mass demonstrate that distinct members of the KRAB-ZFP family are found among genes specifically up- or downregulated in CSCs. Seo and colleagues detected 49 genes that were significantly downregulated and 12 up-regulated in a lung adenocarcinoma cell line subpopulation with stem cell properties. Two KRAB-ZFP transcription factors, namely ZNF567 and ZNF267, were among genes showing higher expression in CSCs.138 The function of ZNF567 is yet unknown, whereas ZNF267 is up-regulated in liver cirrhosis139 and HCC cell lines and tissues140 where it positively correlates with proliferation and migration. ZNF267 might be useful as a prognostic biomarker of HCC development since its expression is also elevated in pre-cancerous tissues.140
Bhat-Nakshatri and colleagues identified a perturbed expression pattern of several KRAB-ZFPs in CSCs subpopulation of the MCF10A cell line. ZNF84, ZNF606, ZNF585A, ZNF420, ZNF41, ZNF25, ZNF226, ZNF222, and ZNF136 were up-regulated, whereas ZNF552 was downregulated in stem cell-like cells.141 Possibly, ZNF84 may function as an oncogene since its expression is elevated in cervical cancer tissues and positively correlates with tumor size and cell proliferation.142 In contrast, ZNF606 might act as a TSG. Its expression is downregulated via promoter hypermethylation in colorectal cancer with mutated BRAF.143 Recently, the CSCs subpopulation of another two breast cancer cell lines was characterized.144,145 CSCs isolated from the basal breast cancer cell line MDA-MB-468 were subjected to whole-transcriptome analysis in order to define stem cell-related gene expression signature. Only one KRAB-ZFP factor, ZNF354A, was significantly downregulated in the CSC population.145 The same factor was detected in our previous studies analyzing KRAB-ZFPs expression in the TCGA datasets.10 The level of ZNF354A was decreased in BRCA, as well as in several other types of cancer samples, i.e., thyroid, prostate, and kidney. Interestingly, the same factor was elevated in other analyzed types, like lung adenocarcinoma, liver cancer, or cholangiocarcinoma.10 The CSC transcriptomic profiling of the MDA-MB-231 BRCA cell line utilizing a single-cell sequencing approach identified only one differentially expressed KRAB-ZFP, namely ZNF566.144 Its expression was found downregulated in CSC-like cells compared to non-CSCs. However, its role in cancer remains unknown.
A single-cell approach demonstrated stem-cell-related features of subpopulations within glioblastoma146 and leukemia cells.147 In the case of patients with chronic myeloid leukemia, only one KRAB-ZFP was detected among stem cell-related genes (ZNF302). The expression of ZNF302 was decreased in CSCs in comparison to normal HSCs, but its role was not explored further in the cancer setting.147 Furthermore, Zhao and colleagues analyzed transcriptomes of two neural cell lines: a glioblastoma CSC line and a neural stem cell line (NSCs). Three KRAB-ZFPs (ZNF92, ZNF480, and ZNF3) were found within genes with increased expression that distinguished CSCs from NSCs.146 ZNF92 was previously found to be highly expressed in multiple cancers, i.e., breast, lung, liver, and bladder.10 ZNF3 is one of the genes that were proposed in a multigene predictor set of metastatic outcome in BRCA patients148 and as an epigenetic marker for gastric cancer risk development in patients with H. pylori infection.149 The function of ZNF480 in carcinogenesis remains unknown.
The role of individual KRAB-ZFP factors in CSCs remains poorly characterized, although certain aspects of their potential engagement in CSCs biology may be pinpointed as reviewed below. The known function of specific KRAB-ZFPs in both cancer and embryonic cells is summarized in Table 2 and Figure 1.
Table 2.
The function of specific KRAB-ZFPs in both cancer and embryonic cells.
| KRAB-ZFP factor/TRIM28 | Function in cancer cells | Function in stem cells | References |
|---|---|---|---|
| TRIM28 | impact on lower survival rate in gastric cancer; positive effect on tumor formation | maintenance of pluripotency, self-renewal, and OCT3/4 and NANOG gene expression | 7,114,128,129 |
| ZFP57 | positive impact of overexpression on metastasis; positive association with NANOG expression; negative impact on patient outcome | maintenance of pluripotency; proper imprinting pattern within ICRs; influence on anchorage-independent growth | 7,137,150, 151, 152 |
| ZNF114 | impact on lower cell survival | maintenance of pluripotency and stemness; repression of differentiation gene DPYSL4 | 7,153 |
| ZNF331 | inhibition of cell migration and invasion; inhibition of c-Myc and proliferation genes – DDX5, EIF5A, GARS, STAM, UQCRFS1, and SET | possible influence on breast CSCs by negative regulation of c-Myc expression | 154, 155, 156 |
| ZNF382 | negative impact on EMT and metastasis | negative impact on OCT4, SOX2, and NANOG expression in CSCs | 157, 158, 159 |
| ZNF398/ZER6 | positive impact on proliferation; impact on p53 ubiquitination | maintenance of pluripotency and inhibition of cell differentiation; influence on expression of TGF-β targeted genes | 160,161 |
| ZNF471 | reduction of cell viability, migration and invasion; induction of apoptosis; inhibition of AKT and Wnt/β-catenin pathways | reduction of CD44, NANOG2, OCT4, SOX2, and KLF4 expression in CSCs | 120,162 |
| ZNF695 | upregulation of cell cycle genes | maintenance of pluripotency, andOCT3/4 and NANOG gene expression in human ESCs | 7,163 |
ZNF695
ZNF695, tested for the effect on stemness in hESCs as described above,7 also seems to affect CSCs. For example, Li and colleagues focused on the master regulator genes in breast cancer that together may form a biomarker panel for the identification of specific molecular tumor subtype.163 Using three cohorts of human breast cancer data, they showed that one of the master regulator genes is ZNF695. Its expression differs depending on the tumor subtype – it is high in ER-negative subtypes (HER2-enriched and basal-like) and low in ER-positive (luminal tumors). Interestingly, ZNF695 was found to be associated with the upregulation of genes involved in cell cycle in basal-like and HER2-enriched subtypes. Also, ZNF695 overexpression was linked to mutations in the TP53 gene, which frequently appear in ER-negative neoplasms.163 These features indicate greater aggressiveness of ER-negative neoplasms, especially in patients' survival from the first metastases. The differences between ER-negative and ER-positive tumors were described in the study by Kennecke and colleagues.2 ER-negative neoplasms (HER-2 enriched and basal-like subtypes) are less common than the luminal A subtype but are characterized by a lower survival rate. ER-negative tumors recur faster than luminal tumors, and median survival from the first distant metastasis is also lower.2 Noteworthy, the marker CD133, which is detected in many types of CSCs, has greater expression in ER-negative neoplasms than in ER-positive ones.164 Moreover, the CD44+/CD24− phenotype is most common in basal-like breast cancer, which may indicate that this type of cancer has a higher percentage of CSCs.165 Considering the increased expression of ZNF695 in ER-negative tumors, it is tempting to speculate that its role in the cell cycle upregulation may be related to the aggressiveness of these tumor subtypes.
ZFP57
ZFP57, a gene involved in imprinting maintenance, is positively regulated by NANOG. In both mESCs and human cancer cells, overexpression of NANOG resulted in increased expression of ZFP57, while its silencing caused ZFP57 down-regulation. Zfp57 was also demonstrated to influence the anchorage-independent growth of mESCs – knock-down of Zfp57 with simultaneous Nanog overexpression resulted in a reduction in the number of cell colonies.152 Higher expression of ZFP57 is thus associated with a greater proliferation potential of ESCs. Moreover, an increased expression of ZFP57 was also observed in grade IV compared to grade II glioblastoma.150 High-grade glioblastomas contain a higher content of CSCs, as evidenced by positive CD133 marker staining in over 60% of grade IV glioblastomas.166 Furthermore, ZFP57 overexpression resulted in higher metastatic potential in the mouse model of colorectal cancer, as reported by Shoji and colleagues.151 In the clinical samples, high levels of ZFP57 were shown to be positively associated with NANOG expression, as well as nodal and liver metastasis in colorectal cancer.151 Indeed, Nanog may participate in the upregulation of Zfp57 as demonstrated in mESCs.152 Moreover, ZFP57 upregulation correlated with poorer patient outcome.151 Altogether, these observations suggest ZFP57 may play a role in the maintenance and proliferation of CSCs.
ZER6/ZNF398
ZER6/ZNF398 exists in two isoforms: p52-ZER6, which has a shorter version of the KRAB domain (truncated KRAB), and p71-ZER6 with an additional HUB-1 domain.160,167 ZER6 may act as a positive factor for the development of tumors, as shown in colorectal160 and breast cancers.168 In colorectal cancer, the p52-ZER6 isoform positively influences the interaction of p53 with MDM2, which leads to the ubiquitination of p53, and lowered expression of its target genes participating in cell cycle restriction. As could be expected, p52-ZER6 silencing resulted in increased p21 expression and inhibited the formation of xenografted tumors, indicating a positive effect of p52-ZER6 on the proliferation of cancer cells.160 The p71-ZER6 isoform, despite having a complete KRAB domain, cannot influence the interaction of p53 with MDM2 due to the presence of HUB-1 domain in its structure.160 Breast cancer cells positive for estrogen receptor alpha (ERα) also express both isoforms of the factor.168 p52-ZER6 interacts with ERα in the presence of 17β-estradiol, while p71-ZER6 through the aforementioned HUB-1 domain does not.167 From the molecular perspective, ZER6/ZNF398 was shown to promote the expression of genes that are targets for transforming growth factor β (TGF-β), which is mediated by colocalization with SMAD3 and histone acetyltransferase EP300.161 Owing to its role in cancer and in the regulation of stemness features, the influence of ZER6/ZNF398 on the phenotype of CSCs would be an interesting research direction. Noteworthy, ZER6/ZNF398 affects human pluripotent SCs by maintaining pluripotency and inhibiting cell differentiation. The factor is also required for the induction of pluripotency in fibroblasts.
ZNF114
ZNF114 is another factor that contributes to the maintenance of pluripotency. Its expression was shown to become up-regulated during iPSC generation, whereas its knock-down caused a slight loss of stemness markers in hESCs. Additionally, ZNF114 repressed the DPYSL4 gene involved in differentiation.7 It also affects the outcome in patients with clear cell renal cell carcinoma, as its higher expression was associated with shorter overall survival.153 Noteworthy, in gastric cancer cell line AZ-521, the silencing of pluripotency factor, SOX2, resulted in the upregulation of ZNF114.169 These observations may indicate that ZNF114 have a different functions in pluripotency-related processes depending on the cell type.
ZNF471
Several KRAB-ZFP factors have a negative effect on the functioning of CSCs. An example is ZNF471, which is a TSG methylated in esophageal squamous cell carcinoma (ESCC)120 and breast cancer cells.162 Tao and colleagues investigated the effects of ZNF471 overexpression in MDA-MB-231 and YCC-B1 cell lines, demonstrating reduced cell viability, induction of apoptosis, and reduction of invasion and migration of cancer cells.162 Analogously, knock-down of ZNF471 increased cell growth, inhibited apoptosis, and promoted metastasis. These aspects may be related to the observed inhibitory effects of ZNF471 on AKT and Wnt/β-catenin pathways that influence tumor growth. In the case of the CSC subpopulation of tested cell lines, cells with higher ZNF471 levels had reduced expression of stemness markers, e.g., CD44, NANOG2, OCT4, SOX2, and KLF4.162 Thus, the inhibition of ZNF471 positively influences the development and growth of CSCs.
ZNF382
ZNF382 also has a suppressor function against CSCs. This factor is expressed during embryogenesis mainly in the kidney and brain, while in adult cells, it is restricted to the heart tissue.170 It acts as a TSG – the expression is repressed by promoter methylation in esophageal cancer,159 gastric cancer,158 and HCC.157 In gastric cancer, the repression of ZNF382 resulted in an increase in tumor cell proliferation, migration, and invasion, and the inhibition of apoptosis. Additionally, ZNF382 silencing had a positive impact on EMT and, thus, on cell metastasis. Furthermore, the in vitro data from two gastric cancer cell lines, MKN45, and SGC7901, indicated that ZNF382 repressed the expression of core genes involved in pluripotency maintenance (OCT4, SOX2, and NANOG).158
ZNF331
KRAB-ZFP factors with TSG function also include ZNF331, whose promoter methylation occurs in cholangiocarcinoma,171 colorectal,154 gastric, and pancreatic172 cancers. Additionally, methylation of the ZNF331 promoter was associated with weaker overall survival of colorectal cancer patients.154 ZNF331 was shown to have a negative effect on cancer features, as it was demonstrated to reduce proliferation, arrest the cell cycle in the G1/S phase,154 and inhibit migratory and invasive properties.155 Also, in mouse xenografts from colorectal cancer cells, high expression of ZNF331, compared to gene silencing, resulted in a smaller tumor volume.154 Differential proteomics studies suggested that ZNF331 may exert its TSG role through suppression of genes involved in migration and invasion (DSTN, and ACTR3), as well as proliferation (DDX5, EIF5A, GARS, STAM, UQCRFS1, and SET).155 Moreover, ZNF331 was demonstrated to reduce the expression of c-Myc oncogene, which is particularly important in terms of CSCs functioning.173 Of note, in the case of breast cancer cells, c-Myc overexpression resulted in the acquisition of stemness traits, including the expression of ALDH1 marker.156 Unlike in breast cancer cells, a decrease in c-Myc expression due to ZNF331 was reported in gastric cancer.155 Further studies are needed to fully understand the role of ZNF331 in different tumors and its role in stemness-related processes.
Conclusion
KRAB-ZFPs are transcription factors involved in many biological processes. In cancer cells, several members of the family were identified as oncogenes, whereas others act as TSGs.11 Still, little is known about their function in CSCs. Certain studies identified the role of specific KRAB-ZFPs in cells with stemness properties, i.e., ESCs or iPSCs. The results shed some light on KRAB-ZFPs' involvement in proliferation, differentiation, and stemness maintenance.7,134 Because physiological stem cells share many features with CSCs, it is possible that similar events are regulated by the same factors also within the cancer subpopulation. CSCs are responsible for tumor metastasis and resistance to chemotherapy.104,174 Understanding the mechanisms responsible for their formation and maintenance in the body may be beneficial for cancer treatment. Unraveling the influence of specific KRAB-ZFPs on CSCs may allow their usage in anti-cancer therapies. However, more research is still needed to further characterize this issue.
Author contributions
Conceptualization: UO, MM; Visualization: AO; Writing – original draft: AO, MM; Writing – review & editing: UO, MM.
Conflict of interests
Authors declare no conflict of interest.
Funding
This work was supported by the National Science Centre, Poland: 2015/17/B/NZ2/03689 to Urszula Oleksiewicz and 2018/31/D/NZ3/03790 to Marta Machnik.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Urszula Oleksiewicz, Email: oleksiewiczu@ump.edu.pl.
Marta Machnik, Email: marta1machnik@gmail.com.
References
- 1.Flavahan W.A., Gaskell E., Bernstein B.E. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357(6348):eaal2380. doi: 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennecke H., Yerushalmi R., Woods R., et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28(20):3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 3.Lathia J., Liu H., Matei D. The clinical impact of cancer stem cells. Oncologist. 2020;25(2):123–131. doi: 10.1634/theoncologist.2019-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oleksiewicz U., Machnik M. Causes, effects, and clinical implications of perturbed patterns within the cancer epigenome. Semin Cancer Biol. 2022;83:15–35. doi: 10.1016/j.semcancer.2020.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Shah M.A., Denton E.L., Arrowsmith C.H., Lupien M., Schapira M. A global assessment of cancer genomic alterations in epigenetic mechanisms. Epigenetics Chromatin. 2014;7(1):29. doi: 10.1186/1756-8935-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ecco G., Imbeault M., Trono D. KRAB zinc finger proteins. Dev Camb Engl. 2017;144(15):2719–2729. doi: 10.1242/dev.132605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oleksiewicz U., Gładych M., Raman A.T., et al. TRIM28 and interacting KRAB-ZNFs control self-renewal of human pluripotent stem cells through epigenetic repression of pro-differentiation genes. Stem Cell Rep. 2017;9(6):2065–2080. doi: 10.1016/j.stemcr.2017.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quenneville S., Verde G., Corsinotti A., et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44(3):361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cylwa R., Kiełczewski K., Machnik M., Oleksiewicz U., Biecek P. KRAB ZNF explorer-the online tool for the exploration of the transcriptomic profiles of KRAB-ZNF factors in the Cancer Genome Atlas. Bioinformatics. 2020;36(3):980–981. doi: 10.1093/bioinformatics/btz624. [DOI] [PubMed] [Google Scholar]
- 10.Machnik M., Cylwa R., Kiełczewski K., et al. The expression signature of cancer-associated KRAB-ZNF factors identified in TCGA pan-cancer transcriptomic data. Mol Oncol. 2019;13(4):701–724. doi: 10.1002/1878-0261.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobocińska J., Molenda S., Machnik M., Oleksiewicz U. KRAB-ZFP transcriptional regulators acting as oncogenes and tumor suppressors: an overview. Int J Mol Sci. 2021;22(4):2212. doi: 10.3390/ijms22042212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 15.Alexandrov L.B., Stratton M.R. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenman C., Stephens P., Smith R., et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International Cancer Genome Consortium. Hudson T.J., Anderson W., et al. International network of cancer genome projects. Nature. 2010;464(7291):993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beerman I., Rossi D.J. Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell. 2015;16(6):613–625. doi: 10.1016/j.stem.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortada I., Mortada R. Epigenetic changes in mesenchymal stem cells differentiation. Eur J Med Genet. 2018;61(2):114–118. doi: 10.1016/j.ejmg.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J., Benavente C.A., McEvoy J., et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481(7381):329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hovestadt V., Jones D.T.W., Picelli S., et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510(7506):537–541. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- 22.Pugh T.J., Weeraratne S.D., Archer T.C., et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baylin S.B., Jones P.A. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8(9):a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You J.S., Jones P.A. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22(1):9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saghafinia S., Mina M., Riggi N., Hanahan D., Ciriello G. Pan-cancer landscape of aberrant DNA methylation across human tumors. Cell Rep. 2018;25(4):1066–1080. doi: 10.1016/j.celrep.2018.09.082. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence M.S., Stojanov P., Mermel C.H., et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Kinzler K.W. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurrion C., Uriostegui M., Zurita M. Heterochromatin reduction correlates with the increase of the KDM4B and KDM6A demethylases and the expression of pericentromeric DNA during the acquisition of a transformed phenotype. J Cancer. 2017;8(14):2866–2875. doi: 10.7150/jca.19477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller-Tidow C., Klein H.U., Hascher A., et al. Profiling of histone H3 lysine 9 trimethylation levels predicts transcription factor activity and survival in acute myeloid leukemia. Blood. 2010;116(18):3564–3571. doi: 10.1182/blood-2009-09-240978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li K., Xu C., Du Y., Junaid M., Kaushik A.C., Wei D.Q. Comprehensive epigenetic analyses reveal master regulators driving lung metastasis of breast cancer. J Cell Mol Med. 2019;23(8):5415–5431. doi: 10.1111/jcmm.14424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ando M., Saito Y., Xu G., et al. Chromatin dysregulation and DNA methylation at transcription start sites associated with transcriptional repression in cancers. Nat Commun. 2019;10(1):2188. doi: 10.1038/s41467-019-09937-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torrano J., Al Emran A., Hammerlindl H., Schaider H. Emerging roles of H3K9me3, SETDB1 and SETDB2 in therapy-induced cellular reprogramming. Clin Epigenetics. 2019;11(1):43. doi: 10.1186/s13148-019-0644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen B., Wu H., Shinkai Y., Irizarry R.A., Feinberg A.P. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41(2):246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q.L., Lei P.J., Zhao Q.Y., Li L., Wei G., Wu M. Epigenomic analysis in a cell-based model reveals the roles of H3K9me3 in breast cancer transformation. Epigenomics. 2017;9(8):1077–1092. doi: 10.2217/epi-2016-0183. [DOI] [PubMed] [Google Scholar]
- 35.Slee R.B., Steiner C.M., Herbert B.S., et al. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene. 2012;31(27):3244–3253. doi: 10.1038/onc.2011.502. [DOI] [PubMed] [Google Scholar]
- 36.Xiang Y., Yan K., Zheng Q., et al. Histone demethylase KDM4B promotes DNA damage by activating long interspersed nuclear element-1. Cancer Res. 2019;79(1):86–98. doi: 10.1158/0008-5472.CAN-18-1310. [DOI] [PubMed] [Google Scholar]
- 37.Al Emran A., Marzese D.M., Menon D.R., et al. Distinct histone modifications denote early stress-induced drug tolerance in cancer. Oncotarget. 2018;9(9):8206–8222. doi: 10.18632/oncotarget.23654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guler G.D., Tindell C.A., Pitti R., et al. Repression of stress-induced LINE-1 expression protects cancer cell subpopulations from lethal drug exposure. Cancer Cell. 2017;32(2):221–237. doi: 10.1016/j.ccell.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Aryee M.J., Liu W., Engelmann J.C., et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med. 2013;5(169):169ra10. doi: 10.1126/scitranslmed.3005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brocks D., Assenov Y., Minner S., et al. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep. 2014;8(3):798–806. doi: 10.1016/j.celrep.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 41.Dietz S., Lifshitz A., Kazdal D., et al. Global DNA methylation reflects spatial heterogeneity and molecular evolution of lung adenocarcinomas. Int J Cancer. 2019;144(5):1061–1072. doi: 10.1002/ijc.31939. [DOI] [PubMed] [Google Scholar]
- 42.Sigalotti L., Fratta E., Coral S., et al. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2′-deoxycytidine. Cancer Res. 2004;64(24):9167–9171. doi: 10.1158/0008-5472.CAN-04-1442. [DOI] [PubMed] [Google Scholar]
- 43.Feinberg A.P., Koldobskiy M.A., Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17(5):284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marjanovic N.D., Weinberg R.A., Chaffer C.L. Cell plasticity and heterogeneity in cancer. Clin Chem. 2013;59(1):168–179. doi: 10.1373/clinchem.2012.184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald O.G., Li X., Saunders T., et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017;49(3):367–376. doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun L., Fang J. Epigenetic regulation of epithelial-mesenchymal transition. Cell Mol Life Sci. 2016;73(23):4493–4515. doi: 10.1007/s00018-016-2303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dawood S., Austin L., Cristofanilli M. Cancer stem cells: implications for cancer therapy. Oncology (Williston Park) 2014;28(12):1101–1107. 1110. [PubMed] [Google Scholar]
- 48.Dick J.E. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 49.Wang J.C.Y., Dick J.E. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15(9):494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 50.Dou J., Gu N. Emerging strategies for the identification and targeting of cancer stem cells. Tumour Biol. 2010;31(4):243–253. doi: 10.1007/s13277-010-0023-y. [DOI] [PubMed] [Google Scholar]
- 51.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim C.F.B., Jackson E.L., Woolfenden A.E., et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121(6):823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 53.Piccirillo S.G.M., Reynolds B.A., Zanetti N., et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444(7120):761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 54.Ponti D., Costa A., Zaffaroni N., et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 55.Wicha M.S., Liu S., Dontu G. Cancer stem cells: an old idea – a paradigm shift. Cancer Res. 2006;66(4):1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 56.Zhou J., Wulfkuhle J., Zhang H., et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci U S A. 2007;104(41):16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuda T., Nakamura T., Nakao K., et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18(15):4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rattigan Y., Hsu J.M., Mishra P.J., Glod J., Banerjee D. Interleukin 6 mediated recruitment of mesenchymal stem cells to the hypoxic tumor milieu. Exp Cell Res. 2010;316(20):3417–3424. doi: 10.1016/j.yexcr.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Mukaida N., Tanabe Y., Baba T. Cancer non-stem cells as a potent regulator of tumor microenvironment: a lesson from chronic myeloid leukemia. Mol Biomed. 2021;2(1):7. doi: 10.1186/s43556-021-00030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akincilar S.C., Unal B., Tergaonkar V. Reactivation of telomerase in cancer. Cell Mol Life Sci. 2016;73(8):1659–1670. doi: 10.1007/s00018-016-2146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swain N., Thakur M., Pathak J., Swain B. SOX2, OCT4 and NANOG: the core embryonic stem cell pluripotency regulators in oral carcinogenesis. J Oral Maxillofac Pathol. 2020;24(2):368–373. doi: 10.4103/jomfp.JOMFP_22_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J., Tam W.L., Tong G.Q., et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8(10):1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 63.Raz R., Lee C.K., Cannizzaro L.A., d'Eustachio P., Levy D.E. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 1999;96(6):2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng H., Ying H., Yan H., et al. Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harb Symp Quant Biol. 2008;73:427–437. doi: 10.1101/sqb.2008.73.047. [DOI] [PubMed] [Google Scholar]
- 65.Nakatake Y., Fukui N., Iwamatsu Y., et al. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol. 2006;26(20):7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohan A., Raj Rajan R., Mohan G., Kollenchery Puthenveettil P., Maliekal T.T. Markers and reporters to reveal the hierarchy in heterogeneous cancer stem cells. Front Cell Dev Biol. 2021;9:668851. doi: 10.3389/fcell.2021.668851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Tomaso T., Mazzoleni S., Wang E., et al. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin Cancer Res. 2010;16(3):800–813. doi: 10.1158/1078-0432.CCR-09-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sherry M.M., Reeves A., Wu J.K., Cochran B.H. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27(10):2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu F., Li J., Chen H., et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30(18):2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kobayashi D., Kuribayshi K., Tanaka M., Watanabe N. SALL4 is essential for cancer cell proliferation and is overexpressed at early clinical stages in breast cancer. Int J Oncol. 2011;38(4):933–939. doi: 10.3892/ijo.2011.929. [DOI] [PubMed] [Google Scholar]
- 71.Gritsko T., Williams A., Turkson J., et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12(1):11–19. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- 72.Gurel B., Iwata T., Koh C.M., et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21(9):1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hall P.E., Lathia J.D., Miller N.G.A., Caldwell M.A., ffrench-Constant C. Integrins are markers of human neural stem cells. Stem Cells. 2006;24(9):2078–2084. doi: 10.1634/stemcells.2005-0595. [DOI] [PubMed] [Google Scholar]
- 74.Osada M., Singh V.J., Wu K., Sant’Angelo D.B., Pezzano M. Label retention identifies a multipotent mesenchymal stem cell-like population in the postnatal thymus. PLoS One. 2013;8(12):e83024. doi: 10.1371/journal.pone.0083024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trempus C.S., Morris R.J., Bortner C.D., et al. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Investig Dermatol. 2003;120(4):501–511. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- 76.Chaurasia P., Gajzer D.C., Schaniel C., D'Souza S., Hoffman R. Epigenetic reprogramming induces the expansion of cord blood stem cells. J Clin Investig. 2014;124(6):2378–2395. doi: 10.1172/JCI70313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woo S.H., Stumpfova M., Jensen U.B., Lumpkin E.A., Owens D.M. Identification of epidermal progenitors for the Merkel cell lineage. Development. 2010;137(23):3965–3971. doi: 10.1242/dev.055970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ortiz-Sánchez E., Santiago-López L., Cruz-Domínguez V.B., et al. Characterization of cervical cancer stem cell-like cells: phenotyping, stemness, and human papilloma virus co-receptor expression. Oncotarget. 2016;7(22):31943–31954. doi: 10.18632/oncotarget.8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schober M., Fuchs E. Tumor-initiating stem cells of squamous cell carcinomas and their control by TGF-β and integrin/focal adhesion kinase (FAK) signaling. Proc Natl Acad Sci U S A. 2011;108(26):10544–10549. doi: 10.1073/pnas.1107807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.López J., Poitevin A., Mendoza-Martínez V., Pérez-Plasencia C., García-Carrancá A. Cancer-initiating cells derived from established cervical cell lines exhibit stem-cell markers and increased radioresistance. BMC Cancer. 2012;12:48. doi: 10.1186/1471-2407-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lathia J.D., Gallagher J., Heddleston J.M., et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6(5):421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rountree C.B., Senadheera S., Mato J.M., Crooks G.M., Lu S.C. Expansion of liver cancer stem cells during aging in methionine adenosyltransferase 1A-deficient mice. Hepatology. 2008;47(4):1288–1297. doi: 10.1002/hep.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O'Brien C.A., Pollett A., Gallinger S., Dick J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 84.Müller A., Homey B., Soto H., et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 85.Mori M., Mimori K., Shiraishi T., et al. Motility related protein 1 (MRP1/CD9) expression in colon cancer. Clin Cancer Res. 1998;4(6):1507–1510. [PubMed] [Google Scholar]
- 86.Hotta H., Ross A.H., Huebner K., et al. Molecular cloning and characterization of an antigen associated with early stages of melanoma tumor progression. Cancer Res. 1988;48(11):2955–2962. [PubMed] [Google Scholar]
- 87.Dong J.T., Lamb P.W., Rinker-Schaeffer C.W., et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science. 1995;268(5212):884–886. doi: 10.1126/science.7754374. [DOI] [PubMed] [Google Scholar]
- 88.Tokuhara T., Hasegawa H., Hattori N., et al. Clinical significance of CD151 gene expression in non-small cell lung cancer. Clin Cancer Res. 2001;7(12):4109–4114. [PubMed] [Google Scholar]
- 89.Saigusa S., Toiyama Y., Tanaka K., et al. Stromal CXCR4 and CXCL12 expression is associated with distant recurrence and poor prognosis in rectal cancer after chemoradiotherapy. Ann Surg Oncol. 2010;17(8):2051–2058. doi: 10.1245/s10434-010-0970-y. [DOI] [PubMed] [Google Scholar]
- 90.Matsui W.H. Cancer stem cell signaling pathways. Medicine. 2016;95(1 suppl 1):S8–S19. doi: 10.1097/MD.0000000000004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sanchez-Vega F., Mina M., Armenia J., et al. Oncogenic signaling pathways in the cancer genome Atlas. Cell. 2018;173(2):321–337. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krebsbach P.H., Villa-Diaz L.G. The role of integrin α6 (CD49f) in stem cells: more than a conserved biomarker. Stem Cells Dev. 2017;26(15):1090–1099. doi: 10.1089/scd.2016.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Z. CD133: a stem cell biomarker and beyond. Exp Hematol Oncol. 2013;2(1):17. doi: 10.1186/2162-3619-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hermann P.C., Huber S.L., Herrler T., et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 95.Ma S., Chan K.W., Hu L., et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132(7):2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 96.Singh S.K., Clarke I.D., Terasaki M., et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 97.Ma I., Allan A.L. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev Rep. 2011;7(2):292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 98.Liu G., Yuan X., Zeng Z., et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ginestier C., Hur M.H., Charafe-Jauffret E., et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li T., Su Y., Mei Y., et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients' outcome. Lab Investig. 2010;90(2):234–244. doi: 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Su Y., Qiu Q., Zhang X., et al. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomark Prev. 2010;19(2):327–337. doi: 10.1158/1055-9965.EPI-09-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heiden K.B., Williamson A.J., Doscas M.E., et al. The sonic hedgehog signaling pathway maintains the cancer stem cell self-renewal of anaplastic thyroid cancer by inducing snail expression. J Clin Endocrinol Metab. 2014;99(11):E2178–E2187. doi: 10.1210/jc.2014-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tirino V., Desiderio V., Paino F., et al. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J. 2013;27(1):13–24. doi: 10.1096/fj.12-218222. [DOI] [PubMed] [Google Scholar]
- 104.Wang D., Lu P., Zhang H., et al. Oct-4 and Nanog promote the epithelial-mesenchymal transition of breast cancer stem cells and are associated with poor prognosis in breast cancer patients. Oncotarget. 2014;5(21):10803–10815. doi: 10.18632/oncotarget.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chiou S.H., Wang M.L., Chou Y.T., et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010;70(24):10433–10444. doi: 10.1158/0008-5472.CAN-10-2638. [DOI] [PubMed] [Google Scholar]
- 106.Xu F., Dai C., Zhang R., Zhao Y., Peng S., Jia C. Nanog: a potential biomarker for liver metastasis of colorectal cancer. Dig Dis Sci. 2012;57(9):2340–2346. doi: 10.1007/s10620-012-2182-8. [DOI] [PubMed] [Google Scholar]
- 107.Lu X., Mazur S.J., Lin T., Appella E., Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene. 2014;33(20):2655–2664. doi: 10.1038/onc.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shen L., Qin K., Wang D., et al. Overexpression of Oct4 suppresses the metastatic potential of breast cancer cells via Rnd1 downregulation. Biochim Biophys Acta. 2014;1842(11):2087–2095. doi: 10.1016/j.bbadis.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 109.Imbeault M., Helleboid P.Y., Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017;543(7646):550–554. doi: 10.1038/nature21683. [DOI] [PubMed] [Google Scholar]
- 110.Huntley S., Baggott D.M., Hamilton A.T., et al. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16(5):669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Friedman J.R., Fredericks W.J., Jensen D.E., et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10(16):2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 112.Lupo A., Cesaro E., Montano G., Zurlo D., Izzo P., Costanzo P. KRAB-Zinc finger proteins: a repressor family displaying multiple biological functions. Curr Genom. 2013;14(4):268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Margolin J.F., Friedman J.R., Meyer W.K., Vissing H., Thiesen H.J., Rauscher F.J., 3rd Krüppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci U S A. 1994;91(10):4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Czerwińska P., Shah P.K., Tomczak K., et al. TRIM28 multi-domain protein regulates cancer stem cell population in breast tumor development. Oncotarget. 2017;8(1):863–882. doi: 10.18632/oncotarget.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iyengar S., Farnham P.J. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem. 2011;286(30):26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ryan R.F., Schultz D.C., Ayyanathan K., et al. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19(6):4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sripathy S.P., Stevens J., Schultz D.C. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26(22):8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheng Y., Geng H., Cheng S.H., et al. KRAB zinc finger protein ZNF382 is a proapoptotic tumor suppressor that represses multiple oncogenes and is commonly silenced in multiple carcinomas. Cancer Res. 2010;70(16):6516–6526. doi: 10.1158/0008-5472.CAN-09-4566. [DOI] [PubMed] [Google Scholar]
- 119.Huang C., Jia Y., Yang S., et al. Characterization of ZNF23, a KRAB-containing protein that is downregulated in human cancers and inhibits cell cycle progression. Exp Cell Res. 2007;313(2):254–263. doi: 10.1016/j.yexcr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 120.Sun R., Xiang T., Tang J., et al. 19q13 KRAB zinc-finger protein ZNF471 activates MAPK10/JNK3 signaling but is frequently silenced by promoter CpG methylation in esophageal cancer. Theranostics. 2020;10(5):2243–2259. doi: 10.7150/thno.35861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xiang S., Xiang T., Xiao Q., Li Y., Shao B., Luo T. Zinc-finger protein 545 is inactivated due to promoter methylation and functions as a tumor suppressor through the Wnt/β-catenin, PI3K/AKT and MAPK/ERK signaling pathways in colorectal cancer. Int J Oncol. 2017;51(3):801–811. doi: 10.3892/ijo.2017.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang T., Wang X.G., Xu J.H., et al. Overexpression of the human ZNF300 gene enhances growth and metastasis of cancer cells through activating NF-kB pathway. J Cell Mol Med. 2012;16(5):1134–1145. doi: 10.1111/j.1582-4934.2011.01388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang L., Hamilton S.R., Sood A., et al. The previously undescribed ZKSCAN3 (ZNF306) is a novel “driver” of colorectal cancer progression. Cancer Res. 2008;68(11):4321–4330. doi: 10.1158/0008-5472.CAN-08-0407. [DOI] [PubMed] [Google Scholar]
- 124.Serra R.W., Fang M., Park S.M., Hutchinson L., Green M.R. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. Elife. 2014;3:e02313. doi: 10.7554/eLife.02313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Busiello T., Ciano M., Romano S., et al. Role of ZNF224 in cell growth and chemoresistance of chronic lymphocitic leukemia. Hum Mol Genet. 2017;26(2):344–353. doi: 10.1093/hmg/ddw427. [DOI] [PubMed] [Google Scholar]
- 126.Chen K., Yu G., Gumireddy K., et al. ZBRK1, a novel tumor suppressor, activates VHL gene transcription through formation of a complex with VHL and p300 in renal cancer. Oncotarget. 2015;6(9):6959–6976. doi: 10.18632/oncotarget.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Montano G., Ullmark T., Jernmark-Nilsson H., et al. The hematopoietic tumor suppressor interferon regulatory factor 8 (IRF8) is upregulated by the antimetabolite cytarabine in leukemic cells involving the zinc finger protein ZNF224, acting as a cofactor of the Wilms' tumor gene 1 (WT1) protein. Leuk Res. 2016;40:60–67. doi: 10.1016/j.leukres.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 128.Seki Y., Kurisaki A., Watanabe-Susaki K., et al. TIF1beta regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc Natl Acad Sci U S A. 2010;107(24):10926–10931. doi: 10.1073/pnas.0907601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hu G., Kim J., Xu Q., Leng Y., Orkin S.H., Elledge S.J. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23(7):837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cammas F., Mark M., Dollé P., Dierich A., Chambon P., Losson R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Dev Camb Engl. 2000;127(13):2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- 131.Messerschmidt D.M., de Vries W., Ito M., Solter D., Ferguson-Smith A., Knowles B.B. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335(6075):1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- 132.Czerwińska P., Mazurek S., Wiznerowicz M. The complexity of TRIM28 contribution to cancer. J Biomed Sci. 2017;24(1):63. doi: 10.1186/s12929-017-0374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Salomonis N., Dexheimer P.J., Omberg L., et al. Integrated genomic analysis of diverse induced pluripotent stem cells from the progenitor cell biology consortium. Stem Cell Rep. 2016;7(1):110–125. doi: 10.1016/j.stemcr.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pontis J., Planet E., Offner S., et al. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell. 2019;24(5):724–735. doi: 10.1016/j.stem.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Turelli P., Playfoot C., Grun D., et al. Primate-restricted KRAB zinc finger proteins and target retrotransposons control gene expression in human neurons. Sci Adv. 2020;6(35):eaba3200. doi: 10.1126/sciadv.aba3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bourque G., Leong B., Vega V.B., et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18(11):1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Takahashi N., Coluccio A., Thorball C.W., et al. ZNF445 is a primary regulator of genomic imprinting. Genes Dev. 2019;33(1–2):49–54. doi: 10.1101/gad.320069.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Seo D.C., Sung J.M., Cho H.J., et al. Gene expression profiling of cancer stem cell in human lung adenocarcinoma A549 cells. Mol Cancer. 2007;6:75. doi: 10.1186/1476-4598-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schnabl B., Hu K., Mühlbauer M., et al. Zinc finger protein 267 is up-regulated during the activation process of human hepatic stellate cells and functions as a negative transcriptional regulator of MMP-10. Biochem Biophys Res Commun. 2005;335(1):87–96. doi: 10.1016/j.bbrc.2005.07.043. [DOI] [PubMed] [Google Scholar]
- 140.Schnabl B., Valletta D., Kirovski G., Hellerbrand C. Zinc finger protein 267 is up-regulated in hepatocellular carcinoma and promotes tumor cell proliferation and migration. Exp Mol Pathol. 2011;91(3):695–701. doi: 10.1016/j.yexmp.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bhat-Nakshatri P., Appaiah H., Ballas C., et al. SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24- phenotype. BMC Cancer. 2010;10(1) doi: 10.1186/1471-2407-10-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li P., Guo H., Zhou G., et al. Increased ZNF84 expression in cervical cancer. Arch Gynecol Obstet. 2018;297(6):1525–1532. doi: 10.1007/s00404-018-4770-0. [DOI] [PubMed] [Google Scholar]
- 143.Boot A., Oosting J., van Eendenburg J.D.H., Kuppen P.J.K., Morreau H., van Wezel T. Methylation associated transcriptional repression of ELOVL5 in novel colorectal cancer cell lines. PLoS One. 2017;12(9):e0184900. doi: 10.1371/journal.pone.0184900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jonasson E., Ghannoum S., Persson E., et al. Identification of breast cancer stem cell related genes using functional cellular assays combined with single-cell RNA sequencing in MDA-MB-231 cells. Front Genet. 2019;10:500. doi: 10.3389/fgene.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schwarz-Cruz Y. Celis A., Ceballos-Cancino G., Vazquez-Santillan K., et al. Basal-type breast cancer stem cells over-express chromosomal passenger complex proteins. Cells. 2020;9(3):709. doi: 10.3390/cells9030709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao Y., Carter R., Natarajan S., et al. Single-cell RNA sequencing reveals the impact of chromosomal instability on glioblastoma cancer stem cells. BMC Med Genom. 2019;12(1):79. doi: 10.1186/s12920-019-0532-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Giustacchini A., Thongjuea S., Barkas N., et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med. 2017;23(6):692–702. doi: 10.1038/nm.4336. [DOI] [PubMed] [Google Scholar]
- 148.Yau C., Esserman L., Moore D.H., Waldman F., Sninsky J., Benz C.C. A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast Cancer Res. 2010;12(5):R85. doi: 10.1186/bcr2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Maeda M., Yamashita S., Shimazu T., et al. Novel epigenetic markers for gastric cancer risk stratification in individuals after Helicobacter pylori eradication. Gastric Cancer. 2018;21(5):745–755. doi: 10.1007/s10120-018-0803-4. [DOI] [PubMed] [Google Scholar]
- 150.Cirillo A., Di Salle A., Petillo O., et al. High grade glioblastoma is associated with aberrant expression of ZFP57, a protein involved in gene imprinting, and of CPT1A and CPT1C that regulate fatty acid metabolism. Cancer Biol Ther. 2014;15(6):735–741. doi: 10.4161/cbt.28408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shoji Y., Takamura H., Ninomiya I., et al. The embryonic stem cell-specific transcription factor ZFP57 promotes liver metastasis of colorectal cancer. J Surg Res. 2019;237:22–29. doi: 10.1016/j.jss.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 152.Yamaguchi Y., Takamura H., Tada Y., et al. Nanog positively regulates Zfp57 expression in mouse embryonic stem cells. Biochem Biophys Res Commun. 2014;453(4):817–820. doi: 10.1016/j.bbrc.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 153.Pan Q., Wang L., Zhang H., Liang C., Li B. Identification of a 5-gene signature predicting progression and prognosis of clear cell renal cell carcinoma. Med Sci Monit. 2019;25:4401–4413. doi: 10.12659/MSM.917399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang Y., He T., Herman J.G., et al. Methylation of ZNF331 is an independent prognostic marker of colorectal cancer and promotes colorectal cancer growth. Clin Epigenetics. 2017;9:115. doi: 10.1186/s13148-017-0417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yu J., Liang Q.Y., Wang J., et al. Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene. 2013;32(3):307–317. doi: 10.1038/onc.2012.54. [DOI] [PubMed] [Google Scholar]
- 156.Poli V., Fagnocchi L., Fasciani A., et al. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat Commun. 2018;9(1):1024. doi: 10.1038/s41467-018-03264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dang S., Zhou J., Chen Y., et al. Dynamic expression of ZNF382 and its tumor-suppressor role in hepatitis B virus-related hepatocellular carcinogenesis. Oncogene. 2019;38(24):4804–4819. doi: 10.1038/s41388-019-0759-9. [DOI] [PubMed] [Google Scholar]
- 158.Pei L., He X., Li S., et al. KRAB zinc-finger protein 382 regulates epithelial-mesenchymal transition and functions as a tumor suppressor, but is silenced by CpG methylation in gastric cancer. Int J Oncol. 2018;53(3):961–972. doi: 10.3892/ijo.2018.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang C., Xiang T., Li S., et al. The novel 19q13 KRAB zinc-finger tumour suppressor ZNF382 is frequently methylated in oesophageal squamous cell carcinoma and antagonises Wnt/β-catenin signalling. Cell Death Dis. 2018;9(5):573. doi: 10.1038/s41419-018-0604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Huang C., Wu S., Li W., et al. Zinc-finger protein p52-ZER6 accelerates colorectal cancer cell proliferation and tumour progression through promoting p53 ubiquitination. EBioMedicine. 2019;48:248–263. doi: 10.1016/j.ebiom.2019.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zorzan I., Pellegrini M., Arboit M., et al. The transcriptional regulator ZNF398 mediates pluripotency and epithelial character downstream of TGF-beta in human PSCs. Nat Commun. 2020;11(1):2364. doi: 10.1038/s41467-020-16205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tao C., Luo J., Tang J., et al. The tumor suppressor zinc finger protein 471 suppresses breast cancer growth and metastasis through inhibiting AKT and Wnt/β-catenin signaling. Clin Epigenetics. 2020;12(1):173. doi: 10.1186/s13148-020-00959-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li R., Campos J., Iida J. A gene regulatory program in human breast cancer. Genetics. 2015;201(4):1341–1348. doi: 10.1534/genetics.115.180125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xia P. CD133 mRNA may be a suitable prognostic marker for human breast cancer. Stem Cell Investig. 2017;4:87. doi: 10.21037/sci.2017.10.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Honeth G., Bendahl P.O., Ringnér M., et al. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10(3):R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tamura K., Aoyagi M., Ando N., et al. Expansion of CD133-positive glioma cells in recurrent de novo glioblastomas after radiotherapy and chemotherapy. J Neurosurg. 2013;119(5):1145–1155. doi: 10.3171/2013.7.JNS122417. [DOI] [PubMed] [Google Scholar]
- 167.Conroy A.T., Sharma M., Holtz A.E., Wu C., Sun Z., Weigel R.J. A novel zinc finger transcription factor with two isoforms that are differentially repressed by estrogen receptor-alpha. J Biol Chem. 2002;277(11):9326–9334. doi: 10.1074/jbc.M107702200. [DOI] [PubMed] [Google Scholar]
- 168.Stabach P.R., Thiyagarajan M.M., Weigel R.J. Expression of ZER6 in ERalpha-positive breast cancer. J Surg Res. 2005;126(1):86–91. doi: 10.1016/j.jss.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 169.Hütz K., Mejías-Luque R., Farsakova K., et al. The stem cell factor SOX2 regulates the tumorigenic potential in human gastric cancer cells. Carcinogenesis. 2014;35(4):942–950. doi: 10.1093/carcin/bgt410. [DOI] [PubMed] [Google Scholar]
- 170.Luo K., Yuan W., Zhu C., et al. Expression of a novel Krüpple-like zinc-finger gene, ZNF382, in human heart. Biochem Biophys Res Commun. 2002;299(4):606–612. doi: 10.1016/s0006-291x(02)02700-6. [DOI] [PubMed] [Google Scholar]
- 171.Andresen K., Boberg K.M., Vedeld H.M., et al. Novel target genes and a valid biomarker panel identified for cholangiocarcinoma. Epigenetics. 2012;7(11):1249–1257. doi: 10.4161/epi.22191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vedeld H.M., Andresen K., Eilertsen I.A., et al. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int J Cancer. 2015;136(4):844–853. doi: 10.1002/ijc.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yoshida G.J. Emerging roles of Myc in stem cell biology and novel tumor therapies. J Exp Clin Cancer Res. 2018;37(1):173. doi: 10.1186/s13046-018-0835-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kim J.K., Jeon H.Y., Kim H. The molecular mechanisms underlying the therapeutic resistance of cancer stem cells. Arch Pharm Res. 2015;38(3):389–401. doi: 10.1007/s12272-014-0531-1. [DOI] [PubMed] [Google Scholar]