Abstract
This study aims to identify the inflammatory factor-related genes which help to predict the prognosis of patients with colorectal cancer. GSEA (Gene Set Enrichment Analysis) was used to acquire inflammation-related genes and the corresponding expression information was collected from TCGA database to determine the DEGs (differentially-expressed genes) in CRC patients. We conducted enrichment analysis and PPI (protein–protein interaction) of these DEGs. Besides, key genes that are both differentially-expressed and prognosis-related were screened out, which were used to establish the prognostic model. We obtained 79 DEGs and 19 prognostic genes, 10 prognostic-related differential genes were eventually screened. These genes were used to construct the prognostic model. We also identified that the immune infiltration score of macrophages between different risk groups was significantly different and similar distinction was witnessed in immune function score of APC (antigen-presenting cell) co-stimulation and type I IFN (interferon) response.
Keywords: Biomarker, Colorectal cancer (CRC), Immune infiltration, Inflammatory factors, Predicting model
Introduction
Colorectal cancer (CRC) is the third most common cancer globally, with 147,950 new cases and 53,200 deaths identified in 2020.1 CRC is considered to be associated with systemic inflammation, which presents in 20%–40% of CRC patients and plays an essential role in promoting the tumor progression through releasing inflammatory factors such as cytokines and growth factors.2,3 Besides, inflammation is an independent factor in predicting a poor prognosis of CRC patients and numerous cases suggest that the transformation from inflammation to tumor is commonly seen in CRC patients.4,5 Therefore, it is urgent to search for inflammatory-related biomarkers to project the cancer development and patients’ survival.
In recent decades, there are multiple studies investigating the mechanism of transformation from inflammation to cancer in CRC patients. Terzić et al demonstrated that chronic inflammation induces the occurrence of CAC (colitis-associated cancer), a subtype of CRC, through promoting mutations in oncogenes and tumor suppressor genes such as p53 and APC (adenomatous polyposis coli) by releasing proinflammatory cytokines.5 Thus, inflammation fosters the activation of anti-apoptotic features of pre-cancer cell and cell proliferation, eventually leading to tumor formation.5 Inflammation can also induce colon carcinogenesis via increasing the production of oxidative stress which may influence genes controlling colonocyte homeostasis, such as p53.6
Furthermore, accumulating research has explored the roles of various inflammatory factors in the progression of CRC. For instance, by increasing DNMT1 (DNA methyltransferase 1) expression in chronically inflamed colon, IL-6 is produced and can inhibit the expression of SOCS 3 (suppressor of cytokine signaling 3), a molecule that negatively modulates the cytokine-induced STAT3 signaling7 Further, the silencing of SOCS3 facilitates the malignant transformation in colons.7 Inflammatory factors such as TNF-α can increase DNA damage and ROS (reactive oxygen species) production in patients with inflammatory bowel disease, which ultimately contributes to intestinal cancer initiation.8 LncRNA CASC9 performs oncogenic activity in CRC cells via promoting the expression of TGF-β2 thus activating TGF-β signaling.9 FOXO3 expression is significantly declined in CRC tissues, which drives inflammation and fosters cell proliferation, associated with poor prognosis.10 Increased level of IL-17A presenting in chronic inflammation of colon tissues is related to poor prognosis and its blockade hinders the transformation from inflammation to cancer.11 As one of the regulators of IL-17 pathway, IL-1 signaling promotes early tumor growth and ablation of IL-1 receptor in epithelial cells can hamper tumor progression.11
Here, we downloaded data of CRC patients in TCGA database and extracted genes linked to inflammatory factors. We further obtained the inflammatory factor-related genes set in GESA and conducted differential expression analysis of these genes. 10 differentially-expressed genes which are prognosis-related were eventually screened out and they were used to establish our prognostic model whose efficacy was verified by GSE39582. Also, the difference of immune-related function in high and low-risk groups were compared. To investigate the relationship between immune infiltration and CRC, we further divided TCGA patients into four types based on immune cell infiltration and difference of risk scores were compared. Besides, stem cell correlation analysis, enrichment analysis and drug-sensitivity test were conducted in different risk groups.
Materials and methods
Data obtainment
Genes related to inflammatory factors were found in GSEA (Gene Set Enrichment Analysis) database. We then extracted the expression information of these genes in tumor and normal tissues of 379 CRC patients from TCGA (The Cancer Genome Atlas) database to determine the DEGs (differentially-expressed genes) of CRC patients. Clinical data encompassing age, gender and stage were collected.
Protein–protein interaction and enrichment analysis of genes
GO (Gene Ontology) enrichment analysis of genes that are differentially expressed in normal and CRC tissues was conducted to identify the biological processes, molecular function and cell components related to these genes. We also performed KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis to obtain the molecular pathways these genes mainly involve. Besides, PPI analysis of these genes was conducted, which showed the correlation between various genes. Based on PPI, we also carried out topology analysis to see whether some of the genes could construct a core interaction model.
Screening of inflammatory genes related to prognosis by in CRC
In order to obtain which genes were significantly associated with CRC patients’ prognosis, univariable regression analysis was performed. Then, we obtained genes from intersection between differentially expressed genes and prognostic genes. Besides, heatmap was created to compared the differential expression of these genes in tumor and normal tissues. We also preformed univariable cox regression analysis of these genes that are both differentially-expressed and prognosis-related to see which of them can serve as indicators for predicting prognosis of CRC patients. According to the expression of these genes, correlation analysis was carried out.
Construction and validation of a prognostic model for CRC patients
We conducted LASSO (Least Absolute Shrinkage and Selection Operator) regression analysis of genes that are differentially-expressed and prognosis-related, which further confirmed the association between these genes and prognosis of CRC patients in the training set. The risk score is calculated according to the following formula: Risk score = coef 1∗gene 1 + coef 2∗gene 2 + coef 3∗gene 3 + … + coefN∗gene. Clinical samples were further divided into high and low-risk groups, based on which the survival time and survival status in these risk groups were investigated. Dimension Reduction analysis was performed, which includes PCA (Principal Component Analysis) and tSNE2 (t-distributed stochastic neighbor embedding) analysis, to determine whether the two risk groups can be clearly distinguished. We implemented Kaplan–Meier analysis to explore the difference in survival probability between high and low-risk groups. To evaluate the accuracy of the model, receiver operating characteristic (ROC) curve analysis was conducted. Similarly, the above process was repeated using the test set to validate the risk model.
Independence analysis of the risk score combined with clinical characteristics
Univariable and multivariate cox regression analyses were implemented in both training and test sets, which aimed to investigate the significance of correlation between the prognosis of CRC patients and various factors including age, gender, stage and risk score. Further, box plots were created, which were used to compare the risk score of patients with different clinical features including age, gender and stage.
Analysis of immune cell infiltration and immune function
Cibersort was used to explore and compare the immune infiltration fraction of 22 types of immune cells (activated dendritic cells, B cells, CD8+ T cells, dendritic cells, interdigitating dendritic cell, macrophages, mast cells, neutrophils, NK cells, Plasmacytoid dendritic cells, T helper cells, T follicular helper cells, T helper type 1 cells, T helper type 2 cell, tumor-infiltrating lymphocytes and regulatory T cells) between high and low-risk groups in training set. Moreover, we implemented immune function analysis which included antigen-presenting cell co-inhibition, antigen-presenting cell co-stimulation, C–C chemokine receptor, check-point, cytolytic activity, human leukocyte antigen, inflammation-promoting, MHC class I, para-inflammation, T cell co-inhibition, T cell co-stimulation, type I interferon response and type II interferon response to compare the corresponding immune score between high and low-risk groups. The same experiments were performed in the test set for verification.
Stemness and PD-L1 expression analysis of the risk score and enrichment analysis
We divided patients into four subgroups based on divergent immune cell infiltration, with the purpose to explore whether there is significant distinction among their risk scores. In addition, to explore the relationship between stemness and the risk score of our model, correlation analysis was preformed, which involved the relation between DNA methylation and risk score, and relation between RNA expression and risk score. Besides, other correlation analyses were also implemented to investigate the relationship of risk score with immune sore and stromal score. Due to the extensive participation of PD-L1 in cancer progression, we further compared the PD-L1 expression level in high and low-risk groups, which was created in box plots. Also, correlation analysis about risk score and PD-L1 expression was performed.
Hallmark enrichment analysis of DEGs (differentially expressed genes) was conducted to obtain the main signaling pathways in which these genes in the high-risk group enriched compared with those in the low-risk group. Additionally, we performed KEGG analysis to explore the biological processes in which prognostic genes in high-risk group involve in comparison with those in low-risk group.
Sensitivity of common drugs analysis
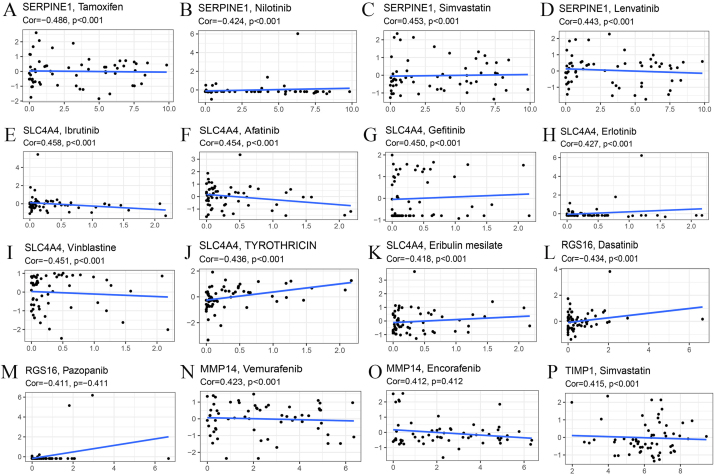
To further explore the association between our model genes and drug sensitivity, correlation analysis was conducted, involving genes such as SERPINE1, SLC4A4, RGS16, MMP14 and TIMP1. For SERPINE1, we analyzed its correlation with the sensitivity of Tamoxifen, Nilotinib, Simvastatin and Lenvatinib. We also investigated the association between SLC4A4 and the sensitivity of drugs including Ibrutinib, Afatinib, Gefitinib, Erlotinib, Vinblastine, TYROTHRICIN and Eribulin mesylate. In terms of RGS16, its expression level was analyzed combined with the sensitivity of Dasatinib and Pazopanib while MMP14 expression was analyzed blended with the sensitivity of Vemurafenib and Encorafenib. In addition, the relation between TIMP1 expression and Simvastatin sensitivity was studied.
Statistical analysis
R software (version number v3.5.2) was used for statistical analysis and Kaplan–Meier method was used to explore the overall survival of CRC patients. We used log-rank to analyze the significant difference and Wilcoxon signed rank-sum test to compare the immune cell infiltration in the two risk groups. P < 0.05 was regarded as the significant threshold in all the correlation analyses.
Results
Enrichment and correlation analysis of 79 inflammation-related genes
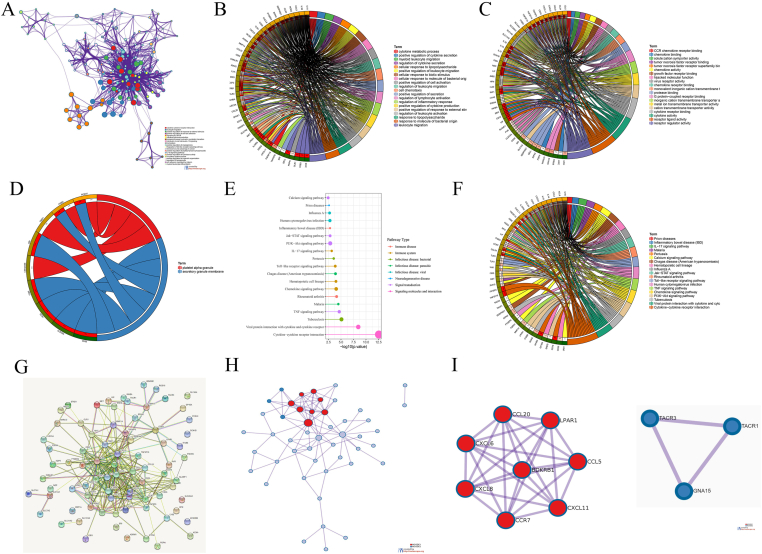
The results of GO (Gene Ontology) enrichment analysis of 79 inflammation-related genes are revealed in Figure 1A. The results of biological processes, molecular function and cell components of GO enrichment analysis are shown in Figure 1B–D respectively. KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis found that 79 genes were mainly enriched in cytokine–cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor and other pathways (Fig. 1E). The circle indicates the result of KEGG analysis of the 79 genes (Fig. 1F). The outcome of PPI (protein–protein interaction) analysis is presented in Figure 1G. We conducted topology analysis based on the results of PPI and found two core interaction models (Fig. 1H, I).
Figure 1.
Enrichment analysis of inflammation-related genes. (A) GO enrichment analysis showing 79 inflammation-related genes. (B–D) Biological processes, molecular function and cell components of GO enrichment analysis. (E, F) KEGG enrichment analysis indicating the main pathways in which the 79 genes participate. (G) PPI analysis of the 79 genes. (H) Topology analysis. (I) Two core models.
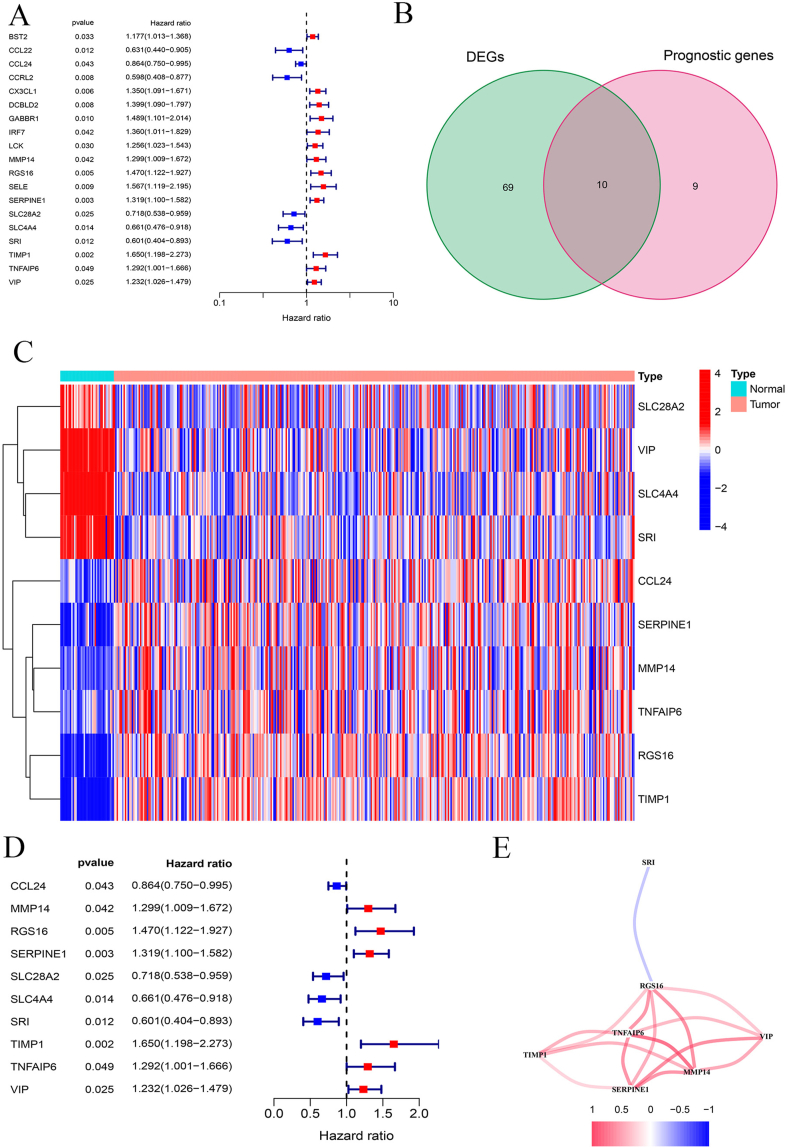
Screening of ten inflammatory-related and prognostic genes
Figure 2A indicates the result of univariable cox regression analysis of 19 prognosis-related genes (BST2, CCL22, CCL24, CCRL2, CX3CL1, DCBLD2, GABBR1, IRF7, LCK, MMP14, RGS16, SELE, SERPINE1, SLC28A2, SLC4A4, SRI, TIMP1, TNFAIP6 and VIP), showing that each gene is associated with CRC patients’ prognosis (Table 1). Based on the intersection of prognostic related genes and differentially expressed genes, a total of 10 overlapping genes were obtained, according to which we drew the corresponding Venn diagram (Fig. 2B). Heatmap presents the differential expression of these ten genes in tumor and normal tissues respectively (Fig. 2C). The result of univariable cox regression analysis of these 10 genes shows that they can act as independent variable for predicting prognosis of CRC patients (Fig. 2D). Among all the ten genes, seven of them (TIMP1, SERPINE1, MMP14, VIP, TNFAIP6, RGS16, and SRI) constitute an interaction module which has strong correlation in expression (Fig. 2E).
Figure 2.
Prognostic genes screening. (A) Univariable cox regression analysis of 19 prognostic genes. (B) Venn diagram presenting the ten genes which are both differentially-expressed and prognosis-related. (C) Heatmap of differential expression of the ten genes in normal and tumor tissues. (D) Univariable cox regression analysis of the ten genes. (E) The interaction network established by seven genes.
Table 1.
The result of univariable cox regression analysis of 19 prognosis-related genes.
| ID | HR | HR.95 L | HR.95H | p value |
|---|---|---|---|---|
| BST2 | 1.177273 | 1.013386 | 1.367664 | 0.032857 |
| CCL22 | 0.630952 | 0.439911 | 0.904955 | 0.012325 |
| CCL24 | 0.863931 | 0.749762 | 0.995485 | 0.04312 |
| CCRL2 | 0.598152 | 0.407977 | 0.876976 | 0.008478 |
| CX3CL1 | 1.350284 | 1.090901 | 1.67134 | 0.005791 |
| DCBLD2 | 1.399249 | 1.089801 | 1.796564 | 0.008431 |
| GABBR1 | 1.489199 | 1.101224 | 2.013863 | 0.009706 |
| IRF7 | 1.35994 | 1.01106 | 1.829206 | 0.042084 |
| LCK | 1.256013 | 1.022597 | 1.542708 | 0.029782 |
| MMP14 | 1.299296 | 1.009472 | 1.672329 | 0.042035 |
| RGS16 | 1.469973 | 1.12159 | 1.926569 | 0.005248 |
| SELE | 1.567228 | 1.118988 | 2.195023 | 0.008948 |
| SERPINE1 | 1.318966 | 1.099802 | 1.581803 | 0.002826 |
| SLC28A2 | 0.718008 | 0.537812 | 0.958579 | 0.024647 |
| SLC4A4 | 0.661031 | 0.475762 | 0.918447 | 0.013627 |
| SRI | 0.600509 | 0.403916 | 0.892787 | 0.01172 |
| TIMP1 | 1.650093 | 1.197812 | 2.27315 | 0.002182 |
| TNFAIP6 | 1.291875 | 1.001471 | 1.666489 | 0.048692 |
| VIP | 1.231964 | 1.025968 | 1.479321 | 0.025445 |
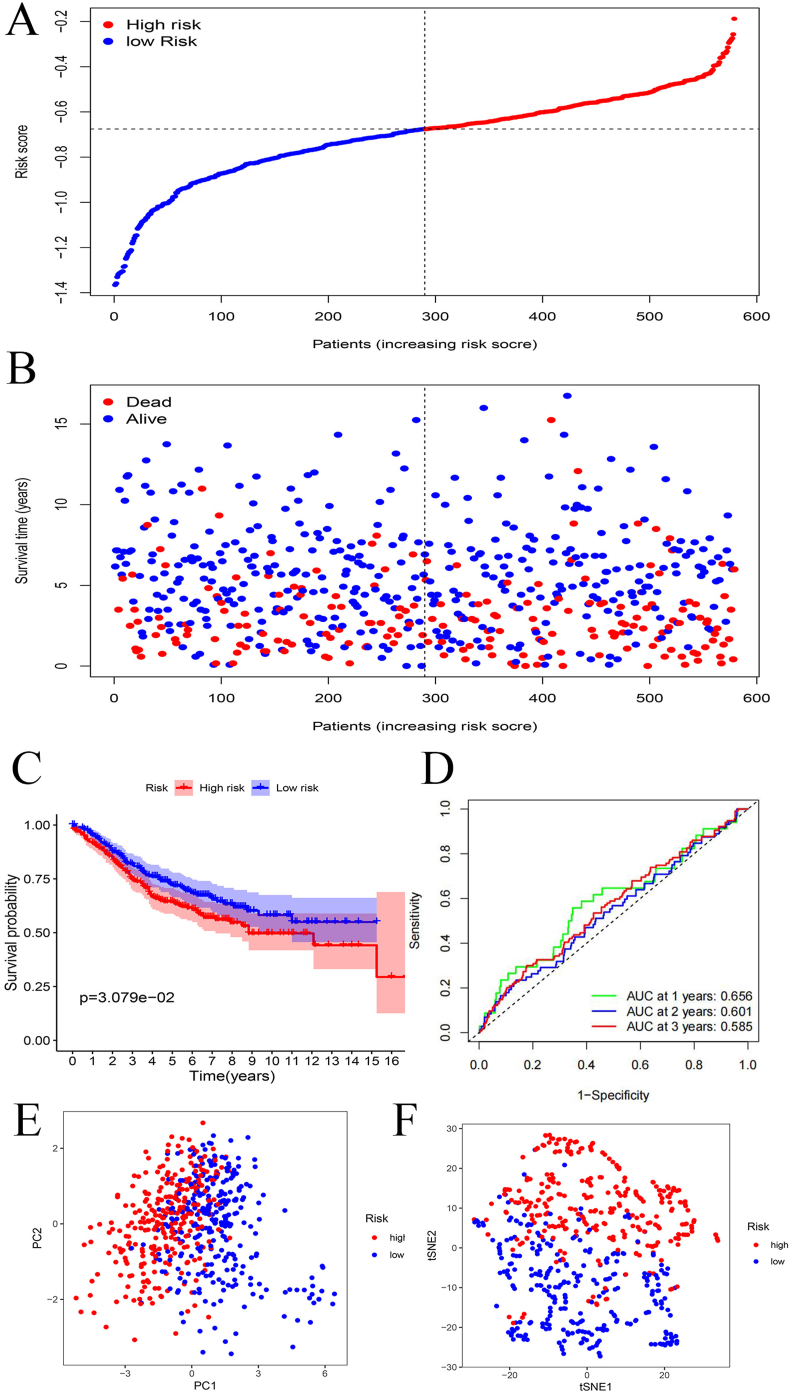
Construction and verification of a prognosis-related model based on the ten genes
LASSO regression analysis found that these ten genes (CCL24, MMP14, RGS16, SERPINE1, SLC28A2, SLC4A4, SRI, TIMP1, TNFAIP6 and VIP) are closely associated with CRC patients’ prognosis (Fig. 3A, B). The ten genes were then used to build our prognostic model. We used samples in TCGA database as training set, which were divided into high and low-risk groups based on LASSO results. The result of PCA (Principal Component Analysis) indicates that samples can be clearly divided into high and low-risk groups (Fig. 3C). Figure 3D suggests the distribution of risk score between high and low-risk groups. Survival status of CRC patients in the two risk groups are shown in Figure 3E. tSNE2 (t-distributed stochastic neighbor embedding) analysis also suggests that the high and low-risk groups can be well distinguished (Fig. 3F). Compared with low-risk groups, patients in high-risk groups have significantly poor survival outcomes, with p < 0.01 (Fig. 3G). Figure 3H indicates the AUC (area under the curve), showing that the 1-year, 2-year and 3-year survival rates are all above 0.7, which confirms that our model has excellent accuracy.
Figure 3.
Construction of the prognostic risk model. (A) Genes screened to achieve the best cut-off value. (B) Coefficients of the ten genes. (C) Principal Component Analysis of high and low-risk groups. (D) Risk score distribution between high and low-risk groups. (E) Survival status of patients in different risk groups. (F) tSNE2 analysis showing the distinguishment of different risk groups. (G) Kaplan–Meier survival analysis of high and low-risk groups. (H) AUC showing the accuracy of the model.
GSE39582 dataset was used as test set and Figure 4A shows the risk score distribution of test samples in high and low-risk groups. The survival status of test samples in two risk groups are suggested in Figure 4B. Kaplan–Meier survival analysis shows that the overall survival of low-risk group is significantly lower than that of high-risk group (Fig. 4C). AUC shows that 1-year survival rate in test set is above 0.65, suggesting that our model has good applicability and good prediction effect in the test set (Fig. 4D). Both PCA and tSNE2 analysis indicate that samples in high and low-risk groups have good distinguishment (Fig. 4E, F).
Figure 4.
Validation of the prognostic model. (A) Risk score distribution of test samples. (B) Survival status of patients in test samples. (C) Kaplan–Meier survival analysis of different risk groups. (D) AUC showing the model applicability. (E, F) PCA and tSNE2 analyses showing the distinguishment of different risk groups.
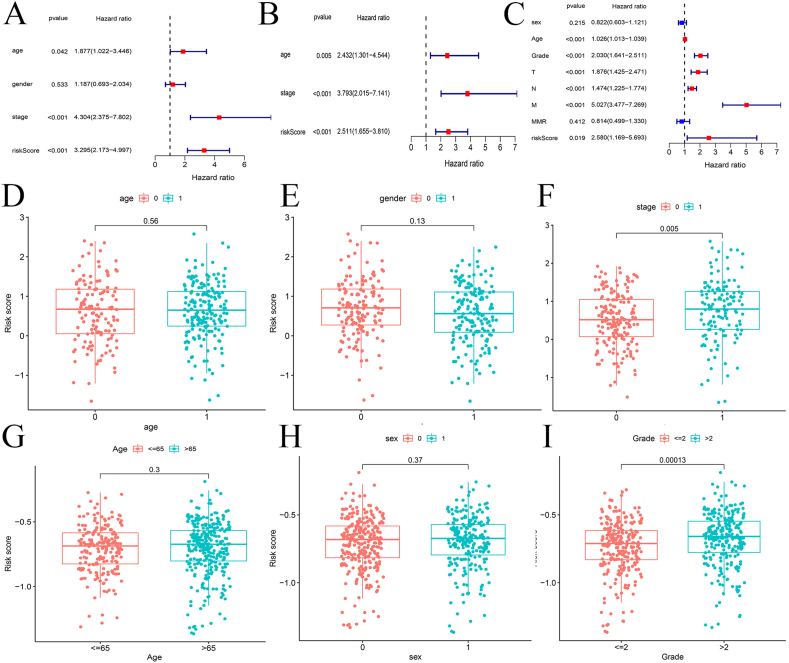
Analysis of risk score and other clinical features
Univariable and multivariate cox regression analysis of patients in training set show that age, stage and risk score are significantly correlated with prognosis (P < 0.05) (Fig. 5A, B). The same results are observed in the test set. Besides, T (tumor), N (nodes) and M (metastasis) are all significantly associated with prognosis (P < 0.001) (Fig. 5C). In training set, no significant correlation is witnessed between age and risk score or between gender and risk score (Fig. 5D, E). However, risk score is found to be significantly related to stage (P < 0.01) (Fig. 5F). As for GEO database, the result is similar to that in TCGA dataset (Fig. 5G–I).
Figure 5.
Comprehensive analysis of the risk score. (A, B) Univariable and multivariate cox regression analysis of age, gender, stage and risk scores of samples in the training group. (C) Univariable cox regression analysis of age, gender, stage and risk scores of samples in the test group. (D–F) Box plots of correlation between risk score and age, gender, stage in the training set, respectively. (G–I) Box plots of correlation between risk score and age, gender, stage in the test set, respectively.
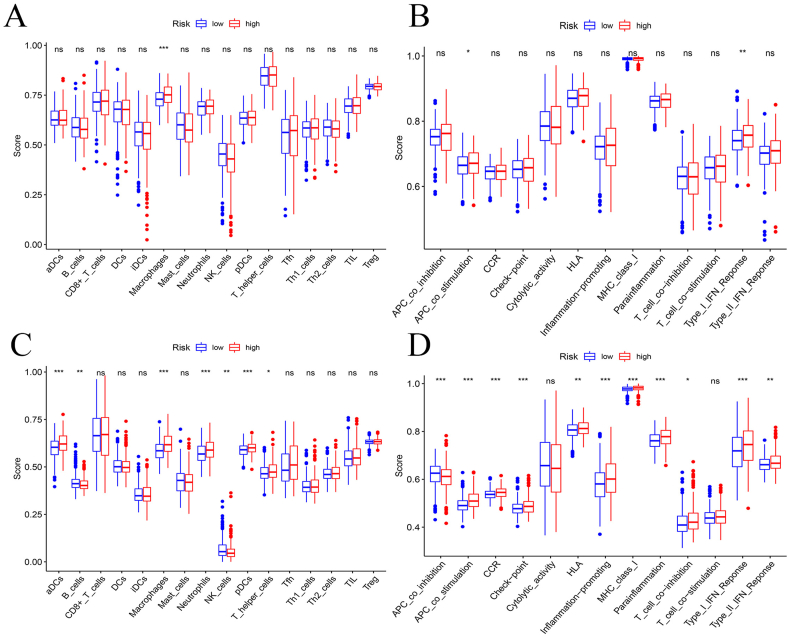
Immune cell infiltration and immune function analysis
We used cibersort to analyze the infiltration of 22 kinds of immune cells in high and low risk groups and found that there is significant difference in immune infiltration score of macrophages between low and high-risk groups while no remarkable distinction can be found in other immune cells between two risk groups (Fig. 6A). Moreover, immune function analysis was conducted and the result shows that there are significant differences in score of APC (antigen-presenting cell) co-stimulation and type I IFN (interferon) response between two risk groups (Fig. 6B). In terms of GEO database, cibersort was used to explore the immune infiltration of the same 22 types of cells and significant difference can be found in immune infiltration score of macrophages and other immune cells between high and low-risk groups (Fig. 6C). The result of immune function analysis shows that compared with low-risk group, the high-risk group has higher score of APC co-stimulation, type I IFN response and other immune function (Fig. 6D).
Figure 6.
Immune infiltration and function. (A, B) Box plots showed the immune infiltration of 22 types of immune cells and the immune function analysis in the high and low risk groups of the training set. (C, D) Box plots showed the immune infiltration of 22 types of immune cells and the immune function analysis in the high and low-risk groups of the test set.
Immune and stemness analysis of risk score
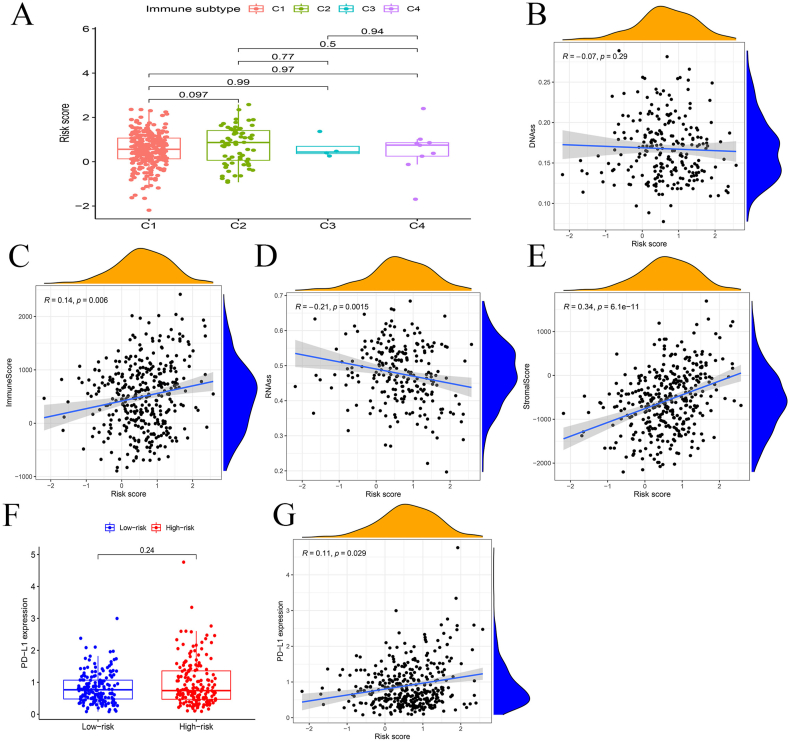
According to different immune cell infiltration, patients were further divided into four subgroups but no remarkable distinction can be found among their risk scores (Fig. 7A). No significant correlation can be observed between DNAss and the risk score (Fig. 7B) while Figure 7C suggests that immune score is positively associated with the risk score. In addition, RNAss is negatively related to the risk score whereas stromal score is positively correlated with the risk score (Fig. 7D, E). Further, PD-L1 expression levels in low and high-risk groups were compared but no significant distinction was found (Fig. 7F). Besides, Figure 7G indicates the positive correlation between PD-L1 expression and the risk score (P < 0.05).
Figure 7.
Immune and stemness analysis. (A) Box plots presented samples divided into four groups based on different immune subtypes. (B–E) Correlation analysis of risk with DNAss, immune score, RNAss and stromal score, respectively. (F) Box plot showed the comparison of PD-L1 expression in high and low-risk groups. (G) Correlation analysis of PD-L1 expression with the risk score.
Enrichment analysis of DEGs and prognostic genes
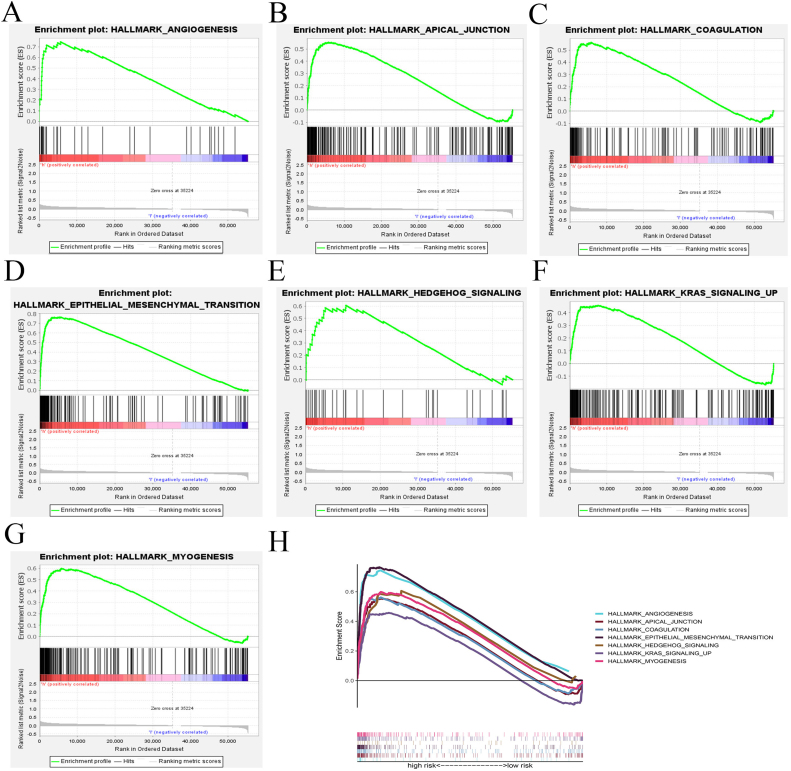
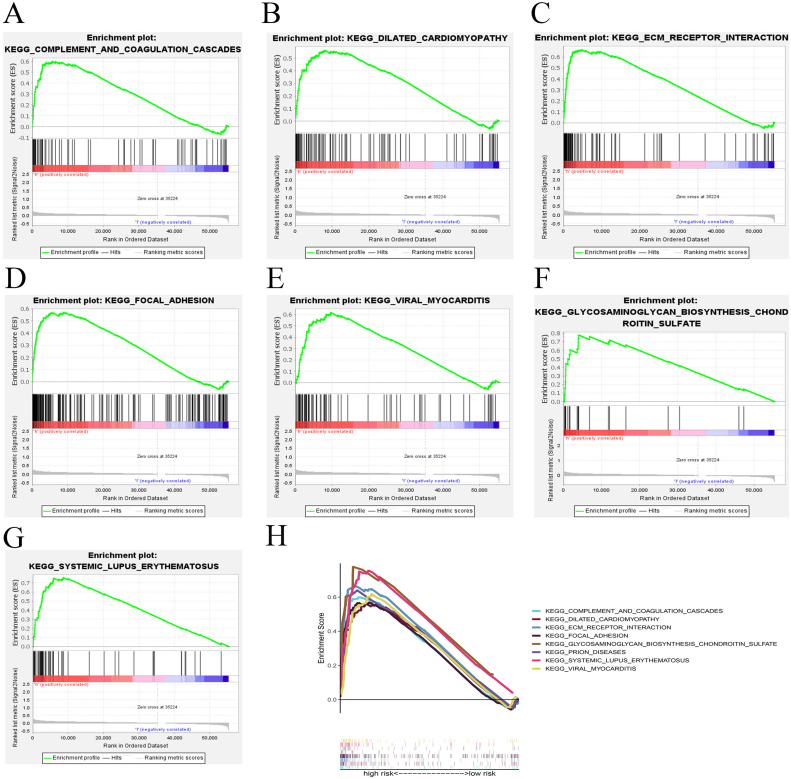
Hallmark enrichment analysis of DEGs (differentially expressed genes) shows that compared with that of low-risk group, DEGs in high-risk group are mainly enriched in signaling pathways such as angiogenesis, apical junction, coagulation, epithelial mesenchymal transition, hedgehog signaling, KARS signaling up and myogenesis (Fig. 8A–H). Besides, KEGG analysis of prognostic genes reveals that biological processes such as complement and coagulation cascades, dilated cardiomyopathy, ECM receptor interaction, focal adhesion, glycosaminoglycan biosynthesis chondroitin sulfate, prion diseases, systemic lupus erythematosus and viral myocarditis are significantly enriched in high-risk group compared with that in low-risk group (Fig. 9A–H).
Figure 8.
Enrichment analysis of DEGs. (A–H) Hallmark enrichment analysis showing the signaling pathways in which DEGs in high-risk group mainly involve.
Figure 9.
Enrichment analysis of prognostic genes. (A–H) KEGG analysis indicating the biological processes in which the prognostic genes in high-risk group mainly enriched.
Correlation analysis of expression of prognostic genes and sensitivity of common drugs
SERPINE1 expression is negatively related to the sensitivity of Tamoxifen and Nilotinib while its expression is positively associated with the sensitivity of Simvastatin, Lenvatinib (Fig. 10A–D). Besides, the expression of SLC4A4 has positive correlation with sensitivity of Ibrutinib, Afatinib, Gefitinib and Erlotinib while it is negatively related to the sensitivity of Vinblastine, TYROTHRICIN and Eribulin mesilate (Fig. 10E–K). In terms of RGS16, the expression level is negatively associated with the sensitivity of Dasatinib and Pazopanib (Fig. 10L, M). In contrast, the expression of MMP14 is positively related to the sensitivity of Vemurafenib and Encorafenib (Fig. 10N, O). Plus, TIMP1 expression is positively correlated with the sensitivity of Simvastatin (Fig. 10P).
Figure 10.
Drug sensitivity analysis. (A–D) Correlation analysis of SERPINE1 expression with sensitivity of Tamoxifen, Nilotinib, Simvastatin and Lenvatinib. (E–K) Correlation analysis of SLC4A4 expression with sensitivity of Ibrutinib, Afatinib, Gefitinib, Erlotinib, Vinblastine, TYROTHRICIN and Eribulin mesylate. (L, M) Correlation analysis of RGS16 expression with sensitivity of Dasatinib and Pazopanib. (N, O) Correlation analysis of MMP14 expression with sensitivity of Vemurafenib and Encorafenib. (P) Correlation analysis of TIMP1 expression with sensitivity of Simvastatin.
Discussion
As one of the most common cancer in world, CRC accounts for 10% of total diagnosed cancer cases and causes 9.4% cancer death worldwide.12 In recent decades, studies have found that long-term inflammatory response plays an essential role in various carcinogenic factors of colorectal cancer. For instance, the bacteria-caused CRC mechanism involves the reduction of helpful bacterial-produced metabolites, the change of intestinal microbiota and the disruption of intestinal epithelial barrier, all of which results in chronic inflammation along with abnormal activation of the immune system, eventually contributing to the development of CRC.13 AT (adipose tissue) inflammation also plays an essential role in obesity-related CRC, with the finding that BMI (body mass index) of CRC patients significantly influences the adipocytes transcriptome, which may foster cancer progression, leading to poor prognosis.14 Compared with the ordinary population, patients suffering from IBD (inflammatory bowel disease) such as ulcerative colitis, have more than twice risk of developing CRC, which further confirms the importance of chronic inflammation in carcinogenesis.15
In recent decades, scientists have focused on the underlying biological processes of various inflammatory factors in promoting CRC tumorigenesis and progression. Becker et al discovered that human colon tumor tissues have distinctively downregulated membrane-bound IL-6 receptors but higher level of tumor-derived ones, which enhances IL-6 signaling because this pathway requires soluble IL-6 receptor made by tumors instead of membrane-bound IL-6 receptor.16 Besides, through IL-6, TGF-β signaling in tumor infiltrating T lymphocytes can modulate the STAT-3 activation in cancer cells, hence regulating the growth of dysplastic colon epithelial cells, which leads to CRC progression.16 Another pro-inflammatory cytokine in CRC is TNFα and Popivanova et al identified that wide type mice treated with AOM (azoxymethane) and DSS (dextran sulfate sodium) had upregulated level of TNFα, which was followed by growth of many colonic tumors.17 De Simone et al discovered that transformation from normal colon mucosa to the neoplastic tissues is characterized by an accumulation of IL-22, IL-17A, IL-6 and TNF-α, which results in the activation of NF-κB and STAT3 to sustain CRC cell growth.18 Moreover, simultaneous neutralization of IL-22 and IL-6 can abolish STAT3 signaling, and synchronous inhibition of IL-17 A and TNF-α can abrogates NF-κB signaling while individual neutralization of these factors still leads to signaling activation.18 IL-22, a cytokine that involves in immune response during inflammation, can promote STAT3 phosphorylation in Lgr5+ crypt base columnar ISCs (intestinal stem cell), which plays an essential role in organoid formation and intestinal-stem-cell-mediated epithelial regeneration.19 Although IL-22 exerts a protective role during intestinal damage, it can also induce tumor initiation when uncontrolled.20 From above, we can see that inflammatory response and various inflammatory factors have an important impact on the occurrence and development of colorectal cancer. However, it is not clear which inflammation-related gene targets are involved in regulating the progression of colorectal cancer and affecting the prognosis of patients.
In our study, we screened out 10 genes (SLC28A2, VIP, SLC4A4, SRI, CCL24, SERPINE1, MMP14, TNFAIP6, RGS16, and TIMP1) related to CRC patients' prognosis and inflammation. These genes are reported to be closely related to the initiation and progression of digestive cancers. For instance, Li et al found that SLC28A2 can affect immune infiltration of Th17 cell and was also significantly related to prognosis of patients with COAD (colorectal adenocarcinoma).21 VIP (vasoactive intestinal polypeptide)/PACAP (pituitary adenylate cyclase-activating peptide) receptors were expressed in multiple human cancers, such as colon adenocarcinoma (96%), stomach carcinoma (54%) and liver carcinoma (49%).22 Moreover, previous study showed that TRPV-1 (transient receptor potential vanilloid type 1)-deficient mice had a downregulated level of VIP, PACAP and other anti-inflammatory neuropeptides, resulting in a proinflammatory condition, which ultimately drives an increased incidence and number of colonic tumors.23 This further confirms the anti-inflammatory function of VIP, which is consistent with our result. SLC4A4 was previously indicated to have an anti-tumor effect in clear cell renal cell carcinoma as miR-223–3p can promote tumor metastasis by downregulating this gene.24 Gao et al identified that SLC4A4 was downregulated in CRC patients and this gene was also remarkably correlated with patients' overall survivals, with high expression of SLC4A4 associated with a reduced risk of death in patients with CRC,25 which also confirms our presumption. The 18-kDa isoform of Ca2+-binding protein sorcin expressed by SRI gene can interact with TRAP1 through a calcium-dependent manner, which is imperative for anti-apoptotic role of TRAP1 in CRC.26 Nevertheless, the 22-kDa sorcin isoform was expressed at a higher level in patients with CRC and by regulating calcium homeostasis, this molecule can also induce resistance to irinotecan, 5-fluorouracil and other chemotherapeutic drugs in sensitized CRC cells.26 CCL24, also known as eotaxin-2 (eosinophil chemotactic protein 2), was upregulated in samples of primary colorectal cancer and liver metastases of colorectal origin.27 Besides, this chemokine may be specific to the cancer type due to its far less presence in adjacent non-neoplastic tissues and this research also demonstrated that carcinoembryonic antigen-positive tumor cells were the cardinal source of CCL24.27 Olsen et al discovered that high tertile level of CCL24 was related to a worse prognosis in CRC patients after operation and an increased cancer specific mortality.28 SERPINE1 (Serpin Family E Member 1) is closely related to CRC patients’ prognosis and its expression is also significantly higher in CRC samples compared with the surrounding tissues.29 In addition, the upregulation of SERPINE1 was particularly found in cancer stroma instead of epithelial tissues, which was related to reduced relapse-free intervals.30 MMP14 has an increased expression level in CRC samples, which is associated with advanced cancer stage and a lower survival time.31 By binding to miR-22-3 P, H19 can promote EMT (epithelial–mesenchymal transition) and raise the expression level of MMP14 while this effect can be inhibited by HDAC2 (histone deacetylase 2) which acts as a metastasis suppressor.32 Previous research indicated that TNFAIP6 was a biomarker of patients with IBD (inflammatory bowel disease), which may contribute to the development of CRC.33 Compared with normal tissues, the expression level of TNFAIP6 was distinctively increased in CRC samples, which was also correlated with TIMP1 expression and poor prognosis.34,35 RGS16, an independent prognostic factor in predicting CRC patients prognosis, was upregulated in CRC tissues in comparison with normal ones and this high expression was correlated with a lower overall survival rate.36 The continual overexpression of TIMP1 was found during the malignant transformation of UC (ulcerative colitis)-associated CRC and negatively related to the CRC patients’ prognosis.37 Also, this molecule may promote tumor progression through modulating cell cycle and cell migration.37
Our study suggested that macrophages, APC co-stimulation and type I interferon response may link to CRC progression. There are also several studies confirming our discovery. For instance, Zhao et al identified that miR-934, which was overexpressed abnormally in CRC, can foster macrophage M2 polarization through activating the PI3K/AKT signaling pathway and inhibiting PTEN expression.38 Then, polarized M2 macrophages can secret CXCL13 thus activating CXCL13/CXCR5/NFκB/p65/miR-934 positive feedback loop in colonic tumor cells to enhance the formation of premetastatic niche and CRLM (colorectal cancer liver metastasis).38 Additionally, macrophages contribute to immune escape of CRC cells though secreting CCL5 (C–C motif chemokine ligand 5) as this molecule can stabilize PD-L1.39 Macrophage-derived CCL5 also hinders T cell-mediated killing of HT29 cells, hence promoting tumor growth.39 STING signaling activation can trigger a robust type I interferon response.40 More importantly, CRC patients with upregulation of STING had early stage tumor with less lymphovascular invasion and more intra-tumoral CD8+ T cell infiltration, and they tended to have longer survival time.41
Enrichment analysis showed that DEGs of CRC were mainly concentrated in angiogenesis, apical junction and other signaling pathways. Deng et al demonstrated that dickkopf2 derived from CRC cells can facilitate aerobic glycolysis and angiogenesis, eventually leading to increased metastasis of tumor cells.42 Alteration of the expression level of various components in apical junction complex was identified in CRC and as one of the dominant elements in tight junction, claudin-3 was upregulated in CRC tissues, which was correlated with an increased malignancy of tumor cells.43 In addition, our research also indicates that the prognostic genes in CRC were mainly enriched in signaling pathways such as ECM receptor interaction and focal adhesion. One study suggested that CRC cells had increased FAK (focal adhesion kinase) expression during early stage of tumorigenesis and this upregulation enhanced adhesive properties and local invasion of tumor cells.44 Besides, miR2155p exerts tumor suppression function through modulating several molecular pathways including ECM-receptor interaction and focal adhesion, which can diminish invasiveness of CRC cells and liver metastasis.45
We established a prognostic risk model based on 10 inflammatory-related genes which may serve as biomarkers for predicting the prognosis of CRC patients. However, the underlying mechanism of these inflammation-related genes involving in colorectal carcinogenesis needs further investigation through cell and animal experiments.
Conclusions
In summary, we constructed a prognostic risk model based on ten genes associated with inflammatory factors, which provides an evaluation system for patients with CRC and can accurately predict their prognosis. Besides, the differences in immune cell infiltration and immune function were identified between the high and low-risk groups, thus offering novel insight into the development of potential immunotherapeutic targets in CRC.
Ethical declaration
This article does not contain any studies with animals performed by any of the authors. All methods are carried out in accordance with relevant guidelines and regulations.
Author contributions
(i) Guarantor of the article: X. Wang.
(ii) Specific author contributions: J. Hu, research design and drafting the manuscript; Y. He, K. Liao, Q. Yang, Y. Xu, and G. Cao, literature searching and helping to draft the manuscript; X. Wang, review and revision of the manuscript and writing guidance; S. Fan, helping to check and revise the manuscript.
(iii) All the authors listed have approved the manuscript that is enclosed.
Conflict of interests
There are no conflict of interests in this study.
Funding
This study was funded by the Natural Science Foundation of China (No.81860034 to WXZ).
Data availability
All data and materials are available. Please contact us to access if it is needed.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Tuomisto A.E., Mäkinen M.J., Väyrynen J.P. Systemic inflammation in colorectal cancer: underlying factors, effects, and prognostic significance. World J Gastroenterol. 2019;25(31):4383–4404. doi: 10.3748/wjg.v25.i31.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J.H., Zhai E.T., Yuan Y.J., et al. Systemic immune-inflammation index for predicting prognosis of colorectal cancer. World J Gastroenterol. 2017;23(34):6261–6272. doi: 10.3748/wjg.v23.i34.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMillan D.C. The systemic inflammation-based Glasgow Prognostic Score: a decade of experience in patients with cancer. Cancer Treat Rev. 2013;39(5):534–540. doi: 10.1016/j.ctrv.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Terzić J., Grivennikov S., Karin E., Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138(6):2101–2114. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 6.Ullman T.A., Itzkowitz S.H. Intestinal inflammation and cancer. Gastroenterology. 2011;140(6):1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 7.Li Y., Deuring J., Peppelenbosch M.P., Kuipers E.J., de Haar C., van der Woude C.J. IL-6-induced DNMT1 activity mediates SOCS3 promoter hypermethylation in ulcerative colitis-related colorectal cancer. Carcinogenesis. 2012;33(10):1889–1896. doi: 10.1093/carcin/bgs214. [DOI] [PubMed] [Google Scholar]
- 8.O'Connor P.M., Lapointe T.K., Beck P.L., Buret A.G. Mechanisms by which inflammation may increase intestinal cancer risk in inflammatory bowel disease. Inflamm Bowel Dis. 2010;16(8):1411–1420. doi: 10.1002/ibd.21217. [DOI] [PubMed] [Google Scholar]
- 9.Luo K., Geng J., Zhang Q., et al. LncRNA CASC9 interacts with CPSF3 to regulate TGF-β signaling in colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):249. doi: 10.1186/s13046-019-1263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullock M.D., Bruce A., Sreekumar R., et al. FOXO3 expression during colorectal cancer progression: biomarker potential reflects a tumour suppressor role. Br J Cancer. 2013;109(2):387–394. doi: 10.1038/bjc.2013.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dmitrieva-Posocco O., Dzutsev A., Posocco D.F., et al. Cell-type-specific responses to interleukin-1 control microbial invasion and tumor-elicited inflammation in colorectal cancer. Immunity. 2019;50(1):166–180. doi: 10.1016/j.immuni.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sung H., Ferlay J., Siegel R.L., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 13.Schwabe R.F., Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13(11):800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Cornò M., Baldassarre A., Calura E., et al. Transcriptome profiles of human visceral adipocytes in obesity and colorectal cancer unravel the effects of body mass index and polyunsaturated fatty acids on genes and biological processes related to tumorigenesis. Front Immunol. 2019;10:265. doi: 10.3389/fimmu.2019.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jess T., Rungoe C., Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol. 2012;10(6):639–645. doi: 10.1016/j.cgh.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Becker C., Fantini M.C., Wirtz S., et al. IL-6 signaling promotes tumor growth in colorectal cancer. Cell Cycle. 2005;4(2):217–220. [PubMed] [Google Scholar]
- 17.Popivanova B.K., Kitamura K., Wu Y., et al. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J Clin Invest. 2008;118(2):560–570. doi: 10.1172/JCI32453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Simone V., Franzè E., Ronchetti G., et al. Th17-type cytokines, IL-6 and TNF-α synergistically activate STAT3 and NF-kB to promote colorectal cancer cell growth. Oncogene. 2015;34(27):3493–3503. doi: 10.1038/onc.2014.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindemans C.A., Calafiore M., Mertelsmann A.M., et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528(7583):560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber S., Gagliani N., Zenewicz L.A., et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 2012;491(7423):259–263. doi: 10.1038/nature11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F., Zhou J., Li Z., Zhang L. Screening of immunosuppressive cells from colorectal adenocarcinoma and identification of prognostic markers. Biosci Rep. 2021;41(4) doi: 10.1042/BSR20203496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reubi J.C., Läderach U., Waser B., Gebbers J.O., Robberecht P., Laissue J.A. Vasoactive intestinal peptide/pituitary adenylate cyclase-activating peptide receptor subtypes in human tumors and their tissues of origin. Cancer Res. 2000;60(11):3105–3112. [PubMed] [Google Scholar]
- 23.Vinuesa A.G., Sancho R., García-Limones C., et al. Vanilloid receptor-1 regulates neurogenic inflammation in colon and protects mice from colon cancer. Cancer Res. 2012;72(7):1705–1716. doi: 10.1158/0008-5472.CAN-11-3693. [DOI] [PubMed] [Google Scholar]
- 24.Xiao W., Wang X., Wang T., Xing J. miR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma. Aging. 2019;11(2):615–633. doi: 10.18632/aging.101763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao X., Yang J. Identification of genes related to clinicopathological characteristics and prognosis of patients with colorectal cancer. DNA Cell Biol. 2020;39(4):690–699. doi: 10.1089/dna.2019.5088. [DOI] [PubMed] [Google Scholar]
- 26.Maddalena F., Laudiero G., Piscazzi A., et al. Sorcin induces a drug-resistant phenotype in human colorectal cancer by modulating Ca(2+) homeostasis. Cancer Res. 2011;71(24):7659–7669. doi: 10.1158/0008-5472.CAN-11-2172. [DOI] [PubMed] [Google Scholar]
- 27.Cheadle E.J., Riyad K., Subar D., et al. Eotaxin-2 and colorectal cancer: a potential target for immune therapy. Clin Cancer Res. 2007;13(19):5719–5728. doi: 10.1158/1078-0432.CCR-07-1145. [DOI] [PubMed] [Google Scholar]
- 28.Olsen R.S., Nijm J., Andersson R.E., Dimberg J., Wågsäter D. Circulating inflammatory factors associated with worse long-term prognosis in colorectal cancer. World J Gastroenterol. 2017;23(34):6212–6219. doi: 10.3748/wjg.v23.i34.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y., Li C., Dong L., Chen X., Fan R. Identification and verification of three key genes associated with survival and prognosis of COAD patients via integrated bioinformatics analysis. Biosci Rep. 2020;40(9) doi: 10.1042/BSR20200141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chida S., Okayama H., Noda M., et al. Stromal VCAN expression as a potential prognostic biomarker for disease recurrence in stage II-III colon cancer. Carcinogenesis. 2016;37(9):878–887. doi: 10.1093/carcin/bgw069. [DOI] [PubMed] [Google Scholar]
- 31.Cui G., Cai F., Ding Z., Gao L. MMP14 predicts a poor prognosis in patients with colorectal cancer. Hum Pathol. 2019;83:36–42. doi: 10.1016/j.humpath.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Hu X.T., Xing W., Zhao R.S., et al. HDAC2 inhibits EMT-mediated cancer metastasis by downregulating the long noncoding RNA H19 in colorectal cancer. J Exp Clin Cancer Res. 2020;39(1):270. doi: 10.1186/s13046-020-01783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Q., Zhang S., Wang H., et al. TNFAIP6 is a potential biomarker of disease activity in inflammatory bowel disease. Biomarkers Med. 2016;10(5):473–483. doi: 10.2217/bmm.16.9. [DOI] [PubMed] [Google Scholar]
- 34.Offenberg H., Brünner N., Mansilla F., Orntoft Torben F., Birkenkamp-Demtroder K. TIMP-1 expression in human colorectal cancer is associated with TGF-B1, LOXL2, INHBA1, TNF-AIP6 and TIMP-2 transcript profiles. Mol Oncol. 2008;2(3):233–240. doi: 10.1016/j.molonc.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Cheng L., Li C., Zhang C., Wang L., Zhang J. Identification of tumor microenvironment-related prognostic genes in colorectal cancer based on bioinformatic methods. Sci Rep. 2021;11(1):15040. doi: 10.1038/s41598-021-94541-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyoshi N., Ishii H., Sekimoto M., Doki Y., Mori M. RGS16 is a marker for prognosis in colorectal cancer. Ann Surg Oncol. 2009;16(12):3507–3514. doi: 10.1245/s10434-009-0690-3. [DOI] [PubMed] [Google Scholar]
- 37.Huang R., Wang K., Gao L., Gao W. TIMP1 is a potential key gene associated with the pathogenesis and prognosis of ulcerative colitis-associated colorectal cancer. OncoTargets Ther. 2019;12:8895–8904. doi: 10.2147/OTT.S222608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao S., Mi Y., Guan B., et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. 2020;13(1):156. doi: 10.1186/s13045-020-00991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C., Yao Z., Wang J., et al. Macrophage-derived CCL5 facilitates immune escape of colorectal cancer cells via the p65/STAT3-CSN5-PD-L1 pathway. Cell Death Differ. 2020;27(6):1765–1781. doi: 10.1038/s41418-019-0460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corrales L., Gajewski T.F. Molecular pathways: targeting the Stimulator of Interferon Genes (STING) in the immunotherapy of cancer. Clin Cancer Res. 2015;21(21):4774–4779. doi: 10.1158/1078-0432.CCR-15-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chon H.J., Kim H., Noh J.H., et al. STING signaling is a potential immunotherapeutic target in colorectal cancer. J Cancer. 2019;10(20):4932–4938. doi: 10.7150/jca.32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng F., Zhou R., Lin C., et al. Tumor-secreted dickkopf2 accelerates aerobic glycolysis and promotes angiogenesis in colorectal cancer. Theranostics. 2019;9(4):1001–1014. doi: 10.7150/thno.30056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pérez A.G., Andrade-Da-Costa J., De Souza W.F., et al. N-glycosylation and receptor tyrosine kinase signaling affect claudin-3 levels in colorectal cancer cells. Oncol Rep. 2020;44(4):1649–1661. doi: 10.3892/or.2020.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeong K.Y. Inhibiting focal adhesion kinase: a potential target for enhancing therapeutic efficacy in colorectal cancer therapy. World J Gastrointest Oncol. 2018;10(10):290–292. doi: 10.4251/wjgo.v10.i10.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machackova T., Vychytilova-Faltejskova P., Souckova K., et al. miR-215-5p reduces liver metastasis in an experimental model of colorectal cancer through regulation of ECM-receptor interactions and focal adhesion. Cancers. 2020;12(12):3518. doi: 10.3390/cancers12123518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials are available. Please contact us to access if it is needed.