Abstract
Aptamers, short single DNA or RNA oligonucleotides, have shown immense application potential as molecular probes for the early diagnosis and therapy of cancer. However, conventional cell-SELEX technologies for aptamer discovery are time-consuming and laborious. Here we discovered a new aptamer BC-3 by using an improved rapid X-Aptamer selection process for human bladder carcinoma, for which there is no specific molecular probe yet. We show that BC-3 exhibited excellent affinity in bladder cancer cells but not normal cells. We demonstrate that BC-3 displayed high selectivity for tumor cells over their normal counterparts in vitro, in mice, and in patient tumor tissue specimens. Further endocytosis pathway analysis revealed that BC-3 internalized into bladder cancer cells via clathrin-mediated endocytosis. Importantly, we identified ribosomal protein S7 (RPS7) as the binding target of BC-3 via an integrated methodology (mass spectrometry, colocalization assay, and immunoblotting). Together, we report that a novel aptamer BC-3 is discovered for bladder cancer and its properties in the disease are unearthed. Our findings will facilitate the discovery of novel diagnostic and therapeutic strategies for bladder cancer.
Keywords: Bladder cancer, Cell-SELEX, Clathrin-mediated endocytosis, Intracellular colocalization, Ribosomal protein S7, X-aptamer selections
Introduction
Bladder carcinoma (BC) is one of the most common severe diseases that occur in human urinary systems. According to global cancer statistics in 2020, bladder cancer is the 10th most diagnosed cancer worldwide, with approximately 573,000 new cases and 213,000 deaths.1 In bladder cancer, the morbidity and the mortality of male to female patients are 4 and 3, respectively. The early clinical manifestations of bladder cancer are not obvious and are easily confused with other urinary diseases. If bladder cancer is suspected, it is most often diagnosed via a cystoscopy. This procedure is sometimes combined with a biopsy.2 Thus, the development of molecular probes and tools for bladder cancer is particularly important to its early diagnosis and treatment.3
Aptamers, short single stranded (ss) DNA or RNA evolved from random oligonucleotide libraries, manifest high binding specificity and affinity due to their specific three-dimensional shapes.4 Compared to antibodies, aptamers have numerous advantages including high biocompatibility, low immunogenicity, low toxicity, and ease of synthesis and modification.5,6 The conventional procedures for cell-SELEX are inefficient and time-consuming, with multiple repetitive rounds of separation and enzymatic amplification.7 In contrast, X-Aptamer selection (X-As) only consists of a two-round process, without any other specialized instruments.8 This rapid selection method is based on a high affinity modified aptamer Library, and coupling beads with multiple copies of sequences can effectively improve their binding opportunities to targets.9 X-As has only been used in protein-SELEX before,7,10,11 and we applied and improved it in cell-SELEX against bladder cancer cells.
Aptamers are internalized from cell membranes by clathrin-mediated endocytosis, caveolae-mediated endocytosis, micropinocytosis, or phagocytosis.12 After that, aptamers are further degraded by lysosomes, sorted for recycling back to the plasma membrane, escape from early endosomes and are delivered to certain subcellular compartments.13 Therefore, lysosomal degradation is the main obstacle for aptamer-based approaches to be used in imaging and intracellular therapy.14,15 For example, in previous research, aptamer S11e was found to be located in mitochondria but not lysosomes through nonlysosomal transport approaches.16 As an important member of the 40S ribosomal protein family, ribosomal protein S7 (RPS7) is correlated with the occurrence and development of many diseases.17, 18, 19 RPS7 can impact the proliferation and metastasis of ovarian cancer through the PI3K/AKT and MAPK signaling pathways and knock down of RPS7 activates p53 and p21.20 RPS7 can regulate tumor progression by deregulating MDM2-p53 signaling in the human lung cancer cell line H1299.21 Moreover, RPS7 is a newly verified tumor promoter in prostate cancer. RPS7 is overexpressed in prostate cancer, and promotes cell migration and growth.22 However, in bladder carcinoma, the mechanism of RPS7 in bladder cancer remains unclear, especially the expression level. Molecular probes or targeted delivery of RPS7 can thus provide an important approach to inhibit the occurrence and development of different cancers via the p53-related signaling pathway.
In this study, we have improved the strategy of X-As and have rapidly obtained a novel aptamer BC-3 which can specifically recognize BC 5637 cells and clinical BC tissues but not normal bladder epithelial cells SV-HUC-1 cells or paracarcinoma tissues. BC-3 accumulated at tumor sites in 5637 xenograft nude mouse models by fluorescence imaging and had no toxicity in vitro according to the CCK-8 assay. Interestingly, BC-3 can escape lysosomal degradation and relocate to the endoplasmic reticulum (ER) by targeting RPS7, which can be developed as a unique tool for intracellular imaging or targeted therapy for bladder cancer.
Materials and methods
Cell culture
All cell lines used in this work and their culture conditions are summarized in Table S1.
X-As kit, ssDNA Library, primers, and buffers
The X-As Selection Kit (AM Biotechnologies, Houston, TX, USA) was purchased from Sangon Biotech Co. Ltd. (Shanghai, China). All DNA sequences used in the X-As were purchased from Hippo Biological Technology Co. Ltd (Huzhou, China). Sequence information can be found in Table S2.
Washing buffer was prepared with Dulbecco's phosphate buffered saline (D-PBS) supplemented with 4.5 g/L glucose and 5 mM MgCl2. Binding buffer was prepared with D-PBS supplemented with 4.5 g/L glucose, 5 mM MgCl2, 0.1 mg/mL yeast tRNA, and 1 mg/mL BSA. Buffer A was prepared with phosphate buffered saline (PBS) supplemented with 1 mM of MgCl2, 0.5 μL/mL Tween 20 and 2 mg/mL BSA. Buffer B was prepared with phosphate buffered saline (PBS) supplemented with 1 mM MgCl2 and 0.5 μL/mL Tween 20.
Apparatus
All flow cytometry experiments were analyzed by the Cytek DxP Athena system, USA. The confocal microscopy graphs were imaged by a confocal laser scanning microscope (Carl Zeiss, Germany). CD spectra were analyzed by Bio-Logic MOS-500 CD spectrophotometer (Claix, France). The fluorescence images of mice in vivo were collected by an IVIS Lumina II imaging system (Caliper LifeSicence, USA). The fluorescence signals of the human bladder tissue array were detected using a Panoramic MIDI (3DHISTECH Ltd., Hungary). All PCR-amplified products were high-throughput sequenced using Illumina MiSeq (Sangon Biotech Co., Ltd. Shanghai, China). DNA gel electrophoresis analysis was performed using a ChemiDoc™ XRS Imager (Bio-Rad, USA).
X-As
Firstly, the X-As bead Library was heated at 95 °C for 5 min and cooled to room temperature. After washing buffer treatment, SV-HUC-1 cells were incubated with the pretreated X-As bead Library with slight shocking for 1 h. After the incubation, unbound Library beads were collected in the EP tube (615601, NEST Biological Technology Co., Ltd) and prepared for the following positive selection. SV-HUC-1 cells were then collected and resuspended in 50 μL buffer A.
Secondly, unbound ssDNA Library beads were incubated with 5637 cells for 90 min at room temperature with rotation. cells were then collected and resuspended in 50 μL buffer B. The above two 50 μL resuspensions, one containing the positively selected bound Library beads and the other containing beads that bound in the negative selection, were added to 50 μL of 1 N NaOH at 65 °C for 30 min to cleave the ssDNA from Library beads. The reaction was neutralized by adding 40 μL 2 M Tris–Cl. X-As was then enriched after the centrifugation step to remove the beads.
Thirdly, 15 μL of the cleaved X-As pool from positive selection was aliquoted into each of 2 tubes (tube #1, #2). Similarly, 15 μL of the cleaved ssDNA pool of negative selection was aliquoted into each of 2 tubes (tube #3, #4). A total of 135 μL Buffer B containing the 5637 cell suspension was added to tubes #2 and #3, as well as 135 μL Buffer B containing SV-HUC-1 cell suspension was added to tubes #1 and #4. All tubes were incubated with rotation for 30 min. Cellular pellets in tubes #1, #2, #3, #4 were recovered by centrifugation.
Finally, each fraction (tubes #1–#4) was subjected to PCR using the corresponding barcoded primer pairs. Then, a clear and definitive band was obtained after PCR for 20 cycles. Finally, 80 μL of each of the above PCR products (tube #1–#4) without particles or cells was sequenced by Sangon Biotechnology Co., Ltd.
The sequencing data of tube #3 were processed with the elimination of data from tubes #1, #2, and #4 to form a candidate aptamer pool. After sequence analysis of their homologous similarity using Mega 7.0 software, six representative candidates were chosen and synthesized for further investigation. Six ssDNA aptamers were selected with a high frequency of occurrence from the candidate pool.
Flow cytometry analysis
The binding ability of candidate aptamers to cells was analyzed by flow cytometry. Each cell line (2 × 105 cells) was incubated with 300 nM candidate aptamers or Library in 200 μL of binding buffer at 4 °C for 1 h. These cells were then washed with washing buffer and resuspended in 400 μL of D-PBS for cytometric analysis.
To measure the dissociation constant (Kd) of aptamers, 5637 cells (3 × 105) were incubated with different concentrations of aptamers in 200 μL of binding buffer at 4 °C for 1 h. After incubation, each sample was washed three times with washing buffer and then resuspended in 200 μL of D-PBS for flow cytometry analysis. These experiments for the binding assay were repeated three times. Using GraphPad Prism 8.0 software, the Kd of aptamers was calculated by the equation BmaxX/(Kd + X) (Y: relative fluorescence intensity; X: aptamer concentration), fitting the dependence of the fluorescence intensity of the cell/aptamer complex on the aptamer concentration.
Confocal microscopy imaging
Target cells were seeded at 1 × 105 in a 35-mm glass bottom dish (801004, NEST Biological Technology Co., Ltd) and cultured for 24 h. After washing with cold washing buffer, the cells were incubated with 300 nM FAM-labeled aptamer in 1 mL of binding buffer at 4 °C for 1 h. After washing twice, the cells were imaged by confocal laser scanning microscopy.
CD analysis
CD spectra were obtained in the range of 220–340 nm at 25 °C in a quartz cuvette. A background CD spectrum of D-PBS was subtracted from the average scan.
Aptamer stability
FAM-labeled aptamers 3 μM were incubated in DMEM with 10% FBS (ABW BIOSCIENCE) for different time periods (0, 1, 2, 3, 4, 6, 8, 10, 12, 18, and 24 h) at 37 °C. At each time point, samples were flash frozen in a dry ice/ethanol bath and then stored at −80 °C. All samples were thawed on ice and run on 3% agarose gels.
Analyzing internalization and uptake mechanism
First, each sample of the 5637 cell line (2 × 105 cells) was washed three times with washing buffer. To analyze the internalization and uptake mechanism, 5637 cells were pretreated with 10 mM M-β-CD for 0.5 h, 3 mM amiloride for 0.5 h, 10 mg/mL chlorpromazine for 0.5 h, and 5 mM cytochalasin D for 0.5 h. Finally the 5637 cells were incubated with 250 nM FAM-labeled BC-3 at 4 °C or 37 °C for 1 h. After incubation with BC-3, the samples were tested by flow cytometry and confocal microscopy.
In vivo fluorescence imaging
Male BALB/c-nude mice (5–6 weeks old) were purchased from Changsha SLAC Animal Laboratory and raised in SPF conditions. All animal experiments were performed with the approval of the Laboratory Animal Center of Hunan University. A total of 5 × 106 5637 cells were implanted in the subcutaneous tissue of the mouse backside. After tumors reached 0.5–1.5 cm in diameter at a time period of 15–20 days, these tumor-bearing mice were anesthetized with isoflurane and intravenously injected with 100 μL of 4.5 nM Cy5-labeled BC-3 or Library (three mice for each group). After fluorescence imaging analysis at 0.5 h postinjection, tumor-bearing mice injected with Cy5-labeled BC-3 or Library were sacrificed and dissected. Their tumor tissues and visceral organs, including the kidneys, liver, stomach, intestine, heart, testis, and prostate, were then imaged.
Fluorescence imaging of clinical pathological section slides
A bladder tissue array (HBlaU050CS01, Shanghai OUTDO Biotech Co., Ltd) was preheated at 60 °C for 2 h and then deparaffinized in xylene (15 min, twice). Tissue sections were then immersed in decreasing ethanol concentrations (100%, 95%, 90%, 80%, and 70%) at 5-min intervals. The hydrated tissues were pretreated in 0.01 M citrate buffer at pH 6.0 and heated in a pressure cooker for 20 min. Afterward, tissue sections were blocked with precooled binding buffer, 20% FBS, and 0.1 mg/mL salmon sperm DNA for 1 h at room temperature and incubated with a 300 nM Cy5-labeled Library or BC-3 for 1 h. The fluorescence intensity of every case was calculated and evaluated by Image J (Version 1.53c) as negative (−, <25.0), weak (+, 25–28.5), moderate (++, 28.5–32.0), or strong (+++, >32.0).
Target protein identification by mass spectrometry (MS) analysis
The binding target of BC-3 was investigated with a similar protocol that was reported in a previous study.23 The plasma membrane proteins of 5637 cells were extracted by the Membrane and Cytosol Protein Extraction Kit (P0033, Beyotime, China). The different protein bands between the BC-3 sample lane and Library sample lane were excised for digestion and analyzed by liquid chromatography (LC)-MS/MS (APTBIO Co., Ltd, China).
Western blot analysis of RPS7 pulled down by aptamer
Total Proteins were extracted by RIPA buffer (WB3100, New Cell & Molecular Biotech Co. Ltd) with Protease Inhibitor Cocktail (4,693,159,001, Roche). Protein samples were separated using 10% SDS-PAGE (P2012, New Cell & Molecular Biotech Co. Ltd) and then transferred to PVDF membranes (Merck Millipore Ltd, ISEQ00010). PVDF membranes were then separately incubated with anti-RPS7 rabbit polyclonal antibody (D127523, Sangon Biotechnology Co., Ltd.) and anti-β-Actin antibody (MA5-11869, Thermo Fisher) at 4 °C overnight. After washing three times with Tris-buffered saline with 0.1% Tween 20 (TBST), the membrane was incubated with goat anti-rabbit secondary antibody (D111018, Sangon Biotechnology Co., Ltd.). The signals were detected with Western Bright™ ECL (K-12045-D50, Advansta) and imaged by a ChemiDoc™ XRS Imager (Bio-Rad, USA).
Colocalization experiment
5637 cells (3 × 105) were seeded into a 35-mm optical vessel. After incubation at 37 °C for 24 h, 5637 cells were treated with 250 nM FAM-labeled aptamer BC-3. The supernatant was removed after incubation for 1 h and 5637 cells was added 200 nM of ER-tracker (C1041, Beyotime) solution or lysosome-tracker (C1046, Beyotime) separately at 37 °C for 30 min. Fluorescence signals were detected by Zeiss LSM510. All experiments were repeated three times.
Molecular docking
Firstly, the protein structure of RPS7 was downloaded from the RCSB PDB databank (PDB ID: 6YBW) with a resolution of 3.1 Å.24 Water molecules and ions in the protein complex were removed from the protein structure. The three-dimensional structure of aptamer BC-3 with the best predicted energy was constructed using the RNA composer software.25,26 After nucleic acid bases T were changed to U, one position-constrained molecular dynamics simulation was performed to eliminate the steric conflicts within the aptamer system.
Secondly, docking simulations were performed with AutoDock software.27,28 The docking protocol began with rigid-body blind docking where all heavy atoms of protein and aptamer systems were strictly restrained. In this step, we randomly placed the protein and aptamer roughly facing each other, and obtained at least 500 docking conformations for further analysis.
Finally, precise docking site prediction was carried out by PyMOL software,29,30 based on the best docking conformation of the RPS7-BC-3 complex. Amino acids forming hydrogen bonds between the protein and aptamer in each docking conformation were collected. The amino acid with a high ratio (> 0.4) that participated in H-bonds was selected as a potential binding site. Binding spots with distances beyond 4 Å were manually deleted and thus simplified to final binding spots.
Bioinformatic analysis
The analysis of the expression or methylation levels of RPS7 in BC was performed using data from the TCGA database (UALCAN, http://ualcan.path.uab.edu).31 The filter indices were set as differential analysis between cancer and normal specimens, individual cancer stages and sexes.31
Statistics
The statistical analysis was implemented by GraphPad Prism 8.0. Statistical significance was determined via t-tests; ∗P < 0.05 was considered significant.
Results
Generation and characterization of a new aptamer
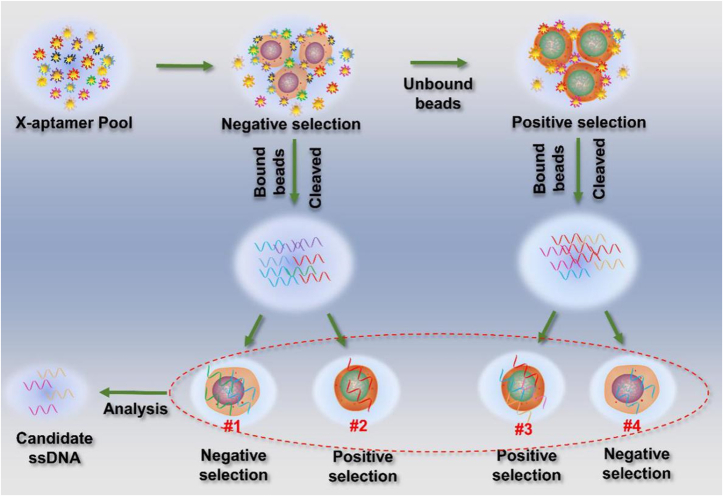
In general, ssDNA aptamers are generated by a selection process known as SELEX. The selection process begins with a synthesized random ssDNA library. After several rounds of selection and amplification, target-specific aptamers are enriched and identified by high-throughput sequencing analysis. However, to rapidly and accurately obtain specific aptamers against human bladder cancer cells, we adopted a novel two-round strategy to obtain specific aptamers from an optimized sequence pool, X-As. The detailed selection process for a new aptamer is illustrated in Figure 1. The gel electrophoresis analysis of different PCR samples before sequencing is shown in Figure S1A. The detailed optimization of the cycle number for PCR is shown in Figure S1B. Six sequences with the highest frequency were selected from different families after bioinformatic analysis (Fig. S2). The analyses from flow cytometry and confocal microscopy imaging for each sequence indicate that BC-3 has the strongest binding ability to 5637 cells among them (Fig. 2A, B), and no sequences, including BC-3, bind to SV-HUC-1 cells (Fig. 2C, D). From the geometric mean fluorescence (GMF) analysis, FITC-labeled BC-3 showed significant binding signals to 5637 cells, but not to SV-HUC-1 cells (Fig. 2E), which also confirmed that BC-3 can distinguish 5637 cells from SV-HUC-1 cells.
Figure 1.
The overall synthetic approach of the X-As process.
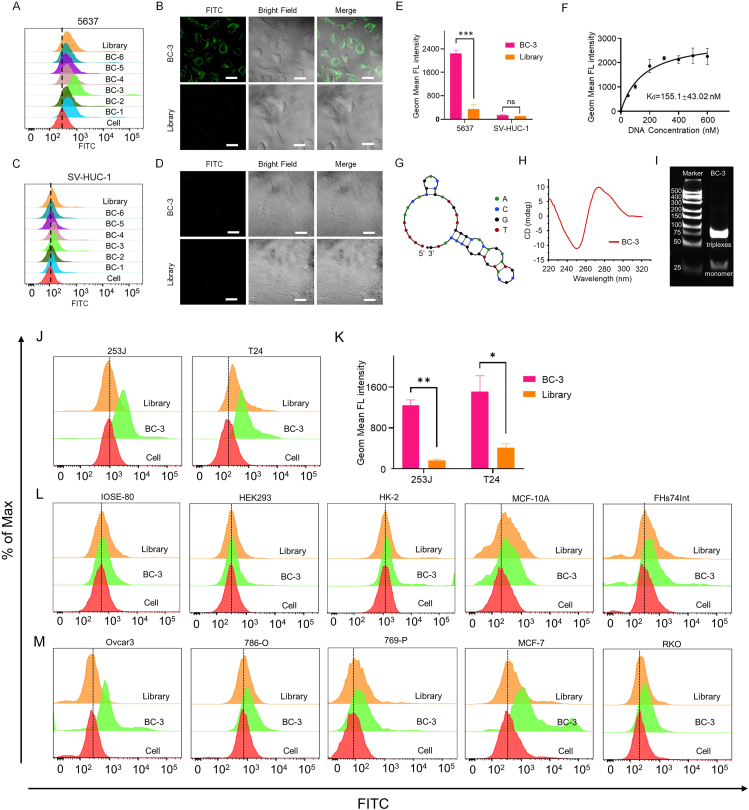
Figure 2.
Characterization and specific binding of aptamer BC-3. 5637 cells (A) and SV-HUC-1 cells (C) were incubated with FITC-labeled BC-1 to BC-6 at 4 °C, with Library or cells only used as a control. The binding ability was analyzed by flow cytometry. The FITC-labeled BC-3 (250 nM) binding to 5637 cells (B) and SV-HUC-1 (D) was displayed by confocal microscopy, with Library used as a control. Images were captured by confocal microscopy. (E) Statistical evaluation of flow cytometry results (∗∗∗P < 0.005; ns, not significant). (F) The dissociation constant of BC-3 for 5637 cells was analyzed by flow cytometry. (G) Predicted secondary structure of aptamer BC-3. (H) CD spectra of BC-3. (I) 12% PAGE gel electrophoresis analysis of BC-3 (scale bar = 40 nm). (J) Affinity of BC-3 to cell lines 253J and T24 was analyzed by flow cytometry. (K) Statistical evaluation of flow cytometry results (Comparison: BC-3 vs. Library: ∗P < 0.05, ∗∗P < 0.01). (L) BC-3 binding to various normal cell lines was analyzed by flow cytometry. (M) BC-3 cells recognizing with the corresponding cancer cell lines were analyzed by flow cytometry.
The equilibrium dissociation constants (Kd) were obtained using equation Y = BmaxX/(Kd + X), fitting the dependence of the fluorescence intensity of samples on the concentration of the aptamers. As shown in Figure 2F, the Kd of BC-3 for 5637 cells was 155.1 ± 43.02 nM. The secondary structure of BC-3 was predicted by the Nucleic Acid Package (NUPACK),32 with four stem loops and one hairpin (Fig. 2G). Circular dichroism (CD) spectral analysis showed that the main positive band of BC-3 was at 275 nm, while the negative band was at 250 nm, as shown in Figure 2H. Consistent with the CD spectrum features of oligonucleotides triplex,33,34 BC-3 may form Pu∗PuPy triplexes in D-PBS solution. Furthermore, gel electrophoresis analysis showed a bright band at 75 bp and a weak band at ∼30 bp, which indicated the triplexes and the monomer of BC-3, respectively (Fig. 2I).
Binding affinity and specificity to other cancer cells
To validate the binding specificity of BC-3 against tumor cells, we incubated various cell lines with FITC-labeled BC-3 separately, and then measured fluorescence signals by flow cytometry individually. By using flow cytometry and statistical analysis of GMF intensity, BC-3 also had high affinity for other BC cell lines such as 253J and T24 (Fig. 2J, K). BC-3 could not recognize normal cell lines, including IOSE-80, HEK293, HK-2, MCF-10A, and FHs74Int, as shown in Figure 2L. BC-3 had a high binding ability to ovarian cancer cells Ovcar3 and breast cancer cells MCF-7, while had low binding ability to 786-O, and 769-P renal cancer cells and RKO colon cancer cells (Fig. 2M). This suggests that BC-3 can be used for detecting bladder cancer cells using clinical urine samples, as it does not bind to kidney cancer cells which may interfere with urine cytology.35,36
Internalization and endocytosis pathways of BC-3
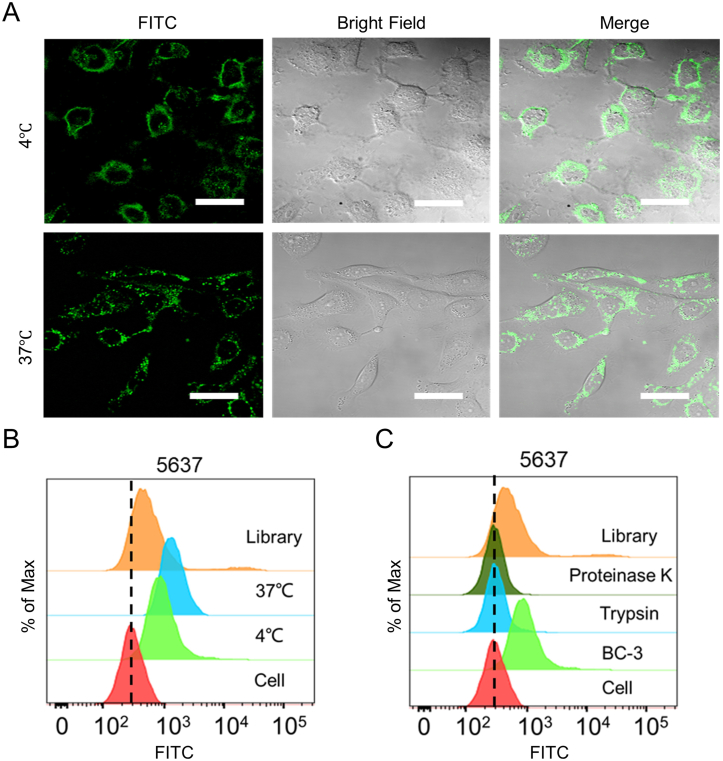
We next used confocal microscope imaging to examine the internalization of BC-3. After 1 h of incubation, fluorescence signals from FITC-labeled BC-3 were mainly displayed inside the membrane of 5637 cells at 4 °C, but dispersed in the cytoplasm at 37 °C (Fig. 3A). The results of flow cytometry analysis also showed that fluorescence signals were stronger at 37 °C than at 4 °C (Fig. 3B), which suggests that BC-3 may be internalized into cells energy dependently via surface receptors.37 Moreover, BC-3 completely lost its binding ability to 5637 cells after individual treatment with trypsin or proteinase K for 2 min (Fig. 3C), suggesting that the receptor type of BC-3 is a membrane protein.
Figure 3.
Internalization of BC-3 in 5637 cells. 5637 cells were incubated with FITC-labeled BC-3 at 4 °C or 37 °C, and images were captured by (A) confocal microscopy and (B) flow cytometry (C) After treatment with proteinase K, trypsin, or EDTA, the binding of BC-3 to 5637 was analyzed by flow cytometry (scale bar = 40 μm).
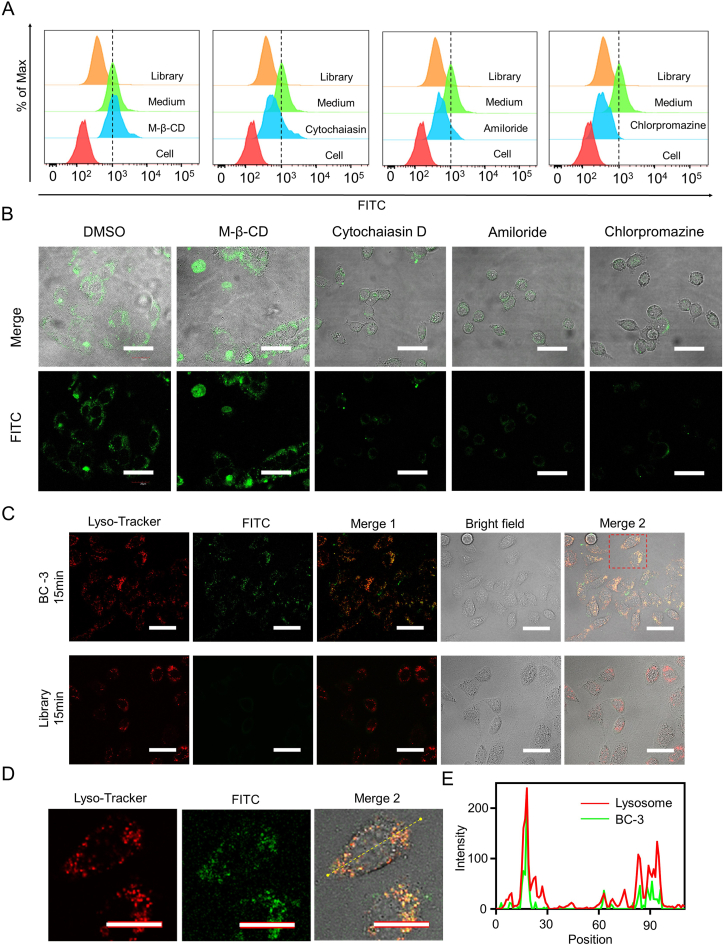
Aptamers can be internalized into cells via receptor-mediated endocytosis pathways,38 including micropinocytosis, clathrin-mediated endocytosis and caveolae-mediated endocytosis. To explore the internalization mechanism of BC-3, we pretreated 5637 cells with M-β-CD to block caveolae function and then incubated them with BC-3 but detected no significant change in fluorescence signals by flow cytometry. After pretreatment with cytochalasin D to inhibit actin polymerization, 5637 cells were incubated with BC-3 and showed decrease in fluorescence signals, demonstrating that actin is involved in the internalization of BC-3. Furthermore, after pretreatment with amiloride or chlorpromazine and incubation with BC-3, 5637 cells displayed a significant decrease in fluorescence signals, indicating that the blockage of either micropinocytosis or the clathrin-mediated pathway can inhibit the cellular uptake of BC-3 (Fig. 4A). These results were further confirmed by confocal images (Fig. 4B).
Figure 4.
Endocytosis pathway analysis of BC-3 in 5637 cells. After 5637 cells were separately treated with DMSO, M-β-CD, cytochalasin D, amiloride, or chlorpromazine, fluorescent signals from the uptake level of BC-3 to 5637 were analyzed by (A) flow cytometry and (B) confocal microscopy. (C) After incubating in 5637 cells for 15 min at 37 °C, co-localization (yellow) of LysoTracker (red) and FITC (green, FITC-labeled BC-3 or FITC-labeled Library) was observed, Library as a control. (D) Enlarged view of intracellular fluorescence signals in colocalization. (E) Fluorescence intensity profile of yellow arrow regions in 5637 cells (Scale bar = 40 μm).
In general, aptamers can form aptamer–receptor complexes and fuse with lysosomes after micropinocytosis or clathrin-mediated endocytosis.39 After LysoTracke pretreatment, 5637 cells were incubated with FITC-labeled BC-3 or FITC-labeled Library for 15 min, and the former displayed overlapping yellow fluorescent signals but the latter did not (Fig. 4C). Intensity profiles with FITC-labeled BC-3 and LysoTracker overlapping were amplified (Fig. 4D), and their Pearson's coefficient (PC) was 0.81 (Fig. 4E). Taken together, BC-3 can be internalized to lysosomes via micropinocytosis or clathrin-mediated endocytosis (Fig. S3).
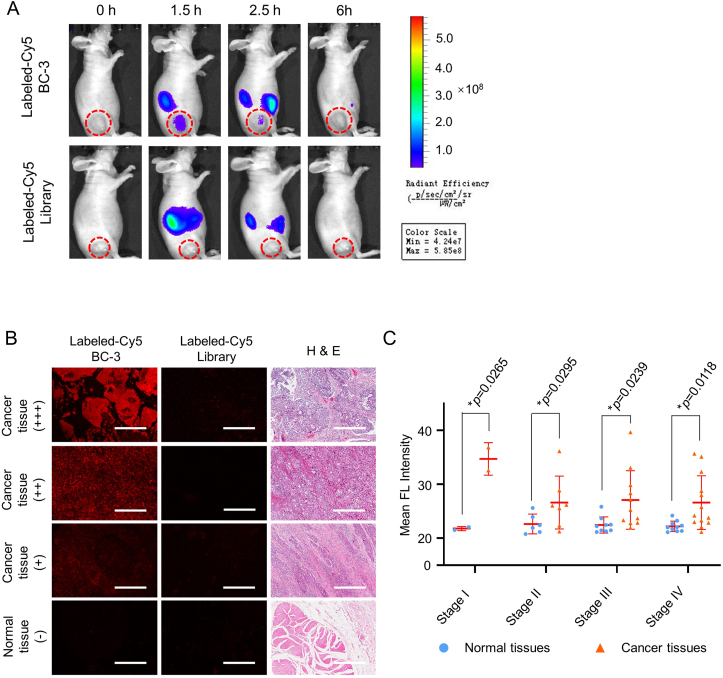
In vivo fluorescence imaging of BC-3
The stability of the aptamer in blood circulation is important for in vivo application. We evaluated the stability of BC-3 by incubating it in pure serum aliquots at 37°C for different time points (Fig. S4A). By using agarose gel electrophoresis, the half-life of BC-3 was calculated to be 5.7 ± 3.5 h (Fig. S4B). To explore the in vivo recognition of BC-3 for BC, we performed tumor imaging with the tail vein injection of Cy5-labeled BC-3 or Cy5-labeled Library into xenografted mice. As shown in Figure 5A, fluorescence signals were accordingly captured at 0, 1.5, 2.5, and 6 h postinjection. After the injection of Cy5-labeled BC-3, fluorescence signals reached the peak in the tumor site at 1.5 h postinjection, then decreased at 2.5 h postinjection and disappeared at 6 h postinjection, indicating that BC-3 can accumulate at the tumor site within its half-life and be further cleared via metabolism (Fig. 5A, upper). In contrast, fluorescence signals were not detected at the tumor site after the injection of Cy5-labeled Library (Fig. 5A, lower), suggesting that Library cannot target tumor sites.
Figure 5.
Fluorescence imaging of BC-3 in xenografted mouse models and clinical BC tissue samples. (A) BC-bearing xenografted mice were intravenously injected with Cy5-labeled BC-3 (upper panel) or Cy5-labeled Library (lower panel). Fluorescence images were taken at different time points by an in vivo imaging system. (B) BC tissues and normal tissues at different stages were incubated with Cy5-labeled BC-3 or Library. Fluorescence signals were detected with panoramic scans (scale bar = 400 μm). (C) Summary statistics of the relative fluorescence intensity of normal bladder and BC tissues (Comparison: Normal tissues vs. Cancer tissues: ∗P < 0.05).
The biodistribution of Cy5-labeled BC-3 or Cy5-labeled Library in xenografted mice was also examined 0.5 h post-injection. Compared to Cy5-labeled Library injection, the fluorescence signals from the mice injected with Cy5-labeled BC-3 accumulated more in tumor and less in the kidneys, suggesting that BC-3 can be finally metabolized and eliminated by the kidneys after accumulating at the tumor site (Fig. S5). Moreover, CCK-8 assays for the cytotoxicity of the BC-3 to BC cell line 5637 or the bladder normal cell line SV-HUC-1 indicated that BC-3 had good biocompatibility (Fig. S6).
Clinical tissue imaging of bladder cancer with BC-3
To test the clinical application potential of BC-3, we stained 60-core tissue arrays, containing 33 BC cases and 27 normal bladder cases, with Cy5-labeled BC-3 or Cy5-labeled Library respectively. As shown in Figure 5B, dividing the fluorescence values into four levels, 57.6% of total bladder cancer tissues had positive fluorescence signals (Table S3), consisting of 100% in stage I, 71.4% in stage II, 54.6% in stage III, 46.2% in stage IV, and 7.4% in normal bladder tissues (Table S4). We also performed a statistical analysis of the mean fluorescence intensity from normal bladder and BC tissues at different stages and found that four intragroup comparisons of the P value were below 0.05 (Fig. 5C). These results indicate that BC-3 can effectively distinguish BC clinical specimens from normal bladder.
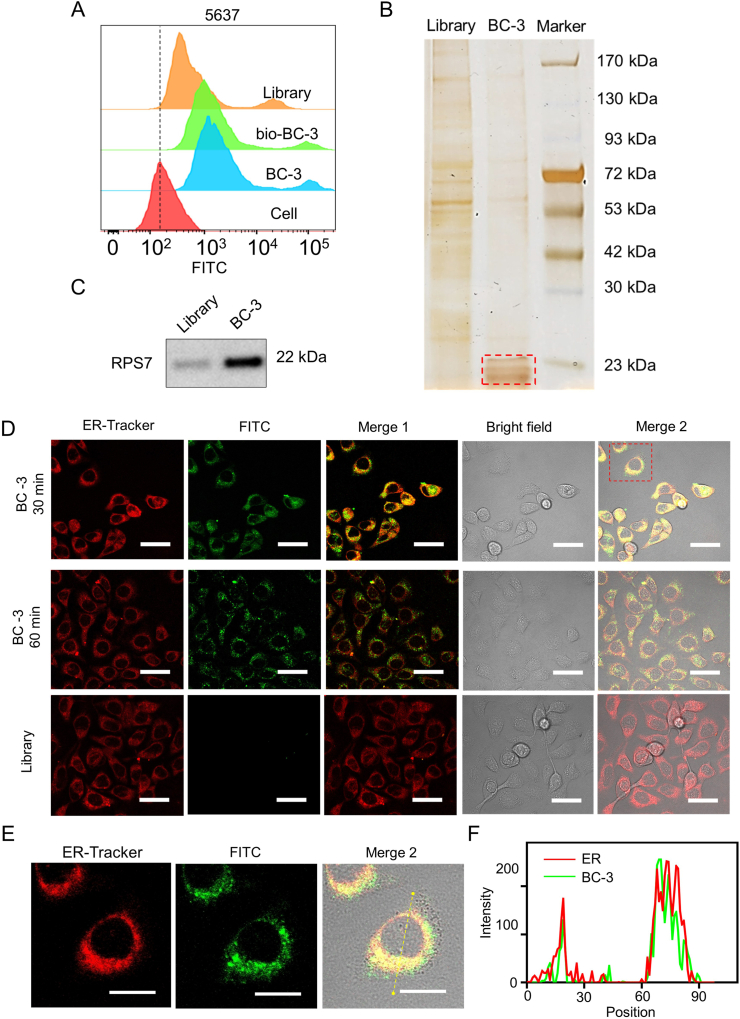
Target identification of BC-3
To investigate the target protein of BC-3, we used quantitative proteomic analysis as previously reported.23 We confirmed that biotin modification of BC-3 did not affect its binding to 5637 cells via a flow cytometry assay (Fig. 6A). After we incubated total membrane proteins with biotin-labeled BC-3 or biotin-labeled Library separately, potential target proteins in 5637 cells were captured by streptavidin-coated Sepharose beads. Protein bands with a molecular weight of ∼23 kDa were obvious in BC-3 treated cells, but not in Library treated cells (Fig. 6B). After those bands were cut, trypsin-digested and identified by LC-MS/MS analysis, we collected representative peptides of these proteins (Fig. S7) and a list of protein hits (Table S5). According to the ratio of proteins captured by aptamer BC-3 to Library, the membrane protein RPS7 is the top-ranked target hit for BC-3 with a relatively high score and coverage (Fig. S8). To confirm this speculation, we carried out Western blot assays with the anti-RPS7 antibody and found that one capture from total protein samples of 5637 cells by BC-3 was specifically recognized by the anti-RPS7 antibody, but a relatively weak band was detected in the other capture by Library (Fig. 6C). The results showed that RPS7 is likely the binding target of aptamer BC-3.
Figure 6.
BC-3 target proteins analyzed by mass spectrometry. (A) The binding of biotin- and FITC-labeled BC-3 to 5637 cells was measured by flow cytometry, with biotin- and FITC-labeled Library as a control. (B) SDS-PAGE with silver staining was used to identify the aptamer target. Lane library, protein captured with biotin-labeled library treated sample; Lane BC-3, protein captured with biotin-labeled BC-3 treated sample. (C) Western blot analysis of the concentrations of RPS7 and β-actin in the preparations captured via Library and BC-3. (D) After incubating the ER fluorescence probe ER-tracker (red) for 0.5 h and FITC-labeled BC-3 or Library (green) for 1 h, 5637 cells displayed the colocalization (yellow) of ER-tracker and FITC-labeled BC-3. (E) Enlarged view of intracellular fluorescence signals in colocalization. (F) Fluorescence intensity profile of yellow arrow regions in 5637 cells (scale bar = 40 μm).
Considering that RPS7 is located mainly in the rough endoplasmic reticulum (ER) and partially in the plasma membrane, we then examined whether BC-3 was localized at the ER. After incubation with ER-tracker (red) for 30 min and FITC-labeled BC-3 (green) for 1 h, 5637 cells displayed overlapping yellow fluorescent signals (Fig. 6D). Intensity profiles with FITC-labeled BC-3 and ER-tracker signal overlap were amplified (Fig. 6E), and their Pearson's coefficient (PC) was 0.83 (Fig. 6F). Taken together, these results indicate that BC-3 can be located at the ER in 5637 cells (Fig. S3).
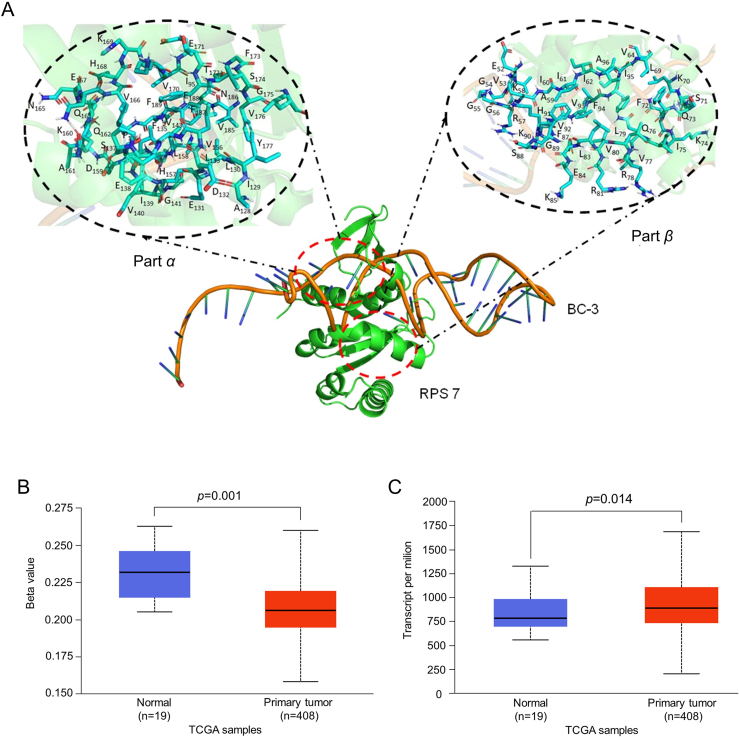
Computational modeling of the binding between RPS7 and BC-3
In our preliminary docking process, a total of 500 binding conformations between RPS7 and BC-3 were established and ranked by docking energy. Among them, 10 conformations with relatively low binding energy were used to analyze their binding area, considering the amino acid residues in RPS7 that the linked with BC-3 conformations as binding sites. Multiple conformation superposition analysis indicated that binding sites between RPS7 and BC-3 overlapped in certain areas. Their binding interface was mainly located at two regions, area α and area β (Fig. 7A). Detailed binding spot analysis showed that BC-3 may bind with all 81 residues in RPS7 (41 residues in part α and 38 residues in part β). Polar interactions were correlated with stabilizing the formation by nonbonded electrostatic interactions. Consequently, 58 binding sites (27 residues in part α and 31 residues in part β) with a relatively high hydrogen bond ratio (r > 0.4) were selected as BC-3 binding residues for further precise docking (Fig. S9). Taken together, computational analysis of the RPS7-binding mode of BC-3 provided in-depth molecular insights into their spatial binding areas.
Figure 7.
Computational analysis of the Binding Model between RPS7 and BC-3. (A) Interaction model between RPS7 and BC-3. Close view of the RPS7 and BC-3 interaction region in part α and part β with key amino acid residues marked. Promoter methylation (B) and the expression level (C) of RPS7 in bladder cancer and normal bladder samples from TCGA.
External dataset analysis of RPS7 expression levels in BC patients
To determine the clinical expression status of RPS7 in BC, we analyzed its expression in BC tumor tissues and normal bladder tissues, using data from The Cancer Genome Atlas (TCGA), a comprehensive “atlas” of cancer genomic profiles.40, 41, 42 After data mining and analysis from TCGA datasets of 408 BC tissues and 19 normal bladder tissues, we found that RPS7 showed a significantly lower methylation level (P = 0.001, Fig. 7B) and a higher expression level (P = 0.014, Fig. 7C) in BC than in normal bladder tissues. These results indicated that RPS7 is a potential biomarker for bladder cancer diagnosis.
Discussion
Here, we identify a novel DNA aptamer BC-3 that can specifically bind bladder cancer cells by a two-round X-As strategy. Aptamer BC-3 can distinguish bladder cancer from renal cancer, which makes BC-3 a potential molecular probe for urine cytology detection. Additionally, BC-3 can specifically recognize BC clinical patient tissues and accumulate at tumor sites in xenograft mouse models. Furthermore, BC-3 can fold into Pu∗PuPy triplexes in D-PBS solution. Different from the base-paired duplex structures, BC-3's triplex forming oligonucleotide (TFO) may be formed by the interaction of an auxiliary single strand with double-helix DNA through Hoogsteen hydrogen bond interactions. BC-3 triplex structures can be developed in DNA nanotechnology and nucleic acid-based functional materials.
Currently, the correlation of ribosomal proteins (RP) with the early diagnosis and development of tumors is attracting more attention. Abnormal expression of ribosomal proteins can induce the occurrence of diseases such as tumors.43 The knockout of highly expressed ribosomal protein S6K (RPS6KB1) in prostatic cancer (PCa) cells was found to increase PCa sensitivity to radiation, which indicated RPS6KB1 to be a new target for PCa drug-assisted radiotherapy.44 We revealed RPS7 as the cellular target of BC-3 via mass spectrometry analysis. A recent study showed that RPS7 plays an important role in the migration of cancer cells through the P53 pathway or other mechanisms.45 We further established a binding model for RPS7 and BC-3 with predicted binding sites via computational simulation docking. The analysis of TCGA datasets on promoter methylation and the expression level of RPS7 showed that RPS7 is expressed at higher levels in BC patient tumor tissues than in normal tissues, which suggests the application potential of RPS7 as a diagnostic biomarker and a therapeutic target for bladder cancer.
Interestingly, BC-3 can quickly avoid being degraded by lysosomes after its intracellular internalization, while most other aptamers are fused with lysosomes, and then sorted for recycling back to the plasma membrane or delivered to late endosomes and lysosomes for degradation. BC-3 can further relocate to ER with the assistance of its target RPS7, which gives the uniqueness for the application of BC-3 in the field of chemo-drug target delivery with an improved therapeutic efficiency and reduced systematic side effects. Considering the high expression level of RPS7 in 5637 bladder cancer and TCGA data based on patient sex and individual cancer stages, further investigation of the mechanism and function of RPS7 in the occurrence and development of bladder cancer will be facilitated. Therefore, our findings also provide new opportunities to accelerate the discovery of novel therapeutic and diagnostic strategies for bladder cancer.
Conflict of interests
The authors have no competing interests to declare.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31970692) (X. Hu), and the Corbett Estate Fund for Cancer Research (USA) (No. 62285-531021-41800, 62285-531021-51800, 62285-531021-61800, and 62285-531021-71800) (E. Wu).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.07.002.
Contributor Information
Xiaoxiao Hu, Email: xxhu@hnu.edu.cn.
Erxi Wu, Email: erxi.wu@bswhealth.org.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Wu S., Chen X., Pan J., et al. An artificial intelligence system for the detection of bladder cancer via cystoscopy: a multicenter diagnostic study. J Natl Cancer Inst. 2022;114(2):220–227. doi: 10.1093/jnci/djab179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamat A.M., Hahn N.M., Efstathiou J.A., et al. Bladder cancer. Lancet. 2016;388(10061):2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhou G., Latchoumanin O., Hebbard L., et al. Aptamers as targeting ligands and therapeutic molecules for overcoming drug resistance in cancers. Adv Drug Deliv Rev. 2018;134:107–121. doi: 10.1016/j.addr.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Madsen M., Gothelf K.V. Chemistries for DNA nanotechnology. Chem Rev. 2019;119(10):6384–6458. doi: 10.1021/acs.chemrev.8b00570. [DOI] [PubMed] [Google Scholar]
- 6.Röthlisberger P., Hollenstein M. Aptamer chemistry. Adv Drug Delivery Rev. 2018;134:3–21. doi: 10.1016/j.addr.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Wang J., Zhang Y., Chen Y., et al. In vitro selection of DNA aptamers against renal cell carcinoma using living cell-SELEX. Talanta. 2017;175:235–242. doi: 10.1016/j.talanta.2017.07.049. [DOI] [PubMed] [Google Scholar]
- 8.Wang H., Li X., Lai L.A., et al. X-aptamers targeting Thy-1 membrane glycoprotein in pancreatic ductal adenocarcinoma. Biochimie. 2021;181:25–33. doi: 10.1016/j.biochi.2020.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu E., Elizondo-Riojas M.A., Chang J.T., Volk D.E. Aptaligner: automated software for aligning pseudorandom DNA X-aptamers from next-generation sequencing data. Biochemistry. 2014;53(22):3523–3525. doi: 10.1021/bi500443e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diego I., Suntravat M., Lucena S.E., et al. The design of an X-aptamer against snake venom disintegrins. Toxicon. 2019;158:S51. [Google Scholar]
- 11.Walss-Bass C., Lokesh G.L.R., Dyukova E., et al. X-aptamer technology identifies C4A and ApoB in blood as potential markers for schizophrenia. Mol Neuropsychiatry. 2019;5(1):52–59. doi: 10.1159/000492331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H., Wang Z., Xie L., et al. Molecular recognition and in-vitro-targeted inhibition of renal cell carcinoma using a DNA aptamer. Mol Ther Nucleic Acids. 2018;12:758–768. doi: 10.1016/j.omtn.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nimjee S.M., White R.R., Becker R.C., Sullenger B.A. Aptamers as therapeutics. Annu Rev Pharmacol Toxicol. 2017;57:61–79. doi: 10.1146/annurev-pharmtox-010716-104558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon S., Rossi J.J. Aptamers: uptake mechanisms and intracellular applications. Adv Drug Deliv Rev. 2018;134:22–35. doi: 10.1016/j.addr.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., Ou H., Pei X., et al. Chemo-drug controlled-release strategies of nanocarrier in the development of cancer therapeutics. Curr Med Chem. 2021;28(31):6307–6322. doi: 10.2174/0929867327666200605153919. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y., Peng C., Zhang H., et al. DNA aptamer S11e recognizes fibrosarcoma and acts as a tumor suppressor. Bioact Mater. 2021;12:278–291. doi: 10.1016/j.bioactmat.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen Y., An Z., Qiao B., Zhang C., Zhang Z. RPS7 promotes cell migration through targeting epithelial-mesenchymal transition in prostate cancer. Urol Oncol. 2019;37(5):297. doi: 10.1016/j.urolonc.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Wu L., Kou F., Ji Z., et al. SMYD2 promotes tumorigenesis and metastasis of lung adenocarcinoma through RPS7. Cell Death Dis. 2021;12(5):439. doi: 10.1038/s41419-021-03720-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C., Qie Y., Yang T., et al. Kinase PIM1 promotes prostate cancer cell growth via c-Myc-RPS7-driven ribosomal stress. Carcinogenesis. 2019;40(1):202. doi: 10.1093/carcin/bgz030. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z., Hou J., Lu L., et al. Small ribosomal protein subunit S7 suppresses ovarian tumorigenesis through regulation of the PI3K/AKT and MAPK pathways. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0079117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y., Poyurovsky M.V., Li Y., et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35(3):316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen Y., An Z., Qiao B., Zhang C., Zhang Z. RPS7 promotes cell migration through targeting epithelial-mesenchymal transition in prostate cancer. Urol Oncol. 2019;37(5):297. doi: 10.1016/j.urolonc.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Wu X., Liu H., Han D., et al. Elucidation and structural modeling of CD71 as a molecular target for cell-specific aptamer binding. J Am Chem Soc. 2019;141(27):10760–10769. doi: 10.1021/jacs.9b03720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brito Querido J., Sokabe M., Kraatz S., et al. Structure of a human 48 S translational initiation complex. Science. 2020;369(6508):1220–1227. doi: 10.1126/science.aba4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popenda M., Szachniuk M., Antczak M., et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40(14):e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antczak M., Popenda M., Zok T., et al. New functionality of RNAComposer: an application to shape the axis of miR 160 precursor structure. Acta Biochim Pol. 2016;63(4):737–744. doi: 10.18388/abp.2016_1329. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Sanner M.F. AutoDock CrankPep: combining folding and docking to predict protein-peptide complexes. Bioinformatics. 2019;35(24):5121–5127. doi: 10.1093/bioinformatics/btz459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodsell D.S., Sanner M.F., Olson A.J., Forli S. The AutoDock suite at 30. Protein Sci. 2021;30(1):31–43. doi: 10.1002/pro.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan S., Chan H.C.S., Hu Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip Rev Comput Mol Sci. 2017;7(2):e1298. [Google Scholar]
- 30.Lu X.J. DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL. Nucleic Acids Res. 2020;48(13):e74. doi: 10.1093/nar/gkaa426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zadeh J.N., Steenberg C.D., Bois J.S., et al. NUPACK: analysis and design of nucleic acid systems. J Comput Chem. 2011;32(1):170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 33.Sun X.G., Cao E.H., He Y.J., Qin J.F. Spectroscopic comparison of different DNA structures formed by oligonucleotides. J Biomol Struct Dyn. 1999;16(4):863–872. doi: 10.1080/07391102.1999.10508298. [DOI] [PubMed] [Google Scholar]
- 34.He Y., Scaria P.V., Shafer R.H. Studies on formation and stability of the d[G(AG)5]∗ d[G(AG)5]. d[C(TC)5] and d[G(TG)5]∗ d[G(AG)5]. d[C(TC)5] triple helices. Biopolymers. 1997;41(4):431–441. doi: 10.1002/(SICI)1097-0282(19970405)41:4<431::AID-BIP7>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 35.Vap L.M., Shropshire S.B. Urine cytology: collection, film preparation, and evaluation. Vet Clin North Am Small Anim Pract. 2017;47(1):135–149. doi: 10.1016/j.cvsm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Becker G.J., Garigali G., Fogazzi G.B. Advances in urine microscopy. Am J Kidney Dis. 2016;67(6):954–964. doi: 10.1053/j.ajkd.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 37.Reyes-Reyes E.M., Šalipur F.R., Shams M., Forsthoefel M.K., Bates P.J. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac 1 activation. Mol Oncol. 2015;9(7):1392–1405. doi: 10.1016/j.molonc.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K., Sefah K., Tang L., et al. A novel aptamer developed for breast cancer cell internalization. ChemMedChem. 2012;7(1):79–84. doi: 10.1002/cmdc.201100457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reyes-Reyes E.M., Teng Y., Bates P.J. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70(21):8617–8629. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blum A., Wang P., Zenklusen J.C. SnapShot: TCGA-analyzed tumors. Cell. 2018;173(2):530. doi: 10.1016/j.cell.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 41.Travaglino A., Raffone A., Mascolo M., et al. TCGA molecular subgroups in endometrial undifferentiated/dedifferentiated carcinoma. Pathol Oncol Res. 2020;26(3):1411–1416. doi: 10.1007/s12253-019-00784-0. [DOI] [PubMed] [Google Scholar]
- 42.Tomczak K., Czerwińska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. 2015;19(1A):A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W., Nag S., Zhang X., et al. Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med Res Rev. 2015;35(2):225–285. doi: 10.1002/med.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussain S.S., Huang S.B., Bedolla R.G., et al. Suppression of ribosomal protein RPS6KB1 by Nexrutine increases sensitivity of prostate tumors to radiation. Cancer Lett. 2018;433:232–241. doi: 10.1016/j.canlet.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 45.Kang J., Brajanovski N., Chan K.T., Xuan J., Pearson R.B., Sanij E. Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Signal Transduct Target Ther. 2021;6(1):323. doi: 10.1038/s41392-021-00728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.