Abstract
Three asparagine synthetase genes, asnB, asnH, and asnO (yisO), were predicted from the sequence of the Bacillus subtilis genome. We show here that the three genes are expressed differentially during cell growth. In a rich sporulation medium, expression of asnB was detected only during exponential growth, that of asnH was drastically elevated at the transition between exponential growth and stationary phase, and that of asnO was seen only later in sporulation. In a minimal medium, both asnB and asnH were expressed constitutively during exponential growth and in stationary phase, while the expression of asnO was not detected in either phase. However, when the minimal medium was supplemented with asparagine, only the expression of asnH was partially repressed. Transcription analyses revealed that asnB was possibly cotranscribed with a downstream gene, ytnA, while the asnH gene was transcribed as the fourth gene of an operon comprising yxbB, yxbA, yxnB, asnH, and yxaM. The asnO gene is a monocistronic operon, the expression of which was dependent on one of the sporulation sigma factors, sigma-E. Each of the three genes, carried on a low-copy-number plasmid, complemented the asparagine deficiency of an Escherichia coli strain lacking asparagine synthetases, indicating that all encode an asparagine synthetase. In B. subtilis, deletion of asnO or asnH, singly or in combination, had essentially no effect on growth rates in media with or without asparagine. In contrast, deletion of asnB led to a slow-growth phenotype, even in the presence of asparagine. A strain lacking all three genes still grew without asparagine, albeit very slowly, implying that B. subtilis might have yet another asparagine synthetase, not recognized by sequence analysis. The strains lacking asnO failed to sporulate, indicating an involvement of this gene in sporulation.
Asparagine biosynthesis in the gram-positive bacteria has not been studied extensively. We chose Bacillus subtilis as a convenient bacterium for such study, since it is able to grow well in minimal media without asparagine, implying that it possesses efficient asparagine biosynthesis pathways. In addition, the completion of the genome sequencing of this organism (10) should allow the identification of genes which could be involved in asparagine biosynthesis.
The reactions that are catalyzed by asparagine synthetase use either glutamine or ammonia as a nitrogen source, as follows: l-Asp + ATP + NH3 → l-Asn + AMP + PPi (reaction 1) and l-Asp + ATP + l-Gln → l-Asn + AMP + PPi + l-Glu (reaction 2). To our knowledge, two families of asparagine synthetase have been reported. One is the AsnA family, represented by AsnA of Escherichia coli, whose members were found in prokaryotes such as E. coli and Klebsiella aerogenes (8, 15). Members of the AsnA family are able to use only ammonia as the amino group donor, as in reaction 1. The other is the AsnB family, represented by AsnB of E. coli, whose members were found in both prokaryotes and eukaryotes (7, 18, 20). Members of the AsnB family are able to use both glutamine and ammonia as the nitrogen donor, but glutamine is preferred. E. coli and K. aerogenes have two asparagine synthetase genes, asnA and asnB, and the presence of either ensures sufficient asparagine biosynthesis, while inactivation of both causes asparagine auxotrophy (8, 15).
Analysis of the genome sequence of B. subtilis predicted three genes encoding glutamine-dependent AsnB-type enzymes but no gene for an ammonia-dependent AsnA-type enzyme. The three genes were designated asnB, asnH, and yisO (10); the last gene is referred to as asnO in this paper. We report here that each of the three genes encodes an asparagine synthetase and describe their expression pattern as well as the study of mutants lacking the three genes individually or in combination, revealing a physiological role for asnB in vegetative cells and for asnO in sporulating cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains used in this study are listed in Table 1. Plasmids pOU71 (11), pBEST-4F and pBEST513, pIC156, and pUC19 (23) were provided by Seiichi Yasuda (Cloning Vector Collection, National Institute of Genetics, Mishima, Japan), Mitsuo Itaya (Mitsubishi Kasei Institute of Life Sciences, Tokyo, Japan), Rozenn Dervyn (Institut National de la Recherche Agronomique, Jouy-en-Josas, France), and Takara Shuzo Co., Ltd. (Ohtsu, Japan), respectively. Plasmid pMUTIN2mcs (19) was provided by Valérie Vagner (Institut National de la Recherche Agronomique, Jouy-en-Josas, France). E. coli cells harboring plasmids were grown on following media containing ampicillin (50 μg/ml): Luria broth (LB) (16) and M9 minimal medium (16) supplemented with asparagine-free Casamino Acids (2 mg/ml) (Difco), thiamine (50 μg/ml), thymine (5 μg/ml), and, when required, asparagine (50 μg/ml). B. subtilis cells were grown on the following media containing appropriate antibiotics when needed (see below): tryptose blood agar base (Difco) supplemented with 0.18% glucose (referred as TBABG), DSM (17), and S6 minimal medium (4) supplemented with tryptophan (50 μg/ml), 0.02% Casamino Acids, and, when required, asparagine (S6 plates were prepared by adding 2.0% Noble agar [Difco] containing no nitrogen source).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype | Source or referencea |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | Our standard strain |
| BFS41 | trpC2 ytnA::pMUTIN2mcs | This work (pYTNA1 → 168) |
| BFS55 | trpC2 asnH::pMUTIN2mcs | This work (pASNH1 → 168) |
| BFS56 | trpC2 PasnB::pMUTIN2mcs | This work (pASNB5D → 168) |
| FU339 | trpC2 PasnO::pMUTIN2mcs | This work (pMASNO → 168) |
| FU340 | trpC2 ΔasnB::neo | This work |
| FU341 | trpC2 ΔasnH::spc | This work |
| FU342 | trpC2 ΔasnO::cat | This work |
| FU343 | trpC2 ΔasnB::neo ΔasnH::spc | This work |
| FU344 | trpC2 ΔasnB::neo ΔasnO::cat | This work |
| FU345 | trpC2 ΔasnH::spc ΔasnO::cat | This work |
| FU346 | trpC2 ΔasnB::neo ΔasnH::spc ΔasnO::cat | This work |
| FU347 | trpC2 ytnA::pMUTIN2mcs ΔasnH::spc | This work |
| FU348 | trpC2 ytnA::pMUTIN2mcs ΔasnH::spc ΔasnO::cat | This work |
| ASK201 | trpC2 spoOH::erm | Kei Asai (spoOH::erm → 168) |
| ASK202 | trpC2 spoIIAC::kan | Kei Asai (spoIIAC::kan → 168) |
| ASK203 | trpC2 spoIIGAB::kan | Kei Asai (spoIIGAB::kan → 168) |
| ASK204 | trpC2 spoIIIG::kan | Kei Asai (spoIIIG::kan → 168) |
| ASK205 | trpC2 spoIVCB::erm | Kei Asai (spoIVCB::erm → 168) |
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′[traD36 proAB+ lacIqlacZΔM15] | 23 |
| ME6279 | F−asnA asnB thi thy str recA | 13 |
| C600 | supE44 hsdR thi-1 thr-1 leuB6 lacY1 tonA21 | 24 |
Arrows indicate transformation from donor DNA to recipient strain. Strain 168 is the strain chosen for the international B. subtilis genome functional analysis project. Strains ASK201 to -205 were kind gifts from Kei Asai (Nara Institute of Science and Technology, Ikoma, Japan); each of the mutations of sigma factor genes was transferred to strain 168 from the original mutant strains established by him or by Patrick Stragier (Institut de Biologie Physico-Chimique, Paris, France). The original strains are MO1781, MO719, MO718, UOT-1850 (1), and MO1027, for the sigma-E (spoIIGAB::kan), -F (spoIIAC::kan), -G (spoIIIG::kan), -H (spoOH::erm), and -K (spoIVCB::erm) mutations, respectively.
Construction of recombinant plasmids.
E. coli plasmids pASNB, pASNH, pASNO, and pYXBB, carrying asnB, asnH, asnO, and the genes from yxbB to asnH of B. subtilis, respectively (Fig. 1), were constructed as follows. DNA fragments carrying the entire coding and 5′ flanking regions of the genes were amplified by PCR with specific primer pairs and chromosomal DNA of B. subtilis 168 as a template (Fig. 1). All PCR was done with a GeneAmp XL PCR kit (Perkin-Elmer). The specific primer pairs used were as follows (restriction sites are underlined): for pASNB, asnBupB (5′-CGCGGATCCATAGCCGCTTACTGGTTAAG-3′) and asnBdnB (5′-CGCGGATCCTGGGTAAATCAATGATGATGG-3′); for pASNH, asnHupE (5′-CCGGAATTCTCGTAAATACCCACACTTGG-3′) and asnHdnB (5′-CGCGGATCCATTGCTAATCCCCTAAGTGC-3′); for pASNO, asnOupE (5′-CCGGAATTCTTTCCGTTTCATCCATGCTG-3′) and asnOdnB (5′-CGCGGATCCTCTTATTGAAGGAATGCGGG-3′); and for pYXBB, yxbBupE (5′-CCGGAATTCTACAAGGAAGGAGGGAAAAG-3′) and asnHdnB (5′-CGCGGATCCATTGCTAATCCCCTAAGTGC-3′). The PCR product for the pASNB construction was trimmed with BamHI and then ligated with pOU71 previously digested with BamHI, and each of the other three products was cleaved with EcoRI and BamHI and then ligated with pOU71 previously digested with EcoRI and BamHI. The ligated DNAs were introduced into E. coli JM109 by transformation to give ampicillin resistance on LB plates. Plasmids in the transformants were extracted, and the identity of each of the PCR products cloned into pOU71 was verified by digesting them with various restriction enzymes.
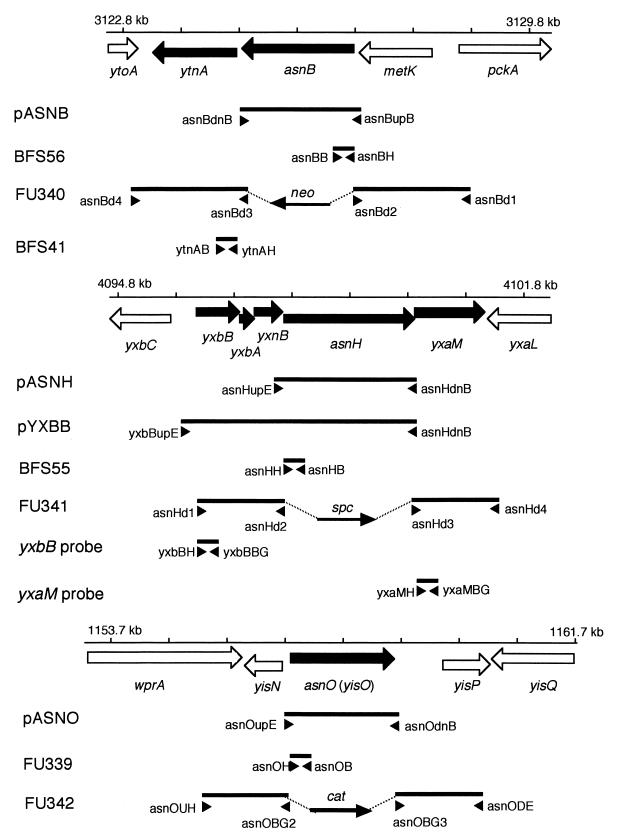
FIG. 1.
Genetic organization of the asnB, asnH, and asnO (yisO) regions and DNA stretches amplified by PCR for plasmid and mutant constructions and probe preparations. The genetic organization of the asnB (top), asnH (middle), and asnO (bottom) regions is shown schematically, and a scale bar with nucleotide positions within the whole genome sequence (10) is given for each of the regions. Genes are shown as thick arrows; solid arrows indicate genes within possible transcription units that contain one of the asparagine synthetase genes. DNA stretches amplified by PCR are shown as solid lines, and names of plasmids, mutants, and probes prepared with the respective PCR products are given on the left. The position and orientation of each specific primer are shown with small arrowheads. The length and orientation of each of antibiotic resistance gene cassette ligated with the PCR products are shown as thinner arrows. The antibiotic resistance genes are abbreviated as follows: cat, chloramphenicol acetyltransferase gene; neo, neomycin resistance gene; spc, spectinomycin resistance gene.
Construction of B. subtilis mutant strains.
B. subtilis BFS41, BFS55, BFS56, and FU339, carrying transcriptional fusions of ytnA, asnH, asnB, and asnO to lacZ, respectively, were constructed as follows. DNA fragments (approximately 300 bp) corresponding to initial parts of each of the genes were amplified by PCR with specific primer pairs and chromosomal DNA of B. subtilis 168 as a template (Fig. 1). The specific primer pairs used for the constructions were as follows (restriction sites are underlined): for BFS41, ytnAH (5′-CCCAAGCTTTAGGGGAGAAGAAGCATG-3′) and ytnAB (5′-CGCGGATCCACCAGTAGTTCCAACCTG-3′); for BFS55, asnHH (5′-CCCAAGCTTCAATAACGCTATTGGGAG-3′) and asnHB (5′-CGCGGATCCTGCTGTCCATTTACAAGG-3′); for BFS56, asnBH (5′-CCCAAGCTTTAGGGGTTCAATGATGAC-3′) and asnBB (5′-CGCGGATCCTCTCTCAGTTCGATATAG-3′); and for FU339, asnOH (5′-CCCAAGCTTGATTGGAGCTGATGTCAC-3′) and asnOB (5′-CGCGGATCCAATGGTGTAGCTATCGCC-3′). Each of the PCR products was trimmed with HindIII and BamHI and was then ligated with pMUTIN2mcs previously digested with HindIII and BamHI. Plasmid pMUTIN2mcs (lacZ lacI amp erm) replicates in E. coli but not in B. subtilis and carries an erythromycin resistance gene that is active in B. subtilis (19). In addition, pMUTIN2mcs carries a promoterless lacZ gene derived from E. coli that can be used as a reporter gene (19). The ligated DNAs were introduced into E. coli C600 by transformation to give ampicillin resistance on LB plates. The identity of each of the PCR products cloned into pMUTIN2mcs was verified by DNA sequencing. The resulting four plasmids, pYTNA1, pASNH1, pASNB5D, and pMASNO, were used to transform B. subtilis 168 to erythromycin (0.3 μg/ml) resistance on TBABG, providing B. subtilis BFS41, BFS55, BFS56, and FU339, respectively. Correct integration of a single copy of each plasmid into the respective genes through a single-crossover event was confirmed by Southern blot analysis. In these strains, each of the target genes was inactivated, and instead lacZ was expressed under the regulation of its upstream sequence.
B. subtilis FU340, FU341, and FU342, lacking asnB, asnH, and asnO, respectively, were constructed as follows. For the construction of strain FU340, two DNA fragments (approximately 2.0 kb) were amplified by PCR with specific primer pairs and chromosomal DNA of B. subtilis 168 as a template; one fragment corresponded to an upstream flanking stretch of the asnB coding region, and the other corresponded to a downstream one (Fig. 1). All PCR was done with a GeneAmp XL PCR kit (Perkin-Elmer). The specific primer pairs used were as follows (restriction sites are underlined): for the upstream fragment, asnBd1 (5′-ATGCCTTCGTTTCGGGAGAG-3′) and asnBd2 (5′-CCGGAATTCCCCTATTTATAGACGCTGTG-3′), and for the downstream fragment, asnBd3 (5′-CGCGGATCCGAGCCATCAGCCTAAAGAAG-3′) and asnBd4 (5′-GGCTCAATCATTTTAGACGG-3′). The upstream and downstream fragments were trimmed with EcoRI and BamHI, respectively, and then ligated with a neomycin resistance cassette derived from pBEST513 previously trimmed with EcoRI and BamHI. DNAs contained in the ligation mixture were used as templates in PCR with a primer pair of asnBd1 and asnBd4 to amplify a tripartitely ligated fragment. The amplified DNA fragment (approximately 5.0 kb) was purified after agarose gel electrophoresis and then used to transform B. subtilis 168 to neomycin (15 μg/ml) resistance on TBABG. In such transformants, the asnB coding region between the upstream and downstream stretches was expected to be deleted and replaced with the cassette through a double-crossover event. The correct replacement of the asnB locus was confirmed by PCR analysis with the primer pair of asnBd1 and asnBd4 and chromosomal DNA of the transformant as a template, which showed that the PCR product obtained was shorter than that from the wild-type locus because of the difference in length between the neomycin cassette and the deleted region (Fig. 1). After this confirmation, one of the transformants was termed FU340.
For the construction of strain FU341, DNA fragments (approximately 1.5 kb) corresponding to upstream and downstream flanking stretches of the asnH coding region were amplified by PCR with specific primer pairs and chromosomal DNA of B. subtilis 168 as a template essentially as described above (Fig. 1). The specific primer pairs used were as follows (restriction sites are underlined): for the upstream fragment of asnH, asnHd1 (5′-GAATGCAGAACGTACAAAG-3′) and asnHd2 (5′-TGCTCTAGACTCCCAATAGCGTTATTG-3′), and for the downstream fragment of asnH, asnHd3 (5′-GACATGCATGCGCCAGAAGGAGCATATAG-3′) and asnHd4 (5′-GGATATGAACTGGTCATTC-3′). The upstream and downstream fragments of asnH were trimmed with XbaI and SphI, respectively, and then ligated with a spectinomycin resistance cassette derived from pIC156 previously trimmed with XbaI and SphI. A tripartitely ligated fragment (approximately 4.2 kb) was amplified with the primer pair of asnHd1 and asnHd4, purified, and then used to transform B. subtilis 168 to spectinomycin (100 μg/ml) resistance to provide strain FU341, the correct construction of which was confirmed by PCR analysis with the primer pair of asnHd1 and asnHd4 as described above.
For the construction of strain FU342, DNA fragments (approximately 1.5 kb) corresponding to upstream and downstream flanking stretches of the asnO coding region were amplified by PCR with specific primer pairs and chromosomal DNA of B. subtilis 168 as a template (Fig. 1). The specific primer pairs used were as follows (restriction sites are underlined): for the upstream fragment of asnO, asnOUH (5′-CCCAAGCTTATGTCACGTACATGAGCG-3′) and asnOBG2 (5′-GGAAGATCTGTGACATCAGCTCCAATC-3′), and for the downstream fragment of asnO, asnOBG3 (5′-GGAAGATCTTTGACGAGAGGTAGGTTC-3′) and asnODE (5′-CCGGAATTCGGCTTCTGCTTCAAAAGC-3′). The upstream and downstream fragments of asnO were trimmed with HindIII and BglII and with BglII and EcoRI, respectively, and then ligated together with plasmid pUC19 DNA previously cleaved with HindIII and EcoRI. The tripartitely ligated pUC19 derivative was introduced into E. coli JM109 by transformation to give ampicillin resistance on LB plates. The plasmid DNA in the transformant cells was extracted, cleaved with BglII, and then ligated with a chloramphenicol resistance cassette derived from pBEST-4F previously trimmed with BamHI. The ligated DNA was introduced again into JM109 by transformation to give chloramphenicol (5 μg/ml) resistance on LB. The resulting plasmid DNAs were extracted, cleaved with EcoRI, and then used to transform B. subtilis 168 to chloramphenicol (5 μg/ml) resistance on TBABG to provide strain FU342, the correct construction of which was confirmed by PCR analysis with the primer pair of asnOUH and asnODE as described above.
B. subtilis FU345, lacking the asnH and asnO genes, was constructed as follows. The chromosomal DNA of strain FU342 was used to transform strain FU341 to chloramphenicol resistance in addition to the original spectinomycin resistance on TBABG. Correct marker replacements in the transformants were confirmed by PCR analysis as described above to provide strain FU345. Similarly, the DNA of strain FU340 was used to transform strains FU341 and FU342 to provide strains FU343 (lacking asnB and asnH) and FU344 (lacking asnB and asnO), respectively. Strain FU346, lacking asnB, asnH, and asnO, was constructed by transforming strain FU345 with the DNA of strain FU340. The ytnA gene of strains FU341 and FU345 was inactivated by pMUTIN2mcs integration. For that, chromosomal DNA of strain BFS41 was used to transform FU341 and FU345 to erythromycin resistance in addition to the original antibiotic resistance on TBABG, providing strains FU347 and FU348, respectively.
RNA techniques.
Cells of B. subtilis strains were inoculated into DSM to an optical density at 600 nm (OD600) of about 0.05 and allowed to grow at 37°C with shaking. The cells were harvested at 1.5 h (in exponential growth), 3 h (at the transition between exponential growth and stationary phase), 4 h (defined as the beginning of sporulation), 5 h (1 h after the beginning of sporulation), 7 h (3 h after the beginning of sporulation), and 9 h (5 h after the beginning of sporulation) after inoculation, at OD600s of approximately 0.4, 1.5, 2.1, 2.3, 2.0, and 2.3, respectively. Total cellular RNA was extracted by mixing the cells with glass beads, phenol, and cetyltrimethylammonium bromide and then purified as described previously (22). Alternatively, cells were inoculated into S6 medium and grown as described above to extract RNAs at 3 h (earlier in exponential growth), 5 h (later in exponential growth), and 8 h (in stationary phase) after inoculation, at OD600s of approximately 0.5, 1.5, and 3.7, respectively.
For Northern blot analysis, RNA (15 μg) was electrophoresed in glyoxal gels, transferred to a Hybond-N membrane (Amersham), and then hybridized with probe DNAs as described previously (22). Probe DNAs for asnB, asnH, asnO, and ytnA were the same DNA fragments cloned into the pMUTIN2mcs vector for the respective mutant constructions described above (Fig. 1). Probe DNAs for yxbB and yxaM (Fig. 1) were prepared by PCR with chromosomal DNA of B. subtilis 168 as a template and specific primer pairs as follows (tag sequences are underlined): for yxbB, yxbBH (5′-GCCGAAGCTTAGACTGTCCCGGATGTATTC-3′) and yxbBBG (5′-GCGCAGATCTGCCAGAACTCTATAGCACTC-3′), and for yxaM, yxaMH (5′-GCCGAAGCTTGATTGGATTAATGGGAGCGG-3′) and yxaMBG (5′-GCGCAGATCTATACCCGCTGGCAATACTTC-3′). Each of the probe DNAs was labeled by using a Bca BEST labeling kit (Takara Shuzo) and [α-32P]dCTP (ICN Biomedicals).
For primer extension analysis, 50 μg of each RNA was annealed to a primer (5′-GTTTTTCCTGGACGAGCTGC-3′) that had been labeled at the 5′ end by using a MEGALABEL kit (Takara Shuzo) and [γ-32P]ATP (Amersham). Primer extension reactions were performed as described previously (22).
RESULTS
Three asparagine synthetase homologs are encoded in the B. subtilis genome.
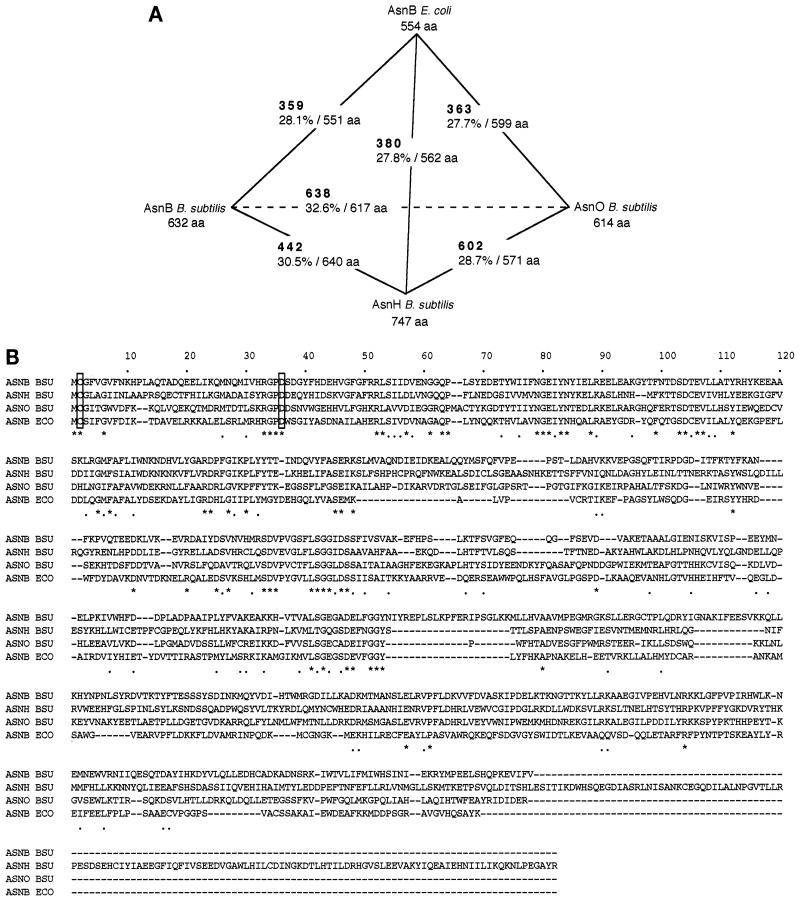
Three genes, asnB, asnH, and asnO (originally named yisO), that encode asparagine synthetase homologs are present in the B. subtilis genome and are located at kb 3126.80, 4097.70, and 1156.80, respectively (10) (Fig. 1). Similarity comparisons among the three gene products and AsnB of E. coli (18) indicated that the three products were paralogous to each other, with the highest similarity between AsnB and AsnO, and also orthologous to AsnB of E. coli to almost the same extent (Fig. 2A). As shown in a multiple alignment of the amino acid sequences of the four proteins (Fig. 2B), they had higher similarity in their N-terminal parts, while the C-terminal parts were less conserved with respect to both sequence and length.
FIG. 2.
Similarity among the putative products of asnB, asnH, and asnO of B. subtilis and asnB of E. coli. (A) Similarity among the four gene products. Similarity was calculated by using the FASTA program (14) for each pairing among the four gene products. The FASTA optimized score (boldface) and sequence identity (percentage of overlapping amino acid residues) are shown. The size of each of the gene products is given as amino acid residues (aa). (B) Alignment of the amino acid sequences of the gene products. The amino acid sequence alignment of AsnB, AsnH, and AsnO of B. subtilis (ASNB BSU, ASNH BSU, and ASNO BSU, respectively) and AsnB of E. coli (ASNB ECO) was performed with the CLUSTAL W program (6). Conserved and related amino acid residues are marked with asterisks and dots beneath the sequences, respectively. Gaps introduced within the sequences to optimize the alignment are shown by hyphens. Conserved Cys and Asp residues are boxed (see text).
AsnB of E. coli has a glutamine amide transfer domain in its N terminus, which is similar to that of PurF-type glutamine amidotransferases (18). This domain of PurF-type glutamine amidotransferases is reported to comprise approximately 194 amino acid residues in the N terminus, containing conserved residues Cys2, Asp29 and His101 for glutamine amide transfer function (12), while that of AsnB of E. coli contains two residues corresponding to Cys2 and Asp29 (18). The Cys2 residue is known to be essential for the glutamine amide transfer function of the AsnB family (21). As shown in the alignment (Fig. 2B), residues corresponding to Cys2 and Asp29 are also conserved in the three AsnB homologs of B. subtilis. The amino acid sequence of AsnO is longer in the N terminus than that of YisO reported originally, since another N terminus, 23 codons upstream, was chosen within the same yisO open reading frame, which is more plausible because it thus contains the essential Cys2.
Expression of the asparagine synthetase genes in B. subtilis.
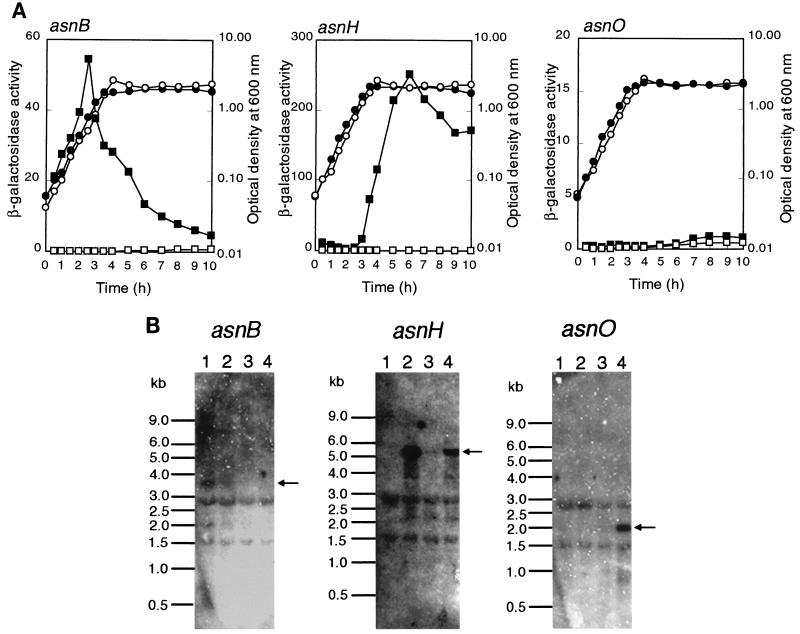
To analyze expression of each of the three asparagine synthetase homolog genes, we constructed transcriptional fusions of each gene with the E. coli lacZ reporter by integration of pMUTIN2mcs into the respective loci on the chromosome via a single-crossover event (see Materials and Methods). β-Galactosidase activity in extracts of the constructs grown in a rich sporulation medium, DSM, was then measured (Fig. 3A). The asnB-lacZ fusion was expressed during exponential growth and repressed in stationary phase. Expression of the asnH-lacZ fusion was dramatically elevated at the transition between exponential growth and the stationary phase. The asnO-lacZ fusion was expressed less well, but the expression was nevertheless significant during sporulation. Thus, the three genes in B. subtilis were likely to be expressed differentially during growth in DSM.
FIG. 3.
Expression analysis of asnB, asnH, and asnO in B. subtilis cells grown in a rich sporulation medium. (A) Expression of lacZ reporters fused with each of the asparagine synthetase homolog genes. Cells of B. subtilis 168 (wild type), BFS55 (asnH::pMUTIN2mcs), BFS56 (asnB::pMUTIN2mcs), and FU339 (asnO::pMUTIN2mcs) were cultured in DSM. At various intervals, cells in 1 ml of the cultures were harvested, and β-galactosidase activity (nanomoles of 2-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per OD600 unit) in cell extracts was determined as described previously (22). Activities of the asnB-lacZ fusion in cells of strain BFS56 (left), asnH-lacZ in cells of strain BFS55 (middle), and asnO-lacZ in cells of FU339 (right) are shown as solid squares, and that of endogenous lacZ in cells of strain 168 are shown as open squares. The OD600 for cells is shown as solid and open circles for each of the mutants and the wild type, respectively. (B) Northern analysis. Results of Northern analyses for the asnB (left), asnH (middle), and asnO (right) transcriptions are shown. RNAs were prepared from cells of strain 168 grown in DSM. The RNA samples were taken during exponential growth (lane 1), at the time of transition between exponential growth and stationary phase (lane 2), and at 1 h (lane 3) and 5 h (lane 4) after the beginning of sporulation. Positions of size marker RNAs (Millennium markers; Ambion) are given on the left of each panel. Positions of major transcripts are indicated with arrows.
In the lacZ fusion analysis described above, each of the asn genes was inactivated by the plasmid integration, which could have influenced its own expression. To confirm the differential expression of the three genes, we analyzed their transcripts in wild-type cells by means of Northern blotting (Fig. 3B). The asnB transcript (3.8 kb) was detected only in exponentially growing cells, as expected from the lacZ fusion analysis. The asnH transcript (5.5 kb) appeared at the transition between exponential growth and stationary phase as expected from the lacZ fusion analysis, decreased early in sporulation, and increased later on. The asnO transcript (2.0 kb) was clearly found only in the sporulating cells, confirming the faint expression of the fused lacZ reporter. Northern analyses with the respective probes for ytnA, yxbB, and yxaM revealed that asnB was possibly cotranscribed with the downstream ytnA gene and that asnH was the fourth gene of an operon comprising yxbB, yxbA, yxnB, asnH, and yxaM (data not shown).
Complementation of E. coli asparagine auxotrophy by B. subtilis asparagine synthetase genes.
It was reported that asnB of E. coli was not able to be cloned into a high-copy-number plasmid in E. coli (18), implying that the same problem might occur in cloning the asparagine synthetase homolog genes of B. subtilis in E. coli. Indeed, asnB of B. subtilis could not be cloned into plasmid pUC19 (data not shown). For cloning of the three genes, therefore, we used a low-copy-number plasmid vector, pOU71, which replicates in E. coli cells at one copy per chromosome (11). A DNA fragment containing the entire asnB coding region as well as its 5′ flanking region, probably carrying a promoter (Fig. 1), was cloned into plasmid pOU71 to provide plasmid pASNB. Similarly a DNA fragment containing asnO (Fig. 1) was cloned to provide plasmid pASNO. In the case of asnH cloning, two DNA fragments, which covered only (i) asnH and (ii) the four genes from yxbB to asnH and the 5′ flanking region of yxbB (Fig. 1), were cloned, to provide plasmids pASNH and pYXBB, respectively, because the Northern analyses described above suggested that a promoter for the operon containing asnH might be located upstream of yxbB.
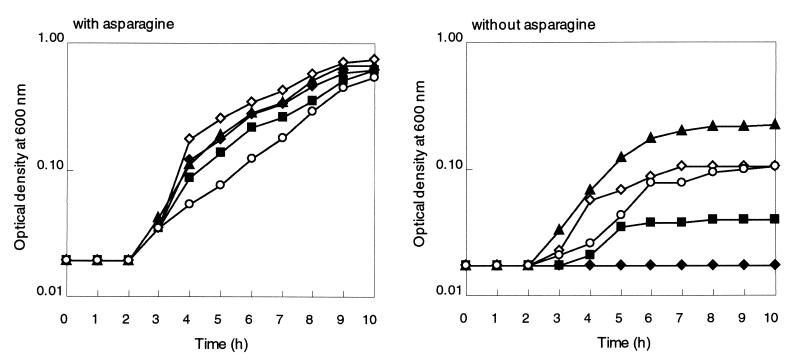
Each of the four recombinant plasmids was introduced into cells of an E. coli asparagine auxotroph (strain ME6279) lacking both the asnA and asnB genes. ME6279 cells harboring pOU71 did not grow in a minimal medium without asparagine, while the cells harboring the recombinant plasmids were able to grow (Fig. 4). In asparagine-supplemented medium all strains grew at essentially equal rates. Therefore, asnB, asnH, and asnO of B. subtilis complemented the asparagine-auxotrophic phenotype of ME6279, suggesting that all three gene products likely functioned as asparagine synthetase. Since the vector has no specific promoter to allow expression of the cloned genes, each of the cloned fragments possibly had some promoter activity in E. coli that allowed expression of the asn genes. However, the extents of the growth supported by different plasmids were not the same, which might suggest a difference in expression levels of the genes and/or in the enzyme activities of the gene products. Plasmid pASNB supported the growth without asparagine more efficiently than the other plasmids. Of the two plasmids carrying asnH, pASNH did support growth without asparagine but much less efficiently than pYXBB, implying that the DNA fragment carried by pASNH might possess only a very weak promoter which led the expression of asnH in E. coli.
FIG. 4.
Complementation of the asparagine-auxotrophic phenotype of E. coli ME6279 by expression of asparagine synthetase genes of B. subtilis. E. coli ME6279 cells harboring plasmid pASNB (solid triangles), pASNH (solid squares), pASNO (open circles), pOU71 (solid diamonds), or pYXBB (open diamonds) were precultured in LB with asparagine (50 μg/ml) for 16 h at 37°C with shaking. The cells in each of the precultures were washed once and then inoculated into M9 medium with (left) and without (right) asparagine (50 μg/ml) and allowed to grow at 37°C with shaking. The OD600s for cells were monitored at 1-h intervals. Data from a single experiment are presented. The same experiments were repeated at least three times with similar results.
Deletion analysis of the asparagine synthetase genes.
To investigate whether the three genes were involved in asparagine biosynthesis in B. subtilis, the coding regions of three asn genes were deleted via double-crossover events, resulting in marker replacement. A series of mutant strains lacking one, two, or all three genes was constructed (see Materials and Methods). As summarized in Table 2, the resulting strains were cultured in S6 liquid minimal medium to compare their growth rates in the absence and presence of low (50 μg/ml) and high (5 mg/ml) levels of asparagine. It is known that some amino acid-auxotrophic phenotypes (for example, tryptophan- and methionine-auxotrophic phenotypes) can be restored by adding the required amino acid at the low level of 50 μg/ml. Among strains lacking one of the three genes, strains FU341 and FU342, lacking asnH and asnO, respectively, grew as well as the wild-type strain even without asparagine, while strain FU340, lacking asnB, grew more slowly than the wild type. Among strains lacking two of the three genes, strain FU345, lacking both of asnH and asnO, grew as well as the wild type, while strains FU343 (lacking asnB and asnH) and FU344 (lacking asnB and asnO) grew more slowly. In addition, the slow growth of strain FU340 was not well restored even in the presence of the high level of asparagine, and strain FU343 exhibited a tendency to grow more slowly than FU340 in the presence of asparagine. Finally, the growth of strain FU346, lacking all three genes, was even slower, in particular when asparagine was present in the medium.
TABLE 2.
Effect on growth of inactivation of asnB, asnH, asnO, and ytnA in B. subtilisa
| Strain (relevant genotype) | Doubling time (h)b in S6 medium with:
|
||
|---|---|---|---|
| No supplement | Asparagine at:
|
||
| 50 μg/ml | 5 mg/ml | ||
| 168 (wild type) | 1.39 ± 0.05 | 1.46 ± 0.08 | 0.95 ± 0.02 |
| FU340 (ΔasnB::neo) | 2.70 ± 0.96 | 2.30 ± 0.78 | 1.97 ± 0.26 |
| FU341 (ΔasnH::spc) | 1.50 ± 0.10 | 1.42 ± 0.08 | 0.95 ± 0.04 |
| FU342 (ΔasnO::cat) | 1.33 ± 0.02 | 1.41 ± 0.05 | 1.13 ± 0.01 |
| FU343 (ΔasnB::neo ΔasnH::spc) | 2.88 ± 0.82 | 3.05 ± 0.39 | 2.46 ± 1.09 |
| FU344 (ΔasnB::neo ΔasnO::cat) | 2.45 ± 0.43 | 2.03 ± 0.40 | 1.88 ± 0.45 |
| FU345 (ΔasnH::spc ΔasnO::cat) | 1.33 ± 0.2 | 1.62 ± 0.17 | 1.41 ± 0.01 |
| FU346 (ΔasnB::neo ΔasnH::spc ΔasnO::cat) | 5.34 ± 0.96 | 3.56 ± 0.37 | 2.54 ± 0.40 |
| BFS41 (ytnA::pMUTIN2mcs) | 1.25 ± 0.17 | 1.26 ± 0.06 | 1.22 ± 0.20 |
| FU347 (ytnA::pMUTIN2mcs ΔasnH::spc) | 1.30 ± 0.20 | 1.20 ± 0.09 | 1.11 ± 0.04 |
| FU348 (ytnA::pMUTIN2mcs ΔasnH::spc ΔasnO::cat) | 1.52 ± 0.07 | 1.60 ± 0.18 | 1.33 ± 0.09 |
Cells of B. subtilis strains were grown in S6 medium with or without two levels (50 μg/ml and 5 mg/ml) of asparagine, and their doubling times were calculated.
Results are means and standard deviations calculated from four independent experiments.
Since different growth rates were obtained with different cultures of each of the asnB mutants for unknown reasons, their average growth rates had considerable deviations (Table 2). To clarify their asparagine dependence, colony formation of the mutants and the wild type was compared on plates with and without asparagine (Fig. 5), and in this test the asparagine dependence of mutants lacking asnB was also observed.
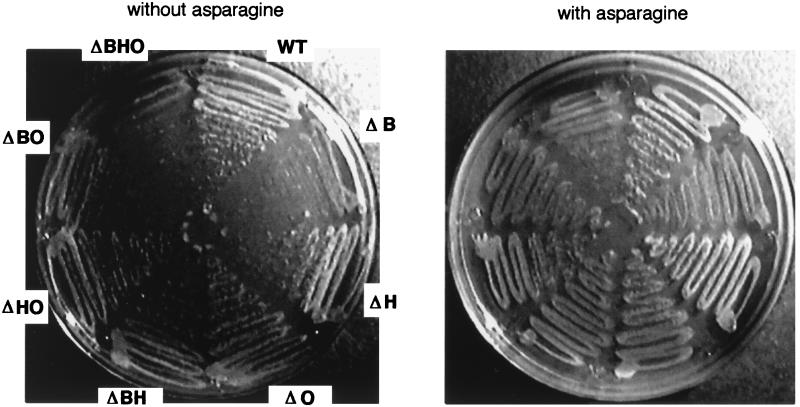
FIG. 5.
Effect on growth of inactivation of asnB, asnH, asnO, and ytnA in B. subtilis. A single colony each of strains 168 (wild type [WT]), FU340 (ΔB), FU341 (ΔH), FU342 (ΔO), FU343 (ΔBH), FU344 (ΔBO), FU345 (ΔHO), and FU346 (ΔBHO) was taken from overnight cultures on TBABG plates containing appropriate antibiotics and spread onto S6 plates with (right) and without (left) asparagine (50 μg/ml). The plates were incubated at 37°C for 36 h, and then colony formation was observed. Data from a single experiment are presented. The same experiments were repeated three times independently with similar results.
Judging from the results of transcription analysis, the asnB deletion possibly also led to inactivation of ytnA. In order to rule out the possibility that the slow growth of asnB mutants resulted from the inactivation of ytnA, we disrupted this gene by pMUTIN2mcs integration. Three strains, BFS41 (without active ytnA), FU347 (without ytnA and asnH), and FU348 (without ytnA, asnH, and asnO) were constructed and cultured in the liquid minimal medium (Table 2). BFS41, FU347, and FU348 exhibited essentially the same growth rates as 168, FU341, and FU345, respectively, indicating that the inactivation of ytnA was not involved in the growth defect of the asnB mutants.
Taken together, the results suggest that asnB may be the main gene involved in asparagine biosynthesis, able to support the normal growth of B. subtilis in the absence of asparagine. However, even if all three asparagine synthetase genes were deleted, cells were still able to grow without asparagine, although almost 3.5 times more slowly than the wild-type counterpart, implying that B. subtilis might possess another, minor asparagine biosynthesis pathway independent of the three genes revealed by sequence analysis.
Expression of each of the three asparagine synthetase genes in the wild-type cells grown in minimal S6 medium with and without asparagine was examined by Northern analysis (Fig. 6). In the absence of asparagine, not only asnB but also asnH was transcribed essentially constitutively during exponential growth and in stationary phase, while transcription of asnO was not detected. Even in the presence of asparagine, both asnB and asnH were still transcribed constitutively, but the transcription of asnH was partially repressed. The result suggested that asnB was expressed and needed even when asparagine was abundant in the medium. In cells grown in a rich medium (DSM), asnH was induced only at the transition between exponential growth and stationary phase, as described above (Fig. 3), implying that the expression of asnH might be repressed by rich nutrients and partially by asparagine. However, the expression of asnH (and possibly asnO) appears to be insufficient to support normal growth in the absence of asparagine (Table 2 and Fig. 5).
FIG. 6.
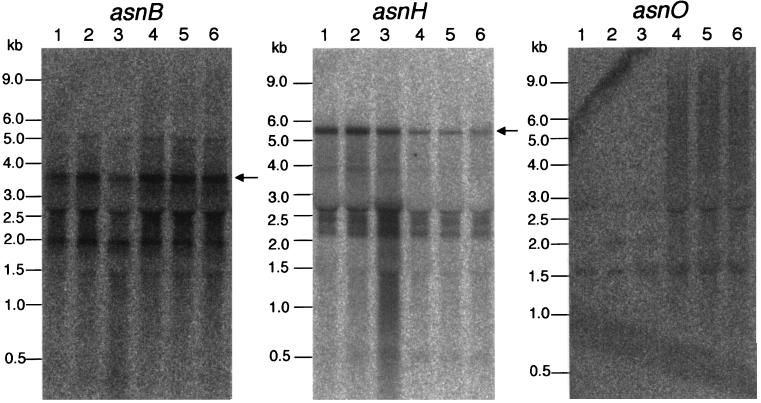
Transcription of asnB, asnH, and asnO in cells of strain 168 grown in minimal medium. Results of Northern analysis of the asnB (left), asnH (middle), and asnO (right) transcriptions are shown. The same specific probes as for the previous experiments (Fig. 3B) were used. RNAs were prepared from cells of strain 168 grown in S6 minimal medium without (lanes 1 to 3) and with (lanes 4 to 6) 5 mg of asparagine per ml. The RNA samples were taken earlier (lanes 1 and 4) and later (lanes 2 and 5) in exponential growth and in stationary phase (lanes 3 and 6). Positions of size marker RNAs are given on the left of each panel. Positions of the detected major transcripts covering each of the entire transcriptional units are indicated with arrows.
The asnO gene is indispensable for sporulation.
Since asnO is expressed only during sporulation (Fig. 3A), we examined whether its transcription depended on sporulation sigma factors. For this purpose RNA samples were prepared from sporulating cells harboring different sigma factor mutations (Table 1) and from the wild-type cells, at stages when the respective sigma factors were activated. The samples were then subjected to Northern analysis, targeting the asnO transcript (Fig. 7A). At the stage when sigma-H was activated, the asnO transcript was not detected even in the wild-type cells, indicating that asnO was not transcribed at the initiation of sporulation. At the stage when sigma-E and -F were activated, the transcript was detected in the wild-type cells but not in the sigma-F or -E mutants. At the stage when sigma-K and -G were activated, the transcript was detectable in both mutants and in the wild-type cells. The sigma-F mutant lacks both sigma-F and -E, but the sigma-E mutant must have active sigma-F. We therefore conclude that asnO transcription depends on sigma-E. In addition, primer extension analysis mapped a 5′ end of the asnO transcript (Fig. 7B) and allowed us to identify a corresponding promoter sequence as −10 (CATAAACT [the −10 position is underlined]) and −35 (TCAACTA) regions, separated by 14-bp spacer. This promoter is likely recognized by sigma-E RNA polymerase (5).
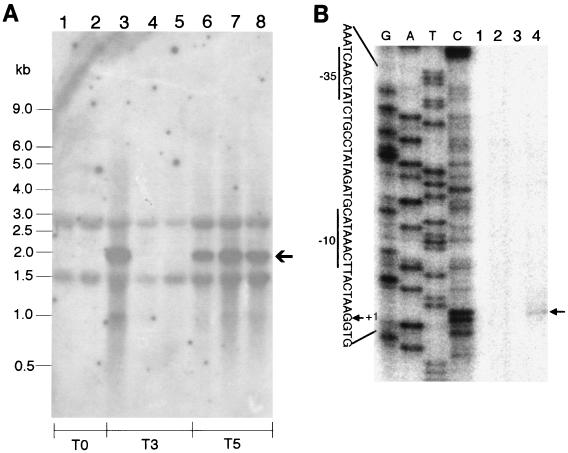
FIG. 7.
Expression of asnO depends on sigma-E. (A) Northern analysis of asnO transcription in sporulating cells of B. subtilis strains. The same specific asnO probe as for the previous experiments (Figs. 3B and 6) was used. RNAs were prepared from cells of B. subtilis strains grown in DSM. The RNA samples were taken from cells of strains 168 and ASK201 (without sigma-H) at the beginning of sporulation (T0) (lanes 1 and 2, respectively); strains 168, ASK203 (without sigma-E), and ASK202 (without sigma-F) at 3 h after the beginning of sporulation (T3) (lanes 3, 4, and 5, respectively); and strains 168, ASK205 (without sigma-K), and ASK204 (without sigma-G) at 5 h after the beginning of sporulation (T5) (lanes 6, 7, and 8, respectively). Positions of size marker RNAs and the asnO transcript (arrow) are given on the left and right, respectively. (B) Primer extension mapping of a 5′ end of the asnO transcript. The end-labeled primer (see Materials and Methods) was hybridized to RNA samples prepared from cells of strain 168 grown in DSM and was then extended with Moloney murine leukemia virus reverse transcriptase (Gibco BRL). The RNA samples were taken in exponential growth (lane 1), at the time of transition between exponential growth and stationary phase (lane 2), and at 1 h (lane 3) and 3 h (lane 4) after the beginning of sporulation. A known DNA sequence ladder (lanes G, A, T, and C) made by dideoxy sequencing reactions with the same end-labeled primer was loaded to estimate the size of the extended product. The position of the extended product is indicated by an arrow on the right. Part of the nucleotide sequence (noncoding strand) of the asnO promoter region is shown on the left. The −10 and −35 promoter regions are underlined, and the 5′ end corresponding to the transcription start site (+1) is indicated.
We also examined the formation of heat-resistant spores of some of the deletion mutants (Table 3). Strains FU340 and FU341, lacking asnB and asnH, respectively, produced heat-resistant spores as well as the wild-type strain. In contrast, strains FU342 and FU346, lacking only asnO and all the three genes, respectively, failed to produce spores, even in the presence of asparagine in the medium. Consequently, asnO was indispensable for sporulation, but the failure in sporulation resulting from its deletion was not corrected by asparagine addition.
TABLE 3.
Heat-resistant spore formation by strains of B. subtilisa
| Strain (relevant genotype) | Asparagine | CFU (108)b
|
Ratio (%)c | |
|---|---|---|---|---|
| With heating | Without heating | |||
| 168 (wild type) | − | 4.4 | 5.3 | 83 |
| + | 3.4 | 4.3 | 79 | |
| FU340 (ΔasnB::neo) | − | 2.4 | 2.9 | 83 |
| + | 2.6 | 3.1 | 84 | |
| FU341 (ΔasnH::spc) | − | 3.5 | 4.4 | 80 |
| + | 2.8 | 3.7 | 76 | |
| FU342 (ΔasnO::cat) | − | NDd | 3.3 | ND |
| + | ND | 2.6 | ND | |
| FU346 (ΔasnB::neo ΔasnH::spc ΔasnO::cat) | − | ND | 2.6 | ND |
| + | ND | 2.4 | ND | |
Cells of each of the B. subtilis strains were inoculated into DSM with (+) and without (−) asparagine (50 μg/ml) and cultivated at 37°C for 24 h with shaking. The cultures were diluted appropriately and divided into two tubes; one was heated at 70°C for 15 min, and the other was not heated. Cells in each of the tubes were then spread onto TBABG plates containing antibiotics when needed, and the plates were incubated at 37°C for 16 h. Colonies that appeared on the plates were counted to calculate CFU.
Data from a single experiment are shown. The same experiments were repeated at least twice with similar results.
Ratio of CFU with heating to CFU without heating.
ND, not determined (<105 CFU).
DISCUSSION
Analysis of the genomic sequence of B. subtilis allowed us to predict three asparagine synthetase gene homologs. These homologs were designated asnB, asnH, and asnO, encoding glutamine-dependent AsnB-type enzymes (Fig. 2). No ammonia-dependent AsnA-type enzyme gene has been detected within the genome. Enteric bacteria, such as E. coli and K. aerogenes, have the two types of asparagine synthetases, and ammonia is their optimal nitrogen source. In contrast, the fastest growth of B. subtilis occurs in medium containing glutamine as the sole nitrogen source (3). Ammonia assimilation in B. subtilis depends on successive reactions involving glutamine synthetase and glutamate synthase, as mutations in either enzyme result in an inability to grow with ammonia as a nitrogen source (2), and glutamate dehydrogenase functions only as a catabolic, not an assimilatory, enzyme in B. subtilis. This indicates that B. subtilis has no other efficient mechanism for ammonia assimilation. These facts might be related to the finding that B. subtilis has no ammonia-dependent AsnA-type enzyme but rather only three glutamine-dependent AsnB-type ones.
Each of the three asparagine synthetase homologs of B. subtilis likely functions as asparagine synthetase, as shown by their complementation of the asparagine-auxotrophic phenotype of E. coli ME6279 (Fig. 4). The analysis of deletion mutants of B. subtilis indicated that asnB is the main gene involved in asparagine biosynthesis in vegetative cells (Table 2 and Fig. 5). It may be relevant that pASNB, carrying asnB, complemented the E. coli auxotrophy most efficiently (Fig. 4). We have no explanation for the fact that the growth rates of B. subtilis mutants lacking asnB fluctuated to a great extent (Table 2). In addition, even a high level of asparagine (5 mg/ml) did not restore the growth of asnB mutants completely (Table 2), and a saturating concentration of asparagine (25 mg/ml) could not either (data not shown). Furthermore, even in the presence of abundant asparagine in the medium, asnB was expressed as constitutively as in the absence of asparagine (Fig. 6). It is possible that not only asparagine itself but also the function of the major asparagine synthetase of the asnB product might be needed to support normal growth in the minimal medium.
The asnB and asnH genes are part of possible longer operons and are followed by ytnA and yxaM, respectively (Fig. 1). These downstream genes could have been inactivated by a possible polar effect of the marker replacements in the asnB and asnH mutants. That possibility was not investigated for yxaM, since there was no discernible phenotype associated with the insertion in the asnH gene. Inactivation of ytnA had no significant effect when asnB was functional (Table 2). However, it is interesting that both ytnA and yxaM are homologous to some of the known amino acid transporter genes (10). If the former was involved in asparagine uptake, the putative polar effect of asnB inactivation might be one of the reasons for the lack of growth restoration by asparagine.
In B. subtilis cells grown in the rich medium, asnH was induced at the transition between exponential growth and stationary phase (Fig. 3). In the minimal medium, not only asnB but also asnH was expressed constitutively, but when the medium was supplemented with asparagine, the expression of asnH was partially repressed (Fig. 6). These results imply that asnH may be repressed by rich nutrients and partially repressed by asparagine. Moreover, in cells grown in the rich medium, the amount of asnH transcript was decreased early in sporulation and then increased again later, suggesting that its regulation is complex. Although homology searches did not suggest any functions for yxbB, yxbA, and yxnB, genes which may be transcribed together with asnH, the three genes might be involved in asnH regulation. However, the expression of asnH alone did not support normal growth in minimal medium without asparagine (Table 2 and Fig. 5). Recently, it was observed that another asnH mutant strain, ASNHd, which was constructed within the framework of the international B. subtilis genome functional analysis project, gave abnormal colonies on high-salt plates and a penicillin-sensitive phenotype (9). The asnH gene might not be as efficient in asparagine biosynthesis as asnB but might be responsible for some function involved in cell surface organization.
The asnO gene was indispensable for sporulation (Table 3). Its transcription depends on sigma-E (Fig. 7), suggesting that the gene was expressed in sporulating mother cells, and the failure of sporulation of the asnO mutants was not restored by adding asparagine to the medium (Table 3). We cannot eliminate the possibility that sporulating mother cells were less efficient in asparagine uptake, but it is also possible that asnO might have an unknown specific role in sporulation besides asparagine biosynthesis.
Finally, the deletion analysis suggested that B. subtilis could have another, minor asparagine biosynthesis pathway independent of the three asparagine synthetases. Because it was impossible to predict candidates for asparagine synthetase genes other than the three described in this paper, the minor biosynthesis might be supported by a novel and unique asparagine synthetase or by some amidotransferases with broad specificity. Further work is required to test these possibilities.
ACKNOWLEDGMENTS
We thank Petar Pujic for his excellent protocols for RNA preparation and Northern analysis with valuable advice and suggestions; Kouji Anami, Hiroki Doyama, Masataka Irie, Mihoko Kaichi, Hiroshi Kataoka, and Hironobu Katsumata for their technical assistance; Rozenn Dervyn, Mitsuo Itaya, Valérie Vagner, and Seiichi Yasuda for providing the plasmids; Akiko Nishimura for E. coli ME6279; Kei Asai and Patrick Stragier for the B. subtilis sporulation sigma mutant strains; Kazuo Kobayashi and Naotake Ogasawara for their personal communication as to the phenotype of an asnH mutant; and Choong-Min Kang for critical reading of the manuscript.
This work was supported by grants JSPS-RFTF96L00105 from the Japan Society for the Promotion of Science and BIO4-CT95-0278 from the EU.
REFERENCES
- 1.Asai K, Kawamura F, Yoshikawa H, Takahashi H. Expression of kinA and accumulation of ςH at the onset of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:6679–6683. doi: 10.1128/jb.177.22.6679-6683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dean D R, Aronson A L. Selection of Bacillus subtilis mutants impaired in ammonia assimilation. J Bacteriol. 1980;141:985–988. doi: 10.1128/jb.141.2.985-988.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher S, Sonenshein A L. Control of carbon and nitrogen metabolism in Bacillus subtilis. Annu Rev Microbiol. 1991;45:107–135. doi: 10.1146/annurev.mi.45.100191.000543. [DOI] [PubMed] [Google Scholar]
- 4.Fujita Y, Freese E. Isolation and properties of a Bacillus subtilis mutant unable to produce fructose-biphosphatase. J Bacteriol. 1981;145:760–767. doi: 10.1128/jb.145.2.760-767.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 7.Hughes C A, Beard H S, Matthews B F. Molecular cloning and expression of two cDNAs encoding asparagine synthetase in soybean. Plant Mol Biol. 1997;33:301–311. doi: 10.1023/a:1005784202450. [DOI] [PubMed] [Google Scholar]
- 8.Humbert R, Simoni R D. Genetic and biochemical studies demonstrating a second gene coding for asparagine synthetase in Escherichia coli. J Bacteriol. 1980;142:212–220. doi: 10.1128/jb.142.1.212-220.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi, K., and N. Ogasawara. Personal communication.
- 10.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 11.Larsen J E, Gerdes K, Light J, Molin S. Low-copy-number plasmid-cloning vectors amplifiable by derepression of an inserted foreign promoter. Gene. 1984;28:45–54. doi: 10.1016/0378-1119(84)90086-6. [DOI] [PubMed] [Google Scholar]
- 12.Mei B, Zalkin H. A cysteine-histidine-aspartate catalytic triad is involved in glutamine amide transfer function in purF-type glutamine amidotransferases. J Biol Chem. 1989;264:16613–16619. [PubMed] [Google Scholar]
- 13.Nakamura M, Yamada M, Hirota Y, Sugimoto K, Oka A, Takanami M. Nucleotide sequence of the asnA gene coding for asparagine synthetase of E. coli K-12. Nucleic Acids Res. 1981;9:4669–4676. doi: 10.1093/nar/9.18.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reitzer L J, Magasanik B. Asparagine synthetase of Klebsiella aerogenes: properties and regulation of synthesis. J Bacteriol. 1982;151:1299–1313. doi: 10.1128/jb.151.3.1299-1313.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Schaeffer P, Millet J, Aubert J P. Catabolite repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scofield M A, Lewis W, Schuster S M. Nucleotide sequence of Escherichia coli asnB and deduced amino acid sequence of asparagine synthase B. J Biol Chem. 1990;265:12895–12902. [PubMed] [Google Scholar]
- 19.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 20.Van Heeke G, Schuster S M. Expression of human asparagine synthetase in Escherichia coli. J Biol Chem. 1989;264:5503–5509. [PubMed] [Google Scholar]
- 21.Van Heeke G, Schuster S M. The N-terminal cysteine of human asparagine synthetase is essential for glutamine-dependent activity. J Biol Chem. 1989;264:19475–19477. [PubMed] [Google Scholar]
- 22.Winstedt L, Yoshida K, Fujita Y, von Wachenfeldt C. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J Bacteriol. 1998;180:6571–6580. doi: 10.1128/jb.180.24.6571-6580.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 24.Young R A, Davis R W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci USA. 1983;80:1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]