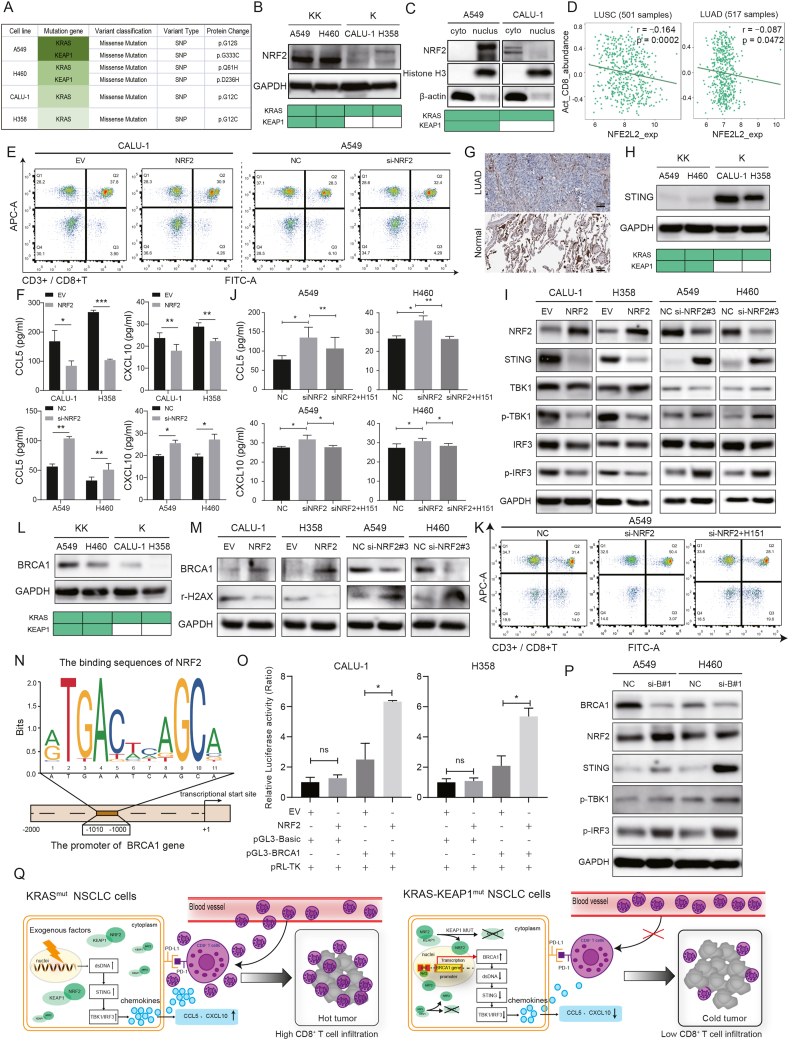
Kirsten rat sarcoma (KRAS) mutant non-small cell lung cancer (NSCLC) has distinct co-mutational profiles. Approximately 20% of KRAS mutant NSCLC cases underwent loss-of-function mutation of Kelch-like ECH-associated protein 1 (KEAP1) gene, namely KRAS-KEAP1 co-mutation (KK) type, which exhibited a poorer immune checkpoint inhibitor (ICI) response compared with individual KRAS mutation (K) type tumors.1 Recent studies suggest that K-type tumors are referred to as “hot tumors” while KK-type tumors are as “cold tumors”. Therefore, clarifying the molecular mechanism of the poor ICI-responsive phenotype of KK-type tumors is the key to improving the clinical efficacy and breaking through the therapy bottleneck. Co-mutations could regulate the immune contexture to reshape the tumor immune microenvironment (TIME), which has been identified as a major influence factor of ICI response. KK-type tumors were confirmed to contain few tumor-infiltrating lymphocytes, especially CD8+ T cells, leading to a suppressive TIME, which is also defined as a “cold tumor".2 Here, we demonstrated that NRF2, the target of KEAP1, exerted its role as a negative regulator of CD8+ T cells recruitment by decreasing CCL5 and CXCL10 chemokines in KK-type NSCLC. Mechanistically, NRF2 promoted the transcription and expression of BRCA1 to repair DNA damage, resulting in STING pathway inactivation. We propose the combination of NRF2 inhibitor or STING agonist with ICI may be a promising therapeutic approach for patients with KK-type NSCLC.
Firstly, A549 and H460 cell lines were identified as KK- type, and CALU-1 and H358 cell lines were as K- type through the CCLE database (Fig. 1A and Table S1). KEAP1 mutation would lead to overexpression and activation of NRF2. As expected, both protein and mRNA expressions of NRF2 were higher in KK-type cells than those in K-type cells (Fig. 1B; Fig. S1A). The nuclear and cytoplasm isolation assay and immunofluorescence staining showed that NRF2 was mainly located in the nuclei of KK-type cells while located in the cytoplasm of K-type cells, suggesting an activated status of NRF2 in KK-type cells (Fig. 1C; Fig. S1B). Collectively, these results demonstrated the potential role of NRF2 in the development of KK-type tumors.
Figure 1.
NRF2 participates in the suppressive tumor immune microenvironment of KRAS/KEAP1 co-mutant non-small cell lung cancer by inhibiting the STING pathway. (A) The mutation characters of cell lines used in this study were identified through CCLE database. (B) The protein expression of NRF2 were determined by Western blot. (C) Nuclear and cytoplasm isolation assay was performed to detect NRF2 localization in both KK- and K-type NSCLC cell lines. (D) The expression of NRF2 was negatively correlated with CD8+ T cell abundance based on TISIDB database in lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). (E) NRF2 was upregulated in K-type cells and knocked down in KK-type cells. CD3+/CD8+ T cell recruitment was evaluated by the chemotaxis assay; cells that migrated to the lower chamber were stained and recognized by a flow cytometer, and analyzed by Flowjo software. (F) ELISA was performed to determine the secretion levels of CCL5 and CXCL10 after upregulating NRF2 in K-type cells and downregulating NRF2 in KK-type cells. (G) The protein expression of STING in LUAD and normal tissues were confirmed by immunohistochemistry results from the HPA database. Scale bar = 50 μm. (H) The expression of STING in both KK- and K-type cells were determined by Western blot. (I) Changes of key protein levels in STING pathway were tested by Western blot after up- or down-regulating NRF2 in K- and KK-type cells, respectively. (J) After knockdown of NRF2 and addition of H151 in A549 and H460 cells, the secretion levels of CCL5 and CXCL10 were tested by ELISA. (K) CD3+/CD8+ T cell recruitment was evaluated by the chemotaxis assay; cells that migrated to the lower chamber were stained and recognized by a flow cytometer, and analyzed by Flowjo software. (L) The expression of BRCA1 in both KK- and K-type cells were determined by Western blot. (M) Changes of BRCA1 and DNA damage marker γ-H2AX levels were tested by Western blot after up- or down-regulating NRF2 in K- and KK-type cells, respectively. (N) Binding sites between the BRCA1 promoter sequence and NRF2 were predicted by JASPAR database. (O) Luciferase activities following transfection with NRF2 expression vector and pGL3-Basic or BRCA1 vector were detected by dual luciferase reporter assay. pRL-TK vectors were transfected as the internal control. (P) Changes of BRCA1, NRF2 and key protein levels in STING pathway were tested by Western blot after down-regulating BRCA1 in A549 and H460 cells. (Q) Schematic representation of the potential mechanisms of “hot” and “cold” tumors in K- and KK-type NSCLC. (Left) In K-type cells, exogenous risks lead to DNA damage, increase the cytoplasmic double-stranded DNA (dsDNA), and activate the STING pathway, resulting in the recruitment of CD8+ T cells by promoting the chemokines CCL5 and CXCL10, leading to a “hot tumor”; (Right) In KK-type cells, when KEAP1 mutation (KEAP1 MUT) occurred, NRF2 translocated into the nucleus and bond to the ARE sequence in the promoter region of BRCA1 gene, to promote BRCA1 transcription and expression, and DNA damage repair, resulting in STING pathway inactivation, chemokines synthesis and secretion reduction, and CD8+ T cell recruitment inhibition, leading to a suppressive TIME (cold tumor). EV, empty vector. NC, negative control. si-NRF2, small interfering RNA that targets NRF2. si-B, small interfering RNA that targets BRCA1. Data are shown as mean ± standard deviation of three independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. ns, not significant.
Bioinformatic analysis results from the TISIDB database showed that the expression of NRF2 was negatively correlated with CD8+ T cell abundance in 501 cases of lung squamous cell carcinoma (LUSC) and 517 cases of lung adenocarcinoma (LUAD), confirming the expectation that NRF2 contributed to the low CD8+ T cell abundance (P = 0.0002 in LUSC and P = 0.0472 in LUAD; Fig. 1D). Then, we screened chemokines that are known to be able to recruit CD8+ T cells, including CCL2/4/5/11 and CXCL9/10/11/133 under comprehensive considering the relationship between these chemokines and the expression of NRF2 and the abundance of CD8+ T cells by GEPIA and TISIDB database, respectively. Only CCL5 and CXCL10 are negatively related to NRF2 (Fig. S2A) and have a significant positive correlation with the abundance of CD8+ T cells (Fig. S2B) in both LUSC and LUAD. Hence, CCL5 and CXCL10 were selected as the analyzed targets in the subsequent research. Overexpression of NRF2 attenuated CD3+/CD8+ T cells recruitment, and the mRNA expression and secretion of CCL5 and CXCL10, whereas the opposite effects were observed in NRF2 knockdown groups (Fig. 1E, F; Fig. S2C–F). Interestingly, the change of NRF2 had no significant effect on the abundance of CD3+/CD8- T cells (Fig. S2C). Taken together, NRF2 regulated CD8+ T cell recruitment and its chemokines negatively.
The role of STING in NSCLC was further studied, for it is able to boost CD8+ T cell recruitment by upregulating CCL5 and CXCL10.4 In most solid tumors, including NSCLC, the mRNA expression of STING was significantly lower in tumors than that in the normal tissues based on GEPIA database (Fig. S3A, B). The lower protein expressions of STING in tumor tissues and cell lines than those in normal ones were confirmed by immunohistochemistry and Western blot, respectively (Fig. 1G; Fig. S3C). Furthermore, the lower STING expression level was associated with worse overall survival and progress-free survival based on the Kaplan–Meier Plotter database, suggesting a favorable prognostic value of STING (Fig. S3D, E). Taken together, these results identified a tumor suppressor role of STING in NSCLC.
A significant negative correlation between STING and NRF2 was identified by the GEPIA database (Fig. S4A). The oppositive protein and mRNA expression levels between them were confirmed as well (Fig. 1H; Fig. S4B). To confirm the hypothesis that NRF2 may be a negative regulator of STING, NRF2 expression was knocked down in KK-type cells using the No.3 siRNA sequence, which exerted the highest transfection efficiency (Fig. S4C), and NRF2 expression was upregulated in K-type cells. The expressions of TBK1 and IRF3, which are key target molecules of STING, remained unchanged regardless of NRF2 levels, whereas the expressions of STING, phospho-TBK1 (p-TBK1), and p-IRF3 were significantly affected by NRF2, indicating an increasing activity of STING pathway (Fig. 1I; Fig. S4D). These results revealed that the STING pathway was negatively regulated by NRF2.
Rescue experiments were performed in KK-type cells, in which NRF2 exhibited high-level expression. H151, a chemical compound specifically inhibiting STING,5 was applied after down-regulating NRF2 in KK-type cells. As shown in Figure S5A, the activity of the STING pathway was lowest after 3 h of exposure to 0.5 μM H151 and increased again by 5 h; hence, this treatment condition was selected for the subsequent experiments. As proved above, the mRNA expression and secretion level of CCL5 and CXCL10 were both increased upon downregulation of NRF2. However, the application of H151 reversed these effects (Fig. 1J; Fig. S5B). A similar rescue effect of H151 on CD3+/CD8+ T cell abundance was observed as well (Fig. 1K; Fig. S5C, D). Besides, the supplement of recombinant human CCL5 and CXCL10 proteins reversed the inhibition effect on CD8+ T cell migration of H151 (Fig. S5E). Overall, these results revealed that the STING pathway was crucial for NRF2-mediated suppressive TIME of KK-type NSCLC.
The possibility of direct interaction between NRF2 and STING was excluded by the String database (Fig. S6A), suggesting the presence of intermediate mediators. DNA damage is a direct activator of the STING pathway and BRCA1 is an important DNA damage repair gene. The mRNA level of BRCA1 was negatively associated with STING whereas positively related with NRF2 based on the GEPIA database (Fig. S6B). The protein expression of BRCA1 was higher in KK-type cells than that in K-type cells (Fig. 1L). Overexpression of NRF2 resulted in upregulation of BRCA1, whereas decreased the level of γ-H2AX, the marker of DNA damage. The opposite effects were observed upon NRF2 knockdown (Fig. 1M; Fig. S6C). Since NRF2 also functions as a transcription factor, whether NRF2 could bind to the promoter of BRCA1 was verified. Based on the JASPAR database, the upstream of the BRCA1 gene transcriptional start site possesses the same sequences as the antioxidant response element, which is an important binding region of NRF2 (Fig. 1N). As expected, the cells transfected with pGL3-BRCA1 exhibited a clearly increase luciferase activity compared to the pGL3-Basic transfected cells by dual-luciferase reporter assay upon NRF2 overexpression (Fig. 1O). Rescue experiments were performed by down-regulating BRCA1 via transfecting No.1 siRNA sequence, which exerted the highest transfection efficiency (Fig. S6D), in KK-type cells. Interestingly, the down-regulation of BRCA1 exerted no obvious effect on the levels of NRF2, implying NRF2 may function as an upstream factor of BRCA1. However, the inactivated STING pathway in KK-type cells was re-activated in response to BRCA1 knockdown (Fig. 1P; Fig. S6E). Taken together, NRF2 inhibited the STING pathway by promoting transcription and expression of BRCA1, resulting in the reduction of DNA damage.
Altogether, we clarified the effects of NRF2 in the suppressive TIME of KK-type NSCLC and revealed a novel regulatory mechanism underlying its effect on the STING pathway (Fig. 1Q). Unfortunately, there is a lack of in vivo experiments to confirm our in vitro observations. In addition, we failed to clarify the specific domain of BRCA1 to which NRF2 bound. Moreover, since BRCA1 is not a major DNA damage repair protein in NSCLC, there are still some other molecules that mediate the effect of NRF2 on the STING pathway, and the effects of NRF2 on other immune cells are not studied here. In addition, further studies are warranted to explore the therapeutic efficacy of NRF2 inhibitors through patient-derived xenograft or organoid models.
Funding
This work was financially supported by the project of lung cancer targeted therapy research of Beijing CSCO (No. Y-2021AST/qn-0013) and the National Natural Science Foundation of China (No. 82103343).
Conflict of interests
The authors declare that they have no competing interests.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.10.009.
Appendix ASupplementary data
The following are the Supplementary data to this article:
References
- 1.Skoulidis F., Heymach J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19(9):495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai M.C., Chen M., Ma P., et al. Clinicopathological, microenvironmental and genetic determinants of molecular subtypes in KEAP1/NRF2-mutant lung cancer. Int J Cancer. 2019;144(4):788–801. doi: 10.1002/ijc.31975. [DOI] [PubMed] [Google Scholar]
- 3.Do H.T.T., Lee C.H., Cho J. Chemokines and their receptors: multifaceted roles in cancer progression and potential value as cancer prognostic markers. Cancers. 2020;12(2):287. doi: 10.3390/cancers12020287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sen T., Rodriguez B.L., Chen L., et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 2019;9(5):646–661. doi: 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haag S.M., Gulen M.F., Reymond L., et al. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559(7713):269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.