The m6A modification involves almost all aspects of RNA biology, including the alternative splicing of mRNA precursors, mRNA transport and stability, and miRNA processing and regulation of target genes.1,2 Alternative splicing controlling the information storage and RNA translation involves the regulation of various biological processes.3, 4, 5 Here, we integrated the genomic information of 161 esophageal cancer (EC) samples to comprehensively evaluate the m6A modification patterns and correlated the m6A modification pattern with the prognosis of EC patients, where two distinct m6A modification patterns were proposed. The combined effects of high m6Ascore and low TMB correlated with a better prognosis of EC patients. In addition, we found an inherent correlation between m6A modification and the occurrence of alternative splicing events in EC patients. Altogether, we established a scoring system to quantify the m6A modification pattern with RNA alternative splicing events in individual EC patients (Fig. S1).
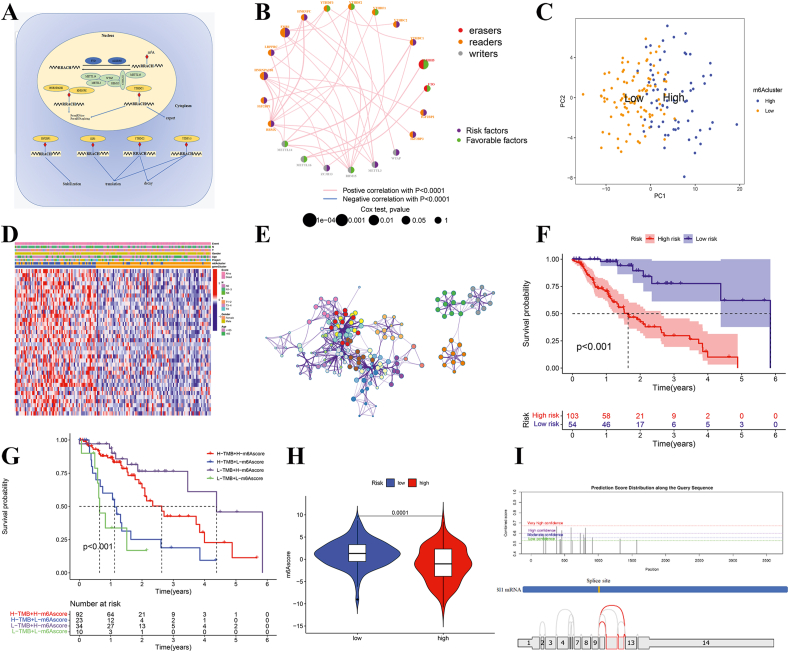
The RNA m6A methylation dynamically regulates different biological functions of RNA (Fig. 1A). Here we identified a total of 22 m6A regulators, including 7 writers, 2 erasers, and 13 readers, and predicted a comprehensive landscape of their interactions, and connections, as well as their prognostic significance for EC patients. We also found that the expression of the 22 m6A regulators was positively correlated with each other (Fig. 1B; Fig. S2, 3).
Figure 1.
Profiling the m6A-regulated RNA expression patterns and alternative splicing features in esophageal carcinoma. (A) Summary of the dynamic reversible process of m6A RNA methylation mediated by regulators “writers”, “erasers”, and “readers” and their potential biological functions for RNA. (B) The interaction between m6A regulators in esophageal cancer. The circle size represented the effect of each regulator on the prognosis, and the range of values calculated by Log-rank test was P < 0.001, P < 0.01, P < 0.05, and P < 0.1, respectively. Green dots in the circle represent risk factors of prognosis. Black dots in the circle represent protective factors of prognosis. The lines linking regulators showed their interactions, and thickness showed the correlation strength between regulators. Negative correlation was marked with blue and positive correlation with red. (C) Principal component analysis for the m6A phenotype-related genes of two m6A modification patterns, showing a remarkable difference on transcriptome between different modification patterns. (D) Unsupervised clustering of 22 m6A regulators in the TCGA-ESCA cohort. The m6Acluster, ESCA molecular subtypes, tumor stage, survival status, and age were used as patient annotations. Red represented high expression of regulators and blue represented low expression. (E) Functional annotation for m6A-related genes using GO enrichment analysis and KEGG enrichment analysis by Metascape. (F) Prognostic signatures based on TMB and m6Ascore in ES for overall survival. (G) DFS-related prognostic model. High-risk and low-risk groups were divided based on the median value of risk score. The upper plot illustrated assignment of patients' survival status and survival times, the middle plot showed the risk score curve, and the bottom heatmap depicted splicing distribution of the AS in compound prognostic models. Color transition from blue to red indicates the increasing PSI value of corresponding AS event from low to high. (H) Differences in m6Ascore among high- and low-risk clusters in TCGA cohort. The Kruskal–Wallis test was used to compare the statistical difference between three gene clusters (P < 0.001). (I) m6A modification site of ABI1 gene is close to the region where ABI1-ES variable editing occurs and the expression of ALKBH5 was correlated with ABI1.
Using unsupervised clustering based on the expression of m6A regulators, two distinct modification patterns were eventually identified, in which 49 cases were classified in pattern A and 112 cases in pattern B (Fig. 1C; Fig. S4). Notably, 22 m6A regulators were highly expressed in pattern A patients who had a worse prognosis. In contrast, the 94 EC patients clustered in gene cluster B had a worse prognosis (Fig. S4).
The following GSVA analyses revealed that the cluster A modification pattern was significantly associated with RNA alternative splicing. The cross-talk among the regulators of writers, readers, and erasers may play critical roles in forming different m6A modification patterns and the prognosis of EC patients.
We also confirmed that DEGs (different expression genes) were characterized by the status of cell cycle and RNA localization (Fig. 1E). Consistent with the above findings, patients with DNA_REPAIR, CELL CYCLE, G2M_CHECKPOINT, and DNA REPLICATION pathways were classified into gene cluster A, which was relevant to the poor survival outcome (Fig. S5, 6 and Table S1–4).
Considering the individual heterogeneity and complexity of m6A modification, we constructed a scoring system based on these phenotype-related genes to quantify the m6A modification pattern of individual EC patients, which was termed as m6Ascore. We also tested the correlation between the known features and the score of m6A, such as the prognosis of patients and tumor staging. The Kruskal–Wallis test revealed that m6A cluster A showed a higher median score than gene cluster B. Moreover, the clustering of EC patients with high m6Ascore has a better prognosis. It suggested that the m6Ascore could better indicate the m6A modification patterns of an individual tumor, and further evaluate the tumor prognosis (Fig. 1F; Fig. S7–10).
Moreover, we found that patients with both low m6Ascore and high TMB score had the poorest prognosis, while patients with both high m6Ascore and low TMB score had the best prognosis. These results suggested that TMB and m6Ascore which are relatively independent (Fig. 1G; Fig. S10), cooperatively affect the prognosis of EC patients. To further construct the risk model of RNA alternative splicing, we used multi-Cox to screen the key RNA alternative editing target molecules. The patients with a higher risk score showed a poor prognosis. Furthermore, we calculated the ROC curves of clinical features, AS prognostic model, and the nomogram in the training group. Further analysis showed a significant negative correlation between risk score and m6Ascore. The multi-Cox analysis established a risk model that had a significant negative correlation with m6Ascore and m6A demethylase ALKBH5 (Fig. 1H; Fig. S11, 12 and Table S6).
Interestingly, we found that the m6A modification site of ABI1 gene is close to its variable editing region, in which the expression of ABI1 was correlated with ALKBH5 (Fig. 1I; Fig. S13–16). According to the above results, the occurrence of ABI1|11037|ES alternative splicing is closely related to the demethylation of ALKBH5, which is conducive to a better prognosis of EC patients.
Two different patterns of m6A methylation modification were identified, which represent distinct types of RNA alternative splicing. Enrichment of m6A regulators leads to a relatively complicated interaction pattern between RNA alternative splicing activation and cardiac activation. We screened the genes closely related to the prognosis from the differential genes in the two clusters, and constructed the m6Ascore model according to the expression of these genes. Interestingly, the high or low m6Ascore model corresponds to the patients in the two clusters. This phenomenon indicates that the m6Ascore model could better evaluate the impact of m6A regulators on the prognosis of patients. The cluster with high TMB and low m6Ascore has a worse prognosis, whereas the cluster with low TMB and high m6Ascore has a better prognosis. This further suggests that the prognosis of tumor patients is determined by many external independent but internal related factors.
The expression of ALKBH5 is positively correlated with a better prognosis in esophageal cancer patients. We further analyzed the RNA alternative splicing in all esophageal cancer samples, screened alternative splicing genes closely related to the prognosis of patients, and constructed a stable risk assessment model through multiple regression analysis. As we know, the probability of ES events is much higher than that of other alternative splicing types. However, the high-risk factor determining the poor prognosis of patients with esophageal cancer is other alternative splicing types such as AT or AA. We speculate that this phenomenon may be caused by the effects of different alternative splicing types on gene function. What's more, we found that there was a close correlation between alternative splicing risk score and m6Ascore (P = 0.0001), which means that m6A modification may directly affect the occurrence of these key alternative splicing events. The results revealed a close correlation between ALKBH5 and ABI1|11037|ES, and the occurrence of ABI1|11037|ES events was closely related to the good prognosis of EC patients, which is further confirmed by the in vitro proliferation and apoptosis assays (Fig. S17, 18).
We constructed a risk model of alternative splicing by multi-Cox analysis, which can predict the prognosis of EC patients. Further studies showed that the alternative splicing risk model is negatively correlated with the m6Ascore model. In addition, the risk model was found to negatively correlate with ALKBH5 expression, and ABI1-ES and SDCBP-ES events. However, only ABI1-ES events are closely related to the poor prognosis of EC patients. These results suggested that the alternative splicing risk model can effectively evaluate the prognosis of EC patients.
Author contributions
YGP and LSK designed this work. CBZ, QYY, FG, FFZ, RX, CFC, WXW, DBH and ZYL integrated and analyzed the data. LSK, YGP and CBZ wrote this manuscript. YGP and LSK edited and revised the manuscript. All authors approved this manuscript.
Conflict of interests
All authors declare that there is no conflict of interests.
Funding
This work was supported by the 2022 Anhui Health Research Project Key Project (China) (No. AHWJ2022a017), the Anhui Provincial Natural Science Foundation of China (No. 2008085MH299). The fundamental Research Funds for the Central Universities (China) (No. WK9110000008, WK9110000090, WK9110000132 and WK9110000086). The Postdoctoral Research Funding of Anhui Province in 2019 (China) (No. 2019B371). The Youth Fund of Anhui Cancer Hospital (China) (No. 2018YJQN017, 2018YJQN004, 2020YJQN003 and 2018YJQN004) and the Youth Technical Backbone Fund of West Branch of the First Affiliated Hospital of USTC granted to CBZ and LSK, respectively.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.12.009.
Contributor Information
Qiyi Yi, Email: yiqiyi@ahmu.edu.cn.
Chunbao Zang, Email: zangchunbao@ustc.edu.cn.
Youguang Pu, Email: pyg@ustc.edu.cn.
Abbreviations
- ABI1
Ablinteractor 1
- ALKBH5
Alkylation repair homolog protein 5
- EC
Esophageal cancer
- FTO
Fat-mass and obesity-associated protein
- GDC
Genomic Data Commons
- GSVA
Gene set variation analysis
- IGF2BP1
Insulin-like growth factor 2 mRNA binding protein 1
- IGF2BP2
Insulin-like growth factor 2 mRNA binding protein 2
- m6A
N6-methyladenosine
- METTL14
Methyltransferase-like 14
- PCA
Principal component analysis
- PSI
Percent spliced-in
- TCGA
Cancer Genome Atlas
- TMB
Tumor mutational burden
- WTAP
Wilms tumor 1 associated protein
- YTHDF1
YTH N6-methyladenosine RNA binding protein 1
- YTHDF2
YTH N6-methyladenosine RNA binding protein 2
- YY1
Yin Yang-1
Appendix A. Supplementary data
The following are the Supplementary data to this article.
.
.
References
- 1.Yue Y., Liu J., He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29(13):1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan D., Batista P.J. RNA post-transcriptional modification speaks to chromatin. Nat Genet. 2020;52(9):868–869. doi: 10.1038/s41588-020-0685-3. [DOI] [PubMed] [Google Scholar]
- 3.Modrek B., Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30(1):13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 4.Stamm S., Ben-Ari S., Rafalska I., et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Ule J., Blencowe B.J. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol Cell. 2019;76(2):329–345. doi: 10.1016/j.molcel.2019.09.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
.
.