Abstract
Circular RNAs (circRNAs) are a special class of single-stranded RNA molecules with covalently closed loops widely expressed in eukaryotic organisms. CircRNAs have long been considered to play important roles in various physiological and pathological processes as non-coding RNAs. However, circRNAs have recently garnered considerable attention due to their ability to be translated into peptides/proteins via internal ribosome entry site- or N6-methyladenosine-mediated pathways or rolling translation mechanisms. Furthermore, dysregulation of translatable circRNAs and their encoded proteins has been associated with developing and progressing diseases such as cancer. This review aims to summarize the driving mechanisms of circRNA translation and the available strategies in circRNA translation research. The main focus is on the emerging biological functions of translatable circRNAs, their regulatory mechanisms, and potential clinical applications in human diseases to provide new perspectives on disease diagnosis, prognosis, and targeted therapy.
Keywords: Circular RNA, Clinical applications, Function, Peptide, Translation
Introduction
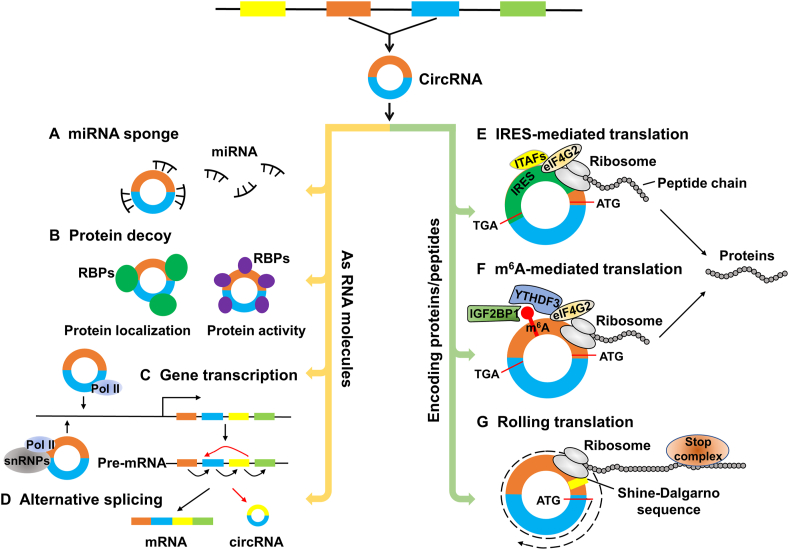
Circular RNAs (circRNAs) are a special class of non-coding RNAs widely present in eukaryotic cells and conserved among different species.1,2 In contrast to traditional linear RNAs, circRNAs have a closed continuous loop structure lacking terminal 5′ caps and 3′ polyadenylated tail.3 CircRNAs are generally expressed at low levels.1,4,5 Therefore, since the first circRNAs were discovered in RNA viruses in 1976,6,7 they have long been considered accidental byproducts or ‘splicing noise’ with little functional potential.8 However, thousands of circRNAs have been discovered recently using high-throughput RNA sequencing (RNA-seq) technology and bioinformatics approaches.9 An increasing number of studies have confirmed multiple biological functions of circRNAs. For instance, circRNAs can act as miRNA sponges or competing endogenous RNAs to combine with miRNA and influence tumorigenesis and metastasis in multiple cancers (Fig. 1A). CircRNAs can also function as protein decoys to regulate their localization and activity10, 11, 12, 13 (Fig. 1B). Additionally, circRNAs can regulate gene transcription or alternative splicing by interacting with RNA polymerase II complex and snRNPs14,15 (Fig. 1C, D).
Figure 1.
The functions and translation mechanisms of circRNAs. (A–D) circRNAs, as RNA molecules, regulate miRNA functions as miRNA sponge (A), protein localization and activity as protein decoy (B), gene transcription (C), and alternative splicing (D). (E–G) circRNAs exert their functions by encoding proteins or peptides through cap-independent mechanisms. (E) IRES-mediated translation. eIF4G2 recognizes and bonds IRES on circRNA to recruit ribosomes and initiate translation with the assistance of IRES-transacting factors (ITAFs). (F) m6A-mediated translation. YTH domain family protein 3 (YTHDF3) or insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1) recognizes m6A modified circRNA and recruits eIF4G2 to m6A to initiate the translation. (G) Rolling translation. CircRNA containing an infinite ORF and start codon (ATG) enables continuous translation. The ribosomal-binding Shine–Dalgarno (SD) sequence on circRNA is associated with the rolling translation initiation, and a stop complex system named “programmed-1 ribosomal frameshifting (-1PRF)-mediated out-of-frame stop codon” can terminate rolling translation in certain natural circRNAs. IRES, internal ribosome entry sites; m6A, N6-methyladenosine; RBPs, RNA-binding proteins; Pol II, polymerase II; snRNP, small nuclear ribonucleoprotein.
In addition to their RNA-based regulatory functions, substantial evidence has revealed that circRNAs can be translated into proteins or peptides.16, 17, 18, 19, 20 For example, circMbl3 was first found to be able to undergo translation by Pamudurti et al in 2017.19 Following this study, more and more endogenous circRNAs were reported to achieve cap-independent protein translation through the internal ribosome entry site (IRES), N6-methyladenosine (m6A), or a unique rolling circle amplification (RCA) method18,21,22 (Fig. 1E–G). Some translatable circRNAs and their encoded proteins have been reported to play important biological functions in human diseases, especially cancers.23, 24, 25, 26 Current research demonstrates that these circRNAs and their encoded proteins are promising biomarkers for human cancer diagnosis, prognosis, and predictive and therapeutic targets.
In this article, we review the latest mechanisms that drive circRNA translation. Moreover, we discuss the biological functions and mechanisms of translatable circRNAs in cancer and other human diseases and their potential clinical applications for cancer diagnosis, prognosis, and targeted therapy. Finally, we summarize the available strategies involved in circRNA translation research, including methods and tools available to predict the coding potential of circRNAs, identify their translation products and study the function of these novel proteins/peptides.
The mechanisms driving circRNA translation
IRES-dependent translation of circRNAs
IRESs are the regulatory RNA elements that can recruit ribosomes to initiate protein translation independent of the 5′ cap structure.27 IRES elements were initially discovered in the viruses of the Picornaviridae family.28,29 Subsequently, some works have reported that IRES elements were also widely distributed in the sequences of eukaryotic mRNAs.30,31 These IRESs can be recognized and bonded by eukaryotic translation initiation factor 4G (eIF4G2). Furthermore, eIF4G2 promotes ribosome assembly and initiates translation with assistance from IRES-transacting factors (ITAFs)32,33(Fig. 1E).
IRES-dependent translation initiation is a widely accepted mechanism of circRNA translation.19,34 Indeed, an earlier study has found that in vitro synthesized circRNA with an IRES element can recruit ribosomes and undergo translation.21 This confirms that circRNAs can be translated in an IRES-mediated manner. Until recently, several studies have revealed that circRNAs containing IRES can also be translated in vivo.19,35,36 For example, Pamudurti et al found that endogenous circMbl was translated into a small protein in an IRES-dependent manner.19 Other circRNAs with ORFs (open reading frames) and IRESs upstream were later reported to be effectively translated by this mechanism in various human diseases (Table 1).
Table 1.
Overview of the translatable circRNAs and their encoded proteins/peptides reported in human diseases.
| Disease | Translatable circRNAs | Expression | Encoded proteins/peptides | Driving models | Biological functions | Action mechanisms | Ref. |
|---|---|---|---|---|---|---|---|
| AD | circAβ-a | Presence in brain | Aβ175 | IRES | Impact the biogenesis of amyloid beta peptides | Aβ175 can be processed to form amyloid beta peptides | 76 |
| BCa | circ-Gprc5a | Up | circGprc5a-peptide (11aa) | Unknown | Promote bladder cancer stem cell self-renewal and invasion | circGprc5a-peptide-Gprc5a- GPCR signaling pathway | 72 |
| CC | HPV16circE7 | Up | E7 oncoprotein-98aa | m6A | Promote cell proliferation and tumor growth | E7 oncoprotein | 48 |
| CRC | circPPP1R12A | Up | circPPP1R12A-73aa | IRES | Promote cell proliferation, migration, invasion, tumor growth, and liver metastasis | circPPP1R12A-73aa-Hippo-YAP signaling pathway | 55 |
| CRC | circFNDC3B | Down | circFNDC3B-218aa | IRES | Inhibit cell proliferation, invasion, migration, tumor growth, and liver metastasis | circFNDC3B-218aa-Snail- FBP1-EMT axis | 64 |
| CRC | circMAPK14 | Down | circMAPK14-175aa | IRES | Inhibit cell proliferation, migration, in vivo tumourigenicity, liver metastasis, and lung metastasis | circMAPK14-175aa-MKK6-MAPK14-FOXC1 axis | 66 |
| CRC | hsa_circ_0006401 | Up | hsa_circ_0006401 peptide (198aa) | Unknown | Promote cell proliferation, migration, tumor growth, and liver metastasis; inhibit cell apoptosis | hsa_circ_0006401-peptide-COL6A3 | 56 |
| CRC | circPLCE1 | Down | circPLCE1-411 | IRES | Inhibit cell proliferation, migration, tumor growth, and liver metastasis | circPLCE1-411-HSP90α-RPS3-NF-κB signaling | 65 |
| CHD | circNlgn | Up | Nlgn173 | Unknown | Increase fibroblast cell cycle and collagen deposition; induce cardiac fibrosis in transgenic mice | Nlgn173-LaminB1-SGK3 and ING4 axis | 77 |
| DMD | circZNF609 | Up | Unknown | IRES | Regulate myoblast proliferation | Unknown | 35 |
| GC | circDIDO1 | Down | DIDO1-529aa | Unknown | Inhibit cell proliferation, migration, invasion, tumor growth, and liver metastasis; promote cell apoptosis | DIDO1-529aa-PARP1 axis and circDIDO1-RBX1-PRDX2 pathways | 69 |
| GC | circMAPK1 | Down | MAPK1-109aa | IRES | Inhibit cell proliferation, migration, tumor growth, and lung metastasis | MAPK1-109aa-MEK1-MAPK pathway | 68 |
| GC | circCOL6A3_030 | Up | circCOL6A3_030_198aa | Unknown | Promote cell migration and liver metastasis | circCOL6A3_030_198aa | 78 |
| GC | circAXIN1 | Up | AXIN1-295aa | IRES | Enhance cell proliferation, migration, invasion, in vivo tumorigenesis, and lung metastasis | AXIN1-295aa-APC-Wnt signaling pathway | 57 |
| GBM | circ-SHPRH | Down | SHPRH-146aa | IRES | Inhibit cell proliferation and in vivo tumorigenicity | SHPRH-146aa-SHPRH-PCNA axis | 36 |
| GBM | circ-AKT3 | Down | AKT3-174aa | IRES | Inhibit cell proliferation, radiation resistance, and in vivo tumorigenicity | AKT3-174aa/p-PDK1-PI3K-AKT pathway | 71 |
| GBM | circ-FBXW7 | Down | FBXW7-185aa | IRES | Induce cell cycle arrest and reduce cell proliferation | FBXW7-185aa-USP28-FBXW7α-c-Myc axis | 67 |
| GBM | circPINTexon2 | Down | PINT87aa | IRES | Inhibit cell proliferation, induce G1 arrest, and enhance IR sensitivity | PINT87aa-PAF1 complex | 17 |
| GBM | circ-SMO | Up | SMO-193a.a. | IRES | Promote the self-renewal, proliferation, and tumorigenicity of brain cancer stem cells | Shh-Gli1-FUS-SMO-193a.a.-Hedgehog signaling | 58 |
| GBM | circ-E-Cad | Up | C-E-Cad | IRES | Promote glioma stem cell self-renewal, sphere-forming frequency, proliferation, invasion, anti-apoptosis, and senescence resistance | C-E-Cad-EGFR-STAT3 signaling | 59 |
| GBM | circ-EGFR | Up | rtEGFR | RCA | Promote tumorigenicity and sensitivity to nimotuzumab | rtEGFR-EGFR signaling | 22 |
| GBM | circHEATR5B | Down | HEATR5B-881aa | IRES | Inhibit the glycolysis, cell proliferation, and tumor growth | HEATR5B-881aa-JMJD5-PKM2 pathway | 24 |
| HCC | circARHGAP35 | Up | circARHGAP35-protein | m6A | Promote tumor cell growth, migration, invasion, and lung metastasis | circARHGAP35 protein-TFII-I | 49 |
| HCC | circMRPS35 | Up | circMRPS35-168aa | IRES | Promote cisplatin resistance | circMRPS35-168aa | 70 |
| HCC | circMAP3K4 | Up | circMAP3K4-455aa | m6A | Promote cell growth, and inhibit cisplatin-induced apoptosis | MIB1-circMAP3K4-455aa-AIF axis | 26 |
| HCC | circβ-catenin | Up | β-catenin-370aa | IRES | Promote cell growth and migration, in vivo tumorigenesis, and liver metastasis | β-catenin-370aa-GSK3β-β-catenin-Wnt pathway | 16 |
| ICC | cGGNBP2 | Up | cGGNBP2-184aa | IRES | Promote cell proliferation, invasion, cell cycle, tumor growth, and lung metastasis | cGGNBP2-184aa-IL-6-STAT3 signaling | 60 |
| LUAD | circASK1 | Down | ASK1-272aa | IRES | Enhance gefitinib sensitivity and gefitinib-induced apoptosis | ASK1-272aa-Akt1-ASK1- JNK-p38 signaling pathway | 50 |
| MM | circBUB1B | Up | circBUB1B_544aa | IRES | Promote cell growth and drug resistance; influence the counterpart cells in the bone marrow microenvironment | Evoke CIN through CEP170 activation | 61 |
| MM | circCHEK1 | Up | circCHEK1_246aa | IRES | Induce CIN and promote osteoclast differentiation | circCHEK1_246aa-CEP170 | 75 |
| MM | circHNRNPU | Up | circHNRNPU_603aa | IRES | Promote cell proliferation and clonal expansion | circHNRNPU_603aa-SKP2-c-Myc axis | 62 |
| NB | ecircCUX1 | Up | p113 | IRES | Promote cell lipid metabolic reprogramming, mitochondrial activity, proliferation, invasion, and lung metastasis | p113-ZRF1-BRD4 axis | 63 |
| TNBC | circ-HER2 | Up | HER2-103 | IRES | Promote cell proliferation, invasion, in vivo tumorigenesis, lung metastasis, and sensitivity to Pertuzumab | HER2-103-EGFR-HER3-AKT signaling pathway | 53 |
| TNBC | circ-FBXW7 | Down | FBXW7-185aa | IRES | Inhibit cell proliferation and migration | FBXW7-185aa-USP28-FBXW7α-c-Myc axis | 109 |
| TNBC | circ-EIF6 | Up | EIF6-224aa | IRES | Promote cell proliferation, cell cycle, migration, invasion, tumor growth, and lung metastasis | EIF6-224aa-MYH9-Wnt/β-catenin pathway | 54 |
Abbreviations: AD, Alzheimer's disease; BCa, bladder cancer; CC, cervical cancer; CIN, chromosomal instability; CRC, colorectal cancer/carcinoma; CHD, congenital heart defects; DMD, Duchenne muscular dystrophy; GC, gastric cancer; GBM, glioblastoma; HCC, hepatocellular carcinoma; ICC, intrahepatic cholangiocarcinoma; IRES, internal ribosome entry site; LUAD, lung adenocarcinoma; m6A, N6-methyladenosine; MM, multiple myeloma; NB, neuroblastoma; RCA, rolling circle amplification; TNBC, triple-negative breast cancer.
Most studies have tested that IRES element is active enough to initiate circRNA translation. Several mechanistic questions remain to be answered. For example, how is IRES-dependent circRNA translation regulated? Only a study by Legnini et al reported that the untranslated regions of circ-ZNF609 could enhance IRES-dependent translation by improving IRES activity.35 Furthermore, Godet et al showed that most IRESs, particularly cellular IRESs, are regulated by ITAFs, exerting their action by at least nine different mechanisms.33 This raises the question of which mechanisms regulate the translation initiation of circRNAs.
Interestingly, recent studies have found that many short IRES-like elements are significantly enriched in endogenous circRNAs and capable of driving the cap-independent translation of circRNAs.37, 38, 39 Any circRNAs sequences longer than 50-nt are expected to contain a short IRES-like element by chance,37, 38, 39 implying that the cap-independent translation of circRNAs driven by short IRES-like elements is pervasive in human cells. Moreover, Fan et al identified that certain RNA binding proteins can specifically recognize these short IRES-like elements and function as a trans-acting factor to promote cap-independent translation of circRNAs, providing a new mechanism for circRNA translation initiation.37
m6A-dependent translation of circRNAs
In addition to the IRES-dependent pathway, the translation of circRNAs can be driven by m6A modification. m6A is the most common and reversible RNA modification in eukaryotic cells.40,41 It has been reported that m6A plays a critical and multifaceted role in regulating mRNA translation. Meyer et al found that m6A in 5′ UTRs (untranslated regions) can initiate cap-independent translation.42 Wang et al showed that the m6A increased translation efficiency in 3′ UTRs.43 Moreover, recent works suggested that m6A in the 5′ UTR of transcripts can influence start codon selection, thereby regulating alternative translation.44,45
Previous transcriptome-wide m6A mapping revealed that the m6A-modified motifs were more enriched in circRNAs than mRNAs.46,47 Given the role of m6A in mRNA translation, it is natural to speculate that m6A may play a similar function in the translation of circRNAs. To test the hypothesis, Yang et al used in vitro translation systems to show that a single m6A site is sufficient to initiate circRNA translation.18 More importantly, when the m6A motif was mutated, circRNA translation was greatly impeded. The authors also noticed that the translation from circRNA is inhibited by m6A demethylate fat mass and obesity-associated protein and enhanced by the methyltransferases METTL3/1418. These observations confirm that the translation of circRNA can be driven by m6A modification. Moreover, a study reported that m6A had been detected and verified to be an essential motif for the translation of circE7.48 In the article by Li et al, the authors reveal that circARHGAP35 exerts its oncogenic functions by translating into a protein in an m6A-dependent manner.49 Recently, Duan et al indicated that circMAP3K4 also could be translated into a peptide via insulin-like growth factor 2 mRNA binding protein 1(IGF2BP1) recognition-mediated m6A modification.26 The mechanism for m6A-mediated circRNA translation is not well known but involves the m6A readers YTH domain family protein 3 (YTHDF3) and IGF2BP1, and the eIF4G2.18 YTHDF3 or IGF2BP1 recognizes m6A-modified circRNAs, recruits eIF4G2 to m6A, and then eIF4G2 initiates the translation (Fig. 1F).
Although many circRNAs with m6A modification have been identified, it has not been established that all of these circRNAs with ORFs can be translated in an m6A-dependent manner. Therefore, carrying m6A modification does not necessarily mean the translation initiation of circRNAs is dependent on them. For example, Wang et al identified a novel protein (ASK1-272a.a) encoded by circASK1 and several m6A sites in circASK1. Nevertheless, they suggested that the m6A modification of circASK1 is responsible for its downregulation in gefitinib-resistant cells instead of its translation initiation.50
Rolling translation of circRNAs
Another important mechanism, called “rolling translation”, drives circRNA translation like DNA rolling circle amplification. Some researchers have demonstrated that circRNAs harboring an infinite ORF and start codon ATG can efficiently translate into proteins in this RCA mechanism.22,51,52 For example, Abe and co-authors reported that synthesized circRNAs are efficiently translated by an RCA mechanism in both cell-free translation systems and living human cells.51,52 Moreover, Liu et al found that circ-EGFR could encode a polymetric protein complex termed rtEGFR (rolling-translated EGFR) through a rolling translation mechanism in glioblastoma, indicating the rolling translation of circRNA also exists endogenously in vivo.22 The mechanism for circRNA rolling translation is not well known but involves the ribosomal-binding Shine–Dalgarno (SD) sequence and a “stop complex”. The ribosomal-binding SD sequence in circRNA was reported to recruit ribosomes and initiate translation.52 Liu et al revealed that the rolling translation of circ-EGFR can be terminated by a stop complex system named “programmed-1 ribosomal frameshifting” (-1PRF)- mediated out-of-frame stop codon” (Fig. 1G).
In summary, it has been demonstrated that IRES- and m6A-mediated translation initiation and rolling translation are important mechanisms for circRNA translation (Fig. 1E–G). However, the current understanding of the translation mechanism of circRNAs is still unsatisfactory.
Roles of translatable circRNAs in cancer
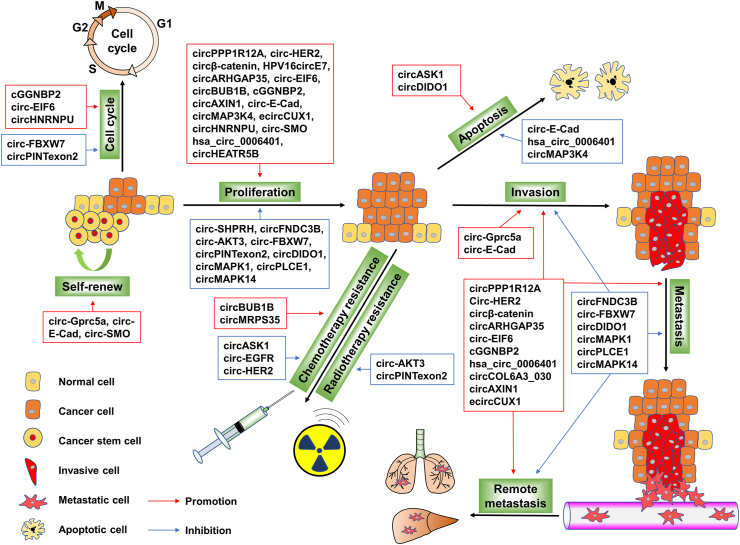
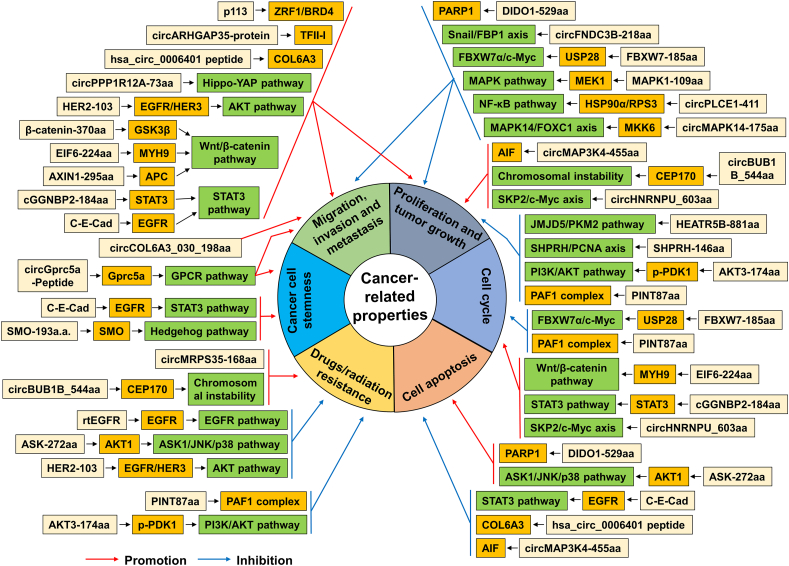
Translatable circRNA disorders are common in human cancers, and the dysregulation of their encoded proteins/peptides is closely associated with the occurrence and progression of cancers. Specifically, translatable circRNAs play important roles in regulating cell proliferation, migration, invasion, metastasis, apoptosis, cell cycle, drug/radiation resistance, and cancer cell stemness by translating into proteins/peptides (depicted and summarized in Fig. 2). Moreover, it has been reported that circRNA-encoded proteins/peptides exert the above-mentioned functions by directly or indirectly regulating the functional proteins or cancer-related signaling pathways (Fig. 3). The biological functions and mechanisms of translatable circRNAs in cancer are also summarized in Table 1.
Figure 2.
Summary of roles of translatable circRNAs in human cancers. Translatable circRNAs play important roles in regulating cell proliferation, migration, invasion, metastasis, apoptosis, cell cycle, drug/radiation resistance, and cancer cell stemness via translation into proteins/peptides. Certain translatable circRNAs tend to affect cancer progression by regulating multiple biological processes.
Figure 3.
Regulatory mechanisms of circRNA-encoded proteins/peptides in cancer-related biological processes. circRNA-encoded proteins/peptides can affect cell proliferation, migration, invasion, metastasis, apoptosis, cell cycle, drug/radiation resistance, and cancer cell stemness by regulating functional proteins or cancer-related signaling pathways directly or indirectly.
Modulating cell proliferation and tumor growth
Major translatable circRNAs identified in human cancers are reported to regulate cell proliferation. The upregulated expression of translatable circRNAs in breast cancer (circ-HER2 and circ-EIF6),53,54 cervical cancer (HPV16circE7),48 colon cancer (circPPP1R12A and hsa_circ_0006401),55,56 gastric cancer (circAXIN1),57 glioblastoma (circ-SMO and circ-E-Cad),58,59 intrahepatic cholangiocarcinoma (cGGNBP2),60 liver cancer (circβ-catenin and circARHGAP35),16,49 multiple myeloma (circBUB1B and circHNRNPU)61,62 or neuroblastoma (ecircCUX1)63 were found to promote cancer cell proliferation both in vitro and in vivo. In-depth studies revealed that these circRNAs could effectively translate into detectable proteins/peptides, promoting cancer cell proliferation via diverse pathways (Fig. 3). For example, Li et al showed that the protein EIF6-224aa encoded by circ-EIF6 promoted the proliferation of breast cancer cells by stabilizing myosin heavy chain 9 (MYH9) and activating the Wnt/β-catenin pathway.54 Likewise, β-catenin-370aa promoted the growth of liver cancer cells, and AXIN1-295aa promoted the proliferation of gastric cancer cells by activating the Wnt/β-catenin pathway.16,57 The signal transducers and activators of the transduction 3 (STAT3) pathway is a key signaling cascade associated with cancer progression. cGGNBP2-184aa activated the STAT3 pathway by interacting with STAT3, thus promoting intrahepatic cholangiocarcinoma cell proliferation and metastasis in vitro and in vivo.60 C-E-Cad promotes glioma stem cell proliferation and invasion by activating epidermal growth factor receptor (EGFR)–STAT3 signaling.59 In addition, circRNA-encoded proteins can exert their functions in cell proliferation by interacting with other functional proteins. Yang et al demonstrated that p113, a protein encoded by ecircCUX1, promotes neuroblastoma cell growth by forming a transcriptional regulatory complex with zuotin-related factor 1 (ZRF1) and bromodomain protein 4 (BRD4).63
Several translatable circRNAs were found to be downregulated in cancer cells and tissues, such as circ-FBXW7 in breast cancer, circFNDC3B in colon cancer, and circDIDO1 in gastric cancer, among others (Table 1). Ectopic overexpression of these circRNAs inhibited cell proliferation and tumor growth. Furthermore, studies have reported that these circRNAs encode proteins/peptides to inhibit cancer cell proliferation and growth via different pathways (Fig. 3). For example, the novel proteins encoded by circFNDC3B, circPLCE1, and circMAPK14 can inhibit the proliferation of colon cancer cells.64, 65, 66 Mechanically, the protein circFNDC3B-218aa can regulate the expression of Snail and fructose-bisphosphatase 1 (FBP1).64 circPLCE1-411 can bind to the HSP90α/RPS3 complex to inhibit NF-κB signaling. Whereas circMAPK14-175aa can competitively bind to MAP kinase kinase 6 (MKK6) to repress MAPK14 phosphorylation, thereby promoting the degradation of forkhead box C1 (FOXC1) via ubiquitination.66 Likewise, studies found that circ-SHPRH, circ-AKT3, circPINTexon2, and circHEATR5B can encode novel proteins in glioblastoma and inhibit the proliferation of glioblastoma cells via different pathways (Fig. 3).
Affecting cell migration, invasion, and metastasis
Regulation of translatable circRNAs expression, in different cancer cells, not only affected their proliferative capacity but also their migratory and invasive abilities. High expression of translatable circRNAs significantly contributed to the capacity for cell migration and invasions in many cancers, such as circ-EIF6 in breast cancer,54 circPPP1R12A in colon cancer,55 and circAXIN1 in gastric cancer,57 among others (Table 1). Conversely, circ-FBXW7, circFNDC3B, circPLCE1, circMAPK14, circDIDO1, and circMAPK1 appeared to be downregulated in cancer cells and tissues. Overexpression of these circRNAs markedly inhibited the migration and invasion of cancer cells.64, 65, 66, 67, 68, 69
Certain translatable circRNAs also play pivotal roles in regulating tumor metastasis in vivo. Most studies used a metastasis animal model created by injecting cancer cells through the tail vein to assess the effects of these circRNAs on tumor metastasis. For example, the injection of cancer cells with circPPP1R12A knockdown into the tail veins of nude mice suppressed tumor liver metastasis compared to those injected with control cells.55 Similarly, downregulation of circ-HER2, circ-EIF6, circAXIN1, cGGNBP2, circβ-catenin, circARHGAP35, or ecircCUX1 significantly inhibited tumor lung metastasis.16,49,53,54,57,60,63 On the contrary, a few translatable circRNAs were shown to have tumor-suppressive effects. circFNDC3B, circPLCE1, circMAPK14, circDIDO1, and circMAPK1 were downregulated in cancer cells, and ectopic overexpression of these circRNAs was able to inhibit tumor metastasis in vivo.64, 65, 66,68,69 The mechanisms by which the above-mentioned translatable circRNAs regulate cancer cell migration, invasion, and metastasis have also been studied. As revealed in Fig. 3, the functional proteins or signaling pathways regulated by circRNA-encoded proteins affect cancer cell proliferation and regulate cancer cell migration, invasion, and metastasis.
Regulating cell cycle and apoptosis
It is well known that dysregulation of the cell cycle and apoptosis play an important role in cancer development and progression. Recently, five translatable circRNAs, cGGNBP2, circ-EIF6, circHNRNPU, circ-FBXW7, and circPINTexon2, were involved in the cell cycle progression of cancer cells. Li et al exhibited that cGGNBP2 encodes a novel protein cGGNBP2-184aa to promote the cell cycle progression of intrahepatic cholangiocarcinoma cells.60 Similarly, circHNRNPU encodes a novel protein circHNRNPU_603aa, which promotes cell cycle progression in multiple myeloma.62 Circ-EIF6 was significantly overexpressed in breast cancer and may promote cell cycle progression by encoding the novel EIF6-224aa peptide.54 On the contrary, circ-FBXW7 and circPINTexon2 stably overexpressing glioblastoma cells exhibited a massive G1 phase arrest compared with their control cells.17,67 As shown in Fig. 3, the mechanisms of these translatable circRNAs exerting functions in cell cycle progression have also been studied.
Regarding cell apoptosis, circMAP3K4, circASK1, circDIDO1, circ-E-Cad, and hsa_circ_0006401 have been associated with the apoptotic process of cancer cells. Specifically, circMAP3K4 encodes a protein circMAP3K4-455aa, which can prevent cisplatin-induced apoptosis in hepatocellular carcinoma cells by interacting with apoptosis-inducing factor mitochondria associated 1 (AIF).26 CircASK1 was downregulated in gefitinib-resistant lung adenocarcinoma cells and could strongly inhibit gefitinib-induced cell apoptosis. Mechanically, circASK1 encodes a novel protein ASK1-272aa to activate apoptosis signal-regulating kinase 1 (ASK1)-mediated apoptotic signaling pathway by competitively binding to Akt1.50 CircDIDO1 overexpression significantly promoted gastric cancer cell apoptosis, whereas circDIDO1 knockdown had the opposite effect, as evaluated by assessing the levels of cleaved caspase 3 and cleaved PARP1.69 Besides, Gao et al reported that circ-E-Cad could decrease cell apoptosis rates of glioma stem cells through its encoded protein.59 Finally, Hsa_circ_0006401 was upregulated in colorectal cancer cells and tissues and could significantly inhibit the apoptotic process of colorectal cancer cells.56
Regulating resistance to therapeutic drugs or radiation
Chemotherapy and radiotherapy are critical cancer treatment strategies, usually administered as preoperative adjuvant or postoperative therapy. Several translatable circRNAs have been reported to play vital roles in drug or radiation resistance (Fig. 2). CircASK1, circ-HER2, circ-EGFR, circBUB1B, and circMRPS35 were associated with drug resistance in different human cancers. CircASK1 enhanced the gefitinib sensitivity of lung adenocarcinoma cells through encoding a novel protein ASK1-272aa, which exerts its function by activating ASK1-dependent apoptosis.50 Likewise, circHER2 can increase the sensitivity of triple-negative breast cancer cells to Pertuzumab via translation into a novel protein HER2-103.53 Liu et al reported that rolling-translated EGFR (rtEGFR) is a polymetric protein complex translated from circ-EGFR. rtEGFR deprivation sensitizes brain tumor-initiating cells to nimotuzumab.22 Tang et al discovered that circBUB1B was significantly upregulated in multiple myeloma and could promote resistance to Bortezomib and Adriamycin in multiple myeloma cells.61 Mechanically, a novel protein (circBUB1B_544aa) encoded by circBUB1B could induce chromosomal instability in multiple myeloma cells by activating CEP170, leading to multiple myeloma drug resistance. In addition, Li et al demonstrated that circMRPS35 encodes a novel peptide (circMRPS35-168aa) upon chemotherapeutic drug treatments, which increased the resistance of hepatocellular carcinoma cells to cisplatin.70
Radiation resistance is also one of the major issues related to treatment failure in patients with malignant tumors. However, the mechanism by which cancer cells become resistant to radiation remains unclear. Recently, two novel circRNA-encoded proteins, AKT3-174aa (encoded by circ-AKT3)71 and PINT87aa (encoded by circPINTexon2),17 were identified as important factors that may be responsible for the resistance to radiation in glioblastoma cells. In glioblastoma cells, AKT3-174aa and PINT87aa were downregulated, and overexpression of the two proteins increased glioblastoma cell sensitivity to radiation. Xia et al further demonstrated that AKT3-174aa inhibits the radiation resistance of glioblastoma cells through binding to phosphorylated pyruvate dehydrogenase kinase 1 (p-PDK1) and thus negatively modulating PI3K/AKT signal intensity.71 In addition, Zhang et al revealed that PINT87aa might contribute to glioblastoma cells' radiation sensitivity by interacting with the polymerase-associated factor 1 (PAF1) complex and blocking cell cycle progression.17
Maintaining the stemness of cancer cells
Translatable circRNAs also affect cancer cell stemness. For example, Wu et al revealed that circ-SMO encodes a novel protein SMO-193a.a. to maintain brain CSC self-renewal via activating Hedgehog signaling. Similarly, Gao et al reported that circ-E-Cad enhanced the stemness properties of glioma stem cells by encoding a protein C-E-Cad, which has been shown to activate EGFR–STAT3 signaling.59 Furthermore, circGprc5a was significantly upregulated in bladder CSCs, mechanically through its involvement in the circGprc5a-Peptide-Gprc5a axis, which drives bladder CSC self-renewal.72
Other potential emerging functions
It is known that the tumor microenvironment (TME) is vital to the progression, metastasis, and treatment of cancer.73,74 Recent studies have reported that several translatable circRNAs or their encoded proteins can be secreted into the TME and might impact the TME. For example, Yang's research team found that multiple myeloma cells could secrete circBUB1B_544aa, circCHEK1_246aa, and circHNRNPU_603aa through exosomes to interfere with various cells in the bone marrow microenvironment.61,62,75 They further demonstrated that these circRNA-encoded proteins could promote multiple myeloma malignant progression and drug resistance by regulating the bone marrow microenvironment. Likewise, the novel protein C-E-Cad encoded by circ-E-Cad was also a secretory protein. It promotes glioblastoma tumorigenicity by directly interacting with EGFR and activating STAT3 signaling.59
A recent study has revealed that a new protein, p113, can promote lipid metabolic reprogramming and mitochondrial activity in neuroblastoma.63 In addition, Song et al showed that a novel protein (HEATR5B-881aa) encoded by zinc finger CCHC-type and RNA-binding motif 1-induced circHEATR5B can suppress aerobic glycolysis in glioblastoma cells.24
Roles of translatable circRNAs in other human diseases
Alzheimer's disease
Mo et al demonstrated that circAβ-a is efficiently translated into a novel polypeptide of 175 amino acids (Aβ175) in cultured cells and the human brain. Aβ175 can be processed into amyloid-beta peptides, hinting at its potential to induce Alzheimer's disease. However, the role and mechanism of Aβ175 in Alzheimer's disease have not been investigated.76
Congenital heart defects
Du et al found that circNlgn is highly expressed in the myocardium of patients with fibrotic heart disease. A novel peptide of 173 amino acids (Nlgn173) translated by circNlgn can function as a transcription factor to promote cardiac remodeling and heart failure.77 Mechanically, Nlgn173 can bind and activate ING4 (inhibitor of growth protein 4) and SGK3 (glucocorticoid-inducible kinase-3) promoters.
Duchenne muscular dystrophy
Circ-ZNF609 is strongly expressed in human myoblasts with Duchenne muscular dystrophy, and its translated peptide is implicated in myoblast proliferation.35 However, the mechanism is still not clear.
Potential clinical applications of translatable circRNAs in cancer
Translatable circRNAs play critical roles in various cancers, and research into their functions and mechanisms suggest promising clinical applications. Based on current research, translatable circRNAs and their encoded proteins might serve as potential diagnostic markers, prognostic markers, predictive markers, or therapeutic targets (Table 2).
Table 2.
The potential clinical applications of translatable circRNAs and their encoded proteins in various cancers.
| Translatable circRNAs or their encoded proteins | Cancer types | Sample size/type | Clinical applications |
Ref. | |||
|---|---|---|---|---|---|---|---|
| Diagnostic biomarker | Prognostic biomarker | Predictive biomarker | Therapeutic target | ||||
| circ-Gprc5a | BCa | 59/tissue | – | Predict survival time | Predict clinical severity and metastasis | Intratumoral injection of circGprc5a antisense oligos impaired tumor growth | 72 |
| circPPP1R12A | CRC | 100/tissue | – | Independent prognostic factor for OS | Predict advanced clinical stage | – | 55 |
| circFNDC3B | CRC | 87/tissue | – | Independent prognostic factor for OS | Predict lymphatic metastasis | – | 64 |
| circMAPK14 | CRC | 72/tissue | – | Predict OS | Predict advanced clinical stage and lymphatic metastasis | – | 66 |
| hsa_circ_0006401 | CRC | 12/tissue | Differentiate CRC patients from healthy individuals | – | Predict lymphatic metastasis | – | 56 |
| circPLCE1/circPLCE1-411 | CRC | 262/tissue; 50/tissue | – | Predict OS | Predict advanced clinical stage | Intratumoral injection of circPLCE1 lentivirus inhibited tumor growths in two CRC PDX models | 65 |
| circDIDO1 | GC | 102/tissue | – | Predict OS | Predict distant metastasis | – | 69 |
| circMAPK1 | GC | 80/tissue | – | Predict OS | Predict advanced tumor stage and LNM | – | 68 |
| MAPK1-109aa | GC | 40/tissue | – | Predict OS | – | – | 68 |
| circCOL6A3_030 | GC | 41/tissue | Predict LNM in GC patients | – | Predict LNM | – | 78 |
| circAXIN1 | GC | 63/tissue | Predict LNM in GC patients | – | Predict advanced tumor stage and LNM | Cholesterol-conjugated siRNA specifically targeting circAXIN1 reduced tumor growth and lung metastasis | 57 |
| circ-FBXW7 | GBM | 60/tissue | – | Predict OS | – | – | 67 |
| circ-SMO/SMO-193a.a. | GBM | 86/tissue | – | Predict OS | – | SMO-193a.a. promoted tumor formation and reduced the overall survival of tumor-bearing mice | 58 |
| circ-EGFR | GBM | 97/tissue | – | Predict OS | – | Circ-EGFR knockdown enhanced the therapeutic effect of nimotuzumab | 22 |
| AKT3-174aa | GBM | 38/tissue | – | Predict OS | – | – | 71 |
| C-E-Cad | GBM | 107/tissue | – | Predict OS | – | Targeting C-E-Cad enhances anti-EGFR therapy for inhibiting GSC tumorigenicity | 59 |
| SHPRH-146aa | GBM | 60/tissue | – | Predict survival time | – | – | 36 |
| circARHGAP35 | HCC | 110/tissue | – | Predict OS, DFS, and recurrence | – | – | 49 |
| circMRPS35 | HCC | 35/tissue | Differentiate HCC patients from healthy individuals | – | Predict LNM and hepatitis B virus infection | – | 70 |
| circMAP3K4 | HCC | 112/tissue | – | Independent prognostic factor for OS and DFS | – | – | 26 |
| cGGNBP2 | ICC | 136/tissue | – | Independent risk factor for OS and RFS | Predict more advanced tumor stage and LNM | – | 60 |
| cGGNBP2-184aa | ICC | – | – | – | – | An auxiliary target for clinical IL-6/STAT3-targeting treatments | 60 |
| circASK1/ASK1-272a.a | LUAD | 48 + 56/tissue | – | Predict OS and PFS | Predict the responsiveness to gefitinib treatment | – | 50 |
| circBUB1B | MM | 48/blood and tissue | – | Predict inferior EFS | – | – | 61 |
| circHNRNPU | MM | 48/blood | – | Predict inferior EFS | – | – | 62 |
| p113 | NB | 42/tissue | – | Predict survival probability | Predict more aggressive clinical course | An inhibitory peptide (ZIP-12) targeted blocking p113-ZRF1 interaction was proved efficient in suppressing NB progression | 63 |
| circ-HER2/HER2-103 | TNBC | 59/tissue | – | Predict OS | Predict the responsiveness to pertuzumab treatment | – | 53 |
| circ-FBXW7 | TNBC | 473/tissue | – | Independent prognostic factor for OS and DFS | Predict LNM | – | 109 |
| circ-EIF6 | TNBC | 98/tissue | – | Independent prognostic factor for OS | Predict distant metastasis | – | 53 |
Abbreviations: BCa, bladder cancer; CRC, colorectal cancer/carcinoma; GC, gastric cancer; GBM, glioblastoma; GSC, glioma stem cells; HCC, hepatocellular carcinoma; ICC, intrahepatic cholangiocarcinoma; LUAD, lung adenocarcinoma; MM, multiple myeloma; NB, neuroblastoma; TNBC, triple-negative breast cancer; OS, overall survival; PFS, progression-free survival; DFS, disease-free survival; RFS, relapse/recurrence-free survival; EFS, event-free survival; LNM, lymph node metastasis; PDX, patient-derived xenograft.
Translatable circRNAs as diagnostic biomarkers
Four translatable circRNAs exhibited distinctive expression patterns in cancer tissues compared with normal tissues and have been reported to have diagnostic values. Hsa circ 0006401 was significantly upregulated in colorectal cancer tissues compared to corresponding para tumor tissues, and its diagnostic value was determined using receiver operating characteristic (ROC) curve analysis.56 Hsa_circ_0006401 could identify colorectal cancer patients among healthy individuals with an area under the ROC curve (AUC) value of 0.77. Similarly, ROC analysis showed that circMRPS35 exhibited excellent diagnostic ability in discriminating between hepatocellular carcinoma patients and normal individuals (AUC = 0.8147).70 In addition, the expression level of circAXIN1 was significantly higher in gastric cancer tissues than the normal control and was associated with advanced tumor stage and lymph node metastasis of gastric cancer. The AUC for circAXIN1 predicting gastric cancer lymph node metastasis was 0.72, with a cutoff value of 1.899.57 Geng et al found that circCOL6A3_030 also yielded diagnostic ability for predicting gastric cancer lymph node metastasis (AUC = 0.675; sensitivity = 92.5%; specificity = 77.5%).78
Translatable circRNAs and their encoded proteins as prognostic biomarkers
Twenty translatable circRNAs and seven circRNA-encoded proteins have been reported to have prognostic values for cancer patients. Their expression level can be used to predict patient survival parameters, such as overall survival (OS), disease-free survival (DFS), progression-free survival (PFS), relapse/recurrence-free survival (RFS), and event-free survival (EFS) (Table 2). The upregulated expression of translatable circRNAs in colon cancer (circPPP1R12A),55 glioblastoma (circ-SMO and circ-EGFR),22,58 hepatocellular carcinoma (circARHGAP35 and circMAP3K4),26,49 intrahepatic cholangiocarcinoma (cGGNBP2),60 and triple-negative breast cancer (circ-HER2 and circ-EIF6)53,54 were reported to predict poor OS. Similarly, several downregulated translatable circRNAs, such as circFNDC3B, circPLCE1, and circMAPK14 in colon cancer,64, 65, 66 circDIDO1, and circMAPK1 in gastric cancer,68,69 circ-FBXW7 in glioblastoma,67 and circASK1 in lung adenocarcinoma,50 have been reported to predict poor OS. Multivariate analysis using Cox proportional hazards model revealed high expression of circPPP1R12A, circMAP3K4, cGGNBP2, and circ-EIF6 and low expression of circFNDC3B and circ-FBXW7 were the independent poor prognostic factors.26,54,55,60,64,67 Furthermore, Kaplan–Meier survival analysis revealed that higher circARHGAP35 and circMAP3K4 expression was associated with shorter DFS,26,49 whereas lower circ-FBXW7 expression was associated with shorter DFS.67 Hepatocellular carcinoma patients with high circARHGAP35 expression and intrahepatic cholangiocarcinoma patients with high cGGNBP2 expression exhibited a shorter RFS.49,60 Wang et al revealed that lung adenocarcinoma patients with low circASK1 expression exhibited shorter PFS times than those with high circASK1 expression.50 In addition, multiple myeloma patients with high circBUB1B and circHNRNPU expression were reported to have significantly inferior EFS survival.61,62
Multiple studies have revealed that the expression levels of circRNA-encoded proteins can also be used to predict cancer patient survival (Table 2). MAPK1-109aa was significantly downregulated in gastric cancer tissues and associated with worse OS.68 Glioblastoma patients with higher SMO-193a.a and C-E-Cad or lower SHPRH-146aa and AKT3-174aa expression were reported to predict poor OS.36,58,59,71 Wang et al found a lower ASK1-272a.a level predicted a shorter PFS in lung adenocarcinoma patients receiving gefitinib treatment.50 In neuroblastoma patients, high p113 expression was associated with poor survival probability.63
Translatable circRNAs and their encoded proteins as predictive biomarkers
Translatable circRNAs and their encoded proteins might be a potential predictive biomarker for cancer progression, metastasis, and therapeutic responses because their expression levels have been reported to be significantly associated with tumor size, grade, differentiation, and stage, lymph node metastasis, distant metastasis, and responsiveness to drug treatment (Table 2). For example, a higher abundance of circPPP1R12A and a lower abundance of circMAPK14, circPLCE1, and its encoded protein circPLCE1-411 were associated with advanced clinical stages in colorectal cancer.55,65,66 Elevated cGGNBP2 expression was significantly associated with a more advanced tumor stage in intrahepatic cholangiocarcinoma; the expression levels of circMAPK1 and circAXIN1 were significantly correlated with tumor stage in gastric cancer.57,60,68 These clinical results suggested that translatable circRNAs and their encoded proteins potentially have predictive value in cancer progression. Furthermore, in terms of metastasis, high expressions of hsa_circ_0006401, circCOL6A3_030, circAXIN1, circMRPS35, cGGNBP2, and circ-EIF6 and low expression of circFNDC3B, circMAPK14, circDIDO1, circMAPK1, and circ-FBXW7 were associated with positive lymph node metastasis or distant metastasis in different cancers, implying that they might act as potential predictors for cancer metastasis (Table 2). Besides, according to Wang and colleagues, ASK1-272a.a might be a biomarker for predicting the responsiveness to gefitinib treatment in patients with advanced lung adenocarcinoma.50 In addition, CircHER2/HER-103 expressing triple-negative breast cancer was sensitive to Pertuzumab, indicating that they have potential values for predicting response to Pertuzumab treatment.53
Translatable circRNAs and their encoded proteins as therapeutic targets
Accumulating studies in patient-derived xenograft (PDX) mouse models and other tumor-bearing mouse models demonstrated that several translatable circRNAs and circRNA-encoded proteins might be potential therapeutic targets for cancer. Liang et al, for example, reported that intratumoral injection of circPLCE1 or circPLCE1-411 lentivirus inhibited tumor growth in two colorectal carcinoma PDX models, implying that circPLCE1 could be a potential therapeutic target for colorectal cancer.65 Similarly, Gu et al claimed that intratumoral injection of circGprc5a antisense oligos significantly suppressed the growth of bladder cancer in tumor-bearing mouse models.72 Furthermore, according to Peng et al, injection of cholesterol-conjugated siRNA specifically targeting circAXIN1 to the subcutaneously formed tumors reduced tumor size in a mouse xenograft model of gastric cancer.57 They also reported that a tail vein injection of circAXIN1 siRNA inhibited the lung metastasis of gastric cancer cells in nude mice. These findings suggest circAXIN1 could be a promising therapeutic target for gastric cancer.57 Interestingly, Liu et al substantiated their results using a mouse xenograft intracranially planted with brain tumor-initiating cells (BTICs) and showed that the overall survival time of mouse-bearing BTICs with stable circ-EGFR knockdown was significantly prolonged after nimotuzumab treatment.22
In addition, circRNA-encoded proteins have received considerable interest for their potential as therapeutic targets (Table 2). For example, Wu et al revealed that SMO-193a.a. promoted self-renewal, proliferation, and tumorigenicity of brain CSCs.58 Deprivation of SMO-193a.a. in the intracranial tumor xenograft model significantly suppressed tumor formation and increased overall survival of mice bearing intracranial brain CSC xenografts, indicating that SMO-193a.a. is a promising target for glioblastoma treatment.58 Likewise, Gao et al confirmed that C-E-Cad is an individual target for combined antibody treatment of glioblastoma because compared to an anti-C-E-Cad antibody or nimotuzumab treatment, a combination treatment with both markedly inhibited the growth of GSC brain tumor xenografts and increased overall survival of the xenograft-bearing mice.59 Besides, Yang et al demonstrated that a novel protein (p113) encoded by ecircCUX1 could facilitate tumorigenesis and the aggressiveness of neuroblastoma (NB) cells via interacting with ZRF1 and BRD4.63 Intravenous administration of an inhibitory peptide (ZIP-12) for targeted blocking of p113-ZRF1/BRD4 interaction led to fewer lung metastatic colonies and prolonged the survival time of mice treated with tail vein injection in NB cells, indicating that p113/ZRF1/BRD4 axis is a potential therapeutic target for NB progression.63 Li et al revealed that cGGNBP2-184aa can facilitate intrahepatic cholangiocarcinoma progression (ICC) by modulating IL-6/STAT3 signaling. Therefore, cGGNBP2-184aa may serve as an auxiliary target for clinical IL-6/STAT3-targeting treatments in ICC.60
Available strategies in circRNA translation research
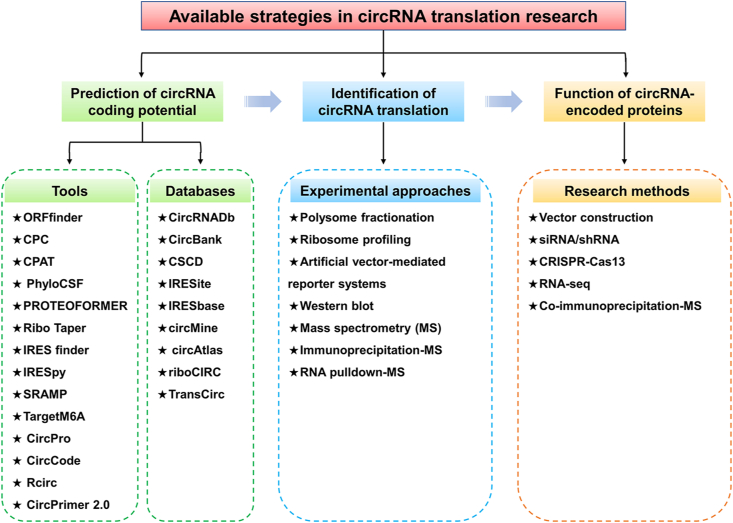
In order to systematically study the translation of circRNAs, numerous research methods and tools have been developed, including bioinformatics tools for predicting circRNA coding potential, experimental approaches for circRNA translation identification, and methods for studying the function of circRNA-encoded proteins/peptides (Fig. 4).
Figure 4.
Strategies of circRNA translation research. These strategies include bioinformatics tools for predicting circRNA coding potential, experimental approaches for circRNA translation identification, and methods for studying the function of circRNA-encoded proteins/peptides.
Prediction of circRNA coding potential
ORFs are nucleic acid sequences beginning with the start codon (AUG in RNA) and ending with one of three stop codons (UAA, UGA, and UAG). ORFs are essential for the translation of RNAs. Therefore, ORFs in circRNA must first be identified to predict whether a circRNA can be translated into peptide/protein. Several tools and databases have been used to predict and identify the ORFs with coding potential in circRNAs. These include sequence analysis tools, methods for identifying coding ORFs, and comprehensive circRNA databases. ORF Finder79 is an easy-to-use sequence analysis tool that can search for all possible ORFs in the user-provided sequence. For an RNA sequence, CPC,80,81 CPAT,82 and PhyloCSF83 can also report the putative ORFs and coding probability. Identifying an ORF's association with ribosomes is required to assess its coding potential. PROTEOFORMER84 and RiboTaper85 are two methods that can identify coding ORFs based on ribosome profiling data. In addition, some comprehensive circRNA databases can be used to predict and identify ORFs with coding potential in circRNAs. For example, CircRNADb,86 CircBank,87 CSCD,88 circAtlas,89 riboCIRC,90 and TransCirc91 can output ORF information and supporting evidence.
In addition to containing ORFs with coding potential, circRNAs require IRESs or m6A modifications to initiate translation. The sequences and structures of many IRESs are well-known. Therefore, many tools and databases have been used to predict IRES. For example, IRES finder92 and IRESpy93 are common tools that can search potential IRES sites on the sequence of circRNAs. IRESite,94 IRESbase,95 circMine,96 circAtlas,89 riboCIRC,90 and TransCirc91 are commonly used databases that allow evaluation of the potential IRES on circRNAs and provide experimental evidence of the IRES region. The m6A is the most abundant base modification of RNAs.18 There are also numerous methods for predicting the m6A modification in circRNAs.23 For example, SRAMP97 and TargetM6A98 are sequence-based tools to predict m6A modification sites on the circRNA sequences of interest. riboCIRC90 and TransCirc91 are two comprehensive databases for translatable circular RNAs, including information about m6A modification sites on circRNAs. In addition, CircPro,99 CircCode,100 Rcirc,101 and CircPrimer 2.0102 are integrated bioinformatics tools for the prediction of circRNAs with protein-coding potential.
Experimental approaches for circRNA translation identification
In general, translatable circRNAs can bind to ribosomes for translation. Polysome fractionation and ribosome profiling can verify whether circRNAs associate with ribosomes.103,104 Polysome fractionation separates polyribosomes by sucrose density gradient centrifugation. After centrifugation, the free RNAs, proteins, and RNAs bound by different ribosomes can be separated in a sucrose solution. Then, circRNA molecules and their active ORFs in the separated solution are analyzed, and the output is used to evaluate the coding potential of circRNAs. Ribosome profiling is a reliable experimental method to sequence ribosome-protected RNA fragments. It uses nuclease to destroy the RNA fragments without ribosome coverage and sequences and analyzes the rest RNA fragments. The output can determine the exact ribosome location and density on circRNAs, ORF location, the starting codon, and other information.105,106
To explore whether the circRNA is indeed translatable, many artificial vector-mediated reporter systems have been developed to test the activity of putative ORFs, IRESs, or m6A in circRNAs. For example, a circRNA overexpressing vector with a Flag tag coding sequence immediately upstream of the stop codon of the predicted ORF can be used to test whether it is translated.55,64 To better exclude the possibility that flag fusion peptides/proteins were translated from an alternative start site inside linear mRNA, another improved circRNA overexpressing vector is often used.53,54,58,67 In this vector, the junction of endogenous circRNA was moved to the stop codon of the predicted ORF, and the Flag tag sequence was separated and inserted reversely on both sides. With this design, the flag fusion peptides/proteins could be produced only upon forming a circular template. Like the Flag tag, green fluorescent protein (GFP) is also a commonly used reporter gene.17 In addition, several reporter systems, such as dual-luciferase reporter system,64,67,71 mCherry/red fluorescent protein (RFP)-GFP dual-cistronic reporter system17,54 and circular vector-based luciferase reporter system,53,58,59 can be used to test the putative IRES activity in circRNAs. To examine the activity of m6A motifs, Yang et al designed a circRNA minigene reporter system that contains a single exon encoding two GFP fragments in a reversed order.19 The schematic structure of the above-mentioned vector-mediated reporter systems was displayed in Figure S1.
Western blot (WB) and mass spectrometry (MS) have been broadly employed to detect and identify putative peptides/proteins encoded by circRNAs. Detecting the protein products of circRNA translation generally requires WB with specifically prepared antibodies, such as those designed against unique amino acid sequences54,59,60 or common amino acid sequences encoded by circRNAs.50,65,69 MS can be used to analyze and identify the amino acid sequence of translated products.
CRISPR/Cas9-mediated gene-editing technology can be used to knock an epitope tag into the predicted ORF of endogenous circRNAs. WB identifies specific bands at the expected molecular weight, indicating the endogenous circRNA was translated. RNA immunoprecipitation and RNA pull-down followed by MS can be performed to verify the interaction of circRNAs with ITAFs or m6A readers to predict whether circRNAs are being translated.
Methods for studying the function of circRNA-encoded proteins
The biological functions of circRNA-encoded protein can be investigated by overexpressing the selected protein. A circRNA expression vector could overexpress circRNA-encoded proteins or a vector carrying the same ORF present in the circRNA but with a linear conformation.53,55,64 However, this approach may lead to the robust overexpression of the artificially constructed protein. To avoid this, Li et al established an inducible SK-Hep-1 cell line expressing circARHGAP35 protein by stably transfecting a Tet-On lentiviral expression vector in which circARHGAP35 ORF sequence was inserted.49 Therefore, a low level of circARHGAP35 protein can be induced to assess its functions.
Another key avenue to investigate the biological function of circRNA-encoded protein is a loss-of-function study by RNA interference. CircRNA knockdown through siRNAs specifically targeting the back–splice junction sequence has been widely used to reduce the expression of its encoded proteins.60,61,71 However, when transfected into cells, siRNAs merely downregulate the targets transiently, and their function relies on high transfection efficiency. The more stable knockdown method uses lentivirus vectors expressing short hairpin RNAs.53,58 Stable circRNA knockdown cell lines can further be obtained by lentiviral infection. The CRISPR-Cas13 RNA targeting system can also be used for circRNA knockdown.107,108
For mechanistic studies, RNA-seq and co-immunoprecipitation (co-IP) analysis are generally carried out to identify the possible signaling pathways by which circRNA-encoded protein exerted its functions.55,59,64 For example, in the study by Zheng et al, RNA-seq was employed to identify the critical signaling pathway regulated by circPPP1R12A-73aa.55 The co-IP assay is a method of investigating putative protein-interacting partners by employing a highly specific antibody against the protein of interest, followed by confirmation by MS. Co-IP/MS has been conducted by many researchers to explore the potential regulatory mechanisms influenced by circRNA-encoded proteins.50,54,60,65
Conclusions and future perspective
Recently, an accumulating number of studies have reported that circRNAs can encode proteins or peptides to participate in the physiology and pathology of human diseases. Despite being early, some circRNAs have been confirmed to achieve protein translation through cap-independent translation mechanisms, including IRES- or m6A-mediated pathways and rolling translation mechanisms. Translatable circRNAs and their encoded proteins have also been demonstrated to exert their biological functions in human diseases through diverse functional proteins or signaling pathways directly or indirectly. Furthermore, the studies described in this review suggest that some translatable circRNAs or their encoded proteins may be used as specific biomarkers or therapeutic targets for human cancers.
Although translatable circRNAs and their encoded proteins have wide application prospects in human diseases, many important issues still need to be addressed in depth. For example, the refined mechanism of natural circRNA translation in eukaryotic cells remains largely unknown. The dysregulation of translatable circRNAs and their encoded proteins is involved in various biological processes of human diseases such as cancer, but the underlying mechanisms have not been fully elucidated. Several translatable circRNAs and the resultant proteins show considerable potential for serving as biomarkers or therapeutic targets in human cancers (Table 2). Nevertheless, whether they can be used in clinical practice has not been established. Moreover, the poor stability, short half-life, and resulting low abundance also significantly limit the clinical application of circRNA-encoded proteins. Therefore, more sensitive approaches are required to detect their expression levels. In addition, it must be confirmed whether functional peptides/small proteins encoded by circRNAs can be developed as new small-molecule peptide drugs to treat current refractory diseases and others. Therefore, more future studies to solve these issues are crucial for the clinical and scientific application of translatable circRNAs and their encoded proteins.
Author contributions
H.L. and C.Z. conceived this manuscript. H.L., W.H., J.Y., and X.W. collected and prepared the related references, drafted the manuscript, and performed data analysis and tabulation. W.H., J.Y., and Y.Z. drew figures. H.L., Y.Z., and C.Z. supervised and revised the manuscript. All authors read and approved the final manuscript.
Conflict of interests
The authors declare no conflict of interests.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81802793 and 82103465), Natural Science Foundation of Shanxi Province (China) (No. 201801D221419), Youth Foundation of First Hospital Affiliated with Shanxi Medical University (China) (No. YQ161701), Science Research Start-up Fund for Doctor of Shanxi Medical University (China) (No. XD1801), Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (China) (STIP; No. 2021-185) and Fund of Shanxi "1331 Project" Key Subjects Construction (China).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.10.015.
Contributor Information
Hongliang Liu, Email: liuhl2018@sxent.org.
Chunming Zhang, Email: zcmsxmu@sxent.org.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Multimedia component 1
References
- 1.Jeck W.R., Sorrentino J.A., Wang K., et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salzman J., Gawad C., Wang P.L., et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo J.U., Agarwal V., Guo H., et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzman J., Chen R.E., Olsen M.N., et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9) doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8(4):547–555. doi: 10.1016/0092-8674(76)90223-3. [DOI] [PubMed] [Google Scholar]
- 7.Sanger H.L., Klotz G., Riesner D., et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocquerelle C., Mascrez B., Hétuin D., et al. Mis-splicing yields circular RNA molecules. Faseb J. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 9.Zheng L.L., Li J.H., Wu J., et al. deepBase v2.0:identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016;44(Database issue):D196–D202. doi: 10.1093/nar/gkv1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashwal-Fluss R., Meyer M., Pamudurti N.R., et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Abdelmohsen K., Panda A.C., Munk R., et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du W.W., Yang W., Liu E., et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q., Wang Y., Wu S., et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metabol. 2019;30(1):157–173. doi: 10.1016/j.cmet.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Li Z., Huang C., Bao C., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Zhang X.O., Chen T., et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Liang W., Wong C.W., Liang P., et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M., Zhao K., Xu X., et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y., Fan X., Mao M., et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pamudurti N.R., Bartok O., Jens M., et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21. doi: 10.1016/j.molcel.2017.02.021. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Wu C., Du Y., et al. Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol Cancer. 2022;21:13. doi: 10.1186/s12943-021-01484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C.Y., Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y., Li Z., Zhang M., et al. Rolling-translated EGFR variants sustain EGFR signaling and promote glioblastoma tumorigenicity. Neuro Oncol. 2021;23(5):743–756. doi: 10.1093/neuonc/noaa279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei M., Zheng G., Ning Q., et al. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J., Zheng J., Liu X., et al. A novel protein encoded by ZCRB1-induced circHEATR5B suppresses aerobic glycolysis of GBM through phosphorylation of JMJD5. J Exp Clin Cancer Res. 2022;41:171. doi: 10.1186/s13046-022-02374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao W., Zhang Y., Zhu Y. Circular RNA circβ-catenin aggravates the malignant phenotype of non-small-cell lung cancer via encoding a peptide. J Clin Lab Anal. 2021;35(9) doi: 10.1002/jcla.23900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan J.L., Chen W., Xie J.J., et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol Cancer. 2022;21:93. doi: 10.1186/s12943-022-01537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Licursi M., Christian S.L., Pongnopparat T., et al. In vitro and in vivo comparison of viral and cellular internal ribosome entry sites for bicistronic vector expression. Gene Ther. 2011;18(6):631–636. doi: 10.1038/gt.2011.11. [DOI] [PubMed] [Google Scholar]
- 28.Jang S.K., Kräusslich H.G., Nicklin M.J., et al. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62(8):2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 30.Jackson R.J. The Current status of vertebrate cellular mRNA IRESs. Cold Spring Harbor Perspect Biol. 2013;5(2):a011569. doi: 10.1101/cshperspect.a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33(20):6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King H.A., Cobbold L.C., Willis A.E. The role of IRES trans-acting factors in regulating translation initiation. Biochem Soc Trans. 2010;38(6):1581–1586. doi: 10.1042/BST0381581. [DOI] [PubMed] [Google Scholar]
- 33.Godet A.C., David F., Hantelys F., et al. IRES trans-acting factors, key actors of the stress response. Int J Mol Sci. 2019;20(4):924. doi: 10.3390/ijms20040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21(2):172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legnini I., Di Timoteo G., Rossi F., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M., Huang N., Yang X., et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 37.Fan X., Yang Y., Chen C., et al. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat Commun. 2022;13(1):3751. doi: 10.1038/s41467-022-31327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y., Wang Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. J Mol Cell Biol. 2019;11(10):911–919. doi: 10.1093/jmcb/mjz091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z. Diverse roles of regulatory non-coding RNAs. J Mol Cell Biol. 2018;10(2):91–92. doi: 10.1093/jmcb/mjy026. [DOI] [PubMed] [Google Scholar]
- 40.Li S., Mason C.E. The pivotal regulatory landscape of RNA modifications. Annu Rev Genom Hum Genet. 2014;15:127–150. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- 41.Wei C.M., Gershowitz A., Moss B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975;4(4):379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 42.Meyer K.D., Patil D.P., Zhou J., et al. 5’ UTR m6A promotes cap-independent translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Zhao B.S., Roundtree I.A., et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J., Wan J., Shu X.E., et al. N6-methyladenosine guides mRNA alternative translation during integrated stress response. Mol Cell. 2018;69(4):636–647. doi: 10.1016/j.molcel.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinnebusch A.G., Ivanov I.P., Sonenberg N. Translational control by 5'-untranslated regions of eukaryotic mRNAs. Science. 2016;352(6292):1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 47.Meyer K.D., Saletore Y., Zumbo P., et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J., Lee E.E., Kim J., et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10:2300. doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y., Chen B., Zhao J., et al. HNRNPL circularizes ARHGAP35 to produce an oncogenic protein. Adv Sci. 2021;8(13) doi: 10.1002/advs.202001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang T., Liu Z., She Y., et al. A novel protein encoded by circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1-dependent apoptosis. Cancer Lett. 2021;520:321–331. doi: 10.1016/j.canlet.2021.08.007. [DOI] [PubMed] [Google Scholar]
- 51.Abe N., Matsumoto K., Nishihara M., et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5 doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abe N., Hiroshima M., Maruyama H., et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem Int Ed Engl. 2013;52(27):7004–7008. doi: 10.1002/anie.201302044. [DOI] [PubMed] [Google Scholar]
- 53.Li J., Ma M., Yang X., et al. Circular HER2 RNA positive triple negative breast cancer is sensitive to Pertuzumab. Mol Cancer. 2020;19(1):142. doi: 10.1186/s12943-020-01259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Wang Z., Su P., et al. circ-EIF6 encodes EIF6-224aa to promote TNBC progression via stabilizing MYH9 and activating the Wnt/beta-catenin pathway. Mol Ther. 2022;30(1):415–430. doi: 10.1016/j.ymthe.2021.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng X., Chen L., Zhou Y., et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang C., Zhou X., Geng X., et al. Circular RNA hsa_circ_0006401 promotes proliferation and metastasis in colorectal carcinoma. Cell Death Dis. 2021;12(5):443. doi: 10.1038/s41419-021-03714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng Y., Xu Y., Zhang X., et al. A novel protein AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin signaling pathway to promote gastric cancer progression. Mol Cancer. 2021;20:158. doi: 10.1186/s12943-021-01457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X., Xiao S., Zhang M., et al. A novel protein encoded by circular SMO RNA is essential for Hedgehog signaling activation and glioblastoma tumorigenicity. Genome Biol. 2021;22:33. doi: 10.1186/s13059-020-02250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao X., Xia X., Li F., et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat Cell Biol. 2021;23(3):278–291. doi: 10.1038/s41556-021-00639-4. [DOI] [PubMed] [Google Scholar]
- 60.Li H., Lan T., Liu H., et al. IL-6-induced cGGNBP2 encodes a protein to promote cell growth and metastasis in intrahepatic cholangiocarcinoma. Hepatology. 2022;75(6):1402–1419. doi: 10.1002/hep.32232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang X., Guo M., Ding P., et al. BUB1B and circBUB1B544aa aggravate multiple myeloma malignancy through evoking chromosomal instability. Signal Transduct Targeted Ther. 2021;6:361. doi: 10.1038/s41392-021-00746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang X., Deng Z., Ding P., et al. A novel protein encoded by circHNRNPU promotes multiple myeloma progression by regulating the bone marrow microenvironment and alternative splicing. J Exp Clin Cancer Res. 2022;41:85. doi: 10.1186/s13046-022-02276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang F., Hu A., Guo Y., et al. p113 isoform encoded by CUX1 circular RNA drives tumor progression via facilitating ZRF1/BRD4 transactivation. Mol Cancer. 2021;20:123. doi: 10.1186/s12943-021-01421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan Z., Cai J., Lin J., et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol Cancer. 2020;19:71. doi: 10.1186/s12943-020-01179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang Z.X., Liu H.S., Xiong L., et al. A novel NF-κB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol Cancer. 2021;20:103. doi: 10.1186/s12943-021-01404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L., Zhou J., Zhang C., et al. A novel tumour suppressor protein encoded by circMAPK14 inhibits progression and metastasis of colorectal cancer by competitively binding to MKK6. Clin Transl Med. 2021;11(10):e613. doi: 10.1002/ctm2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Y., Gao X., Zhang M., et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI J Natl Cancer Inst. 2018;110(3):304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang T., Xia Y., Lv J., et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol Cancer. 2021;20:66. doi: 10.1186/s12943-021-01358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y., Jiang J., Zhang J., et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability. Mol Cancer. 2021;20:101. doi: 10.1186/s12943-021-01390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li P., Song R., Yin F., et al. circMRPS35 promotes malignant progression and cisplatin resistance in hepatocellular carcinoma. Mol Ther. 2022;30(1):431–447. doi: 10.1016/j.ymthe.2021.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xia X., Li X., Li F., et al. Correction to: a novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18:149. doi: 10.1186/s12943-019-1083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu C., Zhou N., Wang Z., et al. circGprc5a promoted bladder oncogenesis and metastasis through Gprc5a-targeting peptide. Mol Ther Nucleic Acids. 2018;13:633–641. doi: 10.1016/j.omtn.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Q., Wang W., Zhou Q., et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19:14. doi: 10.1186/s12943-019-1125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu M., Polyak K. Microenvironmental regulation of cancer development. Curr Opin Genet Dev. 2008;18(1):27–34. doi: 10.1016/j.gde.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gu C., Wang W., Tang X., et al. CHEK1 and circCHEK1_246aa evoke chromosomal instability and induce bone lesion formation in multiple myeloma. Mol Cancer. 2021;20:84. doi: 10.1186/s12943-021-01380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mo D., Li X., Raabe C.A., et al. Circular RNA encoded amyloid beta peptides-a novel putative player in Alzheimer's disease. Cells. 2020;9(10):2196. doi: 10.3390/cells9102196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du W.W., Xu J., Yang W., et al. A neuroligin isoform translated by circNlgn contributes to cardiac remodeling. Circ Res. 2021;129(5):568–582. doi: 10.1161/CIRCRESAHA.120.318364. [DOI] [PubMed] [Google Scholar]
- 78.Geng X., Wang J., Zhang C., et al. Circular RNA circCOL6A3_030 is involved in the metastasis of gastric cancer by encoding polypeptide. Bioengineered. 2021;12(1):8202–8216. doi: 10.1080/21655979.2021.1979915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rombel I.T., Sykes K.F., Rayner S., et al. ORF-FINDER: a vector for high-throughput gene identification. Gene. 2002;282(1–2):33–41. doi: 10.1016/s0378-1119(01)00819-8. [DOI] [PubMed] [Google Scholar]
- 80.Kong L., Zhang Y., Ye Z.Q., et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kang Y.J., Yang D.C., Kong L., et al. CPC2:a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang L., Park H.J., Dasari S., et al. CPAT: coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41(6):e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin M.F., Jungreis I., Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27(13):i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Verbruggen S., Ndah E., van Criekinge W., et al. PROTEOFORMER 2.0:further developments in the ribosome profiling-assisted proteogenomic hunt for new proteoforms. Mol Cell Proteomics. 2019;18:S126–S140. doi: 10.1074/mcp.RA118.001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Calviello L., Mukherjee N., Wyler E., et al. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods. 2016;13(2):165–170. doi: 10.1038/nmeth.3688. [DOI] [PubMed] [Google Scholar]
- 86.Chen X., Han P., Zhou T., et al. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6 doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu M., Wang Q., Shen J., et al. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16(7):899–905. doi: 10.1080/15476286.2019.1600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xia S., Feng J., Chen K., et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu W., Ji P., Zhao F. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020;21(1):101. doi: 10.1186/s13059-020-02018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H., Xie M., Wang Y., et al. riboCIRC: a comprehensive database of translatable circRNAs. Genome Biol. 2021;22(1):79. doi: 10.1186/s13059-021-02300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang W., Ling Y., Zhang S., et al. TransCirc: an interactive database for translatable circular RNAs based on multi-omics evidence. Nucleic Acids Res. 2021;49(D1):D236–D242. doi: 10.1093/nar/gkaa823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao J., Wu J., Xu T., et al. IRESfinder: identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features. J Genet Genomics. 2018;45(7):403–406. doi: 10.1016/j.jgg.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 93.Wang J., Gribskov M. IRESpy: an XGBoost model for prediction of internal ribosome entry sites. BMC Bioinf. 2019;20(1):409. doi: 10.1186/s12859-019-2999-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mokrejs M., Masek T., Vopálensky V., et al. IRESite: a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. 2010;38(Database issue):D131–D136. doi: 10.1093/nar/gkp981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao J., Li Y., Wang C., et al. IRESbase: a comprehensive database of experimentally validated internal ribosome entry sites. Dev Reprod Biol. 2020;18(2):129–139. doi: 10.1016/j.gpb.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang W., Liu Y., Min Z., et al. circMine: a comprehensive database to integrate, analyze and visualize human disease-related circRNA transcriptome. Nucleic Acids Res. 2022;50(D1):D83–D92. doi: 10.1093/nar/gkab809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou Y., Zeng P., Li Y.H., et al. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44(10):e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li G.Q., Liu Z., Shen H.B., et al. TargetM6A: identifying N6-methyladenosine sites from RNA sequences via position-specific nucleotide propensities and a support vector machine. IEEE Trans NanoBioscience. 2016;15(7):674–682. doi: 10.1109/TNB.2016.2599115. [DOI] [PubMed] [Google Scholar]
- 99.Meng X., Chen Q., Zhang P., et al. CircPro: an integrated tool for the identification of circRNAs with protein-coding potential. Bioinformatics. 2017;33(20):3314–3316. doi: 10.1093/bioinformatics/btx446. [DOI] [PubMed] [Google Scholar]
- 100.Sun P., Li G. CircCode: a powerful tool for identifying circRNA coding ability. Front Genet. 2019;10:981. doi: 10.3389/fgene.2019.00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun P., Wang H., Li G. Rcirc: an R package for circRNA analyses and visualization. Front Genet. 2020;11:548. doi: 10.3389/fgene.2020.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhong S., Feng J. CircPrimer 2.0:a software for annotating circRNAs and predicting translation potential of circRNAs. BMC Bioinf. 2022;23(1):215. doi: 10.1186/s12859-022-04705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chassé H., Boulben S., Costache V., et al. Analysis of translation using polysome profiling. Nucleic Acids Res. 2017;45(3):e15. doi: 10.1093/nar/gkw907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ingolia N.T., Ghaemmaghami S., Newman J.R.S., et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ingolia N.T., Brar G.A., Rouskin S., et al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7(8):1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gobet C., Naef F. Ribosome profiling and dynamic regulation of translation in mammals. Curr Opin Genet Dev. 2017;43:120–127. doi: 10.1016/j.gde.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 107.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., et al. RNA targeting with CRISPR-cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., et al. RNA editing with CRISPR-cas13. Science. 2017;358(6366):1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ye F., Gao G., Zou Y., et al. circFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol Ther Nucleic Acids. 2019;18:88–98. doi: 10.1016/j.omtn.2019.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1