Abstract
Osteoarthritis (OA) has been considered non-reversible as articular cartilage wears down with limited repair capacity. Enhanced chondrocyte hypertrophy and increased type X collagen gene (COL10A1) expression have been associated with OA. Therefore, regulators controlling collagen X expression and chondrocyte hypertrophy may play a role in OA intervention. Here, we investigated how Distal-less homeobox 5 (DLX5), the distal-less homeobox family member, controls murine Col10a1 gene expression and chondrocyte hypertrophy in chondrogenic cell models and its role in a murine OA model. Through qRT-PCR and Western blot analyses, we detected significantly increased levels of COL10A1 and DLX5 in hypertrophic MCT and ATDC5 cells compared to their proliferative stage. Forced expression of Dlx5 further increases, while knockdown of Dlx5 decreases COL10A1 expression in hypertrophic MCT cells. We have performed dual-luciferase reporter and ChIP assays and demonstrated that DLX5 promotes reporter activity through direct interaction with Col10a1 cis-enhancer. We established a murine OA model and detected markedly increased COL10A1 and DLX5 in the articular cartilage and subchondral bone of the OA mice compared with the controls. Notably, forced overexpression of DLX5 in hypertrophic MCT cells up-regulates RUNX2, and adjacent DLX5 and RUNX2 binding sites have previously been found within the Col10a1 cis-enhancer. Together, our data suggest that DLX5 may cooperate with RUNX2 to control cell-specific Col10a1 expression and chondrocyte hypertrophy and is involved in OA pathogenesis.
Keywords: Chondrocyte hypertrophy, Col10a1 expression, DLX5, Osteoarthritis, RUNX2
Introduction
Endochondral ossification is the primary path of bone formation for most of the skeleton, especially for long bone development that needs a cartilage intermediate.1,2 In this process, cartilage-forming chondrocytes undergo accumulation, proliferation, hypertrophic differentiation, and apoptosis, and are gradually infiltrated with blood vessels and replaced by bone matrix.3,4 Type X collagen gene (Col10a1), along with hypertrophic chondrocytes where Col10a1 is exclusively expressed, can provide an appropriate environment for blood vessel invasion, mineralization, and modeling of the extracellular matrix that are essential for endochondral ossification.5, 6, 7, 8 Type X collagen is also involved in the process of fracture healing, as increased expression of collagen X was detected in the cartilage callus after injury.9 Human COL10A1 gene mutation is known to cause Schmid-type metaphyseal and other chondrodysplasias that show impaired chondrocyte hypertrophy or maturation.10, 11, 12 Meanwhile, pathological articular chondrocyte hypertrophy with an aberrantly high-level of COL10A1, which mimics the endochondral process, has implicated their roles in osteoarthritis (OA) progression.13, 14, 15, 16, 17, 18
OA is the most common form of arthritis affecting millions of people worldwide. The prominent pathological feature of OA is that the protective articular cartilage wears down over time and this process is progressive and usually non-reversible given its limited repair capacity. However, its association with enhanced chondrocyte hypertrophy and increased collagen X expression makes intervention of OA feasible, as factors that inhibit collagen X expression and chondrocyte hypertrophic differentiation may biologically slow down or reverse the process of OA. These findings above demonstrate that the physiological deposition of collagen X in hypertrophic chondrocytes is essential for endochondral bone formation during skeletal development, while multiple skeletal diseases are associated with aberrant collagen X expression and impaired chondrocyte maturation. Therefore, elucidating the type X collagen gene regulation and identifying its potent regulators during chondrocyte hypertrophy will help with the understanding of the processes of skeletal development, and lead to effective therapeutic strategies aiming to treat bone and cartilage-related disorders, including OA.
For the past 2-to-3 decades, we and others have made significant progress toward understanding the type X collagen gene, especially murine Col10a1 gene regulation. These include the identification of multiple Col10a1 promoters and enhancer elements19 as well as the characterization of many candidate Col10a1 regulators.20,21 We have previously localized murine Col10a1 cis-enhancer to a 150-bp distal promoter and demonstrated that RUNX2 directly interacts with this enhancer and is required but not sufficient for hypertrophic chondrocyte-specific Col10a1 promoter activity in vivo.22, 23, 24 Our further studies indicated that multiple candidate Col10a1 regulators, including DLX5, may bind to this enhancer and drive hypertrophic chondrocyte-specific Col10a1 expression.25,26
The distal-less homeobox (Dlx) gene clusters Dlx1/2, Dlx5/6, and Dlx3/7 constitute one of the pivotal homeobox families in mammalian development.27,28 Specifically, Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development (limb/appendage outgrowth).29 Human DLX5 mutations have been associated with hand-foot deformity syndrome (split hand-foot malformation) that was accompanied by dysplasia of the hands, feet, and phalanges.30,31 DLX5 has also been shown to play an important role in the regulation of osteoblast differentiation by interacting with bone-specific markers RUNX2, bone morphogenetic protein 2 (BMP2), and Msh homeobox 1 (MSX1).32, 33, 34 These findings suggest multiple roles of DLX5 during bone development and possible interaction with RUNX2. Indeed, we have previously identified DLX5 as one of the candidate transcription factors that may interact with the specific murine Col10a1 cis enhancer which locates the RUNX2 binding site.26 RUNX2 is a known essential transcription factor for Col10a1 gene expression, chondrocyte hypertrophy, as well as for osteoblast differentiation, and contributes to OA development.35 In this study, we focused on the roles and mechanisms of DLX5's regulation in cell-specific Col10a1 expression and its effect on chondrocyte hypertrophy in vitro, and possibly in vivo during OA progression.
Materials and methods
Bioinformatics prediction
The identification of the 150-bp Col10al gene enhancer (−4,296 to −4,147) allows us to search for its transcription factor binding sites using web-based software, the TRAP program (http://trap.molgen.mpg.de/cgi-bin/trap_form.cgi) as previously described.26,36 Meanwhile, another software, JASPAR, was also utilized to predict and confirm the transcription factor binding sites of target genes.37 JASPAR (http://jaspar.genereg.net) is an open-access database storing curated, non-redundant transcription factor binding profiles, which document transcription factor binding preferences as position frequency matrices for multiple species in six taxonomic groups. We assessed and calculated the greater possibility of a combination of the scores according to the corresponding algorithm37,38
Cell culture
Three established cell lines were used in this study. The 293T cells were provided by Professor Qixiang Shao at Jiangsu University. These cells were cultured at 37 °C in Dulbecco's Modified Eagle Medium (DMEM, BI, Israel) with 10% fetal bovine serum (FBS, BI, Israel) in a humidified incubator containing 5% CO2. The MCT cells are mouse chondrocytes immortalized with temperature-sensitive simian virus 40 large T antigen. The MCT cell line was originally from Dr. de Crombrugghe's lab at MD Anderson (Houston, USA). These cells proliferate at permissible temperatures (32 °C−33 °C) and express type II collagen and aggrecan but undergo hypertrophic differentiation at 37 °C−39 °C and express type X collagen and osteocalcin, etc.39 MCT cells were maintained in DMEM (BI, Israel) supplemented with 8% FBS (BI, Israel) in a humidified incubator containing 8% CO2 and incubated at 32 °C as per published protocol.39 After being grown until sub-confluence, these MCT cells were further cultured at either 32 °C (proliferative) or 37 °C for additional three days (become hypertrophic) before harvest. The ATDC5 cells were isolated from mouse teratocarcinoma cells and can be treated as a chondrogenic cell model for research on chondrocyte differentiation.40,41 The ATDC5 cells were cultured at 37 °C and maintained in DMEM/F-12(HAM) (1:1) supplemented with 5% FBS (BI, Israel) in a humidified incubator containing 5% CO2. The ATDC5 cell line was kindly provided by Dr. Jiang's lab at Nanjing University Medical School. The ATDC5 cells were treated with 1 × insulin-transferrin-sodium selenite (ITS, Sigma I3146, Germany) to induce hypertrophy.
Plasmids and cell transfection treatment
Small interference RNA (siRNA) sequences for mouse Dlx5 are listed as the following siRNA-Dlx5 sequence (5′-3′CCGUCUCAGGAAUCGCCAATT; 3′-5′UUGGCGAUUCCUGAGACGGTT) and scrambled control sequence (5′-3′UUCUCCGAACGUGUCACGUTT; 3′-5′ACGUGACACGUUCGGAGAATT). All sequences were synthesized by GenePharma (Shanghai, China). Overexpression or interfering with Dlx5 was performed to study its regulatory effect on Col10a1 expression by transient transfection in MCT cells. For the analysis of Col10a1 enhancer activity, the 150-bp enhancer fragment of mouse Col10a1 (ID: 12813) was cloned into pGL4/basic firefly luciferase vector and verified by enzyme digestion and sequencing. Cells were plated into 6-well plates and grown in DMEM supplemented with 8% FBS for 24 h before transfection. Gene knockdown experiment was performed by incubation of 4 μM siRNA with 8 μL of Nano-Trans™ Reagent (Magic science transfer reagent, USA). For plasmids transfection, cells were transfected with lipofectamine 3000 transfection reagent (Catalog: 2092224, Thermo Fisher Scientific, USA) according to the manufacturer's protocol. After 20-min of incubation at room temperature for the mixture of the vectors and lipofectamine 3000 transfection reagent, the complex was then dropped into the media and shaken gently. Cells were cultured at 37 °C in a humidified incubator containing 5% CO2 for 24–48 h according to different experimental requirements.
RNA isolation and qRT- PCR assay
Total RNAs were extracted from cells using Trizol reagent (Ambion, Life Technology, USA), and then quantified using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). 1 μg of total RNA was reversely transcribed into cDNA using the PrimeScript™ RT Reagent Kit (Takara, Osaka, Japan) following the manufacturer's protocol. The complementary DNA product was amplified and analyzed by real-time PCR using SYBR green detection system under the standard protocol. Relative RNA quantities were normalized to levels of β-actin RNA and then calculated using the 2−ΔΔCt method. The primer sequences are listed in Table S1.
Western blotting
The cells in the culture were lysed using the RIPA buffer (Pierce, Rockford, IL, USA) with a protease inhibitor cocktail (Pierce). Total protein was separated by SDS-PAGE electrophoresis using the Novex Nu PAGE system (Invitrogen) and transferred to 0.45-μm PVDF membranes. Membranes were blocked in TBS-T buffer containing 5% nonfat milk for 1 h and incubated overnight at 4 °C with the following primary antibodies: anti-Collagen X (1:1,000, Abcam, #ab182563), anti-DLX5 (1:1,000, Abcam, #ab109737), anti-RUNX2 (1:1,000, Abcam, #ab264077), anti-SOX9 (1:1,000, Abcam, #ab185966), anti-MMP13 (1:1,000, Abcam, #ab51072), and anti-β-ACTIN (1:1,000, Beyotime, China, #AF0003) was used as the internal control. Then the membranes were washed using TBST and hybridized with the horseradish peroxidase (HRP)-linked antibody goat anti-rabbit IgG (1:4,000, Beyotime) or goat anti-mouse IgG for 1 h. Signal detection was carried out with an ECL system (Amersham Pharmacia, Piscataway, NJ, USA).
Transfection and dual-luciferase reporter assay
293T and MCT cells were plated into 24-well plates. After overnight culture, lipofectamine 3000 transfection reagent was used for transfection according to the manufacturer's instructions. Each transfection assay was performed with the addition of either 0.5 μg of the Dlx5 expression vector, or pcDNA3.1 and 0.5 μg of the Col10a1 promoter-luciferase reporter vector, or pGL4 vector, and 0.1 μg pRL-TK was added into each mixed tube. The cells were collected after 48-h incubation, and the firefly and renilla luciferase activities were measured using the dual-luciferase reporter system (Promega, Madison, WI, USA) according to the manufacturer's instructions.
Ethynyldeoxyuridine analysis
Cell proliferation was assessed using a 5-ethynyl-2-deoxyuridine (EdU) labeling and detection kit from RiboBio. Briefly, MCT cells were cultured in 96-well plates at 2 × 104 cells per well and transfected with plasmid DNA or siRNA for 48 h. Then, the cells were incubated with 50 μmol/L EdU labeling media for 4 h at 37 °C under 5% CO2. After treatment with 4% paraformaldehyde and 0.5% Triton X-100, cells were stained with Apollo® fluorescent dye under the manufacturer's protocol. Hoechst 33342 was used to label cell nuclei. The percentage of EdU-positive cells was calculated from five random fields in three wells after analysis with fluorescent microscopy.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using the Pierce™ Agarose ChIP Kit (Thermo Fisher) according to the manufacturer's protocol. In brief, 4 × 106 to 6 × 106 cells were treated with 1% formaldehyde solution for 10 min at room temperature to cross-link proteins to DNA. The sonicated chromatin samples were subjected to immunoprecipitation with anti-DLX5 antibody (ChIP grade) (1:500; Abcam, #ab109737), and these immune complexes were collected by binding with protein A-agarose after overnight incubation at 4 °C. After the complexes were washed with 1 × phosphate-buffered saline, DNA was extracted using phenol/chloroform and ethanol precipitation and then used as a template for PCR amplification of relevant DNA fragments. The specific primer pairs for the 150-bp Col10a1 enhancer sequences are as the following: sense primer, 5′-CCTTCATAAAGTCACAGACCAGT-3'; antisense primer, 5′-ATTGTAGAATCAGAGTATTTGCT-3'.
Destabilization of the medial meniscus (DMM) model and analysis
Animal experiments were performed under the approval of Animal Experimentation of Jiangsu University and according to the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. C57BL/6 male mice at 4 weeks old were purchased from Changzhou Cavens Laboratory Animal Co., Ltd. (Jiangsu, China). After one-week of adaptive feeding, the medial meniscal ligament (MMLT) in the right knee joint was severed to establish the OA model.42,43 For the sham group, a medial incision was operated on the knee joint without MMLT transection. After surgery, the mice were allowed to move and have access to a normal diet and tap water ad libitum. Six weeks after surgery, the mice were euthanized by continuous CO2 inhalation, the right joints were harvested and fixed in 4% paraformaldehyde for 48 h. Whole joints were decalcified in 10% EDTA with rotation at room temperature for 2 weeks, then specimens were embedded in paraffin, and 5-μm frontal sections were taken and subjected to hematoxylin and eosin and safranin O-fast green staining. For immunohistochemistry analysis, sections were incubated with primary antibodies against type X collagen (COL10; ABclonal, Wuhan, China, catalog# A6889; dilution: 1:300) and DLX5 (Abcam, Toronto, ON, Canada, catalog# ab109737; dilution: 1:100).
Statistical analyses
Differences between the groups were determined using Student's t-test. A one-way analysis of variance (ANOVA) test was used to evaluate whether a gross statistically significant change existed. All results were expressed as the mean ± standard error of the mean (SEM). P values < 0.05 or <0.01 were considered statistically significant. All the data were analyzed using GraphPad Prism software version 6.0.
Results
Bioinformatics predicts and basal expression of the candidate transcription factors in MCT cells
We recently reported 48 transcription factor binding sites identified by the TRAP program, here we use another high-quality transcription factor binding profile database JASPAR to further investigate the binding affinity between DLX5 and the Col10a1 distal promoter/enhancer elements.
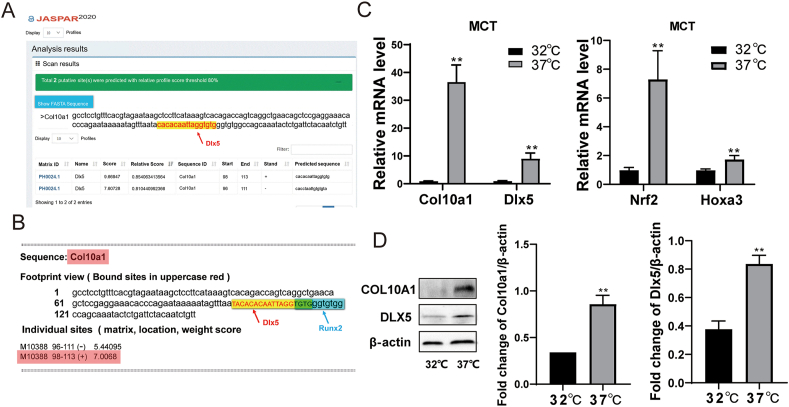
The search criteria were set for vertebrates; after inputting the FASTA-format 150-bp Col10a1 enhancer sequence into the “scan” box, the predicted binding sequences and corresponding scores were listed; the putative DLX5 binding site was at the position of 96–113 of the 150-bp Col10a1 enhancer region (P < 0.05) (Fig. 1A). Meanwhile, the putative DLX5 binding site showed 4 bases overlapping (TGTG) with the tandem-repeat RUNX2 binding sites TGTGGGTGTGGC, which we previously demonstrated to directly interact with the Col10a1 cis-enhancer26 (Fig. 1B).
Figure 1.
Bioinformatics prediction and basal expression of the candidate transcription factors in MCT cells. (A) Two putative and overlapping DLX5 bindings sites within the 150-bp Col10a1 cis-enhancer were predicted by JASPAR. These two sites are at the 3-prime of the enhancers with the relative profile score threshold over 80%. (B) The predicted DLX5 binding site by JASPAR within the Col10a1 enhancer showed four bases overlapping with the previously identified tandem-repeat RUNX2 sites (TGTGGGTGTGGC).26(C) MCT cells were cultured at 32 °C for 3 days (proliferative chondrocytes) and induced to hypertrophy at 37 °C for 24 h (hypertrophic chondrocytes). The mRNA expression of Col10a1, Dlx5, Hoxa3, and Nrf2 was detected by qRT-PCR. Col10a1 mRNA expression was significantly increased in hypertrophic MCT cells as expected. The up-regulation of mRNA levels of Dlx5, as well as two other candidate transcription factors Hoxa3 and Nrf2, were tested and confirmed in hypertrophic MCT cells. (D) MCT cells were cultured at 32 °C for 3 days (proliferative chondrocytes) and induced at 37 °C for 48 h (hypertrophic chondrocytes). The protein expression of COL10A1 and DLX5 was detected by Western blot. Data were expressed as mean ± SEM from three independent experiments. ∗P < 0.05, ∗∗P < 0.01.
MCT cells are mouse chondrocytes, which were immortalized with a temperature-sensitive simian virus 40 large T tumor antigen. According to the change in culture temperature, the chondrocyte differentiation process can be simulated in vitro.39 When cultured at 32 °C, the cells undergo proliferating state, which expresses type II collagen and aggrecan abundantly; while the cells are in a hypertrophic state with increased Col10a1 expression when cultured at 37 °C.39 The expression changes of selected candidate transcription factors at two temperatures were detected by qPCR and Western blot. Compared with proliferating MCT cells (cultured at 32 °C), Col10a1, Dlx5, Hoxa3, and Nrf2 were significantly up-regulated in hypertrophic MCT cells (Fig. 1C). Results of Western blotting also showed that the levels of COL10A1 and DLX5 in hypertrophic MCT cells are significantly higher than that of proliferating MCT cells (Fig. 1D). These results conformed with previous findings and suggested a strong correlation between Dlx5 and Col10a1 expression.37,38
Basal expression of Dlx5 and Col10a1 in ATDC5 cells
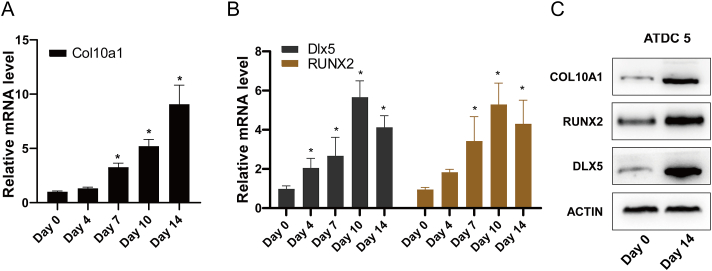
The ATDC5 cell line separated from the mouse teratocarcinoma cells is featured with a chondrocyte phenotype.44, 45, 46 With the chondrogenic potential in the presence of insulin, the ATDC5 cell line is suitable as an in vitro model for exploring cartilage formation. With days of ITS induction, the mRNA expression of Col10a1 was also significantly up-regulated (Fig. 2A). As shown in Figure 2B, the mRNA levels of Dlx5 and Runx2 increased with days and peaked at day 10 after induction. Meanwhile, Western blot results showed that after 14 days of ITS induction, ATDC5 cells underwent hypertrophic differentiation and had highly expressed COL10A1, and the levels of DLX5 and RUNX2 were also significantly up-regulated in hypertrophic ATDC5 cells (Fig. 2C). These results further confirm a strong association between Dlx5 and Col10a1.
Figure 2.
Basal expression of Dlx5 and Col10a1 in ATDC5 cells. (A, B) ATDC5 cells were induced with a complete medium supplemented with 1 × ITS for the corresponding number of days. The mRNA level of Col10a1, Dlx5, and Runx2 were measured by qRT-PCR. (C)Western blotting results showing the protein level of COL10A1, DLX5, and RUNX2 after 14-day ITS induction. Data were expressed as mean ± SEM from three independent experiments, ∗P < 0.05.
Dlx5 up-regulates the expression of Col10a1 in MCT cells
To examine the effect of DLX5 on Col10a1 expression, Dlx5 was either overexpressed or knocked down in MCT cells by transfection with pcDNA3.1/Dlx5 plasmid and Dlx5 small interference fragment siRNA-A respectively.
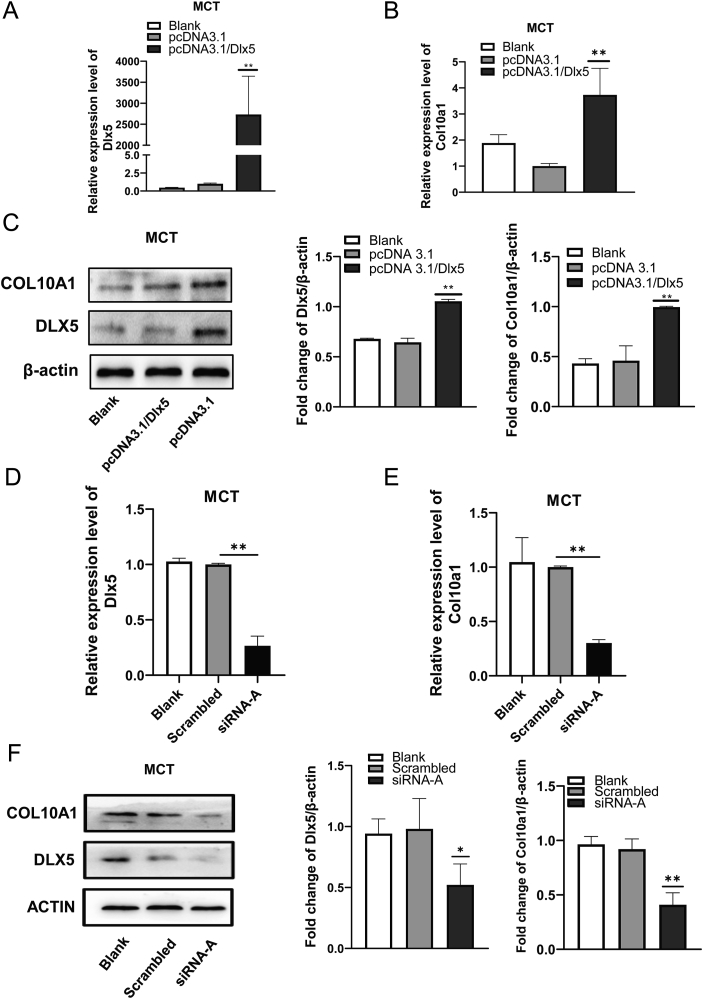
As shown in Figure 3A, C, Dlx5 was over-expressed in MCT cell lines, and the levels of Col10a1 mRNA and protein were significantly increased compared with blank and pcDNA3.1 groups. Meanwhile, the knockdown of Dlx5 by transfection of Dlx5 siRNA-A into MCT cells significantly reduced the expression of Col10a1 gene both at mRNA and protein levels compared with the blank and scrambled control groups (Fig. 3D–F). Together, these results demonstrate that DLX5 is involved in the regulation of Col10a1 and can promote the expression of COL10A1 in hypertrophic chondrocytes.
Figure 3.
Dlx5 promotes the expression of Col10a1 in MCT cells. (A) The mRNA levels of Dlx5 in MCT cells after transfected with pcDNA3.1/Dlx5 and pcDNA3.1 for 24 h. (B) The mRNA levels of Col10a1 after transfected with pcDNA3.1/Dlx5 and pcDNA3.1 for 24 h. (C) Western blotting analysis was used to examine the protein expression of DLX5 and COL10A1 in MCT cells transiently transfected with pcDNA3.1/Dlx5 and pcDNA3.1 for 48 h. β-actin was used as an internal control. (D) The mRNA levels of Dlx5 in MCT cells transfected with Dlx5 siRNA-A and scrambled sequences for 24 h. (E) The mRNA levels of Col10a1 in MCT cells transfected with Dlx5 siRNA-A and scrambled sequences for 24 h. (F) Western blotting analysis was used to examine the protein expression of DLX5 and COL10A1 in MCT cells transiently transfected with Dlx5 siRNA-A and scrambled sequences for 48 h. β-actin was used as an internal control. The results support that forced expression of Dlx5 further increases, while knockdown of Dlx5 decreases Col10a1 expression in hypertrophic MCT cells. Data were expressed as mean ± SEM from three independent experiments. ∗P < 0.05, ∗∗P < 0.01.
Dlx5 strengthens the activity of Col10a1 enhancer
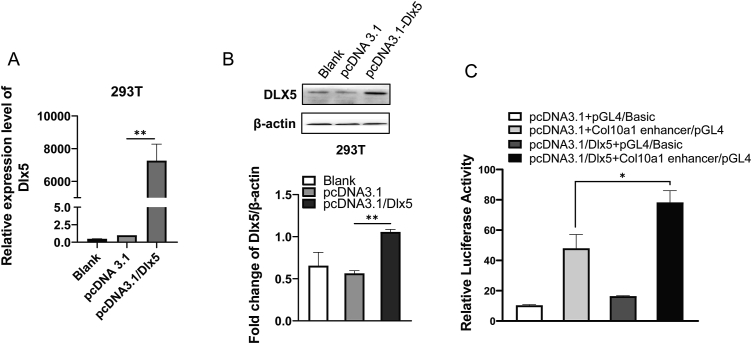
Then we investigated how DLX5 controls Col10a1 expression and the activity of Col10a1 enhancer via dual-luciferase reporter assay. As shown in Figure 4A and B, Dlx5 was weakly expressed in 293T cells as shown in the groups of pcDNA3.1 and blank controls, the protein level of DLX5 was significantly increased after transfection with pcDNA3.1/Dlx5 plasmid. The Col10a1 enhancer activity increased about 1.63 folds in 293T cells co-transfected with pcDNA3.1/Dlx5 and Col10a1 enhancer/pGL4 compared with the cells co-transfected with pcDNA3.1 and Col10a1 enhancer/pGL4, indicating that DLX5 increased Col10a1 enhancer activity (Fig. 4C). Transcription factors or cofactors are known to affect mRNA transcription through influencing the promoter activity of target genes.47 The above results indicate that DLX5 upregulates Col10a1 transcription by enhancing the enhancer activity.
Figure 4.
DLX5 strengthens the activity of the Col10a1 enhancer. (A) 293T cells were transiently transfected with pcDNA3.1/Dlx5 and pcDNA3.1 for 24 h. The relative mRNA level of Dlx5 was detected by qRT-PCR. (B) Western blotting analysis was used to examine the protein expression of DLX5 in 293 T cells transiently transfected with pcDNA3.1/Dlx5 and pcDNA3.1 for 48 h. β-actin was used as an internal control. (C) Dual-Luciferase reporter gene assays were performed to detect relative fluorescence activity in 293T cells. The results demonstrated that DLX5 can promote the activity of the Col10a1 enhancer. Data were expressed as mean ± SEM from three independent experiments. ∗P < 0.05, ∗∗P < 0.01.
Dlx5 directly binds to the 150-bp Col10a1 enhancer
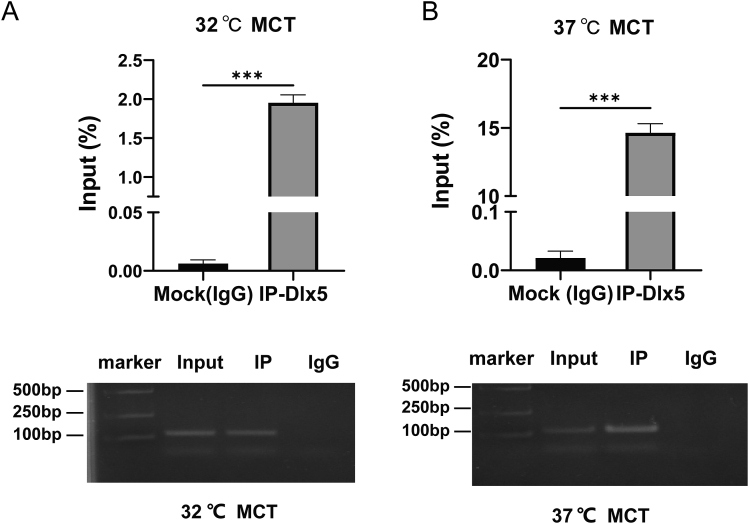
To determine the interaction between DLX5 and Col10a1 promoter/enhancer, we conducted ChIP assays using both proliferative and hypertrophic MCT cells. As shown in Figure 5A and B, immunoprecipitated chromatin samples with anti-DLX5 (Abcam) antibody and input samples were subjected to real-time PCR analysis using a specific primer pair covering the 150-bp Col10a1 enhancer region. The data showed that there is significant enrichment both in proliferative and hypertrophic MCT cells. At the same time, due to the increased expression of Col10a1 and Dlx5 in MCT cells, the fold change of enrichment was higher in hypertrophic than that in proliferative status, suggesting a direct interaction between DLX5 and Col10a1 enhancer.
Figure 5.
DLX5 can directly bind to the 150-bp Col10a1 cis-enhancer. MCT cells cultured at 32 °C (proliferative) (A) and 37 °C (hypertrophy) (B) were subjected to chromatin immunoprecipitation (ChIP) assays and the results demonstrated higher recruitment of DLX5 to the Col10a1 enhancer both in proliferative and in hypertrophic chondrocytes, indicating a direct interaction between DLX5 and Col10a1 ∗∗P < 0.01, ∗∗∗P < 0.001.
Increased Dlx5 and Col10a1 expression during OA progression
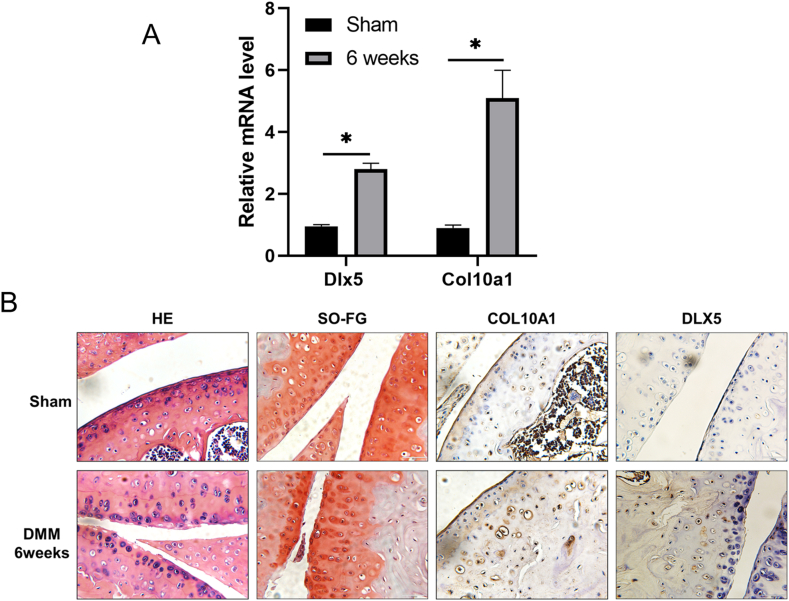
To determine whether DLX5 may play a role in cartilage homeostasis during OA progression, we established a murine OA model by DMM. We then quantified the expression of Dlx5 and Col10a1 in both articular cartilage and subchondral bone tissues of mice with or without surgery. As shown in Figure 6A, the mRNA levels of Dlx5 and Col10a1 in articular cartilage increased significantly six weeks after surgery. Articular surface lesions were visualized in DMM mice by hematoxylin and eosin and safranin O-fast green staining compared with sham controls. Moreover, immunohistochemical analysis detected significantly increased COL10A1 and DLX5 expression in mice receiving DMM surgery compared with the controls (black arrow in Fig. 6B). These data together strongly suggest a correlation between the OA phenotype and the elevated expression of DLX5 and COL10A1.
Figure 6.
Histology and immunohistochemistry analyses of COL10A1 and DLX5 expression in OA mice. (A) The mRNA levels of Col10a1 and Dlx5 in the sham and DMM group. Six weeks after DMM surgery, articular cartilage and part of the subchondral bone tissues were quickly collected and snap-frozen in liquid nitrogen, grounded into powder, and RNA was extracted following the TRizol instructions. As shown in Figure 6A, the mRNA levels of Dlx5 and Col10a1 in articular cartilage increased significantly six weeks after surgery. (B) The hematoxylin and eosin, safranin O-fast green, and immunohistochemistry staining of COL10A1 and DLX5 in the sham and DMM group. Frontal sections of the knee joints were taken and subjected to hematoxylin and eosin and safranin O-fast green staining, smooth and intact knee joint structure could be seen in sham control, while OA mice showed rough surface of the knee joints and joint space narrowing with increased hypertrophic chondrocytes (left two panels). Magnification = 400×. Scale bar = 50 μm. Meanwhile, COL10A1 and DLX5 proteins were stained brown dots after DLX5 and COL10A1 immunostaining in mouse surgery-induced OA cartilage lesions and sham group. Immunohistochemistry staining showed increased hypertrophic chondrocytes with significantly up-regulated COL10A1 and DLX5 expression in the OA mice compared with the sham control (right two panels). Magnification = 400 ×. Scale bar = 50 μm.
Potential effects and mechanism of Dlx5 on chondrocyte differentiation
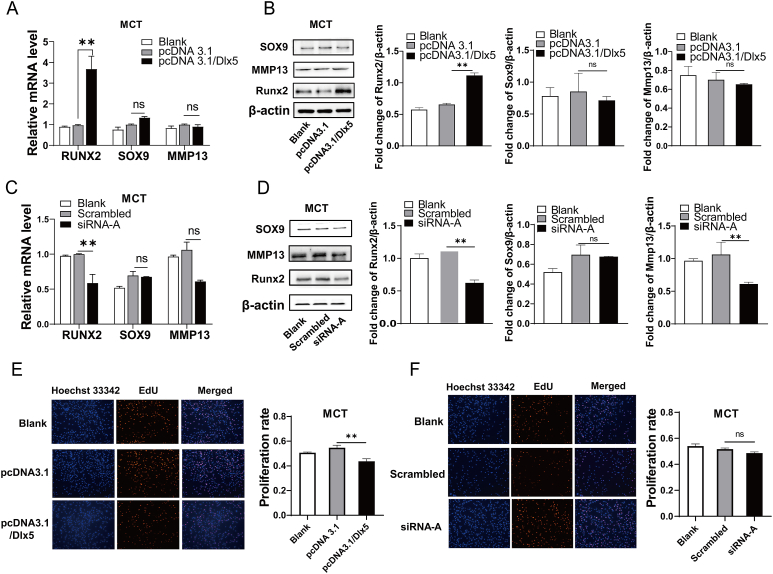
Chondrocyte differentiation and maturation is a complex process involving multiple genes and regulatory factors. Runx2, Sox9, and Mmp13 are all well-known genes closely related to chondrocyte differentiation, maturation, and mineralization. Here, the expression of the above relevant genes in MCT cells and its correlation with DLX5 alteration were evaluated (Fig. 7A–D). The results showed that the protein levels of RUNX2 were significantly increased in pcDNA3.1/Dlx5 transfected MCT cells, while no significant change was observed in the expression levels of SOX9 and MMP13. Meanwhile, the knockdown of Dlx5 in MCT cells by siRNA interference reduced RUNX2 expression. The protein level of MMP13 was also significantly decreased after transfection with siRNA-A of Dlx5. In addition, forced Dlx5 expression in MCT cells suppressed their proliferation, while inhibition of Dlx5 expression had no obvious effect on chondrocyte proliferation (Fig. 7 E, F).
Figure 7.
Effects of DLX5 on chondrocyte differentiation and proliferation. (A) The mRNA levels of Runx2, Sox9, and Mmp13 were detected by qRT-PCR after MCT cells were transiently transfected with pcDNA3.1/Dlx5 for 24 h respectively. (B) Western blotting analysis was used to examine the protein levels of RUNX2, SOX9, and MMP13 in MCT cells transiently transfected with pcDNA3.1/Dlx5 for 48 h respectively. β-actin was used as an internal control. (C) The mRNA levels of Runx2, Sox9, and Mmp13 were detected by qRT-PCR after MCT cells were transiently transfected with Dlx5 siRNA-A for 24 h. (D) Western blotting analysis was used to examine the protein levels of RUNX2, SOX9, and MMP13 in MCT cells transiently transfected with Dlx5 siRNA-A for 48 h. β-actin was used as an internal control. (E) MCT cells were transfected with pcDNA3.1/Dlx5 and pcDNA3.1 for 48 h, and EdU assays were performed to detect cell proliferation ability. (F) MCT cell proliferation ability was detected by EdU after MCT cells were transfected with Dlx5 siRNA-A and scrambled sequences. The results showed that the Runx2 was significantly increased in Dlx5-treated cells, while no significant change in levels of Sox9 and Mmp13 expression. Dlx5 over-expression suppressed MCT cell proliferation, but the knockdown of Dlx5 had no obvious effect on chondrocyte proliferation. Data were expressed as mean ± SEM from three independent experiments. ∗P < 0.05, ∗∗P < 0.01.
Discussion
In this study, we detected increased Dlx5 expression both in hypertrophic MCT cells and ATDC5 cells in which Col10a1 was also increased. We also showed that DLX5 positively up-regulates cell-specific Col10a1 expression in vitro as the forced expression of Dlx5 further increases, while knockdown of Dlx5 decreases Col10a1 expression in hypertrophic MCT cells. Moreover, in a murine OA model, increased expression of DLX5 and COL10A1 was detected in articular cartilage and subchondral bone tissues of OA mice compared to sham controls. These results suggest a positive correlation between DLX5 and Col10a1 expression and that Col10a1 up-regulation by DLX5 may affect chondrocyte hypertrophic differentiation both in vitro and in vivo.
To investigate the underlying mechanism of Dlx5's up-regulation of Col10a1, we analyzed the binding sites within the 150-bp Col10a1 cis-enhancer using JASPAR software.19 The result confirmed the existence of the DLX5 binding site as previously identified by the TRAP program.19 Importantly, we demonstrate that such DLX5 binding site is functional as evidenced by the results of the dual-luciferase reporter and ChIP assays, as well as the results of the co-transfection studies. Not surprisingly, the functional DLX5 binding site is adjacent to the tandem-repeat RUNX2 binding sites within the Col10a1 enhancer as previously described.25 RUNX2 is a known transcription factor that is essential for osteoblast differentiation and chondrocyte maturation during skeletal development.48,49 RUNX2 has also been shown as an indispensable Col10a1 regulator across species.50,51 In our study, we showed that forced expression of Dlx5 up-regulated Runx2 expression in hypertrophic MCT cells while inhibiting chondrocyte proliferation. This suggested that DLX5 may work with Runx2 together to drive the cell-specific Col10a1 expression.19,25 Indeed, it has been reported that DLX5 and myocyte enhancer factor-2C (MEF2C) can directly bind to the 343-bp enhancer of Runx2 and play an important role in directing its expression to osteoblast lineage cells.32
The Dlx gene family, especially Dlx5 and Dlx6 homeobox genes have been extensively studied over the past decade as DLX5 mutation has been associated with hand-foot deformity syndrome (split hand-foot malformation) that was accompanied by dysplasia of the hands, feet, and phalanges. In the meantime, DLX5 is a positive regulator of osteoblastogenesis and is essential for osteoblast-osteoclast coupling. DLX5 has been reported to be specifically expressed in osteogenic cells and may mediate BMP2-induced RUNX2 expression.52 More recently, DLX5 has been shown as a regulator of terminal chondrocyte differentiation and plays a significant role in OA pathogenesis.53 As transiently knockdown of Dlx5 attenuates hypertrophy and decreases apoptotic cell fate in human bone-marrow-derived mesenchymal stem cells, elevated DLX5 expression stimulates cell hypertrophy and apoptosis in cartilage-derived progenitor cells.53 It has been demonstrated that hypertrophic degeneration and undesirable occurrence of calcification remain the major obstacle limiting the application of human mesenchymal stromal cells in cartilage tissue regeneration and repair approaches.54,55 Therefore, a detailed molecular understanding of the hypertrophic process and inhibition of pathological endochondral ossification may offer an alternative strategy for cartilage repair and OA therapeutics.
In our study, we quantified the expression of Dlx5 and Col10a1 in both articular cartilage and subchondral bone tissues of mice with or without the surgery of DMM. The DMM model is a classic micro-surgical technique to mimic the aged spontaneous OA.43,56 During this process, aberrantly hypertrophic chondrocytes exhibit increased synthetic activity which generated matrix degradation products and proinflammatory mediators to stimulate proliferative and proinflammatory response.57,58 Elevated levels of Col10a1 and Dlx5 were observed in the articular cartilage and subchondral bone tissues of the DMM mice compared with the sham group. As the DLX5 binding site is adjacent to the functional RUNX2 sites within the Col10a1 enhancer, we showed that forced overexpression of Dlx5 up-regulates while knockdown of Dlx5 decreases RUNX2 expression in hypertrophic MCT cells. Together, these results suggest that DLX5 may be involved in OA pathogenesis, similar to the role of RUNX2 in OA.35 It has previously been shown that RUNX2 contributes to OA progression as Runx2 heterozygotes are less susceptible to OA.35 However, whether this contribution is related to Dlx5 transactivation remains to be determined.
Taken together, our results suggest that DLX5 positively up-regulates hypertrophic chondrocyte-specific Col10a1 expression via direct interaction with its cis-enhancer. Increased Dlx5 and Col10a1 expression can be seen in the articular cartilage and subchondral bone of OA mice. DLX5 may cooperate with RUNX2 to regulate cell-specific Col10a1 expression and chondrocyte hypertrophy and contribute to OA, although further investigation is needed regarding the role of DLX5 during chondrocyte hypertrophic differentiation and OA progression.
Ethics approval and consent to participate
All experiments and methods were performed in accordance with relevant guidelines and regulations. All animal procedures were approved by the Institutional Animal Care and Use Committee of Jiangsu University.
Author contributions
Conceptualization: Qiping Zheng, Jinnan Chen, Fangzhou Chen, Longwei Qiao; Data curation: Jinnan Chen, Fangzhou Chen, Xuan Wu; Formal analysis: Fangzhou Chen, Qian Wang; Funding acquisition: Qiping Zheng; Methodology: Jinnan Chen, Huiqin Bian, Xiaotong Yang, Haochun Yuan; Software: Xiaojing Zhang, Ruoxuan Hei; Supervision: Qiping Zheng; Writing – original draft: Fangzhou Chen, Jinnan Chen; Writing – review & editing: Qiping Zheng, Qian Wang, Yaojuan Lu.
Conflict of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interests.
Funding
This work was supported by the Jiangsu Provincial Key Research and Development Program (China) (No. BE2020679 to Q. Z.), the Innovation Team (leader) of Jiangsu Province, China (2017, QZ), the National Natural Science Foundation of China (No. 82001576 to L. Q.).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.12.016.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sun M.M., Beier F. Chondrocyte hypertrophy in skeletal development, growth, and disease. Birth Defects Res C Embryo Today. 2014;102(1):74–82. doi: 10.1002/bdrc.21062. [DOI] [PubMed] [Google Scholar]
- 2.Gerstenfeld L.C., Shapiro F.D. Expression of bone-specific genes by hypertrophic chondrocytes: implication of the complex functions of the hypertrophic chondrocyte during endochondral bone development. J Cell Biochem. 1996;62(1):1–9. doi: 10.1002/(SICI)1097-4644(199607)62:1%3C1::AID-JCB1%3E3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 3.Mackie E.J., Ahmed Y.A., Tatarczuch L., et al. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 2008;40(1):46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 5.Chen J., Chen F., Bian H., et al. Hypertrophic chondrocyte-specific Col10a1 controlling elements in Cre recombinase transgenic studies. Am J Transl Res. 2019;11(10):6672–6679. [PMC free article] [PubMed] [Google Scholar]
- 6.Karsenty G., Kronenberg H.M., Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 7.Shen G. The role of type X collagen in facilitating and regulating endochondral ossification of articular cartilage. Orthod Craniofac Res. 2005;8(1):11–17. doi: 10.1111/j.1601-6343.2004.00308.x. [DOI] [PubMed] [Google Scholar]
- 8.Grskovic I., Kutsch A., Frie C., et al. Depletion of annexin A5, annexin A6, and collagen X causes no gross changes in matrix vesicle-mediated mineralization, but lack of collagen X affects hematopoiesis and the Th1/Th2 response. J Bone Miner Res. 2012;27(11):2399–2412. doi: 10.1002/jbmr.1682. [DOI] [PubMed] [Google Scholar]
- 9.Grant W.T., Wang G.J., Balian G. Type X collagen synthesis during endochondral ossification in fracture repair. J Biol Chem. 1987;262(20):9844–9849. [PubMed] [Google Scholar]
- 10.Ikegawa S., Nishimura G., Nagai T., et al. Mutation of the type X collagen gene (COL10A1) causes spondylometaphyseal dysplasia. Am J Hum Genet. 1998;63(6):1659–1662. doi: 10.1086/302158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan J.T., Kremer F., Freddi S., et al. Competency for nonsense-mediated reduction in collagen X mRNA is specified by the 3’ UTR and corresponds to the position of mutations in schmid metaphyseal chondrodysplasia. Am J Hum Genet. 2008;82(3):786–793. doi: 10.1016/j.ajhg.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ain N.U., Makitie O., Naz S. Autosomal recessive chondrodysplasia with severe short stature caused by a biallelic COL10A1 variant. J Med Genet. 2018;55(6):403–407. doi: 10.1136/jmedgenet-2017-104885. [DOI] [PubMed] [Google Scholar]
- 13.van der Kraan P.M., van den Berg W.B. Chondrocyte hypertrophy and osteoarthritis: role in initiation and progression of cartilage degeneration? Osteoarthritis Cartilage. 2012;20(3):223–232. doi: 10.1016/j.joca.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Pullig O., Weseloh G., Ronneberger D., et al. Chondrocyte differentiation in human osteoarthritis: expression of osteocalcin in normal and osteoarthritic cartilage and bone. Calcif Tissue Int. 2000;67(3):230–240. doi: 10.1007/s002230001108. [DOI] [PubMed] [Google Scholar]
- 15.Lamas J.R., Rodríguez-Rodríguez L., Vigo A.G., et al. Large-scale gene expression in bone marrow mesenchymal stem cells: a putative role for COL10A1 in osteoarthritis. Ann Rheum Dis. 2010;69(10):1880–1885. doi: 10.1136/ard.2009.122564. [DOI] [PubMed] [Google Scholar]
- 16.von der Mark K., Kirsch T., Nerlich A., et al. Type X collagen synthesis in human osteoarthritic cartilage. Indication of chondrocyte hypertrophy. Arthritis Rheum. 1992;35(7):806–811. doi: 10.1002/art.1780350715. [DOI] [PubMed] [Google Scholar]
- 17.Dreier R. Hypertrophic differentiation of chondrocytes in osteoarthritis: the developmental aspect of degenerative joint disorders. Arthritis Res Ther. 2010;12(5):216. doi: 10.1186/ar3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von der Mark K., Frischholz S., Aigner T., et al. Upregulation of type X collagen expression in osteoarthritic cartilage. Acta Orthop Scand Suppl. 1995;266:125–129. [PubMed] [Google Scholar]
- 19.Bian H., Zhu T., Liang Y., et al. Expression profiling and functional analysis of candidate Col10a1 regulators identified by the TRAP program. Front Genet. 2021;12 doi: 10.3389/fgene.2021.683939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang L.H., Chen C.H., Wu S.C., et al. Cyclooxygenase-2 regulates PTHrP transcription in human articular chondrocytes and is involved in the pathophysiology of osteoarthritis in rats. J Orthop Translat. 2021;30:16–30. doi: 10.1016/j.jot.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J., Li J., Hu J., et al. Smad4 deficiency impairs chondrocyte hypertrophy via the Runx2 transcription factor in mouse skeletal development. J Biol Chem. 2018;293(24):9162–9175. doi: 10.1074/jbc.RA118.001825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura A., Inose H., Yano F., et al. Runx1 and Runx2 cooperate during sternal morphogenesis. Development. 2010;137(7):1159–1167. doi: 10.1242/dev.045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinoi E., Bialek P., Chen Y.T., et al. Runx2 inhibits chondrocyte proliferation and hypertrophy through its expression in the perichondrium. Genes Dev. 2006;20(21):2937–2942. doi: 10.1101/gad.1482906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Y.F., Soung D.Y., Schwarz E.M., et al. Wnt induction of chondrocyte hypertrophy through the Runx2 transcription factor. J Cell Physiol. 2006;208(1):77–86. doi: 10.1002/jcp.20656. [DOI] [PubMed] [Google Scholar]
- 25.Li F.F., Lu Y.J., Ding M., et al. Runx2 contributes to murine Col10a1 gene regulation through direct interaction with its cis-enhancer. J Bone Miner Res. 2011;26(12):2899–2910. doi: 10.1002/jbmr.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu J., Lu Y., Li F., et al. Identification and characterization of the novel Col10a1 regulatory mechanism during chondrocyte hypertrophic differentiation. Cell Death Dis. 2014;5(10):e1469. doi: 10.1038/cddis.2014.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takechi M., Adachi N., Hirai T., et al. The Dlx genes as clues to vertebrate genomics and craniofacial evolution. Semin Cell Dev Biol. 2013;24(2):110–118. doi: 10.1016/j.semcdb.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Panganiban G., Rubenstein J.L.R. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129(19):4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 29.Robledo R.F., Rajan L., Li X., et al. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 2002;16(9):1089–1101. doi: 10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo Iacono N., Mantero S., Chiarelli A., et al. Regulation of Dlx5 and Dlx6 gene expression by p63 is involved in EEC and SHFM congenital limb defects. Development. 2008;135(7):1377–1388. doi: 10.1242/dev.011759. [DOI] [PubMed] [Google Scholar]
- 31.Kantaputra P.N., Carlson B.M. Genetic regulatory pathways of split-hand/foot malformation. Clin Genet. 2019;95(1):132–139. doi: 10.1111/cge.13434. [DOI] [PubMed] [Google Scholar]
- 32.Kawane T., Komori H., Liu W., et al. Dlx5 and mef2 regulate a novel runx2 enhancer for osteoblast-specific expression. J Bone Miner Res. 2014;29(9):1960–1969. doi: 10.1002/jbmr.2240. [DOI] [PubMed] [Google Scholar]
- 33.Harris S.E., Guo D., Harris M.A., et al. Transcriptional regulation of BMP-2 activated genes in osteoblasts using gene expression microarray analysis: role of Dlx2 and Dlx5 transcription factors. Front Biosci. 2003;8:s1249–s1265. doi: 10.2741/1170. [DOI] [PubMed] [Google Scholar]
- 34.Horie M., Yamaguchi Y., Saito A., et al. Transcriptome analysis of periodontitis-associated fibroblasts by CAGE sequencing identified DLX5 and RUNX2 long variant as novel regulators involved in periodontitis. Sci Rep. 2016;6 doi: 10.1038/srep33666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamekura S., Kawasaki Y., Hoshi K., et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis Rheum. 2006;54(8):2462–2470. doi: 10.1002/art.22041. [DOI] [PubMed] [Google Scholar]
- 36.Zheng Q., Keller B., Zhou G., et al. Localization of the cis-enhancer element for mouse type X collagen expression in hypertrophic chondrocytes in vivo. J Bone Miner Res. 2009;24(6):1022–1032. doi: 10.1359/JBMR.081249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stormo G.D. Modeling the specificity of protein-DNA interactions. Quant Biol. 2013;1(2):115–130. doi: 10.1007/s40484-013-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wasserman W.W., Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5(4):276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 39.Lefebvre V., Garofalo S., de Crombrugghe B. Type X collagen gene expression in mouse chondrocytes immortalized by a temperature-sensitive Simian virus 40 large tumor antigen. J Cell Biol. 1995;128(1–2):239–245. doi: 10.1083/jcb.128.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shukunami C., Ishizeki K., Atsumi T., et al. Cellular hypertrophy and calcification of embryonal carcinoma-derived chondrogenic cell line ATDC5 in vitro. J Bone Miner Res. 1997;12(8):1174–1188. doi: 10.1359/jbmr.1997.12.8.1174. [DOI] [PubMed] [Google Scholar]
- 41.Shukunami C., Shigeno C., Atsumi T., et al. Chondrogenic differentiation of clonal mouse embryonic cell line ATDC5 in vitro: differentiation-dependent gene expression of parathyroid hormone (PTH)/PTH-related peptide receptor. J Cell Biol. 1996;133(2):457–468. doi: 10.1083/jcb.133.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iijima H., Aoyama T., Ito A., et al. Destabilization of the medial meniscus leads to subchondral bone defects and site-specific cartilage degeneration in an experimental rat model. Osteoarthritis Cartilage. 2014;22(7):1036–1043. doi: 10.1016/j.joca.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Glasson S.S., Blanchet T.J., Morris E.A. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15(9):1061–1069. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Atsumi T., Miwa Y., Kimata K., et al. A chondrogenic cell line derived from a differentiating culture of AT805 teratocarcinoma cells. Cell Differ Dev. 1990;30(2):109–116. doi: 10.1016/0922-3371(90)90079-c. [DOI] [PubMed] [Google Scholar]
- 45.Yao Y., Wang Y. ATDC5:an excellent in vitro model cell line for skeletal development. J Cell Biochem. 2013;114(6):1223–1229. doi: 10.1002/jcb.24467. [DOI] [PubMed] [Google Scholar]
- 46.Wilhelm D., Kempf H., Bianchi A., et al. ATDC5 cells as a model of cartilage extracellular matrix neosynthesis, maturation and assembly. J Proteonomics. 2020;219 doi: 10.1016/j.jprot.2020.103718. [DOI] [PubMed] [Google Scholar]
- 47.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 48.Komori T. Runx2, a multifunctional transcription factor in skeletal development. J Cell Biochem. 2002;87(1):1–8. doi: 10.1002/jcb.10276. [DOI] [PubMed] [Google Scholar]
- 49.Komori T. Regulation of proliferation, differentiation and functions of osteoblasts by Runx2. Int J Mol Sci. 2019;20(7):1694. doi: 10.3390/ijms20071694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simões B., Conceição N., Viegas C.S., et al. Identification of a promoter element within the zebrafish colXalpha1 gene responsive to runx2 isoforms Osf2/Cbfa1 and til-1 but not to pebp2alphaA2. Calcif Tissue Int. 2006;79(4):230–244. doi: 10.1007/s00223-006-0111-6. [DOI] [PubMed] [Google Scholar]
- 51.Higashikawa A., Saito T., Ikeda T., et al. Identification of the core element responsive to runt-related transcription factor 2 in the promoter of human type X collagen gene. Arthritis Rheum. 2009;60(1):166–178. doi: 10.1002/art.24243. [DOI] [PubMed] [Google Scholar]
- 52.Heo J.S., Lee S.G., Kim H.O. Distal-less homeobox 5 is a master regulator of the osteogenesis of human mesenchymal stem cells. Int J Mol Med. 2017;40(5):1486–1494. doi: 10.3892/ijmm.2017.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Twomey-Kozak J., Desai S., Liu W., et al. Distal-less homeobox 5 is a therapeutic target for attenuating hypertrophy and apoptosis of mesenchymal progenitor cells. Int J Mol Sci. 2020;21(14):4823. doi: 10.3390/ijms21144823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mueller M.B., Tuan R.S. Functional characterization of hypertrophy in chondrogenesis of human mesenchymal stem cells. Arthritis Rheum. 2008;58(5):1377–1388. doi: 10.1002/art.23370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sekiya I., Vuoristo J.T., Larson B.L., et al. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc Natl Acad Sci U S A. 2002;99(7):4397–4402. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaki S., Blaker C.L., Little C.B. OA foundations - experimental models of osteoarthritis. Osteoarthritis Cartilage. 2022;30(3):357–380. doi: 10.1016/j.joca.2021.03.024. [DOI] [PubMed] [Google Scholar]
- 57.Hunter D.J., Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393(10182):1745–1759. doi: 10.1016/S0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- 58.Sharma L. Osteoarthritis of the knee. N Engl J Med. 2021;384:51–59. doi: 10.1056/NEJMcp1903768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.