Abstract
Alternative splicing (AS) produces the different mRNA splicing bodies, which are then translated into multiple protein isoforms and participate in various biological functions. With a deeper understanding of alternative splicing through the study of transcriptomes using high-throughput sequencing-based methods, the correlation between aberrant AS and diseases triggered a great concern, especially abnormal AS and cancer. Medulloblastoma (MB) is an intracranial tumor in children. Sonic hedgehog MB (SHH-MB) accounted for approximately 30% of MB, which is associated with the activation of SHH signaling. Growing evidence shows that aberrant AS is closely related to the tumorigenesis of MB. Here, we briefly introduced the AS and its mechanism. Next, we described canonical/noncanonical hedgehog signaling and its correlation with MB. The main description focused on AS of various regulators in canonical hedgehog signaling in MB. In addition, we also described AS of various regulators in noncanonical hedgehog signaling. Meanwhile, activated hedgehog signaling also induces AS in MB. Then, we pointed out that aberrant AS of hedgehog signaling is associated with different MB subgroups. Finally, we summarized the therapeutic applications of targeted AS in cancer treatment. In summary, further understanding of AS in SHH-MB could develop therapeutic targets for splicing factors which may be a novel therapeutic strategy.
Keywords: Alternative splicing, Exon, Hedgehog signaling, Medulloblastoma, Tumor suppressor
Introduction
Alternative splicing (AS) is a significant mechanism to generate proteome diversity from limited genes and usually produces transcripts that differ in the coding sequence or untranslated regions (UTR).1 Transcripts from nearly all genes undergo one or more types of AS, which includes exon skip (removal of specific exons, referred to as cassette exons), retained intron (differential retention of introns), switching between alternative 5′ and 3′ splice sites (referred to the alternate donor site and alternate acceptor site), mutually exclusive exons (adjacent inclusive exons cannot coexist), and other patterns (alternate promoter and alternate terminator).2 Different transcripts translate to generate various proteins with multiple functions in life.3
All types of AS require spliceosomes to catalyze RNA splicing reactions.4 The spliceosome consists of small nuclear RNAs (snRNAs) and associated proteins, which comprise a series of small nuclear ribonucleoprotein particles (snRNPs, U1, U2, U4/U6, and U5).5,6 The formation of spliceosome and splicing reactions have been illuminated by some researchers and are not further discussed in this review. However, AS is subject to multiple regulations including cis-acting elements (exon splicing enhancers (ESE), exon splicing silencers (ESSs), intron splicing enhancers (ISEs), and intron splicing silencers (ISSs)) and trans-acting elements (the Ser-Arg-rich (SR) protein family and the heterogeneous nuclear ribonucleoprotein (hnRNP protein family)).2,7 The differentially expressed genes of these splicing factors achieve specificity of splicing as well as flexibility, leading to misregulation of AS and tumorigenesis.8,9
A large number of researchers have elucidated the functions of AS in tumors.10, 11, 12 Firstly, AS is more frequent among cancer cells compared to their normal tissue counterparts.13 There is more mis-splicing in tumors, accompanied by an increase in the rate of transcripts containing premature termination codons (PTCs) in cancer cells. In further data analysis on genes that could be divided into oncogene and tumor suppressor genes (TSGs), PTCs in transcripts encoding TSGs are more frequent than oncogenes. Furthermore, analysis of RNA-seq data across 16 distinct cancer types from The Cancer Genome Atlas (TCGA) revealed that common AS processing differences exist between tumors and their control samples.14 Intron-containing mRNAs are responsible for the transcriptional diversity of multiple cancers. So, what elements caused the diversity of splicing in tumors? Researchers found that somatic, single nucleotide variants (SNVs) triggered intron retention (IR) by integration of RNA-seq analyses of cancer samples with whole exome and whole genome sequencing data from the same patients. SNVs-induced IRs were enriched in TSGs over oncogenes.15 SNVs at intronic splicing donor or acceptor sites are also usually recognized as deleterious. Moreover, mutations interfered with AS through altering splice sites of cancer-associated genes in cis-acting elements or changing the function of trans-acting splicing factors. In addition to mutation, the differentially expressed splicing regulatory proteins also affect splicing. All these researches indicated that cancer cells are associated with aberrant AS.
As for the AS in tumor development, Sciarrillo and colleagues have reviewed the functions of AS in cancer from oncogenesis to drug resistance12 and we no longer further discuss it. We aimed to describe the various types of AS in sonic hedgehog medulloblastoma (SHH-MB), which lacks a comprehensive review.
SHH signaling
Hedgehog (HH) signaling is a highly conserved pathway that is significant in embryonic development and is implicated in numerous birth defects and cancers.16, 17, 18 The mammalian apparatus of the HH pathway includes three kinds of HH ligands (Three HH homologous genes in mammals: Sonic Hedgehog (SHH), Indian Hedgehog (IHH), and Desert Hedgehog (DHH), which encode SHH, IHH, and DHH proteins, respectively), Patched 1 (PTCH1, the 12-pass transmembrane receptor), Smoothened (SMO, the 7-pass transmembrane G protein-coupled receptor), as the obligatory signal transducer across the plasma membrane, and the Gli transcription factors family.19
Canonical SHH signaling
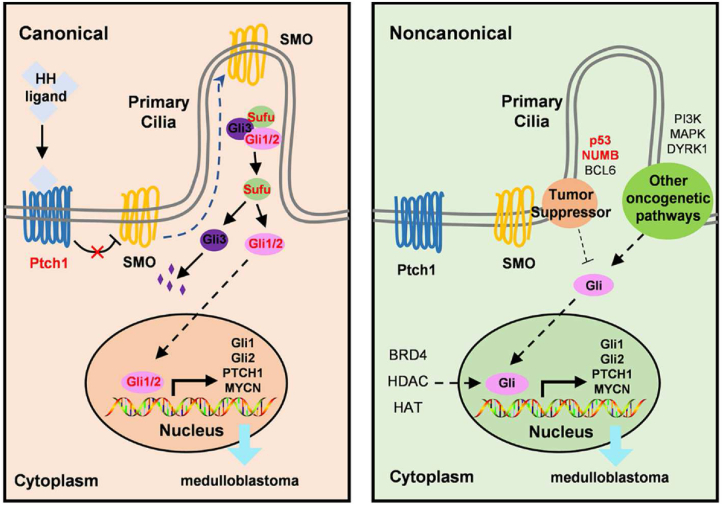
Canonical HH transduction is initiated by HH ligand binding to PTCH1. In absence of an HH ligand, SMO is constitutively repressed by PTCH1, which is located at the base of the primary cilia, an organelle that protrudes from the surface of vertebrate cells and serves as the center of HH signaling.20,21 Thus, the accumulation of SMO in the primary cilia is prevented and its activity is repressed by PTCH1. Once the HH ligand binding to PTCH1, it displaces PTCH1 from the primary cilia by inducing its internalization and lysosomal degradation. Activated SMO translates the HH activating signals into the cytoplasm preventing GLI2 and GLI3 processing and releasing GLI transcription factors from dissociation from the suppressor of fused (SUFU), thereby ultimately promoting GLI to translocate into the nucleus and binds to targets to regulate gene expression (Fig. 1).
Figure 1.
Canonical and noncanonical hedgehog signaling action in MB. Canonical HH signaling: HH ligands (SHH, IHH, and DHH) bind with PTCH1 to relieve suppression of SMO. Active SMO moves to primary cilia and promotes the release of SUFU and Gli proteins. Activated Gli proteins then translocate to the nucleus and promote gene transcription (ie, GLI1, GLI2, PTCH1, and MYCN). Noncanonical HH signaling is activated via other oncogenic pathways (PI3K, DYRK1, MAPK), tumor suppressors (p53, NUMB, BCL6), and other regulators (HAT, HDAC, BRD4).
Noncanonical SHH signaling
Besides the canonical activation of GLI by the HH-PTCH-SMO axis, the noncanonical SHH signaling refers to the cellular and tissue responses to HH ligands that are not dependent on GLI1 isoform transcriptional activity. There is growing evidence pointing out an SMO-independent stimulation of GLI activity.
SHH signaling aberrancy in MB
MB is one of the most malignant brain tumors in children (WHO grade IV).22 As a common embryonal cancer of the cerebellum, MB is speculated to derive from various discrete neuronal stem or progenitor cell populations early in life and is prone to dissemination through the cerebral spinal fluid (CSF).23,24 The peak of incidence is ∼3–4 or 8–10 years of age, although MB sometimes occurs in the first year after birth or adulthood in some individuals.23 MB has four subgroups based on molecular typing, including wingless (WNT), SHH, Group 3, and Group 4.25 Meanwhile, there are several therapeutic options for the treatment of MB. Maximal surgical resection followed by risk-adapted CSI and adjuvant chemotherapy has brought the best survival for MB.26
Among four subgroups of MB, SHH-MB accounted for 30%, together with activated SHH signaling. SHH-MB is usually orientated from the mutations or deletions of key genes in SHH signaling. The majority of patients have either germline or somatic mutations and copy-number alterations in these critical genes, including mutations or deletions in PTCH1 (43% of patients) or SUFU (10%), activating mutations in SMO (9%), GLI1 or GLI2 amplifications (9%), and MYCN amplification (7%).26,27 Nowadays, MYC or MYCN amplification and chromosome 17 imbalances are the signatures of Group 3 MB.28,29 Common SHH-MB cell lines include the Daoy (D324MED) cell line and Ons-76 cell line.30, 31, 32 In WNT-MB, the mutation of CTNNB1 gene is the most significant marker of WNT-MB,33 and the β-catenin protein encoded by CTNNB1 is an important effector of the WNT signaling pathway, common WNT-MB cell lines include BT853 and UW473 cell line.30,34 The molecular mechanism of Group 3 MB remains unclear. MYC amplification is the salient feature of this subgroup MB. Cytogenetically, the main manifestations are the amplification of chromosomes 1q, 7, and 17q, and the deletion of 10q, 11q, 16q, and 17p.35 Common Group 3 MB cell lines include SUMB002, HD-MB03, RCMB40, D425, Med8A, D458, DM283-MED, and D341 cell lines30,36,37; Group 4 MB is the most common molecular subgroup in which MYCN mutations are associated with poorer prognosis. Common Group 4 MB cell lines include ICB299 cell line.30,36
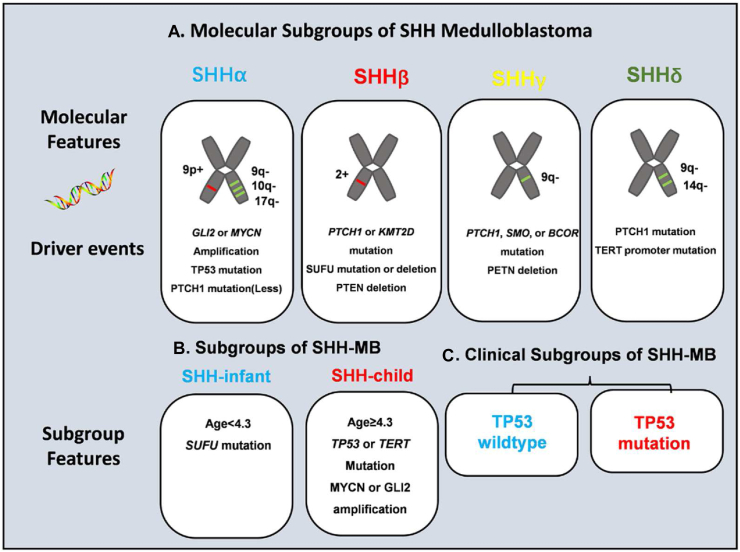
In SHH-MB, comprehensive clustering analysis by Cavalli et al identified four subgroups of SHHα, SHHβ, SHHγ, and SHHδ.38 SHHα has the worst prognosis and is characterized by TP53 mutation, MYCN, or GLI2 amplification. SHHβ is more common in infants, often accompanied by metastasis. SHHγ pathology is mostly MBEN and has a better prognosis. SHHδ is mainly composed of adults and is rich in TERT promoter mutations. Schwalbe et al divided SHH-MB into MBSHH-infant (age <4.3) and MBSHH-child (age ≥4.3) based on age. MBSHH-infant is rich in SUFU mutations and has a good prognosis. MBSHH-infant is characterized by TP53 or TERT mutation, and MYCN or GLI2 amplification.39 Moreover, 21% of SHH-MB have TP53 mutations, which have a poor prognosis and are an important factor for this subgroup of MB.40 In clinical, SHH-MB is further divided into TP53 mutations or TP53 wild type according to whether TP53 is mutant.
In brief, the aberrant activation of the SHH signaling induces aberrant proliferation and tumor growth in MB. The effects of key molecular mutations in the SHH signaling pathway on MB are well known. However, the functions of critical molecular AS in the SHH signaling pathway on MB are unclear.
AS in components of canonical SHH signaling
AS of PTCH1
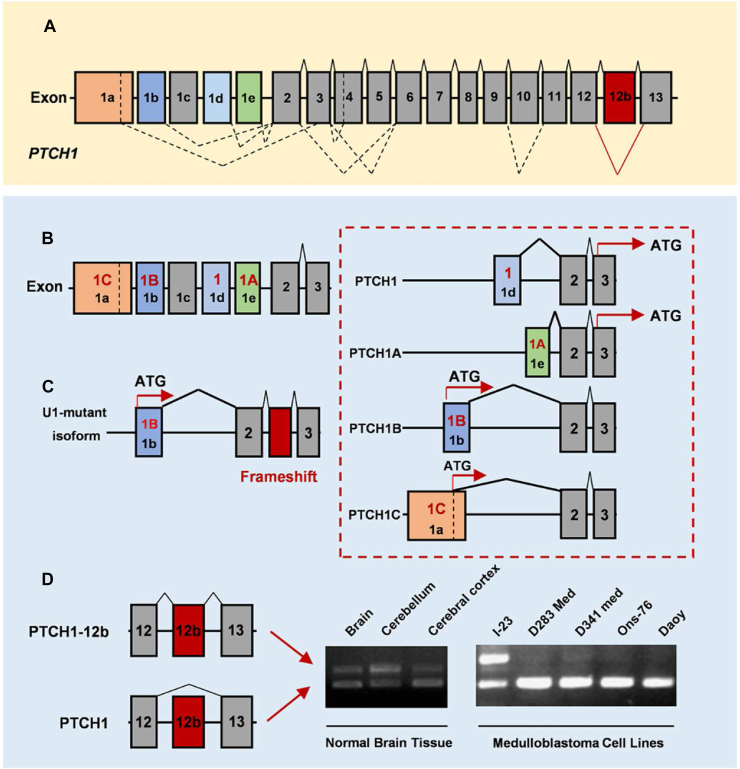
PTCH1 is a significant component of SHH signaling. The binding of SHH to the PTCH1 receptor derepresses SMO, ultimately freeing GLI1 transcription. However, aberrant AS in PTCH1 has been identified in MB patients. Differential PTCH1 transcripts produced through AS have multiple roles in SHH signaling.41 AS of PTCH1 is mainly composed of alternate promoter and exon skip (Fig. 2A). PTCH1 has five alternative first exons involving 1a (also known as 1C), 1b (also known as 1B), 1c, 1d (also known as 1), and 1e (also known as 1A). Alternative first exons of PTCH1 form different transcripts, such as PTCH1, PTCH1A, PTCH1B, and PTCH1C (Fig. 2B). PTCH1 and PTCH1A encode the same short form of PTCH1A, which is translated from an ATG codon within exon 3. PTCH1 and PTCH1A include the alternative first exon-1 and exon 1A, respectively. PTCH1B contains the alternative first exon 1B and encodes a full-length PTCH1 protein. PTCH1C includes the alternative first exon 1C encoding a full-length PTCH1 protein form in which the ATG codon is located in exon 1C. Among the transcripts, the short form of PTCH1A can interact with HH family protein while is incapable of suppressing HH signaling. PTCH1B protein can completely inhibit SMO activity. PTCH1C has a lower capacity for pathway inhibition compared with PTCH1B.42
Figure 2.
Alternative splicing of PTCH1 in MB. (A) All 18 coding exons of the PTCH1 gene are indicated by numbered boxes. (B) Alternative first exons of PTCH1 form different transcripts. (C) U1-snRNA mutations drive cryptic splicing in adults and adolescents with SHH-MB. (D) Alternative splicing of PTCH1 Exon 12b in normal brain tissues and numerous MB cell lines.
As reported previously, PTCH1 transcripts were expressed in numerous human adult tissues, such as the brain, lung, and so on. However, PTCH1 transcripts are specifically expressed in fetal tissues. Fetal brain tissue only expresses exon 1, while adult brain tissue expresses exon 1/1A/1B/1C. Exon 1B/1C expression is lower than that of 1 and 1A. These data indicate the importance of exon 1 variation during early brain development. SHH-MB is associated with the activation of HH signaling that could upregulate exon 1B and 1C.
In addition to the alternate promoter, exon skip also occurs in AS of PTCH1. Exon 12b, an alternative exon, is located between exon 12 and 13 (Fig. 2D). The transcript containing exon 12b is expressed in a brain- and heart-specific fashion, particularly in the cerebellum.43,44 The transcript containing exon 12b encodes a truncated patched-1 protein (PTCH1-12b) since this isoform has an in-frame stop codon, which does not seem to have any functions. PTCH1-12b was found to be expressed in control but not in SHH-MB cell lines (Daoy) (Fig. 2D), thus it is speculated that PTCH1-12b plays a significant suppressor role in the tumorigenesis of MB.44
AS of GLI transcription factors
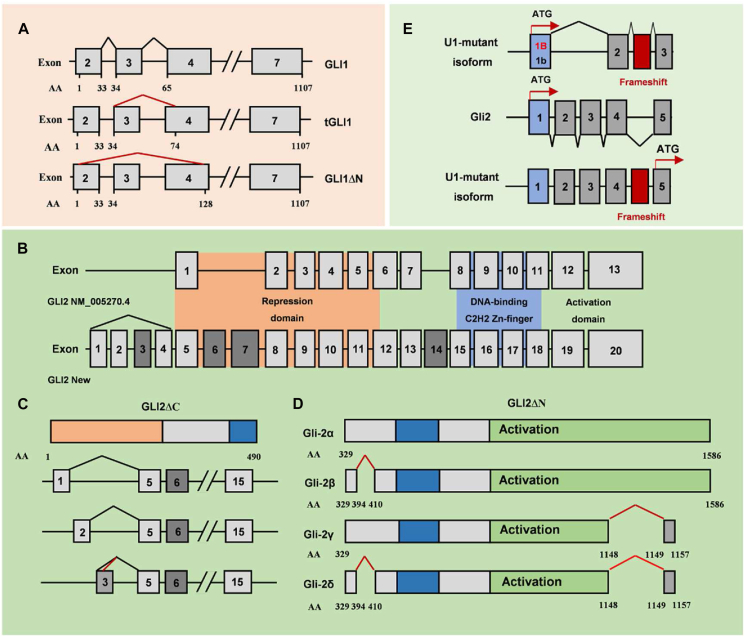
Aberrant splicing of PTCH1 affects the expression of GLI transcription factors. The GLI transcription factors family is composed of GLI1, GLI2, and GLI3. All GLI proteins are members of the GLI-Kruppel family of zinc-finger (ZNF) containing transcription factors. They have a C-terminal transactivation domain, while GLI2/3 also possess an N-terminal repressor domain and function as both transcriptional activators and repressors. GLI1 acts mainly as a transcriptional activator. GLI2FL is a weak activator since the fully activated form (GLI2ΔN) requires the complete removal of its N-terminus.45 GLI3 functions as a repressor in most conditions.46 However, there are also numerous AS in GLI transcription factors. The human GLI1 gene was first identified by Vogelstein and colleagues as an oncogene in glioblastoma.47 Then, Lo and colleagues identified a novel splice variant of GLI1 called tGLI1 in glioblastoma,48 which is generated by exon 3 skipping (Fig. 3A). tGLI1 was identified in D458 cell (a WNT-MB cell line)49; we also found tGLI1 in multiple brain tumors including the Daoy cell (an SHH-MB cell line) (Data are not shown), while tGLI1 doesn't exist in normal cells. The exon 3 skip of GLI1 encodes tGLI1 which upregulates VEGF-A and CD24 gene expression.49 Furthermore, tGLI1 activates metastasis-initiating cancer stem cells by inducing stemness genes CD44, Nanog, Sox2, and OCT4, leading to CSC renewal.50,51 These studies all suggest that tGLI1 has a stronger oncogenic effect than GLI1.
Figure 3.
Alternative splicing of GLI transcription factors in MB. (A) Alternative splicing of GLI1 in MA. (B) All coding exons of the GLI2 gene are indicated by numbered boxes, and the different color represents protein domains. (C) Four mutually exclusive 5′ non-coding exons of GLI2 with different patterns of inclusion in alternatively spliced transcripts. (D) Alternative splicing generates four GLI2 isoforms. (E) U1-snRNA mutations drive the splicing of PTCH1 and GLI2 in adults and adolescents with SHH-MB.
However, the mechanism of AS of tGLI1 remains unknown. tGLI1 is expressed in multiple tumors, and these data indicate AS of tGLI1 is not induced by mutations. We found that hnRNPU expression is associated with tGLI1 in GBM,48 but Lo and colleagues did not explain whether tGLI1 is induced by hnRNPU. Interestingly, Zhao et al found that hnRNPU mediates the long-range regulation of SHH expression during limb development.52 In summary, the AS of tGLI1 is not well understood and requires further research.
Apart from the splicing of Gli1, Gli2 also contains many types of AS in MB. In the past, Gli2 was thought to contain 13 exons (exon 1–13), which encode the N-terminal repressor domain, DNA-binding C2H2 Zn-finger domain, and the C-terminal activation domain. With the development of high-throughput sequencing-based methods, Sadam and colleagues described the revised genomic organization of GLI2 and major ASVs (Fig. 3B).53 They identified four mutually exclusive 5′ non-coding exons of GLI2 with different patterns of inclusion in alternatively spliced transcripts detected in normal versus malignant tissues (Fig. 3C). In SHH-MB, cryptic AS of GLI2 demonstrates the position of a cryptic cassette exon with the 5′ splice site sequence, which encodes Gli2 without N-terminal repressor.54 GLI2ΔN acts as a powerful transcriptional activator, the splicing of it generates four Gli2 isoforms (Gli2α, Gli2β, Gli2γ, and Gli2δ), which all remove the repressor domain at the N terminus (Fig. 3D).
AS of SUFU
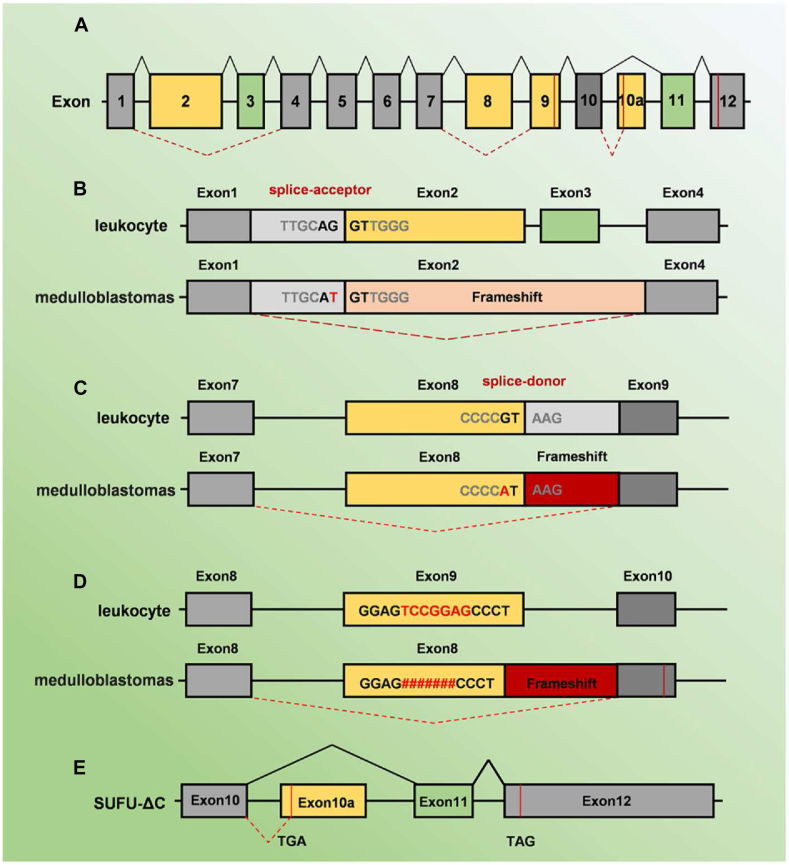
The binding of HH ligand and PTCH1 displaces the suppression of PTCH1 on SMO. Activated SMO translates the HH signals into the cytoplasm and releases GLI transcription factors from dissociation from SUFU. SUFU is a key negative regulator in the HH signaling pathway. Without the HH signal, the GLI2/3-SUFU complex is recruited to cilia, resulting in the processing of GLI2/3FL into GLI2/3R which act as repressors.55 All GLI transcription factors have a SUFU-interacting site at their N-terminus (SIN). GLI2 and GLI3 possess an additional SUFU-interacting site located on their C-terminus (SIC).56 SUFU, as a tumor-suppressor gene, predisposes individuals to MB by modulating the SHH signaling pathway. Growing evidence shows that germline SUFU mutations strongly predispose to MB.57 However, SUFU mutations contain splice acceptor and splice donor mutants, leading to its aberrant splicing in MB (Fig. 4A).58 Specifically, the conserved AG splice-acceptor site of exon 2 mutated to AT, resulting in the splicing of exon 1 to exon 4 with a subsequent frameshift and premature stop codon at the 3′ end of exon 1 (Fig. 4B). Similarly, there was a mutation in the conserved GT splice-donor site consensus sequence of exon 8, leading to splicing of exon 7 to exon 9 with a subsequent frameshift and premature stop (Fig. 4C). Besides, the missing of 7 nucleotides in exon 9 (1129delTCCGGAG) led to a frameshift and premature stop codon in exon 10 (Fig. 4D). These mutations of SUFU in MB all bring about aberrant splicing and loss of its suppressor role, which may lead to carcinogenesis.
Figure 4.
Alternative splicing of SUFU in MB. (A) All 13 coding exons of the SUFU gene are indicated by numbered boxes. (B) The mutation at the splice-acceptor site leads to the splicing of exon 1 to exon 4 with a subsequent frameshift and premature stop codon at the 3′ end of exon 1. (C) The mutation at the splice-acceptor site leads to the splicing of exon 1 to exon 4 with a subsequent frameshift and premature stop codon at the 3′ end of exon 1. (D) Mutation in the conserved GT splice-donor site consensus sequence of exon 8 leads to splicing of exon 7 to exon 9 with a subsequent frameshift and premature stop. (E) SUFU-ΔC harbors an early in-frame translation termination codon exon 10a.
Moreover, there are alternate terminators in SUFU (Fig. 4E), SUFU-ΔC, a carboxy-terminal truncated transcript, harbors an early in-frame translation termination codon exon 10a, which encodes a unique terminal exon. SUFU-ΔC instead of SUFU-FL reduced the protein levels of GLI1FL.59 Furthermore, Xu et al identified SUFUvN, a novel AS transcript variant, contained a new protein-coding exon and had a relationship with lymph node metastasis in pancreatic ductal adenocarcinoma metastasis.60 All the transcripts produced by AS have separate roles in Gli transcription factors and take part in MB.
AS in components of noncanonical SHH signaling
Pietrobono et al summarized the noncanonical SHH signaling in cancer,61 which will not be discussed further. We summarize the recent advances in AS of HH signaling in MB. Firstly, we summarized the upstream regulators of noncanonical HH signaling in MB (Table 1). Among upstream regulators, we focus on the abnormal splicing of tumor suppressor factors, which may lead to a loss of suppression of tumor growth.
Table 1.
Non-canonical regulators of HH-GLI signaling in MB.
| Upstream regulator | Mechanism of action | Reference |
|---|---|---|
| Tumor suppressors | ||
| p53 | Promotes proteasome-dependent degradation of GLI1 (PCAF-dep.) | 62 |
| NUMB | Induces GLI1 ubiquitination and proteasome degradation (ITCH-dep.) | 63 |
| BCL6 | Directly represses Gli1 and Gli2 | 64 |
| PI3K-AKT-mTOR pathway | ||
| mTOR/S6K1 | HH signaling increases SMO translation through a mTORC1/4EBP1-dependent mechanism | 65 |
| MAPK pathway | ||
| MEKK1 | Inhibits GLI1 transcriptional activity | 66 |
| MEKK2/3 | Inhibits GLI1 transcriptional activity and protein stability through SUFU | 67 |
| p38 | The inhibition of p38α causes a decrease in Gli1 and N-myc transcript levels | 68 |
| DYRK family | ||
| DYRK1A | Increase the transcriptional activity of GLI1 | 69 |
| DYRK1B | Enhances GLI1 transcriptional activity | 70 |
| BET proteins | ||
| BRD4 | Increases GLI1/2 transcription | 71 |
| HDAC | ||
| HDAC class I | Increases DNA binding ability of GLI1 (HDAC1) | 72 |
| HDAC class II | Transcriptional control of GLI2 (HDAC6) | 73 |
| HAT | ||
| PCAF | Acts as GLI1 transcriptional cofactor Promotes GLI1 ubiquitination and proteolysis |
74 62 |
| Arbb1/p300 | Suppresses the transcriptional activity of Gli1 | 75 |
AS of TP53
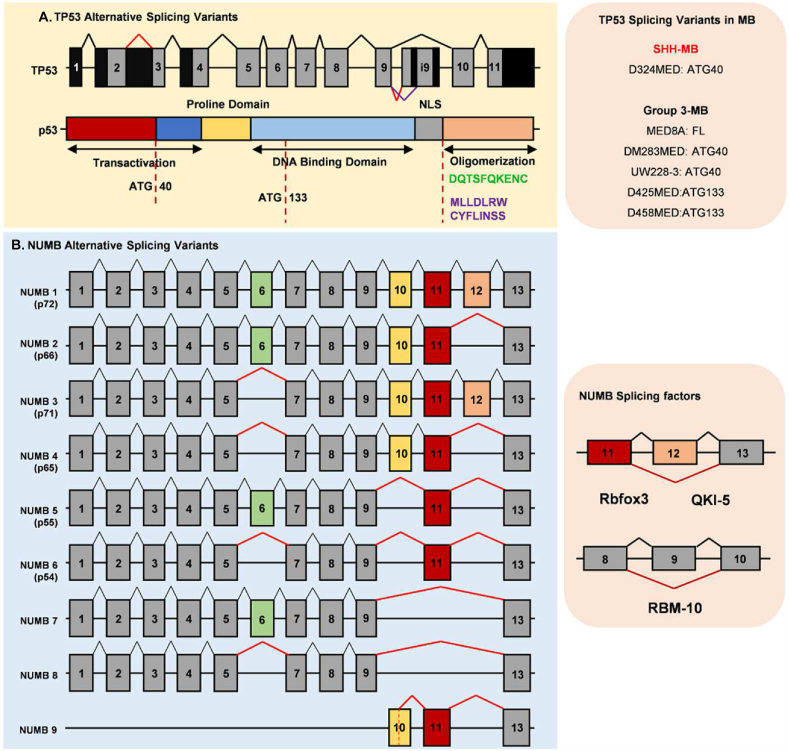
TP53 mutations are present in approximately 30% of SHH-MB, which are frequently germline and form an extremely high-risk group of patients. However, the SA of TP53 in MB is rare. Ghosh and colleagues describe the AS of TP53 (Fig. 5A); they pointed out that N-terminally deleted isoform of p53 (p47) effectively suppresses p53-mediated transcriptional activity and despair p53-mediated tumor growth suppression.76 TP53 has an alternative promoter and transcribes multiple splice variants. P53 isoforms produced by TP53 variants are expressed in different tissue in a tissue-dependent manner.77 P53 and P53β are present in the brain, but the temporal cortex and cerebellum only express P53. As for the AS of TP53 in MB, Philipova and colleagues analyzed nine MB primary biopsy samples and detected full-length P53 and its short form ∼22 kDa by antibody Ab1801, which recognizes amino acids 46–55 in the N-terminus of P53. Thus, the short form is not the N-terminus truncation. Interestingly, the N-terminus truncations of P53 were detected in multiple MB cell lines (Fig. 5A). The Western blot results showed that antibodies Ab1801 and AbD0-7, which recognized separately amino acids 46–55 and 1–45, could not detect the P53 isoforms in D425MED and D458MED cell lines which could be Δ133p53. These results indicate that the translation initiation start of the mRNA of TP53 initiated in intron 4 (Δ133p53). Moreover, D283MED, D324MED, and UW228-3 cell lines detected TP53 transcripts lower than full-length p53 but higher than p47. Although Philipova and colleagues had not explained the certain transcripts in MB cell lines, Ghosh and colleagues consider that this P53 isoform detected by Ab1801 could be p53-338 lacking the C terminus (amino acids 338–393).76 As previously described, Group 3 MB cell lines have various transcripts including TP53-FL and other TP53 transcripts. However, the transcript in the D324MED cell line is TP53 without a transactivation domain; this P53 isoform produced by AS may lose the suppression of HH signaling and play a functional role in MB.
Figure 5.
Alternative splicing of noncanonical regulators in MB. (A) All coding exons of the TP53 gene are indicated by numbered boxes. (B) Structure of p53. Functional domains of p53 are represented by different colored boxes. MB cell lines express different p53 transcripts. (C) Alternative splicing of NUMB forms different NUMB isoforms. (D) Splicing factors in alternative splicing of NUMB.
AS of NUMB
NUMB is another suppressor of HH signaling, and its deregulation is a relevant event in MB.63 Itch, an E3 ligase, regulates a ubiquitin-mediated mechanism of GLI1 degradation. The catalytic activity of Itch could be activated by Numb, which direct recruits and binds with two PPXYs and phospho-serine/proline motifs in the C-terminal region of GLI1. The coordinated action of NUMB and Itch promotes GLI1 degradation. Mutated GLI1 protein, no longer regulated by Itch and Numb, enhanced MB growth, migration, and invasion abilities. Furthermore, different NUMB isoforms are deregulated in MB.78 As shown in Figure 5B, AS forms various subtypes of NUMB. Among the variants, MB is associated with decreased levels of Numb p66 while overexpressed Numb p72. Numb p66 was significantly reduced only in the SHH subgroup, whereas Numb p72 was significantly up-regulated in all MB subgroups.78 Numb p66 lacks exon 12, leading to 366–413 amino acid deletions. The decreased Numb p66 affects MB stem-like cells and cerebellar neuronal stem cells. Choi and colleagues have elaborated on the functions of Numb splicing in cancer stem cells,79 and we do not discuss anything further and pay more attention to the splicing factors of Numb. Rbfox3 promotes neuronal differentiation during development by regulating AS of Numb in the form of exon skip, and it repressed the inclusion of exon 12 (Fig. 5B).80 QKI has a similar function to AS of Numb in lung cancer.81 RBM10 in lung adenocarcinomas also compromises the regulation of NUMB exon 9 and controls cancer cell proliferation.82,83 AS in noncanonical SHH signaling is also associated with other cancer-related signaling such as Notch signaling.
HH signaling induces AS
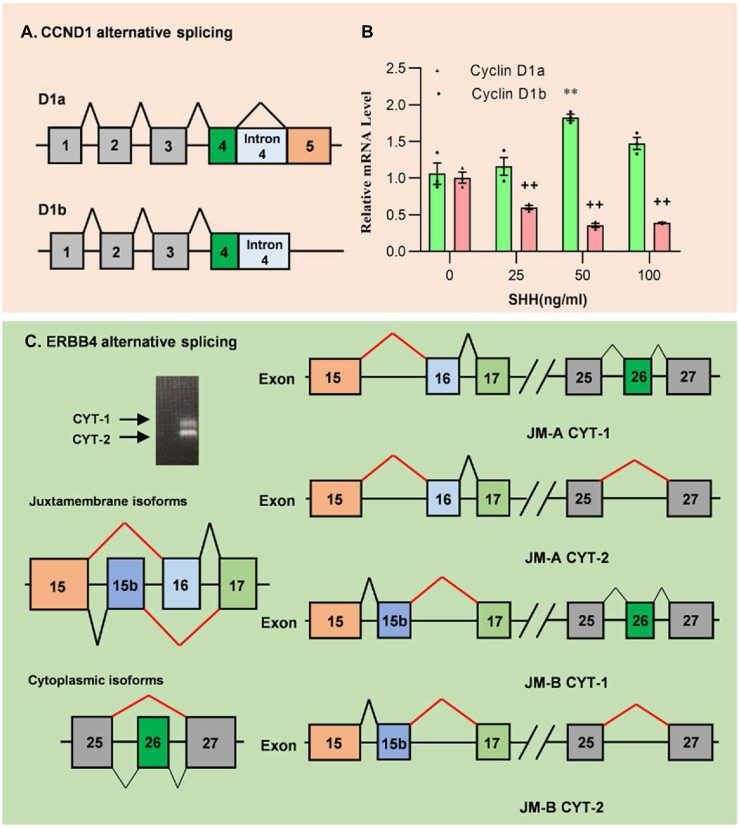
After describing the AS of canonical/noncanonical SHH signaling, HH signaling also induces AS of HH target genes. HH-dependent target genes include Gli transcript factors, Ptch1, Hhip, CCND1, etc. We have shown the AS of Gli transcript factors and Ptch1, and the AS of CCND1 in MB remains unknown. Paronetto and colleagues pointed out that CCND1 is expressed as two transcripts by AS. Compared to Cyclin D1a, Cyclin D1b differ from the inclusion of intron 4 in the mRNA (Fig. 6A). A common polymorphism (G870A) in the exon 4/intron 4 boundary of CCND1 has been related to cyclin D1b, which was detected in various cancers including primary brain tumors.84, 85, 86, 87 Moreover, increased Sam68 stimulates cyclin D1b not D1a expression in human prostate cancers.88 BAF57/SMARCE1 and SRSF1 are also responsible for the mechanical stress-induced AS of cyclin D1.89 To explore the AS of CCND1 in MB, we detected the CCND1 variant in the Daoy cell line and the change of Cyclin D1a/b treated with SHH recombinant protein. The RT-PCR results show that SHH induces the expression of Cyclin D1b while repressing the expression of Cyclin D1a (Fig. 6B). Cyclin D1b could promote tumor growth and invasive migration in various cancers including breast cancer, bladder cancer, and so on.90, 91, 92 However, there is no research indicating the correlation between Cyclin D1b and MB. Our results show that SHH induces cyclin D1b expression in an MB cell line.
Figure 6.
HH signaling induces alternative splicing of genes. (A) Alternative splicing produced two transcripts of CCND1, which differ in the inclusion of intron 4. (B) Daoy cells were treated with SHH for 24h and total RNA was isolated for RT-PCR. (C) Alternative splicing patterns of ERBB4.
ErbbB4, one of the ErbB family of receptors, is another target gene whose expression and AS could be interfaced by HH signaling in MB. Cerebellar ErbB4 in humans and mice is high during early development and decreases in adults.93,94 There are four main transcripts, including JM-A CYT-1, JM-A CYT-2, JM-B CYT-1, and JM-B CYT-2, produced by AS (Fig. 6C). The CYT-1:CYT-2 ratio is inversely correlated with Gli1 expression in human MB and may be an indicator of MB aggressivity.95 Upregulation of ErbB-4 CYT-1 in MB confers a relatively apoptosis-resistant phenotype, whereas the counter-selection of CYT-2 mediates neuregulin-dependent growth suppression. All the evidence suggests that HH signaling induces AS in MB.
Aberrant AS in SHH-driven MB
In addition to mutations or deletions of critical genes in SHH signaling, aberrant AS plays a significant role in MB. To investigate patterns of differential splicing between normal cerebellum and pediatric MB on a genome-wide scale, the Affymetrix Exon arrays were used to analyze 1262 genes identified as potentially generating tumor-associated splice forms. In these analyses, the exon inclusion rate of MB and normal fetal cerebellum samples was significantly lower than that of normal adult cerebellum, and SHH could decrease the cassette exon inclusion rate of multiple genes.96 Moreover, analysis of AS based on 14 normal cerebellar samples and 103 MBs of known subgroups suggest that splicing patterns are distinct and specific in different molecular subgroups.97 Furthermore, the U1 snRNA mutations in adults and adolescents with SHH-MB-mediated AS could activate HH signaling-related genes.54 These data suggest that SHH-MB has specific AS compared to normal tissues. Thus, we try to elucidate the aberrant splicing of molecules in SHH signaling during tumorigenesis.
Growing evidence links AS of regulators in SHH signaling to the occurrence of MB. However, AS of these canonical regulators play different roles in multiple SHH-MB subgroups. According to Schwalbe et al,39 we also divided SHH-MB into MBSHH-infant and MBSHH-child/adult based on age and described the roles of AS of canonical HH regulators in different subgroups of SHH-MB.
AS of SHH signaling in MBSHH-infant subgroup
Growing evidence links AS of regulators in SHH signaling to the occurrence of MB. However, AS of these canonical regulators play different roles in multiple SHH-MB subgroups. According to Schwalbe et al,39 we also divided SHH-MB into MBSHH-infant and MBSHH-child/adult based on age and described the roles of AS of canonical HH regulators in different subgroups of SHH-MB (Fig. 7).
Figure 7.
Subgroups of MB and molecular features. (A) Clinical features, genetic alterations, and gene expression in different subgroups of SHH-MB. (B, C) Subgroups and clinical subgroups of SHH-MB and molecular features. MBSHH-infant is rich in SUFU mutations and has a good prognosis. These mutations of SUFU in MBSHH-infant all bring about aberrant splicing (Fig. 4D, E) and loss of its suppressor role. All the transcripts produced by alternative splicing have separate roles in GLI transcription factors and take parts in MB.
AS of SHH signaling in MBSHH-child/adult subgroup
As we described previously, AS of canonical SHH signaling is associated with the MBSHH-infant subgroup. Similarly, AS of canonical SHH signaling is also represented in MBSHH-child/adult subgroup. In MBSHH-child/adult subgroup, recurrent non-coding U1-snRNA mutations drive cryptic splicing. The mutations occur in 97% of adults (subtype SHHδ) and 25% of adolescents (subtype SHHα) with SHH-MB, but they are largely absent from SHH-MB in infants. The mutations occur in the 5′ splice-site binding region and significantly disrupt RNA splicing. MBSHH-child/adult subgroup has an excess of 5′ cryptic splicing events. Suzuki et al identified cryptic 5′ events with a ‘C’ base at the 6th intronic position and detected cryptic splicing events with high effect sizes in both PTCH1 and GLI2 (Fig. 2C).54 Aberrant AS of PTCH1 leads to the loss of inhibition of SMO and activation of HH signaling.
Apart from splicing of PTCH1 in the MBSHH-child/adult subgroup, recurrent non-coding U1-snRNA mutations also drive splicing GLI2 (Fig. 3E). GLI2 contains many types of AS in the MBSHH-child/adult subgroup MB which is characterized by GLI2 amplification. In SHH-MB, cryptic AS of GLI2 demonstrates the position of a cryptic cassette exon with the 5′ splice site sequence, which encodes Gli2 without an N-terminal repressor.54 GLI2ΔN acts as a powerful transcriptional activator, the splicing of it generates four GLI2 isoforms (GLI2α, GLI2β, GLI2γ, and GLI2δ), which all remove the repressor domain at the N terminus (Fig. 3E).
In SHH signaling, the mutations in snRNAs or splicing sites of molecules result in aberrant AS, which inactivates tumor suppressor genes (PTCH1, SUFU) and activates oncogenes (GLI1, GLI2) in SHH-MB.
Therapeutic applications of targeted AS in cancer treatment
After elucidating the mechanism of AS in SHH signaling and its correlations with MB, we aimed to exploit the potential therapeutic applications of targeted AS in cancer treatment. Along with research in the mechanism of AS, many researchers developed AS-related medicines. Lin concluded the therapeutic applications of targeted AS in cancer treatment.114 Among them, a series of results have been achieved in the development of small-molecule inhibitors targeting core splicing bodies and splicing factors, we further concluded the emerging roles of AS-related drugs (Table 2). In addition, there are oligonucleotide-based cancer splicing modulation therapies.114,115
Table 2.
Small compounds used to alter splicing events.
| Target | Compound | Mechanism | Reference |
|---|---|---|---|
| SF3B1 | Pladienolide E7107 | Abolish the conformation rearrangement of SF3B1 | 98 |
| Spliceostatins | 99 | ||
| Herboxidiene | 100,101 | ||
| U5 snRNP | Brr2 | Interfere with the RNA helicase activity | 102,103 |
| U2 snRNP | Sudemycin K | Inhibitor of spliceosome assembly | 104 |
| hnRNPA1 | VPC-80051 | Prevent AR-V7 generation and reduce c-Myc transcription output | 105 |
| SRSF10 | GPS167 | Increase the interaction of SRSF10 with the CLK1 and CLK4 kinases | 106 |
| SRRK family | SRPIN340 | ATP binding competitor | 107 |
| SRPKIN-1 | Convert the pro-angiogenic VEGF-A165a to the anti-angiogenic VEGF-A165b isoform | 108 | |
| ZINC02154892 (C02) | Binds to targeted ATP binding site and inhibits recruitment of ASF/SF2 by SRPK1 | 109 | |
| CLK family | TG-003; TG-693 | Interfere with ATP binding | 110 |
| CX-4945 | Phosphorylation of serine/arginine-rich proteins | 111 | |
| RECTAS | 112 | ||
| SM08502 | Inhibit SRSF phosphorylation and disrupted spliceosome activity | 113 |
Conclusions and perspectives
With the development of high-throughput sequencing techniques, researchers have found aberrant AS in tumors, including an increase in the rates of AS and the different AS patterns in tumors. The roles of AS in tumors are increasingly clear; for example, specific protein isoforms produced by AS contribute to tumorigenesis. Here, we focused on the effects of AS on SHH-MB. Based on understanding the mutations of critical molecules in the SHH signaling pathway for the development of MB, we comprehensively summarized AS in SHH signaling in MB, which may further expand our understanding of the pathogenesis in MB.
In addition to clarifying the pathogenesis, our review also makes it possible to selectively target AS in MB treatment. Previously, researchers have developed a series of HH inhibitors for the abnormal activation of the HH signaling pathway in various cancers, some inhibitors are approved for marketing, including Vismodegib, Glasdegib, and Erivedge. However, there are still no targeted drugs for MB. Modulation of spliceosome with small molecule inhibitors is perceived to be an anti-cancer strategy. The most commonly applied strategy is that those small molecules directly bind to SF3B1 protein in the U2 snRNP, including spliceostatin A (SSA), meayamycin B (MAMB), sudemycins, E7107, and H3B-8800. There is no evidence that SF3B1 affects the occurrence of MB; aberrant AS of SHH signaling is associated with MB and could be the target. Furthermore, inhibiting specific protein isoforms in SHH signaling produced by AS could potentially halt cancer progression and metastasis. These strategies may have a better efficacy combined with HH inhibitors in MB treatment.
Author contributions
M.Q., Q.H., and X.W. were the main contributors to the conception and design of the review and the manuscript drafting. J.L., T.S., R.G., Y.X., C.X., and M.Q.B. prepared the tables and figures. L.Z. and X.W. supervised the drafted manuscript. All authors read and approved the final manuscript.
Conflict of interests
The authors declare no conflict of interests for this work.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31571493, 81741043, 31871395, 82000046, and 32170841).
Acknowledgements
The authors are grateful to all the staff at the study center who contributed to this work.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Ling-Hui Zeng, Email: zenglh@zucc.edu.cn.
Ximei Wu, Email: xiwu@zju.edu.cn.
Abbreviations
- ACAT1
Acetyl-CoA cholesterol acyltransferase
- AS
Alternative splicing
- BAF57
SWI/SNF complex subunit Brahma-related gene-1-associated factor 57
- CSF
Cerebral spinal fluid
- DHH
Desert Hedgehog
- ESE
Exon splicing enhancers
- ESSs
Exon splicing silencers
- GPCR
G protein-coupled receptor
- hnRNP
heterogeneous nuclear ribonucleoprotein
- IHH
Indian Hedgehog
- IR
Intron retention
- ISEs
Intron splicing enhancers
- ISSs
Intron splicing silencers
- MAMB
Meayamycin B
- MB
Medulloblastoma
- PE
phosphatidylethanolamine
- PS
Phosphatidylserine
- PSS1/2
PS synthase-1/2
- PTCH1
Patched 1
- PTCs
Premature termination codons
- SHH
Sonic Hedgehog
- SMARCE1
SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1
- SMO
Smoothened
- snRNAs
small nuclear RNAs
- SNVs
Single nucleotide variants
- SRSF1
Serine/arginine-rich splicing factor 1
- SSA
Spliceostatin A
- SUFU
Suppressor of fused
- TCGA
The Cancer Genome Atlas
- TSGs
Tumor suppressors genes
- UTR
Untranslated regions
References
- 1.Chen M., Manley J.L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10(11):741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ule J., Blencowe B.J. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol Cell. 2019;76(2):329–345. doi: 10.1016/j.molcel.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Montes M., Sanford B.L., Comiskey D.F., et al. RNA splicing and disease: animal models to therapies. Trends Genet. 2019;35(1):68–87. doi: 10.1016/j.tig.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y., Rio D.C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wan R., Yan C., Bai R., et al. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: insights into spliceosome assembly and catalysis. Science. 2016;351(6272):466–475. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 6.Plaschka C., Lin P.C., Charenton C., et al. Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature. 2018;559(7714):419–422. doi: 10.1038/s41586-018-0323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baralle F.E., Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18(7):437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng X., Peng Q., Wang L., et al. Serine/arginine-rich splicing factors: the bridge linking alternative splicing and cancer. Int J Biol Sci. 2020;16(13):2442–2453. doi: 10.7150/ijbs.46751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kędzierska H., Piekiełko-Witkowska A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 2017;396:53–65. doi: 10.1016/j.canlet.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Lee S.C.W., Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22(9):976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Climente-González H., Porta-Pardo E., Godzik A., et al. The functional impact of alternative splicing in cancer. Cell Rep. 2017;20(9):2215–2226. doi: 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Sciarrillo R., Wojtuszkiewicz A., Assaraf Y.G., et al. The role of alternative splicing in cancer: from oncogenesis to drug resistance. Drug Resist Updates. 2020;53 doi: 10.1016/j.drup.2020.100728. [DOI] [PubMed] [Google Scholar]
- 13.Chen L., Tovar-Corona J.M., Urrutia A.O. Increased levels of noisy splicing in cancers, but not for oncogene-derived transcripts. Hum Mol Genet. 2011;20(22):4422–4429. doi: 10.1093/hmg/ddr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dvinge H., Bradley R.K. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7(1):45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung H., Lee D., Lee J., et al. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. 2015;47(11):1242–1248. doi: 10.1038/ng.3414. [DOI] [PubMed] [Google Scholar]
- 16.Machado M.V., Diehl A.M. Hedgehog signalling in liver pathophysiology. J Hepatol. 2018;68(3):550–562. doi: 10.1016/j.jhep.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimenez-Sanchez M., Menzies F.M., Chang Y.Y., et al. The Hedgehog signalling pathway regulates autophagy. Nat Commun. 2012;3:1200. doi: 10.1038/ncomms2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walton K.D., Gumucio D.L. Hedgehog signaling in intestinal development and homeostasis. Annu Rev Physiol. 2021;83:359–380. doi: 10.1146/annurev-physiol-031620-094324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrov K., Wierbowski B.M., Salic A. Sending and receiving hedgehog signals. Annu Rev Cell Dev Biol. 2017;33:145–168. doi: 10.1146/annurev-cellbio-100616-060847. [DOI] [PubMed] [Google Scholar]
- 20.Bangs F., Anderson K.V. Primary Cilia and mammalian hedgehog signaling. Cold Spring Harb Perspect Biol. 2017;9(5):a028175. doi: 10.1101/cshperspect.a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho E.K., Stearns T. Hedgehog signaling and the primary cilium: implications for spatial and temporal constraints on signaling. Development. 2021;148(9) doi: 10.1242/dev.195552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giangaspero F., Wellek S., Masuoka J., et al. Stratification of medulloblastoma on the basis of histopathological grading. Acta Neuropathol. 2006;112(1):5–12. doi: 10.1007/s00401-006-0064-x. [DOI] [PubMed] [Google Scholar]
- 23.Kieran M.W., Walker D., Frappaz D., et al. Brain tumors: from childhood through adolescence into adulthood. J Clin Oncol. 2010;28(32):4783–4789. doi: 10.1200/JCO.2010.28.3481. [DOI] [PubMed] [Google Scholar]
- 24.Vladoiu M.C., El-Hamamy I., Donovan L.K., et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature. 2019;572(7767):67–73. doi: 10.1038/s41586-019-1158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor M.D., Northcott P.A., Korshunov A., et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Northcott P.A., Robinson G.W., Kratz C.P., et al. Medulloblastoma Nat Rev Dis Primers. 2019;5(1):11. doi: 10.1038/s41572-019-0063-6. [DOI] [PubMed] [Google Scholar]
- 27.Northcott P.A., Buchhalter I., Morrissy A.S., et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellison D.W., Dalton J., Kocak M., et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121(3):381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih D.J.H., Northcott P.A., Remke M., et al. Cytogenetic prognostication within medulloblastoma subgroups. J Clin Oncol. 2014;32(9):886–896. doi: 10.1200/JCO.2013.50.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manoranjan B., Venugopal C., Bakhshinyan D., et al. Wnt activation as a therapeutic strategy in medulloblastoma. Nat Commun. 2020;11(1):4323. doi: 10.1038/s41467-020-17953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higdon R., Kala J., Wilkins D., et al. Integrated proteomic and transcriptomic-based approaches to identifying signature biomarkers and pathways for elucidation of daoy and UW228 subtypes. Proteomes. 2017;5(1):5. doi: 10.3390/proteomes5010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawson C., Ahmed Alta TB., Moschou G., et al. Novel diarylamides and diarylureas with N-substitution dependent activity against medulloblastoma. Eur J Med Chem. 2021;225 doi: 10.1016/j.ejmech.2021.113751. [DOI] [PubMed] [Google Scholar]
- 33.Skowron P., Ramaswamy V., Taylor M.D. Genetic and molecular alterations across medulloblastoma subgroups. J Mol Med. 2015;93(10):1075–1084. doi: 10.1007/s00109-015-1333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geron L., Salomão K.B., Borges K.S., et al. Molecular characterization of Wnt pathway and function of β-catenin overexpression in medulloblastoma cell lines. Cytotechnology. 2018;70(6):1713–1722. doi: 10.1007/s10616-018-0260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Northcott P.A., Jones D.T.W., Kool M., et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12(12):818–834. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pallavicini G., Iegiani G., Berto G.E., et al. CITK loss inhibits growth of group 3 and group 4 medulloblastoma cells and sensitizes them to DNA-damaging agents. Cancers (Basel) 2020;12(3):542. doi: 10.3390/cancers12030542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sreenivasan L., Wang H., Yap S.Q., et al. Autocrine IL-6/STAT3 signaling aids development of acquired drug resistance in Group 3 medulloblastoma. Cell Death Dis. 2020;11(12):1035. doi: 10.1038/s41419-020-03241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavalli F.M.G., Remke M., Rampasek L., et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31(6):737–754. doi: 10.1016/j.ccell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwalbe E.C., Lindsey J.C., Nakjang S., et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 2017;18(7):958–971. doi: 10.1016/S1470-2045(17)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhukova N., Ramaswamy V., Remke M., et al. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J Clin Oncol. 2013;31(23):2927–2935. doi: 10.1200/JCO.2012.48.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kogerman P., Krause D., Rahnama F., et al. Alternative first exons of PTCH1 are differentially regulated in vivo and may confer different functions to the PTCH1 protein. Oncogene. 2002;21(39):6007–6016. doi: 10.1038/sj.onc.1205865. [DOI] [PubMed] [Google Scholar]
- 42.Shimokawa T., Rahnama F., Zaphiropoulos P.G. A novel first exon of the Patched1 gene is upregulated by Hedgehog signaling resulting in a protein with pathway inhibitory functions. FEBS Lett. 2004;578(1–2):157–162. doi: 10.1016/j.febslet.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Nagao K., Togawa N., Fujii K., et al. Detecting tissue-specific alternative splicing and disease-associated aberrant splicing of the PTCH gene with exon junction microarrays. Hum Mol Genet. 2005;14(22):3379–3388. doi: 10.1093/hmg/ddi369. [DOI] [PubMed] [Google Scholar]
- 44.Uchikawa H., Toyoda M., Nagao K., et al. Brain- and heart-specific Patched-1 containing exon 12b is a dominant negative isoform and is expressed in medulloblastomas. Biochem Biophys Res Commun. 2006;349(1):277–283. doi: 10.1016/j.bbrc.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 45.Roessler E., Ermilov A.N., Grange D.K., et al. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet. 2005;14(15):2181–2188. doi: 10.1093/hmg/ddi222. [DOI] [PubMed] [Google Scholar]
- 46.Matissek S.J., Elsawa S.F. GLI3:a mediator of genetic diseases, development and cancer. Cell Commun Signal. 2020;18(1):54. doi: 10.1186/s12964-020-00540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong A.J., Bigner S.H., Bigner D.D., et al. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84(19):6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo H.W., Zhu H., Cao X., et al. A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion. Cancer Res. 2009;69(17):6790–6798. doi: 10.1158/0008-5472.CAN-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao X., Geradts J., Dewhirst M.W., et al. Upregulation of VEGF-A and CD24 gene expression by the tGLI1 transcription factor contributes to the aggressive behavior of breast cancer cells. Oncogene. 2012;31(1):104–115. doi: 10.1038/onc.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rimkus T.K., Carpenter R.L., Sirkisoon S., et al. Truncated glioma-associated oncogene homolog 1 (tGLI1) mediates mesenchymal glioblastoma via transcriptional activation of CD44. Cancer Res. 2018;78(10):2589–2600. doi: 10.1158/0008-5472.CAN-17-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sirkisoon S.R., Carpenter R.L., Rimkus T., et al. TGLI1 transcription factor mediates breast cancer brain metastasis via activating metastasis-initiating cancer stem cells and astrocytes in the tumor microenvironment. Oncogene. 2020;39(1):64–78. doi: 10.1038/s41388-019-0959-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao J., Ding J., Li Y., et al. HnRNP U mediates the long-range regulation of Shh expression during limb development. Hum Mol Genet. 2009;18(16):3090–3097. doi: 10.1093/hmg/ddp250. [DOI] [PubMed] [Google Scholar]
- 53.Sadam H., Liivas U., Kazantseva A., et al. GLI2 cell-specific activity is controlled at the level of transcription and RNA processing: consequences to cancer metastasis. Biochim Biophys Acta BBA Mol Basis Dis. 2016;1862(1):46–55. doi: 10.1016/j.bbadis.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki H., Kumar S.A., Shuai S., et al. Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature. 2019;574(7780):707–711. doi: 10.1038/s41586-019-1650-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cherry A.L., Finta C., Karlström M., et al. Structural basis of SUFU-GLI interaction in human Hedgehog signalling regulation. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 12):2563–2579. doi: 10.1107/S0907444913028473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han Y., Shi Q., Jiang J. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc Natl Acad Sci U S A. 2015;112(20):6383–6388. doi: 10.1073/pnas.1421628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guerrini-Rousseau L., Dufour C., Varlet P., et al. Germline SUFU mutation carriers and medulloblastoma: clinical characteristics, cancer risk, and prognosis. Neuro Oncol. 2018;20(8):1122–1132. doi: 10.1093/neuonc/nox228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor M.D., Liu L., Raffel C., et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31(3):306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 59.Tostar U., Finta C., Rahman M.F., et al. Novel mechanism of action on hedgehog signaling by a suppressor of fused carboxy terminal variant. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0037761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Q., Gao J., Li Z. Identification of a novel alternative splicing transcript variant of the suppressor of fused: relationship with lymph node metastasis in pancreatic ductal adenocarcinoma. Int J Oncol. 2016;49(6):2611–2619. doi: 10.3892/ijo.2016.3753. [DOI] [PubMed] [Google Scholar]
- 61.Pietrobono S., Gagliardi S., Stecca B. Non-canonical hedgehog signaling pathway in cancer: activation of GLI transcription factors beyond smoothened. Front Genet. 2019;10:556. doi: 10.3389/fgene.2019.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mazzà D., Infante P., Colicchia V., et al. PCAF ubiquitin ligase activity inhibits Hedgehog/Gli1 signaling in p53-dependent response to genotoxic stress. Cell Death Differ. 2013;20(12):1688–1697. doi: 10.1038/cdd.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Marcotullio L., Ferretti E., Greco A., et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. 2006;8(12):1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 64.Tiberi L., Bonnefont J., van den Ameele J., et al. A BCL6/BCOR/SIRT1 complex triggers neurogenesis and suppresses medulloblastoma by repressing sonic hedgehog signaling. Cancer Cell. 2014;26(6):797–812. doi: 10.1016/j.ccell.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 65.Wu C.C., Hou S., Orr B.A., et al. mTORC1-mediated inhibition of 4EBP1 is essential for hedgehog signaling-driven translation and medulloblastoma. Dev Cell. 2017;43(6):673–688. doi: 10.1016/j.devcel.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Antonucci L., Di Magno L., D'Amico D., et al. Mitogen-activated kinase kinase kinase 1 inhibits hedgehog signaling and medulloblastoma growth through GLI1 phosphorylation. Int J Oncol. 2019;54(2):505–514. doi: 10.3892/ijo.2018.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu J., Liu L., Zheng M., et al. MEKK2 and MEKK3 suppress Hedgehog pathway-dependent medulloblastoma by inhibiting GLI1 function. Oncogene. 2018;37(28):3864–3878. doi: 10.1038/s41388-018-0249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guldal C.G., Ahmad A., Korshunov A., et al. An essential role for p38 MAPK in cerebellar granule neuron precursor proliferation. Acta Neuropathol. 2012;123(4):573–586. doi: 10.1007/s00401-012-0946-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shimokawa T., Rahman M.F.U., Tostar U., et al. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biol. 2013;10(2):321–333. doi: 10.4161/rna.23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gruber W., Hutzinger M., Elmer D.P., et al. DYRK1B as therapeutic target in Hedgehog/GLI-dependent cancer cells with Smoothened inhibitor resistance. Oncotarget. 2016;7(6):7134–7148. doi: 10.18632/oncotarget.6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Long J., Li B., Rodriguez-Blanco J., et al. The BET bromodomain inhibitor I-BET151 acts downstream of smoothened protein to abrogate the growth of hedgehog protein-driven cancers. J Biol Chem. 2014;289(51):35494–35502. doi: 10.1074/jbc.M114.595348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Canettieri G., Di Marcotullio L., Greco A., et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates hedgehog signalling through gli acetylation. Nat Cell Biol. 2010;12(2):132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 73.Dhanyamraju P.K., Holz P.S., Finkernagel F., et al. Histone deacetylase 6 represents a novel drug target in the oncogenic hedgehog signaling pathway. Mol Cancer Ther. 2015;14(3):727–739. doi: 10.1158/1535-7163.MCT-14-0481. [DOI] [PubMed] [Google Scholar]
- 74.Malatesta M., Steinhauer C., Mohammad F., et al. Histone acetyltransferase PCAF is required for hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res. 2013;73(20):6323–6333. doi: 10.1158/0008-5472.CAN-12-4660. [DOI] [PubMed] [Google Scholar]
- 75.Miele E., Po A., Begalli F., et al. β-arrestin1-mediated acetylation of Gli1 regulates hedgehog/Gli signaling and modulates self-renewal of SHH medulloblastoma cancer stem cells. BMC Cancer. 2017;17(1):488. doi: 10.1186/s12885-017-3477-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghosh A., Stewart D., Matlashewski G. Regulation of human p53 activity and cell localization by alternative splicing. Mol Cell Biol. 2004;24(18):7987–7997. doi: 10.1128/MCB.24.18.7987-7997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bourdon J.C., Fernandes K., Murray-Zmijewski F., et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19(18):2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abballe L., Mastronuzzi A., Miele E., et al. Numb isoforms deregulation in medulloblastoma and role of p66 isoform in cancer and neural stem cells. Front Pediatr. 2018;6:315. doi: 10.3389/fped.2018.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi H.Y., Seok J., Kang G.H., et al. The role of NUMB/NUMB isoforms in cancer stem cells. BMB Rep. 2021;54(7):335–343. doi: 10.5483/BMBRep.2021.54.7.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim K.K., Nam J., Mukouyama Y.S., et al. Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J Cell Biol. 2013;200(4):443–458. doi: 10.1083/jcb.201206146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zong F.Y., Fu X., Wei W.J., et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014;10(4) doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bechara E.G., Sebestyén E., Bernardis I., et al. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52(5):720–733. doi: 10.1016/j.molcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 83.Hernández J., Bechara E., Schlesinger D., et al. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 2016;13(4):466–472. doi: 10.1080/15476286.2016.1144004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ni J., Wang M., Wang M., et al. CCND1 G870A polymorphism and cervical cancer risk: a case-control study and meta-analysis. J Cancer Res Clin Oncol. 2011;137(3):489–494. doi: 10.1007/s00432-010-0904-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liao D., Wu Y., Pu X., et al. Cyclin D1 G870A polymorphism and risk of nasopharyngeal carcinoma: a case-control study and meta-analysis. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0113299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yuan L., Gu X., Shao J., et al. Cyclin D1 G870A polymorphism is associated with risk and clinicopathologic characteristics of bladder cancer. DNA Cell Biol. 2010;29(10):611–617. doi: 10.1089/dna.2010.1018. [DOI] [PubMed] [Google Scholar]
- 87.Zeybek U., Yaylim I., Ozkan N.E., et al. Cyclin D1 gene G870A variants and primary brain tumors. Asian Pac J Cancer Prev. 2013;14(7):4101–4106. doi: 10.7314/apjcp.2013.14.7.4101. [DOI] [PubMed] [Google Scholar]
- 88.Paronetto M.P., Cappellari M., Busà R., et al. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 2010;70(1):229–239. doi: 10.1158/0008-5472.CAN-09-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feng J., Xu X., Fan X., et al. BAF57/SMARCE1 interacting with splicing factor SRSF1 regulates mechanical stress-induced alternative splicing of cyclin D1. Genes. 2021;12(2):306. doi: 10.3390/genes12020306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu F.H., Luo L.Q., Liu Y., et al. Cyclin D1b splice variant promotes αvβ3-mediated adhesion and invasive migration of breast cancer cells. Cancer Lett. 2014;355(1):159–167. doi: 10.1016/j.canlet.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 91.Millar E.K.A., Dean J.L., McNeil C.M., et al. Cyclin D1b protein expression in breast cancer is independent of cyclin D1a and associated with poor disease outcome. Oncogene. 2009;28(15):1812–1820. doi: 10.1038/onc.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burd C.J., Petre C.E., Morey L.M., et al. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci U S A. 2006;103(7):2190–2195. doi: 10.1073/pnas.0506281103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tan W., Dean M., Law A.J. Molecular cloning and characterization of the human ErbB4 gene: identification of novel splice isoforms in the developing and adult brain. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fox I.J., Kornblum H.I. Developmental profile of ErbB receptors in murine central nervous system: implications for functional interactions. J Neurosci Res. 2005;79(5):584–597. doi: 10.1002/jnr.20381. [DOI] [PubMed] [Google Scholar]
- 95.Ferretti E., di Marcotullio L., Gessi M., et al. Alternative splicing of the ErbB-4 cytoplasmic domain and its regulation by hedgehog signaling identify distinct medulloblastoma subsets. Oncogene. 2006;25(55):7267–7273. doi: 10.1038/sj.onc.1209716. [DOI] [PubMed] [Google Scholar]
- 96.Menghi F., Jacques T.S., Barenco M., et al. Genome-wide analysis of alternative splicing in medulloblastoma identifies splicing patterns characteristic of normal cerebellar development. Cancer Res. 2011;71(6):2045–2055. doi: 10.1158/0008-5472.CAN-10-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dubuc A.M., Morrissy A.S., Kloosterhof N.K., et al. Subgroup-specific alternative splicing in medulloblastoma. Acta Neuropathol. 2012;123(4):485–499. doi: 10.1007/s00401-012-0959-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hong D.S., Kurzrock R., Naing A., et al. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs. 2014;32(3):436–444. doi: 10.1007/s10637-013-0046-5. [DOI] [PubMed] [Google Scholar]
- 99.Nakajima H., Sato B., Fujita T., et al. New antitumor substances, FR901463, FR901464 and FR901465. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J Antibiot (Tokyo) 1996;49(12):1196–1203. doi: 10.7164/antibiotics.49.1196. [DOI] [PubMed] [Google Scholar]
- 100.Sakai Y., Tsujita T., Akiyama T., et al. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. I. Taxonomy, production, isolation, physicochemical properties and biological activities. J Antibiot (Tokyo) 2002;55(10):855–862. doi: 10.7164/antibiotics.55.855. [DOI] [PubMed] [Google Scholar]
- 101.Sakai Y., Tsujita T., Akiyama T., et al. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. II. The effects on cell cycle progression and gene expression. J Antibiot (Tokyo) 2002;55(10):863–872. doi: 10.7164/antibiotics.55.863. [DOI] [PubMed] [Google Scholar]
- 102.Hahn D., Kudla G., Tollervey D., et al. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev. 2012;26(21):2408–2421. doi: 10.1101/gad.199307.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Iwatani-Yoshihara M., Ito M., Klein M.G., et al. Discovery of allosteric inhibitors targeting the spliceosomal RNA helicase Brr2. J Med Chem. 2017;60(13):5759–5771. doi: 10.1021/acs.jmedchem.7b00461. [DOI] [PubMed] [Google Scholar]
- 104.Makowski K., Vigevani L., Albericio F., et al. Sudemycin K: a synthetic antitumor splicing inhibitor variant with improved activity and versatile chemistry. ACS Chem Biol. 2017;12(1):163–173. doi: 10.1021/acschembio.6b00562. [DOI] [PubMed] [Google Scholar]
- 105.Carabet L.A., Leblanc E., Lallous N., et al. Computer-aided discovery of small molecules targeting the RNA splicing activity of hnRNP A1 in castration-resistant prostate cancer. Molecules. 2019;24(4):763. doi: 10.3390/molecules24040763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sohail M., Shkreta L., Toutant J., et al. A novel class of inhibitors that target SRSF10 and promote p53-mediated cytotoxicity on human colorectal cancer cells. NAR Cancer. 2021;3(2):zcab019. doi: 10.1093/narcan/zcab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Siqueira R.P., Barbosa É.D.E.A., Polêto M.D., et al. Potential antileukemia effect and structural analyses of SRPK inhibition by N-(2-(piperidin-1-yl)-5-(trifluoromethyl)phenyl)isonicotinamide (SRPIN340) PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0134882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hatcher J.M., Wu G., Zeng C., et al. SRPKIN-1:a covalent SRPK1/2 inhibitor that potently converts VEGF from pro-angiogenic to anti-angiogenic isoform. Cell Chem Biol. 2018;25(4):460–470. doi: 10.1016/j.chembiol.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chandra A., Ananda H., Singh N., et al. Identification of a novel and potent small molecule inhibitor of SRPK1:mechanism of dual inhibition of SRPK1 for the inhibition of cancer progression. Aging. 2020;13(1):163–180. doi: 10.18632/aging.202301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gammons M.V., Lucas R., Dean R., et al. Targeting SRPK1 to control VEGF-mediated tumour angiogenesis in metastatic melanoma. Br J Cancer. 2014;111(3):477–485. doi: 10.1038/bjc.2014.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee J.Y., Yun J.S., Kim W.K., et al. Structural basis for the selective inhibition of Cdc2-like kinases by CX-4945. BioMed Res Int. 2019;2019 doi: 10.1155/2019/6125068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ajiro M., Awaya T., Kim Y.J., et al. Therapeutic manipulation of IKBKAP mis-splicing with a small molecule to cure familial dysautonomia. Nat Commun. 2021;12(1):4507. doi: 10.1038/s41467-021-24705-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tam B.Y., Chiu K., Chung H., et al. The CLK inhibitor SM08502 induces anti-tumor activity and reduces Wnt pathway gene expression in gastrointestinal cancer models. Cancer Lett. 2020;473:186–197. doi: 10.1016/j.canlet.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 114.Lin J.C. Therapeutic applications of targeted alternative splicing to cancer treatment. Int J Mol Sci. 2017;19(1):75. doi: 10.3390/ijms19010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hong D., Kurzrock R., Kim Y., et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med. 2015;7(314):314ra185. doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]