Abstract
Accumulating evidence supports the association of somatic mutations with tumor occurrence and development. We aimed to identify somatic mutations with important implications in hepatocellular carcinoma (HCC) and explore their possible mechanisms. The gene mutation profiles of HCC patients were assessed, and the tumor mutation burden was calculated. Gene mutations closely associated with tumor mutation burden and patient overall survival were identified. In vivo and in vitro experiments were performed to verify the effects of putative genes on proliferation, invasion, drug resistance, and other malignant biological behaviors of tumor cells. Fourteen genes with a high mutation frequency were identified. The mutation status of 12 of these genes was closely related to the mutation burden. Among these 12 genes, LRP1B mutation was closely associated with patient prognosis. Nine genes were associated with immune cell infiltration. The results of in vivo and in vitro experiments showed that the knockdown of LRP1B promotes tumor cell proliferation and migration and enhances the resistance of tumor cells to liposomal doxorubicin. LRP1B could directly bind to NCSTN and affect its protein expression level, thereby regulating the PI3K/AKT pathway. Our mutational analysis revealed complex and orchestrated liposomal alterations linked to doxorubicin resistance that may also render cancers less susceptible to immunotherapy and also provides new treatment alternatives.
Keywords: Doxorubicin, Hepatocellular carcinoma, LRP1B, PI3K/AKT pathway, Tumor mutation burden
Introduction
Liver cancer is the sixth most common malignancy and the fourth leading cause of cancer-related death worldwide.1 Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, accounting for more than 80% of primary liver cancers worldwide.2 Classic clinical treatments for HCC include surgery, radiotherapy, chemotherapy, and immunotherapy. Despite these diverse possible treatments, the mortality rate of HCC remains high; the 10-year overall survival rate is 10%, and the rates of local recurrence and metastasis are often high.3
Tumor mutation burden (TMB) reflects the number of nonsynonymous mutations in the genome and can be used to predict patient prognosis and treatment outcomes. Nonsynonymous mutations can lead to structurally distinct mutant protein products that act as neoantigens, increasing the possibility that cancer cells are recognized by cytotoxic T lymphocytes.4 Therefore, nonsynonymous mutations have clinical significance in predicting the immunotherapy response. Through bioinformatics analysis, we found a close relationship between LDL receptor-related protein 1 B (LRP1B) expression and TMB; in addition, LRP1B mutation is associated with the efficacy of immunotherapy in multiple cancer types.5 Therefore, LRP1B can be used as a biomarker to predict survival and the efficacy of immunotherapy in HCC patients.
We aimed to explore gene mutations associated with the TMB, immune infiltration, and prognosis of liver cancer. Our study showed that mutations in TP53, TTN, MUC16, AHNAK2, OBSCN, FLG, and LRP1B are closely related to TMB. XIRP2, MUC16, HMCN1, and LRP1B are associated with CD8+ T-cell infiltration, and LRP1B mutation is associated with prognosis in patients with liver cancer.
LRP1B functions as a tumor suppressor gene in cancers such as prostate cancer,6 colon cancer,7 thyroid cancer,8 and gastric cancer.9 In addition, the mutation status and expression level of LRP1B are closely related to prognosis in various cancers.10, 11, 12 Multiple miRNAs can promote tumor proliferation, invasion, and migration by targeting LRP1B.6,13 However, the mechanism underlying the role of LRP1B in tumors is unclear. We identified NCSTN as the interacting protein of LRP1B by coimmunoprecipitation (co-IP), mass spectrometry, and other research methods. LRP1B could bind with NCSTN and affect its protein expression level. Furthermore, through in vitro and in vivo experiments, we found that LRP1B exerts a tumor suppressor effect by regulating the PI3K/AKT signaling pathway through NCSTN. In liver cancer, LRP1B could affect lipid metabolism in liver cancer cells and resistance to liposomal doxorubicin.
In conclusion, our bioinformatics analysis revealed that LRP1B has a high mutation rate in HCC and is closely related to tumor mutation burden and prognosis in HCC. Then, we found that LRP1B affects lipid metabolism, proliferation, and drug resistance in cancer cells through the NCSTN/PI3K/AKT signaling axis. Therefore, LRP1B could be used to predict the prognosis of liver cancer patients and the efficacy of doxorubicin treatment and immunotherapy in liver cancer patients and could be used as a potential target for immunotherapy.
Materials and methods
Mutation analysis
Genome sequencing data for 352 and 331 HCC patients were obtained from the International Cancer Genome Consortium (ICGC; daco.icgc.org) database and The Cancer Genome Atlas (TCGA; portal.gdc.cancer.gov) database, respectively. The mutation frequency and mutation type for each gene were quantitatively determined and presented as a waterfall plot. In addition, clinical information of 375 patients with liver cancer, including age, sex, TNM stage, survival time, and survival status, was obtained. Somatic mutation data were analyzed using the “maftools” R package, the TMB of the patients was calculated, and the patients were divided into the low TMB and high TMB groups. Associations between TMB and single-gene mutations were determined by analyzing mutation data from both groups of patients.
Survival analysis based on single-gene mutation status
The mutation data and clinical data of each patient were paired, and the patients were divided into mutant-type and wild-type groups according to the mutation status of every single gene. The survival curves for the two groups of patients were drawn, and the 5-year survival rates were calculated. Univariate and multivariate Cox regression analyses were performed on the mutant genes screened by survival analysis to determine whether each single-gene mutation was an independent prognostic factor.
Immune infiltration analysis
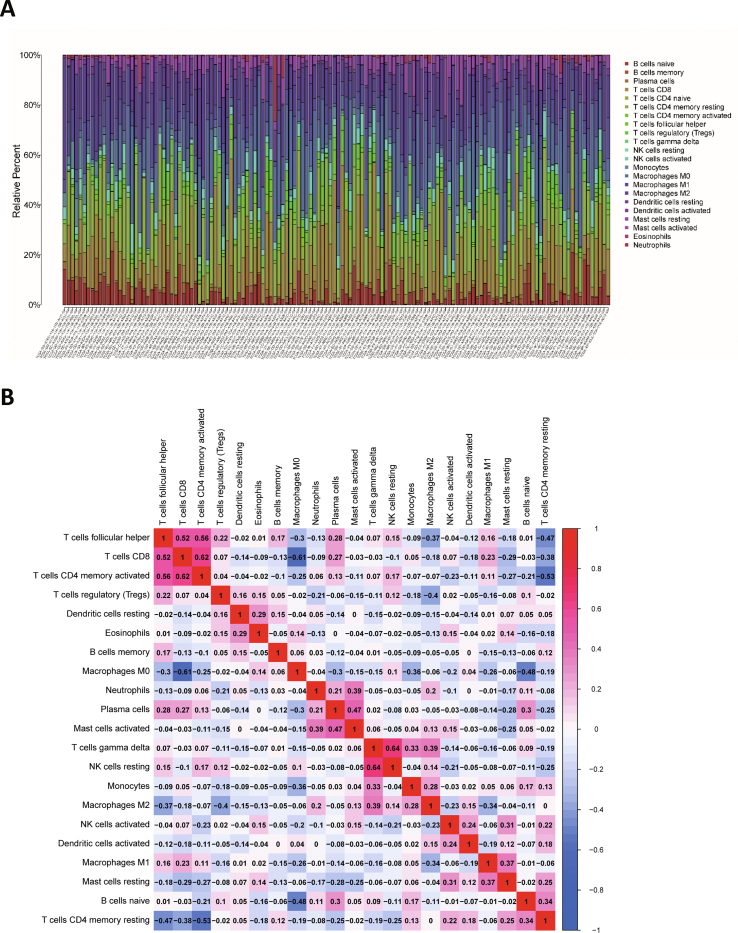
The CIBERSORT algorithm was used to calculate the patients’ immune cell score.14 The analyzed immune cells included B cells (naive B cells and memory B cells), T cells (CD8+ T cells, CD4+ T cells, etc.), natural killer cells, and macrophages. The patients were divided into the wild-type and mutant-type groups according to mutation status. Differences in immune cell infiltration scores between the two groups of patients were analyzed using the “vioplot” package in R software.
Gene ontology (GO) functional enrichment analysis and gene set enrichment analyses (GSEA).
Differences in gene expression between the wild-type and mutant groups of patients were analyzed. Difference analysis, normalization, and visualization of raw transcriptome data were performed with the “limma” package in R software. GO functional enrichment analysis was performed using the “enrichplot”, “org.Hs.eg.db” and “ggplot2” packages in R software. GSEA was performed using software downloaded from “gsea-msigdb.org” (http://www.gsea-msigdb.org/gsea/index.jsp).
Mutation-related gene analysis and identification of potential small molecule drugs
Differentially expressed genes (DEGs) associated with single-gene mutations were analyzed using the online analysis tool muTarget (mutarget.com). DEGs were uploaded to the Connection Map (CMap) database (portals.broadinstitute.org/cmap) to identify drugs that could potentially reverse specific biological effects caused by a single gene mutation in HCC. The structures of potential small molecule drugs were obtained from the PubChem database (pubchem.ncbi.nlm.nih.gov).
Evaluation of cell proliferation ability
Cell proliferation ability was assessed using plate colony formation and Cell Counting Kit-8 (CCK-8) assays. Cells subjected to different treatments were seeded into 6-well plates (approximately 1,000 cells per well). After two weeks of culture, the cells were fixed with formaldehyde, stained with crystal violet, and photographed. A total of 10 μL/well of CCK-8 reagent (Beyotime, Shanghai, China) was added to a 96-well plate containing 2,000 cells per well and incubated at 37 °C for 2 h. The optical density values at 450 nm (OD450) were measured with a 96-well multimode plate reader (Biotech Instruments, Winooski, USA).
Evaluation of cell invasion ability
Cell invasion ability was evaluated using Transwell chambers with 8 μm-pore size polycarbonate membrane filters. After transfection for 24 h, cells were added to the upper compartments of chambers containing membranes coated with Matrigel (BD Bioscience, NJ, USA) and cultured for an additional 24 h. For the wound healing (scratch) assay, cells were seeded in 6-well plates. After the cells were confluent, a scratch was made in the monolayer, and digital images were acquired after 0, 24, and 48 h.
Apoptosis and cell cycle analyses
Transfected cells were examined 48 h post transfection. Apoptosis and the cell cycle status were then assessed using apoptosis and cell cycle kits, respectively (Beyotime, Shanghai, China). Specifically, cells were resuspended in 500 μL of binding buffer and incubated with 5 μL of FITC and 5 μL of PI (apoptosis) or PI/RNase A (cell cycle) for 15 min at room temperature in the dark. Samples were analyzed by flow cytometry (BD Accuri™C6, USA).
Cell immunofluorescence analysis
A total of 3 × 103 cells were seeded on coverslips in each well of a 24-well plate. Cells were fixed with 4% paraformaldehyde for 20 min at room temperature, permeabilized with 0.2% Triton X-100 in PBS for 10 min, blocked in 5% BSA for 1 h at room temperature, and incubated with the primary antibodies overnight at 4 °C. The cells were then incubated with fluorophore-conjugated secondary antibodies (1:1,000; Thermo Fisher Scientific, CA, USA). After washing, sections were incubated with DAPI (Beyotime, Shanghai, China), washed, and mounted. Images were acquired with a ZEISS microscope (Axio Imager A2/AxioCam HRc; Zeiss, Jena, Germany).
Western blotting (WB)
Protein was extracted with 0.5% SDS and quantified using a BCA protein estimation kit (Boster, Wuhan, China). The PVDF membrane was incubated first with the primary antibody overnight at 4 °C and then with the corresponding secondary antibodies at room temperature for 1 h. Immunoreactive bands were visualized using an ECL detection kit (Boster, Wuhan, China) and imaged with a GeneGnome5 Chemiluminescence Series Image Capture system (Syngene, Frederick, USA). Detailed information on all antibodies is shown in Table S1.
Animal model establishment
Male BALB/c nude mice (12–14 g, 3–4 weeks old) were obtained from Hubei Biont Bioscience. Huh7 cells (2 × 106) subjected to different treatments were suspended in 100 μL of PBS and inoculated subcutaneously into the bilateral flanks of nude mice. The tumors were measured every five days, and the volume was calculated according to the following equation: volume = length × width2/2. One month after cell injection, the mice were euthanized, and the tumors were harvested. For immunohistochemical staining, tumor sections were incubated first with the primary antibody and then with the secondary antibody.
Huh7 cells (1 × 106 cells per mouse) subjected to different treatments were injected into BALB/c nude mice via the tail vein. One month after injection, the mice were euthanized, and the lungs were removed. The formation of HCC metastatic foci in the lung was confirmed by hematoxylin and eosin staining.
Co-immunoprecipitation (IP) and ubiquitination assays
The co-IP assay was performed using a Pierce Crosslink Magnetic IP/Co-IP Kit (Thermo Fisher Scientific, CA, USA) according to the manufacturer's instructions. Whole-cell lysates were incubated with the desired antibodies, and the target protein was then pulled down with protein A/G magnetic beads. The microbeads were then washed with a lower-concentration binding buffer, which caused the bound protein to dissociate from the antibody-conjugated beads. The eluate was collected and examined by WB and SDS-PAGE. For the ubiquitination assay, cells co-transfected with the indicated plasmids were lysed in cold IP lysis buffer containing 1% SDS. Subsequently, the lysates were diluted tenfold with IP lysis buffer and subjected to ultrasonic disruption and centrifugation. The next steps were the same as those used for co-IP.
Statistical analysis
The results are presented as the mean ± standard error of the mean (SEM). Statistical analysis was performed using an unpaired t-test. Bioinformatic and statistical analyses were performed using the R package mentioned earlier. DEGs were identified according to the threshold | log2 (fold change) | > 1 and P value < 0.05. GSEA was performed using GSEA version 3.0 (Broad). For the enrichment analysis, the enrichment results were considered meaningful when the false discovery rate (FDR) was <0.05. Statistical analyses of the experimental results were carried out using SPSS version 24.0 (SPSS Inc., Chicago, IL, USA). Two-tailed P values less than 0.05 (P < 0.05) were considered statistically significant.
Results
Tumor mutation burden
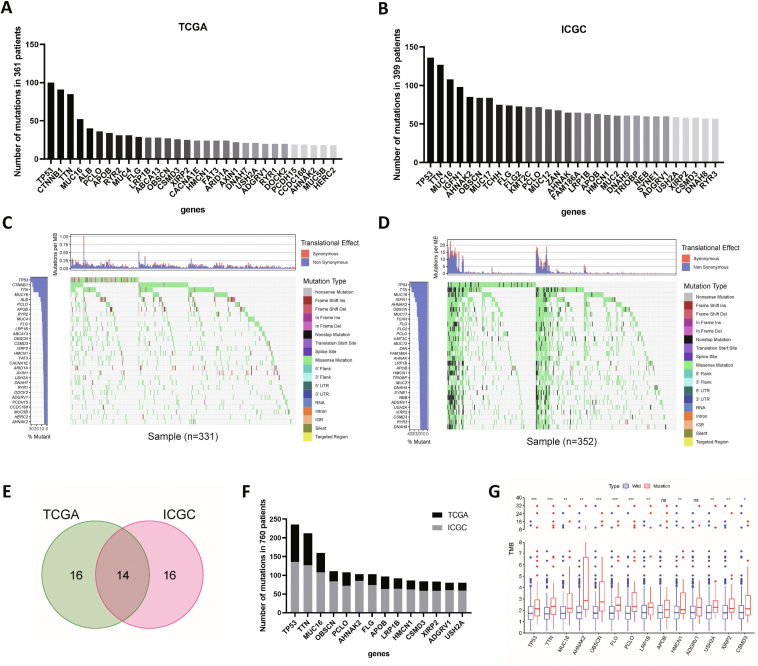
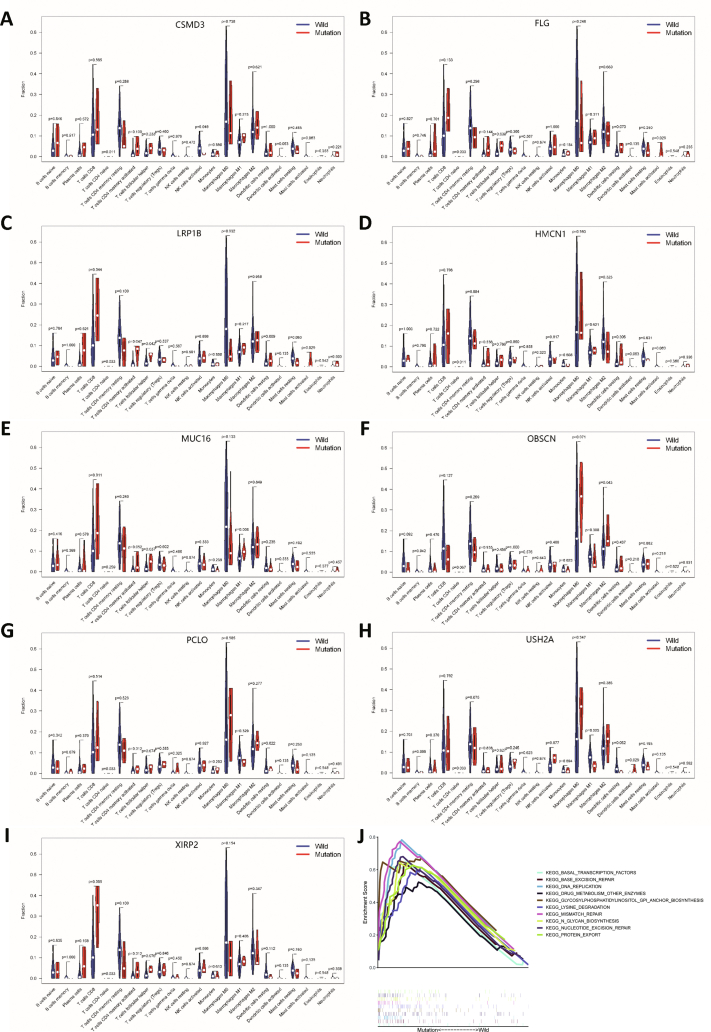
The top 30 high mutation frequency genes from the TCGA database are shown in Figure 1A. In the ICGC database, the top 30 high mutation frequency genes included TP53, TTN, MUC16, IGFN1, and AHNAK2 (Fig. 1B). The mutated genes and types of mutations in each patient were quantitatively determined in the TCGA and ICGC databases, and the most common type of mutation was a missense mutation (Fig. 1C, D). The results obtained in the two databases were intersected to identify 14 genes with high mutation frequencies (Fig. 1E, F). The results of correlation analysis between TMB and gene mutation status showed that the mutation status of TP53, TTN, MUC16, AHNAK2, OBSCN, FLG, PCLO, LRP1B, HMCN1, USH2A, and XIRP2 was closely related to TMB in patients (Fig. 1G). TMB was significantly higher in patients with the above gene mutations (P < 0.01).
Figure 1.
Gene mutation frequency statistics and screening of tumor mutation burden-related mutations. (A) Gene mutation frequencies in the TCGA database. (B) Gene mutation frequencies in the International Cancer Genome Consortium (ICGC) database. (C) Waterfall plot of gene mutation statuses in The Cancer Genome Atlas (TCGA) database. (D) Waterfall plot of gene mutation statuses in the ICGC database. (E) Venn diagram of mutated genes. (F) Mutation frequencies of the overlapping genes. (G) Correlation analysis of TMB and gene mutations.
Prognostic and immune cell infiltration analyses
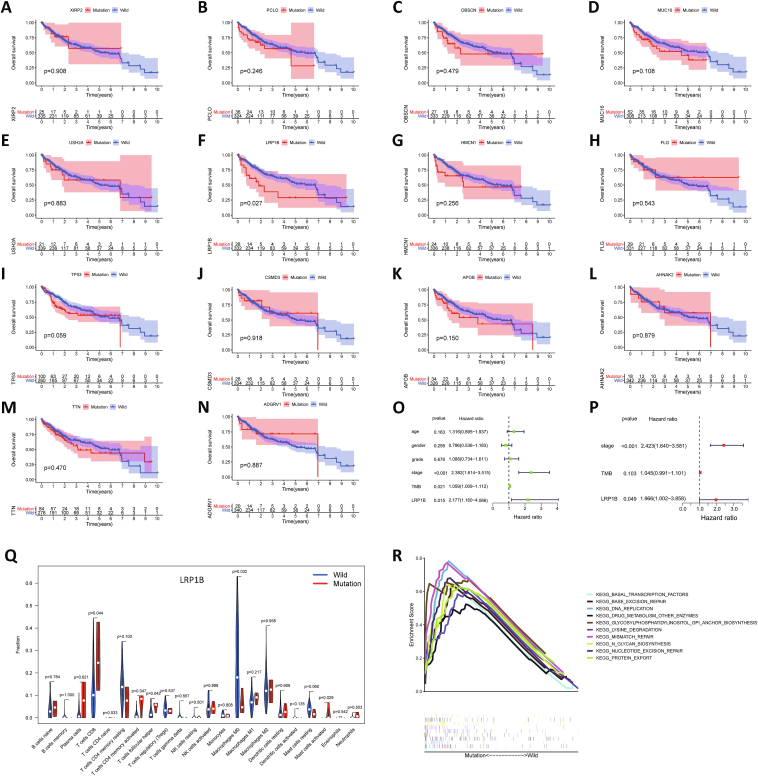
The patients were divided into the wild-type and mutant groups based on the mutation status, and survival analysis was performed using the Kaplan–Meier survival method (Fig. 2A–N). Patients with LRP1B mutation had a worse prognosis than those without LRP1B mutation (Fig. 2F). Univariate Cox regression analysis showed that disease stage and LRP1B mutation status were associated with prognosis (P < 0.05; Fig. 2O). Multivariate Cox regression analysis indicated that disease stage and LRP1B mutation were independent prognostic factors (P < 0.05) and that LRP1B mutation was associated with a poor prognosis (hazard ratio >1, P < 0.05; Fig. 2P). The proportion of immune cells per patient was calculated using CIBERSORT R script v1.03. P < 0.05 was used as the screening threshold, and the results of the immune cell infiltration analysis of 138 patients were considered reliable and used for further analysis (Fig. S1). LRP1B mutation was associated with the infiltration of CD8+ T cells, macrophages, activated mast cells, and activated memory CD4+ T cells (Fig. 2Q). Other genetic mutations were also associated with the infiltration of different immune cells (Fig. S2). GSEA was performed according to the mutation status of LRP1B. Mutation of LRP1B was associated with cellular metabolism and a variety of immune- or inflammation-related pathways, including the nuclear factor kappa-B (NF-κb) signaling pathway and the tumor necrosis factor (TNF) signaling pathway (Fig. 2R).
Figure 2.
Prognostic analysis and immune cell infiltration analysis. (A–N) Survival curves of patients with mutant and wild-type genes: XIRP2 (A), PCLO (B), OBSCN (C), MUC16 (D), USH2A (E), LRP1B (F), HMCN1 (G), FLG (H), TP53 (I), CSMD3 (J), APOB (K), AHNAK2 (L), TTN (M) and ADGRV1 (N). (O) Univariate Cox regression analysis. (P) Multivariate Cox regression analysis. (Q) Differential analysis of immune cell infiltration based on LRP1B mutation status (mutant or wild-type). (R) Gene set enrichment analyses (GSEA) based on LRP1B mutation status.
LRP1B mutation-related gene analysis, functional enrichment analysis, and therapeutic molecule prediction
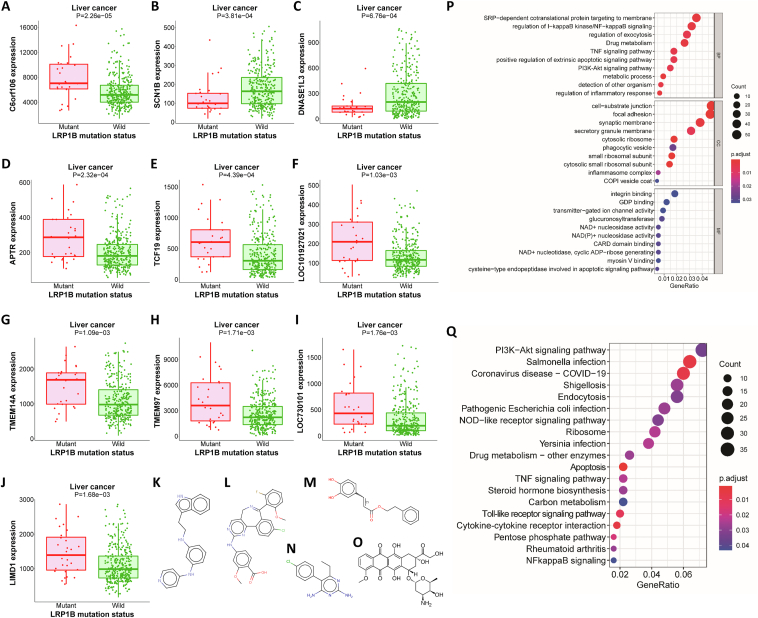
To further analyze the biological function of LRP1B mutation, we identified the genes associated with LRP1B mutation. The expression levels of ILRUN, APTR, TCF19, LOC101927021, TMEM14A, TMEM97, LOC730101, and LIMD1 were significantly higher in patients with LRP1B mutation. Conversely, the expression levels of SCN1B and DNASE1L3 were lower in the mutant group (Fig. 3A–J). Small molecules with potential therapeutic effects against liver cancer were then predicted based on the mutation-related DEG profiles (Table S2). Serdemetan, alisertib, caffeic acid, pyrimethamine, and doxorubicin have strong therapeutic potential (Fig. 3K–O). Among these agents, doxorubicin is widely used in the treatment of liver cancer. GO enrichment analysis showed that LRP1B was associated with various pathways, such as the NF-κB pathway, PI3K/AKT pathway, and TNF pathway (Fig. 3P). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis showed that LRP1B is associated with apoptosis, membrane trafficking, cytokines, drug metabolism, and immune-related pathways (Fig. 3Q).
Figure 3.
LRP1B mutation-related DEGs and functional enrichment analysis. (A–J) Expression levels of f106 (A), SCN1B (B), DNASE1L3 (C), APTR (D), TCF19 (E), LOC101927021 (F), TMEM14A (G), TMEM97 (H), LOC730101 (I) and LIMD1 (J) in the LRP1B mutant and wild-type groups. (K–O) Molecular structure diagrams of serdemetan (K), alisertib (L), caffeic acid (M), pyrimethamine (N), and doxorubicin (O). (P) Gene ontology (GO) enrichment analysis. (Q) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis.
Low expression of LRP1B in hepatocellular carcinoma is associated with cell migration, proliferation, and apoptosis
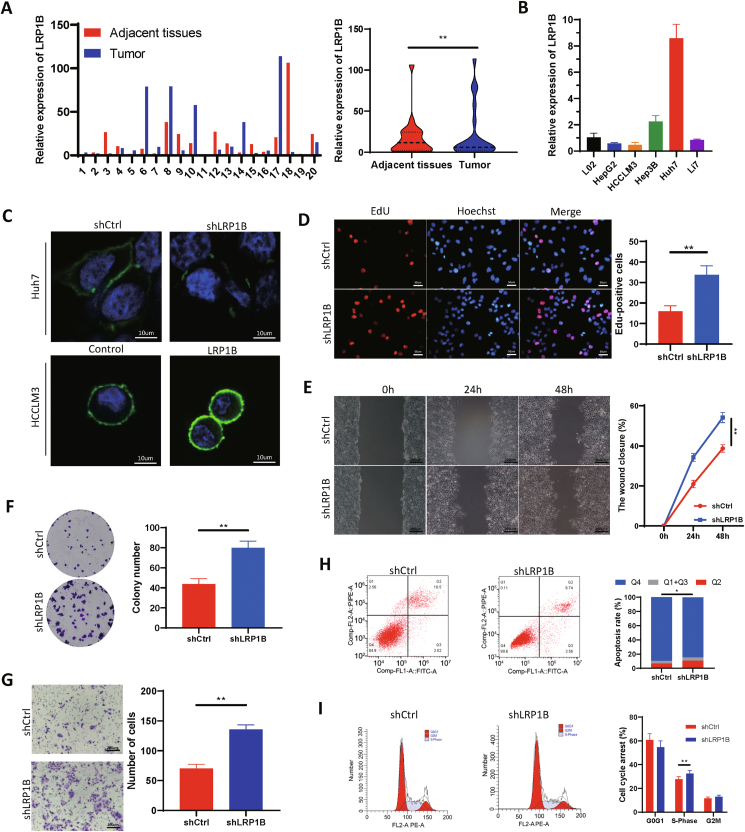
Specimens were collected from 20 patients who underwent resection for liver cancer at the Second Hospital of Shandong University between January 1, 2020, and January 1, 2021, and mRNA was extracted from the tissues for qPCR analysis. In most patients, the expression of LRP1B was lower in tumor tissues than in adjacent tissues (Fig. 4A). The ICGC database indicated that the liver cancer cell line Huh7 was an LRP1B wild-type line. We extracted mRNA from different liver cancer cell lines, and the PCR results showed that the expression level of LRP1B in the Huh7 cell line was significantly higher than that in the other cell lines (Fig. 4B). Therefore, the Huh7 cell line was selected for the functional study of LRP1B. Immunofluorescence analysis showed that the expression level of LRP1B decreased significantly after cells were transfected with shLRP1B. The expression level of LRP1B was obviously increased by LRP1B plasmid transfection (Fig. 4C). We employed 5-ethynyl-2′-deoxyuridine (EdU) incorporation and colony formation assays to detect the effect of LRP1B on cell proliferation. The percentage of EdU-positive cells was significantly higher in the LRP1B knockdown group (Fig. 4D). Knockdown of LRP1B significantly increased the colony number compared with that in the control group (Fig. 4E), suggesting that LRP1B knockdown increases cell proliferation. Huh7 cells with LRP1B knockdown demonstrated an enhanced migratory capacity (Fig. 4F). The Transwell migration assay indicated that the LRP1B knockdown group had a stronger migration ability than the control group (Fig. 4G). LRP1B knockdown led to a decrease in apoptosis upon Nutlin-3 treatment, consistent with the previous results of KEGG enrichment analysis (Fig. 4H). Flow cytometric analysis of the cell cycle showed that after LRP1B knockdown, the proportion of cells in the S phase increased, suggesting that cell proliferation was accelerated (Fig. 4I).
Figure 4.
Low expression of LRP1B in hepatocellular carcinoma is associated with cell migration, proliferation, and apoptosis. (A)LRP1B expression was measured in adjacent and tumor tissues. (B) Relative mRNA expression of LRP1B in different cell lines. (C) Immunofluorescence staining for LRP1B. (D) EdU incorporation assay. (E) Scratch assay of Huh7 cells. (F) Plate colony formation assay. (G) Transwell migration assay. (H) Apoptosis was analyzed by flow cytometry. (I) The cell cycle distribution was analyzed by flow cytometry.
LRP1B affects tumor progression and resistance to liposomal doxorubicin in vivo and in vitro
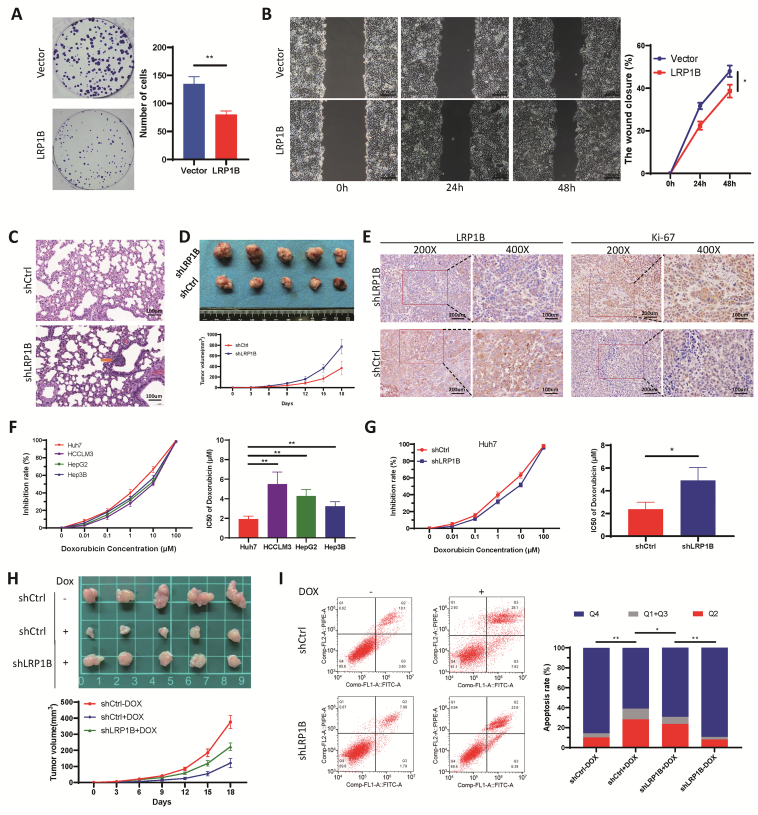
The LRP1B expression plasmid or vector control plasmids were transfected separately into hepatocellular carcinoma cells. The colony formation assays further confirmed that overexpression of LRP1B suppressed cell proliferation (Fig. 5A). The scratch migration assay results showed that overexpression of LRP1B suppressed the migration of HCCLM3 cells compared with that in the control group (Fig. 5B). In an experimental lung metastasis model established by tail vein injection, the number of lung metastases in mice injected with shLRP1B cells was significantly higher than that in mice injected with scramble control cells (Fig. 5C). Mice injected with shLRP1B cells exhibited a larger tumor volume and faster tumor growth (P < 0.05; Fig. 5D). Immunohistochemical analysis of subcutaneous tumors verified the LRP1B knockdown efficiency, and antigen KI-67 (Ki-67) staining was stronger in tumors formed from shLRP1B cells, indicating that the knockdown of LRP1B can promote tumor growth in vivo (Fig. 5E). Under doxorubicin treatment, the inhibition rate was calculated, and a dose–response curve was plotted. The half-maximal inhibitory concentration (IC50) was calculated using GraphPad. The IC50 concentration was lowest in Huh7 cells among the tested cell lines, whereas the expression of LRP1B was the highest (Fig. 5F). The IC50 value in LRP1B knockdown cells was significantly higher than that in control cells (P < 0.05; Fig. 5G). Correspondingly, knockdown of LRP1B was verified to induce the resistance of tumor cells to liposomal doxorubicin in a mouse subcutaneous tumor model (Fig. 5H). Doxorubicin exerts an antitumor effect by inducing apoptosis15 and enrichment analysis showed that LRP1B is related to apoptosis. Therefore, we examined the effect of LRP1B knockout on doxorubicin-induced apoptosis. Indeed, the knockout of LRP1B partially reversed liposomal doxorubicin-induced apoptosis (Fig. 5I).
Figure 5.
Effects of LRP1B on tumor progression and doxorubicin resistance. (A) Plate clone formation assay. (B) Scratch assay of HCCLM3 cells. (C) Lung metastatic foci in the lung metastasis model. (D) Photographs and growth curves of subcutaneous xenografts. (E) Immunohistochemical staining for the expression of LRP1B and Ki67 in subcutaneous xenograft tissues. (F) IC50 values in different cell lines were determined based on 50% growth inhibition using a CCK-8 assay. (G) IC50 values were determined in LRP1B knockdown Huh7 cells and control cells. (H) Growth curves of subcutaneous xenografts treated with doxorubicin. (I) Detection of apoptotic Huh7 cells after doxorubicin and shLRP1B treatment.
LRP1B binds to NCSTN and regulates the PI3K/AKT/mTOR pathway
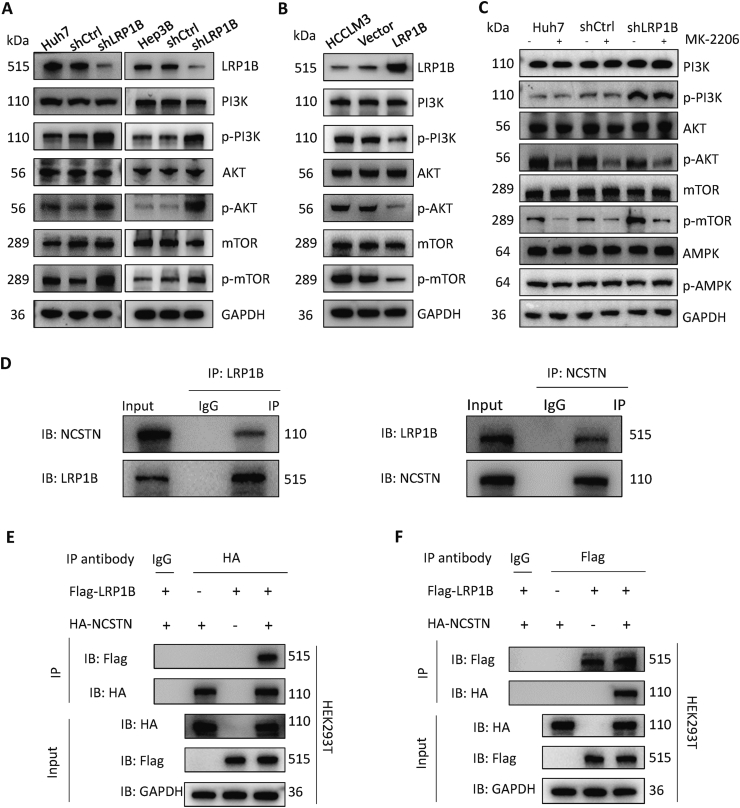
The KEGG enrichment analysis results suggested that LRP1B may be closely related to the PI3K/AKT signaling pathway. Western blot analysis confirmed that the knockdown of LRP1B significantly increased the phosphorylation levels of PI3K, AKT, and mTOR without affecting the corresponding total protein levels (Fig. 6A). Overexpression of LRP1B had the opposite regulatory effects on the PI3K/AKT/mTOR signaling pathway (Fig. 6B). mTOR is regulated by the AMPK/mTOR signaling pathway in addition to the PI3K/AKT signaling pathway; thus, we used the AKT phosphorylation inhibitor MK-2206. The results showed that LRP1B regulates mTOR through the PI3K/AKT signaling pathway but not the AMPK/mTOR signaling pathway (Fig. 6C). Co-IP and mass spectrometry analysis showed that LRP1B may directly bind to NCSTN. We then performed co-IP experiments to verify the formation of the LRP1B/NCSTN protein complex in Huh7 cells (Fig. 6D). Furthermore, we carried out IP using lysates prepared from 293 T cells with exogenous overexpression of LRP1B and NCSTN and proved the reciprocal interaction between NCSTN and LRP1B (Fig. 6E, F).
Figure 6.
LRP1B directly binds to NCSTN and regulates the PI3K/AKT/mTOR pathway. (A)LRP1B knockdown causes upregulation of the PI3K/AKT pathway. (B)LRP1B overexpression inhibited the PI3K/AKT pathway. (C)LRP1B regulates the PI3K/AKT/mTOR pathway. (D) Immunoprecipitation (IP) assay in Huh7 cells. (E) IP assay in 293 T cells using a primary antibody specific for HA. (F) IP was performed with a primary antibody specific for Flag.
LRP1B regulates the ubiquitination and protein level of NCSTN
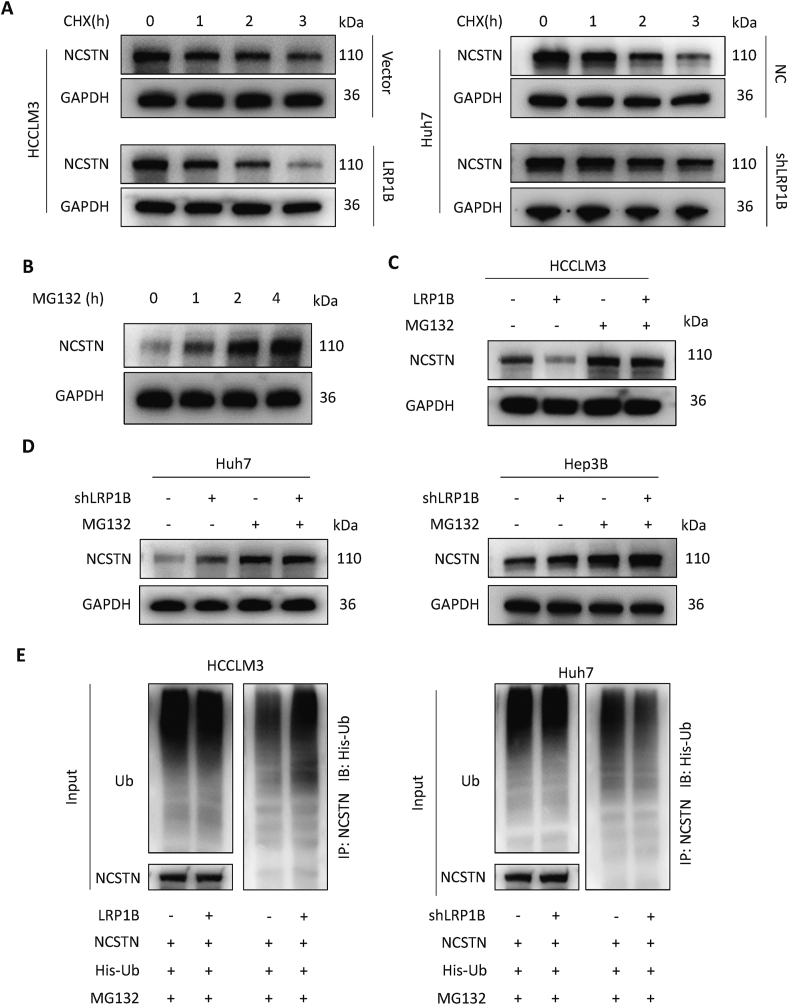
We explored the influence of LRP1B on the degradation of the NCSTN protein by utilizing cycloheximide (CHX), which can inhibit protein synthesis. After overexpression of LRP1B in HCCLM3 cells, the NCSTN protein degradation rate was accelerated compared with that in control cells, indicating that LRP1B promotes the degradation of NCSTN protein (Fig. 7A). To determine whether this clearance involves the proteasome pathway, we treated cells with MG132 to inhibit proteasomal degradation. The protein level of NCSTN increased gradually with prolonged MG132 treatment time, indicating that NCSTN could be degraded via the proteasome pathway (Fig. 7B). Overexpression of LRP1B decreased and knockdown of LRP1B increased the protein level of NCSTN, and these changes were reversed by MG132 treatment (Fig. 7C, D). Overexpression of LRP1B increased the ubiquitination level of NCSTN; similarly, knockdown of LRP1B decreased the ubiquitination level of NCSTN, indicating that LRP1B can regulate the protein level of NCSTN by directly binding to NCSTN and promoting its ubiquitination and degradation (Fig. 7E).
Figure 7.
LRP1B regulates the ubiquitination, degradation, and protein level of NCSTN. (A) The rate of NCSTN degradation after cycloheximide (CHX) inhibition of protein synthesis. (B)NCSTN protein levels were determined after MG132 treatment. (C) The proteasome inhibitor MG132 restored the LRP1B-mediated reduction in the NCSTN protein level in cancer cells. (D)LRP1B regulates the ubiquitination of NCSTN.
LRP1B regulates the malignant behavior of HCC cells through the PI3K/AKT/mTOR signaling pathway
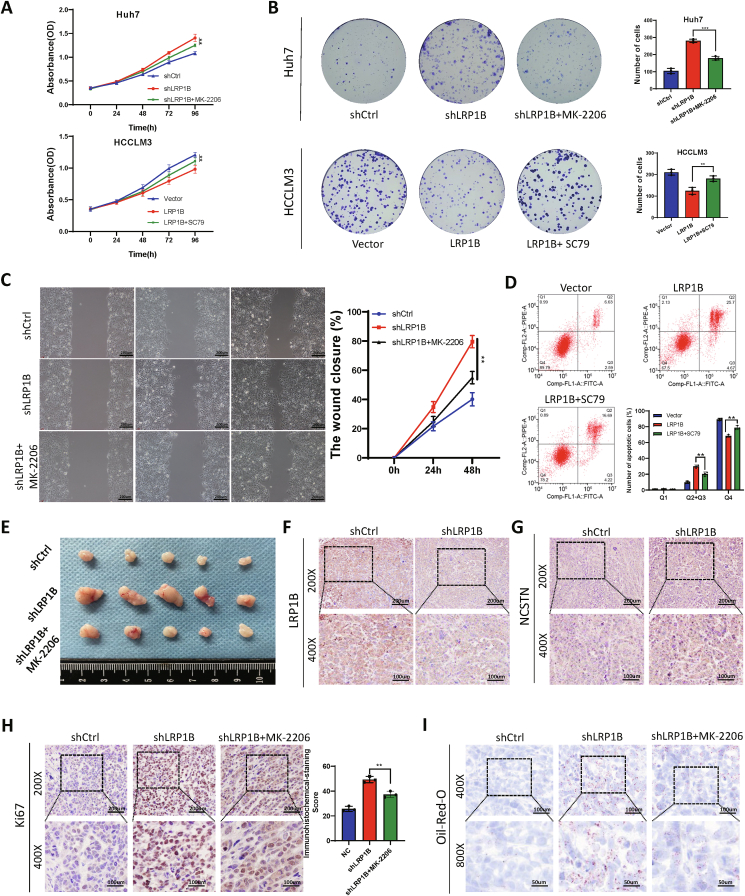
Previous studies have shown that NCSTN has a regulatory effect on the PI3K/AKT signaling pathway.16,17 We performed rescue experiments to demonstrate that LRP1B plays a regulatory role in the malignant behavior of tumor cells through the PI3K/AKT pathway. The PI3K/AKT/mTOR pathway inhibitor MK-2206 and the agonist SC79 reversed the effect of LRP1B on the proliferation of Huh7 and HCCLM3 cells (Fig. 8A, B). The scratch assay showed that the promoting effect of shLRP1B on cell migration was reversed by MK-2206 treatment (Fig. 8C). Overexpression of LRP1B enhanced HCCLM3 cell apoptosis, an effect reversed by SC79 (Fig. 8D). Silencing LRP1B promoted cell proliferation in subcutaneous xenograft tissues in the mouse model, and this effect was reversed by MK-2206 treatment (Fig. 8E). Immunohistochemical assays were used to detect the protein expression levels of LRP1B and NCSTN, and their expression was lower in tumors derived from shLRP1B cells than in tumors derived from control cells (Fig. 8F, G). In addition, the proportion of Ki-67-positive cells was significantly increased in tumors derived from shLRP1B-treated cells, and this increase was partially reversed by MK-2206 treatment (Fig. 8H). Oil Red O staining revealed that knockdown of LRP1B increased lipid droplet formation through the PI3K/AKT pathway and that this effect was reversed by MK-2206 treatment (Fig. 8I).
Figure 8.
LRP1B regulates the malignant behavior of tumor cells through the PI3K/AKT/mTOR pathway. (A) Proliferation was evaluated by a CCK-8 assay. (B) Plate colony formation assays were used to detect cell proliferation. (C) Scratch assay. (D) Flow cytometry was used to detect apoptosis in transfected cells. (E) Subcutaneous xenografts in mice from the indicated groups. (F, G) Immunohistochemical staining for LRP1B (F) and NCSTN (G) in subcutaneous xenograft tissues. (H) Immunohistochemical staining for Ki-67. (I) Oil Red O staining of subcutaneous xenograft tissues.
Discussion
HCC carcinogenesis is a complex multistep process involving multiple genetic mutations. The predominant consequence of mutation accumulation is the activation of proto-oncogenes or silencing of tumor suppressor genes.18 Somatic missense mutations strongly promote the generation of novel tumor epitopes. TMB can reflect the level of mutations in the exon-coding regions of genes in the genome. With the increasing use of immunotherapy in cancer patients, the study of TMB has attracted increasing attention. Generally, the higher the TMB, the greater the difference between tumor cells and normal cells, and the more likely the patient will benefit from immunotherapy. Therefore, the efficacy of immunotherapy in patients can be predicted according to the TMB. Associations between genetic mutations and TMB have previously been reported.19 It is very important to study the relationship between single gene mutations, TMB, tumorigenesis mechanisms, and tumor therapy.
TMB and gene mutations were evaluated as potential biomarkers for immunotherapy and survival. However, studies investigating the relationship between TMB and genes with high mutation frequency in liver cancer are limited. Determining approaches to use effective biomarkers to identify immunotherapy-sensitive people has become a research hotspot. In this study, to further explore TMB-related gene mutations, we (i) counted and screened genes with high mutation frequency in liver cancer patients; (ii) analyzed differences in gene mutations between the high and low TMB groups; and (iii) identified a key mutated gene, LRP1B, associated with liver cancer prognosis. Through these studies, we identified LRP1B mutation as a key genetic mutation associated with TMB and patient prognosis.
LRP1B gene expression is frequently inactivated in numerous human malignancies, suggesting that this gene is a potential tumor suppressor gene. In our study, TP53 was found to have the highest mutation frequency. The mutation frequency of LRP1B was lower than that of TP53, but survival analysis indicated that the LRP1B mutation had a stronger correlation with patient survival. Patients with LRP1B mutations had a significantly worse prognosis, indicating that LRP1B is a tumor suppressor gene with important functions and has important research value. We further analyzed the relationship between LRP1B and tumor immune cell infiltration and found that LRP1B mutation was associated with the infiltration of CD8+ T cells. Inhibition of PD-1/PD-L1 has been reported to increase the infiltration of CD8+ T cells to kill tumor cells.20 This observation suggests that mutations of LRP1B may be closely related to immunotherapy and may become an important immunotherapy target.
LRP1B, a member of the low-density lipoprotein (LDL) receptor family, is localized to the cell membrane, as shown by immunofluorescence. As a member of the LDL receptor family, LRP1B is involved in the uptake of anionic liposomes and drugs.21,22 We identified doxorubicin, which is widely used in the treatment of liver cancer, by predicting the therapeutic effect of small molecule compounds. Based on this finding, we speculated that LRP1B might be involved in the uptake of liposomal doxorubicin. Similarly, previous studies have shown that LRP1B deletion in ovarian cancers is associated with resistance to liposomal doxorubicin.23 We demonstrated that deletion of LRP1B in liver cancer cells is associated with resistance to liposomal doxorubicin by both in vivo and in vitro experiments. Mechanistically, LRP1B may bind to apolipoprotein E (APOE) in liposomes and participate in the uptake of liposomes. When LRP1B is knocked down, the uptake of liposomes is blocked; thus, doxorubicin cannot easily enter cells to exert its effects.
LRP1B participates in liposome uptake and regulates lipid metabolism in cells.24, 25, 26 KEGG enrichment analysis indicated that mTOR, an important regulator of lipid metabolism, may also be regulated by LRP1B.27,28 Therefore, we considered that LRP1B may regulate lipid metabolism in liver cancer cells through mTOR-related pathways. Subsequent experiments also confirmed the regulatory effect of LRP1B on the PI3K/AKT/mTOR pathway and revealed that the knockdown of LRP1B could activate mTOR to promote fat accumulation and liver cancer progression. Whether the LRP1B ectodomain binds to apoE-containing lipoproteins (high-density lipoprotein (HDL) and very-low-density lipoprotein (VLDL)) and is involved in lipoprotein uptake is worth investigating,29,30 but inactivation of LRP1B ultimately leads to accumulation of intracellular lipids. Possible reasons are as follows: (i) lipoproteins and other ligands can induce other regulatory mechanisms mediated by the intracellular tail domain of LRP1B; and (ii) LRP1B and LRP1 antagonize each other's functions by competing for the binding of common ligands, and the of LRP1B enables functional enhancement of LRP1 with increased efficiency of lipoprotein endocytosis, thereby promoting cellular lipid uptake.31 Therefore, the interaction between LRP1B and extracellular ligands and the biological function of the intracellular domain of LRP1B require further study.
LRP1B directly binds to NCSTN and decreases the NCSTN protein level. LRP1B is not a ubiquitinase, but it can increase the ubiquitination level of NCSTN to promote its degradation. Mechanistically, LRP1B may assist in recruiting a ubiquitinase for binding to NCSTN. Alternatively, LRP1B could compete with deubiquitinases for binding to the same binding site on NCSTN. Unfortunately, we have not determined the specific mechanism, and we will study it in depth in subsequent experiments.
This study has limitations. First, the data on gene mutations in liver cancer used in this study came from publicly available data and lacked the support of a large number of independent clinical samples. We plan to include clinical patient samples for gene sequencing in the future to further explore the types of gene mutations and the mutation frequency. Second, although this study showed that the LRP1B mutations are associated with immune cell infiltration in liver cancer, experimental validation is lacking. Finally, the specific site for the binding of LRP1B to NCSTN and the site and type of NCSTN ubiquitination were not determined.
In conclusion, this study demonstrates that mutations in LRP1B in liver cancer are associated with TMB and poor prognosis in patients. LRP1B mutation is associated with the infiltration of CD8+ T cells in the immune microenvironment of liver cancer. Knockdown of LRP1B causes acquired resistance to liposomal doxorubicin in HCC cells. Inactivation of LRP1B can activate the PI3K/AKT/mTOR pathway in tumor cells to promote lipid accumulation, proliferation, invasion, and other malignant biological behaviors. Therefore, our work revealed the important role and molecular mechanism of LRP1B in HCC progression, which may facilitate the development of immunotherapeutic approaches targeting LRP1B.
Author contributions
HZ, XL, and BJ designed the study and conducted the experiments and literature search. XYZ, ZX, GD, XLZ, and TX developed the methodology of analysis. XYZ, XLZ, DM, ZX, and GD helped with the collection and acquisition of data. XYZ, ZX, and GD analyzed the data. XYZ, ZX, XL, HZ, and BJ wrote the manuscript and revised the final version. All authors read and approved the final version of the manuscript.
Conflict of interests
The authors declare no conflict of interests.
Funding
This study was funded by the National Natural Science Foundation of China (No. 81871653), the Natural Science Foundation of Chongqing, China (No. cstc2020jcyj-msxmX0159), Chongqing Science and Health Joint Medical High-end Talent Project (China) (No. 2022GDRC012), Science and Technology Research Program of Chongqing Municipal Education Commission (China) (No. KJZD-K202100402 and KJQN201900449), CQMU Program for Youth Innovation in Future Medicine (China) (No. W0073) and Second Hospital of Shandong University Cultivation Funding(China) (No.2022YP45).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.10.021.
Contributor Information
Xiaosong Li, Email: lixiaosong@cqmu.edu.cn.
Bin Jin, Email: jinbin@sdu.edu.cn.
Hao Zhang, Email: 201735939@mail.sdu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figure S1.
Figure S2.
References
- 1.Yang J.D., Hainaut P., Gores G.J., et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Riley T.P., Keller G.L.J., Smith A.R., et al. Structure based prediction of neoantigen immunogenicity. Front Immunol. 2019;10:2047. doi: 10.3389/fimmu.2019.02047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown L.C., Tucker M.D., Sedhom R., et al. LRP1B mutations are associated with favorable outcomes to immune checkpoint inhibitors across multiple cancer types. J Immunother Cancer. 2021;9(3) doi: 10.1136/jitc-2020-001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng H., Bai L. Hypoxia induced microRNA-301b-3p overexpression promotes proliferation, migration and invasion of prostate cancer cells by targeting LRP1B. Exp Mol Pathol. 2019;111 doi: 10.1016/j.yexmp.2019.104301. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z., Sun P., Gao C., et al. Down-regulation of LRP1B in colon cancer promoted the growth and migration of cancer cells. Exp Cell Res. 2017;357(1):1–8. doi: 10.1016/j.yexcr.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Prazeres H., Torres J., Rodrigues F., et al. Chromosomal, epigenetic and microRNA-mediated inactivation of LRP1B, a modulator of the extracellular environment of thyroid cancer cells. Oncogene. 2011;30(11):1302–1317. doi: 10.1038/onc.2010.512. [DOI] [PubMed] [Google Scholar]
- 9.Lu Y.J., Wu C.S., Li H.P., et al. Aberrant methylation impairs low density lipoprotein receptor-related protein 1B tumor suppressor function in gastric cancer. Genes Chromosomes Cancer. 2010;49(5):412–424. doi: 10.1002/gcc.20752. [DOI] [PubMed] [Google Scholar]
- 10.Hong J.Y., Cho H.J., Kim S.T., et al. Comprehensive molecular profiling to predict clinical outcomes in pancreatic cancer. Ther Adv Med Oncol. 2021;13 doi: 10.1177/17588359211038478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao C.H., Liu R., Lin X.R., et al. LRP1B mutation is associated with tumor HPV status and promotes poor disease outcomes with a higher mutation count in HPV-related cervical carcinoma and head & neck squamous cell carcinoma. Int J Biol Sci. 2021;17(7):1744–1756. doi: 10.7150/ijbs.56970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., Yan K., He X., et al. LRP1B or TP53 mutations are associated with higher tumor mutational burden and worse survival in hepatocellular carcinoma. J Cancer. 2021;12(1):217–223. doi: 10.7150/jca.48983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., Cui R., Li H., et al. miR-500 promotes cell proliferation by directly targetting LRP1B in prostate cancer. Biosci Rep. 2019;39(4) doi: 10.1042/BSR20181854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman A.M., Liu C.L., Green M.R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirzaei S., Zarrabi A., Hashemi F., et al. Nrf2 signaling pathway in chemoprotection and doxorubicin resistance: potential application in drug discovery. Antioxidants. 2021;10(3):349. doi: 10.3390/antiox10030349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao X., He Y., Li C., et al. Nicastrin mutations in familial acne inversa impact keratinocyte proliferation and differentiation through the Notch and phosphoinositide 3-kinase/AKT signalling pathways. Br J Dermatol. 2016;174(3):522–532. doi: 10.1111/bjd.14223. [DOI] [PubMed] [Google Scholar]
- 17.Wang X., Wang X., Xu Y., et al. Effect of nicastrin on hepatocellular carcinoma proliferation and apoptosis through PI3K/AKT signalling pathway modulation. Cancer Cell Int. 2020;20:91. doi: 10.1186/s12935-020-01172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn W.C., Weinberg R.A. Rules for making human tumor cells. N Engl J Med. 2002;347(20):1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 19.Jia Q., Wang J., He N., et al. Titin mutation associated with responsiveness to checkpoint blockades in solid tumors. JCI Insight. 2019;4(10) doi: 10.1172/jci.insight.127901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H.D., Song G.W., Park S., et al. Association between expression level of PD1 by tumor-infiltrating CD8+ T cells and features of hepatocellular carcinoma. Gastroenterology. 2018;155(6):1936–1950. doi: 10.1053/j.gastro.2018.08.030. e17. [DOI] [PubMed] [Google Scholar]
- 21.Chung N.S., Wasan K.M. Potential role of the low-density lipoprotein receptor family as mediators of cellular drug uptake. Adv Drug Deliv Rev. 2004;56(9):1315–1334. doi: 10.1016/j.addr.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Lakkaraju A., Rahman Y.E., Dubinsky J.M. Low-density lipoprotein receptor-related protein mediates the endocytosis of anionic liposomes in neurons. J Biol Chem. 2002;277(17):15085–15092. doi: 10.1074/jbc.M111764200. [DOI] [PubMed] [Google Scholar]
- 23.Cowin P.A., George J., Fereday S., et al. LRP1B deletion in high-grade serous ovarian cancers is associated with acquired chemotherapy resistance to liposomal doxorubicin. Cancer Res. 2012;72(16):4060–4073. doi: 10.1158/0008-5472.CAN-12-0203. [DOI] [PubMed] [Google Scholar]
- 24.May P., Woldt E., Matz R.L., et al. The LDL receptor-related protein (LRP) family: an old family of proteins with new physiological functions. Ann Med. 2007;39(3):219–228. doi: 10.1080/07853890701214881. [DOI] [PubMed] [Google Scholar]
- 25.Masson O., Chavey C., Dray C., et al. LRP1 receptor controls adipogenesis and is up-regulated in human and mouse obese adipose tissue. PLoS One. 2009;4(10) doi: 10.1371/journal.pone.0007422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas J., Beer A.G., Widschwendter P., et al. LRP1b shows restricted expression in human tissues and binds to several extracellular ligands, including fibrinogen and apoE - carrying lipoproteins. Atherosclerosis. 2011;216(2):342–347. doi: 10.1016/j.atherosclerosis.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mossmann D., Park S., Hall M.N. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer. 2018;18(12):744–757. doi: 10.1038/s41568-018-0074-8. [DOI] [PubMed] [Google Scholar]
- 28.Caron A., Richard D., Laplante M. The roles of mTOR complexes in lipid metabolism. Annu Rev Nutr. 2015;35:321–348. doi: 10.1146/annurev-nutr-071714-034355. [DOI] [PubMed] [Google Scholar]
- 29.Strickland D.K., Gonias S.L., Argraves W.S. Diverse roles for the LDL receptor family. Trends Endocrinol Metabol. 2002;13(2):66–74. doi: 10.1016/s1043-2760(01)00526-4. [DOI] [PubMed] [Google Scholar]
- 30.Shiroshima T., Oka C., Kawaichi M. Identification of LRP1B-interacting proteins and inhibition of protein kinase Cα-phosphorylation of LRP1B by association with PICK1. FEBS Lett. 2009;583(1):43–48. doi: 10.1016/j.febslet.2008.11.045. [DOI] [PubMed] [Google Scholar]
- 31.Marzolo M.P., Bu G. Lipoprotein receptors and cholesterol in APP trafficking and proteolytic processing, implications for Alzheimer's disease. Semin Cell Dev Biol. 2009;20(2):191–200. doi: 10.1016/j.semcdb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.