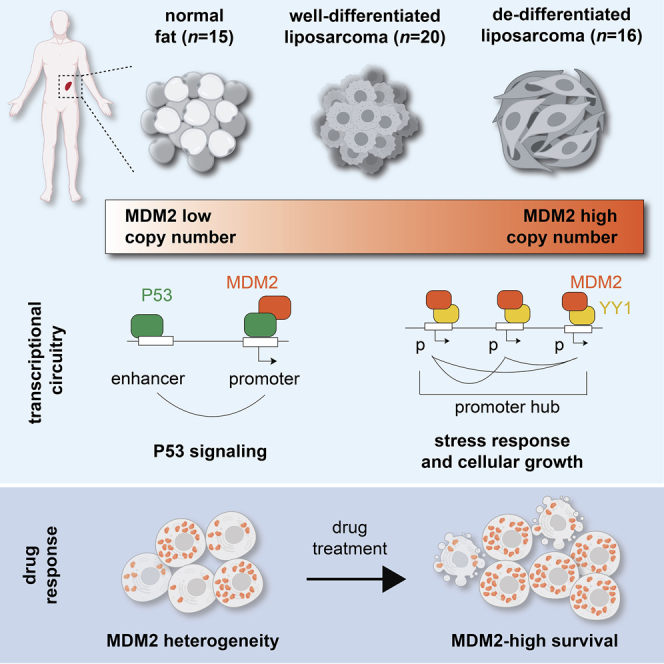
Summary
Amplification of MDM2 on supernumerary chromosomes is a common mechanism of P53 inactivation across tumors. Here, we investigated the impact of MDM2 overexpression on chromatin, gene expression, and cellular phenotypes in liposarcoma. Three independent regulatory circuits predominate in aggressive, dedifferentiated tumors. RUNX and AP-1 family transcription factors bind mesenchymal gene enhancers. P53 and MDM2 co-occupy enhancers and promoters associated with P53 signaling. When highly expressed, MDM2 also binds thousands of P53-independent growth and stress response genes, whose promoters engage in multi-way topological interactions. Overexpressed MDM2 concentrates within nuclear foci that co-localize with PML and YY1 and could also contribute to P53-independent phenotypes associated with supraphysiologic MDM2. Importantly, we observe striking cell-to-cell variability in MDM2 copy number and expression in tumors and models. Whereas liposarcoma cells are generally sensitive to MDM2 inhibitors and their combination with pro-apoptotic drugs, MDM2-high cells tolerate them and may underlie the poor clinical efficacy of these agents.
Keywords: MDM2, liposarcoma, sarcoma, epigenetics, genome topology, P53 independent, therapeutic resistance
Graphical abstract
Highlights
-
•
Genomic and transcriptomic characterization of a liposarcoma clinical cohort
-
•
JUN/RUNX2, P53, and MDM2 underlie dedifferentiated tumor expression programs
-
•
Supraphysiologic MDM2 associates with P53-independent promoter hubs
-
•
Subpopulations with very high MDM2 evade targeted therapy independent of P53
Bevill et al. define transcriptional and regulatory circuits that underlie aggressive tumor states in a clinical cohort of liposarcoma. They find that supraphysiologic MDM2 associates with thousands of growth and stress response genes independent of P53. MDM2 expression is highly variable, and tumor cells with exceptionally high levels tolerate targeted therapies.
Introduction
The TP53 tumor suppressor is the most frequently inactivated gene in human cancers.1 A substantial subset of tumors suppresses its function by overexpressing MDM2, an E3 ubiquitin ligase that targets P53 for degradation.2,3 MDM2 overexpression is often mediated through massive gene amplification on supernumerary rings, rods, and/or extrachromosomal DNA.4,5,6,7 In this context, MDM2 is expressed at supraphysiologic levels many times higher than in wild-type conditions.
The P53 transcription factor (TF) activates its gene targets in response to genotoxicity and cell stress.8,9 MDM2 negatively regulates P53 function by directing its degradation.10 Accordingly, small molecules such as nutlins or HDM201 that abrogate the physical interaction between these proteins reactivate P53 and trigger apoptosis in tumor models.11 However, this model is complicated by reports that MDM2 also localizes to chromatin with P53.12,13 Moreover, MDM2 has multiple additional reported binding partners, many of which are transcriptional regulators or nuclear proteins.13,14,15,16,17,18 Hence, the full scope of MDM2 is likely to include functions in the nucleus and targets other than P53. Furthermore, whether supraphysiologic MDM2 protein levels confer entirely distinct activities remains an open question.
P53 mediates its effects by binding to cognate motifs in promoters and enhancers. In addition to its ∼20,000 genes, the human genome contains a much larger number of enhancers with highly cell-type-specific activities.19 A major challenge in genomics is connecting these distal elements to their target genes. These targets may be predicted on the basis of linear proximity along the DNA and coordinated activities, as well as physical looping interactions.20,21,22
MDM2 is overexpressed in nearly half of all sarcomas, making it an attractive therapeutic target.23,24,25 However, MDM2 inhibitors have largely failed in the clinic, apparently due to either dose-limiting toxicity or tumor cell resistance.26,27 Liposarcoma is a compelling model for investigating MDM2 function and refining therapeutic strategies given that MDM2 amplification is a defining feature of this disease.25,28,29 It is the most common soft-tissue sarcoma that can present as a well-differentiated tumor (WDLPS) or as the much more aggressive dedifferentiated liposarcoma (DDLPS).28,30 Unfortunately, non-surgical therapeutic options for this disease are extremely limited.28 While DDLPS and WDLPS are histologically distinct, no recurrent molecular abnormality distinguishes the two subtypes.31,32 JUN amplifications have been identified in a subset of dedifferentiated tumors, but this feature is not consistently seen in DDLPS.29,31,33 Epigenetically, KLF6 repression and concurrent shifts in H3K9me3 have been associated with the dedifferentiated state, but the underlying mechanism and significance remain unclear.34
Here we present a systematic, functional genomic characterization of WDLPS and DDLPS tumors and models. We distinguish lineage-specific TF circuits from oncogenic programs, and associate MDM2 amplification and overexpression with primitive phenotypes in tumors and cell lines. When expressed at supraphysiologic levels, MDM2 binds hundreds of P53 targets and thousands of additional genes with physically interconnected promoters and functions related to cellular proliferation and stress. MDM2 expression levels vary markedly between and within tumors and cell lines. Whereas cells with moderate MDM2 expression are sensitive to MDM2 inhibitors and pro-apoptotic agents, the highest MDM2 expressers resist these agents and likely confound their clinical application.
Results
Genomic and transcriptional profiles of liposarcoma tumors and models
To investigate genetic and regulatory alterations that underlie the major classes of liposarcoma, we assembled a clinical cohort of primary WDLPS (n = 20) and DDLPS (n = 16) human tumors, and normal adipose tissue (n = 15) (Figure 1A and Table S1). Specimens were reviewed by board-certified pathologists to ensure accurate histological subtyping and appropriate tissue integrity. Whole-genome sequencing defined the genomic landscapes of 21 tumors (Tables S2 and S3). Transcriptional profiles were generated by RNA sequencing (RNA-seq) for 36 samples (Table S4). In addition to our clinical cohort, four liposarcoma cell lines (LPS141, LPS853, T449, and T778) were profiled for transcription (RNA-seq), TF binding, histone modifications (chromatin immunoprecipitation sequencing [ChIP-seq]), and chromatin topology (HiChIP).
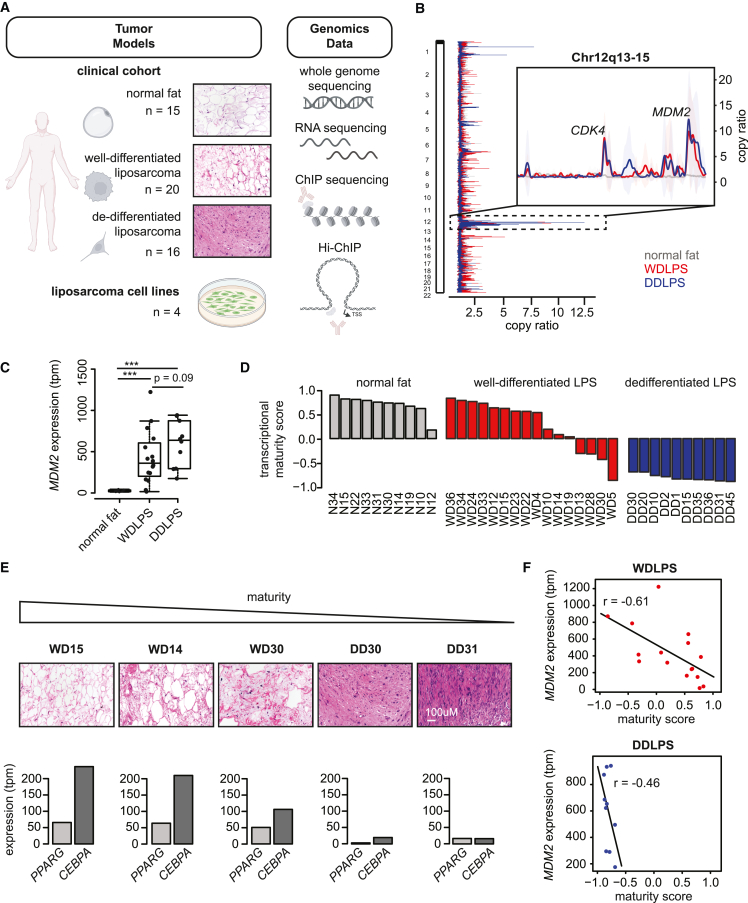
Figure 1.
Liposarcoma maturity score anticorrelates with MDM2 levels
(A) Schematic of study design with indication of primary tumors and cell line models (left) and genome-wide assays (right).
(B) Plot depicts copy number alterations in normal fat (gray), WDLPS (red), and DDLPS (blue) samples from our clinical cohort. Vertical plot at left depicts the average copy number across the genome, while the horizontal view expands the Chr12q locus that contains MDM2.
(C) Box plot shows distribution of MDM2 expression in normal fat and WDLPS and DDLPS. One-tailed t test p values: DDLPS vs. WDLPS = 0.09, NF vs. WDLPS = 8.8e−5, NF vs. DDLPS = 5.5e−5.
(D) Waterfall plots show maturity scores for samples in the clinical cohort.
(E) Hematoxylin and eosin staining for representative well-differentiated (WD) and dedifferentiated (DD) tumors, ranked by maturity score (top) and showing expression of master adipocytic regulators PPARG and CEBPA (bottom).
(F) Scatterplots show correlation between MDM2 expression (y axis) and maturity score (x axis) for WDLPS and DDLPS tumors.
The genomic profiles of tumors revealed low somatic mutational burden (average 1.8 per Mb) but extensive copy number abnormalities (Figure 1B; Tables S2 and S3). Characteristic amplification of MDM2 and CDK4 on the 12q13-15 locus was observed in all WDLPS and DDLPS tumors (Figure 1B and Table S2). Consistent with previous reports, we noted increased genomic amplification centered on the 12q locus in DDLPS, and one DDLPS tumor harboring a JUN amplification (1 out of 10 tumors) (Figure 1B and Table S2).29,31,33,35,36 However, we did not observe any consistent genomic alterations that distinguished DDLPS from WDLPS.
The enhanced genomic complexity in DDLPS prompted us to examine the 12q locus more closely. We found that MDM2 amplification levels were greater in DDLPS (average copy number = 10) compared with WDLPS tumors (average copy number = 7) as has been previously reported (Figures 1B, S1A, and S1B; Table S2). This degree of progressive amplification was not observed for other genes within the 12q amplicon, such as CDK4 (DDLPS = 7, WDLPS = 7), which suggests a specific association between MDM2 copy number and DDLPS (Figures 1B, S1C, and S1D; Table S2). Indeed, we confirmed higher MDM2 mRNA expression in dedifferentiated (mean transcripts per million [tpm] 597) relative to well-differentiated tumors (mean tpm 428) (Figures 1C and S1E; Table S4). The average MDM2 mRNA levels in the liposarcomas were ∼19-fold higher than normal fat (mean tpm 25) (Figure S1E). Expanding this analysis to additional tumors and controls in The Cancer Genome Atlas (TCGA), we defined a distribution of “physiologic” MDM2 expression in normal tissues (normalized log2 expression ranging from 9.44 to 13.22). We found that 88% of DDLPS tumors exceeded the 95th percentile of expression in normal tissues (Figure S1F). While these data support the notion that MDM2 is expressed at supraphysiologic levels in liposarcoma, the WDLPS tumors in our clinical cohort exhibit a surprising range of MDM2 expression, with a subset harboring very high levels comparable with the DDLPS tumors and others exhibiting more moderate overexpression (Figure S1E).
A tumor maturity score inversely correlates with MDM2 expression
The range of MDM2 amplification and expression across tumors prompted us to investigate whether it correlates with specific transcriptional phenotypes. We observed that well-differentiated tumors with high MDM2 levels had lower expression of adipocytic lineage regulators (Figures S1G and S1H). We therefore scored our clinical cohort based on a manually curated set of 30 genes with established roles in adipogenesis and/or specific expression in mature adipocytes (Tables S1 and S5; Figure 1D). This “maturity” score was high across all normal fat samples and consistently low across the DDLPS tumors. The maturity score of WDLPS tumors varied widely, consistent with a spectrum of adipocytic differentiation (Figure 1D). In both WDLPS and DDLPS, it correlated with adipocytic differentiation per histopathology and expression of the master adipocytic regulators PPARG and CEBPA (Figures 1E and S1I). The maturity scores also inversely correlated with MDM2 expression across WDLPS and DDLPS (Figure 1F). Repeating this analysis with an established adipogenesis signature (n = 200)37 confirmed a continuum of maturity across liposarcoma tumors that inversely correlated with MDM2 expression (Figures S1J and S1K).
We used the maturity index as a framework to define additional transcriptional programs associated with liposarcoma cell states by calculating the correlation between the maturity score gene set and all other individual genes across our clinical samples (Figure 2A and Table S6). We identified 1,065 genes whose expression positively correlated (r ≥ 0.5) with the maturity score. As expected, they were enriched for adipogenesis-related programs such as fatty acid metabolism, targets of the master TF PPARG and oxidative phosphorylation. Genes that negatively correlated with the maturity score (r ≤ −0.5, n = 1,298) were enriched for cell-cycle regulators, consistent with the higher proliferative index reported for dedifferentiated tumors.28 We also identified other expression programs that negatively correlated with the maturity score, which could be broadly classified into three functional groups: mesenchymal lineage/skeletal system development, stress response (P53, ER stress), and cellular biosynthetic processes (mRNA metabolism, ribosome biogenesis, translation). Finally, we observed an association between maturity and expression of genes encoded on the mitochondrial genome (Figure 2A).38 These signatures suggest that primitive liposarcoma tumors with higher MDM2 expression adopt a proliferative and biosynthetically active mesenchymal cell state.
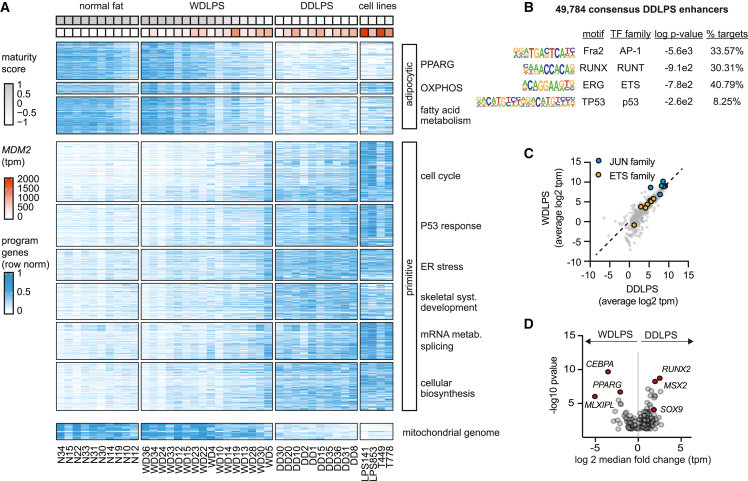
Figure 2.
Transcriptional programs and regulators in dedifferentiated sarcoma
(A) Heatmap depicts expression (blue heat) of differentially expressed genes (rows) across the clinical specimens and cell lines (columns). Genes are grouped into coherent programs (black outlines) ordered by their correlation or anticorrelation with maturity scores. Mitochondrial genome encoded genes are grouped below. Maturity score (gray heat) and MDM2 expression (red heat) are shown above for each sample.
(B) TF binding motifs that are over-represented in DDLPS enhancers marked by H3K27ac.
(C) Scatterplot depicts expression of TFs (points) in WDLPS (y axis) and DDLPS (x axis) tumors. Points along the diagonal represent factors shared between histologies and include multiple AP1-JUN and ETS family members.
(D) Volcano plot depicts fold change and significance (p value ≤ 0.05) of differential expression of TFs (points) between WDLPS and DDLPS. TFs referenced in the text are highlighted.
TF circuits reminiscent of an early mesenchymal progenitor
We sought to define the TF networks that underlie the respective gene expression programs in DDLPS. All four cell lines profiled display primitive morphology and rapid growth in culture despite T449 and T778 originating from WDLPS specimens. Transcriptional analysis showed that programs defined in DDLPS tumors are retained in the four cell lines, supporting their relevance as dedifferentiated disease models (Figure 2A). We therefore charted the regulatory landscape of the two DDLPS-derived lines (LPS141 and LPS853) and four DDLPS tumors (DD10, DD15, DD20, and DD31) by performing ChIP-seq for the enhancer-associated chromatin modification H3 lysine 27 acetylation (H3K27ac). We collated a set of 49,784 putative enhancers marked by H3K27ac in at least two samples (median size 1.8 kb). Scanning the sequences of these putative DDLPS enhancers revealed an over-representation of motifs recognized by AP-1 factors (34% of enhancers), RUNX (30%) and ETS (41%) family TFs, and P53 (8%) (Figure 2B and Table S7). Consistent with the motif enrichments, AP-1 factors (Fos and Jun) and multiple ETS factors (ETS1) were highly expressed in both DDLPS and WDLPS (Figure 2C), suggesting they represent common regulatory TFs, while RUNX2 was selectively induced in DDLPS (Figure 2D).
Established roles for JUN and RUNX2 in mesenchymal development prompted us to map and relate their binding patterns to enhancer circuits in DDLPS (Figure 3A).39,40,41,42,43 ChIP-seq profiles revealed that both TFs preferentially bind to distal elements: 62% and 57% of JUN and RUNX2 target sites, respectively, correspond to putative enhancers, while just 5% and 13% correspond to promoters. The two TFs co-occupy a majority of their enhancer targets (70% of RUNX targets are also bound by JUN), suggestive of shared distal regulatory functions in DDLPS (Figure 3A).
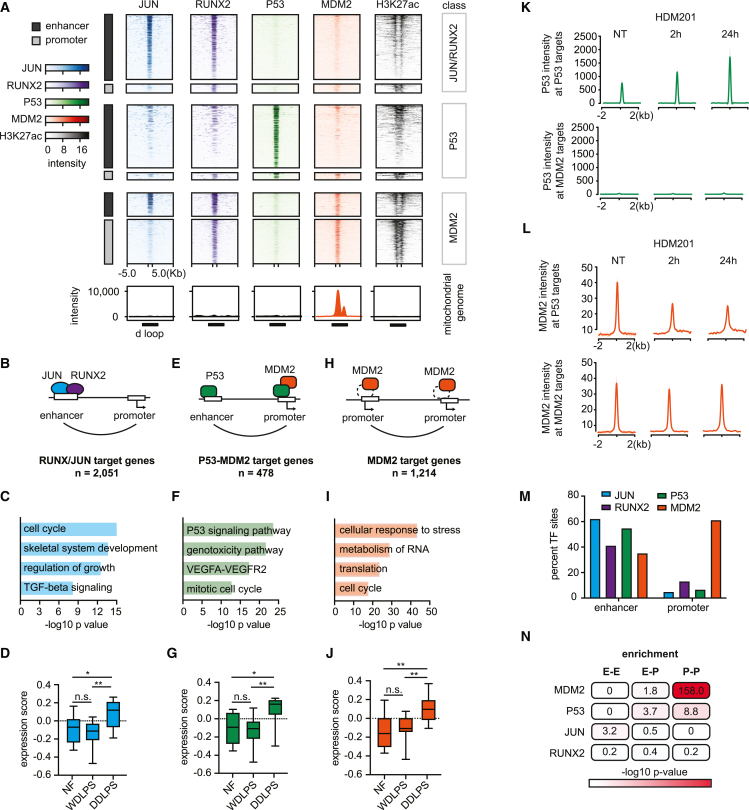
Figure 3.
TF circuits that govern dedifferentiated liposarcoma programs
(A) Heatmap depicts TF binding (color heat) and H3K27ac (grayscale) over candidate enhancers and promoters (rows) of the LPS141 liposarcoma line. Peaks are binned into groups based on combinatorial TF binding, including JUN/RUNX peaks (n = 36,151), P53 peaks (n = 857), and MDM2 peaks without P53 (n = 1,544). Bottom boxes show TF binding on a linearized track of the circular mitochondrial genome centered on the regulatory d loop (black box).
(B, E, and H) Schematics summarize characteristic binding and looping patterns for JUN (blue), RUNX2 (purple), P53 (green), and MDM2 (orange).
(C, F, and I) Bars highlight gene sets enriched among JUN/RUNX2 (blue), P53 (green), and MDM2 (orange) targets, as defined by combination of ChIP-seq and HiChIP.
(D, G, and J) Box plots show expression of TF target genes (defined as in B, E, and H) in normal fat, WDLPS, and DDLPS samples. Two-tailed t test p values: (D) NF vs. DDLPS = 0.017, WDLPS vs. DDLPS = 0.0016; (G) NF vs. DDLPS = 0.012, WDLPS vs. DDLPS = 0.002; (J) NF vs. DDLPS = 0.007, WDLPS vs. DDLPS = 0.002.
(K) Aggregate plots show a P53 binding signal at P53 (top) or MDM2 (bottom) target sites in LPS853 cells with no treatment (NT) or after 2 h or 24 h of HDM201 treatment.
(L) Aggregate plots show MDM2 binding signal at P53 (top) or MDM2 (bottom) target sites in LPS853 cells with no treatment (NT) or after 2 h or 24 h of HDM201 treatment.
(M) Bar plot shows the proportion of TF binding sites that coincide with putative enhancers or promoters.
(N) Table depicts the degree of overlap between TF binding, per ChIP-seq, and sites engaged in enhancer-enhancer (E-E), enhancer-promoter (E-P), or promoter-promoter (P-P) loops, per HiChIP. Red heat represents the significance of overlap between indicated TF and loop pattern calculated with Fisher’s exact test using genome-wide loop patterns as background.
To link the bound enhancers to likely gene targets, we evaluated their physical interactions using H3K27ac HiChIP. This procedure maps chromatin loops by ligating sequence elements in spatial proximity.44 The resulting data revealed physical loops between enhancers and promoters (E-P), as well as enhancer-enhancer (E-E) and promoter-promoter (P-P) loops, indicative of potential regulatory relationships (Figure S2A). Genes whose promoters engaged in physical interactions (E-P or P-P) had higher expression in DDLPS relative to controls (Figure S2B).
Integration of the HiChIP and TF binding data revealed that JUN- and RUNX2-bound elements loop to a largely overlapping set of 2,051 target genes (2,304 JUN, 2,406 RUNX2, ∼87% overlap) (Figure 3B and Table S8). These target genes were highly expressed in DDLPS and enriched for functions related to mesenchymal development and early osteogenic differentiation (Figures 3C and 3D; Table S9). Furthermore, we found that RUNX2 expression levels correlated with these signatures across the DDLPS tumors, consistent with direct regulatory functions (Figures S2C and S2D).
The JUN and RUNX2 gene targets include the mesenchymal lineage TF MSX2. MSX2 and another key mesenchymal TF SOX9 are both preferentially expressed in DDLPS (Figure 2D). JUN, RUNX2, MSX2, and SOX9 are all coordinately expressed in an early developmental window that precedes specification along either an osteogenic or chondrocytic lineage.45,46,47 The expression and interactions among these TFs suggest that DDLPS progenitors may emulate this primitive developmental state. Indeed, histological analysis of DDLPS specimens frequently reveals markers of bone differentiation, in addition to the adipocytic features.48
Supraphysiologic MDM2 restrains tumor-suppressive P53 circuits
We next investigated how this primitive developmental state is corrupted by oncogenic transcriptional circuits in DDLPS. We focused initially on the role of MDM2 in suppressing P53 transcriptional programs. While MDM2 does not directly bind DNA, it physically interacts with and degrades P53 through its E3 ubiquitin ligase activity.49 We therefore mapped genomic occupancy of both MDM2 and P53 in DDLPS cell lines by ChIP-seq (Figures 3A and 3E–3J).
We identified 867 P53 sites and 1,651 MDM2-bound sites at enhancers and promoters in both LPS141 and LPS853 cell lines. P53 primarily bound distal enhancers, which could be linked to putative target genes by the HiChIP data (Figures 3A and 3E). These P53-bound target genes (n = 478) were enriched for canonical P53 programs, including genotoxicity response, vascular endothelial growth factor signaling, and cell cycle (Figure 3F; Tables S8, and S9). Interestingly, these targets showed higher expression in DDLPS relative to WDLPS or normal fat (Figure 3G). This increase may reflect activation of P53 pathways by the proliferative state and genomic instability associated with the more aggressive tumors. We reasoned that it could point to a critical requirement for MDM2 overexpression to hold these programs in check and facilitate malignant progression.
As expected, MDM2 showed overlap with P53 (Figures 3A, S3A, and S3B), most significantly at promoters, and associated with 258 of 478 P53 target genes. To investigate the functional impact of supraphysiologic MDM2 on P53 target genes and circuits we used the inhibitor HDM201, which disrupts the P53-MDM2 interaction and is currently under clinical evaluation.50,51,52 We measured transcriptional changes over a short course of treatment (2, 4, and 6 h) prior to cell death response (Figure S3C) in the LPS141 cell line. We observed rapid upregulation of both MDM2 and P53 protein levels at 2 h followed by a progressive increase of MDM2 up to 6 h (Figures S3D and S3E). In contrast, P53 protein reached maximum levels at 2 h followed by a slow recovery to baseline (Figures S3D and S3E). The rapid increase in P53 is presumably due to its release from MDM2-directed degradation by HDM201. However, MDM2 is also an established P53 target,53 resulting in positive feedback on its expression after the initial induction.
P53 circuit genes were strongly upregulated at early time points (2 and 4 h) of drug treatment (Figure S3F), while JUN/RUNX2 target genes were regulated to a lesser degree (Figure S3G). P53 occupancy at target promoters also increased with HDM201 (Figure 3K, top). However, MDM2 occupancy decreased, consistent with an essential role for the P53 interaction in its recruitment (Figure 3L, top). These analyses show that P53 targets are largely distinct from mesenchymal circuits in DDLPS and support a direct role for MDM2 in their negative regulation.
Supraphysiologic MDM2 binds an expanded promoter repertoire
The ChIP-seq profiles revealed that MDM2 has a much stronger preference for promoters than the other TFs (1,045 bound promoters; 606 enhancers) (Figures 3A, 3M, and 3N). Despite its close association to P53 biology, most MDM2-bound genes in DDLPS are not P53 targets (1,214 out of 1,473; Figures 3A and 3H; Table S8). These P53-independent, putative MDM2 target genes are enriched for functions related to cell growth, biosynthesis, and stress responses (Figure 3I and Table S9), and include previously reported targets involved in serine metabolism (PSPH, SLC1A4, and GSS).14,54 These genes and ontologies are also distinct from those observed for the JUN/RUNX2 circuits (Figure 3C). P53-independent MDM2 targets were expressed at higher levels in DDLPS compared with WDLPS or normal fat (Figure 3J). In contrast to the robust induction of P53 targets, these genes were downregulated after 2 h of HDM201 treatment (Figure S3H). Furthermore, ChIP-seq analysis indicated that MDM2 was retained at these promoters after treatment, while P53 remained absent (Figures 3K and 3L). The impact of MDM2 inhibitor treatment on the transcriptional circuits defined in our study was confirmed using an additional MDM2 inhibitor, Nutlin-3a (Figures S3I–S3K), and by testing MDM2 inhibition in the LPS853 cell line, which sustained P53 induction across the HDM201 time course (Figure S4). These data resolve a set of promoters bound by overexpressed MDM2 that are independent of P53.
In addition to its nuclear genomic targets, the ChIP-seq data reveal that MDM2 associates with the circular mitochondrial genome, which is present at tens to hundreds of copies per cell (Figure 3A).55 MDM2 specifically bound the regulatory d loop, a non-coding region of the mitochondrial genome where transcription and DNA replication originate (Figure 3A).56 In contrast, none of the other TFs in our dataset interacted with the mitochondrial genome (Figure 3A). Although prior studies have localized MDM2 to the mitochondria,15,57,58 these interactions with a key regulatory region of the mitochondrial genome provide additional support for P53-independent localization of MDM2.
MDM2 associates with topological promoter hubs
We observed that many P53-independent MDM2 target promoters were clustered across gene-rich loci. Furthermore, HiChIP data indicated that many of these promoters are connected by promoter-promoter loops (Figure 3N), in some cases forming interconnected hubs (Figures 4A–4C). We used our HiChIP data to define a genome-wide set of 258 promoter-promoter hubs (pp hubs), which involved an average of eight genes each and spanned a median distance of 200 kb (Figures S5A and S5B). The hub promoters are largely contained within the euchromatic compartment and tend to connect highly expressed genes (Figures S5C and S5D). Hub structure was consistent across DDLPS cell lines and three tumors (Figures 4A–4F and S5E–S5G). Promoter hubs appear to represent relatively constitutive structures, as they are evident in published T cell HiChIP (Figures S5H–S5J) and multi-promoter interactions have recently been reported in other cellular contexts.20,59,60,61,62 They may contribute to the coordinated regulation of functionally related genes, as recently observed for physical gene networks in Drosophila.60 Indeed, the expression of genes within a given hub is highly correlated across RNA-seq profiles for DDLPS samples in TCGA (n = 60) (Figures 4G and S5K).
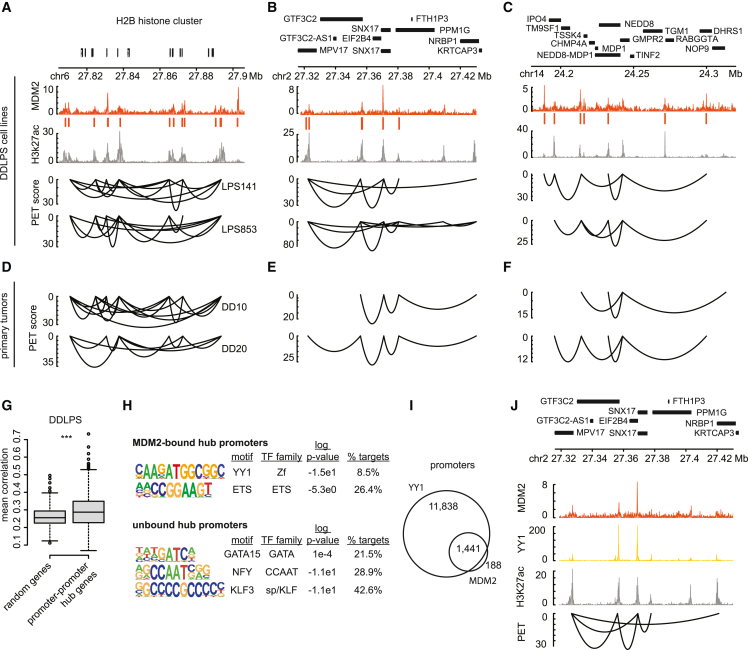
Figure 4.
Supraphysiologic MDM2 associates with promoter hubs
(A–C) Genomic tracks for three genomic regions with representative multi-way promoter hubs. Top tracks represent MDM2 binding intensity and called peaks (orange), and H3K27ac intensity (gray) in LPS141 cells. Bottom tracks depict HiChIP promoter-promoter loops in LPS141 and LPS853 cells. Loop height is proportional to the paired-end tag (PET) score. Genes are shown above.
(D–F) HiChIP promoter-promoter loops in two dedifferentiated tumors (DD10 and DD20) shown for the same loci.
(G) Box plots depict the correlation of the expression of promoter hub genes, compared with random gene sets in DDLPS tumors (n = 60) in the TCGA; two-tailed t test p value = 4.173e−5.
(H) Transcription factor binding motifs that are over-represented in MDM2-bound hub promoters (top) or unbound hub promoters (bottom).
(I) Venn diagram depicting overlap between YY1- and MDM2-bound promoters.
(J) Genomic track for representative promoter hub in the LPS853 cell line. Tracks represent, from top to bottom, MDM2 binding intensity (orange), YY1 binding intensity (yellow), and H3K27ac intensity (gray), and paired-end tag (PET) scores for promoter-promoter loops in H3K27ac HiChIP.
P53-independent MDM2 target promoters are also highly enriched for the binding motif of YY1, a TF implicated in looping interactions (Figure 4H and Table S10).63,64 MDM2 contains a central acidic domain reported to interact with YY1 and other transcriptional regulators.13,16,17,18,65,66 ChIP-seq revealed striking co-localization of YY1 with MDM2-bound promoters (89% of MDM2 sites bound) (Figures 4I, 4J, S6A, and S6B). Given that MDM2 lacks a DNA-binding domain, its association with promoter hubs could reflect pleiotropic interactions with YY1 or other regulators. Moreover, MDM2 occupancy at promoter hubs was considerably more prominent in cell lines with very high MDM2 levels (Figures S6C–S6E). This suggests that P53-independent MDM2 interactions and functions may be enhanced in tumor cells with supraphysiologic expression. Although these P53-independent interactions may facilitate a regulatory role of MDM2 at promoter hubs, further study is needed to define the direct transcriptional impact of MDM2 at these sites.
Heterogeneous MDM2 expression is associated with resistance to targeted therapies
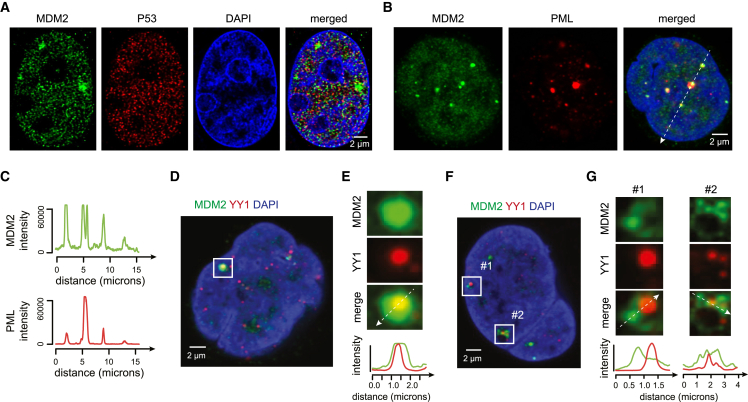
The association of MDM2 with highly interconnected genomic targets prompted us to visualize its localization within nuclei. Immunofluorescence (IF) imaging of liposarcoma cells revealed concentration of MDM2 protein within discrete nuclear foci (Figure 5A). These foci were not co-occupied by P53 but rather co-localized with promyelocytic leukemia protein (PML), a known direct interactor of MDM2 (Figures 5B, 5C, and S7A–S7D).67,68 PML forms nuclear “PML bodies” that sequester client proteins, including MDM2, and have been associated with diverse nuclear functions.68,69,70 PML bodies have also been reported to directly contact transcriptionally active gene-dense loci, including the histone H2B cluster that we identified as a promoter hub co-bound by YY1 and MDM2 (Figures 4A and S6A).71,72,73 Indeed, additional IF analyses of LPS lines revealed that the MDM2 foci were surrounded by smaller YY1 puncta (Figures 5D–5G). These data suggest a role for PML bodies in sequestering MDM2 when it accumulates at high levels, potentially driving its association with specific genomic loci and/or protecting the cell from excessive MDM2 protein.74
Figure 5.
Supraphysiologic MDM2 localizes to nuclear foci
(A) IF images (Airyscan confocal, 63×) for representative LPS853 cell show MDM2 (green), P53 (red), and DAPI (blue).
(B) IF images (confocal, 63×) for representative LPS853 cell show MDM2 (green), PML (red), and DAPI (blue).
(C) Quantification of MDM2 and PML intensity profiles across white dashed line in (B) shows co-localization of MDM2 and PML.
(D and F) IF images (confocal, 63×) for LPS853 cells show MDM2 (green), YY1 (red), and DAPI (blue).
(E and G) High-magnification images shown for white boxed regions in (D) and (F). Bottom plots show quantifications of MDM2 (green) and YY1 (red) intensities across white dashed lines.
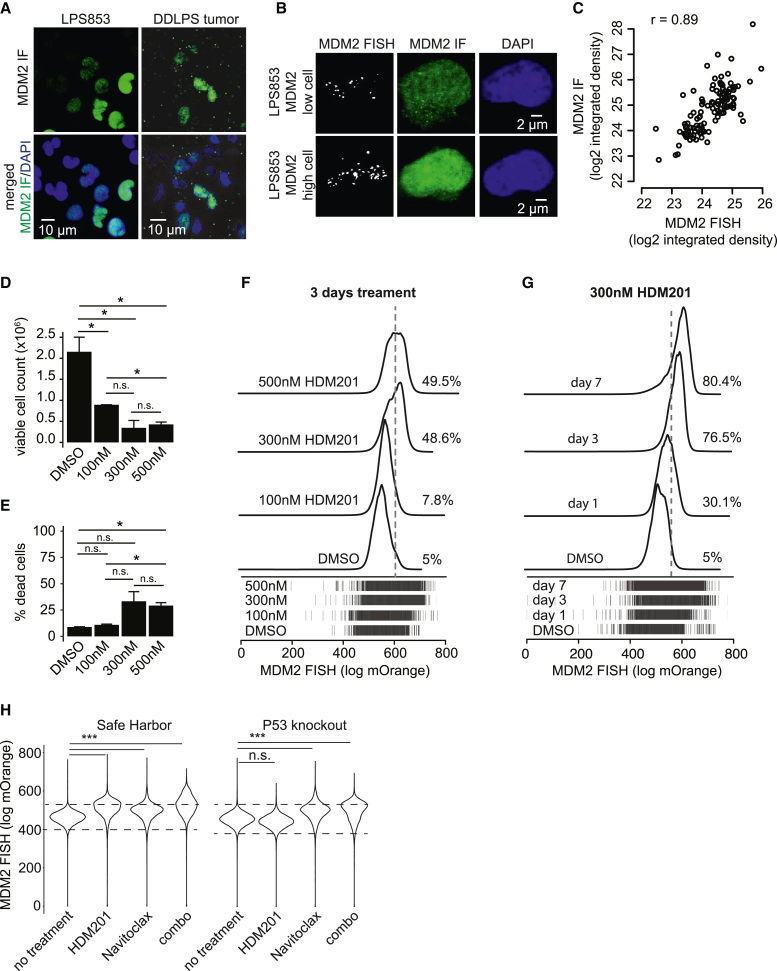
The imaging studies also revealed striking cell-to-cell variability in MDM2 protein levels in cell lines and tumors, with fluorescence signals across individual cells ranging over an order of magnitude (Figures 6A, S8A, and S8B). To relate this protein-level variability to genomic copy number, we combined IF with fluorescence in situ hybridization (FISH) to quantify MDM2 protein and copy numbers in the same single cells. This confirmed a correspondence between protein overexpression and locus amplification levels (Figures 6B and 6C). Metaphase spreads confirmed the presence of MDM2 gene copies on large neochromosomes previously characterized in liposarcoma (Figure S8C).5,6,7 The MDM2-containing neochromosomes were asymmetrically distributed across single cells, similar to non-Mendelian inheritance patterns associated with extrachromosomal DNAs75 and likely underlying the heterogeneous MDM2 protein levels in cell lines and tumors.
Figure 6.
Heterogeneous distribution of MDM2 copy number and expression confound efficacy of targeted therapies
(A) Representative 25× confocal IF images show MDM2 (green) and DAPI (blue) in LPS853 cells and a DDLPS tumor.
(B) Representative maximum-projection 63× confocal z stacks show FISH for the MDM2 locus (gray), IF for MDM2 protein (green), and DAPI (blue).
(C) Scatterplot compares MDM2 FISH and MDM2 IF intensity across 115 single cells.
(D and E) Bar plots comparing counts of viable (D) and dead (E) LPS853 cells following 3 days of HDM201 treatment.
(F) Histograms of MDM2 FISH intensity in LPS853 cells following 3 days of HDM201 treatment (n = 30,000 cells per condition). Dashed line represents the top 5% of the DMSO control. All drug-treated comparisons with the DMSO control displayed two-tailed t test p values < 2.2e−16.
(G) Histograms of MDM2 FISH intensity in LPS853 cells following a time course of 300 nM HDM201 treatment (n = 35,000 cells per condition). Dashed line represents the top 5% of the DMSO control. All drug-treated comparisons with the DMSO control displayed two-tailed t test p values < 2.2e−16.
(H) Violin plots show distribution of MDM2 FISH for safe harbor and P53 knockout LPS853 cells following 3 days treatment with DMSO (control), 100 nM Navitoclax or 300 nM HDM201 as single agents or in combination (n = 30,000 cells per condition). Dashed lines represent the top 5% and bottom 5% of the DMSO control. All comparisons marked as significant displayed two-tailed t test p values < 2.2e−16.
We hypothesized that variable MDM2 expression could impact the response of individual cells to targeted therapies. We therefore tested the sensitivity of liposarcoma cells to the P53-competitive MDM2 inhibitor HDM201. The liposarcoma cells were highly sensitive to submicromolar concentrations of the inhibitor, which led to growth arrest and apoptosis by 24 h of treatment (Figures S8D and S8E). To evaluate the relationship between MDM2 levels and sensitivity, we treated cells with HDM201 for 24 h and then readout MDM2 copy number using FISH coupled to flow cytometry (Figures 6D–6G and S8F). The surviving cells displayed a striking dose- and time-dependent enrichment for the highest MDM2 copies (top 5%; Figures 6F–6G and S8F).
We next evaluated whether the drug tolerance of the highest MDM2 expressers was P53 dependent. To this effect, we established a P53 knockout line from LPS853 using CRISPR-Cas9 (Figure S9A). This derivative maintains a range of MDM2 expression similar to that of a control line with a safe harbor guide RNA or the parental line (Figures S8B, S9A, and S9B). We found that the P53 knockout line was no longer sensitive to the MDM2 inhibitor HDM201 and did not display a shift in MDM2 copy number following treatment, consistent with the known mechanism of action whereby the inhibitor functions by activating P53 (Figures S9C and 6H). We also considered the impact of MDM2 overexpression on other therapeutic modalities. Here we focused on the pro-apoptotic agent Navitoclax, which has been proposed as a combination therapy with MDM2 inhibitors.76 We confirmed that Navitoclax induced cell death and synergized with HDM201 in LPS853 and LPS141 cell lines (Figures S8G–S8I). In contrast to HDM201, Navitoclax induced cell death in the P53 knockout line at an IC50 similar to that of the control line (Figure S9D). Nonetheless, prolonged treatment with Navitoclax enriched cells with high MDM2 copy numbers. Remarkably, this bias toward high MDM2 copy number was evident in both safe harbor and P53 knockout conditions, indicating the high MDM2 can confer resistance to the pro-apoptotic agent independent of P53 (Figure 6H).
These results indicate that cell-to-cell variability in MDM2 copy number and expression contributes to drug-resistant phenotypes in liposarcoma cells via P53-dependent as well as P53-independent mechanisms. While our results highlight the highest MDM2-expressing populations as drivers of drug-resistant phenotypes, remaining liposarcoma cells that also highly express MDM2 may also at least in part share these functional phenotypes. Regardless, our data strongly implicate supraphysiologic MDM2 expression as a mechanism underlying the poor clinical efficacy of the targeted agents against these heterogeneous tumors.
Discussion
We have presented a systematic, functional genomic analysis of liposarcoma, a common soft-tissue sarcoma defined by its overexpression of the MDM2 oncogene.25,28 We define complementary gene-regulatory circuits that underlie the developmental states and malignant phenotypes of well-differentiated and dedifferentiated tumors. We also document striking cell-to-cell variability in MDM2 copy numbers and associate supraphysiologic MDM2 expression with non-canonical genomic targets, nuclear structures, and resistance to targeted therapies. Our findings have broad implications given that MDM2 amplification is a defining feature of these sarcomas and given the high frequency of MDM2 amplification in brain, breast, lung, gastrointestinal, and other tumors.
The developmental state and cell of origin of liposarcoma are not well understood.32,77 While these tumors typically present with primitive mesenchymal and/or adipocytic features, they can also exhibit bone and muscle differentiation.48 Technical challenges include the lack of culture models that recapitulate well-differentiated liposarcomas and difficulties associated with molecular characterization of adipocytic tissues. Genome sequencing studies in primary tumors have tied the various histologies to a shared precursor cell but do not shed light on the underlying developmental programs.32 Our definition of the gene-regulatory circuits and identification of RUNX2, JUN, MSX2, and SOX9 as key TF drivers demonstrate strong parallels between primitive DDLPS tumors and a specific mesenchymal progenitor population.45,47 This progenitor, which coordinately expresses all four TFs, has bipotent potential for bone and cartilage differentiation. We suggest that DDLPS progenitors adopt a closely related state that may be less constrained in its developmental potential. Indeed, we note that the adipocytic signature is subdued in the most primitive DDLPS tumors and is complemented by signatures of skeletal system and muscle development. The exact developmental state and potency of the cells that fuel these tumors is an important question with clinical implications, given the poor prognosis of sarcomas with primitive stem-like features.
Amplification and overexpression of the MDM2 oncogene is a defining feature of liposarcomas that is also pervasive in brain, breast, lung, and stomach carcinomas.1,25,28 The amplicons include MDM2 and CDK431 and are typically contained on supernumerary ring or rod chromosomes, which harbor very high copy numbers.4,5,6,7 A compelling question is why MDM2 is so highly amplified in DDLPS and these other tumors. One likely explanation relates to the inter-relationship between MDM2 and P53. MDM2 oncogenicity is explained by its ability to degrade and thereby abrogate the tumor-suppressive functions of P53. Yet MDM2 is itself a P53 target,53 which creates a feedback loop that was clearly evident in our MDM2 perturbations. While this feedback loop may be important for P53 sensing in normal cells, a corollary is that low-level MDM2 amplification may be insufficient to check P53 activity, as the inherent feedback may simply revert MDM2 levels to their physiologic norm. As such, effective P53 inactivation may require locus amplification to levels that are refractory to feedback control. Furthermore, the most highly MDM2-amplified tumors in our cohort also exhibit signatures of proliferation, DNA damage, and P53 signaling. Thus, the very high copy numbers in these and other tumors could reflect a stringent requirement to offset a robust P53 response to DNA damage and unchecked proliferation in aggressively malignant cells.
One consequence of the supraphysiologic MDM2 protein levels, which are many-fold greater than normal cells, is its physical association with thousands of additional promoters. These promoters, which are distinct from canonical P53-dependent MDM2 targets, tend to be highly active, bound by YY1, and structurally arranged into interactive hubs. Further insight comes from imaging studies showing that supraphysiologic MDM2 concentrates in PML bodies within nuclei. PML bodies have been implicated in the sequestration and degradation of diverse nuclear proteins, including MDM2. Their reported association with transcriptionally active gene-dense loci may explain MDM2’s context-specific interaction with active promoter hubs.73 We note that MDM2 contains, in addition to its E3 ubiquitin ligase functionality, an RNA-binding domain and two intrinsically disordered domains that may promote its association with hubs and nuclear bodies.49 These physical interactions may impact protein degradation within the nuclear bodies and/or perturb the regulation of critical hub genes, with potential impact on cellular fitness and stress responses. Indeed, P53-independent MDM2 functions and PML bodies have both been linked to nutrient sensing and stress responses.14,54,66
The imaging studies also revealed remarkable variability in MDM2 expression across individual cells in liposarcoma lines and tumors, potentially explaining ongoing clinical challenges. Despite some in vitro efficacy, HDM201 and other inhibitors of the P53-MDM2 interaction have lacked clinical efficacy.26,27 Our data suggest that this failure may be due to MDM2-high cells in the populations, which readily tolerate the targeted agents. Although the addition of pro-apoptotic agents, such as Navitoclax, showed synergy in our models, population heterogeneity enabled high expressers to evade both modalities. This ability to rapidly respond to therapeutic or other stresses appears to be a feature of the neo-chromosomal amplifications whose asymmetric segregation during mitosis may result in highly variable copy numbers and protein levels across the population. Resistance to MDM2 inhibitors may simply reflect the capacity of MDM2 to outcompete the small molecule. However, our observation that MDM2-high cells also evade other modalities suggests that supernumerary ring or rod chromosomes containing MDM2 may confer a range of adaptive cell phenotypes. Furthermore, our finding that the resistance of MDM2-high cells to pro-apoptotic agents is retained in P53 knockout cells highlights the potential of supraphysiologic MDM2 to alter phenotypes independent of its canonical P53 target. Our study highlights an urgent need for new therapeutic strategies that exploit vulnerabilities of MDM2 overexpressers, moderate the adaptability of tumors driven by extrachromosomal amplifications, or target intrinsic states that are more constant across the malignant populations.
Limitations of the study
Normal fat, well-differentiated, and dedifferentiated liposarcoma tumors derived in this study were collected from a single institute where patients are administered preoperative irradiation. Additionally, liposarcomas of the extremities or of myxoid or pleomorphic histology were excluded from this study. Alternative cohorts of liposarcoma from other institutions may reveal further insights into liposarcoma biology. We were limited by technical challenges involving processing of well-differentiated tumors and focused our epigenomic and topological analysis on dedifferentiated samples. Therefore, while we define transcriptional circuits in dedifferentiated tumors, it is unclear to what degree these exist in the well-differentiated histology and how they impact the spectrum of maturity that we observed across our cohort. Additional limitations are associated with analysis of bulk datasets, which can be confounded by normal or inflammatory cells that can mask transcriptional circuits specific to tumor cells. Furthermore, all drug results are based on models that cannot fully recapitulate the complex biology of primary tumors.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Actin | Bio-Rad | Cat#12004163, RRID:AB_2861334 |
| H3K27ac | Active Motif | Cat#39133, RRID:AB_2561016 |
| JUN | Cell Signaling | Cat#9165S, RRID:AB_2130165 |
| MDM2 | Abcam | Cat#ab226939, RRID:AB_2754988 |
| P53 | Cell Signaling | Cat#48818, RRID:AB_2713958 |
| PML | Santa Cruz | Cat#sc-966, RRID:AB_628162 |
| RUNX2 | Cell Signaling | Cat#12556S, RRID:AB_2732805 |
| YY1 | Santa Cruz | Cat#sc-7341, RRID:AB_2257497 |
| Drosophila spike-in chromatin antibody | Active Motif | Cat#61686, RRID:AB_2737370 |
| Biological samples | ||
| See Table S1 for a list of patient samples included in the study | N/A | |
| Chemicals, peptides, and recombinant proteins | ||
| HDM201 | SelleckChem | Cat#S8606 |
| Nutlin-3a | SelleckChem | Cat#S8059 |
| Navitoclax | SelleckChem | Cat#S1001 |
| Klenow Fragment (3-5exo-) | NEB | Cat#M0212L |
| Drosophila spike-in chromatin | Active Motif | Cat#53083 |
| MboI | NEB | Cat# R0147M |
| Biotin-dATP | ThermoFisher | Cat#195224016 |
| Streptavidin M280 beads | Invitrogen | Cat#11205D |
| RIPA buffer | Life Technologies | Cat#89900 |
| HALT protease phosphatase inhibitor cocktail | ThermoFisher | Cat#78440 |
| Cell Titer-Glo | Promega | Cat#G7570 |
| 16% Formaldehyde | Life Technologies | Cat#28908 |
| Vectashield | Vector labs | Cat#H-1000-10 |
| Fluoromount G | Invitrogen | Cat#00495802 |
| KaryoMAX colcemid solution | Life Technologies | Cat#15212012 |
| DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher | Cat#D1306 |
| Critical commercial assays | ||
| QIAGEN RNeasy lipid kit | Qiagen | Cat#74804 |
| End-It DNA repair kit | Biosearch Technologies | Cat#ER0720 |
| KAPA HiFi HotStart ReadyMix PCR Kit | Roche | Cat#07958935001 |
| Zymo Clean and Concentrate Kit | Zymo | Cat#DCC-100 |
| Nextera DNA library prep kit | Illumina | Cat#20018704 |
| BCA protein assay kit | ThermoFisher | Cat#23227 |
| Dead cell apoptosis kit | Invitrogen | Cat#V13241 |
| Deposited data | ||
| Sequencing data | This paper | GEO: GSE213300 |
| Human transcriptomic data derived from the TCGA | Xena curated data repository | https://pancanatlas.xenahubs.net |
| H3K27ac HiChIP of normal tissues | Mumbach et al.20 | GEO: GSE101498 |
| Experimental models: Cell lines | ||
| HCT116 | ATCC | Cat#CCL-247, RRID:CVCL_0291 |
| U-2 OS | ATCC | Cat#HTB-96, RRID:CVCL_0042 |
| LPS141 | gift from Dr. Adrian Marino-Enriquez | Brigham and Women’s Hospital |
| LPS853 | gift from Dr. Adrian Marino-Enriquez | Brigham and Women’s Hospital |
| 93T449 | gift from Dr. Adrian Marino-Enriquez | Brigham and Women’s Hospital |
| 94T778 | gift from Dr. Adrian Marino-Enriquez | Brigham and Women’s Hospital |
| Oligonucleotides | ||
| Safe harbor sgRNA | IDT | GGCTAAATTCCTCTTATTCA |
| TP53 sgRNA | IDT | TCGACGCTAGGATCTGACTG |
| PCR of P53 CRISPR targeting region -Forward primer | IDT | CCCAACCCTTGTCCTTACCA |
| PCR of P53 CRISPR targeting region -Reverse primer | IDT | CAACATGCAAAGCCCTGTCT |
| Chr12 FISH probes | Empire Genomics | Cat#CHR12-10-GR |
| MDM2 FISH probes | Empire Genomics | Cat#MDM2-20-OR |
| Recombinant DNA | ||
| pXPR_044, all in one CRISPR-Cas9 vector | gift from John Doench and Dave Root | Broad Institute, Genomics Perturbation Plat-form |
| Software and algorithms | ||
| Code supporting this study | This paper | https://github.com/BernsteinLab/methods_mdm2_sarcoma; https://doi.org/10.5281/zenodo.7814766 |
| STAR | Dobin et al.78 | version 2.7.10 |
| Juicebox | Durand et al.79 | http://aidenlab.org/juicebox/ |
| Hichipper | Lareau et al.80 | version 0.7.9 |
| HiC-Pro | Servant et al.81 | version 2.10.0 |
| Bioconductor | Huber et al.582 | Release 3.13 |
| Homer | Heinz et al.83 | version 4.11 |
| Samtools | Danecek et al.84 | version 1.15.1 |
| Bedtools | Quinlan et al.85 | version 2.29.0 |
| HTSlib | Bonfield et al.86 | version 1.15.1 |
| FIJI | Schindelin et al.87 | version 2.0.0-rc-43/1.5h |
| CellProfiler | Stirling et al.88 | version 4.2.1 |
| SynergyFinder | Ianevski et al.89 | version 1.6.1 |
| FlowJo | BD biosciences | version 10.8.0 |
Resource availability
Lead contact
Further information and request for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Bradley Bernstein (Bradley_Bernstein@dfci.harvard.edu)
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human tumor specimens
All samples were acquired from patients at the Massachusetts General Hospital who had signed written informed consent to provide their cells and tissue collection was given ethical approval by the Institutional Review Board protocol #03–344. When available, paired dedifferentiated and well-differentiated tumor samples were acquired from the same bulk tumor. All tumors originated in the abdomen or retroperitoneum. All specimens underwent review by three independent pathologists to ensure accurate diagnosis, histological subtype and appropriate tissue integrity for downstream analysis. For a subset of these patients, normal fat was taken subcutaneously. Both normal and tumor tissue was snap frozen. Slides for hematoxylin and eosin staining were cut from blocks of frozen tissue sections. A summary of the tissue (normal and tumor) samples is provided in Table S1. Our clinical cohort included 44 tumors and 19 normal subcutaneous fat samples.
Cell lines and cell culture
All studies on human cell lines were given ethical approval by the Institutional Review Board protocol #21–394. All liposarcoma cell lines used in this study (LPS141, LPS853, T449 and T778) were gifts from Dr. Adrian Marino-Enriquez. All liposarcoma cell lines were maintained in RPMI-1640 medium (Gibco, ThermoFisher Scientific) supplemented with 15% FBS, 1% glutaMaX (Gibco), and 1,000 Units/mL Penicillin and 1 mg/mL Streptomycin (Gibco). HCT116 and U-2 OS cell lines were purchased from ATCC. HCT116 and U-2 OS cells were maintained in McCoy’s 5A medium (Gibco, ThermoFisher Scientific) with 10% FBS, 1% glutaMaX (Gibco), 1,000 Units/mL Penicillin and 1 mg/mL Streptomycin (Gibco). All cell lines were regularly screened for mycoplasma.
Method details
RNA-sequencing
For tumors and subcutaneous fat, tissue was dissociated using a blade homogenizer and whole RNA was extracted using the QIAGEN RNeasy lipid kit according to the manufacturer’s protocol. For cell lines, whole RNA was extracted using the QIAGEN RNeasy kit according to the manufacturer’s protocol. For RNA-seq library preparation, Poly(A)+ RNA was enriched using magnetic oligo(dT)-beads (Invitrogen 61011) and then ligated to RNA adaptors for sequencing. RNA-seq was performed with a minimum of two biological replicates per liposarcoma cancer line and in singlicate for tumor and normal fat samples. Libraries were sequenced as 75-base paired-end reads on an Illumina NextSeq500 instrument.
ChIP-sequencing
ChIPseq was performed to generate chromatin state maps (H3K27ac) and binding profiles for transcription factors (P53, MDM2, RUNX2, JUN and YY1). ChIP-seq was performed as described previously unless otherwise specified.90 In brief, crosslinked cells were lysed and DNA was sheared on the Covaris E220 (Fill level 5, Duty Cycle 5, PIP 140, Cycles/burst 200, Time 22–26 min) to between 200 and 2,000 base pair fragments using a covaris water bath sonicator. Antibodies were as follows: H3K27ac (Active Motif #39133), P53 (Cell Signaling #48818), MDM2 (abcam ab226939), JUN (cell signaling 9165S), RUNX2 (cell signaling 12556S), and YY1(cell signaling 63227S). ChIP DNA was used to generate sequencing libraries by end repair (End-It DNA repair kit, Biosearch Technologies ER0720), 30 A base overhang addition via Klenow fragment (NEB M0212L), and ligation of barcoded sequencing adapters. Barcoded fragments were amplified via PCR (KAPA HiFi MM, Roche 07958935001). ChIP-seq was performed with a minimum of two biological replicates per liposarcoma cancer line and in singlicate for DDLPS samples. Libraries were sequenced as 75-base paired-end reads on an Illumina NextSeq500 instrument.
For the HDM201 time course, Drosophila chromatin (Active Motif #53083) and antibody (Active Motif #61686) were spiked-in to samples immediately preceding IP. For transcription factor ChIPs 20μg of spike-in chromatin was added per 25μg of input chromatin. For H3K27ac ChIPs, 50μg of spike-in chromatin was added per 25μg of input chromatin. For all conditions, 2μL of spike-in antibody was added per IP.
HiChIP
HiChIP was performed as previously described91 with the exception that an H3K27ac antibody was used for the immunoprecipitation (Active Motif #39133). In brief, crosslinked chromatin was digested with MboI. Overhangs were filled in and marked with Biotin-dATP (ThermoFisher 19524016) and ends were ligated. Chromatin was then sheared on the Covaris E220 and ChIP was performed overnight for H3K27ac marked chromatin. Resulting DNA was purified using the Zymo clean and concentrate kit (DCC-100), bound to Streptavidin M280 beads (Invitrogen 11205D), and tagmented. Libraries were amplified using the Nextera DNA Library Prep kit (Illumina). HiChIP was performed with a minimum of two biological replicates per liposarcoma cancer line and in singlicate for DDLPS tumors. Libraries were sequenced on a Novaseq S1 (200 cycle kit).
Western blotting
Cells were washed two times with 1x PBS and scraped on ice in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS) supplemented with fresh protease (HALT, Thermofisher) and phosphatase inhibitors (Thermofisher). Protein was quantified and normalized using the BCA Protein Assay kit (Thermo #23227) according to the manufacturer’s protocol. Western blotting was performed according to standard protocols.
Drug treatments
HDM201 drug concentrations were optimized for each cell line in order to achieve comparable activation of P53, determined by western blot. LPS141 were treated with 100nM HDM201 and LPS853 were treated with 300nM HDM201. For Nutlin-3a treatments LPS141 cells were dosed with 1μM drug. Cells were plated and the following day normal growth media was removed and replaced with drug-containing media. Cells were not passaged while on the drug and drug-containing media was changed at least every 2–3 days. For short time courses (2, 4, 6, and 24 h) drug or DMSO containing media was added at staggered timepoints and all plates were harvested at one endpoint. Each treatment was performed across at least two biological replicates.
Dose-response curves
Cells were seeded in 96 well plates at the following densities: LPS53 = 500 cells/well, LPS141 = 750, T449 = 1000 cells/well, T778 = 250 cells/well. The following day regular growth media was replaced with drug-containing media. The optimal dose range for HDM201, or Navitoclax was determined for each cell line and was tested across 10 half-log doses. For synergy analysis, HDM201 and Navitoclax were tested as single agents and in combination across a 5 (HDM201) x 4 (Navitoclax) dose response matrix. Each plate included 0.1% DMSO as a negative control for growth inhibition. Drug media was changed 48 h after initial treatment. Plates were incubated in drug at 37°C for a total of 96 h and lysed by adding 20μL Cell Titer-Glo reagent (Promega, Cat. No. G7570) to 100mL cell media. Luminescence was measured using a Synergy HTX Platereader (BioTek) or GloMax explorer (Promega) and growth inhibition was calculated relative to DMSO treated wells. All experiments were performed in biological duplicates and within a given experiment at least 4 technical replicates were tested per condition.
Drug synergy analysis
Drug synergy scores were generated and plotted using the SynergyFinder package 1.6.1.89 Zero interaction potency (ZIP) scores were calculated without baseline correction and using default parameters with the exception that Emin was specified as 0 and Emax as 100. Synergy scores represent the percent growth inhibition induced by a drug combination which exceeded the expected growth inhibition. Expected growth inhibition was calculated based on the effect of each drug as a single agent.
Flow cytometry
For annexin V/PI staining, ∼1 × 106 cells were trypsinized and washed with PBS. Cells were stained using the Dead Cell Apoptosis Kit (Alexa-fluor 499-AnnexinV/Propidium Iodide) (Invitrogen #V13241) according to manufacturer instructions. Cells were analyzed using a Sony SH880S cell sorter. Cell viability status was analyzed using FlowJo v10.8.0 software. Gates were determined using an unstained control. Experiments were performed with at least two biological replicates.
For MDM2 flow-FISH, drug treated cells were trypsinized to create a single cell suspension, washed in PBS and fixed 12 min in 4% formaldehyde (Life Technologies, #28908) at room temperature. Cells were washed twice in PBS. Approximately 1 million cells were permeabilized for 10 min in 0.05% Tween 20 at room temperature and washed once in 2X SCC. Pelleted cells were resuspended in 10μL MDM2 FISH probes (Empire Genomics, MDM2-20-OR) diluted (1:5) in hybridization buffer (Empire Genomics). DNA was denatured at 80C for 3 min, and probes were hybridized overnight at 37C. Cells were washed quickly with 0.4X SCC, resuspended in 2X SCC and incubated 5 min at 37C to remove non-specific probe. Cells were pelleted, resuspended in PBS +2% FBS and analyzed using a Sony SH880S cell sorter.
Immunofluorescence staining and imaging
For immunocytochemistry cells bound to #1.5 glass coverslips were washed once in PBS and fixed 10 min in 4% formaldehyde (Life Technologies, #28908) at room temperature. Cells were washed three times in PBS and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. Following three PBS washes, coverslips were blocked with 10% normal goat serum (Life Technologies, #50062Z) in PBS for 45 min at room temperature. Primary antibodies were diluted in 10% normal goat serum (1:2000 for MDM2 (Abcam, ab226939), 1:500 for p53 (cell signaling, 48818), 1:500 for YY1 (Santa Cruz, sc-7341), and 1:50 for PML (Santa Cruz, sc-966)) and incubated overnight at 4C in 10% normal goat serum. Coverslips were washed three times in PBS, then secondary antibodies were added at 1:500 dilution for 1 h at room temperature. Coverslips were washed two times in PBS before Hoechst or DAPI (1:10,000) in PBS was added for 5 min at room temperature. Coverslips were mounted on microscope slides using VECTASHIELD (Vector Labs, #H-1000-10), then sealed with nail polish.
For immunohistochemistry of primary tumors, snap frozen samples were post-fixed in 4% formaldehyde (Life Technologies, #28908) at 4°C and then cryoprotected in sucrose solution prior to re-freezing. 10μm sections were prepared on a cryostat and then permeabilized, blocked and stained as above. Sections were mounted with Fluoromount G (Invitrogen, #00495802).
All coverslips were imaged on a Zeiss LSM980 confocal microscope. Concentration of MDM2 in nuclear foci (Figure 5A) was imaged in a single z-plane using a 63× oil objective with Airyscan SR-4Y sampling settings. Post image processing was performed using Airyscan joint deconvolution. Co-stains of MDM2 with PML or YY1 (Figures 5B–5G and S6A–S6D) were imaged in a single z-plane using a 63× oil objective with confocal settings. MDM2 heterogeneity (Figures 6A, 6B, S7A, and S7B) was imaged in a single z-plane using a 25× oil objective with confocal settings. Images were processed using cellprofiler software (v4.2.1).88 For a given cell, masks were generated for each channel to define the nuclear area (Hoechst or DAPI), and area occupied by each protein and fluorescent intensity was measured. The counts for each cell and channel were then plotted using R (v4.1.0). Representative images in all panels were generated using FIJI87 version 2.0.0-rc-43/1.5h.
Dual MDM2 Immunofluorescence-FISH
Cells were plated on coverslips and processed for MDM2 immunofluorescence as above. Following secondary antibody incubation, coverslips were washed three times in PBS +0.05% tween 20. Then coverslips were fixed for 20 min in 300mM disuccinimidyl glutarate (Pierce #20513) at room temperature, washed once in PBS, and equilibrated briefly in 2x SCC buffer. MDM2 FISH probes (Empire Genomics, MDM2-20-OR) were diluted (1:5) in hybridization buffer (Empire Genomics). Coverslips were placed on top of 10μL of diluted probe spotted on a slide. Samples were sealed with rubber cement and denatured at 80C for 20 min and then hybridized at 37C overnight in a humid and dark chamber. Samples were washed once with 0.4x SCC and then twice with 2x SCC +0.1% tween 20. Samples were incubated for 10 min in DAPI (1:2000) diluted in 2x SCC at room temperature. Samples were briefly washed with PBS and mounted on microscope slides using SlowFade Gold Antifade Mountant (Invitrogen, #S36936), then sealed with nail polish.
Coverslips were imaged on a Zeiss LSM980 confocal microscope using a 63× oil objective with confocal settings. Multi-channel Z-stacks were acquired at 0.3 μm spacing. Approximately 35 z-slices were taken per field. Images were processed using FIJI87 version 2.0.0-rc-43/1.5h. The intensity for each channel was summed across the entire z stack. For a given field, nuclear masks were created by thresholding the DAPI signal using the MaxEntropy model. Then fluorescent intensity for MDM2 IF and FISH signal was measured for each nucleus. Paired IF and FISH intensities for each cell were plotted in R. Representative images were generated using FIJI87 version 2.0.0-rc-43/1.5h.
Metaphase spreads
Cells were treated with 0.1 μg/mL KaryoMAX colcemid solution () for 5 h and trypsinized to create a single cell suspension. Cells were resuspended in prewarmed (37C) 75mM potassium chloride for 15 min. Three drops of fresh fixative solution (3:1 methanol/glacial acetic acid) were added with gentle mixing. Cells were pelleted and resuspended in 5mL of fixative solution and stored overnight at 4C. The following day cells were washed 3 times with fresh fixative solution. All but 200μL of fixative solution was removed after final wash. 10μL of cell suspension was dropped on slides that were pre-incubated in HCl for 5 h and rinsed with distilled water. Slides containing cells were exposed to steam for 30 s, dried, and processed for FISH. Slides were equilibrated briefly in 2x SCC buffer. MDM2 FISH probes (Empire Genomics, MDM2-20-OR) and control Chromosome 12 probes (Empire Genomics, CHR12-10-GR) diluted 1:5 in hybridization buffer (Empire Genomics) (1μL MDM2 probe, 1μL Chr12 probe, 8μL hybridization buffer). 10μL of diluted probe were spotted on a slide. Samples were sealed with rubber cement using a coverslip and denatured at 80C for 3 min and then hybridized at 37C overnight in a humid and dark chamber. Slides were washed once with 0.4x SCC and then twice with 2x SCC +0.1% tween 20. Slides were incubated for 10 min in DAPI (1:2000) diluted in 2x SCC at room temperature. Slides were briefly washed with PBS and coverslips were mounted using SlowFade Gold Antifade Mountant (Invitrogen, #S36936), then sealed with nail polish. Slides were imaged on a Zeiss LSM980 confocal microscope using a 63× oil objective with confocal settings.
Generation of LPS853 P53 knockout cell line
TP53 knockout experiments utilized pXPR_044, all-in-one CRISPR-Cas9 vector carrying mCherry and sgRNA (gift from John Doench and Dave Root, Broad GPP). The following CRISPR sgRNAs were cloned into the vector: safe harbor 5′-GGCTAAATTCCTCTTATTCA-3′; TP53 start site 5′-TCGACGCTAGGATCTGACTG-3′. LPS853 cells were transfected with 3 μg/mL of lentivirus plasmid and 5 μL/mL of TransIT-LT1 reagent (MirusBio, MIR2300). The following day cells were selected using fluorescent activated cell sorting (Sony SH800). Gating strategy first involved selecting the live population with forward and side scatter. Untreated cells were used to set the mCherry threshold gate. Cells expressing the highest mCherry (top 50%) were selected. Genomic DNA was isolated and the TP53 start site was amplified using the following primer set: F 5′-CCCAACCCTTGTCCTTACCA-3′; R 5′-CAACATGCAAAGCCCTGTCT-3′. CRISPR-mediated disruption of this amplified region was confirmed via sanger sequencing using the TIDE tool.92
Quantification and statistical analysis
RNA-sequencing analysis
RNA-seq data were mapped to the GRCh38 reference genome using STAR aligner (v2.6.0c).78 Expression was quantified using the –quantMode option and Basic twopassMode in STAR, and counts table was generated in R selecting the column representing the positive-strand counts. Differential expression was calculated using the Bioconductor82 package DESeq2 v(1.32.0).93 In the case of HDM201 treated samples, the 0h time point was used as a reference and contrasts were generated to the samples corresponding to 2h, 4h and 6h of HDM201 treatment. Differentially expressed genes (adjusted p value<0.05) were separated into up- and down-regulated. Genes from each category were further divided in three clusters based on the time-point when they showed their earliest differential expression. Expression heatmaps were calculated using the Fold-change values extracted from DESeq2, and were plotted using the ComplexHeatmap package in R.
Tumor count matrices were subjected to TPM normalization, in which all exons for each gene were combined to determine the gene length. Expression heat maps were plotted with the ComplexHeatmaps R package using a row-normalized TPM expression matrix.
TPM matrices were used to calculate maturity scores for tumors and cell lines using GSVA (v1.40.1)94 with default settings. Genes listed in Table S5 or the hallmark adipogenesis geneset from GSEA37 were used to generate maturity scores. Genes sets associated with maturity were defined by calculating mean Pearson correlations for each gene included in Table S5, a mean correlation value ≥0.5 defined positively correlated genes and a mean correlation value ≤ −0.5 defined negatively correlated genes.
Gene set enrichment for all gene sets was performed using the web-based metascape portal95 using “express analysis” settings.
ChIP-sequencing analysis
ChIP-Seq data were mapped to the GRCh38 reference genome using STAR aligner (v2.6.0c),78 including parameters --alignIntronMax 1 --alignEndsType EndToEnd to make it compatible with genome mapping. Tag directories of the mapped reads were created using homer (v4.11.1 using hg38 v6.4). Peaks were called with the findPeaks function of homer using the histone style parameters for H3k27ac ChIP-seq, and the factor style parameters for MDM2, P53, Jun, RUNX and YY1 ChIP-seqs.
Samples that contained spike-ins (SI) were mapped to the dm6 reference genome assembly. The number of mapped SI reads were used to create normalization factors (NF):
Where represents the maximum number of tags across samples, and is the number of tags for a given sample. Samples were normalized using Spike In normalization factor when available, and using RPGC normalization otherwise for downstream analyses.
For all mitochondrial-related analyses, the ChIP-seq libraries were processed as mentioned above, with the exception of using a mito masked GRCh38 genome assembly for mapping the libraries. In this assembly, the genomic regions that are highly similar to mtDNA sequences are masked.
H3K27ac peaks were used to define active enhancers in dedifferentiated liposarcoma cells. We merged H3K27ac peaks from 8 cell lines and 4 dedifferentiated tumors using mergePeaks in homer,83 and selected H3K27ac merged peaks that were supported by peaks from at least two samples. H3K27ac regions that overlapped promoters were excluded from further enhancer analysis. The set of promoter regions was defined using the 500bp window upstream of Transcription Start Sites in NCBI’s RefSeq GRCh38 assembly.
In order to assess the binding dynamics of the transcription factors in our study, TF peaks from ChIP replicates were merged into an aggregated list of peaks for each cell line. To study the DDLPS programs and create the genomic circuitry, we defined a consensus set of peaks that were present in both DDLPS cell lines (LPS141 and LPS853) for all TFs analyzed. We used bedtools85 (v2.29.0) functions computeMatrix and plotHeatmap to generate binding heatmaps of the transcription factors. TF binding sites were divided into promoter and enhancer regions. We defined three different types of binding sites. P53 sites were defined by P53 binding. MDM2 sites were defined using MDM2-binding sites that were not co-occupied by P53. Finally, Jun and Runx sites were defined using genomic locations that are bound by either of these two factors, and are not co-bound by MDM2 or P53.
For each cell line (HCT116, U-2 OS, LPS141 and LPS853) we obtained a set of peaks that was present in two ChIP replicates, these peak sets were used to study the dynamics of MDM2 and P53 binding sites in supraphysiologic levels of MDM2. The resulting peak files were overlapped with promoters/enhancers lists to determine the proportion of MDM2 and P53 peaks that bind regulatory regions, and how these proportions change between samples with wildtype and supraphysiologic MDM2 levels.
Genomic regions underlying peak binding sites were extracted and subjected to motif enrichment analysis. This analysis was performed using the Homer function findMotifsGenome.pl, using hg38 genome (v6.4) and using the “known motifs” database to extract TF footprints. A random background was used, by setting the -size option to 200bp, 500bp and 1.5kb when analyzing TF peaks, promoters or H3K27ac as input sequences, respectively. Significant enriched motifs (p value ≤0.01) were ordered by the transcription factor family. Enrichment metrics presented in Figures 2B and 4H correspond to the motif that is represented in the highest percent of target sequences per TF family.
SNP analysis from ChIP inputs
Mapped bam files were query sorted using samtools and HTSlib.84,86 The insert size field and mate coordinates were filled in the query-sorted bam files using samtools fixmate. These bam files were then sorted by coordinate. Finally, duplicates were marked using the markdup option of samtools.
SNPs were called using the call option of bcftools with the option -m, to allow for rare-variant calling, and the output was set to vcf. We kept variants that fulfill the following criteria: (1) covered at least 5 times, (2) have a mapping quality score above 39, (3) Have an SNP calling quality score above 19.
Copy number variant analysis
Whole cell extract libraries were mapped to the hg38 genome using Bowtie2. We used the option "--very-sensitive" to obtain the highest possible number of uniquely mapped reads. The output sam files were then converted to bam files and sorted using Samtools. For Copy Number Variant (CNV) calls, we used the Epianalysis software (in preparation). First, the genome was binned into 5kb regions. Then, the number of uniquely mapped reads was measured in each bin. The bin coverage was corrected using mapping quality scores as previously described.96 Finally, the CNAnorm97 package was used to identify underlying ploidy in the binned genome, generating a correction factor corresponding to the FC relative to a diploid coverage. These values were plotted as line plots using ggplot2.
HiChIP analysis
Data were quality and adapter trimmed using seqPrep98 and mapped to the reference genome (hg38) using Bowtie2.99,81 Contact matrices were created with juicebox79 as previously described.91 Chromatin loops were called with hichipper,80 using H3k27ac peaks and hg38 promoters as the predefined peak set to call anchors as in.91 For each pair of potential anchors, hichipper counts the number of read pairs that support their interaction and uses the mango background correction model to estimate the probability of two regions interacting given their genomic distance. We filtered out contacts with an adjusted p value higher than 0.05, and only contacts with at least 10 Paired-End Tags (PETs) were considered for further analyses.
We classified loops in three separate categories depending on the genomic regions where the loop anchors mapped, termed enhancer-enhancer (E-E), enhancer-promoter (E-P), and promoter-promoter (P-P) loops. Each loop type represented a different genomic feature, and contributed to chromatin structure and/or gene regulation. We used bedtools to map transcription factor peaks to regulatory loops. We tested the proportion of loop types associated with each factor, and compared them to the overall proportion of E-E, E-P and P-P loops. We used a Fisher exact test (two-tailed) to determine whether a loop type is significantly associated with any of the TFs tested.
Integrating the HiChIP topology maps with transcription factor binding sites allowed us to map the circuitry associated with each TF. We associated the peaks defined above (section transcription factor circuitry analysis) with the topology maps from HiChIP, and defined a set of target genes associated with each of the transcription factors by connecting the TF binding sites to the gene promoters, using E-P and P-P loops. We defined three different types of transcriptional regulatory circuits. P53 circuits were defined by P53 binding. MDM2 circuits were defined using MDM2-binding sites that were not co-occupied by P53. Finally, Jun and Runx circuits were defined using genomic locations that are bound by either of these two factors, and are not co-bound by MDM2 or P53. In order to isolate the peaks associated with the genes in each regulatory circuit, we used bedtools to extract the peaks that looped to the promoters of the genes of interest. Such peak lists were used for further genomic analysis such as metagene plots across drug treatment samples.
We defined a promoter-promoter hub (pp hub) as a set of 4 or more interconnected HiChIP loops whose anchors span promoter regions. For this study, we defined pp hubs in LPS141 and LPS853 liposarcoma cell lines separately, and overlapped them to create a common list of pp hubs. We visualized pp hubs defined in cell lines in H3K27ac HiChIP datasets of three LPS tumors. We also mined 12 existing H3K27ac datasets20 of normal tissue and processed them in HiC-Pro81 and hichipper,80 using the list of hg38 promoters as the set of reference peaks to calculate the anchors.
The integrative visualization of genomics data was done using the Sushi R package.100 To visualize the topology networks in promoter-promoter hubs, we used cytoscape (v3.8.2).101 We transformed anchors into nodes and created edges between them if they were connected by a loop in the HiChIP data. We added properties to nodes and edges, including gene names and promoter coordinates in nodes, and PET score in the edges. These properties allowed us to filter hubs by PET score or number of anchors.
Hub gene correlation in TCGA datasets
We further tested the significance of the pp hubs by correlating the expression of genes belonging to these hubs in TCGA datasets. We extracted a log-transformed and normalized expression matrix from the TCGA (https://pancanatlas.xenahubs.net) and subsampled a matrix that contained the DDLPS samples. We calculated expression correlation only in hubs that spanned at least three expressed genes. For each hub, we calculated the Pearson correlation between every two genes in the hub, and obtained the mean of the absolute value of all pairwise correlations for a given hub. For each pairwise correlation we selected a pair of genes that were separated by the same genomic distance to be used as a control. The two correlation distributions were compared using a two-tailed t-test in R.
Acknowledgments
We appreciate the contributions of patients and their families. The authors are also grateful for generous support from the Dick Family, Claude Mongeau, the V Foundation for Cancer Research, and the David Liposarcoma Research Initiative at Dana-Farber Cancer Institute supported by KBF Canada via the Rossy Foundation Fund. We publish this work in memory of Spencer Dick. The authors thank all members of the Bernstein laboratory for discussions and Dr. Adrian Marino-Enriquez for providing liposarcoma cell lines and insightful discussions. This work was supported by funds from the NCI/NIH Director’s Fund (DP1CA216873 to B.E.B.). S.E.J. is supported by an NCI K08 career development award from the National Cancer Institute (K08CA259623). G.D.D. is the Quick Family Endowed Chair in Medical Oncology at the Dana-Farber Cancer Institute and is also supported in part by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. B.E.B. is the Richard and Nancy Lubin Family Endowed Chair at the Dana-Farber Cancer Institute and an American Cancer Society Research Professor.
Author contributions
S.M.B., S.E.J., and B.E.B. designed the initial experimental plans. S.M.B., E.G.C., K.A.M., S.E.O., K.J.O., E.H., S.C., M.M.M., F.J.N., and C.A. performed experimental procedures. S.C.-G., C.A.E.F., and S.M.B. performed computational analyses. S.E.J., S.C., J.T.M., G.D.D., and N.R. assisted with the collection and characterization of clinical samples. S.M.B., S.C.G., S.E.J., and B.E.B. drafted the manuscript.
Declaration of interests
B.E.B. declares outside interests in Fulcrum Therapeutics, Arsenal Biosciences, HiFiBio, Cell Signaling Technologies, Design Pharmaceuticals, and Chroma Medicine. G.D.D. reports leadership as co-founder of IDRX; stocks/options/shares in IDRX, Blueprint Medicines, G1 Therapeutics, Caris Life Sciences, Erasca Pharmaceuticals, RELAY Therapeutics, Bessor Pharmaceuticals, CellCarta, IKENA Oncology, and Kojin Therapeutics; paid consulting fees from Bayer, Pfizer, Novartis, Roche/Genentech, GSK, PharmaMar, Daiichi Sankyo, EMD-Serono/Merck KGaA, Mirati, WCG/Arsenal Capital, G1 Therapeutics, Caris Life Sciences, RELAY Therapeutics, CellCarta, IKENA Oncology, Kojin Therapeutics, RAIN Therapeutics, Jazz Pharmaceuticals, Aadi Biosciences, and IDRX; royalties, patents, or licenses from Novartis to Dana-Farber Cancer Institute for “use patent” of imatinib in GIST; and non-financial interests in AACR Science Policy and Government Affairs Committee and Alexandria Real Estate Equities summit conference series.
Published: May 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2023.100321.
Contributor Information
Sarah E. Johnstone, Email: sarah_johnstone@dfci.harvard.edu.
Bradley E. Bernstein, Email: bradley_bernstein@dfci.harvard.edu.
Supplemental information
This table summarizes clinical samples and cell models used in the study and genomic assays performed (related to Figure 1A). Copy number and expression values for commonly amplified genes (related to Figures 1B, 1C, 2A, S1A–S1D, and S1K) and tumor maturity metrics including maturity score and CEBPB/PPARG expression (related to Figures 1D–1F and S1G–S1I) are provided for each sample.
Gene-level copy number calls are provided for each tumor or normal fat sample (related to Figures 1B and S1A–S1D).
Total variants and variants per Mb are provided for each tumor sample (related to section “Genomic and transcriptional profiles of liposarcoma tumors and models”).
Gene expression tpm counts are provided for normal fat, liposarcoma tumors, and cell line models (related to Figures 1C, 1E, 1F, 2A, 2C, 2D, 3D, 3G, 3J, S1G–S1K, S2C, and S2D).
Genes with established roles in adipogenesis and/or specific expression in mature adipocytes that were used to derive maturity scores of tumors and cell lines (related to Figures 1D–1F, 2A, and S1I).
Gene names are provided for genes negatively correlated with tumor maturity (n = 1,298) or positively correlated (n = 1,065) with tumor maturity (related to Figure 2A).
Homer analysis output of transcription factor motifs significantly enriched in enhancers of DDLPS tumors and cell lines (related to Figure 2B).
Target genes defined for each TF regulatory circuit are provided (related to Figures 3B, 3E, and 3H).
Results are provided for functional enrichment analysis of target genes defined for each TF regulatory circuit in Figures 3B, 3E, and 3H (related to Figures 3C, 3F, and 3I).
Homer analysis output for transcription factor motifs significantly enriched in hub promoters either bound or unbound by MDM2 (related to Figure 4H).
Data and code availability
All next generation sequencing data have been deposited at the Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All original code has been deposited at Github: https://github.com/BernsteinLab/methods_mdm2_sarcoma (https://doi.org/10.5281/zenodo.7814766) and is publicly available as of the date of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M., et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haupt Y., Maya R., Kazaz A., Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 4.Kim H., Nguyen N.-P., Turner K., Wu S., Gujar A.D., Luebeck J., Liu J., Deshpande V., Rajkumar U., Namburi S., et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat. Genet. 2020;52:891–897. doi: 10.1038/s41588-020-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedeutour F., Forus A., Coindre J.M., Berner J.M., Nicolo G., Michiels J.F., Terrier P., Ranchere-Vince D., Collin F., Myklebost O., Turc-Carel C. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer. 1999;24:30–41. [PubMed] [Google Scholar]
- 6.Garsed D.W., Marshall O.J., Corbin V.D.A., Hsu A., Di Stefano L., Schröder J., Li J., Feng Z.-P., Kim B.W., Kowarsky M., et al. The architecture and evolution of cancer neochromosomes. Cancer Cell. 2014;26:653–667. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Dal Cin P., Kools P., Sciot R., De Wever I., Van Damme B., Van de Ven W., Van den Berghe H. Cytogenetic and fluorescence in situ hybridization investigation of ring chromosomes characterizing a specific pathologic subgroup of adipose tissue tumors. Cancer Genet. Cytogenet. 1993;68:85–90. doi: 10.1016/0165-4608(93)90001-3. [DOI] [PubMed] [Google Scholar]
- 8.Fields S., Jang S.K. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 9.Kastenhuber E.R., Lowe S.W. Putting p53 in context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubbutat M.H., Jones S.N., Vousden K.H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 11.Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 12.Arva N.C., Gopen T.R., Talbott K.E., Campbell L.E., Chicas A., White D.E., Bond G.L., Levine A.J., Bargonetti J. A chromatin-associated and transcriptionally inactive p53-mdm2 complex occurs in mdm2 SNP309 homozygous cells. J. Biol. Chem. 2005;280:26776–26787. doi: 10.1074/jbc.M505203200. [DOI] [PubMed] [Google Scholar]
- 13.Wienken M., Moll U.M., Dobbelstein M. Mdm2 as a chromatin modifier. J. Mol. Cell Biol. 2017;9:74–80. doi: 10.1093/jmcb/mjw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riscal R., Schrepfer E., Arena G., Cissé M.Y., Bellvert F., Heuillet M., Rambow F., Bonneil E., Sabourdy F., Vincent C., et al. Chromatin-bound MDM2 regulates serine metabolism and redox homeostasis independently of p53. Mol. Cell. 2016;62:890–902. doi: 10.1016/j.molcel.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 15.Arena G., Cissé M.Y., Pyrdziak S., Chatre L., Riscal R., Fuentes M., Arnold J.J., Kastner M., Gayte L., Bertrand-Gaday C., et al. Mitochondrial MDM2 regulates respiratory complex I activity independently of p53. Mol. Cell. 2018;69:594–609.e8. doi: 10.1016/j.molcel.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sui G., Affar E.B., Shi Y., Brignone C., Wall N.R., Yin P., Donohoe M., Luke M.P., Calvo D., Grossman S.R., Shi Y. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Grossman S.R., Perez M., Kung A.L., Joseph M., Mansur C., Xiao Z.-X., Kumar S., Howley P.M., Livingston D.M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol. Cell. 1998;2:405–415. doi: 10.1016/S1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 18.Wienken M., Dickmanns A., Nemajerova A., Kramer D., Najafova Z., Weiss M., Karpiuk O., Kassem M., Zhang Y., Lozano G., et al. MDM2 associates with polycomb repressor complex 2 and enhances stemness-promoting chromatin modifications independent of p53. Mol. Cell. 2016;61:68–83. doi: 10.1016/j.molcel.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ENCODE Project Consortium. Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A., Kaul R., et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mumbach M.R., Satpathy A.T., Boyle E.A., Dai C., Gowen B.G., Cho S.W., Nguyen M.L., Rubin A.J., Granja J.M., Kazane K.R., et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 2017;49:1602–1612. doi: 10.1038/ng.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hariprakash J.M., Ferrari F. Computational biology solutions to identify enhancers-target gene pairs. Comput. Struct. Biotechnol. J. 2019;17:821–831. doi: 10.1016/j.csbj.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popay T.M., Dixon J.R. Coming full circle: on the origin and evolution of the looping model for enhancer-promoter communication. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.102117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordon-Cardo C., Latres E., Drobnjak M., Oliva M.R., Pollack D., Woodruff J.M., Marechal V., Chen J., Brennan M.F., Levine A.J. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994;54:794–799. [PubMed] [Google Scholar]
- 24.Oliner J.D., Kinzler K.W., Meltzer P.S., George D.L., Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 25.Sciot R. MDM2 amplified sarcomas: a literature review. Diagnostics. 2021;11 doi: 10.3390/diagnostics11030496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullard A. p53 programmes plough on. Nat. Rev. Drug Discov. 2020;19:497–500. doi: 10.1038/d41573-020-00130-z. [DOI] [PubMed] [Google Scholar]
- 27.Tisato V., Voltan R., Gonelli A., Secchiero P., Zauli G. MDM2/X inhibitors under clinical evaluation: perspectives for the management of hematological malignancies and pediatric cancer. J. Hematol. Oncol. 2017;10:133. doi: 10.1186/s13045-017-0500-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee A.T.J., Thway K., Huang P.H., Jones R.L. Clinical and molecular spectrum of liposarcoma. J. Clin. Oncol. 2018;36:151–159. doi: 10.1200/JCO.2017.74.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network Electronic address elizabethdemicco@sinaihealthsystemca Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–965.e28. doi: 10.1016/j.cell.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamboa A.C., Gronchi A., Cardona K. Soft-tissue sarcoma in adults: an update on the current state of histiotype-specific management in an era of personalized medicine. CA. Cancer J. Clin. 2020;70:200–229. doi: 10.3322/caac.21605. [DOI] [PubMed] [Google Scholar]
- 31.Hirata M., Asano N., Katayama K., Yoshida A., Tsuda Y., Sekimizu M., Mitani S., Kobayashi E., Komiyama M., Fujimoto H., et al. Integrated exome and RNA sequencing of dedifferentiated liposarcoma. Nat. Commun. 2019;10:5683. doi: 10.1038/s41467-019-13286-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amin-Mansour A., George S., Sioletic S., Carter S.L., Rosenberg M., Taylor-Weiner A., Stewart C., Chevalier A., Seepo S., Tracy A., et al. Genomic evolutionary patterns of leiomyosarcoma and liposarcoma. Clin. Cancer Res. 2019;25:5135–5142. doi: 10.1158/1078-0432.CCR-19-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder E.L., Sandstrom D.J., Law K., Fiore C., Sicinska E., Brito J., Bailey D., Fletcher J.A., Loda M., Rodig S.J., et al. c-Jun amplification and overexpression are oncogenic in liposarcoma but not always sufficient to inhibit the adipocytic differentiation programme. J. Pathol. 2009;218:292–300. doi: 10.1002/path.2564. [DOI] [PubMed] [Google Scholar]
- 34.Keung E.Z., Akdemir K.C., Al Sannaa G.A., Garnett J., Lev D., Torres K.E., Lazar A.J., Rai K., Chin L. Increased H3K9me3 drives dedifferentiated phenotype via KLF6 repression in liposarcoma. J. Clin. Invest. 2015;125:2965–2978. doi: 10.1172/JCI77976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saâda-Bouzid E., Burel-Vandenbos F., Ranchère-Vince D., Birtwisle-Peyrottes I., Chetaille B., Bouvier C., Château M.C., Peoc’h M., Battistella M., Bazin A., et al. Prognostic value of HMGA2, CDK4, and JUN amplification in well-differentiated and dedifferentiated liposarcomas. Mod. Pathol. 2015;28:1404–1414. doi: 10.1038/modpathol.2015.96. [DOI] [PubMed] [Google Scholar]
- 36.Mariani O., Brennetot C., Coindre J.-M., Gruel N., Ganem C., Delattre O., Stern M.-H., Aurias A. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell. 2007;11:361–374. doi: 10.1016/j.ccr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 37.HALLMARK_ADIPOGENESIS https://www.gsea-msigdb.org/gsea/msigdb/human/geneset/HALLMARK_ADIPOGENESIS.html.
- 38.Reznik E., Wang Q., La K., Schultz N., Sander C. Mitochondrial respiratory gene expression is suppressed in many cancers. Elife. 2017;6 doi: 10.7554/eLife.21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu H., Huang Z., Yin X., Zhang J., Gong L., Chen J., Rong K., Xu J., Lu L., Cui L. Role of c-Jun N-terminal kinase in the osteogenic and adipogenic differentiation of human adipose-derived mesenchymal stem cells. Exp. Cell Res. 2015;339:112–121. doi: 10.1016/j.yexcr.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Lee K.S., Kim H.J., Li Q.L., Chi X.Z., Ueta C., Komori T., Wozney J.M., Kim E.G., Choi J.Y., Ryoo H.M., Bae S.C. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol. Cell Biol. 2000;20:8783–8792. doi: 10.1128/MCB.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner E.F. Functions of AP1 (Fos/Jun) in bone development. Ann. Rheum. Dis. 2002;61(Suppl 2):ii40–ii42. doi: 10.1136/ard.61.suppl_2.ii40. ii40–ii42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komori T. Regulation of proliferation, differentiation and functions of osteoblasts by Runx2. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20071694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komori T. Regulation of osteoblast differentiation by Runx2. Adv. Exp. Med. Biol. 2010;658:43–49. doi: 10.1007/978-1-4419-1050-9_5. [DOI] [PubMed] [Google Scholar]
- 44.Mumbach M.R., Rubin A.J., Flynn R.A., Dai C., Khavari P.A., Greenleaf W.J., Chang H.Y. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods. 2016;13:919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karsenty G. Transcriptional control of skeletogenesis. Annu. Rev. Genomics Hum. Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 46.Ono K., Hata K., Nakamura E., Ishihara S., Kobayashi S., Nakanishi M., Yoshida M., Takahata Y., Murakami T., Takenoshita S., et al. Dmrt2 promotes transition of endochondral bone formation by linking Sox9 and Runx2. Commun. Biol. 2021;4:326. doi: 10.1038/s42003-021-01848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donsante S., Palmisano B., Serafini M., Robey P.G., Corsi A., Riminucci M. From stem cells to bone-forming cells. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thway K. Well-differentiated liposarcoma and dedifferentiated liposarcoma: an updated review. Semin. Diagn. Pathol. 2019;36:112–121. doi: 10.1053/j.semdp.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Karni-Schmidt O., Lokshin M., Prives C. The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol. 2016;11:617–644. doi: 10.1146/annurev-pathol-012414-040349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furet P., Masuya K., Kallen J., Stachyra-Valat T., Ruetz S., Guagnano V., Holzer P., Mah R., Stutz S., Vaupel A., et al. Discovery of a novel class of highly potent inhibitors of the p53–MDM2 interaction by structure-based design starting from a conformational argument. Bioorg. Med. Chem. Lett. 2016;26:4837–4841. doi: 10.1016/j.bmcl.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 51.Stein E.M., DeAngelo D.J., Chromik J., Chatterjee M., Bauer S., Lin C.-C., Suarez C., de Vos F., Steeghs N., Cassier P.A., et al. Results from a first-in-human phase I study of siremadlin (HDM201) in patients with advanced wild-type TP53 solid tumors and acute leukemia. Clin. Cancer Res. 2022;28:870–881. doi: 10.1158/1078-0432.CCR-21-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konopleva M., Martinelli G., Daver N., Papayannidis C., Wei A., Higgins B., Ott M., Mascarenhas J., Andreeff M. MDM2 inhibition: an important step forward in cancer therapy. Leukemia. 2020;34:2858–2874. doi: 10.1038/s41375-020-0949-z. [DOI] [PubMed] [Google Scholar]
- 53.Wu X., Bayle J.H., Olson D., Levine A.J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 54.Cissé M.Y., Pyrdziak S., Firmin N., Gayte L., Heuillet M., Bellvert F., Fuentes M., Delpech H., Riscal R., Arena G., et al. Targeting MDM2-dependent serine metabolism as a therapeutic strategy for liposarcoma. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aay2163. [DOI] [PubMed] [Google Scholar]
- 55.Robin E.D., Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell. Physiol. 1988;136:507–513. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- 56.Basu U., Bostwick A.M., Das K., Dittenhafer-Reed K.E., Patel S.S. Structure, mechanism, and regulation of mitochondrial DNA transcription initiation. J. Biol. Chem. 2020;295:18406–18425. doi: 10.1074/jbc.REV120.011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rubio-Patiño C., Trotta A.P., Chipuk J.E. MDM2 and mitochondrial function: one complex intersection. Biochem. Pharmacol. 2019;162:14–20. doi: 10.1016/j.bcp.2018.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elkholi R., Abraham-Enachescu I., Trotta A.P., Rubio-Patiño C., Mohammed J.N., Luna-Vargas M.P.A., Gelles J.D., Kaminetsky J.R., Serasinghe M.N., Zou C., et al. MDM2 integrates cellular respiration and apoptotic signaling through NDUFS1 and the mitochondrial network. Mol. Cell. 2019;74:452–465.e7. doi: 10.1016/j.molcel.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goel V.Y., Huseyin M.K., Hansen A.S. Region Capture Micro-C reveals coalescence of enhancers and promoters into nested microcompartments. bioRxiv. 2022 doi: 10.1101/2022.07.12.499637. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levo M., Raimundo J., Bing X.Y., Sisco Z., Batut P.J., Ryabichko S., Gregor T., Levine M.S. Transcriptional coupling of distant regulatory genes in living embryos. Nature. 2022;605:754–760. doi: 10.1038/s41586-022-04680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deshpande A.S., Ulahannan N., Pendleton M., Dai X., Ly L., Behr J.M., Schwenk S., Liao W., Augello M.A., Tyer C., et al. Identifying synergistic high-order 3D chromatin conformations from genome-scale nanopore concatemer sequencing. Nat. Biotechnol. 2022;40:1488–1499. doi: 10.1038/s41587-022-01289-z. [DOI] [PubMed] [Google Scholar]
- 62.Quinodoz S.A., Ollikainen N., Tabak B., Palla A., Schmidt J.M., Detmar E., Lai M.M., Shishkin A.A., Bhat P., Takei Y., et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018;174:744–757.e24. doi: 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weintraub A.S., Li C.H., Zamudio A.V., Sigova A.A., Hannett N.M., Day D.S., Abraham B.J., Cohen M.A., Nabet B., Buckley D.L., et al. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171:1573–1588.e28. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beagan J.A., Duong M.T., Titus K.R., Zhou L., Cao Z., Ma J., Lachanski C.V., Gillis D.R., Phillips-Cremins J.E. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 2017;27:1139–1152. doi: 10.1101/gr.215160.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bernardi R., Scaglioni P.P., Bergmann S., Horn H.F., Vousden K.H., Pandolfi P.P. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 2004;6:665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 66.Joshi S., Singh A.R., Durden D.L. MDM2 regulates hypoxic hypoxia-inducible factor 1α stability in an E3 ligase, proteasome, and PTEN-phosphatidylinositol 3-kinase-AKT-dependent manner. J. Biol. Chem. 2014;289:22785–22797. doi: 10.1074/jbc.M114.587493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chu Y., Yang X. SUMO E3 ligase activity of TRIM proteins. Oncogene. 2011;30:1108–1116. doi: 10.1038/onc.2010.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei X., Yu Z.K., Ramalingam A., Grossman S.R., Yu J.H., Bloch D.B., Maki C.G. Physical and functional interactions between PML and MDM2. J. Biol. Chem. 2003;278:29288–29297. doi: 10.1074/jbc.M212215200. [DOI] [PubMed] [Google Scholar]
- 69.Zhu H., Wu L., Maki C.G. MDM2 and promyelocytic leukemia antagonize each other through their direct interaction with p53. J. Biol. Chem. 2003;278:49286–49292. doi: 10.1074/jbc.M308302200. [DOI] [PubMed] [Google Scholar]
- 70.Wan J., Block S., Scribano C.M., Thiry R., Esbona K., Audhya A., Weaver B.A. Mad1 destabilizes p53 by preventing PML from sequestering MDM2. Nat. Commun. 2019;10:1540. doi: 10.1038/s41467-019-09471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corpet A., Kleijwegt C., Roubille S., Juillard F., Jacquet K., Texier P., Lomonte P. PML nuclear bodies and chromatin dynamics: catch me if you can. Nucleic Acids Res. 2020;48:11890–11912. doi: 10.1093/nar/gkaa828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shiels C., Islam S.A., Vatcheva R., Sasieni P., Sternberg M.J., Freemont P.S., Sheer D. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 2001;114:3705–3716. doi: 10.1242/jcs.114.20.3705. [DOI] [PubMed] [Google Scholar]
- 73.Wang J., Shiels C., Sasieni P., Wu P.J., Islam S.A., Freemont P.S., Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liebl M.C., Hofmann T.G. Regulating the p53 tumor suppressor network at PML biomolecular condensates. Cancers. 2022;14 doi: 10.3390/cancers14194549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lange J.T., Rose J.C., Chen C.Y., Pichugin Y., Xie L., Tang J., Hung K.L., Yost K.E., Shi Q., Erb M.L., et al. The evolutionary dynamics of extrachromosomal DNA in human cancers. Nat. Genet. 2022;54:1527–1533. doi: 10.1038/s41588-022-01177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeay S., Ferretti S., Holzer P., Fuchs J., Chapeau E.A., Wartmann M., Sterker D., Romanet V., Murakami M., Kerr G., et al. Dose and schedule determine distinct molecular mechanisms underlying the efficacy of the p53-MDM2 inhibitor HDM201. Cancer Res. 2018;78:6257–6267. doi: 10.1158/0008-5472.CAN-18-0338. [DOI] [PubMed] [Google Scholar]
- 77.Matushansky I., Hernando E., Socci N.D., Matos T., Mills J., Edgar M.A., Schwartz G.K., Singer S., Cordon-Cardo C., Maki R.G. A developmental model of sarcomagenesis defines a differentiation-based classification for liposarcomas. Am. J. Pathol. 2008;172:1069–1080. doi: 10.2353/ajpath.2008.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Durand N.C., Robinson J.T., Shamim M.S., Machol I., Mesirov J.P., Lander E.S., Aiden E.L. Juicebox provides a visualization system for hi-C contact maps with unlimited zoom. Cell Syst. 2016;3:99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lareau C.A., Aryee M.J. hichipper: a preprocessing pipeline for calling DNA loops from HiChIP data. Nat. Methods. 2018;15:155–156. doi: 10.1038/nmeth.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.-J., Vert J.-P., Heard E., Dekker J., Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T., et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bonfield J.K., Marshall J., Danecek P., Li H., Ohan V., Whitwham A., Keane T., Davies R.M. HTSlib: C library for reading/writing high-throughput sequencing data. GigaScience. 2021;10 doi: 10.1093/gigascience/giab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stirling D.R., Swain-Bowden M.J., Lucas A.M., Carpenter A.E., Cimini B.A., Goodman A. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinf. 2021;22:433. doi: 10.1186/s12859-021-04344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ianevski A., He L., Aittokallio T., Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics. 2017;33:2413–2415. doi: 10.1093/bioinformatics/btx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liau B.B., Sievers C., Donohue L.K., Gillespie S.M., Flavahan W.A., Miller T.E., Venteicher A.S., Hebert C.H., Carey C.D., Rodig S.J., et al. Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell. 2017;20:233–246.e7. doi: 10.1016/j.stem.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnstone S.E., Reyes A., Qi Y., Adriaens C., Hegazi E., Pelka K., Chen J.H., Zou L.S., Drier Y., Hecht V., et al. Large-Scale topological changes restrain malignant progression in colorectal cancer. Cell. 2020;182:1474–1489.e23. doi: 10.1016/j.cell.2020.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Derrien T., Estellé J., Marco Sola S., Knowles D.G., Raineri E., Guigó R., Ribeca P. Fast computation and applications of genome mappability. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gusnanto A., Wood H.M., Pawitan Y., Rabbitts P., Berri S. Correcting for cancer genome size and tumour cell content enables better estimation of copy number alterations from next-generation sequence data. Bioinformatics. 2012;28:40–47. doi: 10.1093/bioinformatics/btr593. [DOI] [PubMed] [Google Scholar]
- 98.St John, J.S.P. Tool for Stripping Adaptors and/or Merging Paired Reads With Overlap into Single Reads. GitHub https://github.com/jstjohn/SeqPrep.
- 99.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Phanstiel D.H., Boyle A.P., Araya C.L., Snyder M.P. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics. 2014;30:2808–2810. doi: 10.1093/bioinformatics/btu379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table summarizes clinical samples and cell models used in the study and genomic assays performed (related to Figure 1A). Copy number and expression values for commonly amplified genes (related to Figures 1B, 1C, 2A, S1A–S1D, and S1K) and tumor maturity metrics including maturity score and CEBPB/PPARG expression (related to Figures 1D–1F and S1G–S1I) are provided for each sample.
Gene-level copy number calls are provided for each tumor or normal fat sample (related to Figures 1B and S1A–S1D).
Total variants and variants per Mb are provided for each tumor sample (related to section “Genomic and transcriptional profiles of liposarcoma tumors and models”).
Gene expression tpm counts are provided for normal fat, liposarcoma tumors, and cell line models (related to Figures 1C, 1E, 1F, 2A, 2C, 2D, 3D, 3G, 3J, S1G–S1K, S2C, and S2D).
Genes with established roles in adipogenesis and/or specific expression in mature adipocytes that were used to derive maturity scores of tumors and cell lines (related to Figures 1D–1F, 2A, and S1I).
Gene names are provided for genes negatively correlated with tumor maturity (n = 1,298) or positively correlated (n = 1,065) with tumor maturity (related to Figure 2A).
Homer analysis output of transcription factor motifs significantly enriched in enhancers of DDLPS tumors and cell lines (related to Figure 2B).
Target genes defined for each TF regulatory circuit are provided (related to Figures 3B, 3E, and 3H).
Results are provided for functional enrichment analysis of target genes defined for each TF regulatory circuit in Figures 3B, 3E, and 3H (related to Figures 3C, 3F, and 3I).
Homer analysis output for transcription factor motifs significantly enriched in hub promoters either bound or unbound by MDM2 (related to Figure 4H).
Data Availability Statement
All next generation sequencing data have been deposited at the Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All original code has been deposited at Github: https://github.com/BernsteinLab/methods_mdm2_sarcoma (https://doi.org/10.5281/zenodo.7814766) and is publicly available as of the date of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.