Summary
Citrate is a common primary metabolite which often characterizes fruit flavour. The key regulators of citrate accumulation in fruit and vegetables are poorly understood. We systematically analysed the dynamic profiles of organic acid components during the development of kiwifruit (Actinidia spp.). Citrate continuously accumulated so that it became the predominate contributor to total acidity at harvest. Based on a co‐expression network analysis using different kiwifruit cultivars, an Al‐ACTIVATED MALATE TRANSPORTER gene (AcALMT1) was identified as a candidate responsible for citrate accumulation. Electrophysiological assays using expression of this gene in Xenopus oocytes revealed that AcALMT1 functions as a citrate transporter. Additionally, transient overexpression of AcALMT1 in kiwifruit significantly increased citrate content, while tissues showing higher AcALMT1 expression accumulated more citrate. The expression of AcALMT1 was highly correlated with 17 transcription factor candidates. However, dual‐luciferase and EMSA assays indicated that only the NAC transcription factor, AcNAC1, activated AcALMT1 expression via direct binding to its promoter. Targeted CRISPR‐Cas9‐induced mutagenesis of AcNAC1 in kiwifruit resulted in dramatic declines in citrate levels while malate and quinate levels were not substantially affected. Our findings show that transcriptional regulation of a major citrate transporter, by a NAC transcription factor, is responsible for citrate accumulation in kiwifruit, which has broad implications for other fruits and vegetables.
Keywords: Actinidia, kiwifruit, citrate, Al‐activated malate transporters (ALMTs), NAC transcriptional factor
Introduction
Organic acids are important metabolites for plant growth, stress tolerance, as well as fruit flavour (Etienne et al., 2013; Ma et al., 2001). Organic acids are formed within the tricarboxylic acid (TCA) cycle, where they contribute to the biosynthesis of numerous primary metabolites such as amino acids and supply fixed carbon for redox equilibrium and energy balance (Igamberdiev and Eprintsev, 2016). Malate and citrate are the major determinants of fruit acidity representing vital organoleptic quality that contributes to fruit flavour (Etienne et al., 2013). Alterations in the fruit acidity are mainly due to the malate and citrate metabolism influenced by several systems, including synthesis, degradation and vacuole storage (Batista‐Silva et al., 2018; Etienne et al., 2013).
With advances in high‐throughput technologies, the genetic and biochemical basis of flavour traits have been characterized using abundant accessions of tomato (Tieman et al., 2017; Ye et al., 2017), peach (Li et al., 2019; Rawandoozi et al., 2020; Yu, Guan, et al., 2021), grapevine (Liang et al., 2019), pear (Wu et al., 2018), orange (Wang et al., 2021) and apple (Jia et al., 2018; Liao et al., 2021). Although some QTLs and candidate loci have been reported to be associated with fruit flavour traits, only a few genes have been functionally validated. This is often due to the difficulties in stable transformation in many of the aforementioned fruit. Despite this, PpALMT1 was identified to contribute to malate accumulation by using transient overexpression in peach mesocarp tissues (Yu, Guan, et al., 2021). The function of MdALMTII in regulating malate content was verified in transgenic apple calli (Jia et al., 2018). Furthermore, transient repression or overexpression of Ma1 resulted in altered malate and citrate content in apple fruit (Liao et al., 2021). CRISPR‐Cas9‐engineered mutations in SlALMT9 resulted in the reduction of tomato fruit malate content (Ye et al., 2017). Moreover, transcription factors (TFs) have been reported to be involved in organic acid accumulation, including ERF (Li et al., 2016), MYB (Hu et al., 2016, 2017), bHLH (Yu, Gu, et al., 2021), WRKY (Ye et al., 2017; Zhang et al., 2022) and AREB (Bastías et al., 2011) TF families.
Kiwifruit (Actinidia spp.) is an important source of organic acids, sugars and antioxidant nutrients. Quinate metabolism and biosynthesis‐related genes have been investigated in kiwifruit (Marsh et al., 2009; Wang, Shu, et al., 2022), with expression of DAHPS, DHQS and QDH showing correlations with quinate metabolism. It is notable that citrate is a major factor contributing to flavour and makes up a large proportion of organic acid in mature kiwifruit (Marsh et al., 2009). Citrate is accumulated throughout the fruit development, which determines fruit acidity at harvest time and effects consumer preference. Understanding the regulation mechanism of citrate content is an important step forward in improvement of the kiwifruit flavour. Recently, a rapid cycling and continuously flowering kiwifruit has been generated by mutation of kiwifruit CENTRORADIALIS genes (Varkonyi‐Gasic et al., 2019). This has made it more practical to investigate specific gene functions.
In this study, our aim was to investigate the key genes that contribute to kiwifruit citrate alteration. Based on co‐expression network analysis, AcALMT1 expression was highly correlated with citrate profiles during fruit development. We found that AcALMT1 functions as a citrate transporter having an important role in the regulation of citrate content. The AcALMT1 promoter was used as a target to screen transcriptional regulators, which identified AcNAC1 as an activator. A correlation between NAC1 expression and citrate content was found in different tissues and cultivars of kiwifruit, and its homologous gene Solyc07g063420 in tomato also exhibited a significant association with citrate content in tomato accessions. Furthermore, mutagenesis of AcNAC1 resulted in a dramatic reduction in citrate content and AcALMT1 expression during kiwifruit development, suggesting AcNAC1 is a switch that controls citrate accumulation in kiwifruit.
Results
Dynamic profiles of organic acids during development of kiwifruit of two different cultivars
To explore the dynamics of organic acid contents during the development of kiwifruit, nine developmental stages of A. chinensis cv Hongyang (28, 35, 42, 56, 70, 84, 112, 140 and 168 DAFB, Figure 1a) and six developmental stages of A. deliciosa cv Hayward (25, 30, 40, 70, 100 and 150 DAFB) were sampled. The contents of the three predominate organic acid components (malate, citrate and quinate) were measured. As shown in Figure 1b, malate showed the lowest levels in ‘Hongyang’ kiwifruit, which peaked at 42 DAFB (5.75 mg/g) and decreased to 2.07 mg/g at 168 DAFB. Quinate content was high (22.16 mg/g) but decreased during fruit development. In contrast, citrate content was undetectable until 42 DAFB and then exhibited a continuous accumulation pattern from 2.96 mg/g at 56 DAFB to 10.03 mg/g at 168 DAFB (Figure 1b). Similar patterns for organic acids were observed in ‘Hayward’ kiwifruit (Figure 1c), although this cultivar had a significantly higher citrate and quinate content than ‘Hongyang’ fruit.
Figure 1.
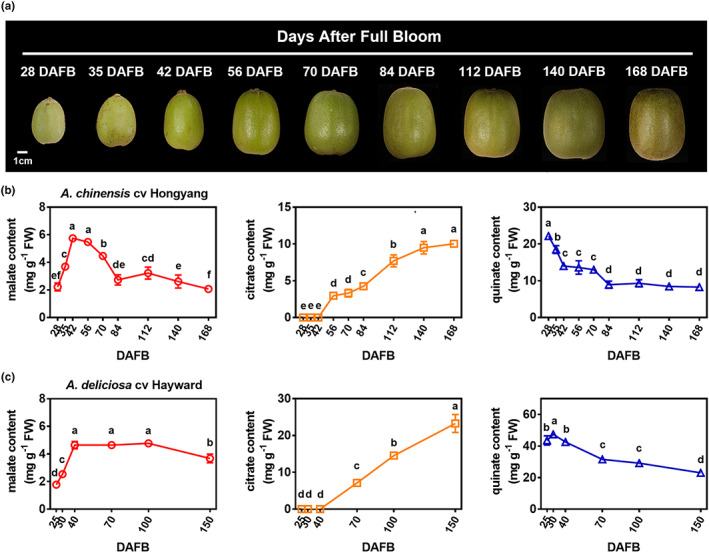
Dynamic profiles of organic acids during the development of kiwifruit. (a) Nine developmental stages of Actinidia chinensis cv Hongyang. DAFB, days after full bloom. Bar = 1 cm. (b) Citrate, malate and quinate content during the developmental stages of Actinidia chinensis cv Hongyang. (c) Citrate, malate and quinate content during the developmental stages of Actinidia deliciosa cv Hayward. Each value represents mean (±SE) of three biological replicates. Different letters indicate significant differences at P < 0.05 determined by one‐way analysis of variance (ANOVA) testing.
Identification of candidate genes associated with citrate accumulation based on RNA‐seq and co‐expression network analysis
In order to discover potential genes that control the accumulation of citrate in these kiwifruit tissues, comparative transcriptome analyses were performed on the two kiwifruit cultivars. An average of 6.31 and 6.94 gigabases (Gb) clean reads were obtained from RNA‐Seq data of ‘Hongyang’ and ‘Hayward’ by removing low‐quality reads, respectively (Dataset S1). A total of 14 680 and 16 611 genes showed differential expression between development stages. Differentially expressed genes (DEGs) with a maximum averaged FPKM from three replicates ≥1 (6470 genes from ‘Hongyang’ and 7728 genes from ‘Hayward’) were selected for WGCNA analysis (Dataset S2, S3).
Co‐expression network analysis showed that genes from ‘Hongyang’ transcriptome were comprised of 12 distinct co‐expression modules (Figure S1a), of which turquoise (R = −0.97, P = 2 × 10−5) and yellow (R = 0.97, P = 2 × 10−5) modules displayed the highest correlation with citrate content (Figure S1b). For ‘Hayward’, a total of 5 co‐expression modules were identified (Figure S1c), and modules with the highest positive and negative correlation were the blue (R = 1, P = 1 × 10−8) and turquoise (R = −0.91, P = 8 × 10−4) (Figure S1d), respectively. Intriguingly, 54 genes were simultaneously present in both kiwifruit cultivars (Figure S1e). Among those, eight genes displayed the strongest up‐ or down‐regulation during 35 DAFB to 56 DAFB with an absolute log2(Fold change) > 4 (P < 0.05). These were Acc30157 (Pleckstrin and lipid‐binding domain protein), Acc23936 (TRANSPARENT TESTA 1, zinc finger protein; involved in flavonoid regulation), Acc11477 (Transmembrane protein 50a), Acc19449 (Cyclic nucleotide‐gated ion channel), Acc19508 (Histone H2A), Acc16834 (Protein RETICULATA‐RELATED 1), Acc23343 (Heat shock transcription factor) and Acc03208 (Aluminium‐activated malate transporter family protein) (Figure S2).
Functional analysis of AcALMT1 in vivo
Based on the gene annotations of the aforementioned eight genes, an Al‐activated malate transporter (Acc03208, named AcALMT1 in the present research) was suggested to encode a malate transporter, and so might have an unexpected role in regulating kiwifruit citrate accumulation. Phylogenetic analysis indicated that AcALMT1 clustered with tomato SlALMT4 and SlALMT9, and showed close homology to Arabidopsis AtALMT3, AtALMT4, AtALMT5, AtALMT6 and AtALMT9 (Figure S3). Several members of the ALMT family have been shown to mediate malate transport (Barbier‐Brygoo et al., 2011). However, the potential function of ALMTs in transporting citrate has not been examined. Here, the electrophysiological properties of AcALMT1 were analysed by heterologous expression in Xenopus laevis oocytes, and transport function measured using the two‐electrode voltage clamp (TEVC) method. In TEVC assays with citrate supplemented in the bath solution, outward currents (anion influx) in the oocytes injected with the AcALMT1‐cRNA were higher than those in the control oocytes injected with water (Figure 2a,b). These results suggest that AcALMT1 functions as a citrate transporter to regulate citrate levels across the membrane.
Figure 2.
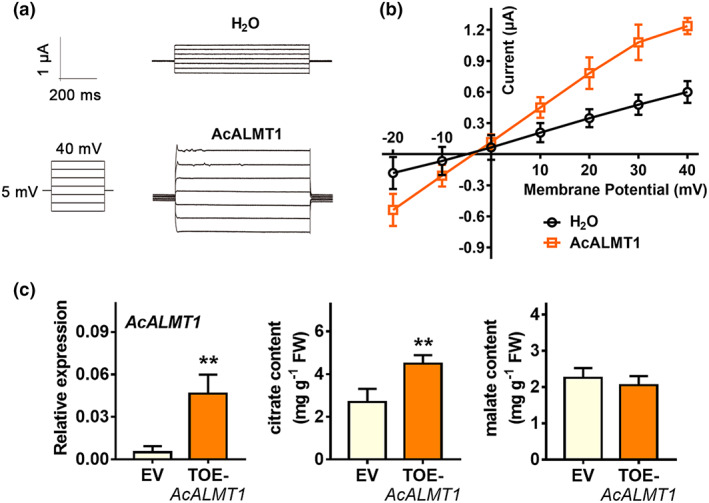
AcALMT1 functions as a citrate transporter. (a) AcALMT1 mediated citrate transport in Xenopus laevis oocyte system. Whole‐cell current recorded in X. laevis oocytes injected with H2O (control) or AcALMT1‐cRNA. The bath solution was added with citrate at a final concentration of 20 mm. The holding potential ranged from 40 to −20 mV in 10 mV increments. The time and current scale bars for the recordings are shown. (b) The current–voltage (I–V) relationship of the steady state currents were recorded in oocytes injected with H2O (control) or AcALMT1‐cRNA. Data are shown as means ± SE (n = 6). (c) A significantly elevated level of citrate content in kiwifruit core tissues by transiently over‐expressing (TOE) AcALMT1. Fruit cores transiently transformed with the empty vector were used as controls (EV). Error bars indicate SE from four biological replicates (**P < 0.01).
The in vivo function of AcALMT1 was then investigated by transient overexpression in the core tissue of A. arguta fruit (Wang et al., 2019). Fruit were infiltrated with pSAK277‐EHA105 (control) and AcALMT1‐pSAK277‐EHA105 at opposite ends of each fruit. AcALMT1 expression level was significantly increased at 2 days after infiltration with AcALMT1‐pSAK277‐EHA105. Citrate contents in the core tissue where AcALMT1 was over‐expressed were higher (1.67‐fold) than those in the empty‐vector control, while there was no significant difference in malate content (Figure 2c).
AcALMT1 expression is regulated by the transcription factor AcNAC1
To screen for possible transcriptional regulation of AcALMT1, 17 TFs were obtained from transcriptome data which showed high correlation (R > 0.9) with the expression pattern of AcALMT1 (Figure 3a). Dual‐luciferase assays of all 17 TFs indicated that the activity of the AcALMT1 promoter was significantly induced by AcNAC1 with 3.3‐fold activation (Figure 3b). Other TFs which caused changes in promoter activity included slight activation by two bHLHs (Acc04806 and Acc19740) and possible repression by a trihelix transcription factor (Acc21389). In plants, NACs were reported to directly target the core binding element, [TA]NN[CT][TCG]TNNNNNNNA[AC]GN[ACT][AT], in the promoters of target genes (Zhong et al., 2010). Four predicted binding elements were present in the AcALMT1 promoter region (P1‐P4, Figure 3c). The purified recombinant AcNAC1 protein was able to bind to probe 3 and 4 (Figure 3d), and this electrophoretic mobility shift was reduced by the addition of increasing concentrations of cold probe. However, AcNAC1 protein did not interact with probe 1 and 2 (Figure S4), which suggested that the sequence context has additional effect on binding activities. These results indicate that AcNAC1 can activate AcALMT1 and has a direct interaction with the AcALMT1 promoter.
Figure 3.
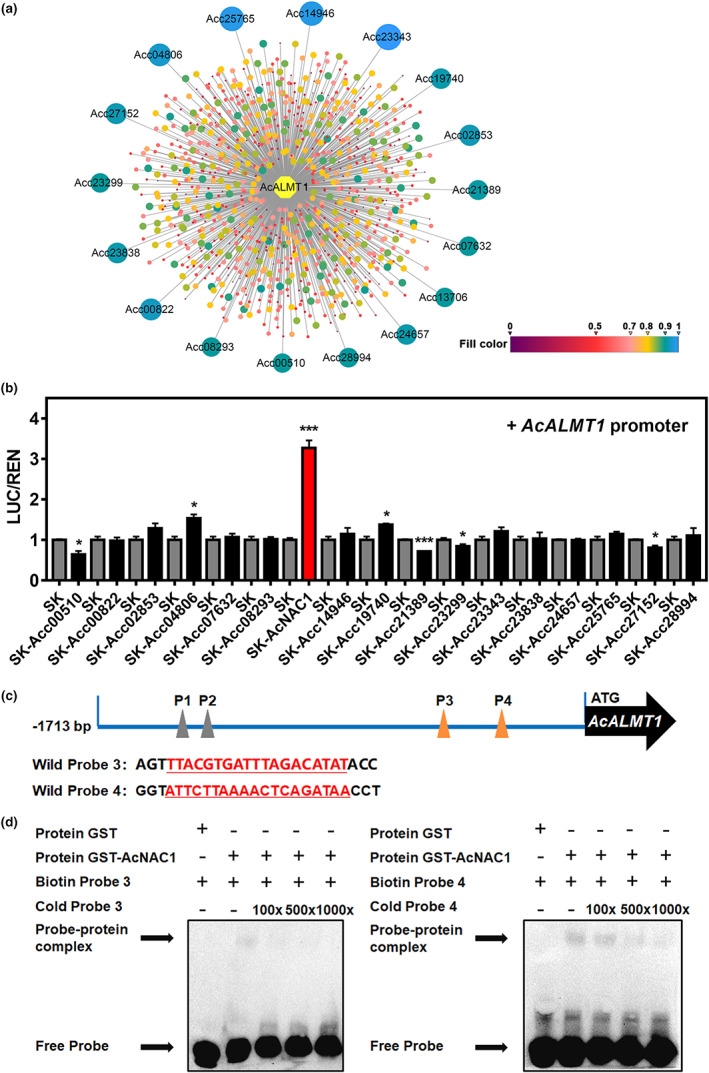
The regulatory effect of AcNAC1 on the AcALMT1 promoter. (a) Transcriptional regulatory network of AcALMT1. The network includes 961 transcription factors (TFs). Circles with different colours represent different TFs, the 17 TFs which labelled with Acc ID (Pilkington et al., 2018) represent correlations higher than 0.9. The colour scale for correlation between AcALMT1 and TFs ranged from 0 to 1. (b) The regulatory effect of AcNAC1 on AcALMT1 promoter using dual‐luciferase assays. The LUC/REN value of empty vector (SK) was set as 1. The red column highlights the effects of AcNAC1. Error bars indicate SE from five biological replicates (*P < 0.05; **P < 0.01; ***P < 0.001). (c) Oligonucleotides used for electrophoretic mobility shift assays (EMSA) with the 19‐bp NAC core sequences are underlined and marked in red. (d) EMSAs of AcNAC1 bindings to the AcALMT1 promoter. The AcNAC1 protein‐DNA complexes were separated on 6% native polyacrylamide gels. ‘−’ and ‘+’ represent absence and presence, respectively. The concentrations of cold probe were set at 100‐fold, 500‐fold and 1000‐fold excess over labelled probes.
Expression pattern of ALMT1 and NAC1 with citrate contents in different fruits
The citrate content and ALMT1/NAC1 expression levels were compared in both outer pericarp and core of A. chinensis cv Hongyang fruit (Figure 4a–d). The citrate content of the outer pericarp was significantly higher than that observed in the core (Figure 4b,d). Expression levels of AcALMT1 and AcNAC1 were up‐regulated at early developmental stages in outer pericarp (Figure 4b), while remaining relatively low in the core (Figure 4d). Moreover, differences in citrate content were also found between cultivars, A. chinensis cv Hongyang and A. deliciosa cv Hayward. In the late developmental stages of the two cultivars, more than 2‐fold more citrate was accumulated in ‘Hayward’ (23.26 mg/g at 150 DAFB) compared to ‘Hongyang’ (10.03 mg/g at 168 DAFB). The expression levels of ALMT1 and NAC1 were significantly higher in ‘Hayward’ than those in ‘Hongyang’ throughout development (Figure 4b,f). AdALMT1 and AdNAC1 exhibited similar expression patterns, with increasing expression from 25 to 70 DAFB, and then maintained at high expression level (Figure 4f). These results suggest that citrate content is highly associated with the expression of ALMT1 and NAC1.
Figure 4.
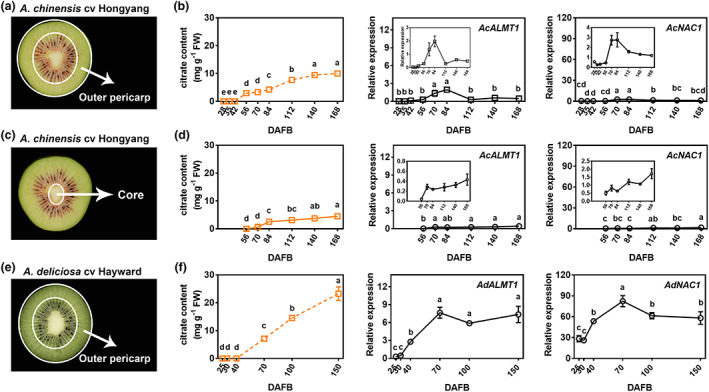
Citrate content and ALMT1/NAC1 expression patterns in different kiwifruit cultivars and tissues. (a) Transverse sections of A. chinensis cv Hongyang fruit. The tissues between the two white oval are defined as outer pericarp. (b) Citrate contents and AcNAC1/AcALMT1 relative expression of outer pericarp during the development of ‘Hongyang’ fruit. DAFB, days after full bloom. (c) Transverse sections of A. chinensis cv Hongyang fruit. The tissues inside the small white oval are defined as core. (d) Citrate contents and AcNAC1/AcALMT1 relative expression of core during the development of ‘Hongyang’ fruit. (e) Transverse sections of A. deliciosa cv Hayward fruit. The tissues between the two white oval are defined as outer pericarp. (f) Citrate contents and AdNAC1/AdALMT1 relative expression levels of outer pericarp during the development of ‘Hayward’ fruit. The dotted line diagram in (b) and (f) indicates that it has been shown in Figure 1. Each value represents mean ± SE of three biological replicates. The statistical analysis was performed using one‐way analysis of variance (ANOVA) testing.
A similar relationship between NAC and citrate content was observed in tomato. 120 accessions were divided into four groups with low malate content (LMC), high malate content (HMC), low citrate content (LCC) and high citrate content (HCC). The expression level of Solyc07g063420, the best match as an AcNAC1 homologue, was significantly associated with citrate content (P = 0.0009) (Figure S5a), while it showed no significance with malate content (Figure S5b). This suggests a correlation between the expression levels of NAC genes and citrate content, and a wider genetic basis for NAC transcription factors to regulate citrate content.
CRISPR‐Cas9 mediated mutagenesis of AcNAC1 greatly reduces citrate accumulation and AcALMT1 expression in kiwifruit
In order to evaluate the regulatory contribution of AcNAC1 on AcALMT1 expression and citrate content, AcNAC1 CRISPR‐Cas9 mutant lines were generated in RF‘DH’ (rapidly‐flowering A. chinensis cv Donghong) kiwifruit via A. tumefaciens‐mediated T‐DNA transformation. Detail of potential editing at the sites of the target guide RNAs in RF‘DH’ acnac1 was analysed using Hi‐TOM platform sequencing (Figure 5a), and showed the presence of indels in AcNAC1 in one or both target sites in three independent lines. Fruit at four developmental stages (6, 9, 13 and 17 WAP) were sampled and used for analysis (Figure 5b). Citrate contents were maintained at low levels during development of the three independent acnac1 lines, from 0.37 mg/g (6 WAP) to 2.39 mg/g (17 WAP), while the wild‐type control (RF‘DH’) showed an increasing trend and reached 11.98 mg/g at 13 WAP (Figure 5c). The levels of malate and quinate did not differ generally between wild‐type and acnac1 (Figure 5d), although at some time points there were differences. This suggests that AcNAC1 regulates citrate biosynthesis, and may indirectly affect other organic acids but to a lesser extent. To exclude the effects of potential off‐target mutations on the citrate contents of CRISPR edited lines, six potential off‐target sites were identified using the web tool CRISPR‐P (Lei et al., 2014), and sequences were obtained by Hi‐TOM platform sequencing. No mutations occurred in the potential off‐target sites in the acnac1 lines (Figure S6), indicating that the target sites are specific mutants of AcNAC1.
Figure 5.
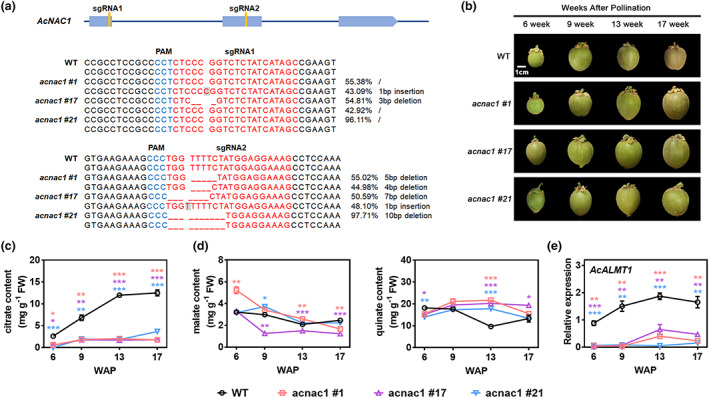
CRISPR‐Cas9 engineered AcNAC1 mutations result in reduced fruit citrate content. (a) Position of two target sites in AcNAC1 and Hi‐TOM sequencing of the CRISPR edited sites in RF‘DH’ acnac1 line #1, #17 and #21. sgRNA‐targeted sequences are highlighted in red and a protospacer adjacent motif (PAM) are shown in blue. Deletion and insertion are represented by dashed line and in grey background, respectively. Each mutation type is represented by the relative percentages. Mutation reads lower than 10% were filtered during data analysis. (b) Four developmental stages of WT (RF‘DH’) and three independent RF‘DH’ acnac1 lines #1, #17 and #21. WAP, weeks after pollination. Bar = 1 cm. (c) Dynamic profiles of citrate during four developmental stages of WT and the AcNAC1 CRIPSR/Cas9 kiwifruits. (d) Dynamic profiles of malate and quinate during four developmental stages of WT and the AcNAC1 CRIPSR/Cas9 kiwifruits. (e) The expression patterns of AcALMT1 in WT and the AcNAC1 CRIPSR/Cas9 kiwifruits. Data are presented as means ± SE of three independent biological replicates. Asterisks above bars indicate significant difference from the WT (*P < 0.05; **P < 0.01; ***P < 0.001) analysed by the Student's t‐test.
Given that mutagenesis of AcNAC1 resulted in the reduction of citrate content in the transgenic kiwifruit, we tested the expression levels of the downstream gene AcALMT1. As expected, AcALMT1 expression has dramatically decreased in the RF‘DH’ acnac1 lines compared to wild‐type (Figure 5e). The expression levels of genes related to citrate metabolism, including citrate synthase (CS), isocitrate dehydrogenase (IDH), aconitase (Aco), succinyl‐CoA synthetase (SCS) and malate dehydrogenase (MDH) were analysed with RT‐qPCR in acnac1 and wild‐type lines (Figure S7). The results showed that only a few genes exhibited significant differences at specific time points. To investigate other downstream genes of AcNAC1, RNA‐seq data was generated for three developmental stages of acnac1 lines and wild‐type. There was a total of 40 upregulated and 49 downregulated DEGs seen throughout fruit development between acnac1 lines compared to wild‐type (Figure S8). However, these genes showed no direct relationship with citrate metabolism, while there were several differentially expressed genes encoding transporters that included AcALMT1. These results suggest that AcALMT1 is the main downstream target gene of AcNAC1, which contributes to citrate accumulation in kiwifruit.
Discussion
AcALMT1 is a citrate transporter, which uncovers regulation of organic acid accumulation
Al‐activated malate transporters (ALMTs) are a family of plant‐specific anion channel proteins that are involved in a range of functions, including abiotic stress resistance, stomatal movement regulation, nutrition and ion homeostasis, fruit acidity and seed germination (Delhaize et al., 2007; Medeiros et al., 2018; Sharma et al., 2016). Over the past decade, several ALMT proteins have been characterized as anion‐selective channels that are responsible for malate influx or efflux at the plasma membrane and tonoplast in Arabidopsis (De Angeli et al., 2013; Eisenach et al., 2017; Imes et al., 2013; Kovermann et al., 2007; Meyer et al., 2010, 2011). Subsequently, apple ALMT‐like genes at the Ma locus on chromosome 16 have been reported to result in different malate content (Bai et al., 2012, 2015; Jia et al., 2018; Khan et al., 2012). In tomato, SlALMT4 and SlALMT5 proteins have been verified in malate transport activities in X. laevis oocytes (Sasaki et al., 2016). Moreover, a genome‐wide association study has characterized a strong peak of SNPs related to natural variation in malate traits near SlALMT9, and a 3‐bp indel in SlALMT9 promoter was linked to high fruit malate content (Ye et al., 2017).
Although ALMTs were generally considered as malate transporters, the accumulative reports on correlations between ALMTs and other organic acids (e.g. citrate) have led to debate on their function. Tieman et al. (2017) conducted a GWAS study using 398 modern, heirloom and wild tomatoes and relatives, and a total of 251 association signals were detected for 20 traits. A locus on LG 6 that has significant effects on the contents of malate and citrate was identified, and corresponded to a ALMT‐like gene (Solyc06g072910). Moreover, another ALMT gene on LG 1 also exhibited a strong significant association for citrate based on the meta‐analysis of 775 tomato accessions (Zhao et al., 2019). In peach, genomic analyses of 548 diverse accessions and molecular‐genetic characterizations revealed PpALMT1 as a candidate gene that contributes to fruit malate accumulation (Yu, Guan, et al., 2021). However, PpALMT9 exhibited highest expression levels in high‐acid peach cultivars which accumulate citrate throughout fruit development (Zheng et al., 2021). Association of ALMT and citrate was also found in citrus fruit (Liu, et al., 2022). Thus, regulation of ALMTs on malate and citrate may be manifested by specific isoforms for different crops.
A role for ALMTs in driving citrate content is suggested, but lacks functional verification. In tomato, overexpression of SlALMT5 markedly increased both malate and citrate content in seeds (Sasaki et al., 2016). Here, the expression level of AcALMT1 was upregulated from 56 DAFB when citrate begun to accumulate (Figure 4b). Given that AcALMT1 mediated large outward currents in X. laevis oocytes (Figure 2a,b), we hypothesized that AcALMT1 mediates citrate influx in kiwifruit, depending on the cytosolic and extracellular citrate concentration. Transient overexpression of AcALMT1 significantly increased citrate rather than malate content in kiwifruit (Figure 2c). Moreover, kiwifruit with mutated AcNAC1, an activator of AcALMT1 expression, showed greatly reduced citrate content (Figure 5c). Taken together, electrophysiological and genetic evidence affirmed the roles of AcALMT1 as an alternative citrate transporter. ALMTs were encoded by a gene family, thus, the specific members for malate and citrate regulation should be discriminated, especially in crops with abundance of different organic acids.
AcNAC1 is a transcriptional regulator of citrate content
In recent years, the regulatory effects of certain TFs on organic acid levels have been studied. The bHLH family TF, AN1, was shown to regulate the expression of PH5, which is associated with alteration in sweet orange acidity (Wang et al., 2021). CitNAC62, CitWRKY1 and CitERF6 trans‐activate the promoters of CitAco3 and CitAclα1 respectively, resulting in fruit citrate degradation (Li et al., 2017, 2020). MdMYB1/10/44/73 can bind to the promoters of vacuolar malate transport and H+‐pumping genes to regulate malate content (Hu et al., 2016, 2017; Jia et al., 2021). In this study, we screened 17 candidate kiwifruit TFs using dual luciferase assays to show that AcNAC1 had the highest activity on the AcAMLT6 promoter (Figure 3a,b). Although several TF families (e.g. WRKY (Ding et al., 2013; Ye et al., 2017), MYB (Hu et al., 2017) and bHLH (Liu, et al., 2022)) have been reported to act as transcription activators or repressors of ALMTs, our results reveal a new transcriptional activator of AcALMT1, which adds to the previously known NAC TF regulatory networks.
NAC TFs share a variety of roles in diverse biological processes, including plant growth and development, ripening and senescence (Kim et al., 2016; Liu, Li, et al., 2022; Nuruzzaman et al., 2013; Olsen et al., 2005). However, a relationship between NACs and organic acid accumulation is not clear. In Arabidopsis, the NAC TF RD26 was identified as a metabolome reprogramming regulator during senescence, and overexpression of RD26 increased higher levels of TCA cycle intermediates including malate, citrate, fumarate and 2‐oxoglutarate compared to wild‐type during extended darkness (Kamranfar et al., 2018). Seven NAC genes were differentially expressed between low‐acid and high‐acid cultivars of peach, related to malate accumulation (Zheng et al., 2021). NAC62 (MD06G1124800) was identified as one of the five genes that showed selection signatures in apple genomic regions and affected fruit acidity (Liao et al., 2021). In this study, we analysed the citrate contents of two main commercial kiwifruit cultivars, A. chinensis cv Hongyang and A. deliciosa cv Hayward. ‘Hayward’ showed significantly higher levels of citrate than that in ‘Hongyang’, and expression of NAC1 was higher (Figure 4b,f). In the core of ‘Hongyang’ fruit, which was lower in citrate content, AcNAC1 expression level was significantly lower than that in outer pericarp (Figure 4d). Moreover, the correlation of NOR‐like Solyc07g063420 expression and citrate content was found in tomato accessions (Figure 5; Tieman et al., 2017; Zhu et al., 2018). This gene is a positive regulator of ripening (Gao et al., 2018). NAC‐NOR genes in tomato has been associated with changes in titratable acidity (Adaskaveg et al., 2021) and differences in citrate content (Kumar et al., 2018), yet the molecular link between this transcription factor and a transporter of organic acids has not been determined.
In conclusion, the present study indicates that AcALMT1 is a citrate transporter in kiwifruit. AcALMT1 expression is regulated by AcNAC1, and mutagenesis of AcNAC1 by CRISPR‐Cas9 switches off kiwifruit citrate accumulation. Moreover, expression of a homologous NAC is positively correlated with citrate content in tomato. Our findings suggest a new regulatory mechanism of NAC‐ALMT in citrate accumulation, and provide a new target to alter acidity in kiwifruit and in other fruits (Figure 6). Ultimately, accumulation of citrate in Citrus will be one such target, yet transformation systems for this genus are problematic. By utilizing fast‐flowering offspring of kiwifruit we achieved functional validation quickly, with a ‘transgene‐free’ low acidity kiwifruit being another potential product.
Figure 6.
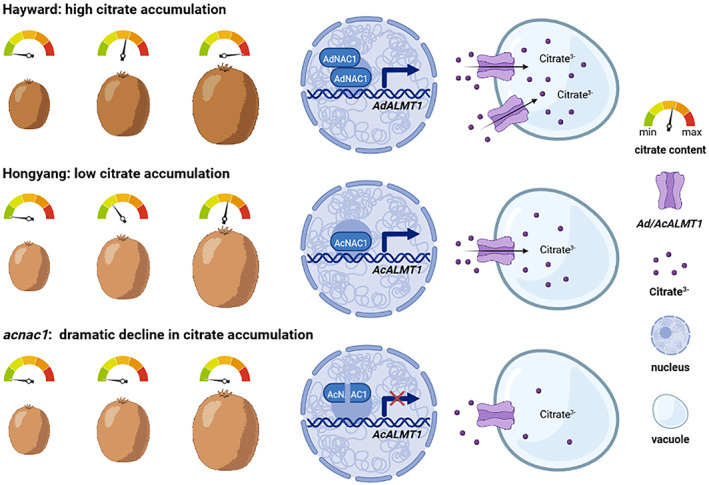
The regulatory mechanism of the transcription factor NAC1 and the citrate transporter ALMT1 during citrate accumulation in fruit. During the development of ‘Hayward’, the expression level of the transcription factor AdNAC1 is higher which activates the transcription of the citrate transporter AdALMT1, thereby increasing the citrate accumulation. Lower expression levels of AcNAC1 and AcALMT1 result in lower citrate accumulation during the development of ‘Hongyang’. After mutagenesis of AcNAC1 by CRISPR‐Cas9, acnac1 lines have a dramatic decline in expression of AcALMT1 and citrate accumulation.
Materials and methods
Plant materials and measurements
Kiwifruit (A. chinensis cv Hongyang) at different developmental stages were collected from a commercial orchard in 2019 in Zhejiang, China. Fruits at different developmental stages were collected from 28 days after full bloom (DAFB) to 168 DAFB, including 28, 35, 42, 56, 70, 84, 112, 140 and 168 DAFB. The kiwifruit (A. deliciosa cv Hayward) at different developmental stages were collected from a commercial orchard in 2018 in Shanxi, China. Fruits at different developmental stages were collected from 25 DAFB to 150 DAFB, including 25, 30, 40, 70, 100 and 150 DAFB. Eighteen fruit with similar sizes and no physical injuries were collected from five individual vines and divided into three groups. Each group containing six fruit were considered to be one biological replicate. The outer pericarp (without skin and seeds) and core of ‘Hongyang’, and outer pericarp of ‘Hayward’ were cut into small pieces, respectively, then rapidly frozen in liquid nitrogen and stored at −80°C for further use.
Determination of organic acids content
Organic acid contents of kiwifruit were measured by gas chromatography–mass spectrometry according to the method described by Wang, Shu, et al. (2022). Each experiment was carried out with three biological replicates.
RNA sequencing and analysis
Total RNA was extracted from frozen kiwifruit flesh by the cetyltrimethylammonium bromide (CTAB) method according to our previous protocol (Wang, Moss, et al., 2022). RNA‐seq were performed using samples of 35, 42 and 56 DAFB (for cv Hongyang fruit) and 30, 40 and 70 DAFB (for cv Hayward fruit), with three biological replicates. Libraries were constructed for each sample and sequenced using Illumina Novaseq 6000 sequencing platform by Biomarker (Beijing, China). The clean reads were filtered from raw data by discarding low‐quality reads (unknown nucleotides >5% or low Q‐value ≤20%) and mapped to the Red5 (A. chinensis) genome (Pilkington et al., 2018). Fragments per kilobase of transcript per million fragments mapped (FPKM) were considered as gene expression levels (Trapnell et al., 2010). The P‐values were corrected using the Benjamini‐Hochberg approach for controlling the false discovery rate. Gene functions were annotated based on the NCBI non‐redundant database (NR), the Swiss‐Prot proteins databases and the pathway databases including COG (http://www.ncbi.nlm.nih.gov/COG/), GO (http://www.geneontology.org/) and KEGG (http://www.genome.jp/kegg/). The differential expression genes (DGEs) between two samples (35 and 42 DAFB, 35 and 56 DAFB, 42 and 56 DAFB, 30 and 40 DAFB, 30 and 70 DAFB, 40 and 70 DAFB) were screened with the standard of the absolute log2(Fold change) ≥ 1. The screening was performed by Excel (Version: 2010).
Weighted gene co‐expression network analysis (WGCNA)
Co‐expression networks were constructed using the WGCNA package in R (Langfelder and Horvath, 2008). The DGEs with a maximum averaged FPKM from three replicates ≥1 were selected for the WGCNA analysis. The modules were obtained using the automatic network construction function blockwise with default settings, except that the soft threshold power was 20 (for ‘Hayward’)/16 (for ‘Hongyang’), minimum module size was 20 and the merge cut height was 0.25. The eigengene value was calculated for each module and used to evaluate the association with the organic acids content during kiwifruit development.
Electrophysiology of AcALMT1 in Xenopus laevis oocytes
For function analysis in Oocytes, full‐length sequence of AcALMT1 was cloned into pGEMHE vector (primers are listed in Table S1). cRNA was synthesized by in vitro transcription using mMESSAGE mMACHINE kit (Invitrogen, Carlsbab, CA, USA). Xenopus laevis oocytes (stage V and VI) were isolated and microinjected with 43 nL of cRNA (0.5 μg/μL) or sterile water (negative control), then incubated at 18 °C in ND96 solution (96 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1 mm CaCl2 and 10 mm HEPES, pH 7.4) supplemented with 5 mg/L gentamicin for 24 h prior to analysis. Electrophysiological measurements were performed by the two‐electrode voltage clamp, oocytes were bathed in the HMg solution (6 mm MgCl2, 1.8 mm CaCl2 and 10 mm MES, pH 6.5) with 20 mm Na‐citrate. The osmolality of all solutions was adjusted to 240–260 mOsmol/kg with D‐mannitol. Voltage test pluses were stepped over a range of −20 to 40 in 10‐mV increments. Experiments were performed three times independently. The current–voltage curves were recorded by Henry's electrophysiological suite (v. 3.5.1, University of Glasgow) and plotted in SigmaPlot (v. 14.0).
Transient overexpression in kiwifruit
Full‐length sequence of AcALMT1 was inserted into pSAK277 vectors, driven by a cauliflower mosaic virus 35S promoter (primers are listed in Table S1). Then all constructs were transformed into A. tumefaciens strain EHA105 and incubated on plate with 100 mg/L spectinomycin and 20 mg/L rifampin. A. tumefaciens cultures were suspended in infiltration buffer (10 mm MgCl2, 150 mm acetosyringone, 10 mm MES, pH 5.6) and grown to an OD600 of 1. A. arguta fruit at 110 DAFB were chosen and harvested from Zhejiang in 2021. The infiltrated procedures were based on the previous report (Zhang et al., 2018). Either 0.2 mL of control (empty pSAK277 vector) or pSAK277‐AcALMT1 were infiltrated separately into two different ends of core tissue within intact fruit, and then fruit were stored in an incubator at 25 °C for 2 days and collected for further analysis with four biological replicates.
cDNA synthesis and RT‐qPCR analysis
PrimeScript RT Reagent Kit with gDNA Eraser (RR047A; Takara, Dalian, China) was used to synthesize the first‐strand cDNA. Three biological replicates with three independent RNA extractions and cDNA synthesis were used for each sampling point. The expression abundance was investigated by RT‐qPCR, using a LightCycler 480 instrument (Roche, Mannheim, Germany) with LightCycler 480 SYBR Green I Master (Roche). The housekeeping gene Actin (GenBank no. EF063572) was chosen as the internal control and the relative expression level was analysed by the 2−∆Ct method (Schmittgen and Livak, 2008; Walton et al., 2009). The sequences of primers (designed with the Primer3 (v.0.4.0) software) are described in Table S1. Each pair of primers was checked by melting curves and product sequencing to ensure their quality and specificity.
Dual‐luciferase assays
Dual‐luciferase assays were used to screen the effective TFs on the AcALMT1 promoter. The full coding DNA sequence of each TF was cloned into the pGreenII 002962‐SK vector, while the promoter fragment of AcALMT1 was cloned into the pGreenII 0800‐LUC vector (primers are listed in Table S1). The above constructs were transferred into A. tumefaciens (strain GV 3101). The prepared A. tumefaciens mixtures of TF and promoter in a ratio of 10: 1 (v/v) were infiltrated into 5‐week‐old N. benthamiana leaves. Luciferase (LUC) and Renilla (REN) luciferase activities were measured after 3 days of infiltration using the dual‐luciferase assay kit (Promega, Wisconsin, USA) on the Promega, Fitchburg, Wisconsin, USA GLOMAX 96 Microplate Luminometer. The regulatory effects of TFs on the AcALMT1 promoter were calculated as the LUC/REN ratio, and the LUC/REN value of the empty vector SK on AcALMT1 promoter was used as a calibrator (set as 1). Three biological replicates were used for each assessment.
Protein extraction and electrophoretic mobility shift assay (EMSA)
The full‐length coding sequence of AcNAC1 was inserted into pGEX‐4T‐1 vector to generate the recombination N‐terminal GST‐tagged protein (primers are listed in Table S1). The construct was sequenced and transformed into Escherichia coli strain Rosetta (DE3). The purification of AcNAC1‐GST was conducted using the GST‐tag Protein Purification Kit (Beyotime, Shanghai, China) for use in further EMSA experiments.
The 25 bp oligonucleotide probes containing NAC‐specific cis‐elements derived from the AcALMT1 promoter were synthesized and labelled with biotin at the 3′ end by HuaGene and dsDNA probes prepared by annealing complementary oligonucleotides. The probes are listed in Table S1. EMSA were performed using a LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer's instructions. The GST protein alone was set as a negative control and the unlabelled probe as the competitor.
Generation of CRISPR‐Cas9 knockout kiwifruit
The construct targeting the first and last exon in CEN and CEN4 was used to generate rapidly flowering A. chinensis cv Donghong (RF‘DH’) (Varkonyi‐Gasic et al., 2019). The simultaneous targeting of AcNAC1 and CEN/CEN4 was followed by the protocol from Varkonyi‐Gasic et al. (2021). Two AcNAC1‐specific guides were combined with two guides from the first and last exon of CEN/CEN4. The Arabidopsis U6‐26 promoter drives the polycistronic transfer RNA (tRNA)‐guide RNA (gRNA) (PTG) cassette, with Gateway recombination sites added on each end was synthesized, and then recombined with the vector pDE‐KRS (Varkonyi‐Gasic et al., 2019). The constructs were transformed into Agrobacterium tumefaciens strain EHA105 by electroporation. A. chinensis cv Donghong tissue cultured leaves were used for transformation (Fu et al., 2021), and grown on media supplemented with 50 μg/mL kanamycin for selection. The rooted transgenic plants were potted and grown in a glasshouse at Zhejiang University, Zhejiang, China (light : dark = 15 h : 9 h, 24 °C). Pollination was carried out at full bloom with stored pollen using a paintbrush. The kiwifruit of RF‘DH’ and RF‘DH’ acnac1 lines were harvest at four different development stages (6, 9, 13 and 17 weeks after pollination) with three biological replicates.
Analysis of target and potential off‐target mutations
The potential off‐target sites of the target sequences were predicted with the web tool CRISPR‐P (Lei et al., 2014). The potential off‐target sites containing 0–4 bp mismatch compared with the target sequence were selected for further analysis. For the detection of target and potential off‐target mutation sites in RF‘DH’ acnac1 lines, the target regions were amplified from genomic DNA of RF‘DH’ and RF‘DH’ acnac1 lines using site‐specific primers (primers are listed in Table S1). PCR products were sequenced using Hi‐TOM platform (China National Rice Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou) (Liu et al., 2019). Mutation reads lower than 10% were filtered during data analysis.
Accession numbers
The transcriptome data generated in this study are available at NCBI database with the BioProject ID PRJNA721028 and PRJNA783998. Sequence data are available at GenBank with the accession numbers AcALMT1, OP178892; AcNAC1, KF319050.
Author contributions
X.Y., B.F. and W.W. conceived and designed the study; B.F. performed most experiments, with help from W.W., T.Q., Q.S., K.L and X.Liu; X.Li analysed the data. B.F. drafted the manuscript and W.W., X.Li, S.L., X.Y. and A.C.A. revised the manuscript.
Supporting information
Figure S1 Weighted correlation network analysis (WGCNA) of two kiwifruit developmental stages.
Figure S2 Identification of eight strongest up‐ or down‐regulated genes during ‘Hongyang’ fruit development.
Figure S3 Phylogenetic analysis of AcALMT1 and ALMTs from Arabidopsis thaliana, Solanum lycopersicum and Vitis vinifera.
Figure S4 Analysis of the binding ability of AcNAC1 to the promoter of AcALMT1.
Figure S5 The correlations between Solyc07g063420 and citrate/malate content in tomato populations.
Figure S6 Analysis of potential off‐target sites in WT (RF‘DH’) and three independent RF‘DH’ acnac1 mutant plants.
Figure S7 Expression profiling of tricarboxylic acid (TCA) cycle genes in WT (RF‘DH’) and acnac1 mutant kiwifruit.
Figure S8 Heatmap of up‐ and down‐regulated genes in WT (RF‘DH’) and acnac1 mutant kiwifruit based on RNA‐seq.
Table S1 Sequences of primers and other oligonucleotides used in this study.
Dataset S1 Throughput and qualities of ‘Hongyang’ and ‘Hayward’ RNA‐Seq data.
Dataset S2 Summary of co‐expression gene modules of ‘Hongyang’ transcriptome.
Dataset S3 Summary of co‐expression gene modules of ‘Hayward’ transcriptome.
Acknowledgements
We thank Prof. Yi‐zhou Wang (Zhejiang University, China) for help with electrophysiological experiments, and thank Prof. Ming‐jun Li (Northwest A&F University, China) for providing the sample of ‘Hayward’ kiwifruit. This research was supported by the National Key Research and Development Program (2018YFD1000200), the National Natural Science Foundation of China (32072635; 32102344), the Key Research and Development Program of Zhejiang Province (2021C02015), China Postdoctoral Science Foundation (2020107), Fruit New Varieties Breeding Project of Zhejiang Province (2021C02066‐8), Fundamental Research Funds for the Central Universities, China (226‐2022‐00152; 226‐2022‐00215).
References
- Adaskaveg, J.A. , Silva, C.J. , Huang, P. and Blanco‐Ulate, B. (2021) Single and double mutations in tomato ripening transcription factors have distinct effects on fruit development and quality traits. Front. Plant Sci. 12, 647035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y. , Dougherty, L. , Li, M. , Fazio, G. , Cheng, L. and Xu, K. (2012) A natural mutation‐led truncation in one of the two aluminum‐activated malate transporter‐like genes at the Ma locus is associated with low fruit acidity in apple. Mol. Gen. Genomics 287, 663–678. [DOI] [PubMed] [Google Scholar]
- Bai, Y. , Dougherty, L. , Cheng, L. , Zhong, G.Y. and Xu, K. (2015) Uncovering co‐expression gene network modules regulating fruit acidity in diverse apples. BMC Genomics 16, 612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier‐Brygoo, H. , De Angeli, A. , Filleur, S. , Frachisse, J.M. , Gambale, F. , Thomine, S. and Wege, S. (2011) Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu. Rev. Plant Biol. 62, 25–51. [DOI] [PubMed] [Google Scholar]
- Bastías, A. , López‐Climent, M. , Valcárcel, M. , Rosello, S. , Gómez‐Cadenas, A. and Casaretto, J.A. (2011) Modulation of organic acids and sugar content in tomato fruits by an abscisic acid‐regulated transcription factor. Physiol. Plant. 141, 215–226. [DOI] [PubMed] [Google Scholar]
- Batista‐Silva, W. , Nascimento, V.L. , Medeiros, D.B. , Nunes‐Nesi, A. , Ribeiro, D.M. , Zsogon, A. and Araujo, W.L. (2018) Modifications in organic acid profiles during fruit development and ripening: correlation or causation? Front. Plant Sci. 9, 1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angeli, A. , Zhang, J. , Meyer, S. and Martinoia, E. (2013) AtALMT9 is a malate‐activated vacuolar chloride channel required for stomatal opening in Arabidopsis . Nat. Commun. 4, 1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize, E. , Gruber, B.D. and Ryan, P.R. (2007) The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Lett. 581, 2255–2262. [DOI] [PubMed] [Google Scholar]
- Ding, Z.J. , Yan, J.Y. , Xu, X.Y. , Li, G.X. and Zheng, S.J. (2013) WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum‐induced malate secretion in Arabidopsis. Plant J. 76, 825–835. [DOI] [PubMed] [Google Scholar]
- Eisenach, C. , Baetz, U. , Huck, N.V. , Zhang, J. , De Angeli, A. , Beckers, G.J.M. and Martinoia, E. (2017) ABA‐induced stomatal closure involves ALMT4, a phosphorylation‐dependent vacuolar anion channel of Arabidopsis. Plant Cell 29, 2552–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne, A. , Genard, M. , Lobit, P. , Mbeguie, A.M.D. and Bugaud, C. (2013) What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J. Exp. Bot. 64, 1451–1469. [DOI] [PubMed] [Google Scholar]
- Fu, B.L. , Wang, W.Q. , Liu, X.F. , Duan, X.W. , Allan, A.C. , Grierson, D. and Yin, X.R. (2021) An ethylene‐hypersensitive methionine sulfoxide reductase regulated by NAC transcription factors increases methionine pool size and ethylene production during kiwifruit ripening. New Phytol. 232, 237–251. [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Wei, W. , Zhao, X. , Tan, X. , Fan, Z. , Zhang, Y. , Jing, Y. et al. (2018) A NAC transcription factor, NOR‐like1, is a new positive regulator of tomato fruit ripening. Hortic Res 5, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, D. , Sun, C. , Ma, Q. , You, C. , Cheng, L. and Hao, Y. (2016) MdMYB1 regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples. Plant Physiol. 170, 1315–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, D.G. , Li, Y.Y. , Zhang, Q.Y. , Li, M. , Sun, C.H. , Yu, J.Q. and Hao, Y.J. (2017) The R2R3‐MYB transcription factor MdMYB73 is involved in malate accumulation and vacuolar acidification in apple. Plant J. 91, 443–454. [DOI] [PubMed] [Google Scholar]
- Igamberdiev, A.U. and Eprintsev, A.T. (2016) Organic acids: The pools of fixed carbon involved in redox regulation and energy balance in higher plants. Front. Plant Sci. 7, 1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imes, D. , Mumm, P. , Bohm, J. , Al‐Rasheid, K.A. , Marten, I. , Geiger, D. and Hedrich, R. (2013) Open stomata 1 (OST1) kinase controls R‐type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 74, 372–382. [DOI] [PubMed] [Google Scholar]
- Jia, D. , Shen, F. , Wang, Y. , Wu, T. , Xu, X. , Zhang, X. and Han, Z. (2018) Apple fruit acidity is genetically diversified by natural variations in three hierarchical epistatic genes: MdSAUR37, MdPP2CH and MdALMTII . Plant J. 95, 427–443. [DOI] [PubMed] [Google Scholar]
- Jia, D. , Wu, P. , Shen, F. , Li, W. , Zheng, X. , Wang, Y. , Yuan, Y. et al. (2021) Genetic variation in the promoter of an R2R3‐MYB transcription factor determines fruit malate content in apple (Malus domestica Borkh.). Plant Physiol. 186, 549–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamranfar, I. , Xue, G.P. , Tohge, T. , Sedaghatmehr, M. , Fernie, A.R. , Balazadeh, S. and Mueller‐Roeber, B. (2018) Transcription factor RD26 is a key regulator of metabolic reprogramming during dark‐induced senescence. New Phytol. 218, 1543–1557. [DOI] [PubMed] [Google Scholar]
- Khan, S.A. , Beekwilder, J. , Schaart, J.G. , Mumm, R. , Soriano, J.M. , Jacobsen, E. and Schouten, H.J. (2012) Differences in acidity of apples are probably mainly caused by a malic acid transporter gene on LG16. Tree Genet. Genomes 9, 475–487. [Google Scholar]
- Kim, H.J. , Nam, H.G. and Lim, P.O. (2016) Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56. [DOI] [PubMed] [Google Scholar]
- Kovermann, P. , Meyer, S. , Hortensteiner, S. , Picco, C. , Scholz‐Starke, J. , Ravera, S. , Lee, Y. et al. (2007) The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 52, 1169–1180. [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Tamboli, V. , Sharma, R. and Sreelakshmi, Y. (2018) NAC‐NOR mutations in tomato Penjar accessions attenuate multiple metabolic processes and prolong the fruit shelf life. Food Chem. 259, 234–244. [DOI] [PubMed] [Google Scholar]
- Langfelder, P. and Horvath, S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Y. , Lu, L. , Liu, H.Y. , Li, S. , Xing, F. and Chen, L.L. (2014) CRISPR‐P: a web tool for synthetic single‐guide RNA design of CRISPR‐system in plants. Mol. Plant 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li, S.J. , Yin, X.R. , Xie, X.L. , Allan, A.C. , Ge, H. , Shen, S.L. and Chen, K.S. (2016) The Citrus transcription factor, CitERF13, regulates citric acid accumulation via a protein‐protein interaction with the vacuolar proton pump, CitVHA‐c4. Sci. Rep. 6, 20151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Yin, X. , Wang, W. , Liu, X. , Zhang, B. and Chen, K. (2017) Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up‐regulation of CitAco3 . J. Exp. Bot. 68, 3419–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Cao, K. , Zhu, G. , Fang, W. , Chen, C. , Wang, X. , Zhao, P. et al. (2019) Genomic analyses of an extensive collection of wild and cultivated accessions provide new insights into peach breeding history. Genome Biol. 20, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.J. , Wang, W.L. , Ma, Y.C. , Liu, S.C. , Grierson, D. , Yin, X.R. and Chen, K.S. (2020) Citrus CitERF6 contributes to citric acid degradation via upregulation of CitAclalpha1, encoding ATP‐citrate lyase subunit alpha . J. Agric. Food Chem. 68, 10081–10087. [DOI] [PubMed] [Google Scholar]
- Liang, Z. , Duan, S. , Sheng, J. , Zhu, S. , Ni, X. , Shao, J. , Liu, C. et al. (2019) Whole‐genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat. Commun. 10, 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, L. , Zhang, W. , Zhang, B. , Fang, T. , Wang, X.F. , Cai, Y. , Ogutu, C. et al. (2021) Unraveling a genetic roadmap for improved taste in the domesticated apple. Mol. Plant 14, 1454–1471. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Wang, C. , Jiao, X. , Zhang, H. , Song, L. , Li, Y. , Gao, C. et al. (2019) Hi‐TOM: a platform for high‐throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7. [DOI] [PubMed] [Google Scholar]
- Liu, G.S. , Li, H.L. , Grierson, D. and Fu, D.Q. (2022) NAC transcription factor family regulation of fruit ripening and quality: A review. Cell 11, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Liu, X. , Gou, B. , Wang, D. , Liu, C. , Sun, J. , Yin, X. et al. (2022) The interaction between CitMYB52 and CitbHLH2 negatively regulates citrate accumulation by activating CitALMT in citrus fruit. Front. Plant Sci. 13, 848869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.F. , Ryan, P.R. and Delhaize, E. (2001) Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 6, 273–278. [DOI] [PubMed] [Google Scholar]
- Marsh, K.B. , Boldingh, H.L. , Shilton, R.S. and Laing, W.A. (2009) Changes in quinic acid metabolism during fruit development in three kiwifruit species. Funct. Plant Biol. 36, 463–470. [DOI] [PubMed] [Google Scholar]
- Medeiros, D.B. , Fernie, A.R. and Araujo, W.L. (2018) Discriminating the function(s) of guard cell ALMT channels. Trends Plant Sci. 23, 649–651. [DOI] [PubMed] [Google Scholar]
- Meyer, S. , Mumm, P. , Imes, D. , Endler, A. , Weder, B. , Al‐Rasheid, K.A. , Geiger, D. et al. (2010) AtALMT12 represents an R‐type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 63, 1054–1062. [DOI] [PubMed] [Google Scholar]
- Meyer, S. , Scholz‐Starke, J. , De Angeli, A. , Kovermann, P. , Burla, B. , Gambale, F. and Martinoia, E. (2011) Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J. 67, 247–257. [DOI] [PubMed] [Google Scholar]
- Nuruzzaman, M. , Sharoni, A.M. and Kikuchi, S. (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, A.N. , Ernst, H.A. , Leggio, L.L. and Skriver, K. (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. [DOI] [PubMed] [Google Scholar]
- Pilkington, S.M. , Crowhurst, R. , Hilario, E. , Nardozza, S. , Fraser, L. , Peng, Y. , Gunaseelan, K. et al. (2018) A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genomics 19, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawandoozi, Z.J. , Hartmann, T.P. , Carpenedo, S. , Gasic, K. , da Silva, L.C. , Cai, L. , Van de Weg, E. et al. (2020) Identification and characterization of QTLs for fruit quality traits in peach through a multi‐family approach. BMC Genomics 21, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, T. , Tsuchiya, Y. , Ariyoshi, M. , Nakano, R. , Ushijima, K. , Kubo, Y. , Mori, I.C. et al. (2016) Two members of the aluminum‐activated malate transporter family, SlALMT4 and SlALMT5, are expressed during fruit development, and the overexpression of SlALMT5 alters organic acid contents in seeds in tomato (Solanum lycopersicum). Plant Cell Physiol. 57, 2367–2379. [DOI] [PubMed] [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Sharma, T. , Dreyer, I. , Kochian, L. and Pineros, M.A. (2016) The ALMT family of organic acid transporters in plants and their involvement in detoxification and nutrient security. Front. Plant Sci. 7, 1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman, D. , Zhu, G. , Resende, M.F., Jr. , Lin, T. , Nguyen, C. , Bies, D. , Rambla, J.L. et al. (2017) A chemical genetic roadmap to improved tomato flavor. Science 355, 391–394. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Williams, B.A. , Pertea, G. , Mortazavi, A. , Kwan, G. , van Baren, M.J. , Salzberg, S.L. et al. (2010) Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi‐Gasic, E. , Wang, T. , Voogd, C. , Jeon, S. , Drummond, R.S.M. , Gleave, A.P. and Allan, A.C. (2019) Mutagenesis of kiwifruit CENTRORADIALIS‐like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnol. J. 17, 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi‐Gasic, E. , Wang, T. , Cooney, J. , Jeon, S. , Voogd, C. , Douglas, M.J. , Pilkington, S.M. et al. (2021) Shy Girl, a kiwifruit suppressor of feminization, restricts gynoecium development via regulation of cytokinin metabolism and signalling. New Phytol. 230, 1461–1475. [DOI] [PubMed] [Google Scholar]
- Walton, E.F. , Wu, R.M. , Richardson, A.C. , Davy, M. , Hellens, R.P. , Thodey, K. , Janssen, B.J. et al. (2009) A rapid transcriptional activation is induced by the dormancy‐breaking chemical hydrogen cyanamide in kiwifruit (Actinidia deliciosa) buds. J. Exp. Bot. 60, 3835–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Tang, W. , Hu, Y. , Zhang, Y. , Sun, J. , Guo, X. , Lu, H. et al. (2019) A MYB/bHLH complex regulates tissue‐specific anthocyanin biosynthesis in the inner pericarp of red‐centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J 99, 359–378. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Huang, Y. , Liu, Z. , He, J. , Jiang, X. , He, F. , Lu, Z. et al. (2021) Somatic variations led to the selection of acidic and acidless orange cultivars. Nat Plants 7, 954–965. [DOI] [PubMed] [Google Scholar]
- Wang, R. , Shu, P. , Zhang, C. , Zhang, J. , Chen, Y. , Zhang, Y. , Du, K. et al. (2022) Integrative analyses of metabolome and genome‐wide transcriptome reveal the regulatory network governing flavor formation in kiwifruit (Actinidia chinensis). New Phytol. 233, 373–389. [DOI] [PubMed] [Google Scholar]
- Wang, W.Q. , Moss, S.M.A. , Zeng, L. , Espley, R.V. , Wang, T. , Lin‐Wang, K. , Fu, B.L. et al. (2022) The red flesh of kiwifruit is differentially controlled by specific activation‐repression systems. New Phytol. 235, 630–645. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Wang, Y. , Xu, J. , Korban, S.S. , Fei, Z. , Tao, S. , Ming, R. et al. (2018) Diversification and independent domestication of Asian and European pears. Genome Biol. 19, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J. , Wang, X. , Hu, T. , Zhang, F. , Wang, B. , Li, C. , Yang, T. et al. (2017) An inDel in the promoter of Al‐ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell 29, 2249–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.Q. , Gu, K.D. , Sun, C.H. , Zhang, Q.Y. , Wang, J.H. , Ma, F.F. , You, C.X. et al. (2021) The apple bHLH transcription factor MdbHLH3 functions in determining the fruit carbohydrates and malate. Plant Biotechnol. J. 19, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Guan, J. , Xu, Y. , Ren, F. , Zhang, Z. , Yan, J. , Fu, J. et al. (2021) Population‐scale peach genome analyses unravel selection patterns and biochemical basis underlying fruit flavor. Nat. Commun. 12, 3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A.D. , Wang, W.Q. , Tong, Y. , Li, M.J. , Grierson, D. , Ferguson, I. , Chen, K.S. et al. (2018) Transcriptome analysis identifies a zinc finger protein regulating starch degradation in kiwifruit. Plant Physiol. 178, 850–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Ma, B. , Wang, C. , Chen, X. , Ruan, Y.L. , Yuan, Y. , Ma, F. et al. (2022) MdWRKY126 modulates malate accumulation in apple fruit by regulating cytosolic malate dehydrogenase (MdMDH5). Plant Physiol. 188, 2059–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Sauvage, C. , Zhao, J. , Bitton, F. , Bauchet, G. , Liu, D. , Huang, S. et al. (2019) Meta‐analysis of genome‐wide association studies provides insights into genetic control of tomato flavor. Nat. Commun. 10, 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, B. , Zhao, L. , Jiang, X. , Cherono, S. , Liu, J. , Ogutu, C. , Ntini, C. et al. (2021) Assessment of organic acid accumulation and its related genes in peach. Food Chem. 334, 127567. [DOI] [PubMed] [Google Scholar]
- Zhong, R. , Lee, C. and Ye, Z.H. (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis . Mol. Plant 3, 1087–1103. [DOI] [PubMed] [Google Scholar]
- Zhu, G. , Wang, S. , Huang, Z. , Zhang, S. , Liao, Q. , Zhang, C. , Lin, T. et al. (2018) Rewiring of the fruit metabolome in tomato breeding. Cell 172, 249–261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Weighted correlation network analysis (WGCNA) of two kiwifruit developmental stages.
Figure S2 Identification of eight strongest up‐ or down‐regulated genes during ‘Hongyang’ fruit development.
Figure S3 Phylogenetic analysis of AcALMT1 and ALMTs from Arabidopsis thaliana, Solanum lycopersicum and Vitis vinifera.
Figure S4 Analysis of the binding ability of AcNAC1 to the promoter of AcALMT1.
Figure S5 The correlations between Solyc07g063420 and citrate/malate content in tomato populations.
Figure S6 Analysis of potential off‐target sites in WT (RF‘DH’) and three independent RF‘DH’ acnac1 mutant plants.
Figure S7 Expression profiling of tricarboxylic acid (TCA) cycle genes in WT (RF‘DH’) and acnac1 mutant kiwifruit.
Figure S8 Heatmap of up‐ and down‐regulated genes in WT (RF‘DH’) and acnac1 mutant kiwifruit based on RNA‐seq.
Table S1 Sequences of primers and other oligonucleotides used in this study.
Dataset S1 Throughput and qualities of ‘Hongyang’ and ‘Hayward’ RNA‐Seq data.
Dataset S2 Summary of co‐expression gene modules of ‘Hongyang’ transcriptome.
Dataset S3 Summary of co‐expression gene modules of ‘Hayward’ transcriptome.


