Summary
Photoperiod‐mediated flowering determines the phenological adaptability of crops including soybean (Glycine max). A genome‐wide association study (GWAS) identified a new flowering time locus, Time of flowering 13 (Tof13), which defined a gene encoding an AP2/ERF transcription factor. This new transcription factor, which we named TOE4b, is localized in the nucleus. TOE4b has been selected for soybean latitude adaptability. The existing natural variant TOE4b H4 was rare in wild soybean accessions but occurred more frequently in landraces and cultivars. Notably, TOE4b H4 improved high‐latitude adaptation of soybean to some extent. The gene‐edited TOE4b knockout mutant exhibited earlier flowering, conversely, TOE4b overexpression delayed flowering time. TOE4b is directly bound to the promoters and gene bodies of the key flowering integration factor genes FT2a and FT5a to inhibit their transcription. Importantly, TOE4b overexpression lines in field trials not only showed late flowering but also altered plant architecture, including shorter internode length, more internodes, more branches and pod number per plant, and finally boosted grain yield per plant by 60% in Guangzhou and 87% in Shijiazhuang. Our findings therefore identified TOE4b as a pleiotropic gene to increase yield potential per plant in soybean, and these results provide a promising option for breeding a soybean variety with an idealized plant architecture that promotes high yields.
Keywords: photoperiodic flowering, yield, adaptability, TOE4b, AP2/ERF transcription factor, soybean
Introduction
Soybean (Glycine max) is one of the most important vegetable oil and protein supply crops in the world, providing more than a quarter of the total protein for human consumption and animal feed (Graham and Vance, 2003). The transition from plant vegetative‐to‐reproductive development determines the length of the plant growth cycle and crop yield, which is especially important for food crops. Soybean is a typical short‐day plant and will be induced flowering when grown under short‐day (SD) photoperiods found at low‐latitude areas, for example near the equator. By contrast, a long‐day (LD) photoperiod common to high latitudes will delay the flowering time and maturity of soybean plants (Lin et al., 2021). Thus, photoperiod‐mediated flowering determines the phenological adaptability of soybean.
Soybean is a typical pod crop with a plant architecture that is unique from that of other grain crops like rice (Oryza sativa), wheat (Triticum aestivum) and maize (Zea mays). In soybean plants, bean pods grow at each node, including the nodes on the main stem and on the branches. The number of nodes and branches thus dictates the final yield. However, increasing the number of nodes along the main stem is not always ideal, as this change in plant architecture would raise plant height and can lead to lodging (bending of the stem towards the soil surface). During the history of crop breeding, the isolation of semi‐dwarf plants, which were later shown to be defective in gibberellin signalling, increased yield, resulting in the success of the Green Revolution, particularly with wheat and rice varieties (Hedden, 2003; Peng et al., 1999; Sasaki et al., 2002). However, the structural components influencing yield in soybean are more complex than those of wheat or rice. Consequently, soybean yield per unit area has yet to experience steep increases as those brought about by the Green Revolution in wheat. As a starting point, the blueprint for an ideal plant architecture has recently been proposed in soybean: optimized plant height, shorter internode length, more internodes, few or no branches, moderate pod number per node, higher podding rate, higher ratio of four seeds per pod, moderate 100‐seed weight, smaller petiole angle and shorter petiole (Liu et al., 2020). However, no genes have been identified that play a role in achieving the phenotypes of the ideal soybean plant type.
Transcription factors are essential during plant growth and development. Members of the APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) family are plant‐specific transcription factors that can be grouped into four major subfamilies: AP2, ERF, DEHYDRATION RESPONSE ELEMENT‐BINDING PROTEIN (DREB) and RELATED TO ABSCISIC ACID‐INSENSITIVE 3/Viviparous (RAV) (Riechmann and Meyerowitz, 1998). These transcription factors play multiple roles during the plant life cycle, such as the regulation of flowering time (Lauter et al., 2005; Zhao et al., 2015), flower and seed development (i.e. seed mass and seed yield) (Jofuku et al., 1994, 2005; Liu et al., 2022; Wang et al., 2022; Wei et al., 2022) and responses to biotic and environmental stress (Zhang et al., 2022; Zlobin et al., 2021). AP2/ERF transcription factors directly bind to the GCC‐box (GCCGCC), G‐box (CACGTG) and dehydration‐responsive element/C‐repeat (DRE/CRT, GCCCAC) cis‐elements to regulate the expression of their target genes (Riechmann and Meyerowitz, 1998; Wei et al., 2022). The Arabidopsis (Arabidopsis thaliana) transcriptional repressor TEMPRANILLO 1 (TEM1) from the RAV subfamily binds to a specific motif (ACACA and CACCTG) in the FLOWERING LOCUS T (FT) promoter to repress floral transition (Hu et al., 2021). Similarly, the AP2 subfamily member TARGET OF EARLY ACTIVATION TAGGED 1 (TOE1) binds to an AT‐rich element in the FT promoter to regulate flowering time (Zhang et al., 2015). AP2/ERF transcription factors also participate in phytohormone signalling pathways like those of ethylene, cytokinin, gibberellin, salicylic acid and jasmonic acid, to regulate plant physiological responses (Gautam and Nandi, 2018; Giri et al., 2014; Liu et al., 2022; Ma et al., 2020; Wang et al., 2022; Xie et al., 2019).
In this study, we identified a major locus regulating photoperiod‐mediated flowering in soybean through a genome‐wide association study (GWAS). We cloned the underlying causal gene, which belongs to the AP2/ERF family and was named TOE4b. Biochemical analysis revealed that TOE4b represses flowering time by binding to the FT2a and FT5a promoters. Furthermore, we established that two main TOE4b haplotypes, H1 and H4, were selected during soybean evolution and spreading process and that TOE4b H4 improves soybean adaptation to high latitudes to some extent. Importantly, TOE4b‐overexpressing soybean raised grain yield per plant by 60% to 87% in field trials through the regulation of yield‐related traits. Thus, our findings identify TOE4b as a new pleiotropic gene in soybean for genetic engineering and crop breeding through plant architecture improvement.
Results
Identification of a new flowering time locus by GWAS
To identify novel genes regulating the flowering time, we conducted GWAS using 449 soybean accessions collected at high latitudes. We detected two significant genomic regions on chromosomes 10 and 13 using data collected at Harbin, China, in both 2017 and 2020 (Figure 1a,b). Further analysis revealed that the strongest association single‐nucleotide polymorphism (SNP) peak on chromosome 10 is the E2 locus (Table S1 and S2), which is known to play a vital role in flowering time, especially at high latitudes (Bernard, 1971; Watanabe et al., 2011). The identification of E2 provided support for the reliability of our phenotypic and GWAS analyses.
Figure 1.
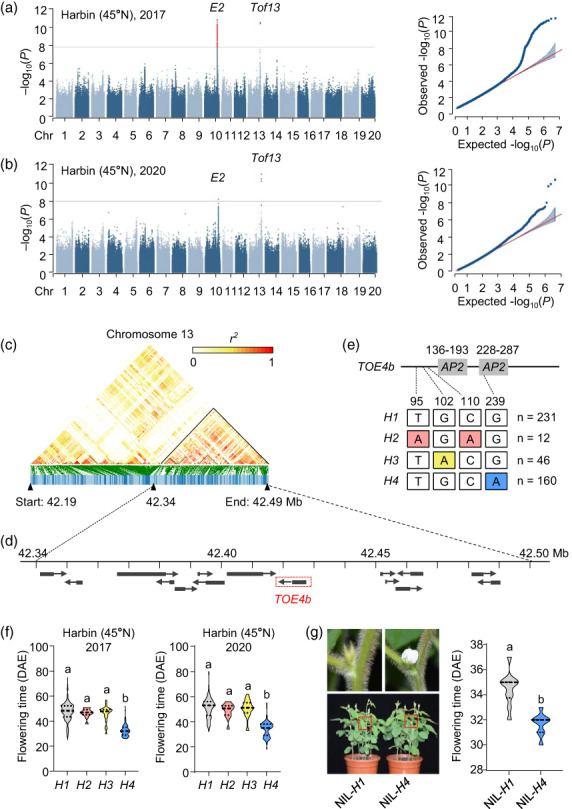
Identification and analysis of major flowering time loci in soybean. (a, b) Manhattan plot for GWAS and quantile–quantile plots using flowering time collected in Harbin, China, in 2017 (a) and 2020 (b). (c) Linkage disequilibrium analysis of SNPs surrounding Tof13. (d) Schematic diagram of the 150‐kb candidate region showing 15 predicted genes. Black boxes represent the location of genes, and arrows represent the direction of transcription. The red box indicates the candidate gene TOE4b. (e) Summary of the main haplotypes for the candidate gene TOE4b (Tof13). The above panel indicates TOE4b gene structure diagram, the numbers on the top indicate the amino acid location of the two AP2 domains. The below panel indicates the position of the amino acid corresponding to the allelic variations and the number of variations in the 449 accessions. (f) Flowering time associated with the four haplotypes at TOE4b. Data are means ± SD, P < 0.05, as determined by multiple comparison testing by one‐way ANOVA. Data are from 449 accessions grown in Harbin (45°55′ N) in 2017 and 2020 (a, b, f). (g) Flowering time phenotypes of TOE4b near isogenic line (NIL) harbouring the H1 or H4 haplotype, grown in the plant growth chamber under LD conditions (16‐h light/8‐h dark). Flowering time, scored at the R1 stage (days from emergence to the appearance of the first open flower on 50% of the plants). Data are means ± SD, n = 10, P < 0.05, two‐tailed Student's t‐test. DAE, days after emergence. Different lowercase letters indicate significant differences.
We also noticed a second strong signal located within a 463 kb region on chromosome 13 (41 879 883–42 343 731 bp) (hereafter referred to as Time of Flowering 13 [Tof13]) (Figure 1a,b; Table S1). Importantly, this region has not been previously associated with flowering time genes. To uncover the underlying causal gene, according to the genome‐wide averaged distance of linkage disequilibrium (LD) decayed (Figure S1), we performed an LD block analysis using the 150‐kb flanking regions upstream and downstream of the peak SNP (chr13: 42343731 bp). We narrowed the candidate region down to a smaller 149.8‐kb linkage region (42 343 631 bp to 42 493 444 bp) containing 15 genes (Figure 1c,d; Table S3).
Generally, plant flowering is the process of leaf‐produced signal that induces floral initiation at the shoot apex (Kong et al., 2010). Thus, we firstly focused on the transcription levels of these 15 genes in the leaves and stem tips of the wild‐type W82, according to FPKM (Fragments Per Kilobase of exon model per Million mapped reads) of the transcriptomic analysis (Table S4), combined with the data from website Phytozome 13 (https://phytozome‐next.jgi.doe.gov/). Only 5 genes (Glyma.13G328900, Glyma.13G329600, Glyma.13G329700, Glyma.13G330100 and Glyma.13G330200) were highly expressed (Figure S2). Next, according to the annotation of gene function, among these 5 genes, we noticed Glyma.13G329700, which encodes a protein belonging to the AP2/ERF transcription factor family that is an ortholog of Arabidopsis TOE1 (AtTOE1) and homologue of soybean TOE4a (Table S3). We thus referred to this gene as TOE4b thereafter. Both AtTOE1 and TOE4a are known to regulate flowering time in plants (Aukerman and Sakai, 2003; Chen, 2004; Zhao et al., 2015). Moreover, AP2‐ and AP2‐like genes perform a conserved role in controlling the vegetative‐to‐reproductive transition in other land plants (Lauter et al., 2005; Martin et al., 2009; Shim et al., 2022). Therefore, we selected TOE4b as a high‐confidence candidate gene for the Tof13 locus.
The TOE4b H4 haplotype confers early flowering
To understand the functional significance of TOE4b, we analysed the association between TOE4b haplotypes and flowering time within the 449‐accession panel. In addition to the W82 reference allele (TOE4b H1 ), we detected three nonsynonymous variants with amino acid changes in the coding region across our panel of 449 accessions (TOE4b H2 , TOE4b H3 and TOE4b H4 ) (Figure 1e). We also determined the TOE4b haplotype harboured by a set of 1744 soybean accession (Lu et al., 2020), which revealed that the H1–H4 haplotypes are by far the most prevalent (1611 out of 1744 accessions) (Figure S3a). Moreover, we established that the reference TOE4b H1 haplotype is likely the ancestral allele from which the other alleles derived (Figure S3b).
The amino acid changes caused by the TOE4b H2 and TOE4b H3 haplotypes occurred upstream of the first AP2 domain, whereas TOE4b H4 was responsible for a single amino acid substitution in the second AP2 domain (Figures 1e and S4a). Interestingly, cultivars with the H4 haplotype flowered significantly earlier than those carrying the other three haplotypes, which flowered at comparable times in both 2017 and 2020 (Figure 1f). Since the two flowering time loci identified by GWAS (Figure 1a,b), we wondered whether another locus E2 would have an effect on Tof13 (TOE4b). Therefore, we divided the accessions used for GWAS into three subgroups according to the main haplotypes of E2 (Wang et al., 2023), to analyse the function of TOE4b. The results showed that TOE4b (H1) and E2 (H1 and H3) functional allele combinations flower latest, on the contrary, both loss‐of‐function allele (TOE4b H4 and E2 H2 ) combinations flower earliest. Moreover, the effect of TOE4b H4 on flowering time is much stronger without e2, but it is not very clear under the E2 background (Figure S5).
To dissect the TOE4b function, we constructed near isogenic line (NIL) by crossing the Harosoy cultivar (harbouring TOE4b H4 ) to W82 (carrying the e2 mutation and TOE4b H1 ). Using F6 lines, we determined that NIL‐H4 flowers were 3 days earlier than NIL‐H1, which was consistent with the population genetic analysis above (Figure 1f,g). Taken together, these results suggested that TOE4b regulates flowering time.
TOE4b H4 improves soybean adaptation to high latitude
Genetic diversity across the whole genome or in specific regions is lower following domestication and crop improvement, and favourable alleles are enriched and possibly fixed for important traits (Doebley et al., 2006). Since flowering time is a key trait behind the domestication syndrome and improvement (Lu et al., 2020), which promotes soybean adaptability, we postulated that TOE4b might have been a target of selection in the process of soybean adaptation to different latitudes. To test this hypothesis, we examined the distributions of the major TOE4b alleles in the 449‐accession panel and 1295 previously sequenced accessions comprising 221 wild soybeans, 516 landraces and 874 cultivars. The TOE4b H4 allele represented only 2%, with the remaining 98% corresponding to TOE4b H1 in the wild soybean accessions. However, the TOE4b H4 frequency increased to 10% and 26% in landraces and modern cultivars, respectively, indicating that TOE4b H4 is enriched in cultivars (Figure 2a). Notably, an exploration of the geographical distribution of TOE4b H1 and TOE4b H4 in the subset of Chinese landraces and cultivars established that accessions with the early flowering allele TOE4b H4 are mainly concentrated at high latitudes, accounting for 29% of all accessions in this region. By contrast, accessions with the early flowering allele TOE4b H4 were less frequent at lower latitudes, indicating that TOE4b H4 contributed to the adaptation of cultivated soybean accessions to high latitudes to some extent (Figure 2b). Meanwhile, we estimated the Tajima's D values and the nucleotide diversity (π) for the TOE4b CDS region. The result showed that Tajima's D values for the TOE4b region were positive in cultivars and landraces but negative in wild soybean accessions (Figure 2c). The nucleotide diversity (π) in the TOE4b region was higher in wild soybean accession than in cultivars and landraces, the latter two having comparable values (Figure 2c). We expanded the 200‐kb region flanking the TOE4b gene and compared the π values between carrying TOE4b H1 and TOE4b H4 accessions. We found that the π values around the TOE4b gene of TOE4b H4 accessions are much lower than that of TOE4b H1 accessions (Figure 2d). F ST analysis showed that TOE4b exhibited a genetic differentiation tendency between TOE4b H1 and TOE4b H4 accessions (Figure 2e). Collectively, these results reveal that TOE4b may have played an important role in the domestication and adaptation of soybean. We thus turned to elucidating the function of TOE4b next.
Figure 2.
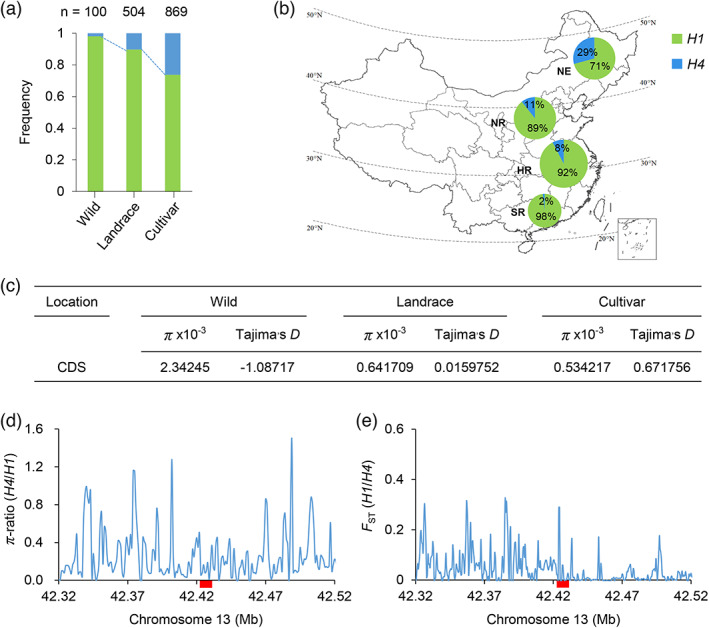
Selection of TOE4b natural variations under soybean adaptation. (a) Frequency of the H1 and H4 alleles of candidate gene TOE4b in 100 wild soybeans, 504 landraces and 869 improved cultivars. (b) Geographical distribution of 439 landraces and 555 cultivars harbouring TOE4b H1 or TOE4b H4 across China. NE, northeastern region of China; NR, northern China; HR, Huanghuai region of China; SR, southern China. The size of each pie chart on the map is proportional to the number of included accessions. (c) Nucleotide diversity (π) and Tajima's D value at TOE4b gene in 221 wild accessions, 516 landraces and 874 cultivars. (d, e) π‐ratio (d) and F ST (e) of TOE4b in 516 landraces and 874 cultivars. The red rectangles indicate the location of TOE4b.
Molecular analysis of TOE4b
To characterize the expression pattern of TOE4b in different tissues, we conducted reverse transcription quantitative PCR (RT‐qPCR) in the cultivar W82. TOE4b was highly expressed in all tested tissues except flowers and pods, suggesting that TOE4b might be involved in multiple developmental and growth stages in soybean (Figure 3a). In particular, TOE4b expression was higher in leaves and stem tips, hinting at a role in photoperiodic flowering and plant height.
Figure 3.
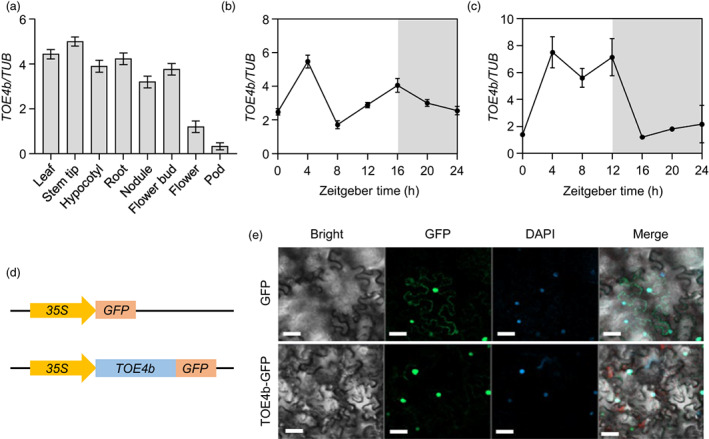
Expression pattern of TOE4b and subcellular localization of TOE4b in soybean. (a) Expression pattern of TOE4b. The plants were grown under SD (12‐h light/12‐h dark) conditions in a plant growth chamber. (b, c) Diurnal expression pattern of TOE4b in leaves under LD (16‐h light/8‐h dark) (b) and SD (12‐h light/12‐h dark) conditions (c). Three leaves were collected from different seedlings at 18 DAE. β‐Tubulin (TUB) was used as a reference transcript. The cultivar Williams 82 (W82) was used. (d) Schematic diagrams of the constructs used for subcellular localization. (e) TOE4b localizes to the nucleus. The vector control (35S:GFP) and 35S:TOE4b‐GFP were introduced into Nicotiana benthamiana leaves via Agrobacterium‐mediated infiltration. Panels from left to right show the bright field, GFP signal channel, the DAPI stain and the merged images. Scale bars, 20 μm.
We also asked whether TOE4b expression responded to photoperiod similar to other flowering‐related genes. To this end, we measured TOE4b transcript levels every 4 h over one diurnal cycle under SD or LD conditions, which showed that TOE4b exhibits a rhythmic pattern with a peak at zeitgeber 4 (ZT4, 4 h after dawn) and another one at dusk (ZT16 for LD, and ZT12 for SD) (Figure 3b,c). This bimodal expression pattern suggests that TOE4b may be involved in the regulation of the soybean photoperiodic flowering pathway.
Next, we assessed the subcellular localization of TOE4b through transient infiltration of Nicotiana benthamiana leaves. Confocal imaging showed that the TOE4b‐GFP (a fusion between TOE4b and the green fluorescent protein) is mainly distributed in the nucleus in N. benthamiana epidermal cells (Figure 3d,e). This nuclear localization was consistent with a role for TOE4b as a transcription factor.
TOE4b delays flowering time
To further characterize the TOE4b function, we generated two lines overexpressing TOE4b‐3Flag (H1 allele) with verified higher expression by RT‐qPCR and protein accumulation by immunoblotting with an anti‐Flag antibody (Figure S6). Compared to the wild‐type (W82), these overexpression lines flowered later by 3–6 days and 2–4 days under LD and SD conditions with the light‐intensity conditions 300 μmol m−2 s−1, respectively (Figure 4a,b,d,e). In particular, line OE7 displayed higher TOE4b expression than line OE2 and flowered later than line OE2. Importantly, overexpression lines also had shorter internodes and overall plant height than W82 (Figures 4a,c,d,f and S7a), which are both beneficial traits for agricultural production. In addition, the number of nodes in transgenic plants decreased slightly under artificial photoperiodic conditions (Figure S7b, d). The results showed that TOE4b could be involved in regulating soybean flowering time and plant architecture.
Figure 4.
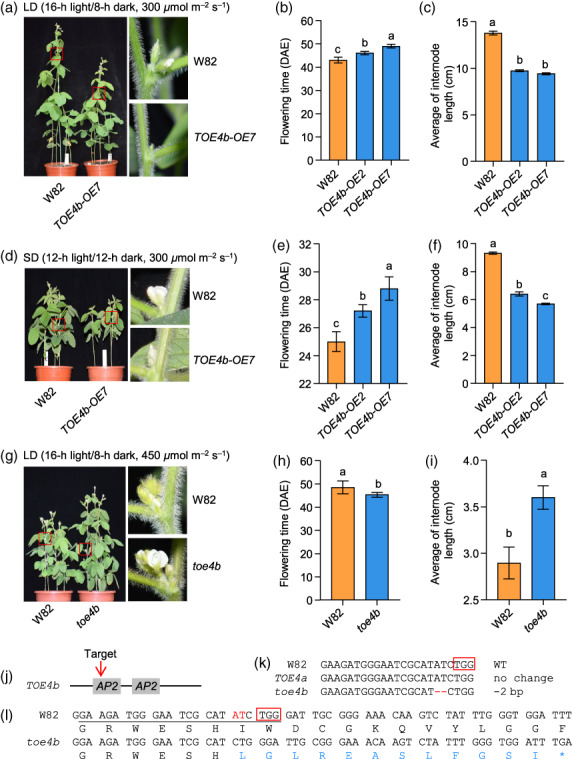
Phenotypes of TOE4b overexpression lines and mutant. (a‐f) Phenotypes of TOE4b overexpression lines. Flowering time (b, e). Average internode length (c, f), calculated as the plant height divided by the number of nodes. The plants were grown in an incubator with an average photon flux of 300 μmol m−2 s−1 under LD (16‐h light/8‐h dark) (a‐c) or SD (12‐h light/12‐h dark) (d‐f) conditions. Phenotypes were recorded at 44 DAE for LD treatment and at 25 DAE for SD treatment. (g‐i) Phenotypes of toe4b mutant under LD (16‐h light/8‐h dark) conditions. The plants were grown in an incubator with an average photon flux of 450 μmol m−2 s−1. W82, wild‐type Williams 82. The red boxes are enlarged to the right and show the axils of trifoliate leaves. Data are means ± SD, n = 5. P < 0.05, as determined by multiple comparison testing by one‐way ANOVA. Different lowercase letters indicate significant differences. (j) Schematic diagram of TOE4b and the selected target site for CRISPR/Cas9‐mediated gene editing. (k) Sequence alignment of the single‐guide RNA (sgRNA) target region in W82 and the homozygous mutant. Red dashes indicate deleted nucleotides. (l) Predicted changes in the amino acid sequence of TOE4b from wild‐type W82 and the toe4b mutant. Underlined nucleotides, sgRNA target sites; red boxes, protospacer‐adjacent motif (PAM); blue letters show the amino acids that are different from W82; asterisk, termination of translation.
To further confirm the gene function, we generated toe4b loss‐of‐function mutants using clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR‐associated nuclease 9 (Cas9)‐mediated gene editing, after testing homologous genes, including TOE4a but also TOE5a and TOE5b, and obtained a single toe4b mutant with a 2‐bp deletion near the target site of the sgRNA, causing a frameshift and premature termination of translation in the first AP2 domain (Figure 4j–l). According to the internode length of TOE4b overexpression lines, we speculated the mutant may have the opposite phenotype, that is longer internode length, which is an agricultural disadvantaged trait. Thus, we examined the mutant phenotypes under stronger light‐intensity conditions (450 μmol m−2 s−1). The results showed that the toe4b‐mutant flowers earlier than the wild‐type W82 still with longer internode lengths and plant height under LD conditions (Figures 4g,i and S7e–f). Taken together, it is revealed that TOE4b indeed delayed soybean flowering time and regulated plant internode length.
TOE4b improves soybean grain yield per plant by altering plant architecture
Given the shorter internodes of the overexpression lines when grown in incubators (Figure 4c,f), we hypothesized that TOE4b may regulate other plant architecture traits or grain yield. We thus designed field experiments in a natural LD field environment in Shijiazhuang (37°27′ N) with a density of 15 cm plant interspace. Compared with the wild‐type W82, TOE4b overexpression lines showed late flowering and moderately decreased plant height (Figure 5a–c), but more nodes (Figure 5d), so that the average internode length was shorter (Figure 5e). Even more striking is that the branches increased as well (Figure 5f), which was not observed in incubators. The increased nodes and branches contributed to the increase of pod number per plant and finally, the seed weight per plant was significantly increased (Figure 5g,h), especially in line OE7, which may be attributed to the TOE4b protein accumulation was much higher than line OE2 (Figure S6).
Figure 5.
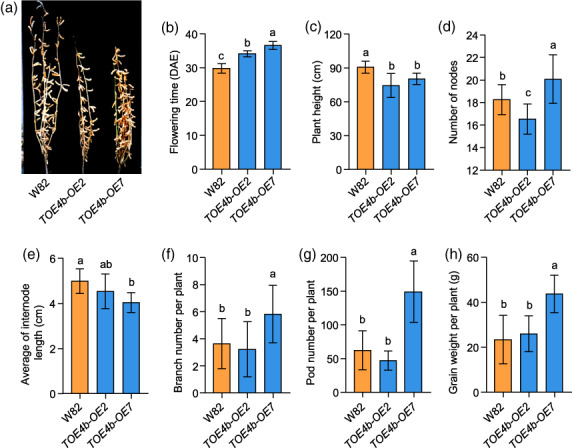
TOE4b regulates flowering time and yield per plant traits under long‐day conditions. (a) Phenotypes of TOE4b overexpression transgenic lines at the maturity stage. (b‐h) Flowering time (b), scored as days after emergence (DAE) and the agronomic traits of plant height (c), measured from the cotyledonary node of the main stem to the apex in centimetres, number of nodes (d), internode length (e), branch number per plant (f), pod number per plant (g) and grain weight per plant (h). The plants were grown in a standard field in Shijiazhuang (37°27′ N) under natural conditions in 2020. W82, wild‐type Williams 82. At least 12 plants were scored for each phenotype; data are means ± SD. P < 0.05, as determined by multiple comparison testing by one‐way ANOVA. Different lowercase letters indicate significant differences.
In 2020, with the same field design, we also tested plants grown in the SD environment of Guangzhou (23°55′ N), where is an induced photoperiod condition for soybean, that is the plants flower and mature so early that yield is extremely low. As expected, the wild‐type W82 exhibited such a phenotype. While we were surprised to discover that both two overexpression lines still flower later and have shorter internodes than wild‐type plants (Figure 6a,b,e), which were stable phenotypes and consistent with incubator‐grown plants. Overexpression lines also produced more nodes, branches and pods per plant in the field, which contributed to higher grain weight per plant compared with control plants (Figure 6d,f–h), the results were consistent with the Shijiazhuang field trial. Importantly, the plant height of overexpression lines did not increase with increasing nodes, which constitutes an ideal plant architecture for agricultural production (Figure 6c,d). Taken together, these results demonstrate that TOE4b can increase soybean grain yield per plant by regulating flowering time and optimizing plant architecture.
Figure 6.
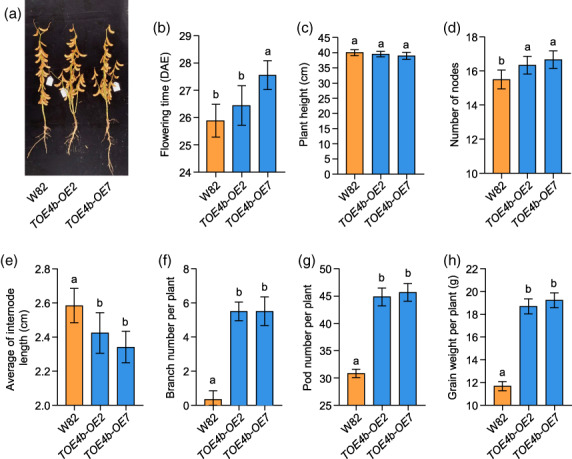
TOE4b is a positive factor in flowering time and yield per plant traits under short‐day conditions. (a) Phenotypes of TOE4b overexpression transgenic lines at the maturity stage. (b‐h) Flowering time (b), scored as days after emergence (DAE); agronomic traits of plant height (c), measured from the cotyledonary node of the main stem to the apex in centimetres; number of nodes (d); internode length (e); branch number per plant (f); pod number per plant (g); and grain weight per plant (h). The plants were grown in a standard field in Guangzhou (23°55′ N) under natural conditions in 2020. W82, wild‐type Williams 82. At least 12 plants were scored for each phenotype; data are means ± SD. P < 0.05, as determined by multiple comparison testing by one‐way ANOVA. Different lowercase letters indicate significant differences.
TOE4b directly binds to the FT2a and FT5a promoters to inhibit soybean flowering
FT is a key flowering integrator in Arabidopsis, and its soybean orthologs FT2a and FT5a coordinately control flowering and have enabled the adaptation of soybean to a wide range of photoperiodic environments (Kong et al., 2010; Li et al., 2021). Considering the late flowering time of TOE4b overexpression lines, we asked whether TOE4b controlled flowering time by regulating the expression of the key flowering integrator genes FT2a and FT5a by RT‐qPCR. We observed that FT2a and FT5a expression levels are much lower in TOE4b overexpression transgenic lines, especially in overexpression line OE7 (Figure 7a,b). Next, we conducted a dual‐luciferase transient expression assay using the FT2a or FT5a promoter cloned upstream of the firefly luciferase (LUC) reporter gene, along with an effector construct consisting of the TOE4b H1 and TOE4b H4 coding sequence driven by the 35S promoter (Figure 7c). When the reporter and TOE4b effector were co‐infiltrated into N. benthamiana leaves, FT2a and FT5a transcription were significantly repressed in the presence of TOE4b, especially the TOE4b H1 haplotype had the stronger inhibitory effect than TOE4b H4 , as evidenced by lower relative LUC activity (Figure 7e). TOE4b‐mediated repression of FT2a and FT5a transcription could explain the observed late flowering phenotypes of TOE4b overexpression lines.
Figure 7.
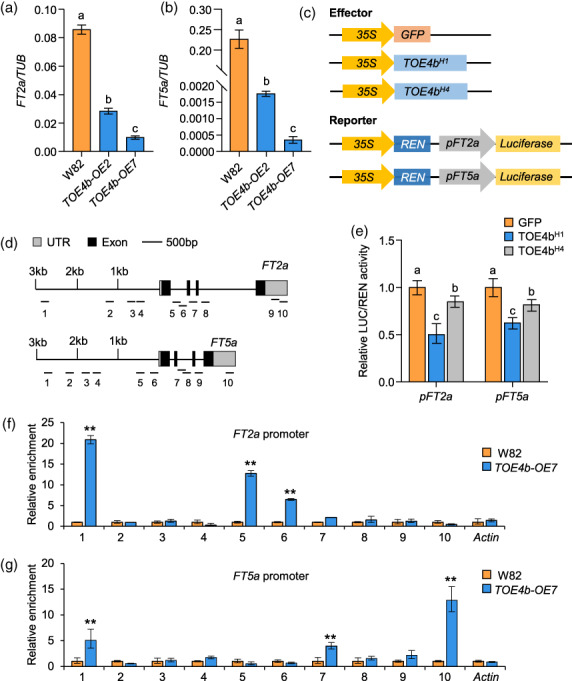
TOE4b delays flowering through direct binding to FT2a and FT5a promoters. (a, b) Relative transcript levels of FT2a (a) and FT5a (b) in wild‐type (W82) and TOE4b overexpression lines. The plants were grown in an incubator under LD conditions (16‐h light/8‐h dark). Leaves were collected at 18 DAE and at ZT4. β‐Tubulin (TUB) was used as a reference transcript. Biological triplicates were averaged. (c) Schematic diagrams of the effector constructs for TOE4b alleles and the reporter construct consisting of the FT2a and FT5a promoter driving the firefly luciferase (LUC) reporter gene for transient infiltration assay in Nicotiana benthamiana leaves. (d) Schematic diagram of the FT2a and FT5a promoter regions. (e) TOE4b alleles repress FT2a and FT5a transcription in a dual‐luciferase reporter assay in N. benthamiana. (f, g) ChIP‐qPCR assay of TOE4b binding to the FT2a (f) or FT5a (g) promoter in wild‐type W82 and the TOE4b‐OE7 line (with TOE4b fused to 3 × FLAG tag). A monoclonal anti‐FLAG antibody was used. Fold enrichment was quantified by normalizing the immunoprecipitation signal to that of the corresponding input. Actin was used as an internal control. Data are means ± SD (n = 3); statistical significance was determined by Student's t‐test (** P < 0.01).
To ascertain whether TOE4b behaved as a transcription factor and repressed FT2a and FT5a transcription directly, we performed a chromatin immunoprecipitation followed by qPCR (ChIP‐qPCR) assay using TOE4b‐OE7 transgenic lines and the wild‐type W82. We detected robust binding by TOE4b upstream of the FT2a promoter region about 3 kb upstream of the translation start site and in the first intron region; TOE4b also showed a significant association with the FT5a promoter region, one intron and the 3′ untranslated region (UTR) (Figure 7d,f,g). Collectively, these results indicate that TOE4b directly binds to the promoters and gene bodies of FT2a and FT5a in vivo and represses their transcription, resulting in delayed flowering time.
Discussion
TOE4b underwent selection in soybean latitude adaptability
We identified the Tof13 locus by GWAS using soybean populations collected from high‐latitude regions. We determined that Tof13 is TOE4b, which has abundant natural variants (Figure S3). Association analysis results showed that the loss‐of‐function allele TOE4b H4 flowered early (Figure 1f), shortened plant height and internode length, decreased nodes, branches and pods, contributing to grain weight per plant decreased, compared with the original allele TOE4b H1 (Figure S8). The results were consistent with our transgenic function verification, except for the plant height and internode length (Figures 4, 5, 6 and S8), which may be due to the difference in genetic background and population structure.
The frequency of TOE4b H4 progressively increased from wild accessions to landraces and to cultivars but was not fixed yet (Figure 2a). The geographical distribution of TOE4b H4 in 439 landraces and 555 cultivars was mostly enriched in high‐latitude regions of northeastern China, and almost absent at low latitudes (Figure 2b). Combined with the π‐ratio and F ST analysis, we propose that TOE4b H4 is a favourable allele for agriculture and may have undergone continuous selection during soybean latitude adaptability. This allele has likely been subjected to natural selection at high latitudes by phenological adaptability, and artificial selection by farmers and breeders. On the other hand, it also indicated that adaptability is selected preferentially during soybean spreading and expanding because excellent adaptability is the prerequisite to survive in nature. Therefore, TOE4b H4 may be much more important for soybean breeding in higher latitude areas, like most of Europe and North America.
Although TOE4b H4 improved high‐latitude adaptation, we speculate that the TOE4b H1 allele can also be used in low‐latitude areas, similar to our TOE4b overexpression lines in Guangzhou field tests, which showed promising higher yields. To facilitate the use of TOE4b in agriculture, we developed an effective molecular marker for this locus (Figure S4b) that can be easily used in breeding.
TOE4b regulates plant architecture and yield potential per plant in soybean
Plant architecture is a complex agronomic trait encompassing plant height, branching patterns, branch and leaf angles, inflorescence morphology and spatial root distribution (Wang and Li, 2008). The Green Revolution resulted in higher yields through the isolation of semi‐dwarf rice and wheat germplasm (Hedden, 2003; Peng et al., 1999; Sasaki et al., 2002). For instance, the ideal plant architecture gene IDEAL PLANT ARCHITECTURE 1 (IPA1) in rice controls tiller number, panicle branching, lodging resistance and grain yield (Jiao et al., 2010; Miura et al., 2010). Maize also underwent a dramatic transformation of both plant and inflorescence architecture, when compared to its wild progenitor teosinte during domestication. Teosinte branched1 (Tb1), a member of the TCP (Teosinte branched1/Cycloidea/Proliferating cell factor) family of transcriptional regulator genes, inhibits branching, resulting in the typical stalk of modern maize plants with only one or two short branches, unlike the highly branched architecture of teosinte (Studer et al., 2011).
We generated toe4b mutants as well as TOE4b overexpression lines in soybean and under low plant density field and growth chamber conditions observed that flowering time and plant architecture were both affected, especially the internode length of overexpressing lines and mutant were still fairly stable even under relatively weak and strong light‐intensity conditions respectively (Figures 4 and S7). Because of supporting stem elongation is a very energy‐intensive process and subsequently leads to lodging that can reduce soybean yield by as much as 21% to 23% (Cooper, 1971). Thus, our particular interest was the plant architecture of overexpression lines, whose later flowering, shorter internodes, more internodes and more branches led to significantly more pods per plant and contributed to a ~60% and ~87% improvement in grain weight per plant in the field experiments of Guangzhou (23°55′ N) and Shijiazhuang (37°27′ N), respectively, compared to wild‐type W82 (Figures 5 and 6). It should be noted that the yield potential per plant presented by our results was obtained at relatively low densities in the field. Our GWAS analysis, association test, NIL, mutant and overexpression lines all supported the notion that TOE4b affects flowering time and plant architecture, and the transcription factor TOE4b plays a pleiotropic role. Plant types resulting from TOE4b‐overexpression could promote lodging resistance and shade avoidance response and are probably well suited to density planting and interplanting cultivation models for which we will perform the field test and mechanism analysis. In addition to regulating plant architecture, TOE4b is the target gene of miR172 (Zhao et al., 2015), which is an important member of the age pathway and is related to reproductive transition, so it is very possible that TOE4b extends the reproductive phase and modulates the structure of the soybean maturity, which need to be investigated in more detail.
Recent studies showed that AP2/ERF family genes in bread wheat (DUO‐B1) and rice (OsDREB1C) both improve grain yield. DUO‐B1 regulates spike inflorescence architecture, and duo‐B1 mutations increase spikelet number in individual spikes, raising grain yield by ~10%/m2 (Wang et al., 2022). Overexpression of OsERF115 significantly increases the grain length, width, thickness and weight by promoting longitudinal elongation and transverse division of spikelet hull cells, as well as enhancing grain‐filling activity (Liu et al., 2022). OsDREB1C overexpression increased secondary branch number, grain length, width, thickness and density, which led to more grain numbers per panicle and increased 1000‐grain weight. Together, OsDREB1C overexpression resulted in a grain yield increase per plant between 45.1% and 67.6% and yield increase per plot from 41.3% to 68.3% compared to wild‐type plants (Wei et al., 2022). In our study, the AP2/ERF family member TOE4b promoted a similar yield per plant increase and phenotype in soybean. Considering that we observed more than a 60% increase in seed yield per plant in TOE4b overexpression lines, it would be interesting to conduct field plot yield assessments to test large‐scale production of soybean.
The targets downstream of TOE4b
In this study, we verified that TOE4b can bind to the promoters of the key flowering integrator genes FT2a and FT5a to repress their transcription, leading to delayed flowering (Figure 7). Some natural variants in the first intron of FT2a and in the 3′ UTR of FT5a have been reported to produce distinct phenotypic changes (Takeshima et al., 2016; Zhao et al., 2016), which proves that the regions are much more important than the others; in agreement, we determined that TOE4b shows the strongest affinity to these regions as well. FT5a was also shown to play major roles in shoot determinacy and post‐flowering stem growth (Takeshima et al., 2019). Given that plant height changed in both TOE4b overexpression and mutant lines, it is likely that FT5a may also contribute to the observed changes in plant architecture.
In our previous study, we had shown that TOE4a, the homologue of TOE4b, inhibits the expression of AP1, LEAFY (LFY) and CONSTANS‐LIKE 1b (COL1b) in shoot apices (Zhao et al., 2015), but we have not yet verified whether TOE4a directly represses the transcription of these genes directly. As TOE4a and TOE4b are functionally partially redundant, and their pleiotropic effects in regulating flowering time and plant architecture, it is likely that TOE4a and TOE4b have more downstream targets outside of FT2a and FT5a. In rice, direct binding of OsDREB1C to the OsRBCS3 (encoding a small subunit of RubisCO) promoter region leads to higher abundance and activity of RubisCO, thus contributing to increased photosynthesis and carbon assimilation rates. Moreover, OsDREB1C acts on its targets NITRATE REDUCTASE2 (OsNR2), NITRATE TRANSPORTER 2.4 (OsNRT2.4) and OsNRT1.1B to promote nitrogen uptake and translocation capacity. OsDREB1C‐mediated activation of another target gene, FT‐LIKE 1 (OsFTL1), conferred a pronounced early flowering phenotype (Wei et al., 2022). The multifaceted functions of OsDREB1C can be attributed to its modulation of several sets of target genes.
Growth and development associated with yield traits are usually related to plant hormones. YIGE1 modulates maize inflorescence, meristem size and ear length to increase grain yield via its involvement in auxin and sugar signalling (Luo et al., 2022). DUO‐B1 regulates spike inflorescence architecture by modulating cytokinin homeostasis to improve bread wheat yield (Wang et al., 2022). In this research, TOE4b played a pleiotropic role, as evidenced by the multiple traits displayed by overexpression lines and mutants, including flowering time, plant height, main stem node number, internode length, branch number, pod number and grain yield per plant (Figures 4, 5, 6), likely to relate with phytohormones. Next, analysing the mechanism of every structural component contributing to yield could help reveal the regulatory network involving TOE4b and its downstream targets and provide a resource for molecular breeding aimed at improving crop yields in soybean. Because TOE4b is a positive regulator, it will be worth exploring its upstream genes as well as gene editing its promoter.
Materials and methods
Plant materials and growth conditions
The 449‐accession panel used in this study was from our previously re‐sequenced accessions (Dong et al., 2022, 2023; Kou et al., 2022; Li et al., 2023; Lu et al., 2020), which consisted of 319 cultivars, 29 landraces and 101 wild accessions (Table S5). These accessions were grown during the cultivation season in Harbin, China (45°55′ N, 126°96′ E) in 2017 and 2020 to determine flowering time. The days after emergence (DAE) from the soil to the first open flower in 50% of the plants were recorded as flowering time, also defined as the R1 stage (Fehr and Cavines, 1977).
The soybean cultivar ‘Harosoy’ (harbouring TOE4b H4 ) was crossed to the e2 mutant previously generated by CRISPR/Cas9‐mediated gene editing in the ‘Williams 82’ (W82) background (which carries TOE4b H1 ) (Wang et al., 2023). Near isogenic line (NIL) was developed for TOE4b with the repetitive heterozygote selection method from F2 plants derived from the above cross. NIL at the F6 generation was used for phenotyping.
TOE4b overexpression transgenic lines and wild‐type W82 were planted in Guangzhou, China and Shijiazhuang, China in 2020 to study flowering time and grain yield‐related traits. Guangzhou (23°55′ N, 113°59′ E) is in Guangdong Province, belonging to the southern China area, and the soybean plant season is from July to September, with naturally short‐day conditions (daylength ~13–12 h) and a subtropical monsoon climate. Shijiazhuang (37°27′ N, 113°30′ E) in Hebei Province is in northern China, and the growing season is from June to October, with naturally long days (daylength >15 h) and a temperate climate. The plants were sown with the same criteria in two different sites, in rows 2 m long, 0.5 m between rows, with a plant interspace of 15 cm. Two rows were planted as biological replications. During the harvest season, at least 6 plants with marginal effects removed in each row were selected to determine the yield structure components per plant, including plant height, number of nodes, branches, pods and seed weight per plant. Data from the two rows were combined for analysis.
For laboratory experiments, the plants grown in the artificial incubators at a consistent air temperature of 25 °C and a relative humidity of 50% (±10%) supplied by Conviron (Canada) with an average photon flux of 300 μmol m−2 s−1or Xunon (China) with an average photon flux of 450 μmol m−2 s−1. Daylength was 12‐h light/12‐h dark for SD and 16‐h light/8‐h dark for LD.
Candidate gene identification by GWAS
Five micrograms of genomic DNA were extracted from each of 449 accessions and used to construct sequencing libraries with an Illumina TruSeq DNA Sample Prep Kit. Libraries were sequenced as paired‐end reads (150 bp in length) on an Illumina HiSeq X Ten instrument with an average coverage depth of 10x for each accession. Burrows‐Wheeler Aligner (BWA) software was used to map paired‐end reads to the soybean W82 reference genome downloaded from Phytozome (https://phytozome.jgi.doe.gov) (Schmutz et al., 2010). SNP calling was performed with GATK (Broad Institute) and SAMtools (Li and Durbin, 2009). After quality control with VCFtools (Wang et al., 2017), 6 540 653 high‐quality SNPs (with minor allele frequency [MAF] > 5%) were used to perform GWAS for flowering time across the 449 accessions with a mixed linear model approach implemented in the rMVP package (https://github.com/XiaoleiLiuBio/rMVP). The threshold for significance was estimated as P = 0.05/total SNPs based on the Bonferroni correction method. LD block analysis was performed within 150‐kb flanking regions of significant loci with LDBlockShow (https://github.com/BGI‐shenzhen/LDBlockShow) (Purcell et al., 2007). Candidate genes were identified from the soybean W82 reference genome (Glycine max W82 a2.v1 version) on Phytozome 13 website (https://phytozome‐next.jgi.doe.gov/).
Detection of selection signals
SNPs with less than 10% missing data and MAF >5% were used for the genetic diversity (π) analysis. The pairwise genomic differentiation values for wild, landrace and cultivated soybean populations were calculated using a—window pi2000—window‐pi‐step 1000 sliding window in VCFtools (Wang et al., 2017). Values for Tajima's D statistics were also calculated using VCFtools as described previously (Lu et al., 2020).
Subcellular localization of TOE4b
The coding sequence of TOE4b was amplified from W82 cDNA and cloned into the pTF101‐GFP vector harbouring the cauliflower mosaic virus (CaMV) 35S promoter to generate the 35S:TOE4b‐GFP construct. The 35S:GFP empty vector was used as a control. Plasmids were introduced into Agrobacterium (Agrobacterium tumefaciens) strain GV3101. Agrobacteria were grown overnight and resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 200 mM Acetosyringone) before transient infiltration into Nicotiana benthamiana leaves. GFP fluorescence was detected with a Zeiss LSM800 confocal laser scanning microscope.
Constructs and plant transformation
To create the 35S:TOE4b‐3Flag construct, the coding sequence of TOE4b was amplified from W82 cDNA and cloned into the pTF101‐3Flag vector. To generate the CRISPR/Cas9‐aided toe4b mutant, a 20‐bp single‐guide RNA (sgRNA) designed to target the sequence corresponding to the first AP2 domain of TOE4b was cloned into the pYLCRISPR/Cas9P35SB vector and placed under the control of the 35S promoter (Ma et al., 2015). The vectors were introduced into Agrobacterium strain EHA101 and used to transform soybean W82 as described previously (Zeng et al., 2004).
RNA extraction and RT‐qPCR
For tissue expression analysis, leaves, stem tips, hypocotyls, roots, nodules, flower buds, flowers, and pod samples were collected from W82 grown under SD conditions. For rhythmic expression analysis, leaves were collected every 4 h from W82 seedlings under LD or SD conditions at 18 DAE. To compare the expression of FT2a and FT5a in wild‐type and TOE4b overexpression seedlings, leaves were collected at 18 DAE from seedlings grown under LD conditions.
Total RNA was extracted from all the above samples using an Ultrapure RNA kit (Cowin Biotech, Beijing, China). First‐strand cDNA synthesis and removal of genomic DNA contaminants were performed using a SuperScript First‐strand cDNA Synthesis System (Takara, Dalian, China). Quantitative PCR (qPCR) was performed using a Roche LightCycle480 system (Roche, Mannheim, Germany) using a qPCR kit (Roche). β‐Tubulin was used as the internal control. Three independent biological replicates were analysed, and three technical replicate reactions were used for each sample. All qPCR primers are listed in Table S6.
Immunoblots
To examine protein abundance in transgenic plants, total proteins were extracted from the leaves of W82 and 35S:TOE4b‐3Flag transgenic lines grown under LD conditions and collected at ZT4 in protein extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% [v/v] Triton X‐100, and protease inhibitor cocktail and PMSF). Proteins were detected with anti‐Flag (A8592, Sigma) antibodies diluted to 1:5000, and the non‐specific protein to Flag antibodies was used as a loading control.
Luciferase activity assay
A ~3‐kb promoter fragment upstream of the translation start site from FT2a and FT5a was amplified from W82 DNA and introduced into the pGreen0800‐LUC/REN vector to generate the 35S:REN‐proFT2a:LUC and 35S:REN‐proFT5a:LUC reporter constructs. The coding sequences of TOE4b H1 and TOE4b H4 were amplified from W82 and Harosoy cDNA, respectively, and then cloned into the pRT107 vector, to generate the 35S:TOE4b effector construct. The appropriate combinations of reporter and effector constructs were co‐infiltrated into N. benthamiana leaves. The firefly luciferase (LUC) and Renilla luciferase (REN) activities were measured using a Luciferase 1000 Assay System (Promega, E4550) and a Renilla Luciferase Assay System (Promega, E2820), respectively. The final transcriptional activity was expressed as the ratio between LUC and REN activities. The primers used in this assay are listed in Table S6.
ChIP assay
The ChIP assay was performed as previously described (Lu et al., 2020). In brief, 2 g of leaves were collected from 18DAE W82 and 35S:TOE4b‐3Flag seedlings at zeitgeber 4 (ZT4, 4 h after lights on) under LD conditions. Leaf samples were fixed in 1% (w/v) formaldehyde for 10 min for protein‐DNA crosslinking. The chromatin complexes were isolated and sonicated to shear DNA into 250–500 bp fragments. The TOE4b‐bound chromatin was immunoprecipitated with anti‐Flag M2 magnetic beads (Sigma, M8823). The coimmunoprecipitated DNA was recovered and analysed by qPCR in triplicates. Relative fold enrichment was calculated by normalizing the amount of a target DNA fragment against input DNA. The enrichment of the Actin genomic fragment was used as a negative control. The primers used for amplification are listed in Table S6.
Development of a cleaved amplified polymorphic sequences (CAPS) marker
The H1 and H4 haplotypes of TOE4b were identified based on the SNP at nucleotide 715 of the coding sequence, based on which one specific CAPS marker was developed. The primers were 5′‐ATTCAGGGGACTTGATGCTGAC‐3′ and 5′‐GTTGGTTGGAACCGTAAAGAGC‐3′. The resulting 744‐bp PCR products were digested with the restriction enzyme HaeIII and separated on 1.5% (w/v) agarose gel. TOE4b H1 was digested into 256‐bp and 488‐bp fragments, whereas TOE4b H4 was not digested and migrated as a 744‐bp band.
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
XZ, FK and BL designed and supervised this study. XZ and HD wrote the manuscript.
HL and ZH performed the experiments and analysed the data. ZH, MH and YZ conducted the genetic experiments. CF, LK, LC and HY investigated agronomic traits in the field. HL performed the bioinformatics analysis. HL, HD and ZH contributed equally.
Supporting information
Figure S1 Genome‐wide averaged distance of linkage disequilibrium decayed. Calculation via squared correlations of allele frequencies (r 2 ) against the distance between polymorphic sites in wild soybeans, landraces and cultivars.
Figure S2 Expression of 15 genes in the linkage disequilibrium block in the leaves and stem tips. (a) Data are from transcriptomic analysis in wild‐type W82. The values represent the RPKM value of genes in transcriptome. Data are presented as means ± SD of two biological replicates. (b) Data are from Phytozome 13 (https://phytozome‐next.jgi.doe.gov/). FPKM (Fragments Per Kilobase of exon model per Million mapped reads).
Figure S3 Natural variation of TOE4b. (a) Summary of TOE4b haplotypes across a diversity panel comprising 1744 soybean accessions. (b) Origin tree of the TOE4b haplotypes. The data are from 1744 accessions, consisting of 221 wild soybeans, 516 landraces and 874 improved cultivars. The red arrows indicate the direction of evolution.
Figure S4 TOE4b H4 variation affects one of the conserved AP2 domains. (a) Alignment of the amino acid sequences of TOE4b with its orthologs and homologues. Amino acid sequences from Glycine max, Phaseolus vulgaris, Medicago truncatula, Arabidopsis thaliana and Oryza sativa were aligned by Clustal W. The red asterisk indicates the amino acid in TOE4bH4 that changes from the conserved G residue to S. (b) CAPS marker for the identification of the TOE4b H1 and TOE4b H4 alleles. PCR products were digested with the restriction enzyme HaeIII. Hetero, heterozygous plant.
Figure S5 Effects of a combination of TOE4b and E2 alleles on soybean flowering time. (a, b) Flowering time of six allelic combinations between TOE4b and E2 across 449 soybean accessions grown in Harbin in 2017 (a) and 2020 (b). The number above the x‐axis represents the number of samples. Different lowercase letters indicate significant differences. The phenotype with two samples was not statistically analysed with other samples.
Figure S6 Generation TOE4b overexpression lines in soybean. (a) Relative transcript levels of TOE4b in wild‐type W82 and T1 overexpression transgenic plants. β‐Tubulin (TUB) was used as a normalization transcript. (b) Abundance of TOE4b‐3Flag in the leaves of T1 transgenic plants, as determined by immunoblotting using anti‐Flag antibodies. The non‐specific band (NS) was used as a loading control.
Figure S7 Phenotypes of TOE4b mutant and overexpression lines. Agronomic traits of plant height (a, c, e), measured from the cotyledonary node of the main stem to the apex in centimetres; number of nodes (b, d, f). The plants were grown in an incubator under LD (16‐h light/8‐h dark) (a‐b) or SD (12‐h light/12‐h dark) conditions with an average photon flux of 300 μmol m−2 s−1 (c–d). For the mutant plants were grown in an incubator under LD (16‐h light/8‐h dark) with an average photon flux of 450 μmol m−2 s−1 (e–f). W82, wild‐type Williams 82. Five plants were scored for each phenotype; data are means ± SD. P < 0.05, as determined by multiple comparison testing by one‐way ANOVA. Different lowercase letters indicate significant differences.
Figure S8 Agronomic traits associated with the major haplotypes of TOE4b. Agronomic trait of plant height (a), measured from the cotyledonary node of the main stem to the apex in centimetres; number of nodes (b); average of internode length (c); branch number per plant (d); pod number per plant (e); and grain weight per plant (f). TOE4b H1 , n = 68; TOE4b H4 , n = 83. The plants were grown in a standard field in Harbin under natural conditions in 2017. At least 6 plants were scored for each phenotype; data are means ± SD. P < 0.05, two‐tailed Student's t‐test. Different lowercase letters indicate significant differences.
Table S1 Significant loci associated with flowering time were identified through GWAS.
Table S2 The –log10(P) values of significant loci associated with flowering time for GWAS.
Table S3 Information of candidate genes within LD block.
Table S4 Transcription levels of the 15 genes in the leaves and stem tips.
Table S5 List of soybean accessions used for GWAS.
Table S6 Primers used in this study.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32090064 to FK), Major Program of Guangdong Basic and Applied Research (2019B030302006 to FK and BL), Science and Technology Projects in Guangzhou (202201020128 to XZ) and the National Key Research and Development Program (2021YFF1001203 to XZ and BL).
Contributor Information
Baohui Liu, Email: liubh@gzhu.edu.cn.
Fanjiang Kong, Email: kongfj@gzhu.edu.cn.
Xiaohui Zhao, Email: zhaoxh@gzhu.edu.cn.
Data Availability Statement
Data supporting the findings of this study are available in the supplementary material of this article. The previously reported re‐sequence data used in this study have been deposited into the NCBI database: 372‐accessions, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA743225.1; 1295‐accessions, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA394629; 349‐accessions, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA776405; 132‐accessions, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA934724; 95‐accessions, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA859249.
References
- Aukerman, M.J. and Sakai, H. (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2‐like target genes. Plant Cell, 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, R.L. (1971) Two major genes for time of flowering and maturity in soybeans. Crop. Sci. 112, 242–244. [Google Scholar]
- Chen, X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science, 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, R.L. (1971) Influence of early lodging on yield of soybean [Glycine max (L.) Merr.]. Agronomy J. 63, 449–450. [Google Scholar]
- Doebley, J.F. , Gaut, B.S. and Smith, B.D. (2006) The molecular genetics of crop domestication. Cell, 127, 1309–1321. [DOI] [PubMed] [Google Scholar]
- Dong, L. , Cheng, Q. , Fang, C. , Kong, L. , Yang, H. , Hou, Z. , Li, Y. et al. (2022) Parallel selection of distinct Tof5 alleles drove the adaptation of cultivated and wild soybean to high latitudes. Mol. Plant, 15, 308–321. [DOI] [PubMed] [Google Scholar]
- Dong, L. , Li, S. , Wang, L. , Su, T. , Zhang, C. , Bi, Y. , Lai, Y. et al. (2023) The genetic basis of high‐latitude adaptation in wild soybean. Curr. Biol. 33, 252–262. [DOI] [PubMed] [Google Scholar]
- Fehr, W.R. and Cavines, C.E. (1977) Stages of soybean development special report. Special Rep. 87, 1–12. [Google Scholar]
- Gautam, J.K. and Nandi, A.K. (2018) APD1, the unique member of Arabidopsis AP2 family influences systemic acquired resistance and ethylene‐jasmonic acid signaling. Plant Physiol. Biochem. 133, 92–99. [DOI] [PubMed] [Google Scholar]
- Giri, M.K. , Swain, S. , Gautam, J.K. , Singh, S. , Singh, N. , Bhattacharjee, L. and Nandi, A.K. (2014) The Arabidopsis thaliana At4g13040 gene, a unique member of the AP2/EREBP family, is a positive regulator for salicylic acid accumulation and basal defense against bacterial pathogens. J. Plant Physiol. 171, 860–867. [DOI] [PubMed] [Google Scholar]
- Graham, P.H. and Vance, C.P. (2003) Legumes: importance and constraints to greater use. Plant Physiol. 131, 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P. (2003) The genes of the green revolution. Trends Genet. 19, 5–9. [DOI] [PubMed] [Google Scholar]
- Hu, H. , Tian, S. , Xie, G. , Liu, R. , Wang, N. , Li, S. , He, Y. et al. (2021) TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis . Proc. Natl. Acad. Sci. U. S. A. 118, e2103895118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y. , Wang, Y. , Xue, D. , Wang, J. , Yan, M. , Liu, G. , Dong, G. et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D. , Den Boer, B.G. , Van Montagu, M. and Okamuro, J.K. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2 . Plant Cell, 6, 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku, K.D. , Omidyar, P.K. , Gee, Z. and Okamuro, J.K. (2005) Control of seed mass and seed yield by the floral homeotic gene APETALA2 . Proc. Natl. Acad. Sci. U. S. A. 102, 3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, F. , Liu, B. , Xia, Z. , Sato, S. , Kim, B.M. , Watanabe, S. , Yamada, T. et al. (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou, K. , Yang, H. , Li, H. , Fang, C. , Chen, L. , Yue, L. , Nan, H. et al. (2022) A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 32, 1728–1742. [DOI] [PubMed] [Google Scholar]
- Lauter, N. , Kampani, A. , Carlson, S. , Goebel, M. and Moose, S.P. (2005) microRNA172 down‐regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. U. S. A. 102, 9412–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. and Durbin, R. (2009) Fast and accurate short read alignment with burrows‐wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Fang, C. , Yang, Y. , Lv, T. , Su, T. , Chen, L. , Nan, H. et al. (2021) Overcoming the genetic compensation response of soybean florigens to improve adaptation and yield at low latitudes. Curr. Biol. 31, 3755–3767. [DOI] [PubMed] [Google Scholar]
- Li, H. , Du, H. , He, M. , Wang, J. , Wang, F. , Yuan, W. , Huang, Z. et al. (2023) Natural variation of FKF1 controls flowering and adaptation during soybean domestication and improvement. New Phytol. 238, 1671–1684. [DOI] [PubMed] [Google Scholar]
- Lin, X. , Liu, B. , Weller, J.L. , Abe, J. and Kong, F. (2021) Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 63, 981–994. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Zhang, M. , Feng, F. and Tian, Z. (2020) Toward a “Green Revolution” for Soybean. Mol. Plant, 13, 688–697. [DOI] [PubMed] [Google Scholar]
- Liu, C. , Ma, T. , Yuan, D. , Zhou, Y. , Long, Y. , Li, Z. , Dong, Z. et al. (2022) The OsEIL1‐OsERF115‐target gene regulatory module controls grain size and weight in rice. Plant Biotechnol. J. 20, 1470–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. , Dong, L. , Fang, C. , Liu, S. , Kong, L. , Cheng, Q. , Chen, L. et al. (2020) Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436. [DOI] [PubMed] [Google Scholar]
- Luo, Y. , Zhang, M. , Liu, Y. , Liu, J. , Li, W. , Chen, G. , Peng, Y. et al. (2022) Genetic variation in YIGE1 contributes to ear length and grain yield in maize. New Phytol. 234, 513–526. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al. (2015) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Ma, Z. , Wu, T. , Huang, K. , Jin, Y.M. , Li, Z. , Chen, M. , Yun, S. et al. (2020) A Novel AP2/ERF transcription factor, OsRPH1, negatively regulates plant height in rice. Front. Plant Sci. 11, 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, A. , Adam, H. , Díaz‐Mendoza, M. , Zurczak, M. , González‐Schain, N.D. and Suárez‐López, P. (2009) Graft‐transmissible induction of potato tuberization by the microRNA miR172. Development, 136, 2873–2881. [DOI] [PubMed] [Google Scholar]
- Miura, K. , Ikeda, M. , Matsubara, A. , Song, X.J. , Ito, M. , Asano, K. , Matsuoka, M. et al. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. [DOI] [PubMed] [Google Scholar]
- Peng, J. , Richards, D.E. , Hartley, N.M. , Murphy, G.P. , Devos, K.M. , Flintham, J.E. , Beales, J. et al. (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature, 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Purcell, S. , Neale, B. , Todd‐Brown, K. , Thomas, L. , Ferreira, M.A. , Bender, D. , Maller, J. et al. (2007) PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am. J. Hum. Genet. 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L. and Meyerowitz, E.M. (1998) The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646. [DOI] [PubMed] [Google Scholar]
- Sasaki, A. , Ashikari, M. , Ueguchi‐Tanaka, M. , Itoh, H. , Nishimura, A. , Swapan, D. , Ishiyama, K. et al. (2002) Green revolution: a mutant gibberellin‐synthesis gene in rice. Nature, 416, 701–702. [DOI] [PubMed] [Google Scholar]
- Schmutz, J. , Cannon, S.B. , Schlueter, J. , Ma, J. , Mitros, T. , Nelson, W. , Hyten, D.L. et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Shim, Y. , Lim, C. , Seong, G. , Choi, Y. , Kang, K. and Paek, N.C. (2022) The AP2/ERF transcription factor LATE FLOWERING SEMI‐DWARF suppresses long‐day‐dependent repression of flowering. Plant Cell Environ. 45, 2446–2459. [DOI] [PubMed] [Google Scholar]
- Studer, A. , Zhao, Q. , Ross‐Ibarra, J. and Doebley, J. (2011) Identification of a functional transposon insertion in the maize domestication gene tb1 . Nat. Genet. 43, 1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima, R. , Hayashi, T. , Zhu, J. , Zhao, C. , Xu, M. , Yamaguchi, N. , Sayama, T. et al. (2016) A soybean quantitative trait locus that promotes flowering under long days is identified as FT5a, a FLOWERING LOCUS T ortholog. J. Exp. Bot. 67, 5247–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima, R. , Nan, H. , Harigai, K. , Dong, L. , Zhu, J. , Lu, S. , Xu, M. et al. (2019) Functional divergence between soybean FLOWERING LOCUS T orthologues FT2a and FT5a in post‐flowering stem growth. J. Exp. Bot. 70, 3941–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. and Li, J. (2008) Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Tu, L. , Lin, M. , Lin, Z. , Wang, P. , Yang, Q. , Ye, Z. et al. (2017) Asymmetric subgenome selection and cis‐regulatory divergence during cotton domestication. Nat. Genet. 49, 579–587. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Du, F. , Wang, J. , Wang, K. , Tian, C. , Qi, X. , Lu, F. et al. (2022) Improving bread wheat yield through modulating an unselected AP2/ERF gene. Nat Plants, 8, 930–939. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Li, H. , He, M. , Dong, L. , Huang, Z. , Chen, L. , Nan, H. et al. (2023) GIGANTEA orthologs, E2 members, redundantly determine photoperiodic flowering and yield in soybean. J. Integr. Plant Biol. 65, 188–202. [DOI] [PubMed] [Google Scholar]
- Watanabe, S. , Xia, Z. , Hideshima, R. , Tsubokura, Y. , Sato, S. , Yamanaka, N. , Takahashi, R. et al. (2011) A map‐based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics, 188, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S. , Li, X. , Lu, Z. , Zhang, H. , Ye, X. , Zhou, Y. , Li, J. et al. (2022) A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science, 377, eabi8455. [DOI] [PubMed] [Google Scholar]
- Xie, Z. , Nolan, T.M. , Jiang, H. and Yin, Y. (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis . Front. Plant Sci. 10, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, P. , Vadnais, D.A. , Zhang, Z. and Polacco, J.C. (2004) Refined glufosinate selection in Agrobacterium‐mediated transformation of soybean [Glycine max (L.) Merrill]. Plant Cell Rep. 22, 478–482. [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Wang, L. , Zeng, L. , Zhang, C. and Ma, H. (2015) Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 29, 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhou, J.F. , Kan, Y. , Shan, J.X. , Ye, W.W. , Dong, N.Q. , Guo, T. et al. (2022) A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science, 376, 1293–1300. [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Cao, D. , Huang, Z. , Wang, J. , Lu, S. , Xu, Y. , Liu, B. et al. (2015) Dual functions of GmTOE4a in the regulation of photoperiod‐mediated flowering and plant morphology in soybean. Plant Mol. Biol. 88, 343–355. [DOI] [PubMed] [Google Scholar]
- Zhao, C. , Takeshima, R. , Zhu, J. , Xu, M. , Sato, M. , Watanabe, S. , Kanazawa, A. et al. (2016) A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 16, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlobin, N. , Lebedeva, M. , Monakhova, Y. , Ustinova, V. and Taranov, V. (2021) An ERF121 transcription factor from Brassica oleracea is a target for the conserved TAL‐effectors from different Xanthomonas campestris pv. campestris strains. Mol. Plant Pathol. 22, 618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Genome‐wide averaged distance of linkage disequilibrium decayed. Calculation via squared correlations of allele frequencies (r 2 ) against the distance between polymorphic sites in wild soybeans, landraces and cultivars.
Figure S2 Expression of 15 genes in the linkage disequilibrium block in the leaves and stem tips. (a) Data are from transcriptomic analysis in wild‐type W82. The values represent the RPKM value of genes in transcriptome. Data are presented as means ± SD of two biological replicates. (b) Data are from Phytozome 13 (https://phytozome‐next.jgi.doe.gov/). FPKM (Fragments Per Kilobase of exon model per Million mapped reads).
Figure S3 Natural variation of TOE4b. (a) Summary of TOE4b haplotypes across a diversity panel comprising 1744 soybean accessions. (b) Origin tree of the TOE4b haplotypes. The data are from 1744 accessions, consisting of 221 wild soybeans, 516 landraces and 874 improved cultivars. The red arrows indicate the direction of evolution.
Figure S4 TOE4b H4 variation affects one of the conserved AP2 domains. (a) Alignment of the amino acid sequences of TOE4b with its orthologs and homologues. Amino acid sequences from Glycine max, Phaseolus vulgaris, Medicago truncatula, Arabidopsis thaliana and Oryza sativa were aligned by Clustal W. The red asterisk indicates the amino acid in TOE4bH4 that changes from the conserved G residue to S. (b) CAPS marker for the identification of the TOE4b H1 and TOE4b H4 alleles. PCR products were digested with the restriction enzyme HaeIII. Hetero, heterozygous plant.
Figure S5 Effects of a combination of TOE4b and E2 alleles on soybean flowering time. (a, b) Flowering time of six allelic combinations between TOE4b and E2 across 449 soybean accessions grown in Harbin in 2017 (a) and 2020 (b). The number above the x‐axis represents the number of samples. Different lowercase letters indicate significant differences. The phenotype with two samples was not statistically analysed with other samples.
Figure S6 Generation TOE4b overexpression lines in soybean. (a) Relative transcript levels of TOE4b in wild‐type W82 and T1 overexpression transgenic plants. β‐Tubulin (TUB) was used as a normalization transcript. (b) Abundance of TOE4b‐3Flag in the leaves of T1 transgenic plants, as determined by immunoblotting using anti‐Flag antibodies. The non‐specific band (NS) was used as a loading control.
Figure S7 Phenotypes of TOE4b mutant and overexpression lines. Agronomic traits of plant height (a, c, e), measured from the cotyledonary node of the main stem to the apex in centimetres; number of nodes (b, d, f). The plants were grown in an incubator under LD (16‐h light/8‐h dark) (a‐b) or SD (12‐h light/12‐h dark) conditions with an average photon flux of 300 μmol m−2 s−1 (c–d). For the mutant plants were grown in an incubator under LD (16‐h light/8‐h dark) with an average photon flux of 450 μmol m−2 s−1 (e–f). W82, wild‐type Williams 82. Five plants were scored for each phenotype; data are means ± SD. P < 0.05, as determined by multiple comparison testing by one‐way ANOVA. Different lowercase letters indicate significant differences.
Figure S8 Agronomic traits associated with the major haplotypes of TOE4b. Agronomic trait of plant height (a), measured from the cotyledonary node of the main stem to the apex in centimetres; number of nodes (b); average of internode length (c); branch number per plant (d); pod number per plant (e); and grain weight per plant (f). TOE4b H1 , n = 68; TOE4b H4 , n = 83. The plants were grown in a standard field in Harbin under natural conditions in 2017. At least 6 plants were scored for each phenotype; data are means ± SD. P < 0.05, two‐tailed Student's t‐test. Different lowercase letters indicate significant differences.
Table S1 Significant loci associated with flowering time were identified through GWAS.
Table S2 The –log10(P) values of significant loci associated with flowering time for GWAS.
Table S3 Information of candidate genes within LD block.
Table S4 Transcription levels of the 15 genes in the leaves and stem tips.
Table S5 List of soybean accessions used for GWAS.
Table S6 Primers used in this study.
Data Availability Statement
Data supporting the findings of this study are available in the supplementary material of this article. The previously reported re‐sequence data used in this study have been deposited into the NCBI database: 372‐accessions, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA743225.1; 1295‐accessions, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA394629; 349‐accessions, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA776405; 132‐accessions, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA934724; 95‐accessions, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA859249.


