Abstract
The Bacillus thuringiensis insecticidal δ-endotoxins have a three-domain structure, with the seven amphipathic helices which comprise domain I being essential for toxicity. To better define the function of these helices in membrane insertion and toxicity, either site-directed or random mutagenesis of two regions was performed. Thirty-nucleotide segments in the B. thuringiensis cry1Ac1 gene, encoding parts of helix α4 and the loop connecting helices α4 and α5, were randomly mutagenized. This hydrophobic region of the toxin probably inserts into the membrane as a hairpin. Site-directed mutations were also created in specific surface residues of helix α3 in order to increase its hydrophobicity. Among 12 random mutations in helix α4, 5 resulted in the total loss of toxicity for Manduca sexta and Heliothis virescens, another caused a significant increase in toxicity, and one resulted in decreased toxicity. None of the nontoxic mutants was altered in toxin stability, binding of toxin to a membrane protein, or the ability of the toxin to aggregate in the membrane. Mutations in the loop connecting helices α4 and α5 did not affect toxicity, nor did mutations in α3, which should have enhanced the hydrophobic properties of this helix. In contrast to mutations in helix α5, those in helix α4 which inactivated the toxin did not affect its capacity to oligomerize in the membrane. Despite the formation of oligomers, there was no ion flow as measured by light scattering. Helix α5 is important for oligomerization and perhaps has other functions, whereas helix α4 must have a more direct role in establishing the properties of the channel.
Bacillus thuringiensis is unique in its capacity to produce a variety of insecticidal δ-endotoxins, which are arranged in different classes (7, 13). The structure of these toxins appears to be highly conserved (10, 13, 19), especially the seven amphipathic α helices which comprise domain I. Following binding of the toxin to specific receptors on cells lining the larval midgut (12, 15), one or more of these helices insert into the membrane and participate in the formation of an ion channel (8, 10, 16, 22). The mode of killing is believed to be colloid osmotic lysis (17), although more subtle and/or more rapid effects have not been ruled out.
Previously we had mutagenized regions of the cry1Ac1 gene encoding residues within three of these helices, i.e., α2, α5, and α6, and found that helix α5 was the only one in which many of the mutations abolished toxicity (2, 27). As an extension of the previous studies, we have investigated the role of helix α4 and the loop connecting helices α4 and α5. Thirty-nucleotide mutagenic oligonucleotides were used to obtain random mutations in regions of the cry1Ac1 gene encoding residues in helix α4 and the loop. Four site-specific mutations which should have altered the hydrophobic properties of helix α3 were also examined. Many mutations in helix α4 resulted in either the loss of toxicity or toxin instability, and one mutant toxin had enhanced activity. Mutations in the loop connecting helices α4 and α5 or within helix α3, however, had little effect on stability or toxicity. The nontoxic α4 mutant toxins oligomerized in the membrane as well as the wild-type toxin but did not form functional ion channels. Helices α4 and α5, which comprise a very hydrophobic loop within domain I, are both important for toxicity but have different roles in toxin aggregation and probably ion channel formation and/or function.
MATERIALS AND METHODS
Bacterial strains and media.
Propagation of phage M13 clones was carried out in Escherichia coli JM101 at 37°C in Luria-Bertani medium (21). Other subclones were propagated in E. coli DH5α, using the same medium, in the presence of 50 μg of ampicillin ml−1. The acrystalliferous derivative of Bacillus thuringiensis subsp. kurstaki HD1, strain CryB (24), was grown at 30°C on a rotary shaker in G-Tris medium (3) with or without erythromycin at 25 μg ml−1.
Mutagenesis and subcloning.
The cry1Ac1 gene in M13mp19 was mutagenized as described previously (27). Thirty-nucleotide mutagenic oligonucleotides encoding residues 129 to 137 within helix α4 (5′ CATGTCATTGAATTGAATACGCATCTC 3′ with 90% as specified plus 3% of each of the other 3 bases) and residues 145 to 155 within the carboxyl terminus of helix α4 and the loop connecting helices α4 and α5 (5′ AACTTGATAATTTTGAACTGCAAAAAGAGGAAT 3′ with 90% as specified plus 3% of each of the other 3 bases) were used to generate random mutations in each of these regions.
Four different single-amino-acid substitutions, i.e., replacement of the asparagine at position 94 by valine (N94V) (5′ TCTAGAAATGGCTTGGACCCTAGCGAATTC 3′), N94F (5′ TCTAGAAATGGCTTGGAACCTAGCGATTC 3′), N105V (5′ GTAAATTTGATAAAGCACGCTTAGTCCTTC 3′), and N105F (5′ GTAAATTTGATAAAGGAAGCTTAGTCCTTC 3′), were created within helix α3 by site-specific mutagenesis (27). Double-stranded DNA was propagated in E. coli JM101, and clones were picked at random for sequencing of single-stranded DNA. Clones with one or two substitutions in helix α4 or the loop and those with specific substitutions in helix α3 were selected for further analyses.
Immunoblotting and bioassays.
E. coli JM101, at a density of 1 × 108 to 2 × 108 in Luria-Bertani medium, was infected with the M13 clones, and the cultures were incubated on a rotary shaker at 37°C for 6 h. To test the stability of the mutant toxins, crude extracts of the infected cells, prepared as described previously (2), were incubated with tolylsulfonyl phenylalanylchloromethyl ketone (TPCK)-trypsin at a ratio of 1 μg of extract to 20 μg of TPCK-trypsin in 0.03 M NaHCO3, pH 8.6, for 2 h. The trypsin-treated toxins were tested for stability by separation by sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE) and immunoblotting with a polyclonal rabbit antibody against the Cry1Ac1 toxin (20).
Bioassays were done, as previously described (2), by spreading various dilutions of cells (100 μl each) expressing stable toxins onto insect diet in bioassay cups. One second- to third-instar larva of Manduca sexta or Heliothis virescens was placed on each of the diet cups, which were incubated for 7 days in an insectary. Ten replicas of each dilution (five to six per assay) were tested for toxicity with cells infected with M13 containing the wild-type cry1Ac1 gene as a control. A portion of the cells used for the bioassays was suspended in 50 μl of 6 M urea–1% SDS–50 mM dithiothreitol–2 mM phenylmethylsulfonyl fluoride, pH 9.6, and lysed by heating in boiling water for 3 min. Ten-microliter aliquots were then electrophoresed and immunoblotted as described above to determine the amount of toxin applied to the diet. Each bioassay was repeated at least three times, and the 50% lethal concentrations (LC50s) and 95% confidence limits were calculated by employing a SAS probit program (SAS Institute, Inc.) as previously described (2). These values were corrected for any differences in the amount of toxin applied to the diet cup.
Internal fragments from all promising mutant genes were subcloned as XhoI-SphI fragments into shuttle vector pHT3101 (1) containing the wild-type cry1Ac1 gene (2), digested with the same enzymes. Toxin stability and alterations in toxicity (LC50s) were confirmed by performing immunoblotting and bioassays of the E. coli DH5α clones expressing the mutated cry1Ac1 genes, as described above.
Toxin purification.
The pHT3101-cry1Ac1 plasmids containing the various mutations were electroporated into B. thuringiensis CryB as described previously (2). Clones were spread on G-Tris-erythromycin agar and incubated for 72 h at 30°C. The confluent plates of spores plus inclusions were harvested in 1 M KCl–5 mM EDTA, pH 7.0. Following centrifugation at 8,000 rpm for 8 min, the pellets were each resuspended in 2 to 3 ml of deionized water and the suspensions were incubated at 65°C for 2 min to inactivate residual proteases. The cells were recentrifuged at 7,700 × g for 8 min, and the pellets were suspended in a minimal volume of solubilization buffer (0.03 M NaHCO3–0.02% α-mercaptoethanol, pH 9.6). Suspensions in this buffer were incubated at 37°C for 20 min and then centrifuged at 7,700 × g, and the supernatants were saved. This extraction was repeated twice, and the pooled supernatants were dialyzed overnight at 4°C against 2 liters of 1 mM Tris, pH 8.5. After dialysis, the solubilized protoxin was incubated with TPCK-trypsin at a ratio of 1 μg of extract to 20 μg of TPCK-trypsin at 37°C for 1 h. This trypsin treatment was repeated with an additional 1-h incubation. The digested toxin was dialyzed in Spectropor dialysis tubing with a 50,000-Da-molecular-size cutoff against 1,000 volumes of 0.03 M Tris-HCl, pH 8.5, followed by dialysis against 0.03 M NaHCO3–0.25 M NaCl, pH 9.6.
Further purification of the toxin was carried out with a 1-ml Mono Q cartridge (Pharmacia). Initially, the column was washed with 5 ml of 0.03 M NaHCO3, pH 9.6, followed by 2 ml of 0.03 M NaHCO3–0.25 M NaCl, pH 9.6, and 5 ml of 0.03 M NaHCO3, pH 9.6. The toxin sample was added to the column, and 1-ml fractions were collected. The bound toxin was eluted with a linear gradient of 0.25 to 0.4 M NaCl in 0.03 M NaHCO3, pH 9.6. Each fraction was assayed for protein content by the use of the bicinchoninic acid reagent (Pierce Chemical), and peak fractions were pooled. Toxin purity and concentration were determined by SDS–10% PAGE, staining the gels with Coomassie blue (18), and comparing stain intensities with those of known concentrations of bovine serum albumin (BSA).
Ligand blotting and membrane insertion studies.
Brush border membrane vesicles (BBMV) were prepared from fifth-instar larvae of M. sexta according to the method of Wolfersberger et al. (26), and the protein concentration of the preparation was determined with the bicinchoninic acid reagent. Ligand blotting was performed by the method of Mohammed et al. (20). Twenty micrograms of BBMV protein (solubilized in the loading buffer) was loaded onto an SDS–8% polyacrylamide gel, and each lane was blotted separately onto a polyvinylidene difluoride membrane (Immobilon P; Millipore) strip. After nonspecific groups were blocked with 5% milk powder in Tris-buffered saline, pH 7.5, the membrane strips were incubated with either the wild-type or mutant toxin. The blots were developed after treatment with rabbit anti-Cry1Ac1 antibody followed by an anti-rabbit antibody–alkaline phosphatase conjugate.
For membrane insertion studies, the nontoxic helix α4 mutant toxins were purified from the transformed B. thuringiensis CryB strain as described above. A nontoxic mutant toxin with a single-amino-acid substitution in helix α5 (A164P) (27) had been previously found to be incapable of inserting into the brush border membrane of M. sexta (4) and served as a negative control. BBMV (20 μg of protein) were first washed with 0.1 M NaHCO3–0.25 M NaCl, pH 9.6, and then incubated with 60 ng of each of the toxins at 30°C for 1 h. The BBMV were centrifuged at 7,700 × g for 8 min, and the pellets were washed twice with 1 ml of 0.1 M NaHCO3–0.1 M NaCl, pH 9.6, and once with 1 ml of 0.1 M NaHCO3–0.25 M NaCl, pH 9.6. The washed pellets were finally each suspended in 10 μl of the latter buffer supplemented with 0.5% SDS and incubated at 65°C for 15 min. Following centrifugation, the supernatants were subjected to SDS–6% PAGE. Immunoblotting of the extracts was performed as described above.
Light scattering assays.
The solute permeability of BBMV containing the wild-type and nontoxic-mutant toxins was analyzed by a light scattering assay as described by Carroll et al. (5) with minor modifications. BBMV (0.2 mg/ml) equilibrated with 10 mM 2-(cyclohexylamino)ethanesulfonic acid (CHES)–KOH (pH 9.0)–1% (wt/vol) BSA were incubated with Cry1Ac1 toxin (36 pmol/mg of BBMV) for 60 min at 21°C. The treated BBMV were mixed with an equal volume of 10 mM CHES–KOH–0.1% (wt/vol) BSA containing 150 mM KCl, pH 9.0, at 21°C. Reswelling was measured by using an SpectraKinetic stopped-flow spectrophotometer (Applied Physics) with 90°C light scattering at 450 nm. All measurements were the averages of at least three replicas with errors as in Table 3.
TABLE 3.
Light scattering measurements of the toxin-induced permeability change in M. sexta BBMV
| Sample tested | Rate constant of light scattering (S−1)a |
|---|---|
| Wild-type toxin | 0.81 ± 0.02 |
| Q133R | 0.12 ± 0.01 |
| I132V | 0.11 ± 0.01 |
| I132N | 0.11 ± 0.01 |
| I132L | 0.11 ± 0.01 |
| I132S | 0.10 ± 0.01 |
| BBMV buffer only | 0.00 |
Values are initial rates of decrease of light scattering as determined from the data in Fig. 4 and are averages of at least three measurements ± 1 standard deviation.
RESULTS
Mutations within helix α4 affect toxicity.
Twelve different single-amino-acid substitutions and four different double-amino-acid substitutions were generated within helix α4 (Table 1). Five of the single mutants (Q133R, I132S, I132L, I132V, and I132N) were nontoxic. The R131L change and the double mutation (I132V-D136Y) resulted in an approximately 10-fold reduction in toxicity. The F134L mutant, on the other hand, showed an approximately threefold increase in toxicity compared to the wild type. Extracts of the M13 clones were tested for toxin stability, and they all produced stable toxins, as shown for three of them in Fig. 1. Five other single-amino-acid substitutions in this region, i.e., R131C, R131S, M130I, M137T, and Q133H, did not affect toxicity. Among the four mutant toxins with double-amino-acid substitutions, M130I-R131L, I132F-N135S, and F134A-M137I were unstable. Those helix α4 mutant toxins which exhibited significant differences from the wild type in terms of toxicity against M. sexta were further tested for toxicity against larvae of H. virescens. All of the mutant toxins which were nontoxic for M. sexta were also nontoxic for H. virescens.
TABLE 1.
Mutations in helix α4 and their effects on toxin stability and toxicity
| Mutation | Trypsin stabilitya | Toxicityb | BBMV protein bindingc |
|---|---|---|---|
| Q133R | Stable | Nontoxic | + |
| I132S | Stable | Nontoxic | + |
| I132L | Stable | Nontoxic | + |
| I132V | Stable | Nontoxic | + |
| I132N | Stable | Nontoxic | + |
| R131L | Stable | Reduced | + |
| R131C | Stable | No change | NT |
| R131S | Stable | No change | NT |
| M130I | Stable | No change | NT |
| M137T | Stable | No change | NT |
| F134L | Stable | Enhancedd | NT |
| Q133H | Stable | No change | NT |
| I132V-D136Y | Stable | Reduced | NT |
| M130I-R131L | Unstable | ||
| I132F-N135S | Unstable | ||
| F134A-M137I | Unstable |
As demonstrated in Fig. 1.
LC50 were determined with infected E. coli cells as described in Materials and Methods. Nontoxic means no toxicity was evident with the most concentrated suspension of either M13-infected cells or of E. coli clones (undiluted cells at 2 × 108 to 3 × 108 ml−1). No change means that the LC50 was within the same range as that of cells infected with the wild type. Reduced means that there was a >10-fold increase in the LC50.
As shown in Fig. 2. NT, not tested; +, protein binding evident.
The LC50 was 106 cells (6.6 × 105 to 1.3 × 106 for 95% confidence limits) for the F134L mutant versus 1.9 × 106 (1.3 × 106 to 3.3 × 106 for 95% confidence limits) for the wild-type infected cells.
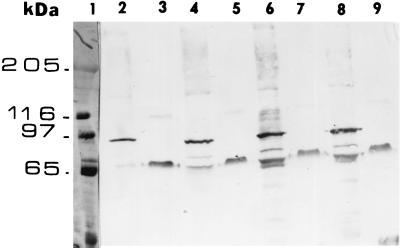
FIG. 1.
Crude extracts of M13 clones digested with trypsin were electrophoresed in an 10% SDS–polyacrylamide gel and transferred to an Immobilon-P membrane for immunoblotting with the Cry1Ac1 antibody. Lane 1, molecular mass standards; lane 2, wild-type M13 crude extract; lane 3, wild-type M13 crude extract, trypsin treated; lane 4, Q133R M13 crude extract; lane 5, Q133R M13 crude extract, trypsin treated; lane 6, I132S M13 crude extract; lane 7, I132S M13 crude extract, trypsin treated; lane 8, I132L M13 crude extract; lane 9, I132L M13 crude extract, trypsin treated. All other nontoxic helix α4 mutants were also stable to trypsin digestion (data not shown).
Mutations within the α4-α5 loop do not affect toxicity.
Seven different single-amino-acid substitutions were generated either within the carboxyl end of helix α4 or within the loop connecting helices α4 and α5 (Table 2). Only the Q154R substitution resulted in instability to trypsin. All of the others produced stable, fully active toxins.
TABLE 2.
Mutations in helix α3 and the loop connecting helices α4 and α5 do not affect toxicity
| Mutation | Trypsin stabilitya | Toxicityb |
|---|---|---|
| Q154R | Unstable | |
| I145D | Stable | No change |
| A149V | Stable | No change |
| F148I | Stable | No change |
| V150D | Stable | No change |
| I145T | Stable | No change |
| I145V | Stable | No change |
| N94V (helix α3) | Stable | No change |
| N94F (helix α3) | Stable | No change |
| N105V (helix α3) | Stable | No change |
| N105F (helix α3) | Stable | No change |
Increasing the hydrophobicity of helix α3 does not affect toxicity.
Among the seven α-helix peptides, the synthetic peptide of helix α3 exhibited the lowest level of binding to phospholipid vesicles (9). It was thought that binding could be enhanced by converting hydrophilic surface residues to hydrophobic ones, thereby increasing toxicity. Asparagines 94 and 105 were identified as being solvent exposed (14) and were mutated to either V or F (Table 2). None of any of the four single-amino-acid substitutions had any effect on toxicity or toxin stability.
Binding to a toxin receptor and oligomerization within the membrane.
To test whether the lack of toxicity of the helix α4 mutants was due to a loss of receptor binding, ligand blot and membrane insertion studies were performed. The nontoxic helix α4 mutant toxins bound as well as the wild-type toxin to a single 120-kDa protein in M. sexta BBMV (Fig. 2). Nontoxic helix α4 mutant toxins were recovered from BBMV as oligomers of ca. 200 and 130 kDa to about the same extent as the wild-type toxin (Fig. 3). Results like those shown for mutants Q133R and I132S in Fig. 3 were obtained with all of the helix α4 mutant toxins. In contrast, a nontoxic helix α5 mutant toxin, A164P, did not insert efficiently into the membrane or oligomerize as previously reported for this mutant and other nontoxic helix α5 mutants (4).
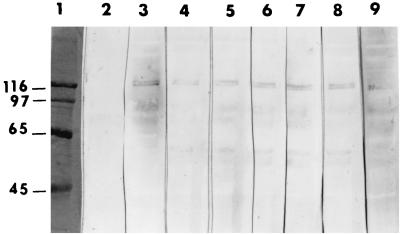
FIG. 2.
Immunoblot of M. sexta BBMV solubilized proteins fractionated by SDS–8% PAGE and incubated with Cry1Ac1 wild-type and mutant toxins. The blots were developed after treatment with rabbit anti-Cry1Ac1 antibody followed by an anti-rabbit antibody–alkaline phosphatase conjugate. Lane 1, molecular mass standards (in kilodaltons); lane 2, BBMV not incubated with toxin; lane 3, BBMV incubated with wild-type toxin; lane 4, BBMV incubated with mutant toxin Q133R; lane 5, BBMV incubated with mutant toxin I132S; lane 6, BBMV incubated with mutant toxin I132L; lane 7, BBMV incubated with mutant toxin I132V; lane 8, BBMV incubated with mutant toxin I132N; lane 9, BBMV incubated with mutant toxin A164P. Mutant toxins R131L and I132V-D136Y, with reduced toxicity, also bound to the 120-kDa BBMV protein (data not shown).
FIG. 3.
Binding and oligomerization of toxins in M. sexta BBMV. BBMV (20 μg of protein) were incubated with 60 ng of toxin at 30°C for 1 h. The vesicles were centrifuged, washed, and extracted as described in Materials and Methods. The extracts were subjected to electrophoresis (SDS–6% polyacrylamide gels) and western blotting, and the immunoblots were treated with rabbit anti-Cry1Ac1 antibody and then an anti-rabbit antibody conjugated with alkaline phosphatase. Lane 1, molecular mass standards; lane 2, wild-type toxin; lane 3, wild-type toxin extracted from BBMV; lane 4, mutant toxin A164P; lane 5, mutant toxin A164P extracted from BBMV; lane 6, mutant toxin Q133R; lane 7, mutant toxin Q133R extracted from BBMV; lane 8, mutant toxin I132S; lane 9, mutant toxin I132S extracted from BBMV; lane 10, BBMV. All other nontoxic helix α4 mutants also oligomerized in the membrane (data not shown).
Nontoxic helix α4 mutant toxins are affected in BBMV permeability.
Toxin-induced changes in BBMV permeability were measured by a light scattering assay (Fig. 4). The rate of decrease for the wild-type toxin was much higher than that for the I132L mutant, with the latter being close to the buffer control value. Similar analyses were done for all of the nontoxic helix α4 mutant toxins, and in all cases there was a seven- to eightfold difference in the initial rate compared to that of the wild type (Table 3).
FIG. 4.
Toxin-induced permeability changes in M. sexta BBMV as measured by a decrease in light scattering with time (5). x, wild-type toxin (w.t.); o, I132L mutant toxin; +, BBMV with buffer only.
DISCUSSION
Six of 12 single-amino-acid changes within helix α4 resulted in either a total loss of or greatly reduced toxicity. This frequency is comparable to that found for random mutations in helix α5 (27). No nontoxic mutants had been isolated following random mutagenesis in the regions of the cry1Ac1 gene encoding helices α2 and α6 (2, 27). As discussed below, mutations of surface residues within helix α3 did not affect toxicity (Table 2). Helix α1 does not bind to synthetic phospholipid vesicles (9), and it is the only part of the toxin that is susceptible to protease K after binding of the toxin to BBMV (4). It is unlikely, therefore, to play a critical role in toxin function within the membrane.
Since only mutations in helices α4 and α5, among six of the seven helices comprising domain I, affected toxicity, this portion of the toxin must contribute significantly to the formation and function of an ion channel. The α4-loop-α5 is the most hydrophobic region of the Cry1A toxins (10), and it probably inserts into the membrane, as indicated by studies with synthetic peptides of these helices (9). Helix cross-linking studies of the Cry1Aa1 toxin indicate the importance of this region of domain I in toxicity (23). Studies of mutants with single proline substitutions in helix α4 or α3 of the Cry4Ba1 toxin also suggested the importance of helix α4 in toxicity (25).
Single substitutions for 6 of the 10 residues encoded by the mutagenic oligonucleotide were identified, 4 within the hydrophobic face and 2 within the hydrophilic face of the amphipathic helix (Fig. 5). Among the former, only one in five mutations (Q133R) resulted in the loss of toxicity. Interestingly, the Q133H mutant was fully toxic, as were the other endotoxins with mutations in this region, which were largely hydrophobic-to-hydrophobic changes. It was also interesting that F134L was about threefold more toxic than the wild type. Five mutations (affecting only two residues) in the hydrophilic face resulted in the loss of toxicity. One, R131L, involved the loss of a charge; the other four were relatively conserved changes of I132. None of the nontoxic mutants was affected in terms of binding to the receptor. It appears that I132, which is a hydrophobic residue within the hydrophilic face of the helix, and Q133, which is within the hydrophobic face, have critical functions in the properties of the ion channel. Since the F134L mutant showed a toxicity increase versus the wild type for two different insects, further studies are under way to characterize this mutant and to study the effect of other mutations of this residue.
FIG. 5.
Helical wheel of 18 residues in helix α4, including the 10 mutagenized residues (E129 to N138). Mutations which resulted in the loss of toxicity (including one double mutant) are indicated by solid arrows. Changes with no resultant loss of toxicity are indicated by dashed arrows. The asterisk indicates an increase in toxicity. Parentheses indicate double mutations. The hydrophobic and hydrophilic faces are demarcated by the internal lines.
None of six mutations among four residues either at the carboxyl end of helix α4 or within the loop connecting helices α4 and α5 resulted in the loss of toxicity (Table 2). Some of these changes were relatively conserved, but others involved the substitution of a charged residue (I145D or V150D) or replacement of a large residue (F148I). Following toxin insertion into the membrane, this loop could be close to the cytoplasmic side of the membrane or might even project into the cytoplasm. It was conceivable, therefore, that there was a specific interaction of this loop with a cytoplasmic component which was important for toxicity. If there were such an interaction, it is not likely that A149 and V150 would be involved.
Site-directed mutagenesis of two N residues in helix α3 was undertaken to enhance the hydrophobic properties of this helix, since a synthetic peptide of helix α3 bound very poorly to phospholipid vesicles (9). Residues N94 and N105 are surface exposed (14), so the mutations should have increased the hydrophobicity of this helix and, thus, its affinity for BBMV. Since there was no change in toxicity for any of the four mutants (Table 2), binding studies were not done. It appears that the surface properties of this helix are not critical for toxicity. An unexpected result was the ability of the nontoxic helix α4 mutants to insert into BBMV and oligomerize as well as the wild-type toxin (Fig. 3). It should be noted, however, that the relative rates of insertion were not determined. An inactive helix α5 mutant toxin, A164P, did not remain bound to the membrane, nor did other nontoxic helix α5 mutant toxins (4), although they all bound to a 120-kDa protein from BBMV in immunoblots (as in Fig. 2). All helix α5 mutant toxins which retained toxicity, except for H168R, did oligomerize (4).
Some very large (>200-kDa) toxin oligomers have been found in purified Cry1Ac1 and other toxins, and this capacity to aggregate may be important for toxin insertion into the membrane after binding to the receptor (11). There is some aggregation of purified Cry1Ac1, but not Cry1Ab3, toxin in solution, but in both cases the formation of ca. 200-kDa oligomers was enhanced by incubating purified toxin with BBMV (4). In addition, this oligomer is not a complex of a toxin molecule and the 120-kDa aminopeptidase N receptor (15), since antibody to the latter did not react with this oligomer (4). While interaction with other membrane components has not been ruled out, the formation of a toxin trimer is likely. The lower 130-kDa band could represent toxin dimers.
Helix α5 seems to be very important for oligomerization, perhaps among its other functions in toxicity. In contrast, mutations within helix α4 did not affect the capacity of the toxin to oligomerize in BBMV, despite the lack of permeability to ions in light scattering experiments. A different role for this helix, most likely in the function of the ion channel, is indicated. It was recently reported that a nontoxic helix α4 mutant toxin (N135Q) of Cry1Ac1 was altered in a second phase of binding, as measured in a BIOCORE biosensor instrument with aminopeptidase N anchored in synthetic phospholipid (6). The inability to form an irreversible association implied a lack of membrane penetration by this helix α4 mutant, in contrast to the results with other helix α4 mutants reported here. The difference in results may be due to the specific mutation or, more likely, the use of BBMV, rather than synthetic phospholipids, in the present experiments.
ACKNOWLEDGMENTS
This research was supported by a grant from the USDA BARD program.
Jeffrey Bolin provided the program and expertise for determining solvent-exposed residues. Jeffrey Lucas provided the SAS program and expertise for the probit analysis. William Cramer and Stanislav Zakharov were most helpful in the light scattering measurements. The technical assistance of Lan Wu in BBMV preparations is gratefully acknowledged.
REFERENCES
- 1.Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 2.Aronson A I, Wu D, Zhang C. Mutagenesis of specificity and toxicity regions of a Bacillus thuringiensis prototoxin gene. J Bacteriol. 1995;177:4059–4065. doi: 10.1128/jb.177.14.4059-4065.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson A I, Han E-S, McGaughey W, Johnson D. The solubility of inclusion proteins from Bacillus thuringiensis is dependent upon protoxin composition and is a factor in toxicity to insects. Appl Environ Microbiol. 1991;57:981–986. doi: 10.1128/aem.57.4.981-986.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronson A I, Geng C, Wu L. Aggregation of Bacillus thuringiensis Cry1A toxins upon binding to target insect larval midgut vesicles. Appl Environ Microbiol. 1999;65:2503–2507. doi: 10.1128/aem.65.6.2503-2507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll J, Wolfesberger M G, Ellar D J. The Bacillus thuringiensis CryIAc toxin-induced permeability change in Manduca sexta midgut brush border membrane vesicles proceeds by more than one mechanism. J Cell Sci. 1997;110:3099–3104. doi: 10.1242/jcs.110.24.3099. [DOI] [PubMed] [Google Scholar]
- 6.Cooper A M, Carroll J, Travis E R, Williams D H, Ellar D J. Bacillus thuringiensis CryIAc toxin interaction with Manduca sexta aminopeptidase N in a model membrane environment. Biochem J. 1998;333:677–683. doi: 10.1042/bj3330677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crickmore N, Zeigler D R, Feitelson J, Schnepf E, Van Rie J, Lereclus D, Baum J, Dean D H. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.English L, Robbins H L, Van Tersch M A, Kulesza C A, Ave D, Coyl D, Jany S C, Slain S L. Mode of action of Cry IIA: a Bacillus thuringiensis delta endotoxin. Insect Biochem Mol Biol. 1994;24:1025–1035. [Google Scholar]
- 9.Gazit E, La Rocca P, Sansom M S P, Shai Y. The structure and organization within the membrane of the helices composing the pore-forming domain of Bacillus thuringiensis δ-endotoxin are consistent with an “umbrella-like” structure of the pore. Proc Natl Acad Sci USA. 1998;95:12289–12294. doi: 10.1073/pnas.95.21.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grochulski P, Masson L, Borisova S, Pusztai-Carey M, Schwartz J L, Brousseau R, Cygler M. Bacillus thuringiensis CryIA(a) insecticidal toxin: crystal structure and channel formation. J Mol Biol. 1995;254:1–18. doi: 10.1006/jmbi.1995.0630. [DOI] [PubMed] [Google Scholar]
- 11.Guereca L, Bravo A. The oligomeric state of Bacillus thuringiensis Cry toxins in solution. Biochim Biophys Acta. 1999;1429:342–350. doi: 10.1016/s0167-4838(98)00241-6. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann C, Vanderbruggen H, Hofte H, Van Rie J, Jansens S, Van Mellaert H. Specificity of Bacillus thuringiensis delta-endotoxins is correlated with the presence of high affinity binding sites in the brushborder membrane of target insect midguts. Proc Natl Acad Sci USA. 1988;85:7844–7848. doi: 10.1073/pnas.85.21.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Höfte H, Whiteley H R. Insecticidal crystal protein of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbard S J, Thornton J M. NACCESS computer program. London, United Kingdom: Department of Biochemistry and Molecular Biology, University College; 1993. [Google Scholar]
- 15.Knight J K, Crickmore N, Ellar D J. The receptor of Bacillus thuringiensis CryIA(c) delta-endotoxin in the brushborder membrane of the lepidopteran Manduca sexta is aminopeptidase N. Mol Microbiol. 1994;11:429–436. doi: 10.1111/j.1365-2958.1994.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowles B H. Mechanism of action of Bacillus thuringiensis insecticidal δ-endotoxins. Adv Insect Physiol. 1994;24:275–308. [Google Scholar]
- 17.Knowles B H, Ellar D J. Colloid osmotic lysis is a general feature of the mechanism of action of Bacillus thuringiensis delta-endotoxins with different insect specificities. Biochim Biophys Acta. 1987;924:509–518. [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Carroll J, Ellar D J. Crystal structure of an insecticidal protein. The delta-endotoxin of Bacillus thuringiensis subsp. tenebrionis at 2.5 Å resolution. Nature. 1991;353:815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- 20.Mohammed S I, Johnson D E, Aronson A I. Altered binding of the Cry1Ac toxin to larval membranes but not to the toxin-binding protein in Plodia interpunctella selected for resistance to different Bacillus thuringiensis isolates. Appl Environ Microbiol. 1996;62:4168–4173. doi: 10.1128/aem.62.11.4168-4173.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler D R, Dean D H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz J L, Juteau M, Grochulski P, Cygler M, Prefontaine G, Brosseau R, Masson L. Restriction of intramolecular movements within the CryIA(a) toxin molecule of Bacillus thuringiensis through disulphide bond engineering. FEBS Lett. 1997;410:397–402. doi: 10.1016/s0014-5793(97)00626-1. [DOI] [PubMed] [Google Scholar]
- 24.Stahly C P, Dingman D W, Bulla L A, Jr, Aronson A I. Possible origin and function of the parasporal crystals in Bacillus thuringiensis. Biochem Biophys Res Commun. 1978;84:581–585. doi: 10.1016/0006-291x(78)90745-3. [DOI] [PubMed] [Google Scholar]
- 25.Uawithya P, Tuntitippawan T, Katzenmeier G, Panyim S, Angsuthanasombat C. Effects of larvicidal activity of single proline substitutions in α3 or α4 of the Bacillus thuringiensis Cry4B toxin. Biochem Mol Biol Int. 1998;44:825–832. doi: 10.1080/15216549800201872. [DOI] [PubMed] [Google Scholar]
- 26.Wolfersberger M G, Luthy P, Mauer A, Parenti P, Sacchi V F, Giordano B, Hanozet G M. Preparation and partial characterization of amino acid transporting brushborder membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae) Comp Biochem Physiol. 1987;A86:301–312. [Google Scholar]
- 27.Wu D, Aronson A I. Localized mutagenesis defines regions of the Bacillus thuringiensis delta endotoxin involved in toxicity and specificity. J Biol Chem. 1992;267:2311–2317. [PubMed] [Google Scholar]