Summary
RAD23 (RADIATION SENSITIVE23) proteins are a group of UBL‐UBA (ubiquitin‐like‐ubiquitin‐associated) proteins that shuttle ubiquitylated proteins to the 26S proteasome for breakdown. Drought stress is a major environmental constraint that limits plant growth and production, but whether RAD23 proteins are involved in this process is unclear. Here, we demonstrated that a shuttle protein, MdRAD23D1, mediated drought response in apple plants (Malus domestica). MdRAD23D1 levels increased under drought stress, and its suppression resulted in decreased stress tolerance in apple plants. Through in vitro and in vivo assays, we demonstrated that MdRAD23D1 interacted with a proline‐rich protein MdPRP6, resulting in the degradation of MdPRP6 by the 26S proteasome. And MdRAD23D1 accelerated the degradation of MdPRP6 under drought stress. Suppression of MdPRP6 resulted in enhanced drought tolerance in apple plants, mainly because the free proline accumulation is changed. And the free proline is also involved in MdRAD23D1‐mediated drought response. Taken together, these findings demonstrated that MdRAD23D1 and MdPRP6 oppositely regulated drought response. MdRAD23D1 levels increased under drought, accelerating the degradation of MdPRP6. MdPRP6 negatively regulated drought response, probably by regulating proline accumulation. Thus, “MdRAD23D1‐MdPRP6” conferred drought stress tolerance in apple plants.
Keywords: apple, UBL‐UBA protein, drought stress, proline‐rich protein, degradation via ubiquitination, free proline accumulation
Introduction
Because plants are unable to move, they encounter diverse environmental stressors, such as drought, high salinity, and extreme temperatures, which inhibit their growth and development (Zhu, 2016). Drought stress is a major environmental constraint on agricultural production, it affects many physiological processes, including photosynthesis, stomatal opening and closing, and the detoxification of reactive oxygen species (ROS). To cope with drought stress, plants have evolved many morphological, physiological, and molecular mechanisms to minimize damage (Farooq et al., 2009; Liu et al., 2019; Ogura et al., 2019; Zhao et al., 2020). For example, ubiquitination, which is a vital way to regulate protein activities and levels at post‐translational level in cells, has been shown to mediate drought response in plants. Wheat (Triticum aestivum) proteins TaPUB2 and TaPUB3 are E3 ligases that positively regulate Arabidopsis response to drought stress (Kim et al., 2022). The UBC27‐AIRP3 complex modulates the ABA signalling by promoting ABI1 degradation through ubiquitination in Arabidopsis (Pan et al., 2020).
The UBL‐UBA (ubiquitin‐like‐ubiquitin‐associated) proteins are ubiquitin receptors and transporters in the ubiquitin‐proteasome system (UPS). The UBL domain in UBL‐UBA proteins can physically interact with the proteasome receptors Rpn1, Rpn10, and Rpn13 and the UBA domain is linked with ubiquitinated substrates (Finley, 2009; Tsuchiya et al., 2017; Zhang et al., 2009). There are possibly five different types of UBL‐UBA proteins, including RAD23 (RADIATION SENSITIVE23; Farmer et al., 2010; Lambertson et al., 1999; Schultz and Quatrano, 1997; Sturm and Lienhard, 1998), DSK2 (DOMINANT SUPPRESSOR OF KAR2; Funakoshi et al., 2002; Kang et al., 2006; Wang et al., 2020), DDI1 (DNA DAMAGE‐INDUCIBLE1; Gabriely et al., 2008), NUB1 (NEDD8 ultimate buster 1; Kito et al., 2001), and possibly UBL7 (the Ub‐like 7; Liu et al., 2003). After being labelled by the poly‐Ub chains, the target protein is recognized and delivered to the 26S proteasome for degradation, and this process is precisely controlled by the UBL‐UBA proteins. The vital role of UBL‐UBA proteins in protein degradation through the UPS pathway makes them important regulators of growth and stress response in plants. The Arabidopsis protein RAD23B regulates pollen development by mediating degradation of KRP1 (KIP‐related protein 1; Li et al., 2020a). The rice (Oryza sativa) protein OsDSK2a mediates seedling growth and salt response by regulating the degradation of its substrate protein EUI, thus regulating gibberellin metabolism (Wang et al., 2020).
The RAD23 proteins are first known for their role in repairing the damaged DNA caused by UV (Guzder et al., 1998). Plants' RAD23 isoforms can rescue the UV‐sensitive phenotype of rad23D in yeast, they also contribute to UV tolerance in plants (Lahari et al., 2018; Sturm and Lienhard, 1998). As the UBL‐UBA proteins, RAD23 proteins have been proven to be essential in the cell cycle, morphology, and fertility of plants through their delivery of UPS substrates to the 26S proteasome, as evidenced by the severely dwarfed plants or reproductive lethality in different Arabidopsis mutants (Farmer et al., 2010; Li et al., 2020a). Proteomic analysis points to the role of them in apical dominance in Pinus sylvestris (Ning et al., 2013). Besides, RAD23 proteins appear to be the key factors in pathogen‐plant interaction. Arabidopsis RAD23C and RAD23D function in insect colonization via interacting with a phytoplasma virulence protein SAP54 (secreted AY‐WB protein), which can degrade MADS‐box proteins to hijack plant reproduction (MacLean et al., 2014). A recent study shows RAD23 proteins interact with bacterium effectors Lso‐HPE1 (Lso hypothetical protein effector 1) and CLIBASIA_00460 from Candidatus Liberibacter solanacearum and Candidatus Liberibacter asiaticus, respectively, thus to promote infection (Oh et al., 2022).
Proline‐rich proteins (PRPs), a type of cell wall proteins rich in proline and hydroxyproline, play important roles in the drought stress response in plants (Gujjar et al., 2018; Liu et al., 2018). The proline is an excellent osmolyte, its accumulation helps the cell to maintain the water content and swelling potential, thereby preserving normal cellular function under stresses. It has been clearly demonstrated that plants accumulate proline via the P5CS pathway under stresses (Perez‐Arellano et al., 2010). And studies show that the accumulation of proline can also be achieved independent of the P5CS pathway, through the degradation of PRP proteins under stress conditions (Barthakur et al., 2001; Gujjar et al., 2019; Stines et al., 1999). The tomato proline‐rich protein SlPRP is rapidly down‐regulated while the concentration of proline increases in cells under drought stress, helping plants improve the drought resistance (Gujjar et al., 2018). Elevated cellular proline content, in turn, may promote PRP levels (Stein et al., 2011). Besides the temporary pool of proline, PRPs are also implicated in cell wall signal transduction cascades (Kishor et al., 2015). The hybrid PRPs may facilitate signal molecule transport at specific sites in cells (Banday et al., 2022). The phosphorylated AZTI (azelaic acid induced 1), a hybrid PRP, may function as a membrane signalling protein in the response to stresses in plants (Pitzschke et al., 2014a,b, 2016).
Apple trees are perennial woody fruit trees, and the apple is an economically important fruit around the world. However, drought stress imposes limitations on the development of apple trees and apple production (Mihaljevic et al., 2021; Sun et al., 2018). The UBL‐UBA proteins have been demonstrated to function in various stress responses in different plant species; however, little is known about them in apple plants. In the present study, we identified a UBL‐UBA protein MdRAD23D1 from the ‘Golden delicious’ apple. We demonstrated that MdRAD23D1 positively regulated the drought response in apple plants by regulating the levels of MdPRP6, which is a PRP (Zhang et al., 2021a). MdRAD23D1 can physically interact with MdPRP6, resulting in the degradation of MdPRP6 through the 26S proteasome. In contrast to MdRAD23D1, MdPRP6 negatively regulated the drought response in apple plants. The accumulation of free proline increased in MdPRP6‐Ri plants and decreased in MdRAD23D1‐Ri plants under drought, and exogenous proline alleviated drought stress in both MdPRP6‐cOE calli and MdRAD23D1‐Ri plants. Thus, we suggested that the MdRAD23D1‐MdPRP6 module regulated the drought response of plants, and this process was probably linked to the accumulation of free proline in cells.
Results
The UBL‐UBA protein MdRAD23D1 plays a positive role in apples responding to drought stress
We previously identified MdRAD23 family members in the apple genome (Wang et al., 2017), like Arabidopsis RAD23 proteins, MdRAD23s also contained the signature UBL domain at the N terminal and UBA domain at the C terminal (Figure S1a). The evolutionary relationship between Arabidopsis and apples was analysed, and we found that C and D type RAD23s were close in the phylogenetic tree (Figure S1b). MdRAD23D1, MdRAD23D2, and MdRAD23A were, respectively, collinear with AtRAD23C, AtRAD23D, and AtRAD23A (Figure S1c). Among these three genes, the expression level of MdRAD23D1 was induced by both ABA treatment and drought stress (Wang et al., 2017, Figure 1a). We also examined the MdRAD23D1 protein levels under drought using ‘Orin’ apple calli. 200 mm mannitol was applied to apple calli to monitor osmotic stress caused by drought, and MdRAD23D1 protein obviously accumulated under mannitol treatment (Figure 1b). To explore the biological function of MdRAD23D1 in response to drought stress, we obtained three transgenic apple plants with suppressed expression of MdRAD23D1, including Ri18, Ri22, and Ri23 (Figure S2a). qPCR analyses showed that the relative expression levels of MdRAD23D1 decreased by 50%–70% in Ri plants compared with wild type (WT; Figure S2b), without suppression in the expression levels of the other MdRAD23 members (Figure S2d). The MdRAD23D1 protein levels were also obviously lower in Ri plants when detected using anti‐MdRAD23D1 antibody (Figure S2c). After being treated with drought for 8 days, Ri plants exhibited severe leaf wilting, whereas WT was less affected (Figure 1c). Ri leaves drooped away from the stem, to form bigger leaf angles than in WT (Figure 1d). Lower relative water content (RWC) and higher EL were detected in Ri plants than in WT (Figure 1e,f). Furthermore, values for Pn and Fv/Fm were lower in Ri than in WT plants under drought treatment (Figure 1g,h), consistent with weaker fluorescence observed in Ri leaves (Figure 1i). The MDA content was higher in Ri plants (Figure S3a), the stains of 3,3‐diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) produced darker colour in Ri than in WT plants (Figure S3b), and higher ROS accumulation was observed in Ri plants by quantitative analyses (Figure S3c,d). Taken together, these results suggest that MdRAD23D1 plays a positive role in the drought stress response in apple plants.
Figure 1.
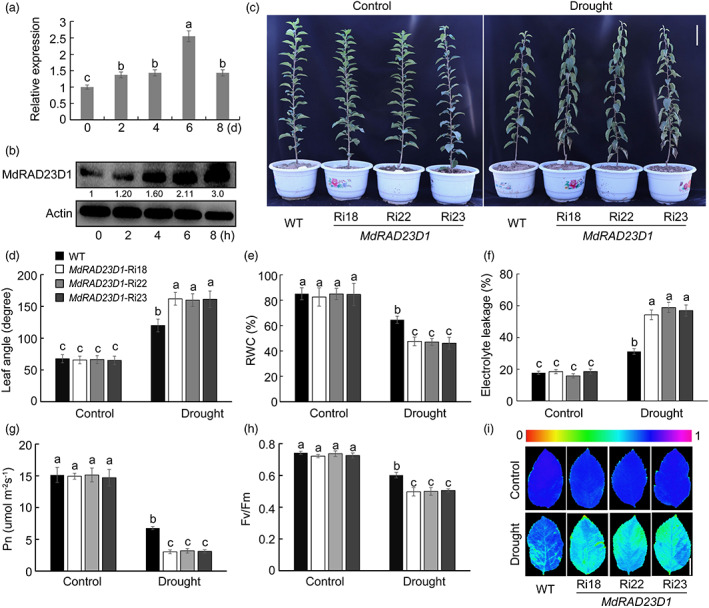
Expression patterns of MdRAD23D1 and response of MdRAD23D1 transgenic plants to drought. (a) The expression level of MdRAD23D1 under drought stress. qPCR was carried out using the GL‐3 apple (Malus domestica) leaves after drought treatment. (b) MdRAD23D1 levels under drought stress. 200 mm mannitol was applied to the ‘Orin’ apple (M. domestica) calli to monitor drought stress. The calli samples were collected at 0, 2, 4, 6, and 8 h, and a specifically synthesized anti‐MdRAD23D1 antibody was used to detect MdRAD23D1 protein levels. MdActin was used as the loading control. The amount of MdRAD23D1 was quantified using an Ultra‐sensitive multifunctional imager (Uvitec), and the protein level at 0 h was set to 1. (c) Morphology differences of WT and transgenic apple plants under control and drought conditions. Forty‐five‐day‐old WT and MdRAD23D1‐Ri plants in the greenhouse were withheld from water for 8 days. The scale bar = 8 cm. (d–i) Leaf angle (d), relative water content (e), electrolyte leakage (f), Net photosynthesis (Pn) (g), Fv/Fm ratio (h), and chlorophyll fluorescence images (i) in WT and transgenic plants with and without drought treatment. The scale bar in (i) = 3 cm. WT, wild type, here we used GL‐3 apple (M. domestic), which was also used as explants in generating transgenic apple plants; MdRAD23D1‐Ri18/22/23: transgenic apple plants with suppressed expression of MdRAD23D1 via RNA‐interference. Data are shown as the means ± SD. Different letters indicate significant differences according to the one‐way ANOVA followed by Tukey's multiple range test (P < 0.05).
AtRAD23 proteins have proven to interact with multiple components of the UPS, so we also investigated whether the apple protein MdRAD23D1 has similar interaction. The purified MdRAD23D1‐GST protein bound to anti‐GST magnetic beads was co‐incubated with polyubiquitin chains assembled via K48 or K63 linkages (Ub2‐6), and the results showed that MdRAD23D1 bound to both K48‐ and K63‐linked polyubiquitin chains well (Figure 2a). In addition, MdRAD23D1 can interact with MdRPN13, which is a ubiquitin receptor of the 26S proteasome subunit (Husnjak et al., 2008), in vitro and in vivo (Figure 2b–e). Thus, we conclude that MdRAD23D1 associates with the 26S proteasome and is a receptor for ubiquitinated proteins, serving as a shuttle factor in protein degradation via ubiquitination.
Figure 2.
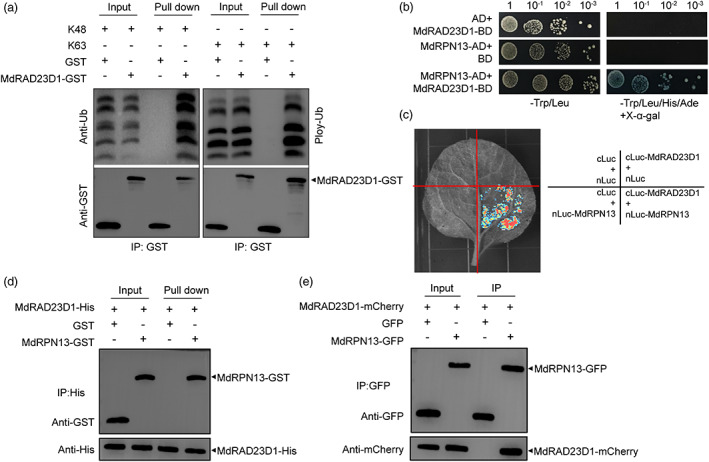
MdRAD23D1 binds poly‐ubiquitin (poly‐Ub) chains and interacts with the 26S proteasome. (a) Binding of MdRAD23D1 to poly‐Ub chains in vitro. The Escherichia coli–expressed MdRAD23D1‐GST fusion protein and GST protein were first incubated with anti‐GST magnetic beads and then the mixture was incubated with poly‐Ub chains linked via K48 or K63. The bound proteins were eluted and separated by SDS‐PAGE and the amount of protein levels were determined by immunoblotting with anti‐GST and anti‐Ub antibodies. Arrowhead shows the MdRAD23D1‐GST protein. (b–e) Interaction analysis of MdRAD23D1 and MdRPN13, a subunit of 26S proteasome via yeast two‐hybrid (b), Split‐Luc assay (c), Pull‐down (d), and Co‐IP (e). In yeast two‐hybrid assay, the full‐length CDS of MdRAD23D1 and MdRPN13 was cloned into the pGBKT7 and pGADT7 vectors, respectively. After co‐transformation, yeast cells were cultured on SD base/‐Leu/‐Trp (left) and SD base/‐Leu/‐Trp/‐His/‐Ade+x‐α‐gal (right). In the Split‐Luc assay, the full‐length CDS of MdRAD23D1 and MdRPN13 were cloned into pRI‐101‐cLuc and pRI‐101‐nLuc, respectively. The fusion proteins MdRAD23D1‐cLuc and MdRPN13‐nLuc were transiently co‐expressed in Nicotiana benthamiana leaves, and the fluorescence signal was observed by an Ultra‐sensitive multifunctional imager (Uvitec). In the Pull‐down assay, Escherichia coli–expressed MdRAD23D1‐His protein was first incubated with anti‐His magnetic beads and then the mixture was incubated with E. coli‐expressed MdRPN13‐GST or GST. The bound proteins were eluted and detected using anti‐GST and anti‐His antibodies. In the Co‐IP assay, the fusion proteins MdRAD23D1‐mCherry and MdRPN13‐GFP were transiently co‐expressed in N. benthamiana leaves, and the total proteins were extracted and immunoprecipitated with anti‐GFP magnetic beads. Immunoblotting was performed with anti‐GFP and anti‐mCherry antibodies.
MdRAD23D1 interacts with a proline‐rich protein, MdPRP6
As mentioned above, MdRAD23D1 was a positive regulator in drought stress response in apples. As a first approach, we screened an apple cDNA library by the yeast‐two hybrid using the CDS of MdRAD23D1 as the bait. Multiple prey cDNAs identified by the screen expressed a putative 14 KDa PRP, which corresponded to previously named MdPRP6 (Zhang et al., 2021a) and was selected for further study. MdPRP6 has a transmembrane domain and a signal peptide is located in the cell membranes (Zhang et al., 2021a, Figure S4a,b), and MdRAD23D1 is located in the nucleus and the cell membranes (Figure S4a,c), the same location in the cell also suggested the potential relation between MdPRP6 and MdRAD23D1. To further determine the interaction between MdRAD23D1 and MdPRP6, the full‐length CDS of MdPRP6 was cloned using ‘Golden Delicious’ apple cDNA and fused to the pGADT7 vector. Yeast cells co‐transformed with both MdRAD23D1 and MdPRP6 grew well and exhibited blue colour on ‐Trp/‐Leu/‐His/‐Ade+x‐α‐gal medium (Figure 3a). When MdRAD23D1 and MdPRP6 were co‐expressed in Nicotiana benthamiana leaves, the strong luciferase signal was detected with an Ultra‐sensitive multifunctional imager (Figure 3b). In the pull‐down assay, we incubated purified MdRAD23D1‐GST with MdPRP6‐His, and the interaction between them was detected after pull‐down (Figure 3c). We also performed Co‐IP assay using N. benthamiana leaves co‐expressing MdRAD23D1‐GFP and MdPRP6‐Flag, and the immunoblot analyses showed that MdRAD23D1 interacted with MdPRP6 (Figure 3d). Together, these results demonstrate that MdRAD23D1 interacts with MdPRP6 in vitro and in vivo.
Figure 3.
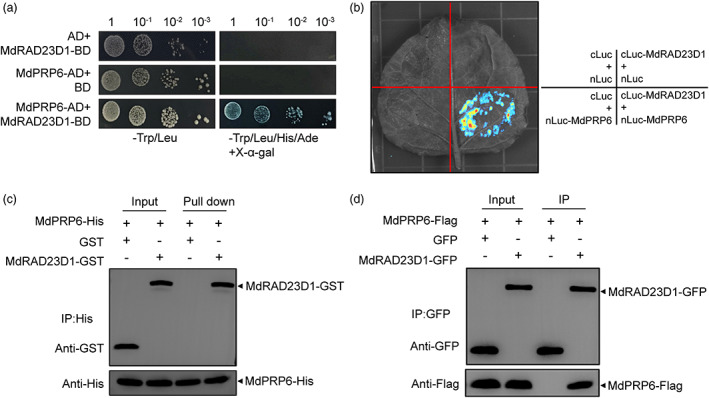
MdRAD23D1 interacts with MdPRP6 in vivo and in vitro. (a) Yeast two‐hybrid assays. The full‐length CDS of MdRAD23D1 and MdPRP6 were cloned into pGBKT7 and pGADT7 vectors, respectively. Co‐transformed yeast cells were grown on SD‐Trp/‐Leu and SD‐Trp/‐Leu/‐His/‐Ade+X‐α‐gal medium. (b) The Split‐Luc assay. The full‐length CDS of MdRAD23D1 and MdPRP6 were cloned into pRI‐101‐cLuc and pRI‐101‐nLuc, respectively. The fusion proteins MdRAD23D1‐cLuc and MdPRP6‐nLuc were transiently co‐expressed in Nicotiana benthamiana leaves, and the fluorescence signal was observed by an Ultra‐sensitive multifunctional imager (Uvitec). (c) Pull‐down assays. Escherichia coli–expressed MdPRP6‐His protein was first incubated with anti‐His magnetic beads and then the mixture was incubated with E. coli‐expressed GST or MdRAD23D1‐GST. The bound proteins were eluted and detected using anti‐GST and anti‐His antibodies. (d) Co‐IP assay. The fusion proteins MdRAD23D1‐GFP and MdPRP6‐Flag were transiently co‐expressed in N. benthamiana leaves, and the total proteins were extracted and immunoprecipitated with anti‐GFP magnetic beads. Immunoblotting was performed with anti‐GFP and anti‐Flag antibodies.
MdPRP6 negatively regulates the drought response of apple
Previously we demonstrated that MdPRP6 was expressed in leaves, stems, and roots of apple plants, and it played a role in plants responding to heat, salt, and low nitrogen stresses (Zhang et al., 2021a, 2022, 2023). Here, we further examined the spatial expression patterns of MdPRP6 by in situ hybridization in different tissues. Strong signals of MdPRP6 were detected in vascular tissues, such as xylem and phloem, implying that MdRPR6 might affect the water uptake of apple plants (Figure 4a). In contrast to MdRAD23D1, the expression levels of MdPRP6 were inhibited by drought stress (Figure 4b). We examined the MdPRP6 protein levels under drought using MdPRP6‐Flag transgenic apple calli (MdPRP6‐cOE in Figure S5a–c). MdPRP6 protein levels strongly decreased after 8 h of 200 mm mannitol treatment (Figure 4c), without a corresponding decrease in MdPRP6 expression levels (Figure S5d).
Figure 4.
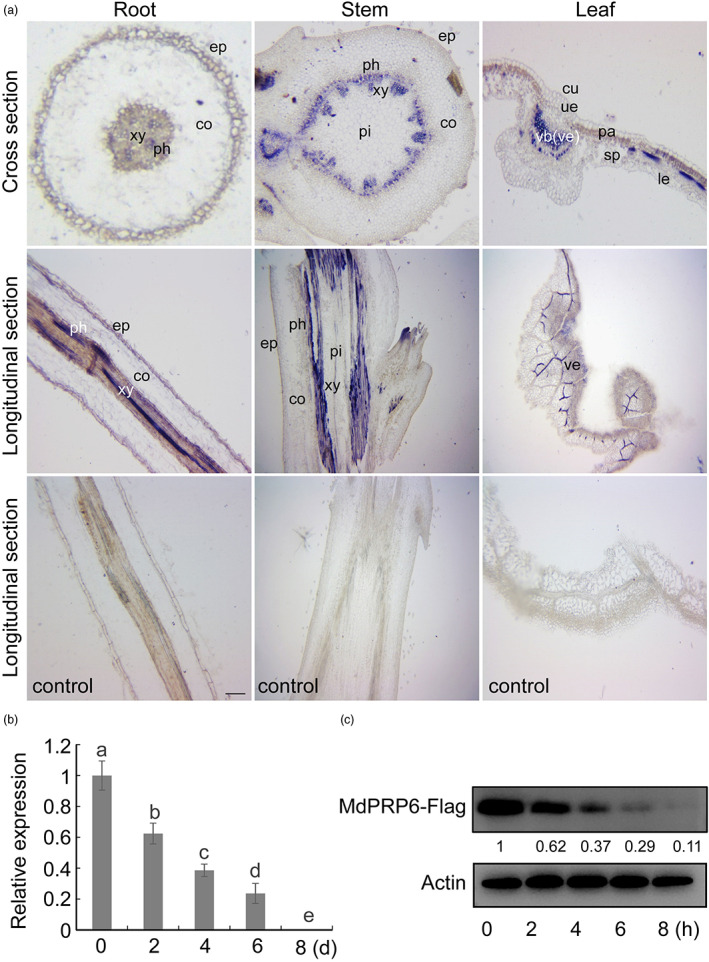
Expression patterns of MdPRP6. (a) In situ hybridization of MdPRP6 in roots, stems, and leaves of GL‐3 apple (Malus domestica). ep, epidermis; xy, xylem; ph, phloem; pi, pith; co, cortex; cu, cuticle; ue, upper epidermis; le, lower epidermis; pa, palisade mesophyll cells; sp, spongy mesophyll cells; vb, vascular bundle; ve, vein. The scale bar = 100 μm. (b) The expression level of MdPRP6 under drought stress. qPCR was carried out using the GL‐3 apple leaves after drought treatment. (c) MdPRP6 levels under drought stress. 200 mm mannitol was applied to the transgenic apple calli (‘Orin’ calli, M. domestica) expressed 35S::MdPRP6‐Flag to monitor drought stress. The MdPRP6‐Flag calli were cultured on MS medium containing 200 mm mannitol and 75 μm CHX. The calli sample was collected at 0, 2, 4, 6 and 8 h and immunoblotting was performed with anti‐Flag antibody. MdActin was used as the loading control. The amount of MdPRP6 was quantified using an Ultra‐sensitive multifunctional imager (Uvitec), and the protein level at 0 h was set to 1. Data in (b) are shown as the means ± SD. Different letters indicate significant differences according to a one‐way ANOVA followed by Tukey's multiple range test (P < 0.05).
To gain a better understanding of the role of MdPRP6 under drought stress, we generated another transgenic line with suppressed expression of MdPRP6 in GL‐3 via RNA interference silencing (Ri3 in Figure S6a,b), without a decrease in expression levels of the other MdPRP members (Figure S6c, Ri1 and Ri2 were previously obtained in Zhang et al., 2022). After 8 days of plant exposure to drought stress, leaves of WT were substantially more wilting than those of Ri plants (Figure 5a,b). Values for leaf angles and EL were lower in Ri plants than in WT plants (Figure 5c,e), whereas values for RWC were higher in Ri plants (Figure 5d). Under drought treatment, Ri plants had higher Pn and Fv/Fm values than WT plants, as well as stronger chlorophyll fluorescence (Figure 5f–h). We also noticed that the MDA content was higher in WT plants than in Ri plants (Figure S7a). The leaves of WT plants were stained more densely with DAB and NBT compared with Ri leaves (Figure S7b). Ri plants under drought stress also had consistently lower H2O2 and contents (Figure S7c,d). These results demonstrate that MdPRP6 negatively regulates the drought stress response in apple plants.
Figure 5.
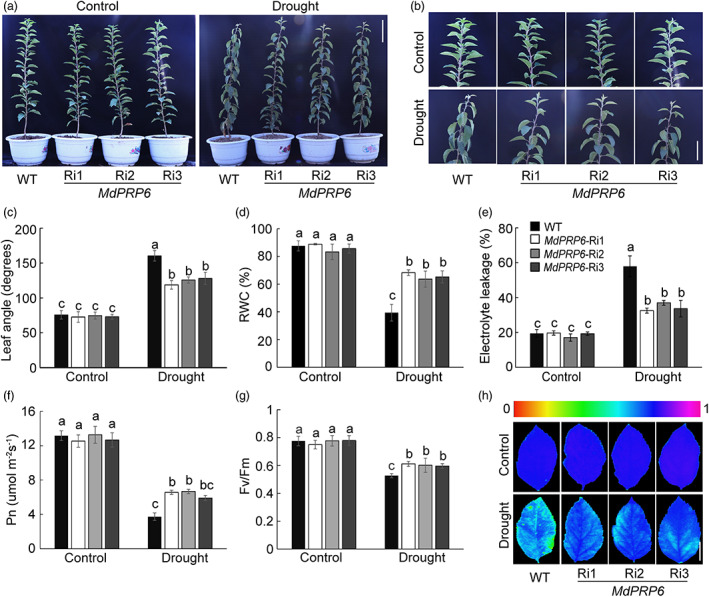
MdPRP6 negatively regulates the drought tolerance of apple plants. (a, b) Morphology differences in whole plants (a) and top enlarged view (b) of WT and transgenic apple plants under control and drought conditions. Forty‐five‐day‐old WT and MdPRP6‐Ri plants in the greenhouse were withheld from water for 8 days. The Scale bars in (a) and (b) were 15 cm and 7 cm, respectively. (c–h) Leaf angle (c), relative water content (d), electrolyte leakage (e), net photosynthesis (Pn) (f), Fv/Fm ratio (g), and chlorophyll fluorescence images (h) in WT and MdPRP6‐Ri plants with and without drought treatment. The Scale bar in (h) = 3 cm. Data are shown as the means ± SD. Different letters indicate significant differences according to the one‐way ANOVA followed by Tukey's multiple range test (P < 0.05). WT, wild type, here we used GL‐3 apple (Malus domestic), which was also used as explants in generating transgenic apple plants; MdPRP6‐Ri1/2/3, transgenic apple plants with suppressed expression of MdPRP6 via RNA‐interference.
MdRAD23D1 affects the protein levels of MdPRP6
We analysed the protein levels of MdRAD23D1 in MdPRP6‐cOE and MdPRP6‐cRi (Figure S5e,f) calli, using the anti‐MdRAD23D1 antibody. We found that the altered expression of MdPRP6 did not affect MdRAD23D1 levels (Figure S8a). To analyse whether MdRAD23D1 affects MdPRP6 levels, we generated MdRAD23D1‐cOE (Figure S9a–c) and MdRAD23D1‐cRi (Figure S9d–f) transgenic apple calli, and then expressed MdPRP6‐Flag in MdRAD23D1‐cOE and MdRAD23D1‐cRi calli (MdRAD23D1‐cOE/MdPRP6‐cOE and in MdRAD23D1‐cRi/MdPRP6‐cOE Figure S10a–d). We found that when MdRAD23D1 levels increased, MdPRP6‐Flag levels decreased, and vice versa (Figure S8b), indicating that MdRAD23D1 might function upstream of MdPRP6.
Given the shuttle function of MdRAD23D1, it probably transports ubiquitylated MdPRP6 to the 26S proteasome. As a first approach, we detected the MdPRP6 levels in MdPRP6‐Flag transgenic calli supplied with or without MG132. Without MG132, MdPRP6‐Flag levels decreased over time. In contrast, when MG132 was added, MdPRP6‐Flag was more stable (Figure 6a). When we incubated MdPRP6‐His with protein extracts from WT or MdRAD23D1‐Ri plants, MdPRP6‐His protein levels decreased more slowly in MdRAD23D1‐Ri plants than in WT, and MG132 inhibited the decrease in MdPRP6‐His levels in both two (Figure 6b). We also employed different co‐transgenic calli in the assay. At each time point, MdPRP6‐Flag was most abundant in MdRAD23D1‐cRi/MdPRP6‐cOE calli and least abundant in MdRAD23D1‐cOE/MdPRP6‐cOE calli (Figure 6c). We used anti‐Flag and anti‐Ubi antibodies to detect the ubiquitination levels of MdPRP6 in the co‐transgenic calli and found that changes in MdRAD23D1 levels did not affect the ubiquitination levels of MdPRP6‐Flag (Figure 6d). In summary, these data demonstrate that MdPRP6 would be degraded by the UPS, and MdRAD23D1 promotes its degradation.
Figure 6.
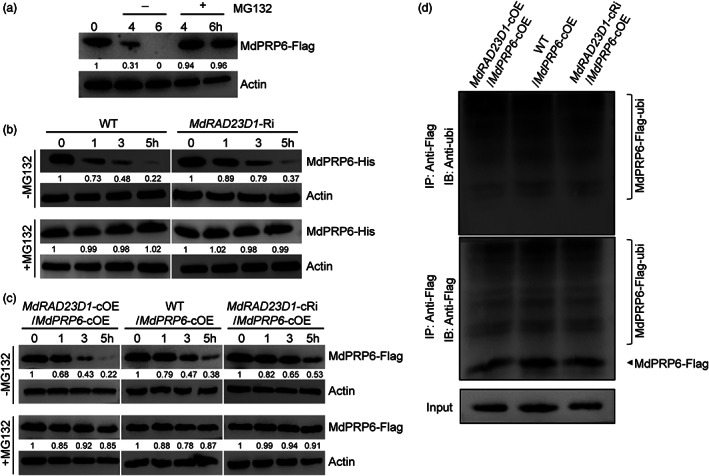
MdRAD23D1 promotes the degradation of MdPRP6. (a) Effects of MG132 on MdPRP6 degradation. The transgenic apple calli expressed 35S::MdPRP6‐Flag were treated with 75 μm CHX with (+) or without (−) 50 μm MG132 for the indicated time, and the amount of MdPRP6‐Flag was determined using anti‐Flag antibody. (b) Cell‐free degradation assays in apple plants. The Escherichia coli–expressed MdPRP6‐His protein was incubated with the protein extracts from WT or MdRAD23D1‐Ri apple plants with or without MG132, and then immunoblotting was performed using anti‐His antibody at the indicated time points. (c) MdRAD23D1 affects the degradation of MdPRP6 in vivo. Total proteins separately were extracted from the WT/MdPRP6‐cOE, MdRAD23D1‐cOE/MdPRP6‐cOE, and MdRAD23D1‐cRi/MdPRP6‐cOE transgenic apple calli after treated with 75 μm CHX with or without MG132. The immunoblotting was performed with anti‐Flag antibody. (d) Ubiquitination of MdPRP6 in vivo. The total proteins were separately extracted from the same three calli in (c) and immunoprecipitated using anti‐Flag magnetic beads. The MdPRP6‐Flag protein was detected with anti‐ubi antibody and anti‐Flag antibody. WT in (b), wild type, here we used GL‐3 apple (Malus domestica), which was also used as explants in generating transgenic apple plants; MdRAD23D1‐Ri, transgenic apple plants with suppressed expression of MdRAD23D1 via RNA interference; MdRAD23D1‐cOE/MdPRP6‐cOE, transgenic apple calli co‐expressed 35S::MdRAD23D1‐HA and 35S::MdPRP6‐Flag; MdRAD23D1‐cRi/MdPRP6‐cOE, transgenic apple calli with suppressed expression of MdRAD23D1 via RNA‐interference, and co‐expressed with 35S::MdPRP6‐Flag; WT/MdPRP6‐cOE, transgenic apple calli expressed 35S::MdPRP6‐Flag. All the transgenic apple calli was generated using ‘Orin’ calli (M. domestica). MdActin was used as the loading control (a–c) or input control (d). The amounts of proteins were quantified using an Ultra‐sensitive multifunctional imager (Uvitec), and the protein levels at 0 h were set to 1.
MdRAD23D1 promotes MdPRP6 degradation under drought stress
MdRAD23D1 promoted the degradation of MdPRP6, and drought stress also inhibited the expression of MdPRP6. To further investigate the role of their association in regulating apple drought response, a cell‐free degradation assay and in vivo protein degradation assay were carried out to measure MdPRP6 levels. When MdPRP6‐His fusion protein was incubated with protein extracts from WT or MdRAD23D1‐Ri plants under drought stress, MdPRP6‐His protein levels significantly decreased after 3 h of drought stress treatment in WT plants but after 5 h of drought stress treatment in MdRAD23D1‐Ri plants (Figure 7a). We also detected the levels of MdPRP6‐Flag in co‐transgenic apple calli treated with 200 mm mannitol. MdPRP6‐Flag band was difficult to detect at 3 h in the MdRAD23D1‐cOE/MdPRP6‐cOE calli, and at 5 h in the WT/MdPRP6‐cOE calli, but it was still detectable at 5 h in the MdRAD23D1‐cRi/ MdPRP6‐cOE calli (Figure 7b).
Figure 7.
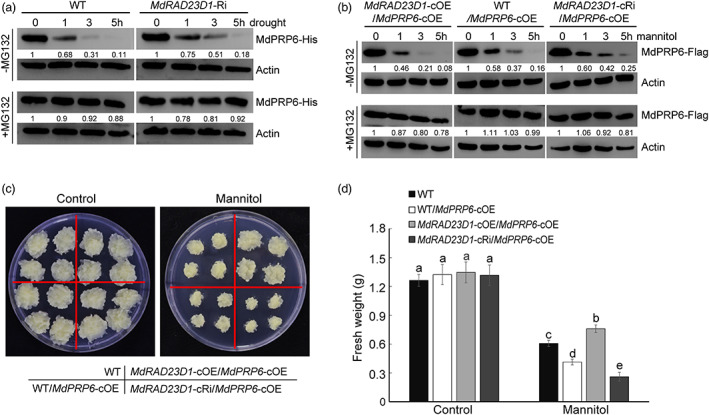
MdRAD23D1‐promoted MdPRP6 degradation affects drought tolerance. (a) Promotion of MdPRP6 degradation by MdRAD23D1 under drought in vitro. The Escherichia coli–expressed MdPRP6‐His protein was incubated with the total proteins extracted from WT or MdRAD23D1‐Ri apple plants with or without MG132 under drought treatment. The amount of MdPRP6‐His was determined with anti‐His antibody. (b) Promotion of MdPRP6 degradation by MdRAD23D1 under drought in vivo. 200 mm mannitol was applied to apple calli to monitor osmatic stress caused by drought. Total proteins were extracted from WT/MdPRP6‐cOE, MdRAD23D1‐cOE/MdPRP6‐cOE, and MdRAD23D1‐cRi/MdPRP6‐cOE transgenic apple calli treated with 75 μm CHX with or without MG132. The immunoblotting was performed with anti‐Flag antibody. (c) Morphology differences of apple calli grown on MS medium with or without 200 mm mannitol for 20 days. (d) The fresh weights of apple calli. WT in (a), wild type, here we used GL‐3 apple (Malus domestica), which was also used as explants in generating transgenic apple plants; WT in (b–d), wild type, here we used ‘Orin’ apple calli (Malus domestica), which was also used as explants in generating transgenic apple calli; MdRAD23D1‐Ri, transgenic apple plants with suppressed expression of MdRAD23D1 via RNA‐interference; MdRAD23D1‐cOE/MdPRP6‐cOE, transgenic apple calli co‐expressed 35S::MdRAD23D1‐HA and 35S::MdPRP6‐Flag; MdRAD23D1‐cRi/MdPRP6‐cOE, transgenic apple calli with suppressed expression of MdRAD23D1 via RNA‐interference, and co‐expressed with 35S::MdPRP6‐Flag; WT/MdPRP6‐cOE, transgenic apple calli expressed 35S::MdPRP6‐Flag. MdActin was used as the loading control in (a) and (b). The amounts of proteins were quantified using an Ultra‐sensitive multifunctional imager (Uvitec), and the protein levels at 0 h were set to 1. Data in (d) are shown as the means ± SD. Different letters indicate significant differences according to a one‐way ANOVA followed by Tukey's multiple range test (P < 0.05).
We examined the drought sensitivity of different transgenic calli. The size of MdRAD23D1‐cOE calli was larger than the size of WT calli and MdRAD23D1‐cRi calli under 200 mm mannitol treatment (Figure S11a,b). We then expressed MdPRP6‐Flag in the above three transgenic calli and treated the co‐trangenic calli with 200 mm mannitol (Figure 7c). When MdPRP6 was overexpressed alone (WT/MdPRP6‐cOE), the fresh weight of transgenic calli decreased upon treatment with 200 mm mannitol. However, the fresh weight of transgenic calli was higher when MdRAD23D1 was also overexpressed (MdRAD23D1‐cOE/MdPRP6‐cOE), and it was lowest when the expression of MdRAD23D1 was suppressed (MdRAD23D1‐cRi/MdPRP6‐cOE; Figure 7d). These results suggest that MdRAD23D1 affects the drought tolerance of MdPRP6.
The free proline contributes to the drought stress response in MdPRP6 transgenic apple plants
Previously, we found that MdPRP6 affects the free proline accumulation under salt stress (Zhang et al., 2023), and the same physiological changes were also detected under drought stress. The MdPRP6‐Ri plants contained higher levels of free proline than WT plants under drought (Figure 8a). To verify whether the proline is implicated in MdPRP6‐mediated drought response, we tested the stress response of MdPRP6 transgenic calli after being treated with proline. The results showed that when MdPRP6 was overexpressed, the free proline content decreased in transgenic calli under 200 mm mannitol treatment, and vice versa (Figure 8b). After treated with exogenous proline, the proline content was almost the same in MdPRP6‐cOE calli and WT under 200 mm mannitol treatment (Figure 8c), and no difference in calli size and fresh weight were observed between MdPRP6‐cOE calli and WT (Figure 8d,e). These results indicate that the proline in MdPRP6 transgenic plants is implicated with MdPRP6‐mediated drought response.
Figure 8.
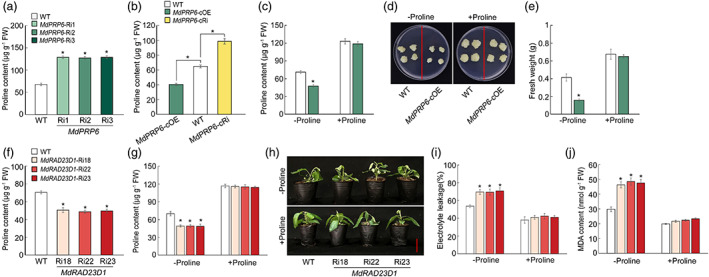
Effects of proline metabolism on the drought response of WT, MdPRP6, and MdRAD23D1 transgenic apple plants. (a) The free proline content in WT and MdPRP6‐Ri apple plants under drought stress. (b) The free proline content in WT and MdPRP6 transgenic apple calli. (c–e) The free proline content (c), phenotypes analyses (d), and fresh weight (e) in WT and MdPRP6‐cRi calli supplied with or without proline under 200 mm mannitol treatment. (f) The free proline content in WT and MdRAD23D1‐Ri apple plants under drought stress. (g–j) The free proline content (g), phenotypes analyses (h), electrolyte leakage (i), and MDA content (j) in WT and MdRAD23D1‐Ri plants supplied with or without proline under drought stress. The scale bar = 5 cm in (h). WT in (a) and (f–j), wild type, here we used GL‐3 apple (Malus domestic), which was also used as explants in generating transgenic apple plants; MdPRP6‐Ri1/2/3, transgenic apple plants with suppressed expression of MdPRP6 via RNA interference; MdRAD23D1‐Ri18/22/23: transgenic apple plants with suppressed expression of MdRAD23D1 via RNA‐interference. WT in (b–e), wild type, here we used ‘Orin’ apple calli (Malus domestica), which was also used as explants in generating transgenic apple calli; MdPRP6‐cOE, transgenic apple calli expressed 35S::MdPRP6‐Flag; MdPRP6‐cRi, transgenic apple calli with suppressed expression of MdPRP6 via RNA‐interference. Data are shown as the means ± SD. Asterisks indicate significant differences between WT and transgenic materials exposed to the same treatment (*, P < 0.05).
Given the promoted MdPRP6 degradation by MdRAD23D1 under drought stress, we reasoned that the proline content would also change in MdRAD23D1 transgenic materials. So we measured it and found that the proline content was less in MdRAD23D1‐Ri plants than in WT under drought (Figure 8f). When we applied exogenous proline to MdRAD23D1‐Ri plants under drought treatment, the proline content was similar between MdRAD23D1‐Ri plants and WT (Figure 8g). And the proline‐treated MdRAD23D1‐Ri plants were as tolerant as the proline‐treated WT plants, no difference was observed in the performance, EL, and MDA content between them under drought treatment (Figure 8h–j). We also examined the proline content in MdRAD23D1 transgenic calli and tested their stress response treated with proline. The results showed that when MdRAD23D1 was overexpressed, the proline content increased in transgenic calli under 200 mm mannitol treatment, and vice versa (Figure S12a). After treated with exogenous proline, the proline content was almost the same in MdRAD23D1‐cRi calli and WT under 200 mm mannitol treatment (Figure S12b), and no difference in calli size and fresh weight were observed between MdRAD23D1‐cRi calli and WT (Figure S12c,d). In addition, we measured the free proline content in co‐transgenic calli. When MdRAD23D1 was overexpressed (MdRAD23D1‐cOE/MdPRP6‐cOE), the free proline content significantly increased under mannitol treatment, much higher than in WT and WT/MdPRP6‐cOE calli. When it was inhibited (MdRAD23D1‐cRi/MdPRP6‐cOE), the free proline content decreased, lower than in WT/MdPRP6‐cOE calli (Figure S12e). All these results demonstrate that the free proline is also involved in MdRAD23D1‐mediated drought response.
Discussion
Drought is an emerging global problem that limits plant growth and crop yield (Iglesias et al., 2011). The apple is an economically important fruit worldwide, and drought stress seriously reduces the yield and quality of apples in China and around the world. Plants have evolved multiple physiological mechanisms to cope with drought stress. For example, increasing root branching and density to promote water uptake (Henry et al., 2011), decreasing stomatal density in leaves to reduce water loss (Hughes et al., 2017), enhancing autophagic activity to improve water use efficiency (Jia et al., 2021b). They also changed at the molecular level in response to drought. For example, the E3 ligase MdMIEL1 degrades MdBBX7, thus conferring drought tolerance in apples (Chen et al., 2022). Apple SERRATE directly interacts with MdMYB88 and MdMYB124 to regulate their expression, it also regulates the microRNA biogenesis, thus regulating drought response (Li et al., 2020b). In this study, we uncovered a new regulatory module, MdRAD23D1‐MdPRP6, in the response to drought stress in apple plants. Our results suggest that MdRAD23D1 could interact with MdPRP6 and promote its degradation via the UPS under drought stress, which modulates stress response in apple plants.
RAD23s are UBL‐UBA proteins, and they are involved in plant growth and development as well as the responses to various stresses in different species. Arabidopsis RAD23B regulates pollen development by mediating degradation of KRP1 (Li et al., 2020a). Dsk2 interacts with Irc22 and confers salt stress tolerance in yeast (Ishii et al., 2014). Recombinant Rad23 protein can protect HeLa cells from UV irradiation and inhibits apoptosis after exposure to UV irradiation (Li et al., 2013). Our previous research revealed that the six MdRAD23s were induced by various stresses in apples (Wang et al., 2017). Here, we found that the expression of MdRAD23D1 was strongly induced by drought and osmotic stress (Figure 1), implying that MdRAD23D1 may play a role in the response to drought in apples. Using different transgenic materials, we observed that MdRAD23D1 positively regulated drought stress response of apple plants. The MdRAD23D1‐Ri plants were much more sensitive to drought, as evidenced by their wilted leaves, higher EL values, MDA content, and ROS accumulation, as well as their lower RWC and Photosynthesis efficiency compared with WT plants (Figure 1, Figure S3). In addition, the MdRAD23D1‐cRi calli were less tolerant to osmotic stress, but MdRAD23D1‐cOE calli were more tolerant, as revealed by the changed size and fresh weight under mannitol treatments (Figure S11).
Expression of MdRAD23D1 improved the drought resistance of apple plants. In addition to the physiological changes, we also examined this phenomenon at the molecular level. We performed a yeast two‐hybrid screening assay to identify proteins that interacted with MdRAD23D1 and identified a proline‐rich protein MdPRP6, which was further confirmed by in vivo and in vitro experiments (Figure 3). RAD proteins are the ubiquitin shuttles of the UPS, and they are connected to the 26S proteasome by their UBL domains and to the ubiquitinated protein by the UBA domains (Farmer et al., 2010; Ishii et al., 2006; Liang et al., 2014). MdRAD23D1 was the apple protein homologous to RAD proteins and also functioned as a transporter (Figure 2). Proteins that interact with the shuttles might be ubiquitinated and can be degraded via UPS (Hara et al., 2005; Zhang et al., 2021b, c). We found that when MdRAD23D1 levels increased, MdPRP6 levels decreased (Figure S8). MdPRP6 was degraded via UPS and MdRAD23D1 promoted its degradation without affecting its ubiquitination levels (Figure 6a–d). In addition, MdRAD23D1‐promoted MdPRP6 degradation affected the stress response of apple calli treated with mannitol (Figure 7). Together, we hypothesized that this was a possible molecular mechanism for how MdRAD23D1 promoted drought response in apple plants.
When we considered the degradation of MdPRP6 as a potential reason to explain the regulation of MdRAD23D1 under drought stress, we met with a new problem: what was the role of MdPRP6 in apple plants experiencing drought stress? It is known that PRPs are a type of cell wall structural protein that play important roles in coping with various stresses in different species (Li et al., 2016; Mellacheruvu et al., 2016; Priyanka et al., 2010; Seo et al., 2011). HyPRP1 negatively regulates salt and oxidative stress responses in tomatoes (Solanum lycopersicum) by modulating sulfite metabolism (Li et al., 2016). The expression of GhHyPRP4 was down‐regulated in response to drought, salt, and ABA treatment (Huang et al., 2011). In this study, we found that MdPRP6 also played a negative role in the response to drought stress in apple plants. Its expression was inhibited by drought stress (Figure 4b,c), and when we suppressed the expression of MdPRP6 in apple plants, we found that MdPRP6‐Ri plants were much more tolerant to drought stress than WT plants (Figure 5, Figure S7).
The proline‐rich secreted protein FOCL1 involves in the stomata development in the drought response in Arabidopsis (Hunt et al., 2017). The SlPRP in tomatoes is down‐regulated by drought stress and may regulate the free proline content in the cellular response to drought stress (Gujjar et al., 2018). MdPRP6 negatively regulated drought response, so how it works? Proline is an important osmolyte in plants responding to drought stress (Huang et al., 2021; Wang et al., 2021). We found that the cellular free proline is implicated with MdPRP6‐mediated drought response (Figure 8a–e). As previously observed under salt stress (Zhang et al., 2023), we found that the P5CS activity was also higher in MdPRP6‐Ri plants under drought (Figure S13a,b), suggesting that MdPRP6 might affect the P5CS pathway in accumulating proline under stresses. Besides the P5CS pathway, the proline can also be produced from degradation of the PRPs to counteract stress (Barthakur et al., 2001; Gujjar et al., 2018, 2019; Raymond and Smirnoff, 2002; Saikia et al., 2020). We also tested this possibility by silencing MdP5CS via VIGS in MdPRP6‐Ri plants (Figure S14), and then we found it failed to totally destroy the proline accumulation and drought resistance in MdPRP6‐Ri plants (Figure S13c–f). So, the increased proline contents in MdPRP6‐Ri plants might be derived from both the P5CS pathway and MdPRP6 degradation.
Drought stress quickly induces the synthesis of ABA, the increased ABA then activates its signal transduction, thus activating cellular response (Furihata et al., 2006; Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009). MdRAD23D1 is induced by ABA treatment (Wang et al., 2017), and MdPRP6 is also induced by ABA in 24 h (Zhang et al., 2021a), but it was inhibited since day two (Figure S15a). So what would happen if blocking the ABA signalling? We treated MdPRP6‐Ri plants with NDGA (Nordihydroguaiaretic acid), one of the ABA biosynthesis inhibitors, to block the synthesis of ABA, thus blocking its signalling (Cutler et al., 2010; Lin et al., 2021; Figure S15b,c). The results showed that NDGA eliminated response differences between WT and MdPRP6‐Ri plants (Figure S15d–g). It seems that MdPRP6 also worked in an ABA‐dependent manner under drought, just like many other drought‐responsive genes (Collin et al., 2020; Joo et al., 2019). However, although decreased ABA level would inhibit its signal, NDGA is not a specific inhibitor to block its signal, so MdPRP6 might also function upstream of the ABA biosynthesis. How does ABA or ABA signalling interact with MdPRP6? More work is needed to clarify these findings.
In conclusion, we demonstrated that MdRAD23D1 and MdPRP6 function oppositely in the response of apple plants to drought. MdRAD23D1 positively regulates drought response and promotes the degradation of MdPRP6 via UPS under drought stress. MdPRP6 negatively regulates drought response. It can affect the accumulation of cellular free proline, possibly via both the P5CS pathway and its degradation under drought. The increased free proline contents then contribute to the improved drought resistance of apple plants. In addition, the ABA pathway might also function in MdPRP6‐mediated drought response (Figure 9). Our findings suggest that “MdRAD23D1‐MdPRP6” is a new type of regulon in the response to drought in apple plants.
Figure 9.
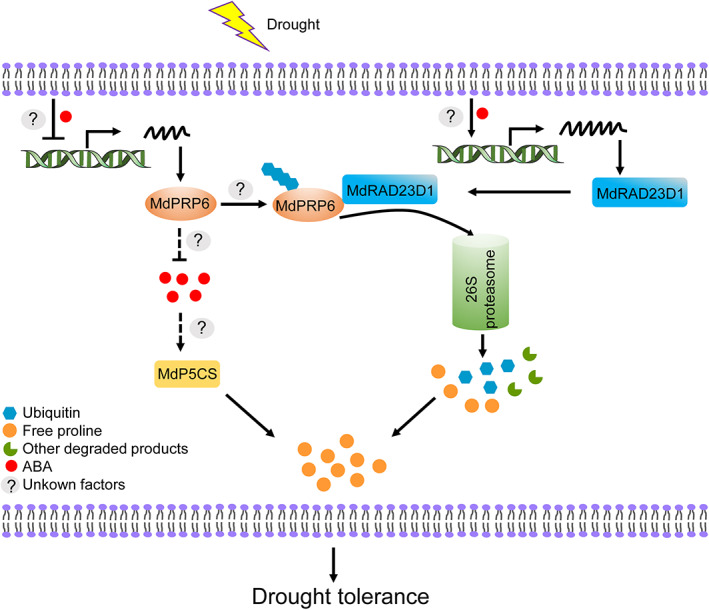
A proposed mechanism model of ‘MdRAD23D1‐MdPRP6’ regulon in apples under drought. Under drought stress, MdRAD23D1 protein levels increase, accelerating MdPRP6 degradation via 26S proteasome, there by the MdPRP6 protein levels decrease under drought. The free proline from MdPRP6 degradation might contribute to build up a high cellular free proline content. In addition, MdPRP6 affects the P5CS activity, might via the ABA pathway, to regulate proline synthesis. The increased free proline then makes apple plants resist drought stress.
Experimental procedures
Analysis of phylogenetic relationships
RAD23 proteins were identified from the apple (GDR; https://www.rosaceae.org/) and Arabidopsis genomes (Tair; https://www.arabidopsis.org/). All amino acid sequences were analysed to identify domains using Pfam (http://pfam.xfam.org/) and SMART (http://smart.embl‐heidelberg.de/) with the default parameters. DNAMAN software was used to compare the amino acid sequences of RAD23 from Arabidopsis and apples. A phylogenetic tree of RAD23s was then constructed by MEGA 6.06 software according to the neighbour‐joining approach, and the test parameter bootstrap was repeated 1000 times. We then constructed a homologous relationship map between the RAD23 genes of apple and Arabidopsis using Dual Synteny Plotter software (Chen et al., 2020).
Plant materials and growth conditions
Tissue‐cultured GL‐3 apple (Malus domestica) plants and ‘Orin’ apple (M. domestica) calli were used in this study. GL‐3 and transgenic apple plants were cultured as described by Sun et al. (2018) and Zhou et al. (2019). Briefly, after rooting, tissue‐cultured plants were transferred to the plastic pots (10 × 10 × 10 cm) filled with a mixture of organic substrate/vermiculite/perlite (3 : 1 : 1, v : v : v) and placed in a light incubator (23 °C, 16/8 h light/dark). Thirty days later, they were transferred to larger plastic pots (30 × 18 cm) with the same weight mixture of soil/sand/organic matter (v : v : v, 5 : 1 : 1) and placed in a greenhouse.
‘Orin’ apple calli were cultured on MS medium with 1.5 mg L−1 2,4‐D (2,4‐Dichlorophenoxyacetic acid) and 0.4 mg L−1 6‐BA (N‐(Phenylmethyl)‐9H‐purin‐6‐amine) in continuous darkness.
Vector construction and genetic transformation
To construct overexpression (OE) vectors, the full‐length CDS of MdRAD23D1 was separately inserted into three vectors: pRI‐101 (with a GFP tag), pRI‐101 (with a mCherry tag), and pGWB415 (with a HA tag). The full‐length CDS of MdPRP6 was separately cloned into pCambia2300 (with a Flag tag) and pCambia2300 (with a GFP tag). The full‐length CDS of MdRPN13 was cloned into the vector pCambia2300 (with a GFP tag). To generate RNA interference (Ri) vectors, a 295 bp sequence from the 5′ end of MdRAD23D1 (218–512 bp) was cloned into the pHellsgate2 vector, a 200 bp sequence at the 5′ end of MdPRP6 (1–200 bp) was cloned into the pK7GWIWG2D vector. All constructed OE and Ri vectors were introduced into Agrobacterium tumefaciens strain EHA105 by the freeze and thaw method.
Genetic transformation of apple plants (using GL‐3 as explants) and calli (using ‘Orin’ as explants) were carried out as described by Dai et al. (2013) and Li et al. (2012). The transgenic lines were screened on a medium containing 50 mg/L kanamycin or 15 mg/L hygromycin B (only for apple calli). The resistant materials were verified by PCR. Expression levels of MdRAD23D1 and MdPRP6 in the positive transgenic materials were analysed by qPCR, and MdRAD23D1 levels were detected using a specifically synthesized antibody, MdPRP6 levels were detected using an anti‐Flag antibody. All the primers are listed in Table S1.
RNA extraction and gene expression analysis
The expression levels of MdRAD23D1 and MdPRP6 under drought stress were determined using the leaves of GL‐3, and the protein levels of MdRAD23D1 and MdPRP6 under drought stress were detected using WT (‘Orin’) calli and MdPRP6‐cOE calli (expressed 35S::MdPRP6‐Flag), respectively. For qPCR, total RNA was extracted from apple leaves or calli as described by Huo et al. (2020). The synthesis of the first‐strand cDNA was performed using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA). qPCR was conducted on a LightCycler 96 system (Roche, Basel, Switzerland). MDH (encoding malate dehydrogenase) was used as an internal reference to calculate the relative expression levels for each gene. Three biological replicates and three technical replicates were performed for each experiment. All the primers used for qPCR are listed in Table S1. For separately analysing the protein levels of MdRAD23D1 and MdPRP6 under drought stress, the total protein was extracted from different calli samples treated with 200 mm mannitol at 0, 2, 4, 6, and 8 h, and then immunoblot analyses were performed using anti‐MdRAD23D1 antibody and anti‐Flag antibody, respectively.
In situ hybridization assay
In situ hybridizations of MdPRP6 in apple plants were conducted as previously described by Geng et al. (2018) with slight modification. Briefly, the roots, stems, and leaves of GL‐3 were collected and immediately fixed in formalin‐aceto‐alcohol (FAA) solution (4% formaldehyde, 10% acetic acid, and 50% ethanol). The tissues were decolorized, embedded in Paraffin, and sliced. The glass slides were then placed at 37 °C for 8‐12 h. Xylene was used to remove all the wax, and gradient ethanol and DEPC water were used to dehydrate the samples. Finally, the glass slides were digested in 1 μg/mL protease K at 37 °C. The glass slides were incubated with the probe at 42 °C overnight and then rinsed with washing buffer at 23 and 65 °C before being stored in TE buffer. The probes are listed in Table S1.
Drought stress treatments
The transgenic and WT (here we used GL‐3) apple plants were cultured as described above. After growth in the greenhouse for 45 days, all apple plants were fully irrigated and the drought treatment was performed as previously described (Jia et al., 2021a). The apple plants of uniform size were selected and randomly divided into two groups as follows: (1) the control group, which was watered every 2 days; (2) the drought group, which received no water for 8 days. Samples were then collected at 0, 2, 4, 6, and 8 d and stored at −80 °C for subsequent experiments. There were three independent biological replicates for each group, containing at least 60 plants in total.
The transgenic and WT (here we used ‘Orin’) apple calli were cultured as described above. To monitor drought stress, 200 mm mannitol was added to the medium, and 2‐week‐old WT and transgenic apple calli of equal weight (0.1 g) were used in the treatments. Samples were then collected at 20 days and stored at −80 °C for subsequent experiments. There were four independent replicates for each experiment.
VIGS (virus‐induced gene silencing)‐mediated silencing of MdP5CS in apple plants was performed according to previous methods (Sasaki et al., 2011; Zhu et al., 2022) with minor modification (the detailed method is included in Figure S14). After confirmation, all the positive plants were treated with drought stress for 15 days by stopping watering.
For the proline or NDGA treatments, 1‐month‐old MdRAD23D1‐Ri apple plants or MdPRP6‐Ri apple plants were, respectively, applied with 5 mm proline or 10 μm NDGA and treated with drought stress for 15 days. Exogenous proline or NDGA was sprayed on leaves every 2 days. In addition, the transgenic and WT apple calli (0.1 g) were grown on MS medium treated with 200 mm mannitol with or without 5 mm proline for 20 days. The samples were harvested at the end of treatment and stored at −80 °C for subsequent experiments. There were four independent replicates for each experiment.
Measurement of physiological traits and photosynthetic parameters
Electrolyte leakage (EL) was measured according to Sun et al. (2018). The total chlorophyll was extracted with 80% acetone, and a UV‐2250 spectrophotometer (Shimadzu, Kyoto, Japan) was used to measure the chlorophyll content according to Liu et al. (2021). RWC was calculated following the method of Jia et al. (2021b). The leaf angle was the angle between the straight part of the leaf and the stem. A CIRAS‐3 portable photosynthesis system (CIRAS‐3; PP Systems, Amesbury, MA) was used to measure photosynthetic parameters. The chlorophyll fluorescence was detected using a chlorophyll fluorescence imaging system (IMAGING‐PAM, WALZ, Bavaria, Germany).
NBT and DAB were used to detect the accumulation of superoxide radical () and hydrogen peroxide (H2O2), respectively (Fryer et al., 2002). Free proline content, as well as MDA, H2O2, and P5CS activity were detected using their corresponding detection kits (Suzhou Comin Biotechnology Co., Ltd., Suzhou, China).
Screening of the cDNA library via the yeast two‐hybrid (Y2H) assay
To identify proteins that interact with MdRAD23D1, the CDS of MdRAD23D1 was cloned into the pGBKT7 to construct a MdRAD23D1‐BD vector. The cDNA library was constructed using mixed samples of apple roots, stems, and leaves, according to the protocol of Clontech. Then MdRAD23D1‐BD was expressed in the Y2H Gold (Clontech) strain to test self‐activation. Screening of the cDNA library and yeast transformation were performed according to the protocol. When analysing the interaction between MdRAD23D1 and MdRPN13, MdRAD23D1 and MdPRP6, the CDS of MdPRP6 and MdRPN13 were cloned into the pGADT7 to generate the MdPRP6‐AD and MdRPN13‐AD constructs. And then MdRAD23D1‐BD and MdPRP6‐AD or MdRPN13‐AD were co‐transformed into yeast strain Y2H Gold according to the protocol. All positive clones were selected on SD base/‐Leu/‐Trp/‐His/‐Ade + x‐α gal (5‐Bromo‐4‐chloro‐3‐indoxyl‐α‐d galactopyranoside) to confirm the interactions.
Subcellular localization of MdRAD23D1 and MdPRP6
MdRAD23D1 was transiently expressed in tobacco (N. benthamiana) leaves and Arabidopsis protoplasts to analyse its localization in cells, MdPRP6 was transiently expressed in onion epidermal cells and Arabidopsis protoplasts to analyse its localization in cells. The detailed method is included in Figure S4.
Pull‐down assays
The CDS of MdRAD23D1, MdPRP6, and MdRPN13 were cloned into the pET28a (with a His tag) and pGEX4T‐1(with a GST tag) vectors to generate the recombined constructs pET28a‐35S::MdRAD23D1‐His, pGEX‐4 T‐1‐35S::MdRAD23D1‐GST, pET28a‐35S::MdPRP6‐His, and pGEX‐4T‐1‐35S::MdRPN13‐GST. To study protein expression, 100 ng of each plasmid was separately transformed into Escherichia coli BL21 (DE3) and fusion proteins were induced by 1 mm isopropyl β‐d‐1‐ thiogalactopyranoside (IPTG) for 12 h at 16 °C. Then, the fusion proteins bound to His magnetic beads were incubated with MdRPN13‐GST (or MdRAD23D1‐GST) fusion proteins in binding buffer A containing 50 mm Tris–HCl, pH 8.0, 200 mm NaCl, 1 mm EDTA, 1 mm DTT (DL‐Dithiothreitol), 10 mm MgCl2, 1% Nonidet P‐40, 1 mm PMSF (phenylmethylsulfonyl fluoride), and 1× complete protease inhibitor cocktail (P9599; Sigma‐Aldrich, St. Louis, MO, USA) for at least 2 h at 23 °C with gentle shaking. Finally, the proteins were eluted according to the protocol of Beyotime and immunoblotted using anti‐GST and anti‐His antibodies (Yeasen, Shanghai, China).
Split‐luciferase assays
The CDS of MdRAD23D1 was cloned into pRI‐101‐cLuc, and the CDS of MdPRP6 and MdRPN13 were cloned pRI‐101‐nLuc to generate cLuc‐MdRAD23D1, nLuc‐MdPRP6, and nLuc‐MdRPN13. These constructs were transformed into EHA105 and then injected into the leaves of N. benthamiana. The luciferase expression signals were detected with an Ultra‐sensitive multifunctional imager (Uvitec, Cambridge, UK).
Co‐Immunoprecipitation (Co‐IP) assays
To perform the Co‐IP assays, different combinations of proteins were co‐expressed in N. benthamiana leaves via injection, including pRI‐101‐35S::GFP and pCambia2300‐35S::MdPRP6‐Flag, pRI‐101‐35S::MdRAD23D1‐GFP and pCambia2300‐35S::MdPRP6‐Flag, pCambia2300‐35S::GFP and pRI‐101‐35S::MdRAD23D1‐mCherry, and pRI‐101‐35S::MdRAD23D1‐mCherry and pCambia2300‐35S::MdRPN13‐GFP. Total proteins were extracted using an extraction buffer containing 50 mm Tris–HCL, 150 mm NaCl, 5 mm EDTA, 1% NP‐40, 10% glycerol, 1 mm PMSF, 5 mm DTT, and 1× complete protease inhibitor cocktail. Anti‐GFP magnetic beads were used for Co‐IP assays according to the manufacturer's manuals (Beyotime, Shanghai, China). Briefly, 10 μL pre‐prepared anti‐GFP magnetic beads were added to each 500 μL of protein sample and incubated at 4 °C overnight with gentle shaking. The beads were then washed three times with extraction buffer, and the immunoprecipitated proteins were eluted according to the protocol of Beyotime. Protein blot analysis was performed using anti‐GFP, anti‐Flag, or anti‐mCherry antibodies (Yeasen, Shanghai, China).
Ubiquitin chain binding assays
The purified MdRAD23D1‐GST protein was first incubated with anti‐GST magnetic beads at 4 °C overnight according to the manufacturer's manuals (Beyotime, Shanghai, China). Then, the mixture was incubated with 20 μL purified Lys‐48 (K48) or Lys‐63 (K63) linked chains (Ub2‐6) (Ubbiotech, Changchun, China) in buffer A at 23 °C for 2 h. Then, the bound proteins were washed three times in buffer A and eluted according to the protocol of Beyotime. Finally, the eluted proteins were immunoblotted using anti‐GST antibody (Yeasen) and anti‐Ub antibody (CST, Denver, MA, USA).
Protein degradation and in vivo ubiquitination assay
For the cell‐free assay, DMSO or 50 μm MG132 (stock solution prepared with 0.5% DMSO) was added to apple plants 1 h before extraction. The total protein of WT or MdRAD23D1‐Ri transgenic apple plant was extracted using degradation buffer containing 25 mm Tris–HCl (pH 7.5), 10 mm NaCl, 10 mm MgCl2, 4 mm PMSF, 5 mm DTT, and 10 mm ATP (Yang et al., 2021). The protein extracts were incubated with E. coli expressed MdPRP6‐His at 23 °C for specific time points and detected by immunoblotting with an anti‐His antibody (Yeasen).
For the in vivo degradation assay, DMSO or 50 μm MG132 (stock solution prepared with 0.5% DMSO) and 75 μm CHX (Cycloheximide, a protein synthesis inhibitor) were added to MdPRP6 transgenic and co‐transgenic apple calli. The samples were harvested at specific time points and total protein was extracted as described above. The extracted protein was immunoblotted using anti‐Flag antibody.
For the in vivo ubiquitination assay, the co‐transgenic apple calli were treated with 50 μm MG132 for 10 h before extraction. Then, the extracted total proteins were immunoprecipitated with anti‐Flag magnetic beads (Beyotime) and immunoblotted with anti‐Flag antibody (Yeasen) and anti‐Ub antibody (CST).
All the proteins were quantified using an Ultra‐sensitive multifunctional imager (Uvitec), and the protein levels at 0 h were set to 1. MdActin protein was used as the loading control or input control.
Statistical analysis
All data were analysed using IBM SPSS Statistics software (version 20; SPSS Inc., Chicago, IL). One‐way ANOVA and Tukey's multiple‐range tests or Student's t‐test were used to evaluate significant differences (P < 0.05).
Accession numbers
The Arabidopsis sequences were found from the Arabidopsis Information Resource (TAIR) site and the accession numbers were as follows: AtRAD23A (AT1G16190), AtRAD23B (AT1G79650), AtRAD23C (AT3G02540), and AtRAD23D (AT5G38470). The apple sequences data used in this study can be downloaded from Genome Database for Rosaceae (GDR) site and the accession numbers were as follows: MdRAD23A (MD09G1229800), MdRAD23B (MD17G1228500), MdRAD23C1 (MD13G1043000), MdRAD23C2 (MD16G1043900), MdRAD23D1 (MD17G1076200), MdRAD23D2 (MD09G1087400), MdPRP1 (MD07G1116600), MdPRP2 (MD02G1209700), MdPRP3 (MD02G1023200), MdPRP4 (MD04G1086500), MdPRP5 (MD04G1087300), MdPRP6 (MD15G1126700), MdPRP7 (MD06G1062200), MdPRP8 (MD06G1062000), MdPRP9 (MD15G1166100), and MdRPN13 (MD16G1238000).
Conflict of interest
No conflicts of interest were declared.
Author contributions
X‐QG, X‐LZ, and F‐WM conceived and designed the experiments. X‐LZ, X‐JS, and H‐XY performed the research. S‐YC, J‐WH,D‐YL and Z‐LL analysed the data, and X‐LZ, X‐QG, and F‐WM wrote the paper. X‐QG, F‐WM, and M‐JL revised the paper.
Supporting information
Figure S1 Phylogenetic analyses of UBL‐UBA proteins.
Figure S2 Identification of MdRAD23D1 transgenic apple plants.
Figure S3 The ROS accumulation in wild type and MdRAD23D1‐Ri transgenic plants under drought conditions.
Figure S4 Localization analyses of MdRAD23D1 and MdPRP6 using tobacco leaves, Arabidopsis protoplasts, and onion epidermal cells.
Figure S5 Identification of MdPRP6 transgenic apple calli.
Figure S6 Identification of MdPRP6 transgenic apple plants.
Figure S7 The ROS accumulation in wild type and MdPRP6‐Ri transgenic apple plants under drought conditions.
Figure S8 MdRAD23D1 affects the protein abundance of MdPRP6.
Figure S9 Identification of MdRAD23D1 transgenic apple calli.
Figure S10 Identification of co‐transgenic apple calli.
Figure S11 Effects of mannitol treatment on the growth of wild type and MdRAD23D1 transgenic apple calli.
Figure S12 Effects of exogenous proline on the stress response of wild type and MdRAD23D1‐cRi calli.
Figure S13 Effects of P5CS pathway on the proline accumulation and stress response of wild type and MdPRP6‐Ri plants.
Figure S14 Identification of MdP5CS‐VIGS apple plants.
Figure S15 Effects of exogenous NDGA on the stress response of wild type and MdPRP6‐Ri plants.
Table S1 Application of primers and sequences.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFD1000300), the National Natural Science Foundation of China (31972391), the Fundamental Research Funds for the Central Universities (2452022118), the earmarked fund for the China Agricultural Research System (CARS‐27) and the Key S&T Special Projects of Shannxi Province (2020zdzx03‐01‐02). And the authors are grateful to Dr Zhihong Zhang from Shenyang Agricultural University for providing tissue‐cultured GL‐3 apple plants, as well as the Horticulture Science Research Center at the College of Horticulture, NWAFU for their technical support in this work.
Contributor Information
Ming‐jun Li, Email: limingjun@nwsuaf.edu.cn.
Feng‐wang Ma, Email: fwm64@sina.com, Email: fwm64@nwsuaf.edu.cn.
Data availability
Research data are not shared.
References
- Banday, Z.Z. , Cecchini, N.M. , Speed, D.J. , Scott, A.T. , Parent, C. , Hu, C.T. , Filzen, R.C. et al. (2022) Friend or foe: hybrid proline‐rich proteins determine how plants respond to beneficial and pathogenic microbes. Plant Physiol. 190, 860–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthakur, S. , Babu, V. and Bansal, K.C. (2001) Over‐expression of osmotin induces proline accumulation and confers tolerance to osmotic stress in transgenic tobacco. J. Plant Biochem. Biotechnol. 10, 31–37. [Google Scholar]
- Chen, C.J. , Chen, H. , Zhang, Y. , Thomas, H.R. , Frank, M.H. , He, Y.H. and Xia, R. (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant, 13, 1194–1202. [DOI] [PubMed] [Google Scholar]
- Chen, P.X. , Zhi, F. , Li, X.W. , Shen, W.Y. , Yan, M.J. , He, J.Q. , Bao, C.N. et al. (2022) Zinc‐finger protein MdBBX7/MdCOL9, a target of MdMIEL1 E3 ligase, confers drought tolerance in apple. Plant Physiol. 188, 540–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin, A. , Daszkowska‐Golec, A. , Kurowska, M. and Szarejko, I. (2020) BarleyABI5 (abscisic acid insensitive 5) is involved in abscisic acid‐dependent drought response. Front. Plant Sci. 11, 1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R. , Rodriguez, P.L. , Finkelstein, R.R. , Abrams, S.R. , Merchant, S. , Briggs, W.R. and Ort, D. (2010) Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. [DOI] [PubMed] [Google Scholar]
- Dai, H.Y. , Li, W.R. , Han, G.F. , Yang, Y. , Ma, Y. , Li, H. and Zhang, Z.H. (2013) Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci. Hortic. 164, 202–208. [Google Scholar]
- Farmer, L.M. , Book, A.J. , Lee, K.H. , Lin, Y.L. , Fu, H.Y. and Vierstra, R.D. (2010) The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis . Plant Cell, 22, 124–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq, M. , Wahid, A. , Kobayashi, N. , Fujita, D. and Basra, S.M.A. (2009) Plant drought stress: effects, mechanisms and management. Agron. Sustain. Dev. 29, 185–212. [Google Scholar]
- Finley, D. (2009) Recognition and processing of ubiquitin‐protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer, M.J. , Oxborough, K. , Mullineaux, P.M. and Baker, N.R. (2002) Imaging of photo‐oxidative stress responses in leaves. J. Exp. Bot. 53, 1249–1254. [PubMed] [Google Scholar]
- Funakoshi, M. , Sasaki, T. , Nishimoto, T. and Kobayashi, H. (2002) Budding yeast dsk2p is a polyubiquitin‐binding protein that can interact with the proteasome. Proc. Natl Acad. Sci. USA, 99, 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata, T. , Maruyama, K. , Fujita, Y. , Umezawa, T. , Yoshida, R. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2006) Abscisic acid‐dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl Acad. Sci. USA, 103, 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely, G. , Kama, R. , Gelin‐Licht, R. and Gerst, J.E. (2008) Different domains of the UBL‐UBA ubiquitin receptor, Ddi1/Vsm1, are involved in its multiple cellular roles. Mol. Biol. Cell, 19, 3625–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, D.L. , Chen, P.X. , Shen, X.X. , Zhang, Y. , Li, X.W. , Jiang, L.J. , Xie, Y.P. et al. (2018) MdMYB88 and MdMYB124 enhance drought tolerance by modulating root vessels and cell walls in apple. Plant Physiol. 178, 1296–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujjar, R.S. , Karkute, S.G. , Rai, A. , Singh, M. and Singh, B. (2018) Proline‐rich proteins may regulate free cellular proline levels during drought stress in tomato. Curr. Sci. 114, 915–920. [Google Scholar]
- Gujjar, R.S. , Pathak, A.D. , Karkute, S.G. and Supaibulwatana, K. (2019) Multifunctional proline rich proteins and their role in regulating cellular proline content in plants under stress. Biol. Plant, 63, 448–454. [Google Scholar]
- Guzder, S.N. , Sung, P. , Prakash, L. and Prakash, S. (1998) Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J. Biol. Chem. 273, 31541–33154. [DOI] [PubMed] [Google Scholar]
- Hara, T. , Kamura, T. , Kotoshiba, S. , Takahashi, H. , Fujiwara, K. , Onoyam, A.I. , Shirakawa, M. et al. (2005) Role of the UBL‐UBA protein KPC2 in degradation of p27 at G (1) phase of the cell cycle. Mol. Cell. Biol. 25, 9292–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, A. , Gowda, V.R.P. , Torres, R.O. , McNally, K.L. and Serraj, R. (2011) Variation in root system architecture and drought response in rice (Oryza sativa): phenotyping of the OryzaSNP panel in rainfed lowland fields. Field Crop Res. 120, 205–214. [Google Scholar]
- Huang, G.Q. , Gong, S.Y. , Xu, W.L. , Li, P. , Zhang, D.J. , Qin, L.X. , Li, W. et al. (2011) GhHyPRP4, a cotton gene encoding putative hybrid proline‐rich protein, is preferentially expressed in leaves and involved in plant response to cold stress. Acta Bioch. Biophys. Sin. 43, 519–527. [DOI] [PubMed] [Google Scholar]
- Huang, D. , Wang, Q. , Jing, G.Q. , Ma, M.N. , Li, C. and Ma, F.W. (2021) Overexpression of MdIAA24 improves apple drought resistance by positively regulating strigolactone biosynthesis and mycorrhization. Tree Physiol. 41, 134–146. [DOI] [PubMed] [Google Scholar]
- Hughes, J. , Hepworth, C. , Dutton, C. , Dunn, J.A. , Hunt, L. , Stephens, J. , Waugh, R. et al. (2017) Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 174, 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, L. , Amsbury, S. , Baillie, A. , Movahedi, M. , Mitchell, A. , Afsharinafar, M. , Swarup, K. et al. (2017) Formation of the stomatal outer cuticular ledge requires a guard cell wall proline‐rich protein. Plant Physiol. 174, 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo, L.Q. , Guo, Z.J. , Zhang, Z.J. , Jia, X. , Sun, Y.M. , Sun, X. , Wang, P. et al. (2020) The apple autophagy‐related gene MdATG9 confers tolerance to low nitrogen in transgenic apple callus. Front. Plant Sci. 11, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak, K. , Elsasser, S. , Zhang, N.X. , Chen, X. , Randles, L. , Shi, Y. , Hofmann, K. et al. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias, A. , Quiroga, S. and Diz, A. (2011) Looking into the future of agriculture in a changing climate. Eur. Rev. Agric. Econ. 38, 427–447. [Google Scholar]
- Ishii, T. , Funakoshi, M. and Kobayashi, H. (2006) Yeast Pth2 is a UBL domain‐binding protein that participates in the ubiquitin‐proteasome pathway. EMBO J. 25, 5492–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, T. , Funakoshi, M. , Kobayashi, H. and Sekiguchi, T. (2014) Yeast Irc22 is a novel Dsk2‐interacting protein that is involved in salt tolerance. Cell, 3, 180–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, X. , Jia, X.M. , Li, T.T. , Wang, Y. , Sun, X. , Huo, L.Q. , Wang, P. et al. (2021a) MdATG5a induces drought tolerance by improving the antioxidant defenses and promoting starch degradation in apple. Plant Sci. 312, 111052. [DOI] [PubMed] [Google Scholar]
- Jia, X. , Mao, K. , Wang, P. , Wang, Y. , Jia, X.M. , Huo, L.Q. , Sun, X. et al. (2021b) Overexpression of MdATG8i improves water use efficiency in transgenic apple by modulating photosynthesis, osmotic balance, and autophagic activity under moderate water deficit. Hortic Res. 8, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo, J. , Lee, Y.H. and Song, S.I. (2019) OsbZIP42 is a positive regulator of ABA signaling and confers drought tolerance to rice. Planta, 249, 1521–1533. [DOI] [PubMed] [Google Scholar]
- Kang, Y. , Vossler, R.A. , Diaz‐Martinez, L.A. , Winter, N.S. , Clarke, D.J. and Walters, K.J. (2006) UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J. Mol. Biol. 356, 1027–1035. [DOI] [PubMed] [Google Scholar]
- Kim, J.H. , Kim, M.S. , Kim, D.Y. , Amoah, J.N. and Seo, Y.W. (2022) Molecular characterization of U‐box E3 ubiquitin ligases (TaPUB2 and TaPUB3) involved in the positive regulation of drought stress response in Arabidopsis. Int. J. Mol. Sci. 22, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishor, P.B.K. , Kumari, P.H. , Sunita, M.S.L. and Sreenivasulu, N. (2015) Role of proline in cell wall synthesis and plant development and its implications in plant ontogeny. Front. Plant Sci. 6, 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kito, K. , Yeh, E.T.H. and Kamitani, T. (2001) NUB1, a NEDD8‐interacting protein, is induced by interferon and down‐regulates the NEDD8 expression. J. Biol. Chem. 276, 20603–20609. [DOI] [PubMed] [Google Scholar]
- Lahari, T. , Lazaro, J. and Schroeder, D.F. (2018) RAD4 and RAD23/HMR contribute to Arabidopsis UV tolerance. Genes, 9, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertson, D. , Chen, L. and Madura, K. (1999) Pleiotropic defects caused by loss of the proteasome‐interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae . Genetics, 153, 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.Y. , Mao, K. , Zhao, C. , Zhao, X.Y. , Zhang, H.L. , Shu, H.R. and Hao, Y.J. (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light‐induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.S. , Zhang, J.H. , Zhao, X.Y. and Liu, X.M. (2013) Protective effects of Rad23 protein on ultraviolet damage to HeLa cells. J. Cent. South Univ. 20, 2974–2980. [Google Scholar]
- Li, J.H. , Ouyang, B. , Wang, T.T. , Luo, Z.D. , Yang, C.X. , Li, H.X. , Sima, W. et al. (2016) HyPRP1 gene suppressed by multiple stresses plays a negative role in abiotic stress tolerance in tomato. Front. Plant Sci. 7, 967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Li, B. , Xie, C. , Zhang, T. , Borassi, C. , Estevez, J.M. , Li, X.S. et al. (2020a) Arabidopsis RAD23B regulates pollen development by mediating degradation of KRP1. J. Exp. Bot. 71, 4010–4019. [DOI] [PubMed] [Google Scholar]
- Li, X.W. , Chen, P.X. , Xie, Y.P. , Yan, Y. , Wang, L.P. , Dang, H. , Zhang, J. et al. (2020b) Apple SERRATE negatively mediates drought resistance by regulating MdMYB88 and MdMYB124 and microRNA biogenesis. Hortic. Res. 7, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, R.Y. , Chen, L. , Ko, B.T. , Shen, Y.H. , Li, Y.T. , Chen, B.R. , Lin, K.T. et al. (2014) Rad23 interaction with the proteasome is regulated by phosphorylation of its ubiquitin‐like (UbL) domain. J. Mol. Biol. 426, 4049–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Z. , Li, Y. , Wang, Y. , Liu, X. , Ma, L. , Zhang, Z. , Mu, C. et al. (2021) Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 12, 2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.X. , Yu, Y.Z. , An, H.Z. , Li, N. , Lin, N.S. , Wang, W.Y. , Zhang, W.P. et al. (2003) Cloning and identification of a novel ubiquitin‐like protein, BMSC‐UbP, from human bone marrow stromal cells. Immunol. Lett. 86, 169–175. [DOI] [PubMed] [Google Scholar]
- Liu, D.Q. , Han, Q. , Shah, T. , Chen, C.Y. , Wang, Q. , Tang, B.F. and Ge, F. (2018) A hybrid proline‐rich cell‐wall protein gene JsPRP1 from juglans sigillata dode confers both biotic and abiotic stresses in transgenic tobacco plants. Trees‐Struct. Funct. 32, 1199–1209. [Google Scholar]
- Liu, Y. , Yang, T.Y. , Lin, Z.K. , Gu, B.J. , Xing, C.H. , Zhao, L.Y. , Dong, H.Z. et al. (2019) A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 17, 1770–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.M. , Jin, Y.B. , Tan, K.X. , Zheng, J.Z. , Gao, T.T. , Zhang, Z.J. , Zhao, Y.J. et al. (2021) MdTyDc overexpression improves alkalinity tolerance in Malus domestica . Front. Plant Sci. 12, 625890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Szostkiewicz, I. , Korte, A. , Moes, D. , Yang, Y. , Christmann, A. and Grill, E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science, 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- MacLean, A.M. , Orlovskis, Z. , Kowitwanich, K. , Zdziarska, A.M. , Angenent, G.C. , Immink, R.G.H. and Hogenhout, S.A. (2014) Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS‐box proteins and promotes insect colonization in a RAD23‐dependent manner. PLoS Biol. 12, e1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu, S. , Tamirisa, S. , Vudem, D.R. and Khareedu, V.R. (2016) Pigeonpea hybrid‐proline‐rich protein (CcHyPRP) confers biotic and abiotic stress tolerance in transgenic rice. Front. Plant Sci. 6, 1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaljevic, I. , Viljevac Vuletic, M. , Simic, D. , Tomas, V. , Horvat, D. , Josipovic, M. , Zdunic, Z. et al. (2021) Comparative study of drought stress effects on traditional and modern apple cultivars. Plants, 10, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, D.L. , Lu, T.C. , Liu, G.F. , Yang, C.P. and Wang, B.C. (2013) Proteomic analysis points to a role for RAD23 in apical dominance in Pinus sylvestris var. mongolica . Plant Mol. Biol. Rep. 31, 1283–1292. [Google Scholar]
- Ogura, T. , Goeschl, C. , Filiault, D. , Mirea, M. , Slovak, R. , Wolhrab, B. , Satbhai, S.B. et al. (2019) Root system depth in Arabidopsis is shaped by EXOCYST70A3 via the dynamic modulation of auxin transport. Cell, 178, 400–412.e16. [DOI] [PubMed] [Google Scholar]
- Oh, J. , Levy, J.G. , Kan, C.C. , Ibanez‐Carrasco, F. and Tamborindeguy, C. (2022) CLIBASIA 00460 disrupts hypersensitive response and interacts with citrus Rad23 proteins. Int. J. Mol. Sci. 23, 7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, W.B. , Lin, B.Y. , Yang, X.Y. , Liu, L.J. , Xia, R. , Li, J.G. , Wu, Y.R. et al. (2020) The UBC27‐AIRP3 ubiquitination complex modulates ABA signaling by promoting the degradation of ABI1 in Arabidopsis. Proc. Natl. Acad. Sci. USA, 117, 27694–27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.Y. , Fung, P. , Nishimura, N. , Jensen, D.R. , Fujii, H. , Zhao, Y. , Lumba, S. et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Arellano, I. , Carmona‐Alvarez, F. , Martinez, A.I. , Rodriguez‐Diaz, J. and Cervera, J. (2010) Pyrroline‐5‐carboxylate synthase and proline biosynthesis: from osmotolerance to rare metabolic disease. Protein Sci. 19, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke, A. , Datta, S. and Persak, H. (2014a) Salt stress in Arabidopsis: lipid transfer protein AZI1 and its control by mitogen‐activated protein kinase MPK3. Mol. Plant, 7, 722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke, A. , Datta, S. and Persak, H. (2014b) Mitogen‐activated protein kinase‐regulated AZI1‐an attractive candidate for genetic engineering. Plant Signal. Behav. 9, e27764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke, A. , Xue, H. , Persak, H. , Datta, S. and Seifert, G.J. (2016) Post‐translational modification and secretion of Azelaic Acid Induced 1 (AZI1), a hybrid proline‐rich protein from Arabidopsis . Int. J. Mol. Sci. 17, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyanka, B. , Sekhar, K. , Reddy, V.D. and Rao, K.V. (2010) Expression of pigeonpea hybrid‐proline‐rich protein encoding gene (CcHyPRP) in yeast and Arabidopsis affords multiple abiotic stress tolerance. Plant Biotechnol. J. 8, 76–87. [DOI] [PubMed] [Google Scholar]
- Raymond, M.J. and Smirnoff, N. (2002) Proline metabolism and transport in maize seedlings at low water potential. Ann. Bot. 89, 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia, B. , Singh, S. , Debbarma, J. , Velmurugan, N. , Dekaboruah, H. , Arunkumar, K.P. and Chikkaputtaiah, C. (2020) Multigene CRISPR/Cas9 genome editing of hybrid proline rich proteins (HyPRPs) for sustainable multi‐stress tolerance in crops: the review of a promising approach. Physiol. Mol. Biol. Plants, 26, 857–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, S. , Yamagishi, N. and Yoshikawa, N. (2011) Efficient virus‐induced gene silencing in apple, pear and Japanese pear using Apple latent spherical virus vectors. Plant Methods 7, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, T.F. and Quatrano, R.S. (1997) Characterization and expression of a rice RAD23 gene. Plant Mol. Biol. 34, 557–562. [DOI] [PubMed] [Google Scholar]
- Seo, S.G. , Jeon, S.B. , Kim, J.S. , Shin, J.M. , Kim, J.B. , Kang, S.W. , Lee, G.P. et al. (2011) Characterization and expression pattern of IbPRP1 and IbPRP2 stress‐related genes from sweetpotato. Genes Genom. 32, 487–497. [Google Scholar]
- Stein, H. , Honig, A. , Miller, G. , Erster, O. , Eilenberg, H. , Csonka, L.N. , Szabados, L. et al. (2011) Elevation of free proline and proline‐rich protein levels by simultaneous manipulations of proline biosynthesis and degradation in plants. Plant Sci. 181, 140–150. [DOI] [PubMed] [Google Scholar]
- Stines, A.P. , Naylor, D.J. , Hoj, P.B. and van Heeswijck, R. (1999) Proline accumulation in developing grapevine fruit occurs independently of changes in the levels of delta (1)‐pyrroline‐5‐carboxylate synthetase mRNA or protein. Plant Physiol. 120, 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm, A. and Lienhard, S. (1998) Two isoforms of plant RAD23 complement a UV‐sensitive rad23 mutant in yeast. Plant J. 13, 815–821. [DOI] [PubMed] [Google Scholar]
- Sun, X. , Wang, P. , Jia, X. , Huo, L.Q. , Che, R.M. and Ma, F.W. (2018) Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 16, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, H. , Ohtake, F. , Arai, N. , Kaiho, A. , Yasuda, S. , Tanaka, K. and Saeki, Y. (2017) In vivo ubiquitin linkage‐type analysis reveals that the Cdc48‐Rad23/Dsk2 axis contributes to K48‐linked chain specificity of the proteasome. Mol. Cell, 66, 488–502.e7. [DOI] [PubMed] [Google Scholar]
- Umezawa, T. , Sugiyama, N. , Mizoguchi, M. , Hayashi, S. , Myouga, F. , Yamaguchi‐Shinozaki, K. , Ishihama, Y. et al. (2009) Type 2C protein phosphatases directly regulate abscisic acid‐activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA, 106, 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Gong, X.Q. and Ma, F.W. (2017) Genome‐wide identification of the radiation sensitivity protein‐23 (RAD23) family members in apple (Malus × domestica Borkh.) and expression analysis of their stress responsiveness. J. Integr. Agr. 16, 820–827. [Google Scholar]
- Wang, J. , Qin, H. , Zhou, S.R. , Wei, P.C. , Zhang, H.W. , Zhou, Y. , Miao, Y.C. et al. (2020) The ubiquitin‐binding protein OsDSK2a mediates seedling growth and salt responses by regulating gibberellin metabolism in rice. Plant Cell, 32, 414–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.P. , Chen, Q. , Zheng, J.Z. , Zhang, Z.J. , Gao, T.T. , Li, C. and Ma, F.W. (2021) Overexpression of the tyrosine decarboxylase gene MdTyDC in apple enhances long‐term moderate drought tolerance and WUE. Plant Sci. 313, 111064. [DOI] [PubMed] [Google Scholar]
- Yang, K. , An, J.P. , Li, C.Y. , Shen, X.N. , Liu, Y.J. , Wang, D.R. , Ji, X.L. et al. (2021) The apple C2H2‐type zinc finger transcription factor MdZAT10 positively regulates JA‐induced leaf senescence by interacting with MdBT2. Hortic Res. 8, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.N. , Chen, T. , Ziv, I. , Rosenzweig, R. , Matiuhin, Y. , Bronner, V. , Glickman, M.H. et al. (2009) Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain‐length sensor. Mol. Cell, 36, 1018–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.L. , Gong, X.Q. , Li, D.Y. , Yue, H. , Qin, Y. , Liu, Z. , Li, M.J. et al. (2021a) Genome‐wide identification of PRP genes in apple genome and the role of MdPRP6 in response to heat stress. Int. J. Mol. Sci. 22, 5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Xue, Y.F. , Yang, X. , Liu, J. and Liu, Q. (2021b) Toxoplasma gondii UBL‐UBA shuttle proteins regulate several important cellular processes. FASEB J. 35, e21898. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Yang, X. , Ying, Z. , Liu, J. and Liu, Q. (2021c) Toxoplasma gondii UBL‐UBA shuttle protein DSK2s are important for parasite intracellular replication. Int. J. Mol. Sci. 22, 7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.L. , Gong, X.Q. , Cheng, S.Y. , Yu, H.X. , Li, D.Y. , Su, X.J. , Lei, Z.L. et al. (2022) Proline‐rich protein MdPRP6 alters low nitrogen stress tolerance by regulating lateral root formation and anthocyanin accumulation in transgenic apple (Malus domestica). Environ. Exp. Bot. 197, 104841. [Google Scholar]
- Zhang, X.L. , Gong, X.Q. , Yu, H.X. , Su, X.J. , Cheng, S.Y. , Huang, J. , Lei, Z.L. et al. (2023) The proline‐rich protein MdPRP6 confers tolerance to salt stress in transgenic apple (Malus domestica). Sci. Hortic. 308, 111581. [Google Scholar]
- Zhao, S. , Gao, H.B. , Jia, X.M. , Li, X.W. , Mao, K. and Ma, F.W. (2020) The gamma‐clade HD‐Zip I transcription factor MdHB‐7 regulates salt tolerance in transgenic apple (Malus domestica). Plant Soil, 463, 509–522. [Google Scholar]
- Zhou, K. , Hu, L.Y. , Li, Y.T.S. , Chen, X.F. , Zhang, Z.J. , Liu, B.B. , Li, P.M. et al. (2019) MdUGT88F1‐mediated phloridzin biosynthesis regulates apple development and valsa canker resistance. Plant Physiol. 180, 2290–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2016) Abiotic stress signaling and responses in plants. Cell, 167, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L.C. , Li, B.Y. , Wu, L.M. , Li, H.X. , Wang, Z.Y. , Wei, X.Y. , Ma, B.Q. et al. (2022) MdERDL6‐mediated glucose efflux to the cytosol promotes sugar accumulation in the vacuole through up‐regulating TSTs in apple and tomato. Proc. Natl Acad. Sci. USA, 118, e2022788118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phylogenetic analyses of UBL‐UBA proteins.
Figure S2 Identification of MdRAD23D1 transgenic apple plants.
Figure S3 The ROS accumulation in wild type and MdRAD23D1‐Ri transgenic plants under drought conditions.
Figure S4 Localization analyses of MdRAD23D1 and MdPRP6 using tobacco leaves, Arabidopsis protoplasts, and onion epidermal cells.
Figure S5 Identification of MdPRP6 transgenic apple calli.
Figure S6 Identification of MdPRP6 transgenic apple plants.
Figure S7 The ROS accumulation in wild type and MdPRP6‐Ri transgenic apple plants under drought conditions.
Figure S8 MdRAD23D1 affects the protein abundance of MdPRP6.
Figure S9 Identification of MdRAD23D1 transgenic apple calli.
Figure S10 Identification of co‐transgenic apple calli.
Figure S11 Effects of mannitol treatment on the growth of wild type and MdRAD23D1 transgenic apple calli.
Figure S12 Effects of exogenous proline on the stress response of wild type and MdRAD23D1‐cRi calli.
Figure S13 Effects of P5CS pathway on the proline accumulation and stress response of wild type and MdPRP6‐Ri plants.
Figure S14 Identification of MdP5CS‐VIGS apple plants.
Figure S15 Effects of exogenous NDGA on the stress response of wild type and MdPRP6‐Ri plants.
Table S1 Application of primers and sequences.
Data Availability Statement
Research data are not shared.


