Abstract
Analytical techniques with high sensitivity and selectivity are essential to the quantitative analysis of clinical samples. Liquid chromatography (LC) coupled to tandem mass spectrometry (MS/MS) is the gold standard in clinical chemistry. However, tandem mass spectrometers come at high capital expenditure and maintenance costs. We recently showed that it is possible to generate very similar results using a much simpler single MS detector by performing enhanced in-source fragmentation/annotation (EISA) combined with correlated ion monitoring. Here we provide a step-by-step protocol for optimizing the analytical conditions for EISA, so anyone properly trained in LC-MS can follow and apply this technique for any given analyte. We exemplify the approach by using 2-hydroxyglutarate (2-HG) which is a clinically relevant metabolite whose D-enantiomer is considered an “oncometabolite”, characteristic to cancers associated with mutated isocitrate dehydrogenases 1 or 2 (IDH1/2). We include procedures for determining quantitative robustness, and show results of these relating to the analysis of DL-2-HG in cells, as well as in serum samples from acute myeloid leukemia patients that contain the IDH1/2 mutation. This EISA-MS protocol is a broadly applicable and low-cost approach for the quantification of small molecules that has been developed to work well for both single quadrupole and time-of-flight mass analyzers.
INTRODUCTION
Atmospheric pressure ionization (API), especially electrospray ionization (ESI), has significantly enhanced liquid chromatography mass spectrometry (LC-MS) capabilities largely due to its ability to generate intact molecular ions with high sensitivity. LC-API-MS allows the detection of primarily intact molecular ions, and tandem MS (MS/MS) was designed to provide an added dimension of information contained in the fragmentation spectra of each molecular ion. The concept has widely become adapted and is of crucial value in a variety of MS applications, including mixture analysis, molecular structure elucidation and quantitative analysis1–4.
The place where fragments are generated is called the collision cell. In the collision cell, ion fragmentation is performed by collision-induced dissociation (CID), a process in which the ion acquires internal energy by collisions with neutral molecules and/or atoms. CID is widely used in both high- and low-resolution MS instrumentation to generate fragment spectra. However, adding additional tandem MS hardware components such as the collision cell comes at the cost of ion losses during transmission in combination with increased instrumentation and maintenance costs, ultimately limiting an even broader application of MS.
We recently introduced the concept of enhanced in-source fragmentation annotation (EISA) as a simple and efficient alternative approach to traditional tandem MS fragment ion generation, producing ESI in-source fragments5,6. By optimizing in-source fragmentation conditions, fragments generated using EISA on single quadrupole MS machines can mimic the tandem MS fragments produced via CID without compromising ion transmission. Moreover, EISA-generated spectra are highly comparable with CID fragment spectra and hence can be searched using MS/MS spectral libraries such as METLIN5. However, the most significant advantage that EISA allows is in using the fragment ions to perform quantitative analyses using much simpler (single quadrupole) technology. This approach is termed quantitative multiple fragment ion monitoring (Q-MFM) or alternatively Q-MRM referring to single quadrupole multiple reaction monitoring and comes at a fraction of the cost of traditional triple quadrupole (QqQ) mass spectrometers. In principle, the approach for method development and validation described in this protocol can be applied to any given analyte. As an example for this Q-MFM protocol, we chose the clinically relevant DL-2-hydroxyglutarate (DL-2-HG) enantiomers. The analysis of these enantiomers is useful because of the importance of D-2-HG as an oncometabolite. This can be challenging because enantiomers have the same physicochemical properties and therefore cannot be separated in a standard chromatographic approach. For this, the diastereomers must be firstly formed by means of derivatisation with a enantiomerically pure agent.
DEVELOPMENT OF THE PROTOCOL
Initially EISA was investigated as an alternative approach for the generation of fragment ions6. As a logical next step the quantitative nature and applicability of EISA was investigated for small molecule analysis and peptide fragmentation in bottom-up proteomics7. The approach is designated as Q-MFM, referring to the use of a single stage MS in combination with a software tool for correlating fragment ions (correlated ion monitoring, CIM) so that it is possible to see results similar to those obtained using a reaction monitoring approach6.
Thus far quantitative EISA Q-MFM has been applied to several small molecule metabolites in diverse matrices6 and in peptide quantification7. Overall, the metrics for measuring quantitative soundness for these compounds (dynamic range, matrix effects, trueness and precision) have been adequate6. In principle this approach can be adapted for use with any kind of analyte that is ionizable using electrospray ionization.
In this protocol we describe, quantitative Q-MFM analysis of the clinically relevant DL-enantiomers of 2-HG. The ratio between the D- and L-2-HG is clinically meaningful as increased levels of D-2-HG can be used as biomarker for mutated isocitrate dehydrogenases 1 or 2 (IDH1/2)8. For chromatographic separation we applied the method originally developed by Struys et al.9, that transforms the DL-2-HG enantiomers into separable diastereomers by derivatization with diacetyl-L-tartaric anhydride (L-DATAN). Subsequently, we optimized chromatographic conditions as well as Q-MFM-based analysis and validated the assay for several matrices.
COMPARISON WITH OTHER METHODS
The industry standard for quantitative LC-MS analysis of small molecules is isotope dilution-based MS (IDMS) executed in multiple reaction monitoring (MRM) mode on QqQ instrumentation10. As outlined above, this involves fragmenting molecules by CID in the collision cell. Our here described procedure adopts the IDMS and MRM concept, however all molecules are fragmented within the ion source allowing for the removal of the collision cell from the procedure.
The selected ions specific to any particular molecule can provide for quantitative LC-MS methods using single stage MS systems (e.g. quadrupole (Q) or time-of-flight (TOF)) with sensitivities comparable to QqQ instrumentation. The fragmentation of all molecules within the ion source and the removal of an extra mass filter significantly reduces hardware costs, although it limits the selectivity of the method. Nevertheless, modern UHPLC systems and core shell columns allow for superior chromatographic resolutions and advanced separation of analytes11. In turn, Q-MFM coupled to UHPLC is a broadly applicable, cheap alternative for the quantitative analysis of small molecules in complex mixtures. It is important to keep in mind that fragmentation is dependent on the molecule chemical structure. Some molecules might be prone to fragment while others will be more challenging. However, this is not a limitation of EISA, rather a pre-requisite of an analyte chemical nature and it has been shown that EISA and MS/MS fragmentation patterns are very similar5. We have already demonstrated the applicability of EISA in a total of 50 endogenous molecules with a broad range of physicochemical properties and chemical structures, including amino acids, lipids, and fatty acids6. This method worked well for all of these compounds and all of the fragmentation patterns followed those observed in MS/MS. As collision cell is not needed, EISA method enables MRM level quantitative performance by monitoring/correlating precursor and fragment ions on existing single quadrupole instrumentation that are generally inexpensive.
EXPERIMENTAL DESIGN
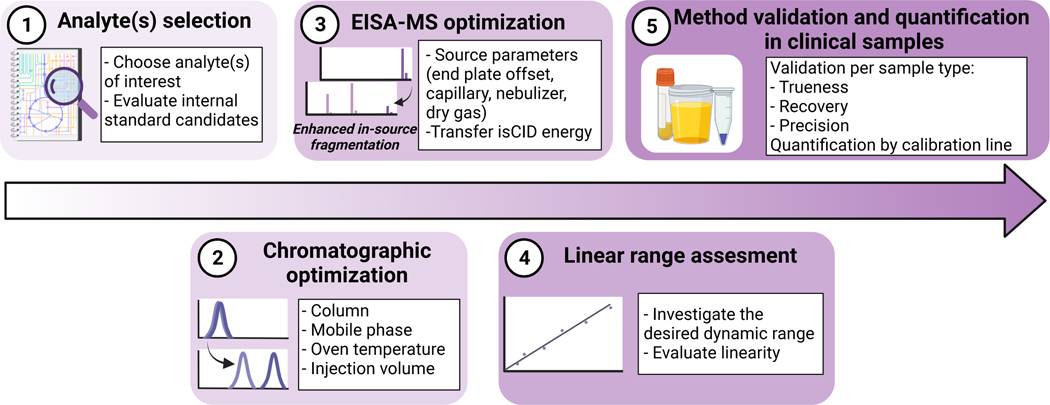
A detailed depiction of the PROCEDURE can be found in Figure 1. Experimentally some steps might be modified or carried out in an alternative fashion, however, in our experience the described PROCEDURE is the most practical and facile approach. Using Figure 1 as guideline, we provide a step-by-step description of the entire protocol.
Figure 1.
Workflow to optimize an EISA method for the analysis of clinical samples. This figure has been created with BioRender.com.
Selection of the analyte(s)
The first step is to select the metabolite(s) of interest. Once the analytes have been chosen, it is important to determine a suitable internal standard (IS). The IS plays a critical role not only in correcting for possible losses during the sample preparation but also to correct for matrix effects12. The ratio calculated by dividing the response of the targeted analyte and that of the IS can then be interpolated onto a calibration line to calculate the unknown concentration of the analyte in the sample. Ideally the IS should be a stable (heavy) isotope-labelled version of the target analyte (deuterated or 13C labelled). If using the isotopic-labeled variant is not feasible (e.g. elevated cost or not commercially available), a surrogate can be used, i.e. a similarly structured molecule. Importantly, the IS should not be present in the biological sample of interest and the retention time must be close to that of the selected analyte.
Optimization of chromatographic separation
Although Q-MFM analysis is highly comparable to QqQ-based MRM analysis in terms of sensitivity6, it does lack an additional mass filter, which translates into a lower selectivity when compared to “traditional” QqQ-based MRM analysis. This is the reason why in Q-MFM analysis, chromatographic separation of target analytes from overlapping components is even more important to take into account. In the here described Protocol we present such an optimization strategy for the derivatized DL-enantiomers of 2-HG.
Q-MFM optimization
Subsequent to the optimization of the chromatographic workflow, the Q-MFM parameters can be optimized taking the eluent composition at analyte specific retention times into account during direct-infusion experiments. As described below, the main goal is the optimization of the in-source fragmentation parameters so that recognizable fragment ions are observed without significantly compromising the intensity of the molecular ion. Several parameters have to be optimized such as the end plate offset, capillary voltage, nebulizer pressure, drying gas and in-source collision induced dissociation (isCID).
Linear range assessment
Once both the LC and Q-MFM methods have been optimized, the next step is to identify what is the expected concentration range for the monitored analyte(s) of interest and thereafter assess method linearity. The calibration line should contain at least six datapoints obtained from solutions with increasing concentrations of the analyte and identical IS concentration. This way, the linear relationship between the analyte concentration and the IS-normalized peak area of the analyte (areaanalyte/areaIS) can be established. Subsequently, the unknown concentration in the samples can be predicted using the least squares regression method. Since in LC-MS low concentrations are subjected to a higher variability, it is common practice is to use weighted least squares linear regression. Using a 1/x2-type of weighting results in a better accuracy for low concentration datapoints. High correlation coefficients (R2) (e.g. higher than 0.99) mean that the calibration curve approaches linearity. Another way of proving that the calibration curve is linear is to perform appropriate statistical method such as analysis of variance (ANOVA).
Validation of the LC-Q-MFM method
As for any quantitative LC-MS assay, a thorough validation of the final assay is mandatory. Standard analytical figures of merit include, linearity, limits of detection (LOD), and quantification (LOQ), trueness (the accuracy performance characteristic refers to systematic and random error, being trueness and precision) , recovery and precision. This should be done for each matrix under investigation. Ultimately, validation will prove the applicability of Q-MFM for a specific target analyte in a given matrix. We also performed an interlaboratory comparison with two different single stage MS instruments and found largely overlapping results.
MATERIAL
REAGENTS
Reagents used in the UHPLC-EISA-TOF system
LC-MS grade acetonitrile (Honeywell, cat. no. 34967).
LC-MS grade methanol (Supelco, cat. no. 1.06035.2500).
-
LC-MS grade 2-propanol (Honeywell, cat. no. 34965).
! CAUTION: Acetonitrile, methanol and 2-propanol are highly flammable.
-
LC-MS grade formic acid ≥ 99% (VWR, cat. no. 84865.180).
! CAUTION: Formic acid is corrosive, flammable, and toxic.
LC-MS grade water (Honeywell, cat. no. 14263).
n-Hexane (Honeywell, cat. no. 32293).
LC-MS calibration standard, for ESI-TOF (Agilent Technologies, cat. no. G1969–85000).
LC-MS grade acetic acid ≥ 99% (Sigma Aldrich, cat. no. A6283).
LC-MS grade ammonium formate (Fluka, cat. no. 55674).
Disodium D-2-hydroxyglutarate (Merck, cat. no. 61382).
Disodium L-2-hydroxyglutarate (Merck, cat. no. 90790).
Disodium DL-2-hydroxyglutarate-d3,OD (CDN isotopes, cat. no. D-7496).
Sodium lactate (Sigma Aldrich, cat. no. L7022).
(+)−Diacetyl-L-tartaric anhydride (L-DATAN) (Aldrich, cat. no. 358924).
Sodium azide (Merck, cat.no. 7290).
Reagents used in the HPLC-EISA-Q system
LC-MS grade acetonitrile (Fisher Chemical, cat. no. A955–4).
LC-MS grade methanol >99.9% (Honeywell, cat. no. LC230–4).
LC-MS grade 2-propanol >99.9% (Honeywell, cat. no. 34965–4X4L).
ACS reagent grade Formic acid ≥ 98% (Sigma-Aldrich, cat. no. 33015–500ML).
LC-MS grade water (Honeywell, cat. no. 365–4).
LC-MS calibration standard, for ESI-TOF (Agilent Technologies, cat. no. G1969–85000).
Acetic acid Reagent Plus ≥ 99% (Sigma-Aldrich, cat. no. A6283–500ML).
Ammonium formate solution BioUltra 10 mol/L in water (Sigma-Aldrich, cat. no. 78314).
Disodium D-2-hydroxyglutarate (Cayman, cat. no. 11605).
Disodium L-2-hydroxyglutarate (MilliporeSigma Supelco, cat. no. 61313-10MG).
Disodium DL-2-hydroxyglutarate-d3,OD (CDN isotopes, cat. no. D-7496).
ACS reagent grade Lactic acid solution ≥ 85% (Sigma-Aldrich, cat. no. 252476–100G).
(+)−Diacetyl-L-tartaric anhydride (L-DATAN) (Aldrich, cat. no. 358924).
BIOLOGICAL MATERIAL
Cell lines
The K562 cell line (https://scicrunch.org/resolver/RRID:CVCL_0004, ATCC, Manassas, VA, USA) was cultured in IMDM with 10% heat-inactivated fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and 1.5% 200 mmol/L L-glutamine (Lonza). The chondrosarcoma cell lines CH2879 (https://scicrunch.org/resolver/RRID:CVCL_9921, IDH1 and IDH2 wildtype)13,14 and JJ012 (https://scicrunch.org/resolver/RRID:CVCL_D605, IDH1 p.R132G)13,15 were cultured in RPMI medium supplemented with 10% heat-inactivated FBS (F524, Sigma-Aldrich, Sant Louis, MO, USA). Cells were trypsinized, put on ice, and counted with a hemocytometer. Cell pellets of 1 × 106 cells were prepared, washed twice with ice-cold PBS, and stored at −80 °C until DL-2-HG extraction. Heat inactivated fetal calf serum (FCS) was obtained from Bodinco B.V. (Alkmaar, the Netherlands).
! CAUTION: The cell lines used should be regularly checked to ensure they are authentic and are not infected with mycoplasma
Biological fluids
Urine from a healthy male volunteer was collected as first urine of the day. To prevent bacterial growth 0.1% (vol/vol) of sodium azide in water was added upon collection. Serum samples obtained from patients at diagnosis of acute myeloid leukemia (AML) with written informed consent were selected from the Hematology Biobank of Leiden University Medical Center with approval by the institutional review board (no. B18.047). In both patients, the IDH1-R132H mutation was detected in diagnostic cell samples by RNA-Seq16. Human plasma was acquired from Sigma-Aldrich (cat. No. P9523–5ML). Identical material was used for Q-MFM analysis on TOF and single quadrupole MS systems.
EQUIPMENT
Equipment used in the UHPLC-EISA-TOF system
Aqua C30 column (3 μm; 2 mm inner diameter (i.d.) × 150 mm length (Develosil, cat. no. RPAR320150W).
HPLC guard holder for 2.1 to 4.6 mm ID columns (Phenomenex, cat. no. AJ0–9000).
C8 2.1 mm ID precolumn (Phenomenex, cat. no. AJ0–8784).
Dionex UltiMate 3000 UHPLC (Thermo Fisher) or generic HPLC.
Bruker Impact II Q-TOF system (Bruker).
Stuart Scientific Block heater (for the derivatization step; Fisher scientific).
Mini Spin centrifuge (Eppendorf).
Fisherbrand vortex mixer (Fisher Scientific).
Centrifuge 1.5–2 mL Eppendorf tubes (Eppendorf).
Heat block.
pH meter (Mettler Toledo).
Ultrasonic bath.
Magnetic stir bar.
Polypropylene microcentrifuge tubes (1.5 mL; Eppendorf, cat. no. 0030 123.328).
Polypropylene microcentrifuge tubes (2.0 mL; Eppendorf, cat. no. 0030 120.094).
HPLC vials with screw (Agilent, cat. no. 5182–0714).
HPLC cap vials (Agilent, cat. no. 5182–0717).
250 μL inserts with polymer feet (Agilent, cat. no. 5181–1270).
Solid phase extraction system (Waters).
C18 Cartridges (Waters, cat. no. WAT054945).
500 μL glass syringe (Hamilton, cat. no. 81265).
Equipment used in the HPLC-EISA-Q system
InfinityLab Poroshell 120 EC-C18 column, 4.6 × 50 mm, 2.7 μm (Agilent, cat.no. 699975–902T).
Agilent 6130 Single Quadrupole LC-MS.
Agilent HPLC 1260 Infinity.
pH meter (Mettler Toledo).
Thermomixer R (Eppendorf).
Vortex mixer (Thermo Scientific).
Centrifuge 1.5–2 mL Eppendorf tubes (Eppendorf).
Refrigerated CentriVap Concentrator (Labconco).
Safe-Lock Eppendorf tubes, 1.5 mL (Eppendorf, cat. no. 022363204).
Safe-Lock Eppendorf tubes, 2.0 mL (Eppendorf, cat. no. 022363344).
HPLC glass vials with screw (Thermo Fisher, cat. no. 4000-S1W).
HPLC vial caps (Agilent, cat. no. 5182–0717).
Vial inserts, 250 μL, deactivated glass with polymer feet (Agilent, cat. no. 5181–8872).
SOFTWARE
Software used in the UHPLC-EISA-TOF system
UPLC system was controlled by Chromeleon version 6.80 (Dionex, USA).
Bruker Impact II Q-TOF system was controlled by otof Control version 3.4 (Bruker Daltonik GmbH, Germany).
LC-MS hyphenation was managed by Compass HyStar (Bruker Daltonik GmbH, Germany).
Peak integration was conducted via Compass DataAnalysis version 4.2 (Bruker Daltonik GmbH, Germany).
Software used in the HPLC-EISA-Q system
Agilent ChemStation Rev. C01.10.
Agilent MassHunter Quantitative Analysis Version B.08.00.
Correlated ion monitoring (CIM) algorithm.
EQUIPMENT SETUP
CIM algorithm.
This algorithm was developed to perform peak picking, alignment and data analysis and can be accessed via https://github.com/ricoderks/eisaCIM (see also ref. 6). Briefly, chromatograms for each individual metabolite are constructed by using each SIM trace (for single Q analyzers) using XCMS (v3.12.0) R-package with an established intensity threshold. This is a crucial step as to obtain clean signals that do not include co-eluting peaks. Finally, SIM traces containing precursor and fragment ions can be aligned and grouped as a CIM chromatogram based on a given retention time window.
REAGENT SETUP
Cells.
For cultured cells, use at least 1 × 106 cells.
Biological fluids.
For biological fluid samples, start with aliquots of 10 μL of fetal calf serum, human serum or plasma or 20 μL of human urine in 1.5-mL polypropylene microcentrifuge tubes.
Methanol (80% (vol/vol)).
To prepare a volume of 100 mL of a 80% (vol/vol) methanol solution for metabolite extraction and protein precipitation, mix 80 mL of LC-MS grade methanol and 20 mL of LC-MS grade water in a glass jar and gently shake for mixing. Store at −20 °C until further use.
2-propanol (50% (vol/vol)).
To prepare a volume of 500 mL of a 50% (vol/vol) 2-propanol solution to clean the needle of the UHPLC system between injections, mix 250 mL of LC-MS grade 2-propanol and 250 mL of LC-MS grade water in a HPLC bottle. Cap and gently shake for mixing. Slightly open the bottle and sonicate for 10 min to eliminate oxygen. The solution can be stored for three weeks at room temperature (~ 20–25 °C).
Formic Acid (50% (vol/vol)).
To prepare a volume of 1 mL of a 50% (vol/vol) formic acid solution to adjust the pH buffer solution A, mix 500 μL of LC-MS grade formic acid and 500 μL of LC-MS grade water in a 1.5-mL polypropylene microcentrifuge tube. Cap and gently shake for mixing. The solution can be stored for three weeks at room temperature (~ 20–25 °C).
Acetonitrile:acetic acid (4:1 (vol/vol)).
To prepare a volume of 1 mL of 4:1 (vol/vol) acetonitrile:acetic acid, to dissolve the L-DATAN, mix 800 μL of LC-MS grade acetonitrile and 200 μL of acetic acid in a 1.5-mL polypropylene microcentrifuge tube. Cap and gently shake for mixing. The solution can be stored for three weeks at room temperature (~ 20–25 °C).
LC-MS grade water:acetic acid (4:1 (vol/vol)).
To prepare a volume of 1 mL of a 4:1 (vol/vol) LC-MS grade water:acetic acid to reconstitute the dry pellet prior LC-MS analysis, mix 800 μL of LC-MS grade water and 200 μL of acetic acid in a 1.5-mL polypropylene microcentrifuge tube. Cap and gently shake for mixing. The solution can be stored for three weeks at room temperature (~ 20–25 °C).
L-DATAN solution (50 mg/mL in (4:1 (vol/vol) acetonitrile:acetic acid).
To prepare 1 mL, add 50 mg of L-DATAN to a 1.5-mL polypropylene microcentrifuge tube and add 1 mL of (4:1 (vol/vol)) acetonitrile:acetic acid. Cap and gently shake for mixing. The solution must be prepared fresh directly before use and cannot be stored.
Sodium L-lactate (10 mmol/L in water).
To prepare 2 mL, add 2.2 mg of sodium lactate to a 2.0-mL polypropylene microcentrifuge tube and add 2 mL of LC-MS grade water. Cap and gently shake for mixing. The solution can be stored for at least six months at −20 °C.
D-2-hydroxyglutarate (D-2-HG) (50 mmol/L in water).
To prepare 1 mL, add 9.6 mg of D-2-HG disodium salt to a 1.5-mL polypropylene microcentrifuge tube and add 1 mL of LC-MS grade water. Cap and gently shake for mixing. The solution can be stored for at least six months at −20 °C.
L-2-hydroxyglutarate (L-2-HG) (50 mmol/L in water).
To prepare 1 mL, add 9.6 mg of L-2-HG disodium salt to a 1.5-mL polypropylene microcentrifuge tube and add 1 mL of LC-MS grade water. Cap and gently shake for mixing. The solution can be stored for at least six months at −20 °C.
DL-2-HG (2 mmol/L in water).
To prepare 1 mL, mix 20 μL of the 50 mmol/L D-2-HG solution, 20 μL of the 50 mmol/L L-2-HG solution and 960 μL of LC-MS grade water in a 1.5-mL polypropylene microcentrifuge tube. Cap and gently shake for mixing. The solution can be stored for at least six months at −20 °C. Please, note that the individual concentration per enantiomer is 1 mmol/L.
DL-2-HG (200 μmol/L in water).
To prepare 1 mL, mix 100 μL of the 2 mmol/L DL-2-HG solution and 900 μL of LC-MS grade water in a 1.5-mL polypropylene microcentrifuge tube. Cap and gently shake for mixing. The solution can be stored for at least six months at −20 °C. Please, note that the individual concentration per enantiomer is 100 μmol/L.
DL-2-HG (2 μmol/L in water).
To prepare 1 mL, mix 10 μL of 200 μmol/L DL-2-HG solution and 990 μL of LC-MS grade water in a 1.5-mL polypropylene microcentrifuge tube. Cap and gently shake for mixing. The solution can be stored for at least six months at −20 °C. Please, note that the individual concentration per enantiomer is 1 μmol/L.
DL-2-HG-d3 (IS) (10 mmol/L in water).
To prepare 1 mL, add 2.0 mg of disodium DL-2-HG-d3 to a 1.5-mL microcentrifuge tube and add 1 mL of LC-MS grade water. Cap and gently shake for mixing. The solution can be stored for at least six months at −20°C. Please, note that the individual concentration per enantiomer is 5 mmol/L.
DL-2-HG-d3 (IS) (1 mmol/L).
To prepare 1 mL, mix 100 μL of 10 mmol/L DL-2-HG-d3 solution and 900 μL of LC-MS grade water in a 1.5-mL polypropylene microcentrifuge tube. Cap and gently shake. The solution can be stored for at least six months at −20 °C. Please, note that the individual concentration per enantiomer is 0.5 mmol/L.
LC mobile phase A (125 mg/L of ammonium formate in water, pH 3.6).
To prepare LC mobile phase A, add 125 mg of ammonium formate to a 1-liter HPLC bottle and add 990 mL of LC-MS grade water. Shake the solution using a magnetic stirrer and add 250 μL of a 50% (vol/vol) formic acid solution. Check the pH to assure it is at 3.6 and complete the volume up to 1 L with LC-MS grade water. Sonicate for 10 min to degas the solution. The solution can be stored for three weeks at room temperature (~ 20–25 °C).
LC mobile phase B (95% (vol/vol) acetonitrile in water).
To prepare LC mobile phase B, mix 950 mL of acetonitrile and add 50 mL of LC-MS grade water to a 1-liter HPLC bottle. Cap and shake the solution. Sonicate for 10 min to degas the solution. The solution can be stored for three weeks at room temperature (~ 20–25 °C).
PROCEDURE
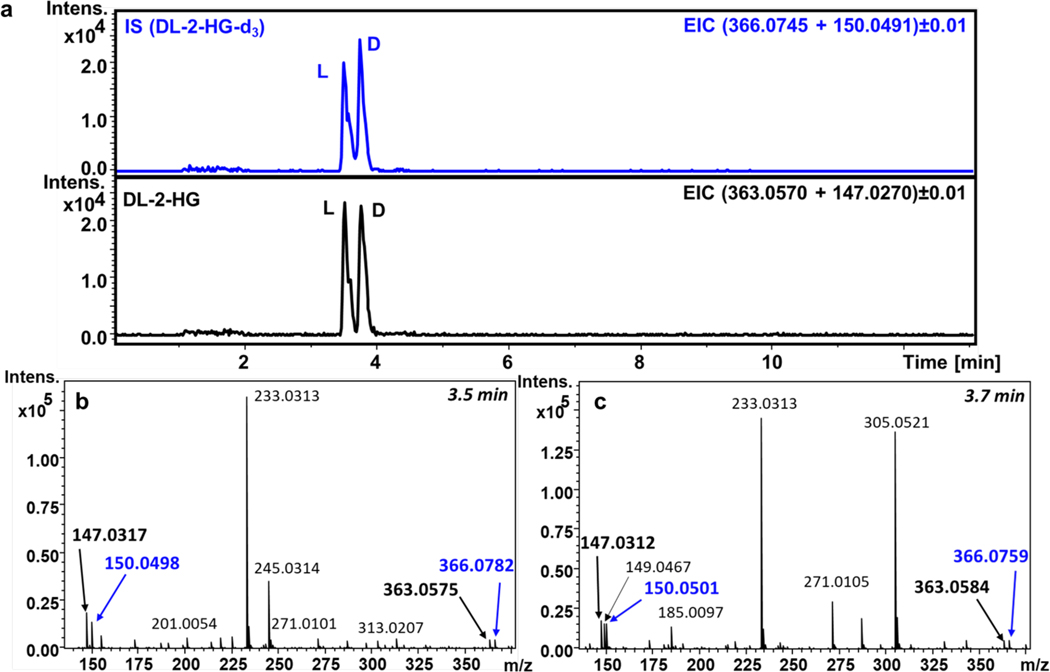
The overall objective of this protocol is to show a practical guide to develop a LC-Q-MFM strategy for quantitative purposes (see Figure 1). We exemplify this process by showing results for the quantification of DL-2-HG (see Figure 2).
Figure 2.
UHPLC-EISA-TOF separation of DL-2-HG (black) and its d3-labelled IS (blue) derivatized with L-DATAN and resulting MS spectra of L- and D-enantiomers at 3.5 and 3.7 min, respectively.
-
1
Select the target molecule/s.
-
2
Prepare the standard solutions.
-
3
Develop the chromatographic method. The development of the LC method for DL-2-HG is out of scope for this protocol but we have included a detailed description in the Supplementary Material (see also Supplementary Figures 1-4). <CRITICAL STEP> It is advisable that the optimization of the EISA method is conducted once a satisfactory chromatographical separation has been obtained. This way, the infused standard solution can exactly resemble subsequent mobile phase composition.
-
4
(optional) Decide whether derivatisation is needed. In Box 1, we provide steps for derivatisation with L-DATAN as an example. However this is highly dependent on the analyte of interest.
Importantly, although we exemplify this protocol for a chiral molecule, researchers interested in incorporating the EISA strategy to their laboratories for non-chiral analysis can skip the steps included in Box 1. The Supplementary Material also includes a more detailed description of the EISA optimization for our particular example (DL-2-HG).
Box 1. Preparation of DL-2-HG standard solution including L-DATAN-derivatization and SPE purification.
This step is required for the optimization of the analysis of DL-2-HG standard and does not always need to be performed. This should be conducted only for the analytes of interest (DL-2-HG in our case) and not to the IS. This step is not relevant for the quantification non-chiral molecules.
SPE purification is performed to minimize interference of non-derivatized L-DATAN during the EISA method optimization via direct infusion and avoid contamination of the system. This process might also be relevant if you have used a different derivatisation method.
Mix 25 μL of 50 mmol/L D-2-HG solution and 25 μL of 50 mmol/L L-2-HG solution in a 1.5-mL polypropylene microcentrifuge tube.
Follow the steps 10–17 from section “Derivatization of the calibration standards and extractant samples e.g. with diacetyl-L-tartaric anhydride (L-DATAN)” and after Step 17, redissolve the dried residue in 1 mL of LC-MS grade water.
- Purify the derivatized compound using a solid phase extraction (SPE) system and a C18 cartridge (Sep-Pak C18 200 mg) as follows:
- Wash with 3 mL of methanol to condition the stationary phase.
- Wash with 3 mL of LC-MS grade-water.
- Add 1 mL of the derivatized DL-2-HG solution.
- Wash with 3 mL of LC-MS grade-water. Discard this fraction.
- Wash with 3 mL of hexane. Discard this fraction.
- Place a glass test tube and add 3 mL of acetonitrile to the SPE cartridge to elute and collect the derivatized compound.
Evaporate the derivatized compound using a N2 evaporator operated at 40 °C using a water bath.
Redissolve the dried residue in 1 mL of the composition of the mobile phase where the analytes elute in the LC-MS system. In our case this was a mixture of 70:30 (v/v) mobile phase A and B, respectively. The final concentration was 1.25 mM for each 2-HG enantiomer.
Load a 500 μL syringe with the SPE-purified (derivatized) compound to optimize the MS parameters by direct infusion.
Optimization of the EISA method.
Timing: 6 h.
<CRITICAL> The procedure requires that the EISA-MS parameters are optimized for in-source fragmentation. Similar to tandem MS/MS method optimisation, different parameters have to be fine-tuned to reach the optimum values for Q-MRM experiments. The particular goal of the EISA optimization is to get as many high abundant fragment ions as possible without significantly compromising the intensity of the molecular ion (precursor). To speed up the process, we advise optimising most of these parameters using direct infusion.
<CRITICAL> When multiple analytes are to be determined each optimization should be done separately. The optimum condition is one where all analytes have some in-source fragmentation while maintaining the highest intensity of both precursor and product ions.
-
5Asses the following parameters using direct infusion:
- End plate offset: from 0 to 2000 V.
- Capillary: from 3000 to 5000 V.
- Nebulizer: from 1 to 2.4 bar.
- Dry Gas: from 1 to 10 L/min.
<CRITICAL STEP> Any parameters related to voltage and heat in the ion source are normally associated with the in-source fragmentation of analytes.
Selection of the parameters are largely dependent on the research purpose and are instrument-dependent so instrument vendor recommendations should be checked to know lower and upper limits. Our optimization was conducted on a Bruker Impact II system.
<CRITICAL STEP> In our lab, we performed step-wise optimization: we optimized the first parameter, e.g. end plate offset, we selected the optimum value (i.e. 10 V) and then continued with the next parameter using this optimized value. There are other ways that this could be done, you might consider using “design of experiments” to find the optimum value among the multiple parameters assayed here.
-
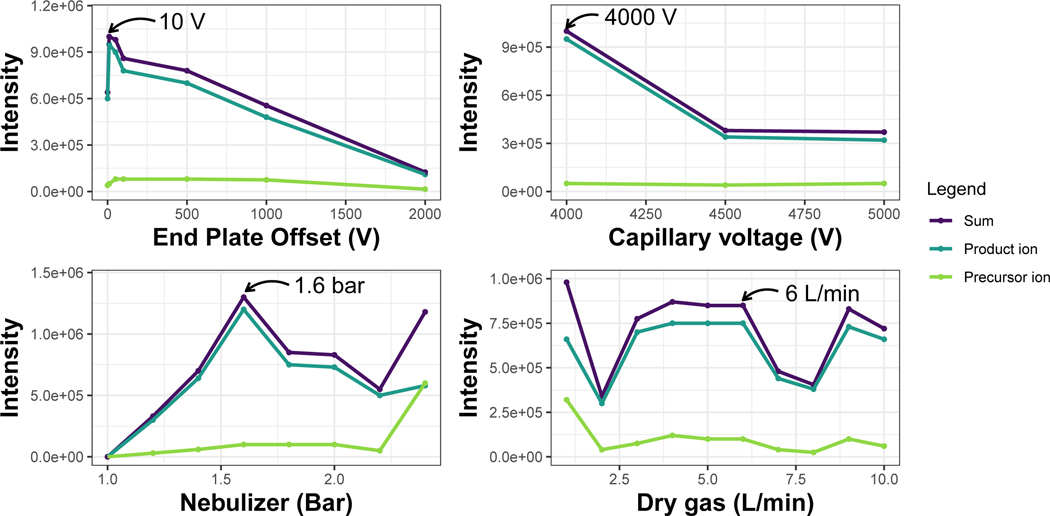
6To choose the optimum value per condition, look for the value that enables the highest intensity of the precursor ion with adequate fragmentation. Create a plot that shows the intensity of the precursor and the product ions. It is also helpful to represent the sum of the intensities for both ions. Figure 3 shows the different intensity points obtained for each of the parameters assayed. From these data we decided on the following parameters:
- End plate offset: 10 V
- Capillary: 4000 V
- Nebulizer: 1.6 bar
- Dry Gas: 6 L/min
Figure 3.
Optimized parameters of the EISA method: a) End plate offset; b) Capillary voltage; c) Nebulizer pressure; d) Dry gas. All parameters were optimized by direct infusion as detailed in Box 1. The optimum values are the ones indicated by the arrow.
<CRITICAL STEP> Although in panel Figure 3d we found that 1 L/min was the optimum value the ionization was too low to be stable, thus, we selected 6 L/min as it also gave high intensity of precursor + product.
-
7(optional) Optimize the in-source collision induced dissociation parameters for the analytes in the conditions they are in after LC separation.
- Under optimum LC-MS conditions assess the isCID voltage between 0 and 80 eV. To do this, prepare your standard mix as described in Step XX and inject it at least twice per isCID voltage assayed.
- Extract the EIC of the product and precursor ions and integrate the resulting peaks. In the example shown in Figure 4, these are m/z 147 and m/z 363 respectively. Determine the S/N ratio using the given software.
- Plot the S/N of each analyte per assayed isCID condition.
- To choose the optimum value, select the one with the highest S/N (see Figure 4).
Figure 4.
Signal-to-noise ratio (S/N) of each 2-HG enantiomer (20 μM) per assessed isCID value. Duplicate analyses are shown.
<CRITICAL STEP> In many ESI sources, it is possible to select a parameter which is exclusively designed for controlling the in-source fragmentation energy. This is vendor dependent, e.g., isCID energy in Bruker mass spectrometers and fragmentor voltage in the Agilent mass spectrometers. In our case we could acquire fragment ions until 40 eV, although the S/N at that isCID was compromised. Complete fragmentation of the precursor ion was achieved at higher isCID energies.
<CRITICAL STEP> If more than one product ion is obtained, under optimum LC-MS conditions you can always evaluate what combination of precursor + product/s ions gives the highest S/N. For the example we have selected, the precursor (m/z 363) + just one product (m/z 147) ion gave the best S/N.
<CRITICAL STEP> When using a single quadrupole mass analyzer it is possible to work in so-called single ion monitoring (SIM) so that each ion (precursor and product) becomes independent channels. Each SIM channel has their own settings (including some parameters controlling the in-source fragmentation depending on the vendor), which can be optimized exclusively for the performance of the target ion. However, these ion channels share many parameters in the ion source, which need to be optimized by taking all target ions into account (see Note 6 of Box 1). By means of the CIM algorithm, it is possible to reconstruct these ion channels into a single molecular trace (https://github.com/ricoderks/eisaCIM (see also ref. 6). The CIM algorithm is designed to align and selectively compile multiple ions within one chromatogram by analyzing the SIM traces within a pre-specified RT window from mzxml files, including peak picking, alignment and data analysis. A CIM chromatogram can be created as a compilation of the individual ion signals only if each signal satisfies pre-set criteria.
Preparation of an internal standard solution for calibration.
Timing: 30 min. <CRITICAL> Since the overall aim is to quantify the selected metabolite (in this example, DL-2-HG) in samples of clinical interest through a simplified strategy, it is important to evaluate the dynamic range and linearity of the method. It is also important to incorporate an IS. In this particular case, we used the deuterated (d3) version of DL-2-HG.
-
8
To prepare the calibration line, design a pipetting scheme like that shown in Table 1. Each solution must include the same amount of IS (for area correction), and any supporting reagents required for derivatization (e.g. sodium lactate (to facilitate the derivatization of low concentrations of DL-2-HG17 and methanol (to aid in the drying step prior derivatization)). In order to calculate the actual standard concentration, take into account the final reconstitution volume used for your samples. In our case this is 100 μL.
Table 1.
Pipetting scheme to prepare the calibration line of the DL-2-HG for the EISA method.
| Calibration points | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|
| |||||||||
| Concentration (μmol/L) per DL-2-HG enantiomer | 0.5 | 1.25 | 2.5 | 5 | 7.5 | 10 | 20 | 50 | 100 |
|
| |||||||||
| Concentration (μmol/L) of the stock solutions used for spikinga | 2 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 |
|
| |||||||||
| Volume (μL) of stock solution addedb | 50 | 1.25 | 2.5 | 5 | 7.5 | 10 | 20 | 50 | 100 |
|
| |||||||||
| Volume (μL) of 1 mmol/L IS solution addeda,b | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
|
| |||||||||
| Volume (μL) of 10 mmol/L sodium lactate solution addedb | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
|
| |||||||||
| Volume (μL) of methanol added | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 |
The concentration depicted here is the sum of both enantiomers.
In case volumes lower than 20 μL cannot be accurately taken, use less concentrated stock solutions so that larger volumes of the corresponding aliquots can be used.
Preparation of biological samples: DL-2-HG extraction
-
9
The extraction of DL-2-HG from the different biological samples is the first step in the analysis and quantification process. Here it is important to take into account the type of sample and the needed solvent to achieve a successful extraction that will lead to protein precipitation, necessary before any LC analysis. To carry out the extraction from cells follow option A (see Figure 5a). Here we exemplify the sample preparation by using myelogenous leukemia cells (K562) and chondrosarcoma cells (CH2879-IDH1wt and JJ012-IDH1R132G), but it can be used for any other mammalian cell line.
To conduct the extraction from biofluids (fetal calf serum, human serum, plasma and urine) follow option B (see Figure 5b).
Make sure to keep samples on ice at all times to prevent sample degradation and minimize residual enzymatic activity.
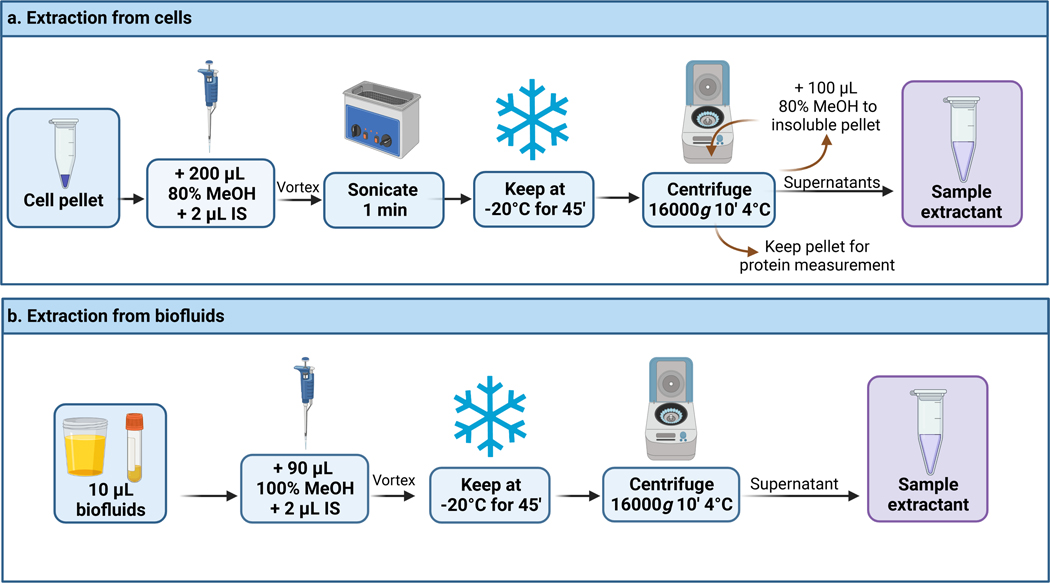
Figure 5.
Extraction protocol for the analysis of DL-2-HG from cell pellets or biofluids. This figure has been created with BioRender.com.
(A) Extraction from cell pellets.
Timing: 1.5 h.
Add 200 μL of ice-cold (−20 °C) 80% (vol/vol) methanol to a frozen cell pellet in a 1.5 or 2.0 mL-polypropylene microcentrifuge tube. We typically expect the cell pellet to contain ~ 1 × 106 cells. This number could be optimized depending on the sensitivity of the method and the expected concentration of the desired analytes.
-
Add 2 μL of IS stock solution (1 mmol/L DL-2-HG-d3).
CRITICAL STEP. You must ensure that the IS solution is added at this point to account for errors during sample preparation and analysis.
(optional) If needed during method validation, add at this step the desired concentration of DL-2-HG standard according to the validation of the method section.
Vortex for 15 s and sonicate for 1 min at room temperature.
Keep at −20 °C for 30 min to allow for protein precipitation.
Centrifuge at 16,000g for 10 min at 4 °C.
Transfer the supernatant to a new 1.5 mL-polypropylene microcentrifuge tube. Note: make sure the insoluble pellet is not taken.
Add an additional 100 μL of ice-cold 80% (vol/vol) methanol to the insoluble pellet to ensure everything is extracted.
Centrifuge at 16,000g for 10 min, 4 °C.
Transfer the supernatant to the previous 1.5 mL-polypropylene microcentrifuge tube. If you are performing derivatisation, then perform this reaction immediately (follow all steps in this Protocol) if you are not performing derivatisation, skip Steps 12–17.
If needed, keep the insoluble pellet for bicinchoninic acid (BCA) protein quantification for normalization purposes. For details on protein analysis, refer to the manufacturer’s instructions.
<PAUSE POINT> If needed samples can be stored at −20 °C for maximum one week.
(B) Extraction from biofluids: fetal calf serum, human serum, plasma and urine.
Timing: 1 h.
Add 9 volumes of ice-cold LC-MS grade methanol (cooled to −20 °C) to 1 volume of biofluid. In our case, we added 90 μL of methanol to 10 μL of fetal calf serum, human serum or plasma and 180 μL of methanol to 20 μL of human urine in a 1.5 or 2 mL-polypropylene microcentrifuge tube.
Add 2 μL of IS stock solution (1 mmol/L DL-2-HG-d3).
(optional) If needed during method validation, add at this step the desired concentration of DL-2-HG standard according to the validation of the method section.
Vortex for 15 s at room temperature.
Keep at −20 °C for 30 min to allow for protein precipitation.
Centrifuge at 16,000g for 10 min at 4 °C.
Transfer the supernatant to a new 1.5 mL-polypropylene microcentrifuge tube. Take care to not transfer any of the insoluble pellet. If you are performing derivatisation, then perform this reaction immediately (follow all steps in this Protocol) if you are not performing derivatisation, skip Steps 12–17.
(optional) Derivatization of the calibration standards and extractant samples e.g. with diacetyl-L-tartaric anhydride (L-DATAN).
Timing: 4 h.
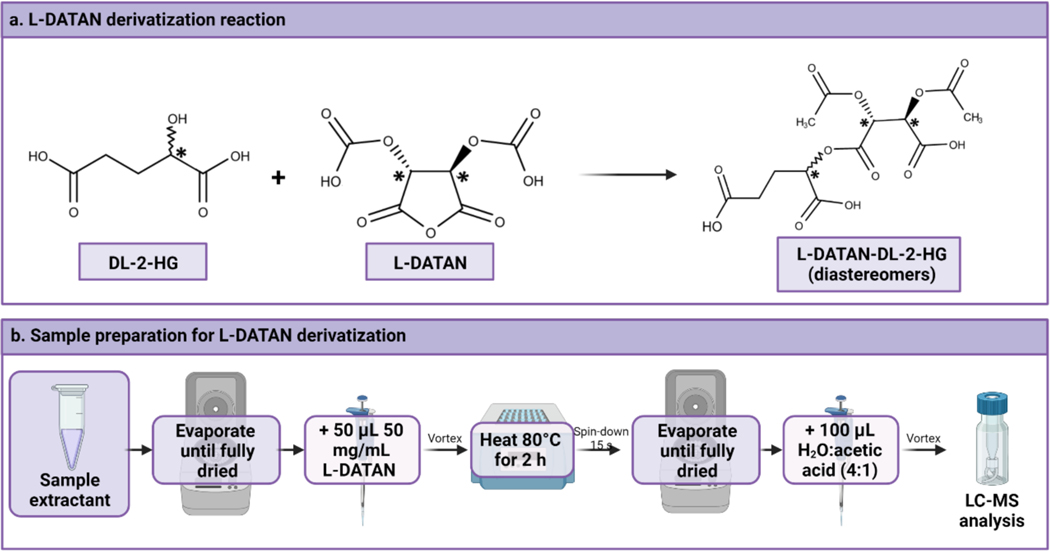
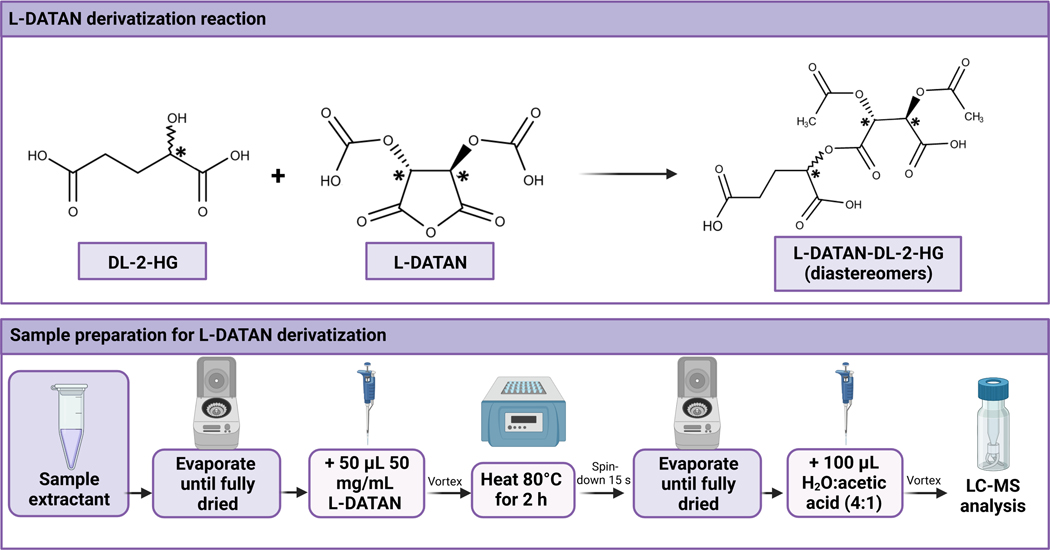
<CRITICAL> In order to carry out the chiral separation of DL-2-HG, derivatization with L-DATAN is necessary to obtain the corresponding diastereomers that will be then separated on a standard reverse phase column (see Figure 6). This section can be left out if your method does not require derivatisation of the analyte compound. It can also be replaced with the derivatisation method that is most appropriate for your analyte; much of the advice we have included here will be relevant to other derivatisation methods.
Figure 6.
Derivatization reaction scheme and sample protocol to achieve the derivatization of DL-2-HG with L-DATAN. This figure has been created with BioRender.com.
-
10
Evaporate samples until total dryness using a SpeedVac for 45 min at 60 °C.
<PAUSE POINT> If needed samples can be stored at this point for later analysis. Dried samples could be kept at −20 °C if analyzed in the same week or at −80 °C if analysis cannot be initiated in a week. However we recommend to prepare and analyze the samples as soon as possible.
-
11
Add 100 μL of LC-MS grade methanol to the microcentrifuge tubes to help any remaining droplets of water to evaporate. Dry in the SpeedVac for an additional 20 min at 60 °C.
CRITICAL STEP. You must ensure that all water is evaporated and that the sample has completely dried, otherwise the derivatization will not take place.
-
12
Prepare a fresh solution of 50 mg/mL L-DATAN in 4:1 (vol/vol) acetonitrile:acetic acid. Prepare immediately before use to avoid unnecessary evaporation.
-
13
Add 50 μL of the L-DATAN solution.
-
14
Vortex for 15 s.
-
15
Heat at 80 °C in a heat block for 2 h. Depending on the concentration, samples may turn into a dark yellow or brownish color.
CRITICAL STEP. Make sure that the polypropylene microcentrifuge tubes are tightly closed. The heat will generate vapors inside the tubes.
-
16
Keep at room temperature for 2 min and spin-down for 15 s at room temperature. This will enable that all the content returns to the bottom of the polypropylene microcentrifuge tube and will not be lost.
-
17
Dry samples using a SpeedVac for 60 min until completely dry.
-
18
Redissolve the dried residue in 100 μL of a 4:1 (vol/vol) LC-MS grade water:acetic acid solution and vortex.
CRITICAL STEP. Make sure that the dried pellet resulting from the drying step is completely redissolved. This might need vortexing for several minutes.
-
19
If a turbid solution is observed, centrifuge at 16,000g for 10 min at room temperature.
-
20
Transfer 100 μL in a HPLC vial with insert for LC-MS analysis.
Preparing the UHPLC-MS for sample analysis and data acquisition.
Timing: 10 min (calibration of the MS instrument) + 10 min (setting the method parameters) + 10 min (conditioning of the column) + 13 min (analysis time per sample).
-
21
Calibrate the MS instrument according to the manufacturer’s instructions using a Tuning Mix solution.
-
22
Create an instrument method in MS scan mode following the instructions of the acquisition software. Work in negative ion mode, use a mass range from m/z 30 to 370 and a spectra rate of 1.00 Hz.
-
23
Set up the ion source conditions. These are the optimized values obtained by performing Steps 5–7. In our case these values are included in the Supplementary Material’s “EISA optimization” section. The rest of the MS-related parameters are also included in this section.
-
24
Before starting the analytical run, condition the Aqua C30 column with the initial gradient conditions for 10 min until reaching a stable pressure, in our case, 100% mobile phase A at 0.4 mL/min resulted in a backpressure of ~ 260 bar.
-
25
Set up the parameters and the gradient for the UHPLC system. In our case, these parameters and the gradient can be found in the Supplementary Information “Optimization of the LC conditions” section.
-
26
Include necessary blanks at the beginning, at the end and during the run to monitor sample carryover. When the run has finished, wash the column following manufacturer’s recommendations. In our case we washed the Aqua C30 column with acetonitrile for 10 min, LC-MS grade water for 10 min and 60% (vol/vol) acetonitrile:water for 10 min at a flow of 0.4 mL/min.
Evaluating the analytical characteristics of the method
<CRITICAL> To demonstrate the suitability of the method for quantification purposes, its analytical characteristics must be validated. In this example we assess its fit-for-purpose according to linearity, LOD, LOQ, trueness (accuracy), recovery, and precision.
Linearity.
Timing: 5 h (analysis time) + 2 h (data analysis).
-
27
Construct a calibration line for each of your chosen analytes (e.g. using the information in Table 1 where the concentration range is 0.5–100 μmol/L for each enantiomer). Refer to the Step 8.
-
28
Inject the nine calibration line solutions and analyse these using the optimized EISA method. Refer to the section: “Preparing the UHPLC-MS for sample analysis and data acquisition”. Integrate the peak areas of both the external and the internal standard (e.g. for both DL-2-HG and deuterated DL-2-HG).
-
29
For each enantiomer, plot the area ratio versus the nine calibration line. A weighted linear regression is often recommended due to the fact that variance over the entire concentration range is not equal, rather the variance increases as concentration increases. For this reason, 1/x2 is the most commonly used weighting factor that improves accuracy in quantifying low concentrations standards.
-
30
To assess the linearity, calculate the R2 of the regression model which should be as high as possible (ideally > 0.99). As recommended by the United States Food and Drug Administration (FDA), the linearity should also be evaluated by appropriate statistical methods such as ANOVA18.
Determination of the LOD.
Timing: 5 min.
-
31
Once the linearity is assessed, determine the LOD as 3.3 × (standard deviation of the intercept of the calibration line) / (slope of the calibration line).
Determination of the LOQ.
Timing: 15 min.
-
32
Once the linearity is assessed, determine the LOQ as the lowest concentration from the calibration line that can be quantified with acceptable accuracy (between 70–130 %) and precision (coefficient of variation <30%).
Trueness.
Timing: 12.5 h (analysis time) + 2 h (data analysis).
<CRITICAL> Here we asses trueness expressed in terms of “bias” by recovery experiments using spiked samples. In this context, spiked samples are biological samples to which different concentrations of external standard have been added. The external standard is added together with the IS before sample extraction (and before the derivatization when needed). The samples themselves may also contain some of the compound of interest. Thus it is important having a control sample (without adding the external standard) functioning as reference of the endogenous concentration. Considering that a sample with no detectable endogenous analyte was used, the analysis of a spiked sample should reveal a quantified concentration equal to the concentration used (within a 80–120% range).
-
33
Prepare a calibration series (see e.g. Table 1) for each analyte of interest. Refer to the Step 8.
-
34
Prepare the different biological samples according to section “Preparation of biological samples: DL-2-HG extraction” and on step iii) add the required volume of the external standard stock solution to assess the trueness at four different levels. For the DL-2-HG example these would be: 0 (non-spiked), 10, 50 and 70 μmol/L (see Table 2 for pipetting scheme).
-
35
Inject both the calibration line solutions and the samples (spiked or non-spiked) using the EISA method as detailed in “Preparing the UHPLC-MS for sample analysis and data acquisition”. Integrate the peak areas of both the external and the internal standard (in this example, DL-2-HG and the IS).
-
36
Using the calibration line to calculate the concentration of each analyte of interest in each spiked sample as well as in the non-spiked sample.
-
37
Evaluate the trueness of the method by subtracting the amount of the analyte of interest (e.g. DL-2-HG) in the non-spiked solutions from the amount detected in the three different spiked levels (10, 50 and 70 μmol/L) (this needs to be done if there were endogenous levels of the compound in the sample). Divide the resulting concentration by the value of the known added concentration and multiply by 100. The values should be in the 80–120% range.
Table 2.
Pipetting scheme to assess the trueness of the DL-2-HG EISA method
| Spiked concentration (μmol/L) per DL-2-HG enantiomer | ||||
|---|---|---|---|---|
|
| ||||
| 0 (non-spiked) | 10 | 50 | 70 | |
|
| ||||
| Concentration (μmol/L) of the stock solutions used for spikinga | - | 2 | 2 | 2 |
|
| ||||
| Volume (μL) of stock solution addedb | - | 1 | 5 | 7 |
|
| ||||
| Volume (μL) of 1 mmol/L IS solution addeda,b | 2 | 2 | 2 | 2 |
|
| ||||
| Volume (μL) of methanol added | 90 | 90 | 90 | 90 |
The concentration depicted here is the sum of both enantiomers.
In situations where volumes lower than 20 μL cannot be accurately taken, use less concentrated stock solutions so that larger volumes of the corresponding aliquots can be used.
Recovery.
Timing: 6.5 h (analysis time) + 1 h (data analysis).
-
38
Prepare the different biological samples according to section “Preparation of biological samples: DL-2-HG extraction”, but for each experimental replicate create two samples: one where the external standard is added either before and one where it is added after the extraction step. In the example shown, add the necessary volume of the stock solution to achieve a final 10 μmol/L per enantiomer (for a detailed pipetting scheme follow the same procedure as in the accuracy, corresponding to the 10 μmol/L column, see Table 2).
-
39
Inject both the spiked samples using the EISA method as detailed in “Preparing the UHPLC-MS for sample analysis and data acquisition”. Integrate the peak areas of both the external and the internal standard (e.g. DL-2-HG and the IS).
-
40
To calculate the recovery, use the external standard area ratio obtained for the sample where the standard was added before extraction and divide this value by the resulting area ratio obtained after extraction and multiply by 100. The resulting value, should be in the 80–120 % range.
Precision.
Timing: 7.5 h over a span of 3 days (2.5 h each day) + 1.5 h (data analysis).
-
41
Prepare the different biological samples as described in the section section “Preparation of biological samples: DL-2-HG extraction” and add the required volume of external standard stock solution (for this example use the necessary volume of the stock solution so that that final concentration is 10 μmol/L per enantiomer). Follow the same procedure as in the accuracy study, corresponding to the 10 μmol/L column (see Table 2). The corresponding samples should be prepared in triplicate during three consecutive days to assess the precision.
-
42
Inject both the spiked samples using the EISA method as described in “Preparing the UHPLC-MS for sample analysis and data acquisition”. Integrate the peak areas of both the external and the internal standard (e.g. DL-2-HG and the IS).
-
43
To calculate the between-injection repeatability, use the area ratio for the external standard obtained in three consecutive injections of the same replicate and calculate the relative standard deviation (RSD) (%).
-
44
To calculate the measurement repeatability, use the area ratio for the external standard obtained in three replicates originating from the same sample, each injected in triplicate in the same day and calculate the RSD (%).
-
45
To calculate the intermediate measurement precision, use the area ratio for the external standard obtained in three replicates originating from the same sample, injected in triplicate on three consecutive days and calculate the RSD (%).
? TROUBLESHOOTING
For a detailed troubleshooting see Table 3.
Table 3.
Troubleshooting of the UHPLC-EISA-TOF method for the quantification of DL-2-HG in biological samples
| Step | Problem | Observation | Possible reason | Solution |
|---|---|---|---|---|
| Derivatization (Step 16) | Derivatization did not work | Standards or samples do not turn into yellow or brownish color | Not all water was evaporated. Alternatively, it could also be possible that the sample is not very concentrated and thus the color is not that noticeable. | Make sure all water has been evaporated before adding L-DATAN solution. |
| Liquid chromatography instrument (Steps 24–26) | High column backpressure | Pressure near 400 bar and/or low reproducibility of the retention time | Column may be clogged | Clean the column using 2-propanol. If the problem persists, replace the column |
| Pressure fluctuation | The pressure is not constant | Valve or pump seal leaks and might be damaged | Replace the check valve and the pump seals | |
| Loss of chiral separation | The resolution of the peaks is lower than expected | Mobile phase A and B are not prepared correctly or column is defective | Make sure mobile phase A and B are well prepared and/or replace the column | |
| Mass spectrometry instrument (Steps 21–23) | No peaks appear | Ions of the compound are not observed | Some parameters are not set correctly setup and/or the derivatization step was not successful | Check the MS parameters and/or make sure the derivatization was conducted in the proper way |
| Poor mass accuracy | Peaks are not observed when they are extracted with a width of m/z ±0.01 | The system is not well calibrated | Calibrate the MS detector according to the manufacturer’s instructions | |
| Loss of sensitivity | Ion counts are too low | The ion source may be contaminated | Clean the ion source, spray shield and the glass capillary following the manufacturer’s instructions |
TIMING
Step 5–7, Optimization of an EISA method: 6 h.
Step 8, Preparation of an internal standard solution for calibration line: 30 min.
Step 9A, Extraction from cells pellets: 1.5 h.
Step 9B, Extraction from biofluids: fetal calf serum, human serum, plasma and urine: 1 h.
(optional) Step 10–20, Derivatization of the calibration standards and extractant samples e.g. with diacetyl-L-tartaric anhydride (L-DATAN). Timing: 4 h.
Step 21–26, Preparing the UHPLC-MS for sample analysis and data acquisition: 10 min (calibration of the MS instrument) + 10 min (setting the method parameters) + 10 min (conditioning of the column) + 13 min (analysis time per sample).
Step 27–30, Linearity. 5 h (analysis time) + 2 h (data analysis).
Step 31, Determination of the LOD: 5 min.
Step 32, Determination of the LOQ: 15 min.
Step 33–37, Trueness: 12.5 h (analysis time) + 2 h (data analysis).
Step 38–40, Recovery: 6.5 h (analysis time) + 1 h (data analysis).
Step 41–45, Precision: 7.5 h in a span of 3 days (2.5 h each day) + 1.5 h (data analysis).
ANTICIPATED RESULTS
EISA takes advantage fragment ions produced in the ion source; these fragment ions are similar to those generated by tandem MS/MS. Among the benefits of EISA we can highlight:
Selective and sensitive results using a single mass analyzer, allowing for broad applicability at low cost.
Fragment ions can be produced over a wide dynamic range of ion intensities.
Precursor ion remains at high intensity when producing recognizable fragments, therefore, increasing the sensitivity.
Robust identification of a wide range of small molecules.
The purpose of this protocol is to show the development of an EISA methodology for quantification purposes. Here we exemplified this by showing the quantitative analysis of DL-2-HG in different biological matrices.
The results of the validation for each sample are illustrated in Table 4. It demonstrates that this protocol allows for the accurate and precise quantification of DL-2-HG in all cases obtaining a good recovery. Moreover, the LOD calculated was 0.08 and 0.09 μmol/L for L-2-HG and D-2-HG, respectively. Here we want to provide readers with a brief discussion of the anticipated results of our Q-MFM assessed in the clinical setting.
Table 4.
Results of assessing the trueness, precision and recovery of the different biological samples by the developed EISA-TOF method
| K562 cells | Urine | FCS | Serum | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| L-2-HG | D-2-HG | L-2-HG | D-2-HG | L-2-HG | D-2-HG | L-2-HG | D-2-HG | |
| Trueness | % Mean recovery ± SD | |||||||
|
| ||||||||
| 10 μmol/L | 97 ± 4 | 90 ± 5 | 98 ± 8 | 105 ± 5 | 105 ± 8 | 101± 4 | 103 ± 2 | 106 ± 6 |
| 50 μmol/L | 95 ± 2 | 87 ± 2 | 100 ± 8 | 113 ± 10 | 100 ± 5 | 96.7 ± 0.3 | 106 ± 3 | 105 ± 5 |
| 70 μmol/L | 97 ± 2 | 83 ± 5 | 98 ± 3 | 105 ± 5 | 99 ± 2 | 96 ± 3 | 105 ± 1 | 105 ± 2 |
| Precision | RSD (%) | |||||||
|
| ||||||||
| Between-injection repeatability (n=3)a | 5.0 | 5.0 | 3.6 | 3.9 | 1.1 | 2.2 | 1.3 | 2.2 |
| Measurement repeatability (n=3)b | 4.8 | 4.8 | 1.8 | 2.4 | 5.3 | 6.8 | 3.5 | 3.1 |
| Intermediate measurement precision (n=9)c | 4.9 | 4.9 | 4.6 | 4.4 | 6.7 | 7.3 | 3.8 | 5.2 |
| Recovery (before and after the extraction) | % Mean recovery ± SD | |||||||
|
| ||||||||
| 104 ± 8 | 104 ± 8 | 99 ± 2 | 94 ± 5 | 99 ± 3 | 94 ± 7 | 95 ± 3 | 99 ± 5 | |
Three consecutive injections of the corresponding biological sample spiked with 10 μmol/L of L-2-HG, 10 μmol/L of D-2-HG, 10 μmol/L of L-2-HG-d3 and 10 μmol/L of D-2-HG-d3.
Three replicates of the corresponding biological sample spiked with 10 μmol/L of L-2-HG, 10 μmol/L of D-2-HG, 10 μmol/L of L-2-HG-d3 and 10 μmol/L of D-2-HG-d3 injected in triplicate on the same day.
Three replicates of the corresponding biological sample spiked with 10 μmol/L of L-2-HG, 10 μmol/L of D-2-HG, 10 μmol/L of L-2-HG-d3 and 10 μmol/L of D-2-HG-d3 injected during three consecutive days.
Assessing the reproducibility of Q-MFM: benchmarking the EISA-TOF against an EISA-Q method
To further assess the reproducibility of the Q-MFM method we transferred our DL-2-HG approach to a single quadrupole system located in a different laboratory. The Pearson correlation between the area ratios from the calibration line (Table 1) of both enantiomers in the Q-MFM methods (TOF and Q) revealed a high level of agreement between the two instruments (Supplementary Figure 5). In both laboratories, the method was validated in terms of trueness (accuracy), recovery and precision using the same commercially available human plasma (Table 5). As can be seen, both strategies were highly comparable, which highlights the high reproducibility of this approach even when performed at two different locations, with two different instruments, different vendors and handled by different laboratory staff.
Table 5.
Results of the calibration line and the analytical characteristics of the plasma analysis for the EISA-TOF and EISA-Q methods.
| EISA-TOF | EISA-Q | |||
|---|---|---|---|---|
|
| ||||
| Internal standard calibration method (0.5–100 μmol/L)a | ||||
|
| ||||
| L-2-HG | D-2-HG | L-2-HG | D-2-HG | |
| Slope ± SD | 0.131 ± 0.002 | 0.116 ± 0.002 | 0.124 ± 0.002 | 0.119 ± 0.003 |
| Intercept ± SD | 0.011 ± 0.003 | 0.021 ± 0.003 | 0.009 ± 0.003 | 0.013 ± 0.004 |
| R2 | 0.998 | 0.997 | 0.998 | 0.995 |
| LOQb | 0.5 μmol/L | 0.5 μmol/L | 0.5 μmol/L | 0.5 μmol/L |
| F-value of ANOVAc | 3268 | 2918 | 4046 | 1668 |
| Plasma | ||||
|
| ||||
| Trueness | % Mean recovery ± SD | |||
|
| ||||
| 10 μmol/L | 98 ± 4 | 100 ± 4 | 85 ± 6 | 96 ± 21 |
| 50 μmol/L | 97 ± 5 | 98.9 ± 0.1 | 99 ± 3 | 104 ± 4 |
| 70 μmol/L | 95 ± 8 | 99 ± 10 | 101 ± 4 | 104 ± 11 |
| Precision | RSD (%) | |||
|
| ||||
| Between injection repeatability (n=3)d | 3.6 | 2.8 | 2.3 | 6.8 |
| Measurement repeatability (n=3)e | 2.6 | 5.5 | 5.4 | 5.6 |
| Intermediate Measurement precision (n=9)f | 4.1 | 6.0 | 4.7 | 6.7 |
| Recovery (before and after the extraction) | % Mean recovery ± SD | |||
|
| ||||
| 106 ± 5 | 97 ± 4 | 86 ± 4 | 88 ± 14 | |
Nine standard solutions at concentration levels from 0.5 to 100 μmol/L for L-2-HG and D-2-HG. Each solution contains 10 μmol/L of L-2-HG-d3 and D-2-HG-d3. The calibration line was established by plotting the area ratio of each enantiomer versus the concentration using a 1/x2 weighting.
LOQ estimated as the lowest concentration in the calibration that can be quantified with acceptable trueness (between 70–130%) and precision (coefficient of variation <30%).
F-value for ANOVA test that confirms that experimental data fit to a linear model. In all cases the p-values were lower than 0.05.
Three consecutive injections of a plasma sample spiked with 10 μmol/L of L-2-HG, 10 μmol/L of D-2-HG, 10 μmol/L of L-2-HG-d3 and 10 μmol/L of D-2-HG-d3.
Three replicates of plasma samples spiked with 10 μmol/L of L-2-HG, 10 μmol/L of D-2-HG, 10 μmol/L of L-2-HG-d3 and 10 μmol/L of D-2-HG-d3 injected in triplicate on the same day.
Three replicates of plasma samples spiked with 10 μmol/L of L-2-HG, 10 μmol/L of D-2-HG, 10 μmol/L of L-2-HG-d3 and 10 μmol/L of D-2-HG-d3 injected during three consecutive days.
Assessing the applicability of Q-MFM in the diagnosis of IDH1 mutation in biological specimens: analysis of cell lines, and serum from acute myeloid leukemia patients
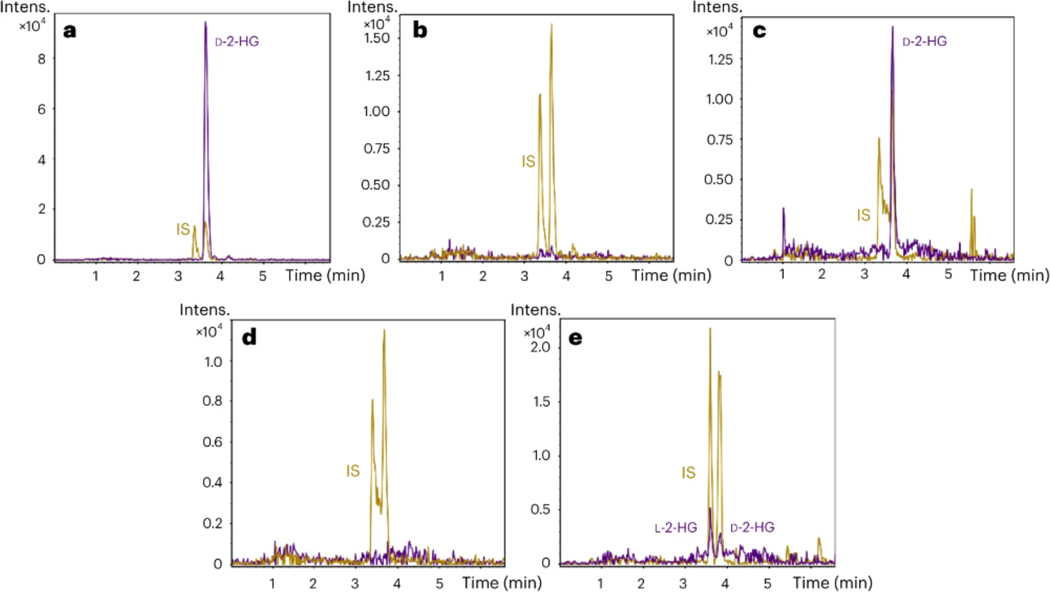
To further demonstrate the applicability of the Q-MFM in a clinical setup, we analyzed several biological specimens suspected to have endogenous D-2-HG. As expected, JJ012 chondrosarcoma cells carrying a mutation in the IDH1 gene accumulated high amounts of the D-enantiomer (62 ± 0.8 μmol/L) whereas the L-counterpart was not detected (Figure 7a). On the other hand, no D-2-HG was detected in CH2879 chondrosarcoma cells carrying wildtype IDH1 and IDH2 genes (Figure 7b).
Figure 7.
UHPLC-EISA-TOF extracted ion chromatograms of different biological samples derivatized with L-DATAN: (a) chondrosarcoma IDH1 mutant cell line; (b) chondrosarcoma IDH1 wild type cell line; (c) serum from an AML patient carrying a IDH1 mutation; (d) serum from a healthy donor; (e) Urine from a healthy donor. In all cases, the black trace corresponds to L-DATAN derivatized D- and/or L-2-HG (m/z 363.0570 + 147.0270) if present endogenously in the samples. The blue trace corresponds to the L-DATAN derivatized internal standard (IS) DL-2-HG-d3 (m/z 366.0745 + 150.0491). Both were extracted with a m/z ± 0.01 window.
Serum samples from two different AML patients carrying IDH1 mutations also showed presence of D-2-HG: 2.6 ± 0.2 and 12.0 ± 0.8 μmol/L (Figure 7c) in a volume of 20 μL whereas L-2-HG levels were below the LOD. On the contrary, serum from a healthy donor did not contain neither D- nor L-2-HG (Figure 7d). This is a clear demonstration that the developed method based on Q-MFM is able to quantify D-2-HG and correlate it to deficiencies in IDH1, being a potential tool for clinical practice and diagnosis.
Assessing the applicability of the Q-MFM in the quantification of D- and L-2-HG in urine
Urine samples of a healthy donor were screened for DL-2-HG analysis and results indicated that using a volume of 20 μL both enantiomers could be quantified at near 1-to-1 ratio, namely, 9.7 ± 0.2 and 6.9 ± 0.1 μmol/L for D- and L-2-HG, respectively (see Figure 7e). It is known that both enantiomers are present at low levels in urine samples from healthy individuals as previously reported9. A striking accumulation of any of these would be indicative of D- and/or L-2-hydroxyglutaric acidurias, a hallmark of encephalopathies and psychomotor retardation19.
Supplementary Material
Acknowledgements:
This work was supported by the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2019ZT08L213), the Guangdong Provincial Key Laboratory Project (2019B121203011), Key-Area Research and Development Program of Guangdong Province (2020B1111380003), the Leiden Center for Computational Oncology (LCCO), the Spanish Ministry of Science and Innovation (PID2019-104913GB-I00 project) and the Spanish Ministry of Economy and Competitiveness (S.B.B.’s predoctoral research contract (BES-2017-082458)).
The authors are grateful to A. Llombart-Bosch (University of Valencia, Spain) for the CH2879 cell line and J.A. Block (Rush University Medical Centre, Chicago, IL, USA) for the JJ012 cell line. Authors would also like to thank Ieva Palubeckaitė and Judith Bovée (Department of Pathology, Leiden University Medical Center, the Netherlands) for providing chondrosarcoma cells and valuable discussion, Hans Dalebout and René van Zeijl (Center for Proteomics and Metabolomics, Leiden University Medical Center, the Netherlands) for their technical assistance and Peter van Balen (Department of Hematology, Leiden University Medical Center, The Netherlands) for providing the serum samples.
Footnotes
Competing interests:
Authors declare no competing interests.
References
- [1].Holcapek M, Jirasko R, Lisa M. Recent developments in liquid chromatography–mass spectrometry and related techniques. J. Chromatogr. A 1259, 3–15 (2012). [DOI] [PubMed] [Google Scholar]
- [2].Leung KS-Y, & Fong BM-W LC-MS/MS in the routine clinical laboratory: has its time come? Anal. Bioanal. Chem 406, 2289–2301 (2014). [DOI] [PubMed] [Google Scholar]
- [3].Wishart D. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug. Discov 15, 473–484 (2016). [DOI] [PubMed] [Google Scholar]
- [4].Blazenovic I, Kind T, Ji J, Fiehn O, Blaženović. Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics. Metabolites 8, 31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Xue J. et al. Enhanced in-Source Fragmentation Annotation Enables Novel Data Independent Acquisition and Autonomous METLIN Molecular Identification. Anal. Chem 92, 6051–6059 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Xue J. et al. Single Quadrupole Multiple Fragment Ion Monitoring Quantitative Mass Spectrometry, Anal. Chem 93, 10879–10889 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xue J, Derks RJE, Hoang L, Giera M, Siuzdak G. Proteomics with Enhanced In-Source Fragmentation/Annotation: Applying XCMS-EISA Informatics and Q-MRM High-Sensitivity Quantification. J. Am. Soc. Mass Spectrom 32, 2644–2654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dang L. et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–743 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Struys EA, Jansen EEW, Verhoeven NM, Jakobs C. Measurement of urinary D- and L-2-hydroxyglutarate enantiomers by stable-isotope-dilution liquid chromatography-tandem mass spectrometry after derivatization with diacetyl-L-tartaric anhydride, Clin. Chem 50, 1391–1395 (2004). [DOI] [PubMed] [Google Scholar]
- [10].Zhou J. & Yin Y. Strategies for large-scale targeted metabolomics quantification by liquid chromatography-mass spectrometry. Analyst 141, 6362 (2016). [DOI] [PubMed] [Google Scholar]
- [11].González-Ruiz V, Olives AI, Martin MA Core-shell particles lead the way to renewing high-performance liquid chromatography, TrAC-Trend. Anal. Chem 64, 17–28 (2015). [Google Scholar]
- [12].Ciccimaro E. & Blair IA Stable-isotope dilution LC-MS for quantitative biomarker analysis, Bioanalysis 2, 311–341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pansuriya TC et al. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat. Genet 43, 1256–1261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gil-Benso R. et al. Establishment and characterization of a continuous human chondrosarcoma cell line, ch-2879: comparative histologic and genetic studies with its tumor of origin. Lab. Invest 83, 877–887 (2003). [DOI] [PubMed] [Google Scholar]
- [15].Scully SP et al. Marshall Urist Award. Interstitial collagenase gene expression correlates with in vitro invasion in human chondrosarcoma. Clin. Orthop. Relat. Res 376, 291–303 (2000). [DOI] [PubMed] [Google Scholar]
- [16].Arindrarto W, et al. Comprehensive diagnostics of acute myeloid leukemia by whole transcriptome RNA sequencing. Leukemia 35, 47–61 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Oldham WM and Loscalzo J. Quantification of 2-hydroxyglutarate enantiomers by liquid chromatography-mass spectrometry. Bio. Protoc 20, e1908 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].FDA, editor. Analytical Procedures and Methods Validation U.S.D.o.H.a.H. Services Rockville, Maryland: Food and Drug Admistration; (2000). [Google Scholar]
- [19].Kranendijk M, Struys EA, Salomons GS, Van der Knaap MS, Jakobs C. Progress in understanding 2-hydroxyglutaric acidurias. J. Inherit. Metab. Dis, 35, 571–587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.