Abstract
We investigated the regulation of the S10 ribosomal protein (r-protein) operon among members of the gamma subdivision of the proteobacteria, which includes Escherichia coli. In E. coli, this 11-gene operon is autogenously controlled by r-protein L4. This regulation requires specific determinants within the untranslated leader of the mRNA. Secondary structure analysis of the S10 leaders of five enterobacteria (Salmonella typhimurium, Citrobacter freundii, Yersinia enterocolitica, Serratia marcescens, and Morganella morganii) and two nonenteric members of the gamma subdivision (Haemophilus influenzae and Vibrio cholerae) shows that these foreign leaders share significant structural homology with the E. coli leader, particularly in the region which is critical for L4-mediated autogenous control in E. coli. Moreover, these heterologous leaders produce a regulatory response to L4 oversynthesis in E. coli. Our results suggest that an E. coli-like L4-mediated regulatory mechanism may operate in all of these species. However, the mechanism is not universally conserved among the gamma subdivision members, since at least one, Pseudomonas aeruginosa, does not contain the required S10 leader features, and its leader cannot provide the signals for regulation by L4 in E. coli. We speculate that L4-mediated autogenous control developed during the evolution of the gamma branch of proteobacteria.
All organisms have evolved mechanisms to regulate ribosome synthesis so that rapidly dividing cells dedicate a larger fraction of their mass and energy to manufacturing ribosomes than do more slowly growing cells. This optimization of metabolism has been particularly well studied in the bacterium Escherichia coli. Regulation of rRNA synthesis depends, at least in part, on an unusual sensitivity of rRNA transcription initiation to the nucleoside triphosphate concentration (9), which increases with growth rate (2). Ribosomal protein (r-protein) synthesis is regulated, in turn, in response to the availability of nascent rRNA molecules. Most r-protein operons contain a gene whose product functions not only as an r-protein but also as a repressor of the expression of the operon (see reference 46 for a review). When r-protein production exceeds rRNA synthesis, free regulatory r-proteins accumulate and each represses expression of its own operon, usually by inhibiting translation of the r-protein mRNA. This autogenous control mechanism is believed to coordinate the production of rRNA and r-protein and to balance the expression of individual r-protein operons (46) (although additional mechanisms also contribute to regulating r-protein production [22, 46]).
The 11-gene S10 operon of E. coli is autogenously regulated by r-protein L4, encoded by the third gene of the operon (Fig. 1A). Unlike other regulatory r-proteins, L4 inhibits not only translation but also transcription. Regulation of transcription is accomplished by L4-mediated termination (attenuation) within the S10 leader, about 140 bases from the start of transcription (Fig. 1B). This process requires the RNA polymerase accessory factor NusA and two hairpins in the nascent leader transcript (35, 36, 47, 51). One of these hairpins is also required for L4-mediated repression of translation, although distinguishable attributes of the leader RNA are required for the two levels of regulation (7, 34).
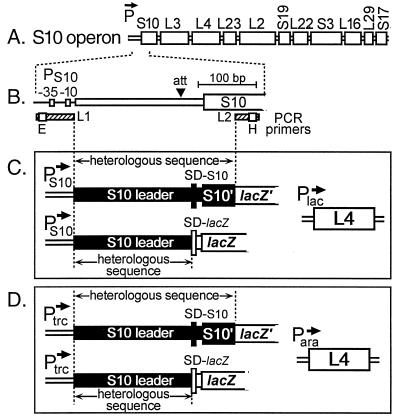
FIG. 1.
Maps of the S10 operon and plasmids used for regulatory studies. (A) Organization of genes in the E. coli S10 operon. (B) Region of the S10 operon amplified by PCR. The site of L4-mediated termination (att) is indicated by the arrowhead. Positions of the primers L1 and L2 are indicated by hatched bars. Open boxes on the primer bars indicate recognition sites for EcoRI (E) and HindIII (H) introduced during the amplification reaction. (C and D). General structures of L4 target and L4 source plasmids. Leader-S10′/lacZ′ or leader-lacZ genes driven by E. coli PS10 were introduced into cells carrying an IPTG-inducible Plac-L4 plasmid (C). Leader-S10′/lacZ′ or leader-lacZ genes driven by the IPTG-inducible Ptrc were introduced into cells carrying an arabinose-inducible Para-L4 plasmid (D). SD-S10 and SD-lacZ refer to the Shine-Dalgarno sequences for the S10 and lacZ structural genes, respectively.
While the molecular mechanisms of autogenous control of r-protein synthesis have been studied extensively in E. coli, knowledge of the control of r-protein synthesis in other organisms remains comparatively sparse. The general organization of r-protein genes is highly conserved. For example, in systems as diverse as gram-positive and gram-negative eubacteria, cyanobacteria, thermophilic eubacteria, archaea, protist cyanelles, chloroplasts, and mitochondria, clusters of r-protein genes corresponding to the S10, spc, and alpha operons of E. coli are strikingly similar (16, 42, 43). Nevertheless, the regulatory circuits identified in E. coli are not universal for all bacteria, since the positions of promoters and transcription terminators are not well conserved. For example, in E. coli, the 28 genes in the S10-spc-alpha region are organized into three consecutive transcription units (21, 27–29, 38). In contrast, the corresponding genes in Bacillus subtilis appear to be organized into a single transcription unit (15, 19, 38). Also, genes encoding r-proteins identified as regulators in E. coli are sometimes dissociated in other organisms from some or all of the r-protein genes that they regulate in E. coli. For example, the gene encoding r-protein S4, the autogenous regulator of the alpha operon in E. coli, maps outside the S10-spc-alpha cluster of B. subtilis (10, 14) and regulates only its own synthesis in this organism (11). Thus, while r-protein gene order in the S10-spc-alpha region is highly conserved, the molecular mechanisms for regulation of these genes are clearly more diverse.
To learn more about the evolution of regulatory mechanisms governing r-protein synthesis, we have focused on the regulation of the genes corresponding to the S10 operon of E. coli. Our earlier experiments suggested that in B. subtilis this gene cluster is regulated by a mechanism different from the E. coli-like L4-mediated control (19). To extend these studies, we inspected the S10 operons in a variety of eubacterial species within the gamma subdivision of the proteobacteria, which includes E. coli. Since the target for L4-mediated autogenous control is contained within the untranslated leader of the E. coli operon (7, 35, 48), we concentrated on this region. Our studies suggest that specific features of the S10 leader structure that are essential for L4-mediated autogenous control in E. coli are conserved in several, but not all, branches of the gamma subdivision, including all of the enterobacteria that we investigated. Moreover, our regulatory studies confirm that foreign leaders containing these features are sensitive to L4-mediated control in E. coli. We speculate that the conservation of these features reflects a conservation of L4-mediated autogenous control and therefore that this regulatory mechanism developed during the evolution of the gamma branch of the proteobacteria.
MATERIALS AND METHODS
Strains, plasmids, and other materials.
The E. coli strain was LL308 (23). Salmonella typhimurium, Citrobacter freundii, Yersinia enterocolitica, Morganella morganii, and Pseudomonas aeruginosa were obtained from the School of Medicine, University of Rochester. We obtained S. typhimurium LT2 and Serratia marcescens from the American Type Culture Collection and S. typhimurium MS1868 from M. Suskind via R. Wolf. Haemophilus influenzae was a gift from G. J. Barcak. Vibrio cholerae was from J. B. Kaper.
Plasmids pLF1 (8), pLF17 (8), pLL235 (20), pSma2R (48), pLL202 (8), pLL226 (48, 51), pLL229 (48), pACYC-Bsu (19), and pAra-L4 (19) have all been described previously. The pBAD-L4 plasmids are analogous to pAra-L4 but are derived from pBAD18 (12).
Restriction enzymes were purchased from New England Biolabs or Promega. Taq DNA polymerase was from Perkin-Elmer/Cetus, Fisher Biotech, or Promega. Vent DNA polymerase was purchased from New England Biolabs. Reagents and enzymes used for sequencing reactions were purchased from U.S. Biochemical.
Cloning and sequencing of PCR-amplified DNA.
Chromosomal DNA was prepared according to standard procedures. PCRs to amplify the S10 leader regions were performed under standard reaction conditions. For genomes for which we had no a priori S10 operon sequence information (S. typhimurium, C. freundii, Y. enterocolitica, and M. morganii), the upstream oligonucleotide was L1 (TTGAATTCCTAGCAATACGCTTGCGTTCGGTGGTTAAGTATGTATA ATG); the underlined sequence corresponds to the −35/−10 region of the E. coli S10 operon (Fig. 1B). The downstream oligonucleotide was L2 (CGAAGCTTTCCGCGGTTGCTTGATCGA), corresponding to the E. coli S10 structural gene (Fig. 1B). Amplified DNA was purified by cutting the band from a low-melting-point agarose gel and passing it through an Elutip column (Schleicher & Schuell) or a Wizard PCR Prep DNA purification system (Promega). The purified DNA was digested with the indicated restriction enzymes, cloned into M13mp18, and sequenced (33). For S. marcescens, the 5′ oligonucleotide, O175 (CGTACTACTGACGTGACTGG), was derived from the upstream tufA gene, and the downstream oligonucleotide was O243 (GCGCTTGGCAGTCTCGACGAT), from the S10 gene. The amplified DNA was sequenced directly. In all cases, two to five independent clones or PCR products were sequenced.
For in vivo regulatory studies, heterologous leader/S10′ sequences were inserted into pLL202 (8), containing a partial lacZ gene (lacking the first eight codons). The resulting constructs consisted of the E. coli S10 promoter (PS10), heterologous leader, and proximal 54 codons of the S10 gene, fused in frame with lacZ′ (Fig. 1C). To analyze only transcriptional control, heterologous leader sequences (without the S10 gene) were inserted upstream of the intact lacZ gene in pLL229 (48) (Fig. 1C). In later experiments, the heterologous leader/S10′ sequences were cloned downstream of the Ptrc promoter by replacing the B. subtilis S10 leader/S10′ fragment in pACYC-Bsu (19; Fig. 1D). For in vitro transcription studies, the E. coli leader in plasmid pLL226 (48, 51) was replaced with the heterologous leader.
Computer analysis.
DNA sequence alignments were determined by first using a multiple-sequence alignment program such as CLUSTAL W (39) and then manually aligning the sequences to obtain maximum identity. The RNA secondary structures were predicted by using Zuker’s MFOLD program on the mfold server (55), using the energy rules described by Walter et al. (41). Searches for S10 genes in other bacteria were performed by using BLAST 2.0 (1) on the NCBI BLAST server (27). The phylogenetic tree was constructed by using the Ribosome Database Project server (24, 31).
Labeling and gel electrophoresis of proteins.
E. coli cells were grown at 37°C in AB minimal medium (3) supplemented with 0.5% glycerol, 1 μg of thiamine per ml, and the appropriate antibiotics. For experiments with cells carrying a Plac-L4 plasmid and a PS10-foreign leader-S10′/lacZ′ or lacZ plasmid, cells were labeled for 2 min with 50 μCi of [35S]methionine (ca. 1,000 Ci/mmol) per ml immediately before or 10 min after addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to induce L4 oversynthesis and then lysed at 95°C in sodium dodecyl sulfate (SDS) sample buffer (17). For cells carrying both pAra-L4 and a Ptrc-foreign leader-S10′/lacZ′ or lacZ plasmid, the synthesis of S10′/β-galactosidase (β-Gal′) or β-Gal was first induced by addition of IPTG (to 2 mM). Immediately before and 15 to 20 min after addition of IPTG, aliquots of the culture were pulse-labeled and harvested by lysing at 95°C in SDS sample buffer (17). Synthesis of L4 protein was then induced by addition of arabinose (to 0.25%), and 10 to 15 min later aliquots were pulse-labeled and harvested. The total extracts were fractionated by gel electrophoresis on an SDS–7.5% or 12% (wt/vol) polyacrylamide gel (17) and visualized by autoradiography. The 12% gel resolves small proteins like L4 (22 kDa) and allowed us to confirm that the synthesis of L4 was increased following arabinose induction; based on earlier studies, we estimate that L4 synthesis was induced at least threefold (52). The 7.5% gel provides better resolution of the S10′/β-Gal′ fusion protein. The radioactivity in the S10′/lacZ′ protein band was quantified by using a Molecular Dynamics PhosphorImager. To control for variable loadings, the radioactivity in S10′/β-Gal′ was normalized to the radioactivity in a protein band whose intensity was not affected by IPTG or arabinose addition.
S. typhimurium MS1868 was grown and pulse-labeled as for E. coli except that the growth medium included 19 amino acids (no methionine). The source of L4 was plasmid pLL235 (Plac-L4) (20), and the target plasmid was pLF1 (PS10-E. coli S10 leader-S10′/lacZ′) (8, 45) or pSma2R (PS10-E. coli S10 leader-lacZ) (48).
In vitro transcription reactions.
Standard 10-μl transcription reaction mixtures contained 20 mM Tris acetate (pH 7.9), 4 mM magnesium acetate, 0.1 mM EDTA, 100 mM potassium glutamate, 20 nM E. coli RNA polymerase, and 20 nM plasmid DNA. Where indicated, r-protein L4 or S7 from E. coli was added to 160 nM, and NusA from E. coli was added to 40 nM. The DNA template was pLL226 (48, 51) or a derivative containing a non-E. coli leader. The reaction components were mixed together with 500 μM each CTP and GTP and incubated at 37°C for 10 min to allow formation of the initiation complex and incorporation of the proximal several nucleotides. A single round of transcription elongation was then started by addition of ATP to 500 μM, UTP to 100 μM, 5 μCi of [32P]UTP, and rifampin to 10 μg/ml. Reactions were terminated at the indicated times by the addition of 8 μl of sequencing stop mix. The RNA products were heated at 95°C for 2 min before loading on a 8% sequencing gel.
Nucleotide sequence accession numbers.
The S10 leader sequences determined in our laboratory have been deposited in the GenBank database under accession no. AF089898 (C. freundii), AF089899 (M. morganii), AF058449 (S. typhimurium), AF058451 (S. marcescens), and AF089900 (Y. enterocolitica). The H. influenzae leader sequence is found in the database under accession no. U32761.
RESULTS
Cloning and sequencing of the S10 leader from gamma proteobacteria.
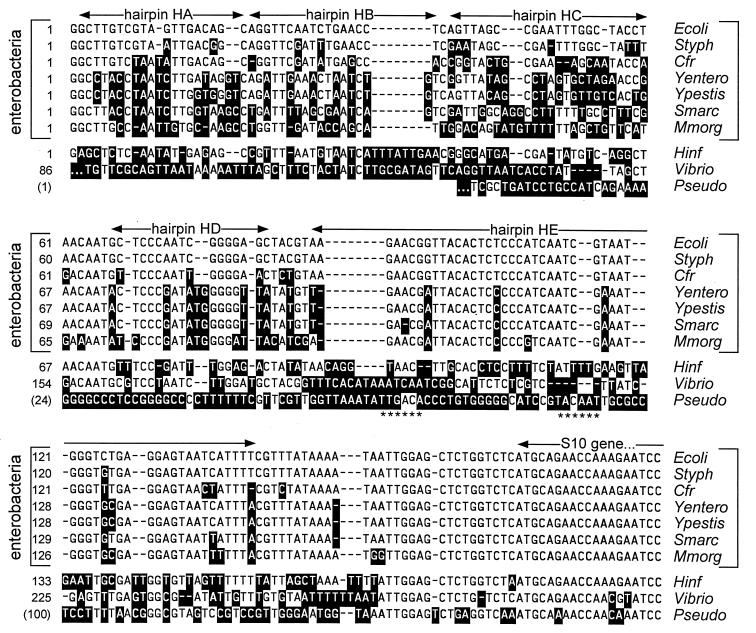
Our Southern analysis of the DNA at the 5′ end of the S. typhimurium S10 operon showed that the sequence immediately upstream of the promoter has no detectable homology with the corresponding E. coli sequence (data not shown). Therefore, to clone the S10 leader of S. typhimurium and other enteric bacteria, we designed an upstream primer that contained the E. coli promoter (as well as an EcoRI site to use for subsequent manipulations [Fig. 1B]), expecting that the promoter regions in the various enterobacteria would be sufficiently similar to hybridize to an oligonucleotide containing the E. coli promoter sequence. We also predicted that the S10 structural gene sequence would be very similar in all enterobacteria, and so the downstream primer contained a sequence from the E. coli S10 gene, as well as a HindIII site for cloning (Fig. 1B). Amplification of chromosomal DNA from four species of enterobacteria, S. typhimurium, C. freundii, Y. enterocolitica, and M. morganii, generated PCR products of the expected ca. 270 bases (not shown); this DNA was then cloned into M13mp18 and sequenced. The S10 leader region from the enterobacterium S. marcescens was also amplified and sequenced directly from the PCR product. Sequences of the five enterobacterial S10 leaders were then aligned to determine their relationship with the E. coli sequence. Recently, we identified the Yersinia pestis S10 leader sequence via a BLAST search of the DNA sequence database at the Sanger Centre (44); this sequence was included in our alignment analysis. As shown in Fig. 2, all six enterobacterial leaders showed significant primary sequence homology with E. coli’s, particularly in the 3′ two-thirds of the leader.
FIG. 2.
Alignment of S10 leader sequences. Bases differing from the E. coli sequence are indicated by white text on black background. The approximate positions of presumed hairpins are also shown. Ecoli, E. coli; Styph, S. typhimurium; Cfr, C. freundii; Yentero, Y. enterocolitica; Ypestis, Y. pestis; Smarc, S. marcescens; Mmorg, M. morganii; Hinf, H. influenzae; Vibrio, V. cholerae; Pseudo, P. aeruginosa. Except for the Pseudomonas sequence (see below), the 5′ ends of the various leaders were presumed from potential promoter sequences. The Vibrio leader probably begins about 80 bases upstream of the indicated sequence. The alignment programs did not identify any significant sequence homology in the Vibrio leader upstream of nucleotide 149, and so only the 3′ two-thirds of the leader sequence is shown. The complete sequence is shown in Fig. 3H. The asterisks below the Pseudomonas sequence refer to likely −35 and −10 sequences for the S10 promoter in this species. Therefore, the leader is probably much shorter than the leaders of other eubacteria. However, for comparison the sequence upstream of the presumptive transcription start site is included in the alignment.
To examine S10 leader sequences in other bacteria, we searched available bacterial databases, using the E. coli S10 protein sequence as the query sequence in BLAST2 (1) and then investigating the sequence upstream of the structural gene. Since the putative S10 leaders of more distant species shared no primary or secondary structure similarity with E. coli (data not shown), we focused on species that, like the enterobacteria, are members of the gamma subdivision of the proteobacteria. In addition to Y. pestis (44), already described, we analyzed H. influenzae, whose complete DNA sequence has been released (5), and V. cholerae (40) and P. aeruginosa (30). The latter two genomes were in the process of being sequenced.
The putative S10 leader sequences of the nonenteric proteobacteria showed very limited sequence similarity with the E. coli sequence (Fig. 2). For P. aeruginosa, the predicted S10 leader is very short, and only the S10 structural gene and 15 or so bases immediately upstream could be aligned with the E. coli sequence. For V. cholerae and H. influenzae, the alignment was somewhat more convincing (Fig. 2). Assuming that we have identified the correct S10 promoter sequences, the V. cholerae leader is considerably longer.
Secondary structure of the S10 leaders.
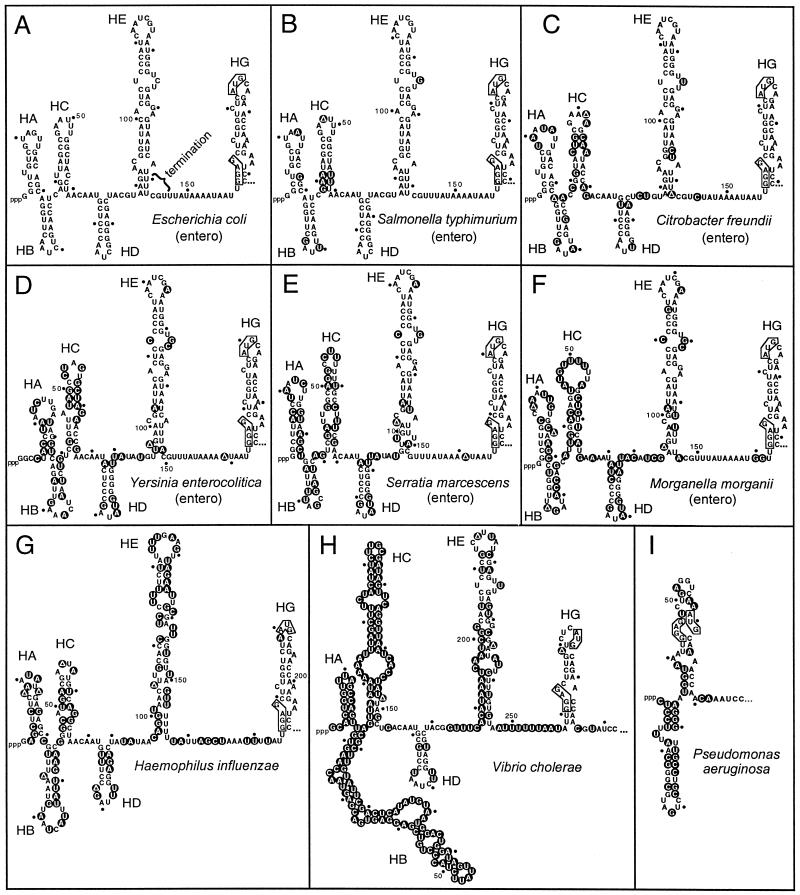
Our previous studies showed that two hairpins within the E. coli S10 leader are required for L4-mediated autogenous control. Hairpin HD (Fig. 3A) is essential for transcriptional, but not translational, control (7, 48). Hairpin HE (Fig. 3A) is required for both levels of control (7). To determine if the S10 leader sequences shown in Fig. 2 can form comparable hairpins, we used the mfold computer algorithm (54) to determine possible secondary structures of each leader.
FIG. 3.
Secondary structure predictions for the S10 leader regions. The secondary structures were predicted by using mfold (55). The computer-predicted E. coli structure has been confirmed by in vitro structure probing analysis (37). The site of L4-mediated transcription termination in the E. coli leader is indicated. Nucleotides differing from the E. coli sequence in the alignments shown in Fig. 2 are indicated by the black circles with white text. The Shine-Dalgarno sequence and the AUG initiation codon for the S10 structural gene are indicated by boxes. entero, enterobacterium.
All of the enterobacterial leaders could in fact generate structures strikingly similar to the structure from E. coli (Fig. 3B to F). Despite the unimpressive primary sequence homology for the H. influenzae and V. cholerae leaders, these two RNAs also generated secondary structures with remarkable similarities to E. coli structures in the region containing the HD, HE, and HG hairpins (Fig. 3G and H). However, the very short P. aeruginosa leader had no obvious structural similarity with E. coli’s (Fig. 3I).
Consistent with the primary sequence comparisons (Fig. 2), the 5′ third of the enterobacterial and H. influenzae and V. cholerae leaders displays limited secondary structure similarity: while all of the species have leader sequences compatible with the formation of hairpins HA, HB, and HC, there are significant variations between species with respect to details such as bulges, helix length, and loop size, particularly for the much longer V. cholerae leader. Variability among these three hairpins is not likely to have a significant effect on L4-mediated regulation of the operon, since their deletion from the E. coli leader has no measurable effect on autogenous control (48).
The structure of the fourth hairpin (HD) is completely preserved among the various enterobacterial species and is indeed an excellent example of a hairpin whose structure is supported by compensatory changes during evolution. That is, there are changes in basepairs in the stem, but none disrupt the helix (Fig. 3C to F). The H. influenzae and V. cholerae leaders also form an HD hairpin with 6 bp, although they both have additional bases in the loop (Fig. 3G and H). These results suggest that hairpin HD, which is required for L4-mediated transcription termination in E. coli (34, 48), is conserved in these proteobacteria.
All of the enterobacterial S10 leader sequences have the potential to form an extended hairpin similar to the HE hairpin of E. coli. There is almost complete sequence conservation in this region of the leader and therefore insufficient substitutions to provide a phylogenetic proof for this structure. Nevertheless, all of the enterobacteria have sequences compatible with the salient features of the E. coli HE hairpin: an upper hairpin consisting of a 5-bp stem and an 8-base loop with changes in only one base, an extended stem with several internal loops, and a U-rich sequence on the distal side in the position corresponding to the site of L4-mediated transcription termination in E. coli.
Despite having very different primary sequences, the leaders from H. influenzae and V. cholerae form HE hairpins similar in size and overall structure to the HE hairpins found in the enterobacteria, including a U-rich sequence at the base of the descending side of the hairpin. However, the closing loop of V. cholerae has six bases, while H. influenzae’s HE hairpin loop has nine bases.
Our previous studies showed that the upper stem-loop structure in hairpin HE is involved in both transcription and translation control by L4 (7, 34) and that the string of U’s near the descending base of the HE hairpin is required for transcription control (34, 35). Again, our phylogenetic results suggest that these structural features are conserved among these proteobacteria.
The most distal hairpin in the E. coli leader, HG, contains the ribosome binding site and initiation codon for the S10 structural gene. Since the sequence of this region of the leader is identical in all enterobacterial species, the structure of the HG hairpin is also preserved. The leaders from H. influenzae and V. cholerae form similar HG hairpin structures. Furthermore, the single-stranded region between HE and HG, which may be an entry site for ribosomes initiating translation of the S10 structural gene (46), is AU rich in all of the species. The P. aeruginosa leader lacks the single-stranded AU-rich sequence.
In vivo regulation in E. coli with non-E. coli S10 leaders.
Given the conservation of the HD and HE hairpin structures in the various bacterial species and their critical role in L4-mediated autogenous control in E. coli, we suspected that these foreign S10 operons might also be regulated autogenously by r-protein L4. We analyze L4 regulation by inducing oversynthesis of the r-protein from a plasmid and then measuring the effect on the expression of a lacZ gene placed downstream of the S10 promoter/leader on a second plasmid (7, 48, 53). Since most of the species analyzed here lack well-developed genetic systems that would allow conditional oversynthesis of L4, we tested the regulation of the heterologous leaders in E. coli. We have shown that L4 proteins from bacteria as divergent from E. coli as Bacillus stearothermophilus (only 42% amino acid identity) maintain the features required for autogenous control of the E. coli leader (53). Therefore, we reasoned that if the S10 operons from closely related species were subject to L4-mediated autogenous control, their leaders should respond to E. coli L4.
We first analyzed the leaders from the enterobacteria S. typhimurium, C. freundii, M. morganella, and Y. enterocolitica, by inserting the heterologous leader/S10′ sequences between the E. coli S10 promoter and lacZ′ (Materials and Methods; Fig. 1C). Each of these plasmids was transformed into an E. coli strain already containing a plasmid with an E. coli L4 gene under control of the lac promoter (Fig. 1C). The effect of L4 was measured by pulse-labeling the cells with [35S]methionine immediately before and 10 min after inducing L4 oversynthesis by addition of IPTG and examining whole-cell extracts of the labeled cells by SDS-polyacrylamide gel electrophoresis (PAGE). All four of the enterobacterial leader constructs were sensitive to L4 regulation, as evidenced by the decreased rate of synthesis of S10′/β-Gal′ after induction of L4. Indeed, expression of S10′/β-Gal′ from the heterologous leader clones was inhibited by L4 to approximately the same extent as the protein expressed from the E. coli leader (data not shown).
Although the foreign leaders appeared to be well regulated by E. coli L4, the plasmids containing these leaders slowed the growth of the E. coli host, resulting in a selection for mutated plasmids with no or reduced synthesis of the S10′/lacZ′ product. Indeed, when we re-sequenced the Y. enterocolitica plasmid, we found a mutation in the −35 region of the S10 promoter. Because the constitutive expression of heterologous leader-S10′/lacZ′ constructs appeared to be toxic to the E. coli host (for unidentified reasons), we modified the system so that the leader-S10′/lacZ′ was expressed from the IPTG-inducible Ptrc promoter and the L4 gene was expressed from an arabinose-inducible promoter (Fig. 1D). Using this system, we analyzed the E. coli and M. morganii leaders, as well as the more divergent H. influenzae and V. cholerae RNAs. For comparison, we also analyzed the P. aeruginosa leader, which shares no obvious structural similarities with the other proteobacterial S10 leaders.
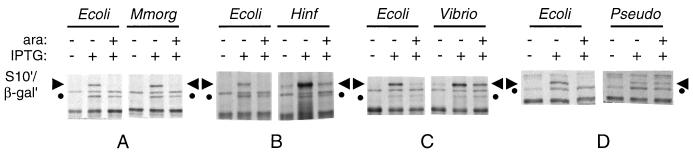
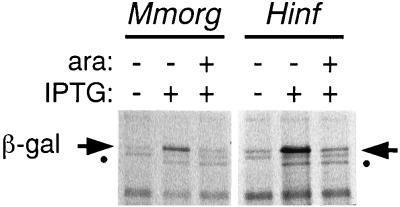
The effect of L4 regulation of these leaders was monitored by first inducing expression of the S10′/lacZ′ gene by addition of IPTG, and then, approximately 20 min later, inducing oversynthesis of L4 by addition of arabinose. The results (Fig. 4; Table 1) indicate that not only the M. morganii leader but even the H. influenzae and V. cholerae leaders, which have little primary sequence conservation relative to E. coli, contain the determinants sufficient for autogenous control by L4. Regulation of the M. morganii and H. influenzae leaders was not significantly different from the regulation of E. coli, but the V. cholerae leader, whose secondary structure is also less similar to E. coli’s, was not regulated as well. Not surprisingly, the P. aeruginosa leader, which has neither primary sequence nor secondary structure homology with the E. coli leader, showed no regulation by E. coli L4 (Fig. 4; Table 1).
FIG. 4.
Effect of L4 on in vivo synthesis of S10′/β-Gal′ from plasmids carrying foreign S10 leaders. Cells carrying a Para-L4 plasmid and a Ptrc-leader-S10′/lacZ′ fusion plasmid (Fig. 1D) with the indicated heterologous leader sequence were grown exponentially. Aliquots of the culture were labeled for 2 min with [35S]-methionine immediately before or 20 min after the addition of IPTG to induce expression of S10′/β-Gal′. Twenty-three minutes after IPTG addition, arabinose (ara) was added to the culture to induce L4 oversynthesis; after another 15 min, an aliquot was labeled with [35S]methionine. Total protein extracts were fractionated by PAGE and analyzed by autoradiography. The protein band corresponding to S10′/β-Gal′ is indicated by the horizontal arrows. Control experiments showed that the synthesis of this protein is dependent on the presence of a plasmid carrying the Ptrc-leader-S10′/lacZ′ construct, and its regulation is dependent on the leader and on the induction of an active L4 protein (references 8, 19, 20, and 48 and data not shown). A second band, smaller than the S10′/β-Gal′ band, is also induced by IPTG but is not affected by L4 induction. This protein (●) is the product of the lacZΔM15 gene carried by the F′ lac plasmid carried in LL308 (23). It is well resolved in the gels shown in panels A to C but coelectrophoreses with another band in panel D. Ecoli, E. coli; Mmorg, M. morganii; Hinf, H. influenzae; Vibrio, V. cholerae; Pseudo, P. aeruginosa.
TABLE 1.
L4-mediated regulation of heterologous S10 leader sequences
| Leader | S10′/β-Gal′ synthesis, +L4/−L4a |
|---|---|
| E. coli | 0.078 (0.046) |
| M. morganii | 0.036 (0.016) |
| H. influenzae | 0.084 (0.030) |
| V. cholerae | 0.32 (0.11) |
| P. aeruginosa | 0.95 (0.16) |
Calculated as the radioactivity in the S10′/β-Gal′ fusion protein after arabinose induction of L4 oversynthesis divided by the radioactivity in S10′/β-Gal′ before arabinose induction (see Materials and Methods for details). Values in parentheses are standard deviations. Each leader was analyzed in at least three different labeling experiments.
Since L4 regulates both transcription and translation of the E. coli S10 operon (7, 47), inhibition of S10′/β-Gal′ synthesis in the E. coli version of the construct analyzed in Fig. 4 is the product of both levels of control. We have previously described a fusion plasmid containing the E. coli S10 leader upstream of an intact lacZ gene with its own Shine-Dalgarno sequence: synthesis of the β-Gal protein from this plasmid is subject to only transcriptional control (7). Therefore, to analyze the level of L4 control mediated by the heterologous leaders, we constructed a similar plasmid containing the Ptrc-regulated S10 leader upstream of the SDlac-lacZ sequence (Fig. 1D). The results, shown in Fig. 5, indicate that both M. morganii and H. influenzae leaders are sensitive to transcription control by E. coli L4. Note that we have not directly assayed the response of these foreign leaders to L4-mediated translation control, because we have no facile way to monitor translation regulation in the absence of transcription regulation.
FIG. 5.
Effect of L4 on in vivo synthesis of β-Gal from plasmids carrying foreign S10 leaders. Cells carrying a Para-L4 plasmid and a Ptrc-leader-lacZ fusion plasmid (Fig. 1D) with the indicated heterologous leader sequence were labeled as described in the legend to Fig. 4. The protein band corresponding to β-Gal is indicated by the horizontal arrows. The smaller band that is induced by IPTG (●) is the product of the F′ lacZΔM15 gene. Mmorg, M. morganii; Hinf, H. influenzae; ara, arabinose.
In vitro transcription regulation with heterologous S10 leaders.
We also tested the ability of the heterologous leaders to support L4-mediated transcription termination in a cell-free transcription reaction with purified E. coli RNA polymerase and NusA protein. We have previously shown that RNA polymerase pauses transiently in vitro at the site in the E. coli leader of in vivo transcription termination (51). Purified L4 stabilizes the paused transcription complex, facilitating termination, but only if NusA is also included in the transcription reaction (36, 49–51). We constructed DNA templates consisting of the E. coli S10 promoter followed by either the E. coli or a heterologous leader, upstream of a strong transcription terminator from the E. coli rrnC operon (Fig. 6A).
FIG. 6.
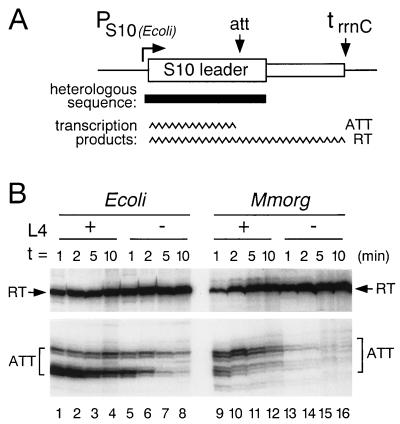
In vitro transcription of E. coli and M. morganii leaders. (A) The general structure of the DNA template, pLL226 or its heterologous derivatives, is shown. The position of the substituted DNA from M. morganii is indicated by the solid bar. PS10, E. coli promoter for the S10 operon; att, site of L4-stimulated transcription termination; trrnC, terminator from rRNA rrnC operon. (B) Transcription reactions were performed in the presence of NusA, and, where indicated, L4. Aliquots were removed at the indicated times after the start of transcription, fractionated on an 8% urea-polyacrylamide gel, and analyzed by autoradiography. Relevant portions of the gel are shown. The RT band contains readthrough transcripts terminated at trrnC; the ATT bands contain attenuated transcripts reflecting RNA polymerases paused or terminated at the attenuation site. Ecoli, E. coli; Mmorg, M. morganii.
E. coli RNA polymerase in the presence of E. coli NusA responded to the foreign templates in a similar way to its response to the native leader. Results with the M. morganii template are shown in Fig. 6B; we observed similar results with C. freundii, S. typhimurium, Y. enterocolitica, and H. influenzae templates (not shown). In the absence of L4 (Fig. 6B, lanes 13-16), the enzyme paused only transiently at the attenuation site (corresponding to the cluster of U’s at the base of the HE hairpin, at nucleotides 140 to 150 in Fig. 3F). In the presence of E. coli L4 (Fig. 6B, lanes 9 to 12), the stability of the paused transcription complex was significantly enhanced. The addition of NusA was required for the L4 effect (not shown).
While pausing occurred at a site in the M. morganii leader that corresponds to the E. coli attenuation site, the pattern of paused transcripts was slightly different, probably a result of the different distribution of U’s around the pause site. Another difference was that, with or without L4, the pause was not as stable with the M. morganii leader as with the E. coli leader (Fig. 6B; compare lanes 1 to 4 with lanes 9 to 12 or lanes 5 to 8 with lanes 13 to 16). Nevertheless, our in vitro studies confirm our conclusion from in vivo experiments that the heterologous S10 leaders contain the determinants sufficient for NusA-dependent, L4-stimulated transcription termination.
L4-mediated autogenous control in S. typhimurium.
Although we could not readily analyze the regulation of the S10 operons within most of the proteobacteria whose leaders we had analyzed, we were able to characterize the response in S. typhimurium to the oversynthesis of L4. Our source of L4 was a previously constructed plasmid, pLL235 (20), which contains the L4 gene from E. coli under control of the lac promoter. Since the Salmonella L4 gene encodes a protein with the same amino acid sequence as the E. coli protein (32), it was not necessary to construct a plasmid carrying the S. typhimurium L4 gene. Similarly, we used target plasmids containing the E. coli leader, since the S. typhimurium leader is nearly identical.
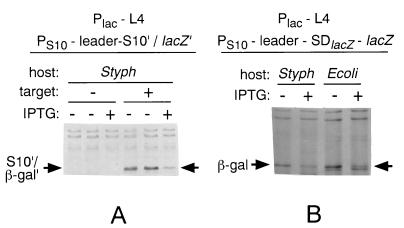
Using the target plasmid pLF1 (PS10-E. coli S10 leader-S10′/lacZ′) (45), we observed that the synthesis of S10′/β-Gal′ was inhibited by oversynthesis of L4 (Fig. 7A) to about the same extent as we observe in E. coli. Plasmid pLF1 is subject to both transcription and translation control by L4. To look specifically at transcription control, we repeated the experiment with pSma2R (PS10-E. coli S10 leader-lacZ) (48). Again, the extent of inhibition by L4 was equal to the inhibition that we observe in E. coli (Fig. 7B). We conclude that S. typhimurium is capable of L4-mediated attenuation control.
FIG. 7.
Autogenous regulation by L4 in S. typhimurium. S. typhimurium (Styph) or E. coli (Ecoli) cells carrying the indicated plasmids were pulse-labeled with [35S]methionine before (−) or 10 min after (+) addition of IPTG to induce oversynthesis of E. coli L4. See Materials and Methods for details. Total protein extracts were fractionated by PAGE and analyzed by autoradiography. The protein band corresponding to S10′/β-Gal′ (A) or β-Gal (B) is indicated by the horizontal arrows. The band just below the β-Gal band in the E. coli lanes (B) is the product of the F′ lacZΔM15 gene. In panel A, “target” refers to the absence (−) or presence (+) of the PS10-E. coli S10 leader-S10′/lacZ′ target plasmid.
DISCUSSION
Because about 75% of a cell’s mass is made of protein, protein synthesis is a major drain on the cell’s resources during growth. This cost includes the biogenesis of ribosomes and auxiliary proteins such as translation initiation, elongation, and termination factors. Not surprisingly, all organisms have evolved mechanisms to regulate the formation of the protein synthesis apparatus. We are interested in understanding the evolution of the regulatory mechanisms for control of ribosome formation. Since the major r-protein gene cluster corresponding to the S10, spc, and alpha operons of E. coli is preserved in many bacteria, archaea, and plastids (16, 42, 43), one might have expected that gene order and regulation have coevolved. Indeed, autogenous regulation, a major principle for control of r-protein production in E. coli, has been observed in diverse microorganisms, including the gram-positive bacterium B. subtilis (11) and the archaeum Methanococcus vannielii (13). Nevertheless, the molecular mechanisms of this regulation are different from the mechanisms described for E. coli.
Since our earlier studies had suggested that the E. coli-like L4-mediated autogenous control mechanism is not the mechanism for regulating the S10 cluster in B. subtilis (19), we investigated the regulation of the S10 operon in bacteria more closely related to E. coli, focusing on the gamma branch of the proteobacteria. All of the enterobacteria we analyzed, including S. typhimurium, S. marcescens, C. freundii, M. morganii, and Y. enterocolitica, contain S10 leaders that apparently form secondary structures that are strikingly similar to the structure that we have described for E. coli (37). In particular, the 3′ region of the S10 leaders contains highly conserved structures corresponding to hairpins HD and HE that we have shown are involved in L4-mediated transcription and translation control (7, 35, 37, 48). Not surprisingly, these leaders all respond to E. coli L4.
Two more distantly related members of the gamma subdivision of the purple bacteria, H. influenzae and V. cholerae, also contain leaders that can form E. coli-like HD and HE hairpins. Furthermore, these leaders also respond in E. coli to r-protein L4. Taken together with the finding that heterologous L4 proteins from M. morganii, Y. pseudotuberculosis, H. influenzae, and B. stearothermophilus can substitute for E. coli L4 in the regulation of the E. coli S10 operon (53), our results suggest that the L4-mediated regulatory mechanism that we have described for E. coli might also govern the regulation of the S10 operons of other enterobacteria and of other closely related members of the gamma subdivision of the proteobacteria such as H. influenzae and V. cholerae.
Interestingly, it appears that not all members of the gamma subdivision utilize an E. coli-type mechanism for regulating the S10 gene cluster. P. aeruginosa has a much shorter untranslated sequence upstream of its S10 gene, and neither the primary sequence nor the computer-predicted secondary structure of the leader suggests any homology with the E. coli leader. Indeed, the P. aeruginosa leader is not sensitive to autogenous control by E. coli L4. Since its leader is also not regulated by the homologous P. aeruginosa L4 (data not shown), it is likely that P. aeruginosa utilizes a very different mechanism for regulating expression of its S10 operon.
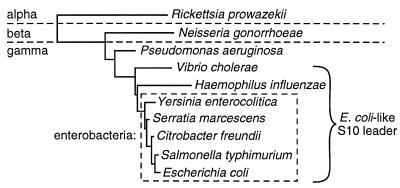
We have also surveyed the available sequences of S10 regions of other members of the proteobacteria, using the NCBI BLAST algorithm (26). So far, we have found no other examples of leader sequences that form an E. coli-like structure. Indeed, for most of these proteobacteria (including Rickettsia prowazekii, Neisseria gonorrhoeae, and Thiobacillus cuprinus), the S10 gene is located less than 100 nucleotides downstream of tufA and often appears to be cotranscribed with the tufA gene. A phylogenetic tree based on the rRNA sequences of relevant proteobacteria and summarizing our S10 leader studies is shown in Fig. 8. Although we are still exploring the S10 operon structures of other members of the proteobacterial branch, our current data suggest that L4-mediated autogenous control was established during the evolution of the gamma subdivision of proteobacteria. Our studies indicate that this control involves L4-stimulated transcription termination. Whether translation control by L4 is also conserved in these other bacteria is under investigation.
FIG. 8.
Phylogenetic tree of relevant proteobacteria based on 16S rRNA sequences.
As already mentioned, previous studies of B. subtilis (11, 15, 19, 38) had suggested that the highly conserved gene order of the major r-protein gene cluster in gram-negative and gram-positive bacteria does not ensure that they utilize the same mechanisms for autogenously regulating the expression of those genes. The present study reveals that this dichotomy between gene order and regulatory mechanism exists even within a more restricted group of the bacterial kingdom.
The evolutionary preservation of r-protein gene order in eubacteria and the archaea is especially remarkable, since in general no other major gene clusters are preserved among the bacteria whose genomes have been sequenced (see, for example, references 25 and 42). The similarities in r-protein gene organization imply that the last common eubacterial/archaebacterial ancestor had already established this gene order (4, 42) and that selective pressure maintained this order despite continual rearrangements of other genes during evolution (42). We do not yet understand the forces behind either the original clustering of the r-protein genes or the subsequent preservation of the gene order. The selfish-operon model (18) accounts for clustering of nonessential genes but cannot explain the formation of clusters of essential genes like those encoding r-proteins. Indeed, the primordial history of these r-protein clusters precludes reliable speculation about their formation (18).
In any case, once formed, the r-protein clusters clearly offered a strong evolutionary advantage. Given that these genes encode proteins that must interact physically, there may have been selective pressure to ensure that modules of r-protein genes were exchanged during a recombination event; a similar argument has been used to explain the functional clusters of bacteriophage genes (6). Moreover, once the r-protein genes had been gathered during evolution, the extremely short intragenic regions in the cluster would make it very difficult to survive transpositions disrupting the cluster, since such events would have a high probability of inactivating these essential genes (although such events clearly occurred in some species, since there are examples of extra or missing genes within the S10-spc-alpha cluster [16, 42]). Finally, the clustering of r-protein genes into operons provides a mechanism for cotranscriptional and coupled translational control. The latter advantage, however, presumably evolved after the primordial organization of the genes, resulting in a plethora of regulatory schemes that remain virtually unexplored.
ACKNOWLEDGMENTS
We thank Mike Noorani for technical assistance, Xiao Li and Yizhong Sha for technical advice, and Knud Nierhaus for purified L4 and S7 proteins.
This work was supported by grants AI-15286 and GM-54876 from the National Institutes of Health.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck C, Ingraham J, Maaløe O, Neuhard J. Relationship between the concentration of nucleoside triphosphates and the rate of synthesis of RNA. J Mol Biol. 1973;78:117–121. doi: 10.1016/0022-2836(73)90431-2. [DOI] [PubMed] [Google Scholar]
- 3.Clark D J, Maaløe O. DNA replication and the division cycle in Escherichia coli. J Mol Biol. 1967;23:99–112. [Google Scholar]
- 4.Doolittle W F, Brown J R. Tempo, mode, the progenote, and the universal root. Proc Natl Acad Sci USA. 1994;91:6721–6728. doi: 10.1073/pnas.91.15.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 6.Ford M E, Sarkis G J, Belanger A E, Hendrix R W, Hatfull G F. Genome structure of mycobacteriophage D29: implications for phage evolution. J Mol Biol. 1998;279:143–164. doi: 10.1006/jmbi.1997.1610. [DOI] [PubMed] [Google Scholar]
- 7.Freedman L P, Zengel J M, Archer R H, Lindahl L. Autogenous control of the S10 ribosomal protein operon of Escherichia coli: genetic dissection of transcriptional and post-transcriptional regulation. Proc Natl Acad Sci USA. 1987;84:6516–6520. doi: 10.1073/pnas.84.18.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freedman L P, Zengel J M, Lindahl L. Genetic dissection of stringent control and nutritional shift-up response of the Escherichia coli S10 ribosomal protein operon. J Mol Biol. 1985;185:701–712. doi: 10.1016/0022-2836(85)90055-5. [DOI] [PubMed] [Google Scholar]
- 9.Gaal T, Bartlett M S, Ross W, Turnbaugh C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 10.Grundy F J, Henkin T M. Cloning and analysis of the Bacillus subtilis rpsD gene, encoding ribosomal protein S4. J Bacteriol. 1990;172:6372–6379. doi: 10.1128/jb.172.11.6372-6379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundy F J, Henkin T M. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J Bacteriol. 1991;173:4595–4602. doi: 10.1128/jb.173.15.4595-4602.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanner M, Mayer C, Köhrer C, Golderer G, Grobner P, Piendl W. Autogenous translational regulation of the ribosomal MvaL1 operon in the archaebacterium Methanococcus vannielii. J Bacteriol. 1994;176:409–418. doi: 10.1128/jb.176.2.409-418.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henkin T M, Chambliss G H. Genetic mapping of a mutation causing an alteration in Bacillus subtilis ribosomal protein S4. Mol Gen Genet. 1984;193:364–369. doi: 10.1007/BF00330694. [DOI] [PubMed] [Google Scholar]
- 15.Henkin T M, Moon S H, Mattheakis L C, Nomura M. Cloning and analysis of the spc ribosomal protein operon of Bacillus subtilis: comparison with the spc operon of Escherichia coli. Nucleic Acids Res. 1989;17:7469–7486. doi: 10.1093/nar/17.18.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koonin E V, Galperin M Y. Prokaryotic genomes: the emerging paradigm of genome-based microbiology. Curr Opin Genet Dev. 1997;7:757–763. doi: 10.1016/s0959-437x(97)80037-8. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence J G, Roth J R. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Lindahl L, Sha Y, Zengel J M. Analysis of the Bacillus subtilis S10 ribosomal protein gene cluster identifies two promoters that may be responsible for transcription of the entire 15-kilobase S10-spc-alpha cluster. J Bacteriol. 1997;179:7046–7054. doi: 10.1128/jb.179.22.7046-7054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Lindahl L, Zengel J M. Ribosomal protein L4 from Escherichia coli utilizes different determinants for its structural and regulatory functions. RNA. 1996;2:24–37. [PMC free article] [PubMed] [Google Scholar]
- 21.Lindahl L, Sor F, Archer R H, Nomura M, Zengel J M. Transcriptional organization of the S10, spc and α operons of Escherichia coli. Biochim Biophys Acta Gene Struct Expr. 1990;1050:337–342. doi: 10.1016/0167-4781(90)90191-4. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl L, Zengel J M. Autogenous control is not sufficient to ensure steady-state growth rate-dependent regulation of the S10 ribosomal protein operon in Escherichia coli. J Bacteriol. 1990;172:305–309. doi: 10.1128/jb.172.1.305-309.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindahl L, Zengel J M. Operon-specific regulation of ribosomal protein synthesis in Escherichia coli. Proc Natl Acad Sci USA. 1979;76:6542–6546. doi: 10.1073/pnas.76.12.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mushegian A R, Koonin E V. Gene order is not conserved in bacterial evolution. Trends Genet. 1996;12:289–290. doi: 10.1016/0168-9525(96)20006-x. [DOI] [PubMed] [Google Scholar]
- 26.NCBI Blast server. revision date. 15 March 1999. http://www.ncbi.nlm.nih.gov/BLAST/ http://www.ncbi.nlm.nih.gov/BLAST/. [Online.] [27 April, last date accessed.] . [Online.] [27 April, last date accessed.] [Google Scholar]
- 27.Olins P O, Nomura M. Regulation of the S10 ribosomal protein operon in E. coli: nucleotide sequence at the start of the operon. Cell. 1981;26:205–211. doi: 10.1016/0092-8674(81)90303-2. [DOI] [PubMed] [Google Scholar]
- 28.Post L E, Arfsten A E, Davis G R, Nomura M. DNA sequence of the promoter region for the α ribosomal protein operon in Escherichia coli. J Biol Chem. 1980;255:4653–4659. [PubMed] [Google Scholar]
- 29.Post L E, Arfsten A E, Reusser F, Nomura M. DNA sequences of promoter regions for the str and spc ribosomal protein operons in Escherichia coli. Cell. 1978;15:215–229. doi: 10.1016/0092-8674(78)90096-x. [DOI] [PubMed] [Google Scholar]
- 30.Pseudomonas Genome Project. 12/15/1998, release date. Contig 65. http://www.pseudomonas.com/. [Online.] [3/2/1999, last date accessed.]
- 31.Ribosomal Database Project.http://www.cme.msu.edu/RDP. [Online.] [13 August 1998, last date accessed.]
- 32.Salmonella typhi Genome Project. 2/22/1999, posting date. Contig 510. The Sanger Center. http://www.sanger.ac.uk/Projects/S_typhi/. [Online] [3/2/1999, last date accessed.]
- 33.Sanger F, Nicklen S, Coulsen A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sha Y. Ph.D. thesis. Rochester, N.Y: University of Rochester; 1997. [Google Scholar]
- 35.Sha Y, Lindahl L, Zengel J M. RNA determinants required for L4-mediated attenuation control of the S10 r-protein operon of Escherichia coli. J Mol Biol. 1995;245:486–498. doi: 10.1006/jmbi.1994.0040. [DOI] [PubMed] [Google Scholar]
- 36.Sha Y, Lindahl L, Zengel J M. Role of NusA in L4-mediated attenuation control of the S10 r-protein operon of Escherichia coli. J Mol Biol. 1995;245:474–485. doi: 10.1006/jmbi.1994.0039. [DOI] [PubMed] [Google Scholar]
- 37.Shen P, Zengel J M, Lindahl L. Secondary structure of the leader transcript from the Escherichia coli S10 ribosomal protein operon. Nucleic Acids Res. 1988;16:8905–8924. doi: 10.1093/nar/16.18.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh J W, Boylan S A, Oh S H, Price C W. Genetic and transcriptional organization of the Bacillus subtilis spc-α region. Gene. 1996;169:17–23. doi: 10.1016/0378-1119(95)00757-1. [DOI] [PubMed] [Google Scholar]
- 39.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vibrio cholerae Genome Project. 2/11/1999, posting date. Contig asm970. The Institute for Genome Research. http://www.tigr.org:Vibrio cholerae. [Online.] [3/2/1999, last date accessed.]
- 41.Walter A E, Turner D H, Kim J, Lyttle M H, Müller P, Mathews D H, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe H, Mori H, Itoh T, Gojobori T. Genome plasticity as a paradigm of eubacteria evolution. J Mol Evol. 1997;44(Suppl. 1):S57–S64. doi: 10.1007/pl00000052. [DOI] [PubMed] [Google Scholar]
- 43.Wittmann-Liebold B, Köpke A K E, Arndt E, Krömer W, Hatakeyama T, Wittmann H-G. Sequence comparison and evolution of ribosomal proteins and their genes. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function and evolution. Washington, D.C.: American Society for Microbiology; 1990. pp. 598–616. [Google Scholar]
- 44.Yersinia pestis Genome Project. 2/15/1999, posting date. Contig 795. 1999. The Sanger Centre http://www.sanger.ac.uk/Projects/Y_pestis/. [Online.] [3/2/1999, last date accessed.]
- 45.Zengel J M, Archer R H, Freedman L P, Lindahl L. Role of attenuation in growth rate-dependent regulation of the S10 r-protein operon of E. coli. EMBO J. 1984;3:1561–1565. doi: 10.1002/j.1460-2075.1984.tb02011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zengel J M, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acid Res Mol Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 47.Zengel J M, Lindahl L. Escherichia coli ribosomal protein L4 stimulates transcription termination at a specific site in the leader of the S10 operon independent of L4-mediated inhibition of translation. J Mol Biol. 1990;213:67–78. doi: 10.1016/S0022-2836(05)80122-6. [DOI] [PubMed] [Google Scholar]
- 48.Zengel J M, Lindahl L. A hairpin structure upstream of the terminator hairpin required for ribosomal protein L4-mediated attenuation control of the S10 operon of Escherichia coli. J Bacteriol. 1996;178:2383–2387. doi: 10.1128/jb.178.8.2383-2387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zengel J M, Lindahl L. Ribosomal protein L4 and transcription factor NusA have separable roles in mediating termination of transcription within the leader of the S10 operon of E. coli. Genes Dev. 1992;6:2655–2662. doi: 10.1101/gad.6.12b.2655. [DOI] [PubMed] [Google Scholar]
- 50.Zengel J M, Lindahl L. Ribosomal protein L4 of Escherichia coli: in vitro analysis of L4-mediated attenuation control. Biochimie. 1991;73:719–727. doi: 10.1016/0300-9084(91)90052-3. [DOI] [PubMed] [Google Scholar]
- 51.Zengel J M, Lindahl L. Ribosomal protein L4 stimulates in vitro termination of transcription at a NusA-dependent terminator in the S10 operon leader. Proc Natl Acad Sci USA. 1990;87:2675–2679. doi: 10.1073/pnas.87.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zengel J M, Mueckl D, Lindahl L. Protein L4 of the E. coli ribosome regulates an eleven gene r-protein operon. Cell. 1980;21:523–535. doi: 10.1016/0092-8674(80)90490-0. [DOI] [PubMed] [Google Scholar]
- 53.Zengel J M, Vorozheikina D, Li X, Lindahl L. Regulation of the Escherichia coli S10 ribosomal protein operon by heterologous L4 ribosomal proteins. Biochem Cell Biol. 1995;73:1105–1112. doi: 10.1139/o95-119. [DOI] [PubMed] [Google Scholar]
- 54.Zuker M. In: Mathematical methods for DNA sequences. Waterman M S, editor. Boca Raton, Fla: CRC Press; 1987. pp. 159–184. [Google Scholar]
- 55.Zuker, M. mfold mserver. http://www.ibc.wustl.edu/∼zuker/rna/form1.cgi. [Online.] [26 April 1999, last date accessed.]