Abstract
Abrupt aggregation of misfolded proteins is the underlying molecular cause of numerous severe pathologies including Alzheimer's and Parkinson's diseases. Protein aggregation yields small oligomers that can later propagate into amyloid fibrils, β‐sheet‐rich structures with a variety of topologies. A growing body of evidence suggests that lipids play an important role in abrupt aggregation of misfolded proteins. In this study, we investigate the roles of length and saturation of fatty acids (FAs) in phosphatidylserine (PS), an anionic lipid that is responsible for the recognition of apoptotic cells by macrophages, in lysozyme aggregation. We found that both the length and saturation of FAs in PS contribute to the aggregation rate of insulin. PS with 14‐carbon‐long FAs (14:0) enabled a much stronger acceleration of protein aggregation compared to PS with 18‐carbon‐long FAs (18:0). Our results demonstrate that the presence of double bonds in FAs accelerated the rate of insulin aggregation relative to PS with fully saturated FAs. Biophysical methods revealed morphological and structural differences in lysozyme aggregates grown in the presence of PS with varying lengths and FA saturation. We also found that such aggregates exerted diverse cell toxicities. These results demonstrate that the length and saturation of FAs in PS can uniquely alter the stability of misfolded proteins on lipid membranes.
Keywords: amyloid fibrils, lysozyme, oligomers, phosphatidylserine, toxicity
1. INTRODUCTION
Pathological aggregation of misfolded proteins is a major contributor to the onset and progression of amyloid diseases, including diabetes type 2, Alzheimer's (AD), and Parkinson's disease (PD) (Chiti & Dobson, 2017; Eisenberg et al., 2006; Nelson et al., 2005). This process results in the formation of cytotoxic oligomers, protein aggregates that exhibit a large variety of forms and shapes (Almeida et al., 2006; Kayed & Lasagna‐Reeves, 2013; Konig et al., 2021; Lasagna‐Reeves et al., 2011). Some of these oligomers can propagate into filaments that braid and intertwine with other filaments forming amyloid fibrils (Sengupta et al., 2016; Vosough & Barth, 2021). Utilization of electron microscopy revealed the presence of both tape‐like and twisted fibril polymorphs in amyloid plaques, pathological formations observed in the brain cortex of patients diagnosed with AD (Kidd, 1963; Kollmer et al., 2019; Paravastu et al., 2009).
Both amyloid plaques and Lewy bodies contain fragments of lipid membranes (Han et al., 2017; Killinger et al., 2019; Shahmoradian et al., 2019). It was also found that lipids could alter rates of protein aggregation (Alza et al., 2019; Galvagnion, 2017; Galvagnion et al., 2016; Matveyenka et al., 2023; Matveyenka, Rizevsky, & Kurouski, 2022b). Specifically, Matveyenka and co‐workers reported that zwitterionic lipids strongly inhibit aggregation of insulin, a small peptide hormone that can aggregate upon injection causing amyloidosis and type 2 diabetes (Matveyenka et al., 2023). Anionic lipids, such as phosphatidylserine (PS), strongly accelerated insulin aggregation (Matveyenka et al., 2023; Matveyenka, Rizevsky, & Kurouski, 2022b). Similar findings were recently reported by Matveyenka et al. for lysozyme (Matveyenka, Zhaliazka, et al., 2022). However, unlike in the case of insulin, PS did not alter the rate of lysozyme aggregation present at a 1:1 molar ratio. However, an increase in the concentration of PS relative to the concentration of lysozyme to 1:5 and 1:10 resulted in a drastic increase in the rate of protein aggregation. (Zhaliazka, Serada, et al., 2023b) Furthermore, it was found that lipids not only altered the rates of protein aggregation, but uniquely modified the morphology and secondary structure of protein aggregates formed in their presence (Matveyenka et al., 2023; Matveyenka, Rizevsky, & Kurouski, 2022a; Matveyenka, Rizevsky, & Kurouski, 2022b; Matveyenka, Rizevsky, & Kurouski, 2022c; Matveyenka, Rizevsky, & Kurouski, 2022d; Matveyenka, Zhaliazka, et al., 2022). Experimental results reported by Dou et al. suggested that lipids could temper protein aggregation (Dou et al., 2021; Dou & Kurouski, 2022). It was proposed that such protein‐lipid complexes are held together by electrostatic interactions between polar head groups of lipids and charged amino acids (Alza et al., 2019; Galvagnion, 2017; Kiechle et al., 2020). NMR results revealed strong hydrophobic interactions between the aliphatic tails and hydrophobic amino acids of α‐Syn. Similar interactions are thought to stabilize such protein‐lipid aggregates (Alza et al., 2019; Galvagnion, 2017; Giasson et al., 2001; Kiechle et al., 2020; Ueda et al., 1993). Using nano‐Infrared spectroscopy, our group demonstrated that such aggregates possessed lipids in their structure (Zhaliazka et al., 2022; Zhaliazka & Kurouski, 2023).
PS is an anionic lipid that is localized in the inner part of the plasma membrane via ATP‐dependent flippase transport (Alecu & Bennett, 2019; Fitzner et al., 2020). An increase in the concentration of PS on the outer surface of the plasma membrane is recognized by macrophages which can trigger apoptosis (Levental et al., 2020; Matveyenka, Rizevsky, & Kurouski, 2022b). We hypothesized that the presence of PS in the outer membrane can drastically alter the stability of misfolded proteins which can stimulate the aggregation of α‐Syn, insulin, and other membrane‐linked proteins (Dou et al., 2021; Dou & Kurouski, 2022; Matveyenka, Rizevsky, & Kurouski, 2022b; Zhaliazka et al., 2022).
A growing body of evidence suggests that not only the net charge, but also the length and saturation of fatty acids (FAs) in phospholipids can play an important role in protein aggregation (Matveyenka, Rizevsky, & Kurouski, 2022b; Matveyenka, Rizevsky, & Kurouski, 2022c; Matveyenka, Rizevsky, & Kurouski, 2022d). Specifically, Matveyenka and co‐workers found that PS with 14‐carbon‐long FAs, known as 1,2‐dimyristoyl‐sn‐glycero‐3‐phospho‐Lserine (DMPS), accelerated the rate of insulin aggregation compared to PS with 18‐carbon‐long FAs that possess double bonds (1,2‐dioleoyl‐sn‐glycero‐3‐phospho‐L‐serine, DOPS) (Matveyenka, Rizevsky, & Kurouski, 2022b). It remains unclear whether this observed difference stems from the length of the FAs or their degree of unsaturation. Expanding upon this, we investigate rates of lysozyme aggregation in the equimolar concentrations of DMPS, DOPS, 1‐palmitoyl‐2‐oleoyl‐sn‐glycero‐3‐phospho‐L‐serine (POPS), and 1,2‐distearyl‐sn‐glycero‐3‐phospho‐L‐serine (DSPS), Scheme 1.
SCHEME 1.
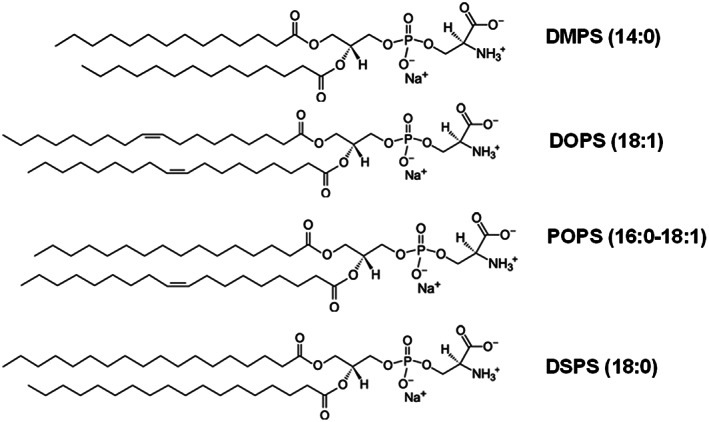
Molecular structures of DMPS, DOPS, POPS, and DSPS.
We also utilize a set of biophysical methods to determine the morphology and secondary structure of lysozyme oligomers and fibrils formed in the lipid‐free environment and in the presence of DMPS, DSPS, DOPS, and POPS. Finally, a lactate dehydrogenase (LDH) based cell toxicity assay was employed to elucidate the extent to which phospholipids altered the toxicity of lysozyme oligomers.
1.1. The effect of length and saturation of FAs in PS on the rate of lysozyme aggregation
We first utilized the thioflavin T (ThT) assay to determine the extent to which structural differences in PS could alter the rate of lysozyme aggregation. For this, 200 μM of lysozyme was incubated at 37°C under constant agitation in the presence of ThT. Our results showed that lysozyme aggregation was observed in the equimolar presence of DMPS already at 12 h ± 0.2 h after the initiation of protein aggregation, Figure 1. This conclusion could be made by the observed increase in the ThT signal. Similar effects on lysozyme aggregation were observed for DOPS and POPS. However, these lipids catalyzed protein aggregation only at 45 ± 0.8 h and 47 ± 1.2 h after the initiation of protein aggregation, respectively. Finally, we observed an increase in the ThT signal for Lys:DSPS only at 110 h ± 0.5 h. It should be noted that in the absence of lipid vesicles, lysozyme aggregation was not observed even by 160 h after the initiation of protein aggregation. These results demonstrated that all tested PS could trigger protein aggregation at pH 3.0, 37°C. Our findings also showed that PS with 14:0 FAs (DMPS) exhibited much greater catalytic properties compared to PS with 18:0 FAs (DSPS). Thus, the length of the FAs plays an important role in lipid‐facilitated lysozyme aggregation. Finally, our results showed that the unsaturation of FAs in PS facilitated lysozyme aggregation compared to the fully saturated FAs with the same number of carbon atoms.
FIGURE 1.
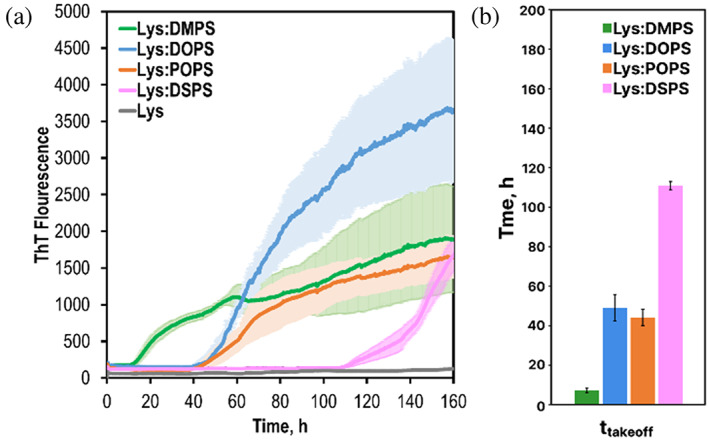
Length and saturation of FAs in PS uniquely alter the rate of lysozyme aggregation. (a) ThT aggregation kinetics of insulin in the lipid‐free environment (gray), and in the presence of DMPS (green), DOPS (blue), POPS (orange), and DSPS (pink). Each kinetic curve is the average of three independent measurements. (b) A histogram of ttakeoff of lysozyme aggregation in the presence of DMPS (green), DOPS (blue), POPS (orange), and DPSP (pink). For Lys, 200 μM of chicken‐egg lysozyme was dissolved in 1xPBS with 2 mM of ThT; pH adjusted to pH 3.0. For Lys:DMPS, Lys:DOPS, Lys:POPS, and Lys:DSPS, 200 μM of chicken‐egg lysozyme was mixed with an equivalent concentration of the corresponding lipid; pH was adjusted to pH 3.0. All samples were kept at 37°C under 510 rpm for 160 h.
1.2. Morphological analysis of lysozyme aggregates formed in the presence of DMPS, DOPS, POPS, and DSPS
We investigated the morphology of lysozyme aggregates formed in the presence of structurally different PS, as well as in the lipid‐free environment. We found that in the absence of PS, after 160 h incubation at 37°C, lysozyme formed small oligomers that had 3–5 nm in height. AFM revealed that in addition to morphologically similar aggregates, Lys:DMPS possessed short fibrillar species, as well as aggregates that looked like a string of interconnected oligomers. These bead‐like aggregates had ~6–8 nm in height and stretched four (or 4) microns (μm) in length, Figure S1. We did not observe such bead‐like aggregates in Lys:DOPS and Lys:POPS. Instead, these samples possessed only small oligomers that had 5–8 nm in height. We also found that most of Lys:DOPS oligomers were surrounded by lipid shells, suggesting that protein aggregation was initiated in lipid microbubbles. AFM imaging of Lys:DSPS revealed the presence of small protein oligomers that had 5–8 nm in height. These results demonstrate that under physiological conditions, in the presence of structurally different PS, lysozyme forms only oligomers that exhibit similar topologies to the aggregates formed by lysozyme in the lipid‐free environment. Although some differences in the morphologies of protein oligomers were observed for Lys:DMPS, very few differences in topology were observed between the other samples.
In agreement with previous findings, our results demonstrated that lysozyme aggregation at 65°C resulted in the formation of fibrils, Figure S2 (Matveyenka, Zhaliazka, et al., 2022; Shashilov et al., 2007; Zhaliazka et al., 2022). Following these findings, we aggregated lysozyme at 65°C in the presence of DMPS, DOPS, POPS, and DSPS, Figure 2. Under these experimental conditions, lysozyme formed long fibril species that had 15–20 nm in height.
FIGURE 2.
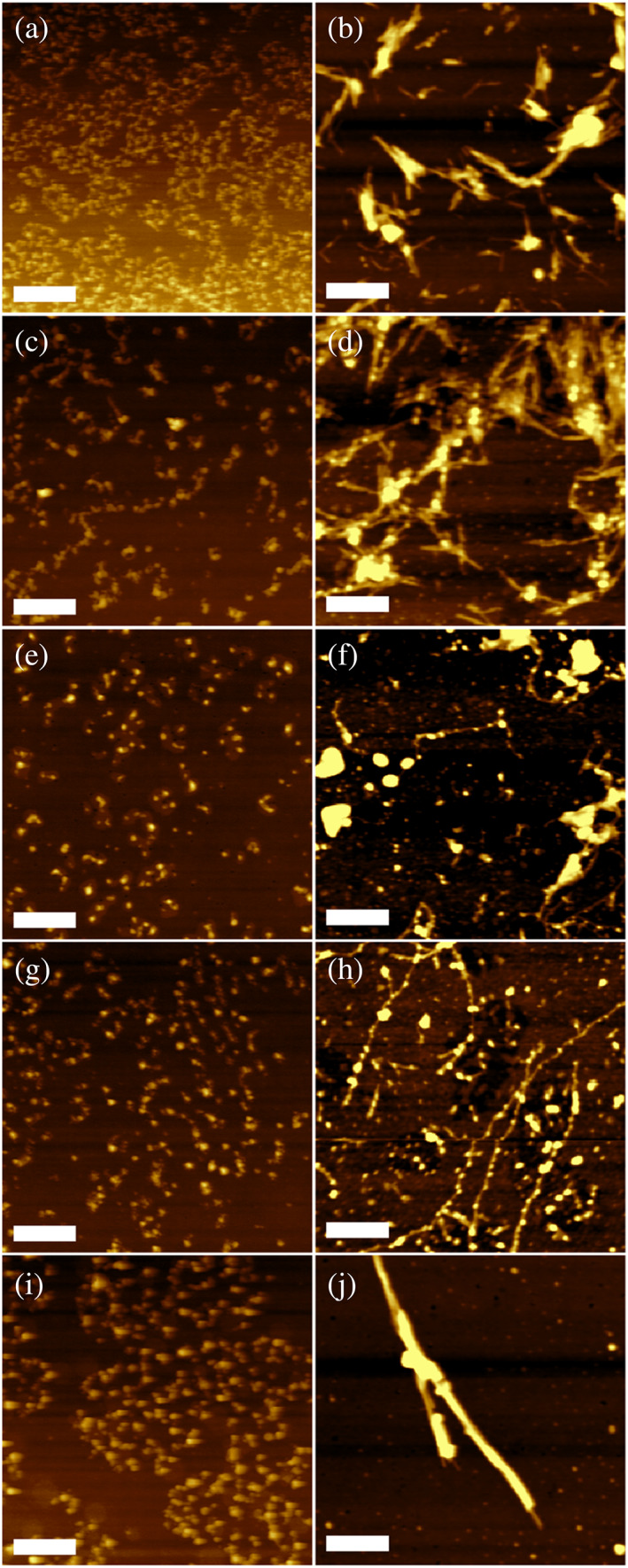
Morphological characterization of protein aggregates formed in the presence of DMPS, POPS, DOPS, and DSPS, as well as in the lipid‐free environment. AFM images of lysozyme oligomers (a, c, e, g, and i) and fibrils (b, d, f, h, and j) formed in the absence of lipids (a and b), as well as in the presence of DMPS (c and d), DOPS (e and f), POPS (g and h) and DSPS (i and j). Scale bars are 1 μm (a, c, e, g, i) and 375 nm (b, d, f, h, and j).
Morphologically similar aggregates were formed by lysozyme in the presence of DMPS. In addition to the straight and uniform fibrils, we observed formations similar to the strings of oligomers discussed above. These strings, however, had substantially longer length compared to those formed in the presence of DMPS at 37°C. Morphologically similar strings of oligomers were observed for Lys:DOPS and Lys:POPS. We also found that some of those aggregates were covered by lipid shells. Finally, in the presence of DSPS, lysozyme formed straight uniform fibrils that were much thicker compared to the aggregates observed in all other samples. These results showed that saturation and length of FAs in PS uniquely alter the morphology of lysozyme fibrils formed in their presence. Specifically, we observed bead‐like fibrils formed in the presence of Lys:DMPS, Lys:DOPS, and Lys:POPS. These fibrils were not observed for Lys:DSPS. We also found significant morphological differences between aggregates grown in the presence of DMPS and DSPS suggesting that the length of FAs in PS plays an important role in lysozyme aggregation. Similar conclusions could be made about the role of saturation of FAs in PS.
1.3. Elucidation of protein secondary structure of lysozyme aggregates grown in the presence of DMPS, POPS, DOPS, and DSPS
We utilized both circular dichroism (CD) and attenuated total internal reflection Fourier‐transformed Infrared (ATR‐FTIR) spectroscopy to examine the secondary structure of lysozyme oligomers and fibrils formed in both the presence of structurally different PS and lipid‐free environments.CD spectra acquired from lysozyme oligomers grown at 37°C exhibited a broad trough, which indicates a mixture of α‐helical and unordered protein secondary structure with insignificant amounts of β‐sheets, Figure 3, A and Figure S3. We also did not observe significant differences between the spectra acquired from Lys:DMPS, Lys:POPS, Lys:DOPS, and Lys:DSPS. At the same time, CD spectra acquired from lysozyme fibrils grown at 65°C point on the predominance of β‐sheets in the secondary structure of these aggregates. This conclusion can be made by the observed shift of the trough in the CD spectra to ~217 nm, which is characteristic of β‐sheet secondary structures. ATR‐FTIR spectra acquired from suspensions of lysozyme oligomers confirmed the discussed above CD results. We observed only one amide I band centered around 1650 cm−1, which indicates the predominance of α‐helical and unordered protein secondary structure in the analyzed samples. We also observed small variations in the position of the amide I band in such spectra. In the ATR‐FTIR spectra of Lys:DMPS and Lys:POPS, the amide I band was centered at 1652 cm−1, whereas in the spectra acquired form Lys:DOPS and Lys:DSPS this band was slightly red‐shifted to ~1560 cm−1. It should be noted that in the ATR‐FTIR spectra of Lys, we observed an amide I band at ~1648 cm−1. We infer that these small spectroscopic changes originate from slight differences in the ratio of α‐helical and unordered protein in these samples.
FIGURE 3.
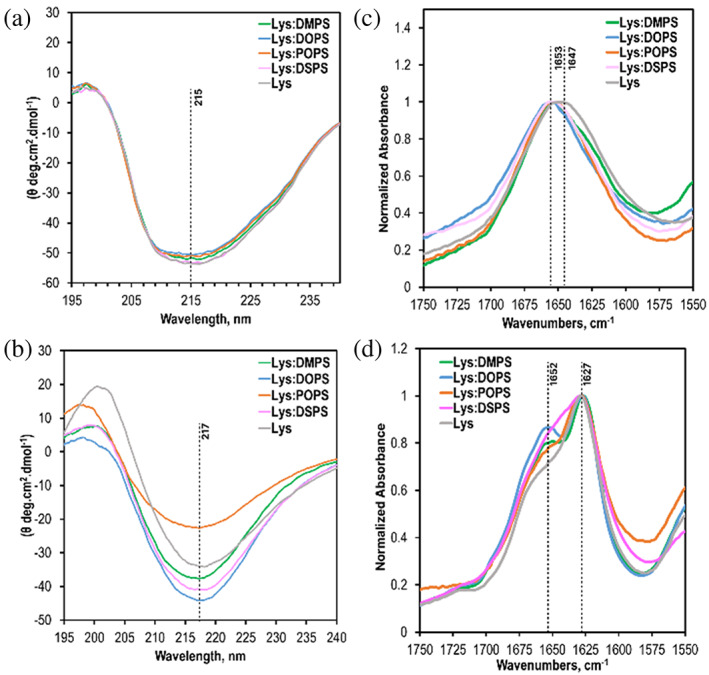
Structural characterization of protein aggregates formed in the presence of DMPS, POPS, DOPS, and DSPS. CD (a and b) and ATR‐FTIR (c and d) spectra acquitted form Lys, Lys:DMPS, Lys:POPS, Lys:DOPS, and Lys:DSPS oligomers (a and c) formed at 37°C after 160 h agitation at 510 rpm (top) and fibrils and fibrils (b and d) grown at 65°C after 24 h agitation at 510 rpm.
ATR‐FTIR spectra acquired from lysozyme fibrils grown at 65°C exhibit two amide I bands at 1625 cm−1 and 1660 cm−1, which could be assigned to parallel β‐sheets and unordered proteins, respectively. These results showed that the secondary structure of lysozyme fibrils was dominated by parallel β‐sheets with some amount of unordered proteins present. We also found that IR spectra acquired from different samples were very similar if not identical. These findings demonstrate that the structural organization of Lys:DMPS, Lys:POPS, Lys:DOPS, and Lys:DSPS, as well as lysozyme aggregates formed in the lipid‐free environment (Lys) were very similar.
1.4. Length and saturation of FAs of PS uniquely alter the toxicity of lysozyme aggregates grown in the presence of lipids
The question is whether lysozyme oligomers and fibrils formed in the presence of DMPS, POPS, DOPS, and DSPS exert different cell toxicity, or not. To answer this question, we utilized N27 rat dopaminergic cell line and lactate dehydrogenase (LDH) assay. We found that lysozyme oligomers grown in the presence of DMPS, POPS, DOPS, and DSPS exerted significantly lower cell toxicity compared to the lysozyme oligomers formed in the lipid‐free environment, Figure 4. Specifically, the lowest cell toxicity was observed for Lys:POPS and Lys:DSPS. Lys:DMPS and Lys:DOPS exerted greater cell toxicity than Lys:POPS and Lys:DSPS, but were significantly lower than the Lys oligomers. These results demonstrate that the length and saturation of FAs in PS uniquely alter the toxicity of lysozyme oligomers. Our results also showed that the length and saturation of FAs in PS uniquely altered the toxicity of lysozyme fibrils. Specifically, Lys:POPS exerted significantly lower cell toxicity, whereas Lys:DOPS and Lys:DPSP were significantly higher than Lys fibrils. We observed no statistically significant difference between the toxicity of Lys:DMPS and Lys aggregates. Based on these results, we can conclude that the length and saturation of FAs in PS uniquely alter the toxicity of both oligomers and fibrils. Finally, it is important to note that lipids themselves do not exert any significant cell toxicity, Figure S4.
FIGURE 4.
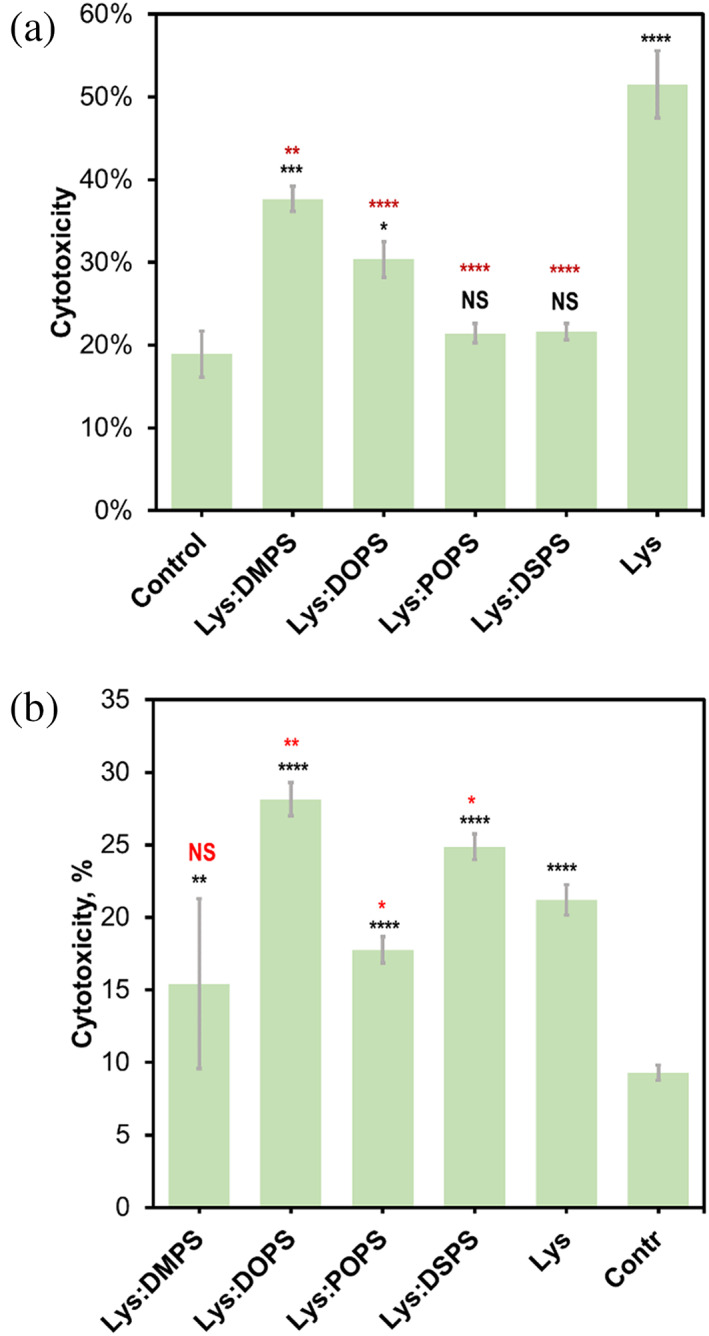
Toxicity of lysozyme aggregates grown in the presence of DMPS, POPS, DOPS, and DSPS, as well as in the lipid‐free environment. Histograms of LDH toxicity assays of Lys:DMPS, Lys:DOPS, Lys:POPS, Lys:DPSP, and Lys oligomers formed at 37°C after 160 h agitation at 510 rpm (top) and fibrils grown at 65°C after 24 h agitation at 510 rpm (bottom). Rat N27 dopaminergic cell line was used for the toxicity assay. For each of the presented results, three independent measurements were made. Black asterisks (*) demonstrate statistical significance between the LDH of protein aggregates and the control; red asterisks show statistical significance between the LDH of Lys and protein aggregates formed in the presence of lipids. ANOVA and Bonferroni corrected Post Hoc T‐test were used to analyze the results of toxicity assays. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; NS, nonsignificant difference.
2. DISCUSSION
Amyloid diseases are caused by the abrupt aggregation of misfolded proteins. For instance, the onset and spread of PD are linked to the aggregation of α‐Syn in the midbrain, hypothalamus, and thalamus. A growing body of evidence indicates AD is linked to the abrupt aggregation of Aβ peptide. Similar to α‐Syn, Aβ can bind lipids. Avdulov et al. demonstrated that lipids preferably interact not with monomeric Aβ but with protein oligomers (Avdulov et al., 1997). It was also found that Aβ exhibited substantially higher affinity to cholesterol compared to phospholipids, and FAs (Avdulov et al., 1997). Our own experimental results showed that lipids present in the structure of Aβ oligomers formed in their presence (Zhaliazka & Kurouski, 2023). We infer that the presence of lipids drastically changes the hydrophobic properties of oligomer surfaces facilitating their property to permeabilize plasma membranes (Matveyenka et al., 2023; Matveyenka, Rizevsky, & Kurouski, 2022a; Matveyenka, Rizevsky, & Kurouski, 2022b; Matveyenka, Zhaliazka, et al., 2022). Our group demonstrated that the toxicity of such lipid‐rich aggregates directly depended on the saturation of FAs in phospholipids (Matveyenka, Rizevsky, & Kurouski, 2022b; Matveyenka, Rizevsky, & Kurouski, 2022c; Matveyenka, Rizevsky, & Kurouski, 2022d). Specifically, we found that amyloid oligomers formed in the presence of unsaturated exerted greater cell toxicity compared to the aggregates formed in the presence of saturated phospholipids (Matveyenka, Rizevsky, & Kurouski, 2022b; Matveyenka, Rizevsky, & Kurouski, 2022c; Matveyenka, Rizevsky, & Kurouski, 2022d).
The reported results indicate that lysozyme strongly interacts with PS, an anionic lipid that is involved in the recognition of malfunctioning cells by macrophages. Our results also show that such interactions are determined by the length and saturation of FAs of PA. Such interactions uniquely alter the rate of lysozyme aggregation. We found that both DMPS drastically accelerated lysozyme aggregation compared to DOPS, POPS, and DSPS. These results are in agreement with the experimental findings reported by Matveyenka et al. for insulin (Matveyenka, Rizevsky, & Kurouski, 2022b). However, Matveyenka et al. found that POPS slightly decelerated insulin aggregation, whereas our results demonstrated that this phospholipid had the opposite effect on lysozyme aggregation. These differences in the aggregation kinetics of insulin and lysozyme in the presence of POPS indicate that the secondary structure and amino acid composition of protein play an important role in such lipid‐protein interactions. Structural and morphological examination of lysozyme aggregates grown in the presence of DMPS, DOPS, POPS, and DSPS demonstrated that the length and saturation of FAs also changed the secondary structure and morphology of lysozyme fibrils formed in the presence of these phospholipids. These results are congruent with the morphological and structural analysis of insulin fibrils formed in the presence of DMPS, POPS, and DOPS reported by Matveyenka, Rizevsky, and Kurouski (2022b). Finally, the LDH assay confirmed that structurally different lysozyme aggregates exert drastically different cell toxicity.
Previously reported results by Matveyenka and co‐workers demonstrated that both oligomers and fibrils could be endocytosed by cells (Matveyenka et al., 2023). Alternatively, protein aggregates may directly permeabilize lipid bilayers (Srinivasan et al., 2013). In the former case, oligomers and fibrils damage endosomes and leak out to the cytosol, where they impair the physiological function of the endoplasmic reticulum and mitochondria. In the latter case, protein aggregates generate ROS, which ultimately leads to cell death. Matveyenka and co‐workers found that protein aggregates formed in the presence of lipids and lipid‐free environments exert drastically different cell toxicity to N27 cells (Matveyenka et al., 2023). It was also reported that insulin and lysozyme aggregates formed in the presence of LUVs possessed lipids in their structure. One can expect that the presence of lipids on the surface of oligomers and fibrils drastically changed their interactions with the cell membranes, which ultimately lead to the observed differences in the oligomers' and fibrils' toxicity. At the same time, Zhaliazka and co‐workers recently demonstrated a direct relationship between the amount of β‐sheet and toxicity of amyloid β1‐42 aggregates (Zhaliazka, Matveyenka, & Kurouski, 2023a). Thus, one can expect that differences in the secondary structure of amyloid aggregates determine their toxicity. Our current results show that likely both factors, the presence of lipids and protein secondary structure determine the toxicity of lysozyme aggregates formed in the presence of DMPS, DOPS, POPS, and DSPS.
3. CONCLUSIONS
In this study, we investigated the effect of the length and saturation of FAs in PS on the aggregation rate of lysozyme. We also determined the structure and toxicity of protein aggregates formed in the presence of these lipids. Our results demonstrate that both length and saturation uniquely alter the rate of lysozyme aggregation. Furthermore, the presence of PS of varying length and FA saturation during protein aggregation results in the formation of structurally and morphologically distinct aggregates, which, in turn, cause different cell toxicity.
4. MATERIALS AND METHODS
4.1. Materials
Chicken‐eff lysozyme was purchased from Sigma‐Aldrich (St. Louis, MO, USA), 1,2‐dioleoyl‐sn‐glycero‐3‐phospho‐L‐serine (DOPS), 1‐palmitoyl‐2‐oleoyl‐sn‐glycero‐3‐phospho‐L‐serine (POPS), 1,2‐dimyristoyl‐sn‐glycero‐3‐phospho‐L‐serine (DMPS) and 1,2‐distearyl‐sn‐glycero‐3‐phospho‐L‐serine (DSPS) was purchased from Avanti (Alabaster, AL, USA).
4.2. Liposome preparation
For the preparation of large unilamellar vesicles (LUVs) of DMPS, POPS, DOPS, and DPSP 0.6 mg of the lipid was dissolved in 2.6 mL of phosphate buffered saline (PBS) pH 7.4. Next, solutions were heated in a water bath to ~50°C for 30 min. After that, the solutions were immediately immersed in liquid nitrogen for 3–5 min. This heating‐thawing cycle was repeated 10 times. Finally, lipid solutions were passed 15 times through a 100 nm membrane that was placed into the extruder (Avanti, Alabaster, AL, USA). LUV sizes were determined by dynamic light scattering.
4.3. Protein aggregation
In the lipid‐free environment, 400 μM of lysozyme was dissolved in PBS; the solution pH was adjusted to pH 3.0 using concentrated HCl. For Lys:DMPS, Lys:POPS, Ins:DOPS, and Lys:DSPS, 75 μL of 200 μM of lysozyme was mixed with the equivalent volume and concentration of the corresponding lipid; the solution pH was adjusted to pH 3.0 using concentrated HCl. Next, the solutions were placed in the plate reader (Tecan, Männedorf, Switzerland) and incubated at 37°C under 510 rpm for 160 h or at 65°C under 510 rpm for 24 h.
4.4. Kinetic measurements
Protein aggregation was monitored using a thioflavin T (ThT) fluorescence assay. For this, samples were mixed with 2 mM of ThT solution and placed in the plate reader (Tecan, Männedorf, Switzerland) where samples were incubated at 37°C under 510 rpm for 160 h. Fluorescence measurements were taken every 10 min using 450 nm excitation. Emission was collected at 488 nm.
4.5. AFM imaging
AFM analysis of protein aggregates was performed using tapping‐mode AFM probes with related parameters force constant 2.7 N/m and resonance frequency 50–80 kHz were purchased from Appnano (Mountain View, CA) on AIST‐NT‐HORIBA system (Edison, NJ). Analysis of collected images was performed using AIST‐NT software (Edison, NJ).
4.6. Circular Dichroism (CD)
CD spectra were collected from solutions of lysozyme aggregates at 25°C on J‐1000 CD spectrometer (Jasco, Easton, MD, USA). Three spectra were collected for each sample within 190–250 nm.
4.7. Attenuated total reflectance Fourier‐transform Infrared (ATR‐FTIR) spectroscopy
Sample aliquots were placed onto ATR crystal and dried at room temperature. Spectra were measured using Spectrum 100 FTIR spectrometer (Perkin‐Elmer, Waltham, MA, USA). Three spectra were collected from each sample.
4.8. Cell toxicity assays
The N27 rat dopaminergic cell line was grown in RPMI 1640 Medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Waltham, MA, USA). The cells were cultured in a 96‐well plate at a density of 10,000 cells per well and incubated at 37°C with 5% CO2. After 24 h, the cells adhered to the wells and reached approximately 70% confluency. Next, 100 μL of the cell culture medium was replaced with 100 μL of RPMI 1640 Medium containing 5% FBS and 10 μL of protein samples. After 24 h of incubation, the lactate dehydrogenase (LDH) assay was performed on the cell medium using the CytoTox 96 nonradioactive cytotoxicity assay (G1781, Promega, Madison, WI, USA). Absorbance measurements were taken at 490 nm using a plate reader (Tecan, Männedorf, Switzerland).
CONFLICT OF INTEREST STATEMENT
The authors declare no competing financial interests.
Supporting information
Data S1. Supporting information.
ACKNOWLEDGMENTS
We are grateful to the National Institute of Health for the provided financial support (R35GM142869).
Frese A, Goode C, Zhaliazka K, Holman AP, Dou T, Kurouski D. Length and saturation of fatty acids in phosphatidylserine determine the rate of lysozyme aggregation simultaneously altering the structure and toxicity of amyloid oligomers and fibrils. Protein Science. 2023;32(8):e4717. 10.1002/pro.4717
Addison Frese and Cody Goode contributed equally.
Review Editor: Aitziber L. Cortajarena
REFERENCES
- Alecu I, Bennett SAL. Dysregulated lipid metabolism and its role in alpha‐synucleinopathy in Parkinson's disease. Front Neurosci. 2019;13:328 PMID: 31031582 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida CG, Takahashi RH, Gouras GK. Beta‐amyloid accumulation impairs multivesicular body sorting by inhibiting the ubiquitin‐proteasome system. J Neurosci. 2006;26:4277–4288. PMID: 16624948 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alza NP, Iglesias Gonzalez PA, Conde MA, Uranga RM, Salvador GA. Lipids at the Crossroad of alpha‐Synuclein Function and Dysfunction: Biological and Pathological Implications. Front Cell Neurosci. 2019;13:175 PMID: 31118888 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdulov NA, Chochina SV, Igbavboa U, Warden CS, Vassiliev AV, Wood WG. Lipid binding to amyloid beta‐peptide aggregates: preferential binding of cholesterol as compared with phosphatidylcholine and fatty acids. J Neurochem. 1997;69:1746–1752. PMID: 9326304 {Medline}. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. PMID: 28498720 {Medline}. [DOI] [PubMed] [Google Scholar]
- Dou T, Kurouski D. Phosphatidylcholine and phosphatidylserine uniquely modify the secondary structure of alpha‐synuclein oligomers formed in their presence at the early stages of protein aggregation. ACS Chem Nerosci. 2022;13:2380–2385. PMID: 35904551 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou T, Zhou L, Kurouski D. Unravelling the structural organization of individual alpha‐synuclein oligomers grown in the presence of phospholipids. J Phys Chem Lett. 2021;12:4407–4414. PMID: 33945282 {Medline}. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Nelson R, Sawaya MR, Balbirnie M, Sambashivan S, Ivanova MI, et al. The structural biology of protein aggregation diseases: fundamental questions and some answers. Acc Chem Res. 2006;39:568–575. PMID: 16981672 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzner D, Bader JM, Penkert H, Bergner CG, Su M, Weil MT, et al. Cell‐type‐ and brain‐region‐resolved mouse brain lipidome. Cell Rep. 2020;32:108132 PMID: 32937123 {Medline}. [DOI] [PubMed] [Google Scholar]
- Galvagnion C. The role of lipids interacting with ‐synuclein in the pathogenesis of Parkinson's disease. J Parkinsons Dis. 2017;7:433–450. [DOI] [PubMed] [Google Scholar]
- Galvagnion C, Brown JW, Ouberai MM, Flagmeier P, Vendruscolo M, Buell AK, et al. Chemical properties of lipids strongly affect the kinetics of the membrane‐induced aggregation of alpha‐synuclein. Proc Natl Acad Sci U S A. 2016;113:7065–7070. PMID: 27298346 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson BI, Murray IV, Trojanowski JQ, Lee VM. A hydrophobic stretch of 12 amino acid residues in the middle of alpha‐synuclein is essential for filament assembly. J Biol Chem. 2001;276:2380–2386. PMID: 11060312 {Medline}. [DOI] [PubMed] [Google Scholar]
- Han S, Kollmer M, Markx D, Claus S, Walther P, Fandrich M. Amyloid plaque structure and cell surface interactions of beta‐amyloid fibrils revealed by electron tomography. Sci Rep. 2017;7:43577 PMID: 28240273 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R, Lasagna‐Reeves CA. Molecular mechanisms of amyloid oligomers toxicity. J Alzheimers Dis. 2013;33 Suppl 1:S67–S78. PMID: 22531422 {Medline}. [DOI] [PubMed] [Google Scholar]
- Kidd M. Paired helical filaments in electron microscopy of Alzheimer's disease. Nature. 1963;197:192–193. PMID: 14032480 {Medline}. [DOI] [PubMed] [Google Scholar]
- Kiechle M, Grozdanov V, Danzer KM. The role of lipids in the initiation of alpha‐synuclein misfolding. Front Cell Dev Biol. 2020;8:562241 PMID: 33042996 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killinger BA, Melki R, Brundin P, Kordower JH. Endogenous alpha‐synuclein monomers, oligomers and resulting pathology: let's talk about the lipids in the room. NPJ Parkinsons Dis. 2019;5:23 PMID: 31728405 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmer M, Close W, Funk L, Rasmussen J, Bsoul A, Schierhorn A, et al. Cryo‐EM structure and polymorphism of Abeta amyloid fibrils purified from Alzheimer's brain tissue. Nat Commun. 2019;10:4760 PMID: 31664019 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig AS, Rosener NS, Gremer L, Tusche M, Flender D, Reinartz E, et al. Structural details of amyloid beta oligomers in complex with human prion protein as revealed by solid‐state MAS NMR spectroscopy. J Biol Chem. 2021;296:100499 PMID: 33667547 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasagna‐Reeves CA, Glabe CG, Kayed R. Amyloid‐beta annular protofibrils evade fibrillar fate in Alzheimer disease brain. J Biol Chem. 2011;286:22122–22130. PMID: 21507938 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I, Levental KR, Heberle FA. Lipid rafts: controversies resolved, mysteries remain. Trends Cell Biol. 2020;30:341–353. PMID: 32302547 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveyenka M, Rizevsky S, Kurouski D. Amyloid aggregates exert cell toxicity causing irreversible damages in the endoplasmic reticulum. Biochim Biophys Acta Mol Basis Dis. 2022a;1868:166485 PMID: 35840040 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveyenka M, Rizevsky S, Kurouski D. The degree of unsaturation of fatty acids in phosphatidylserine alters the rate of insulin aggregation and the structure and toxicity of amyloid aggregates. FEBS Lett. 2022b;596:1424–1433. PMID: 35510803 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveyenka M, Rizevsky S, Kurouski D. Length and unsaturation of fatty acids of phosphatidic acid determines the aggregation rate of insulin and modifies the structure and toxicity of insulin aggregates. ACS Chem Nerosci. 2022c;13:2483–2489. PMID: 35930674 {Medline}. [DOI] [PubMed] [Google Scholar]
- Matveyenka M, Rizevsky S, Kurouski D. Unsaturation in the fatty acids of phospholipids drastically alters the structure and toxicity of insulin aggregates grown in their presence. J Phys Chem Lett. 2022d;13:4563–4569. PMID: 35580189 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveyenka M, Rizevsky S, Pellois JP, Kurouski D. Lipids uniquely alter rates of insulin aggregation and lower toxicity of amyloid aggregates. Biochim Biophys Acta Mol Cell Biol Lipids. 2023;1868:159247 PMID: 36272517 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveyenka M, Zhaliazka K, Rizevsky S, Kurouski D. Lipids uniquely alter secondary structure and toxicity of lysozyme aggregates. FASEB J. 2022;36:e22543. PMID: 36094052 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, et al. Structure of the cross‐beta spine of amyloid‐like fibrils. Nature. 2005;435:773–778. PMID: 15944695 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Seeded growth of beta‐amyloid fibrils from Alzheimer's brain‐derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci U S A. 2009;106:7443–7448. PMID: 19376973 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta U, Nilson AN, Kayed R. The role of amyloid‐beta oligomers in toxicity, propagation, and immunotherapy. EBioMedicine. 2016;6:42–49. PMID: 27211547 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmoradian SH, Lewis AJ, Genoud C, Hench J, Moors TE, Navarro PP, et al. Lewy pathology in Parkinson's disease consists of crowded organelles and lipid membranes. Nat Neurosci. 2019;22:1099–1109. PMID: 31235907 {Medline}. [DOI] [PubMed] [Google Scholar]
- Shashilov V, Xu M, Ermolenkov VV, Fredriksen L, Lednev IK. Probing a fibrillation nucleus directly by deep ultraviolet Raman spectroscopy. J Am Chem Soc. 2007;129:6972–6973. PMID: 17500518 {Medline}. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Patke S, Wang Y, Ye Z, Litt J, Srivastava SK, et al. Pathogenic serum amyloid A 1.1 shows a long oligomer‐rich fibrillation lag phase contrary to the highly amyloidogenic non‐pathogenic SAA2.2. J Biol Chem. 2013;288:2744–2755. PMID: 23223242 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Fukushima H, Masliah E, Xia Y, Iwai A, Yoshimoto M, et al. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:11282–11286. PMID: 8248242 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosough F, Barth A. Characterization of homogeneous and heterogeneous amyloid‐beta42 oligomer preparations with biochemical methods and infrared spectroscopy reveals a correlation between infrared spectrum and oligomer size. ACS Chem Nerosci. 2021;12:473–488. PMID: 33455165 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaliazka K, Kurouski D. Lipids uniquely alter the secondary structure and toxicity of amyloid beta 1‐42 aggregates. FEBS J. 2023;290:3203–3220. 10.1111/febs.16738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaliazka K, Matveyenka M, Kurouski D. Lipids uniquely alter the secondary structure and toxicity of amyloid beta 1‐42 aggregates. FEBS J PMID: 36705524 {Medline}. 2023a;290:3203–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaliazka K, Rizevsky S, Matveyenka M, Serada V, Kurouski D. Charge of phospholipids determines the rate of lysozyme aggregation but not the structure and toxicity of amyloid aggregates. J Phys Chem Lett. 2022;13:8833–8839. PMID: 36111888 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaliazka K, Serada V, Matveyenka M, Rizevsky S, Kurouski D. Protein‐to‐lipid ratio uniquely changes the rate of lysozyme aggregation but does not significantly alter toxicity of mature protein aggregates. Biochim Biophys Acta Mol Cell Biol Lipids. 2023b;1868:159305 PMID: 36907244 {Medline}. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting information.


