Abstract
CRISPR-Cas9-based base editing allows precise base editing to achieve conversion of adenosine to guanine or cytosine to thymidine. In this issue of Cell, McAuley et al. use adenine base editing to correct a single base-pair mutation causing human CD3δ deficiency, demonstrating superior efficiency of genetic correction with reduced undesired genetic alterations compared with standard CRISPR-Cas9 editing.
Severe combined immune deficiency (SCID) comprises a group of genetically determined inborn errors of immunity characterized by profound T cell deficiency leading to serious infections and early death unless treated by hematopoietic stem cell (HSC) transplantation, gene therapy, or enzyme replacement therapy.1 In this issue of Cell, McAuley et al. explored the use of adenine base editing (ABE) to correct human CD3δ deficiency both in vitro and in vivo in an animal model.2 CD3δ deficiency is caused by biallelic loss-of-function mutations of the CD3D gene that compromise expression of the CD3/T cell receptor (TCR) complex necessary for T cell development and function.3 CD3δ deficiency accounts for an ultrarare form of SCID (~1% of all cases of SCID).4 The rarity of the disorder and the severity of the clinical condition limit access to patients’ primary cells, making development of new treatment forms problematic. To circumvent this issue, McAuley et al. used an array of cellular and molecular tools to assess safety and potential efficacy of ABE to attempt correction of a CD3D nonsense mutation (c.202C>T, p.R68X) that results in a lack of CD3δ protein expression, which has been recurrently observed in patients of Mennonite origin.
Using Jurkat T cells engineered to contain the pathogenic c.202C>T variant, McAuley et al. demonstrated that the ABE strategy is superior to the CRISPR-Cas-based homology-directed repair (HDR) method that uses recombination with a single-strand oligodeoxynucleotide donor (ssODN) in achieving gene correction and restoration of surface expression of the CD3 complex and CD3-mediated signaling. The details of CRISPR-Cas9-mediated HDR editing versus base editing are compared in Figure 1.
Figure 1. Comparison of standard CRISPR-Cas9 double-strand cut-mediated homology-directed repair versus CRISPR-Cas9 nickase single-strand cut-mediated base editing.
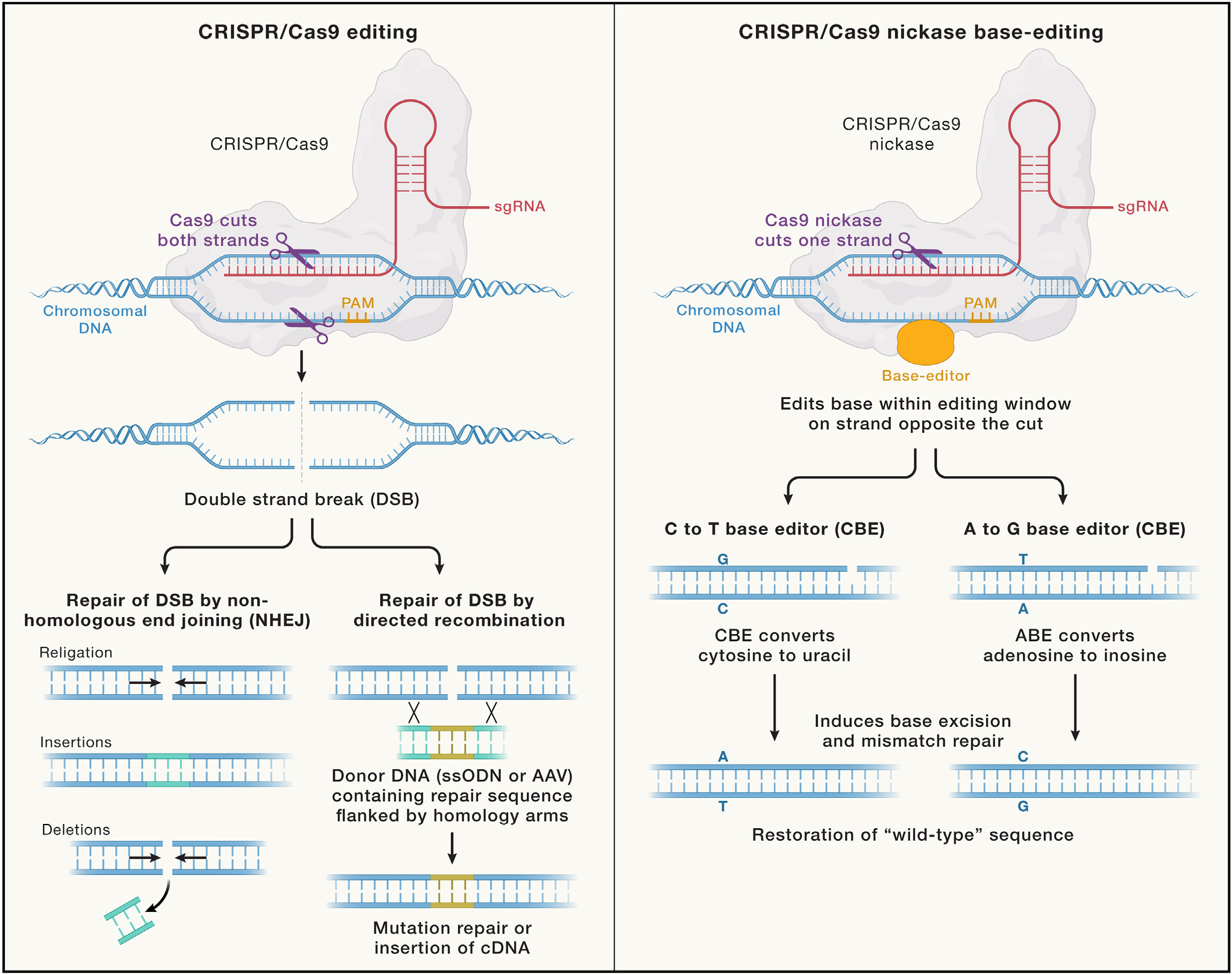
Both methods of editing use CRISPR together with the sgRNA to specify the genomic location for binding to chromosomal DNA. The standard CRISPR-Cas9 editing achieves a double-strand cut, which sets into motion the competing cellular repair mechanisms of NHEJ pathway versus the HDR pathway that can use a donor DNA (generally delivered either as ssODN or within an adenoassociated virus [AAV]) to achieve homology-directed recombination. The CRISPR-Cas9 nickase base editing employs Cas9 that has been mutated to only cut a single strand of the chromosomal DNA and also includes either of two types of highly engineered CBE or ABE enzymes that, respectively, can convert cytosine to uracil or adenosine to inosine within the editing window. Subsequent base excision with mismatch repair converts these to thymidine or guanine, respectively, to achieve correction of an SBPM to the wild-type sequence.
Upon selection of the optimal adenine base editor, minimal levels of local bystander editing and of genome-wide off-site targeting were observed. The authors then used a humanized mouse model to demonstrate that editing of the CD3D mutation did not affect engraftment and multilineage differentiation. Finally, they cultured unedited and gene-edited CD34+ bone marrow cells from a CD3δ-SCID infant in an in vitro artificial thymic organoid (ATO) system5 and demonstrated that patient-derived gene-edited CD34+ cells were able to generate a polyclonal repertoire of mature single-positive (SP) cells expressing the CD3/TCRαβ complex with normal response to CD3 stimulation. In the same experiment, the authors were able to precisely map the T cell differentiation block of unedited, CD3D-mutated CD34+ cells at the immature SP4/double-positive early (DPE) stage. These data expand the fine-mapping of T cell developmental blocks associated with SCID in humans, as defined by studies in the ATO system.6 Moreover, the developmental block observed in CD3δ deficiency was associated with skewing of TCR rearrangements, with reduced usage of 5′ Vα and of 3′ Jα genes, a defect that was corrected in base-edited cells.
This study illustrates the remarkable potential for ex vivo CRISPR base editing of HSCs to achieve high-efficiency gene correction. Importantly, base editing uses a modified Cas nickase mediating a single-strand cut of DNA (Figure 1), thus greatly reducing the risk of introducing the insertions or deletions that more commonly occur by non-homologous end joining (NHEJ) when seeking to achieve CRISPR-Cas9 double-strand break-mediated HDR.7 CRISPR base editing would, at first glance, seem to have a niche role within the broad range of the rapidly developing gene-editing technology or virus vector approaches to gene therapy. However, the Human Gene Mutation Database (http://www.hgmd.org/) indicates that single base-pair mutations (SBPMs) comprise about two-thirds of all disease-causing mutations.8 Moreover, 32% of SBPMs are CG-to-TG or CG-to-CA transitions.9 This fortuitously presaged the utility of one of the only two currently available major classes of base editors: adenine base editors, allowing A>G conversions as used by McAuley et al.,2 and cytidine base editors, allowing C>T conversions (Figure 1). The development of CRISPR base-editor variants with a much wider range of protospacer adjacent motif (PAM) binding sites and an increased range of target editing window distance from the PAM10 has greatly expanded the range of SBPMs amenable to correction, and future innovation is likely to expand that range.
The clinical potential of base-editing correction of each patient’s specific SBPM as quintessential personalized medicine does require that targeting be optimized for each mutation. However, there are a few disorders for which many patients share the same SBPM (sickle cell disease being such an example), and there are many examples of founder SBPMs in defined communities, such as the CD3D c.202C>T variant among the Mennonites. Moreover, for many disorders, there are hot spot SBPMs that comprise 5%–20% of the patients. However, many base-editing correctable mutations may be unique to individual patients. Fortunately, for SBPMs that are amenable to correction in HSCs using the current base-editing tools, the design and optimization of choice of editor and guide RNA (gRNA) to achieve high-efficiency correction with low off-target rates is relatively straightforward. Thus, it can be anticipated that CRISPR base editing will increasingly become the method of choice for correction of suitable SBPMs.
ACKNOWLEDGMENTS
H.L.M. and L.D.N. are supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, Klein C, Morio T, Oksenhendler E, Picard C, et al. (2022). Human inborn errors of immunity: 2022 update on the classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 42, 1473–1507. 10.1007/s10875-022-01289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McAuley GE, Yiu G, Chang PC, Newby GA, Campo-Fernandez B, Fitz-Gibbon ST, Wu X, Kang S-HL, Garibay A, Butler J, et al. (2023). Adenine base editing of hematopoietic stem cells rescues T cell development for CD3d severe combined immune deficiency. Cell 186, 1398–1416. 10.1016/j.cell.2023.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dadi HK, Simon AJ, and Roifman CM (2003). Effect of CD3delta deficiency on maturation of alpha/beta and gamma/delta T-cell lineages in severe combined immunodeficiency. N. Engl. J. Med. 349, 1821–1828. 10.1056/NEJMoa031178. [DOI] [PubMed] [Google Scholar]
- 4.Dvorak CC, Haddad E, Buckley RH, Cowan MJ, Logan B, Griffith LM, Kohn DB, Pai SY, Notarangelo L, Shearer W, et al. (2019). The genetic landscape of severe combined immunodeficiency in the United States and Canada in the current era (2010–2018). J. Allergy Clin. Immunol. 143, 405–407. 10.1016/j.jaci.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seet CS, He C, Bethune MT, Li S, Chick B, Gschweng EH, Zhu Y, Kim K, Kohn DB, Baltimore D, et al. (2017). Generation of mature T cells from human hematopoietic stem and progenitor cells in artificial thymic organoids. Nat. Methods 14, 521–530. 10.1038/nmeth.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosticardo M, Pala F, Calzoni E, Delmonte OM, Dobbs K, Gardner CL, Sacchetti N, Kawai T, Garabedian EK, Draper D, et al. (2020). Artificial thymic organoids represent a reliable tool to study T-cell differentiation in patients with severe T-cell lymphopenia. Blood Adv. 4, 2611–2616. 10.1182/bloodadvances.2020001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, and Liu DR (2017). Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471. 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, and Cooper DN (2017). The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 136, 665–677. 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper DN, and Krawczak M (1990). The mutational spectrum of single base-pair substitutions causing human genetic disease: Patterns and predictions. Hum. Genet. 85, 55–74. 10.1007/BF00276326. [DOI] [PubMed] [Google Scholar]
- 10.Tan J, Zhang F, Karcher D, and Bock R (2020). Expanding the genome-targeting scope and the site selectivity of high-precision base editors. Nat. Commun. 11, 629. 10.1038/s41467-020-14465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]


