Abstract
The biogenesis of the polar flagellum of Caulobacter crescentus is regulated by the cell cycle as well as by a trans-acting regulatory hierarchy that functions to couple flagellum assembly to gene expression. The assembly of early flagellar structures (MS ring, switch, and flagellum-specific secretory system) is required for the transcription of class III genes, which encode the remainder of the basal body and the external hook structure. Similarly, the assembly of class III gene-encoded structures is required for the expression of the class IV flagellins, which are incorporated into the flagellar filament. Here, we demonstrate that mutations in flbT, a flagellar gene of unknown function, can restore flagellin protein synthesis and the expression of fljK::lacZ (25-kDa flagellin) protein fusions in class III flagellar mutants. These results suggest that FlbT functions to negatively regulate flagellin expression in the absence of flagellum assembly. Deletion analysis shows that sequences within the 5′ untranslated region of the fljK transcript are sufficient for FlbT regulation. To determine the mechanism of FlbT-mediated regulation, we assayed the stability of fljK mRNA. The half-life (t1/2) of fljK mRNA in wild-type cells was approximately 11 min and was reduced to less than 1.5 min in a flgE (hook) mutant. A flgE flbT double mutant exhibited an mRNA t1/2 of greater than 30 min. This suggests that the primary effect of FlbT regulation is an increased turnover of flagellin mRNA. The increased t1/2 of fljK mRNA in a flbT mutant has consequences for the temporal expression of fljK. In contrast to the case for wild-type cells, fljK::lacZ protein fusions in the mutant are expressed almost continuously throughout the C. crescentus cell cycle, suggesting that coupling of flagellin gene expression to assembly has a critical influence on regulating cell cycle expression.
Cells of the gram-negative, aquatic bacterium Caulobacter crescentus possess an intrinsic asymmetry, which upon division results in the formation of two distinct daughter cells: a motile swarmer cell and a sessile stalked cell (reviewed in references 10, 26, and 80). These cell types differ in their programs of gene expression and DNA replication. For example, in the stalked cell, replication of chromosomal DNA initiates immediately following cell division, whereas this process is silenced for a defined period in the newly formed swarmer cell. Following this period of repression, the swarmer cell differentiates into a stalked cell. This differentiation event is accompanied by the degradation of flagellar components, stalk growth, the transcription of genes encoding DNA replication proteins, and the initiation of DNA replication (10, 26, 80).
Differentiation into a stalked cell type also initiates a cascade of events that culminate in the assembly of a single polar flagellum at the pole opposite the stalk. Flagellar biogenesis in C. crescentus requires the coordinated expression of approximately 50 genes (17, 20, 39) and is regulated by a complex interplay of both cell cycle and flagellar assembly events. The initiation of DNA replication is required for expression of early class II flagellar genes (13, 75), which encode the MS ring (37, 58), the flagellar switch (58, 83), and a flagellum-specific secretory apparatus (29, 64, 68, 75, 84), as well as the trans-acting factors encoded by flbE (78) and flbD (7, 8, 54, 59, 63, 79, 81) (Fig. 1). Class II promoters are transcribed relatively early in the cell cycle and contain conserved cis-acting sequences located between 35 and 10 bp upstream of the transcription start site (29, 58, 68, 75, 84). The transcription of these early flagellar operons is regulated by the global response regulator CtrA, which has been shown to bind to these conserved sequence elements and to activate the transcription of class II flagellar promoters at a specific time in the C. crescentus cell cycle (15, 50, 62).
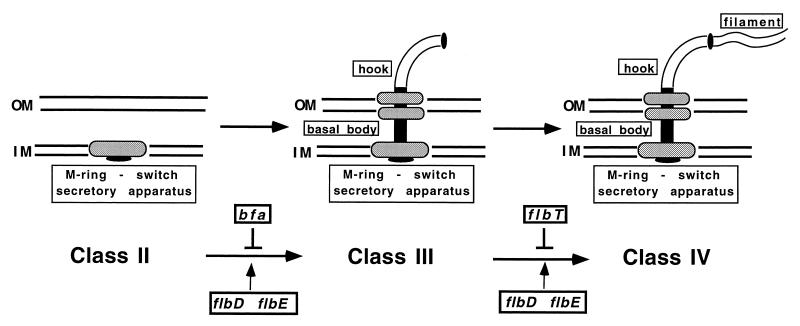
FIG. 1.
The C. crescentus flagellar regulatory hierarchy (reviewed in references 10, 26, and 80). A schematic of the flagellar regulatory hierarchy is shown. The hypothetical structures are depicted in the order in which they are assembled. Below each intermediate structure is indicated the class of flagellar genes that encode it. Flagellar assembly is coupled to gene expression at two distinct levels of the regulatory hierarchy. A cell cycle cue activates the response regulator CtrA, which in turn activates the transcription of the class II subset of flagellar genes. These genes encode regulatory proteins such as FlbD and FlbE, as well as the MS ring, the flagellar switch, and components of the flagellum-specific class III secretory apparatus. The expression and activation of FlbD and FlbE are required for the transcription of class III genes, which encode components of the basal body and hook structure. In addition, in the absence of assembly of the MS ring-switch secretory complex, the transcription of class III genes is negatively regulated by the bfa gene product. The proper assembly of class III genes is, in turn, necessary for the expression of the class IV genes, which encode flagellins. In this report, we describe how FlbT negatively regulates the expression of flagellin in the absence of assembly of the basal body-hook complex. OM, outer membrane; IM, inner membrane.
The expression of class III flagellar genes, which encode the rods and rings of the basal body and hook, follows the assembly of class II gene products (11, 12, 28, 30, 40, 46, 51–53). Class III gene expression is regulated by two distinct cellular events: the progression to a specific stage in the cell division cycle and the assembly of class II gene products into the nascent flagellar structure. The transcription of class III flagellar promoters requires the general transcription factors ς54-containing RNA polymerase (3, 9) and integration host factor (27, 28, 46), as well as the flagellar transcription factor FlbD (64). FlbD is a member of the response regulator family of bacterial two-component regulatory systems (64) and binds to a conserved enhancer element called ftr which is located approximately 100 bp from the transcription start site (7, 8, 52–54, 79, 81). Cell cycle expression of class III promoters is accomplished through the temporal phosphorylation of FlbD (8, 79). The product of the flbE gene is also required for the temporal expression of class III genes and has been shown to be required for FlbD activity (78). Activation of class III gene expression requires localization of FlbE to the mid-cell site in the predivisional cell (78). Therefore, FlbE is thought to couple an early cell division event to the activation of class III gene expression.
The assembly of class II gene products is also required for the expression of class III genes (Fig. 1). For example, a mutation in any one of the class II flagellar genes results in a lack of transcription of class III genes (11, 45, 56, 59, 82). A mutation, in bfa (for bypass of flagellar assembly), that permits expression of class III genes in the absence of an assembled flagellar structure has been isolated (45). Strains containing a mutation in bfa also exhibit an alteration in the cell cycle timing of class III gene transcription, indicating that bfa, and presumably flagellar assembly, has an influence in regulating temporal expression (45). It is hypothesized that bfa encodes a repressor of class III gene expression whose activity or availability is regulated by the assembly of an early flagellar structure. This would be analogous to the anti-sigma factor FlgM, which inhibits ς28 activity in response to a flagellar assembly defect in Salmonella typhimurium (24, 25, 34, 60). The bfa gene has not yet been isolated, and therefore it is not known whether Bfa inhibits FlbE and/or FlbD activity or whether it binds to class III promoters and represses transcription directly.
The incorporation of flagellin monomers into the filament follows the assembly of the hook structure (Fig. 1). C. crescentus possesses six different flagellin genes that map in two clusters at distinct chromosomal locations. The α cluster contains genes encoding 27-kDa (fljL), 25-kDa (fljK), and 29-kDa (fljJ) flagellins (21–23, 49). The genetically unlinked β cluster contains three genes, fljMNO, each encoding a 25-kDa flagellin (18). The transcriptional regulation of fljL and fljK has been examined in some detail (49). Like class III flagellar promoters, fljL and fljK require ς54-containing RNA polymerase (3, 9, 49, 51) and integration host factor (27). In addition, these promoters each contain FlbD binding sites (7, 8, 49, 79). fljL transcription requires the assembly of class II gene products (2, 45) and therefore is regulated by bfa. Interestingly, although fljK transcription requires the same trans-acting factors as fljL expression, it is not regulated by bfa (i.e., the promoter is transcribed in class II flagellar mutants) (2, 79).
Experiments using bfa mutants have revealed another level of this trans-acting regulatory hierarchy (45). bfa mutant strains, which were able to transcribe the 27-kDa flagellin gene, fljL, in the absence of flagellar assembly, synthesized no detectable flagellin protein (45). It has also been shown that although flagellin promoters are transcribed in strains with mutations in class III genes, flagellin protein and flagellin-lacZ protein fusions are not expressed (2). Furthermore, in the case of the 25-kDa flagellin (fljK product), this effect requires the 5′ untranslated region of the mRNA (2). From these experiments, it has been inferred that the assembly of class III flagellar gene products is required for the expression of flagellin, which comprises the final assembled structure of the flagellar filament (2). In contrast to the coupling of class III gene expression to the assembly of class II flagellar structures, which is regulated through the inhibition of transcription, the coupling of the assembly of class III flagellar structures to flagellin synthesis is regulated at the posttranscriptional level.
In this study, we investigated the mechanism of posttranscriptional regulation of fljK expression. We show that mRNA sequences that are 5′ to the translation start codon are sufficient to exert regulation and that fljK mRNA has a dramatically shorter half-life in the absence of flagellar assembly. Mutations in flbT, a well-characterized (16, 68–71) but previously unclassified flagellar gene, can reverse this effect, indicating that the flbT gene product may act as a negative regulator of fljK gene expression in the absence of flagellar assembly. Strains containing mutations in flbT have an altered cell cycle pattern of fljK expression, suggesting that posttranscriptional repression and the progression of flagellar assembly have a critical role in influencing temporal expression of C. crescentus flagellin genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in these experiments are listed in Table 1. C. crescentus NA1000, a motile, synchronizable strain, was used as the wild-type strain. AE8006 is a mutant strain with a Tn5-VB32 (6) insertion in flgE (11). A flbT flgE double mutant strain (JG551) was created by transducing flgE806::Tn5-VB32 into SC276 (flbT650) by using phage φCR30 (19). C. crescentus cells were grown with shaking at 31°C in either PYE medium (61) or minimal M2-glucose medium (38). Escherichia coli strains were grown in Luria-Bertani medium at 37°C (48). To generate a wild-type fljK::lacZ translational fusion, a 505-bp PstI-EcoRI fragment spanning the entire fljK promoter region to codon 23 within the gene was fused to the eighth codon of lacZ in pJBZ282 (45). The fljK::lacZ fusions in pJBZ282 were introduced into C. crescentus by conjugation, using E. coli S17-1 containing the helper plasmid pLVC9 (74). A fljK-lacZ transcription fusion (79) contained this same sequence cloned upstream of a promoterless lacZ in pRK290 (14) (placZ/290) (28). To construct a template for site-directed mutagenesis, the 505-bp PstI-EcoRI fragment was subcloned first into Bluescript KS (Stratagene) and then into M13-BM21 (Boehringer Mannheim Biochemicals) as a 524-bp BamHI-HindIII fragment.
TABLE 1.
Bacterial strains and plasmids used
| Bacterial strain or plasmid | Genotype | Reference or source |
|---|---|---|
| E. coli S17-1 | Rp4-2 Tc::Mu Km::Tn7 | 74 |
| C. crescentus | ||
| NA1000 | syn-1000 | 21 |
| AE8006 | flgE806::Tn5-VB32 | 11 |
| SC276 | flbT650 | 38 |
| SC511 | flgE::IS511 | 38 |
| SC508 | fliQR153 | 38 |
| SC603 | flaF673 | |
| SC604 | fla-674 | |
| JG548 | syn-1000 fljK::lacZ | This work |
| JG549 | flgE806::Tn5-VB32 fljK::lacZ | This work |
| JG550 | flbT650 fljK::lacZ | This work |
| JG551 | flgE806::Tn5-VB32 flbT650 fljK::lacZ | This work |
| JG552 | flgE806::Tn5-VB32 flaF673 fljK::lacZ | This work |
| JG553 | flgE806::Tn5-VB32 fla-674 fljK::lacZ | This work |
| SC1032 | flbD198::Tn5 str-152 | 57 |
| SC1137 | podF199::Tn5 str-152 | 77 |
| SC1054 | fliF173::Tn5 proA103 str-140 (old flaO) | 57 |
| SC1055 | rpoN610::Tn5 proA103 str-140 | 17 |
| SC1060 | flaN176::Tn5 proA103 str-140 | 57 |
| SC1066 | fliL::Tn5 proA103 str-140 | 57 |
| SC1114 | flbE607::Tn5 str-152 | 57 |
| SC1132 | flhA608::Tn5 str-152 | 57 |
| SC2201 | flbS636::Tn5-132 rif-175 | 17 |
| SC2204 | flbX639::Tn5-132 rif-175 | 17 |
| SC3476 | podF199::Tn5-132 str-152 | Conversion of the Tn5 in SC1137 to Tn5-132 |
| SC3730 | fliL::Tn5 str-140 | Transduction of SC1066 |
| SC3799 | flbT650 fliL179::Tn5 | Phage SC3730 × SC276 |
| SC3800 | flbT650 flhA608::Tn5 | Phage SC1132 × SC276 |
| SC3801 | flbT650 flaN176::Tn5 | Phage SC1060 × SC276 |
| SC3802 | flbT650 flbD198::Tn5 | Phage SC1032 × SC276 |
| SC3803 | flbT650 fliF173::Tn5 | Phage SC1054 × SC276 |
| SC3804 | flbT650 flbS636::Tn5 | Phage SC2201 × SC276 |
| SC3805 | flbT650 flbX639::Tn5 | Phage SC2204 × SC276 |
| SC3806 | flbT650 flabE607::Tn5 | Phage SC1114 × SC276 |
| SC3807 | flbT650 rpoN610::Tn5 | Phage SC1055 × SC276 |
| SC3808 | flbT650 podF199::Tn5 | Phage SC3476 × SC276 |
| SC3809 | fliQR153 zzz::Tn5 recA101 | Transduction of SC508 to KanrrecA |
| SC3843 | flbT650 fliQR153 zzz::Tn5 | Phage SC3809 × SC276 |
| Plasmids | ||
| pJBZ282 | lacZ protein fusion vector | M. R. K. Alley |
| pfljK::lacZ | 505-bp PstI-EcoRI fragment containing the fljK promoter and 23 codons inserted in frame to lacZ in pJBZ282 | This work |
| pfljK1::lacZ | 497-bp BamHI-HindIII fragment containing the fljK promoter and codons 1–14 inserted in frame to lacZ in pJBZ282 | This work |
| pfljK2::lacZ | 458-bp BamHI-HindIII fragment containing the fljK promoter and the ATG inserted in frame to lacZ in pJBZ282 | This work |
| pfljK3::lacZ | 471-bp BamHI-HindIII fragment containing the fljK promoter region with the upstream leader deleted and 23 codons inserted in frame to lacZ in pJBZ282 | This work |
| pfljK-lacZ/290 | 505-bp PstI-EcoRI fragment containing the fljK promoter region subcloned upstream of a promoterless lacZ reporter gene | 79 |
Mutagenesis of fljK mRNA.
The 505-bp PstI-EcoRI fljK fragment in M13-BM21 (Boehringer Mannheim Biochemicals) was used as a template for site-directed mutagenesis. Single-stranded DNA was isolated from E. coli CJ236, and mutagenesis was performed as described by Kunkel and Roberts (42). Mutants were identified by DNA sequencing (69). To generate pfljK1::lacZ, a HindIII site was created at codon 14. Likewise, pfljK2::lacZ was constructed by creating a HindIII site after codon 1 of fljK. pfljK3::lacZ was generated by creating a BglII site immediately following the transcription start site and another BglII site before the ribosomal binding site in the upstream leader. The resulting mutant was cloned from M13-BM21 into Bluescript KS by using BamHI-HindIII. The upstream leader was then deleted by digesting with BglII and religating. All products of site-directed mutagenesis were subcloned in frame to pJBZ282. Expression of wild-type and mutant constructs was assayed by β-galactosidase activity (48) with C. crescentus cultures grown to mid-logarithmic phase in PYE medium at 31°C. The reported β-galactosidase values represent mean values from assays performed in triplicate on three separately grown cultures.
Immunoprecipitation of flagellin and β-galactosidase.
C. crescentus cultures were grown in M2-glucose to an optical density at 660 nm of 1.0 to 1.4. An aliquot was removed and pulse-labeled with Tran35S-label (ICN) for 5 min. Labeled protein was immunoprecipitated with either a polyclonal antiflagellin antibody (33) or a monoclonal anti-β-galactosidase antibody (Boehringer Mannheim Biochemicals).
mRNA stability assay by primer extension.
C. crescentus NA1000, AE8006, SC276, and JG551 cells were grown in M2-glucose medium to an optical density at 600 nm of 1.0 to 1.2. To initiate the experiment, rifampin was added to the cultures to a final concentration of 200 μg/ml. Aliquots of cells were removed at various times following the addition of rifampin, and RNA was isolated from each sample (29). Primer extension was performed as described by Ausubel et al. (5) with an oligonucleotide primer complementary to fljK mRNA coding sequences 53 to 71 nucleotides from the start of translation. The 71-nucleotide extension products were resolved by electrophoresis in a denaturing 8% acrylamide–urea sequencing gel. The dried gel was exposed to X-ray film and subjected to phosphorimager analysis.
Cell cycle expression experiments.
C. crescentus cultures were grown in M2-glucose to an optical density at 660 nm of 1.0 to 1.4. Swarmer cells were isolated by centrifugation through a Ludox gradient (21). Swarmer cell populations (greater than 97% pure as assayed by light microscopy) were suspended in fresh M2-glucose medium and allowed to progress through the cell cycle. At various times throughout the cell cycle, samples were removed and proteins were pulse-labeled with Tran35S-label (ICN) for 5 min. Labeled protein was immunoprecipitated with the monoclonal anti-β-galactosidase antibody (Boehringer Mannheim Biochemicals) (33) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the dried gel was subjected to fluorography.
RESULTS
Previous experiments have demonstrated that although the C. crescentus 25-kDa flagellin gene (fljK) is transcribed in strains containing mutations in class II flagellar genes, flagellin protein is not synthesized (79). Furthermore, experiments comparing the expression of flagellin-lacZ transcription fusions and protein fusions have shown that class III flagellar mutant strains have a marked reduction in β-galactosidase activity generated from lacZ protein fusions, whereas transcription is largely unaffected (2). The conclusion from both of these experiments is that flagellin synthesis is subject to negative posttranscriptional regulation in strains that exhibit defects in flagellar assembly.
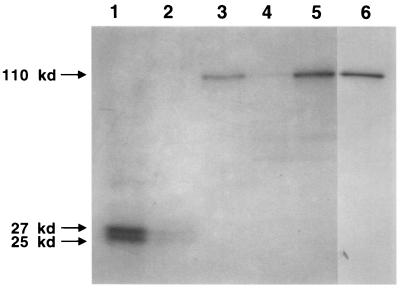
To determine whether the rate of β-galactosidase synthesis generated by reporter fusions was regulated in the same manner as that of flagellin protein, cellular proteins were pulse-labeled with Tran35S-label and immunoprecipitation experiments were performed. Wild-type and hook mutant (SC511) strains were assayed for expression of either a fljK::lacZ protein fusion reporter or a fljK-lacZ transcriptional fusion reporter (Fig. 2). Proteins were immunoprecipitated with a polyclonal antiflagellin antibody or a monoclonal anti-β-galactosidase antibody. Flagellin protein was expressed in wild-type cells (Fig. 2, lane 1), but was poorly expressed in hook mutant cells (Fig. 2, lane 2). Likewise, a fljK::lacZ protein fusion was expressed in wild-type cells (Fig. 2, lane 3) and exhibited a marked decrease in expression in a hook mutant strain (Fig. 2, lane 4). However, a fljK-lacZ transcriptional fusion was expressed in both wild-type cells and a hook mutant background (Fig. 2, lanes 5 and 6). These results suggest that a fljK::lacZ protein fusion is regulated by a posttranscriptional mechanism in the same manner as wild-type flagellin protein.
FIG. 2.
Posttranscriptional regulation of the 25-kDa flagellin gene, fljK. Expression of flagellin protein or fljK-lacZ fusions in either wild-type or hook mutant cells was assayed by immunoprecipitation. Cells were grown in M2 medium to mid-log phase, and an aliquot of cells was removed and labeled with Tran35S-label for 5 min. Labeled protein was immunoprecipitated with either antiflagellin (lanes 1 and 2) or anti-β-galactosidase (lanes 3 to 6) antibody and separated by SDS-PAGE as described in Materials and Methods. Lane 1, NA1000; lane 2, SC511; lane 3, NA1000 containing a fljK::lacZ protein fusion; lane 4, SC511 containing a fljK::lacZ protein fusion; lane 5, NA1000 containing a fljK-lacZ transcription fusion; lane 6, SC511 containing a fljK-lacZ transcription fusion. The apparent molecular masses of the immunoprecipitated proteins are indicated with arrows.
Mutations in flbT restore flagellin protein synthesis in strains bearing mutations in flagellar structural genes.
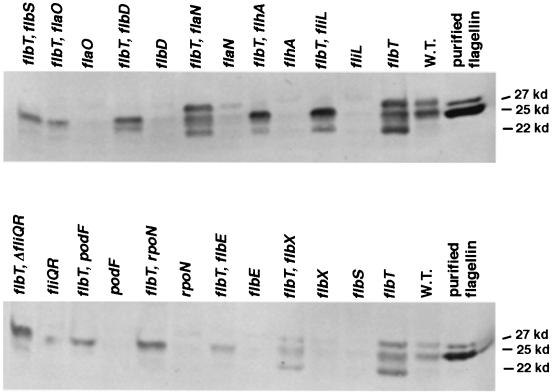
Previous experiments have demonstrated that mutations in flbT, a gene that lies near the α-flagellin gene clusters in C. crescentus, increase the level of flagellin expression (69, 72). Therefore, FlbT is an attractive candidate for a negative regulator of flagellin expression. To test this idea, the levels of flagellin in strains containing a mutation in a flagellar structural gene alone or in combination with a flbT mutation were assayed by immunoblotting (Fig. 3). Compared to wild-type cells, a strain bearing a flbT mutation produced slightly increased levels of the 27-kDa flagellin, encoded by the fljL gene, and the 25-kDa flagellin, encoded by fljK as well as fljMNO of the β cluster. In addition, as previously demonstrated, the mutant flbT strain also produced a variant of the 25-kDa flagellin that migrates at an apparent molecular mass of 22 kDa (70, 73). The effect of a flbT mutation on flagellin expression in class II and class III flagellar mutants was also examined. All of the class II and III mutant strains tested produced little or no flagellin protein (Fig. 3). If these strains also contained a mutation in flbT, 25-kDa flagellin expression was restored in all cases. Therefore, in the case of the 25-kDa flagellins, mutations in flbT bypass the requirement for flagellar assembly. In most cases, the mutation in flbT could not restore expression of the 27-kDa flagellin. Expression was evident in only two strains: a flbT flaN mutant and a flbT flbX mutant. These two strains contain mutations in class III flagellar genes, whereas the remainder of the strains tested contain mutations in class II genes. In contrast to the 25-kDa flagellin gene, fljK, fljL is not transcribed in class II mutants (2, 45) as a consequence of Bfa activity (45) and therefore is not expressed in flbT-class II double mutant strains.
FIG. 3.
A mutation in flbT restores flagellin expression in flagellar mutants. Proteins from whole-cell extracts from mutant strains were separated by SDS-PAGE and subjected to immunoblotting as described in Materials and Methods. Mutations in a single flagellar gene were introduced into the flbT mutant strain SC276 by generalized transduction. The mobilities of flagellins derived from these strains are compared to that of purified flagellin protein. W.T., wild type.
Interestingly, mutations in flbT were also able to restore flagellin expression in strains containing mutations in genes encoding trans-acting factors, such as rpoN (encoding the ς54 subunit of RNA polymerase holoenzyme), flbD (encoding a ς54 transcriptional activator), and flbE (encoding a trans-acting factor required for FlbD activity). Previous experiments have shown that these genes are essential for the expression of fljK and fljL. The simplest interpretation of these results is that the flagellin produced in these double mutant strains derives from fljMNO in the β cluster. Recent evidence indicates that the transcription of fljMNO is not dependent on rpoN or flbD (18). However, the fact that this cluster does not produce flagellin in the absence of flagellar assembly probably indicates that protein synthesis is regulated in a fashion similar to that for fljL and fljK.
Effect of flbT mutations on fljK::lacZ expression.
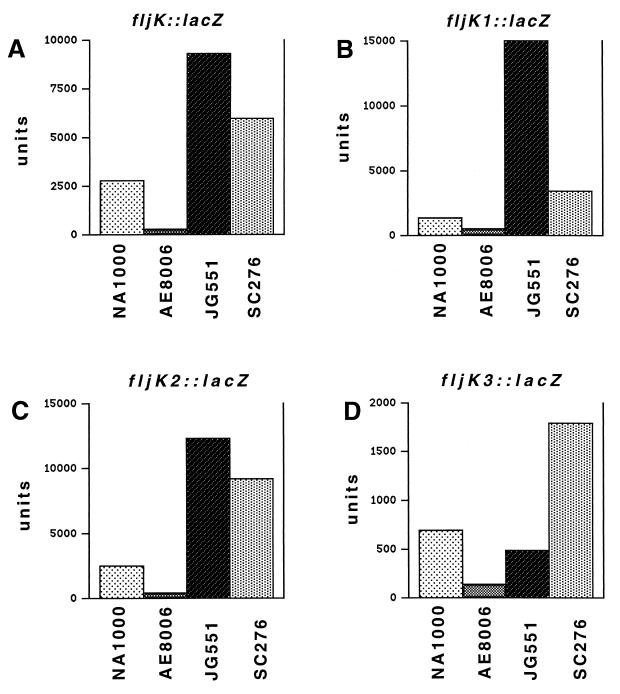
We next tested whether the flbT mutant strain had the same effect on fljK::lacZ reporter expression. To accomplish this, a wild-type fljK::lacZ protein fusion (Fig. 4) was introduced into wild-type cells, a flgE mutant (AE8006), a flbT mutant (SC276), and a flgE flbT double mutant (JG551) (Fig. 5A). This fusion generated 2,772 U of β-galactosidase activity in wild-type cells. In contrast, in the flgE mutant, the fljK::lacZ fusion was expressed at levels 10% of that in wild-type cells, generating 246 U of β-galactosidase activity (Fig. 5A). This result is consistent with the observation that fljK expression is subject to negative posttranscriptional regulation. In a flbT mutant, expression of fljK::lacZ was almost twice that in wild-type cells (5,955 U). This increase in expression is similar to that observed when flagellin protein was assayed in cell extracts derived from wild-type and flbT mutants (Fig. 3). To determine whether a mutation in flbT could restore fljK::lacZ expression in a class III mutant, we constructed a strain, JG551, that contained both the flgE::Tn5-VB32 mutation and the flbT650 mutation and then assayed β-galactosidase activity. The mutation in flbT completely bypassed the requirement for an assembled hook structure; the fljK::lacZ fusion in this strain possessed approximately 40 times the β-galactosidase activity of strain AE8006 which contains only a mutation in flgE (9,253 versus 246 U) (Fig. 5A). This result indicates that flbT has a critical role in negatively regulating fljK expression in the absence of flagellar assembly.
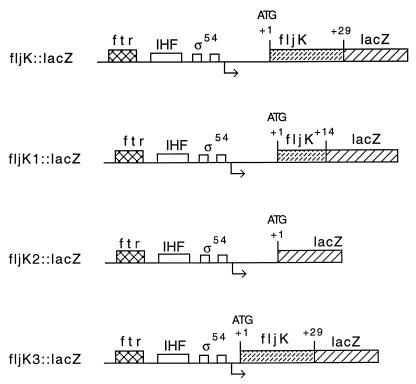
FIG. 4.
Schematic diagram of deletion derivatives of fljK::lacZ translation fusions. Site-directed mutagenesis was used to construct different deletions of fljK. The sequence encoding amino acids 1 to 23 was fused in frame to lacZ to create fljK::lacZ. fljK1::lacZ corresponds to a deletion of amino acids 15 to 23 fused in frame to lacZ. The sequence encoding amino acids 2 to 23 was deleted, resulting in an in-frame fusion of the ATG from fljK to lacZ to create fljK2::lacZ. fljK3::lacZ is a deletion of the upstream leader, leaving only the ribosomal binding site intact, fused in frame to lacZ. IHF, integration host factor.
FIG. 5.
Effect of deletions on the expression of fljK::lacZ translation fusions in wild-type and mutant strains. Wild-type and deleted fljK::lacZ translation fusions were introduced into the mutant strains indicated on the x axis. Values on the y axis represent β-galactosidase activity, in units (48), assayed in triplicate from three different mid-logarithmic-phase cultures. NA1000 is a synchronizable derivative of wild-type C. crescentus CB15. AE8006 contains a Tn5-VB32 insertion in flgE (hook). JG551 contains a Tn5-VB32 insertion in flgE and a flbT650 mutation from strain SC276. SC276 contains the flbT650 allele. (A) Mean β-galactosidase activity generated from pfljK::lacZ, which contains the entire 5′ untranslated region of fljK and the first 23 codons fused in frame to lacZ. The mean β-galactosidase activities were 2,772 U for NA1000, 246 U for AE8006, 9,253 U for JG551, and 5,955 U for SC276. (B) β-Galactosidase activity generated from pfljK1::lacZ, which contains the entire 5′ untranslated region of fljK and the first 14 codons fused in frame to lacZ. The mean β-galactosidase activities were 1,328 U for NA1000, 503 U for AE8006, 14,926 U for JG551, and 3,407 U for SC276. (C) β-Galactosidase activity generated from pfljK2::lacZ, which contains the entire 5′ untranslated region of fljK and the first codon fused in frame to lacZ. The mean β-galactosidase activities were 2,426 U for NA1000, 373 U for AE8006, 12,279 U for JG551, and 9,135 U for SC276. (D) β-Galactosidase activity (note difference in scale) generated from pfljK3::lacZ, in which the entire 5′ untranslated region of fljK except the ribosome binding site was deleted, fused in frame to lacZ. The mean β-galactosidase activities were 685 U for NA1000, 132 U for AE8006, 479 U for JG551, and 1,791 U for SC276. See Fig. 4 for a schematic representation of these fusions.
Since the nature of the flbT mutation in strain SC276 is unknown, we tested whether strains containing deletions in flbT exhibited a similar effect. The flgE::Tn5-VB32 mutation was introduced, by transduction, into SC603 and SC604, which have flmGH flbT flaF and flmGH flbT deleted, respectively (44, 72). flmG and flmH encode proteins that are homologous to O-linked acetylglucosamine transferases and acetyltransferases, respectively, and are regarded to be required for posttranslational modification of flagellins (44). The function of flaF is unknown. Previous experiments have demonstrated that mutations in flmG, flmH, and flaF result in a decrease in the level of flagellin produced (72). In strains SC603 and SC604, the level of β-galactosidase generated by fljK::lacZ was increased approximately twofold over that in wild-type cells (data not shown). The deletion in flbT in both these strains is apparently able to bypass the requirement for a hook structure. When the flgE::Tn5-VB32 allele was introduced into these strains, the level of β-galactosidase generated by fljK::lacZ was increased approximately 23 times over that assayed in AE8006 (5,800 versus 246 U) (data not shown). Since the other genes deleted in both of these strains are probably involved in the posttranslational modification of flagellin (44), the simplest conclusion that can be drawn from these results is that the deletion in flbT is responsible for the restoration of fljK::lacZ expression in the absence of a hook structure.
The restoration of fljK expression observed in flbT and flgE flbT mutants could be attributable to an increase in transcription and/or translation. To distinguish between these two possibilities, we tested the expression of fljK-lacZ transcription fusions in the flbT mutant strain SC276. In contrast to the high levels of β-galactosidase generated by the fljK::lacZ protein fusion, expression of the fljK-lacZ transcription fusion was dramatically decreased compared to that in wild-type cells (135 versus 3,500 U) (data not shown). This result indicates that FlbT probably functions as a posttranscriptional repressor. In addition, the decrease in transcription and the increase in translation in SC276 suggest that transcription of fljK is influenced by the level of translation.
In an attempt to define the region of fljK mRNA that is responsible for the posttranscriptional repression in a class III mutant background, a set of deletions were created and in-frame fusions to lacZ were constructed (Fig. 4). Mutants in which amino acids 15 to 23 (pfljK1::lacZ) or 2 to 23 (pfljK2::lacZ), encoded in the fljK coding region, were deleted (Fig. 4) exhibited a pattern of regulation similar to that for the fljK::lacZ fusion, which encoded all 23 of these amino acids; the fusions were expressed in wild-type cells and had a decreased expression in a hook mutant strain (Fig. 5B and C). The flbT mutation increased the expression of both of these fusions and could restore expression in a hook mutant to levels greater than that measured in wild-type cells (Fig. 5B and C). A final deletion mutant was created, in which the entire upstream leader sequence except the ribosomal binding site had been deleted (pfljK3::lacZ) (Fig. 4). In contrast to fusions with deletions within the coding region, this fusion was expressed poorly in wild-type cells (Fig. 5D [note change in scale]). Expression was reduced an additional fivefold in the hook mutant strain, indicating that this fusion was still subject to posttranscriptional repression. Furthermore, introduction of a flbT mutation into this strain partially restored expression; however, the increase in expression compared to that in the hook mutant was less than that observed for fusions containing a wild-type leader sequence. In this case, when the hook mutant strain also contained a flbT mutation (JG551), the expression of fljK3::lacZ was only 3.6 times higher than that in the strain containing a single mutation in the hook. In strains containing a fljK::lacZ fusion possessing a wild-type leader sequence, a flbT mutation increased expression levels by 37-fold. These results indicate that the sequences within the upstream leader are required for the regulation of expression of fljK in class III flagellar mutants and, in addition, that the likely site of FlbT-mediated regulation lies within this leader region.
flbT regulates fljK mRNA stability.
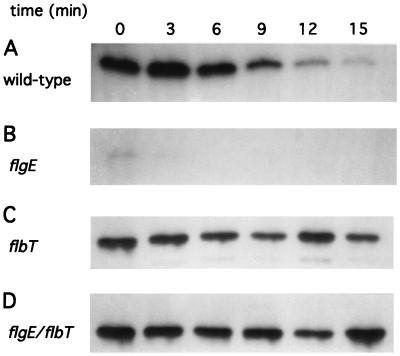
The decreased expression of flagellins and fljK::lacZ protein fusions in class III flagellar mutants may be a consequence of negative regulation of translation. If this is the case, a block in translation should result in a shorter half-life of flagellin mRNA, as has been observed for other bacterial mRNAs whose translation is negatively regulated (32, 35, 55, 65). To test this idea, we assayed the decay of fljK mRNA over time in wild-type cells, and in flgE, flbT, and flgE flbT mutant strains. The cultures were treated with rifampin to inhibit transcription, and total RNA was isolated at 3-min intervals and subjected to primer extension analysis. In wild-type cells, fljK mRNA had an estimated half-life of 11 min (Fig. 6). Previous experiments have demonstrated that the steady-state levels of flgK mRNA were reduced in a class III flagellar mutant (2). Consistent with this observation, in a strain containing a mutation in flgE, the fljK mRNA had a greatly reduced half-life of less than 1.5 min. This result suggests that decreased flagellin synthesis in class III mutant cells may be attributable to a decrease in the stability of fljK mRNA. fljK mRNA stability was increased to greater-than-wild-type levels in a flbT mutant background (half-life of greater than 30 min). The same experiment was performed with a flbT flgE double mutant. Like the flbT mutant background alone, a mutation in flbT was sufficient to increase fljK message stability to greater-than-wild-type levels (half-life of greater than 30 min). We hypothesize that the decrease in mRNA half-life observed in flgE mutant strains is a consequence of inhibition of translation, perhaps mediated through the action of the flbT gene product.
FIG. 6.
Effect of flagellar mutations on fljK::lacZ mRNA stability. C. crescentus NA1000 cells were grown to mid-logarithmic phase in M2 medium. At 0 min, 200 μg of rifampin per ml was added to inhibit transcription. At 3, 6, 9, 12, and 15 min, an aliquot was removed, RNA was isolated from each sample, and primer extension was performed with a 32P-labeled oligonucleotide that hybridized to the coding sequence of lacZ. The primer extension products were subjected to electrophoresis in a denaturing polyacrylamide gel. (A) C. crescentus NA1000 cells. The mRNA half-life is approximately 11 min. (B) Strain AE8006 (flgE::Tn5-VB32). The mRNA half-life is less than 1.5 min. (C) Strain SC276 (flbT650). The mRNA half-life is greater than 30 min. (D) Strain JG551 (flgE::Tn5-VB32 flbT650). The mRNA half-life is greater than 30 min.
A mutation in flbT alters the temporal pattern of fljK expression.
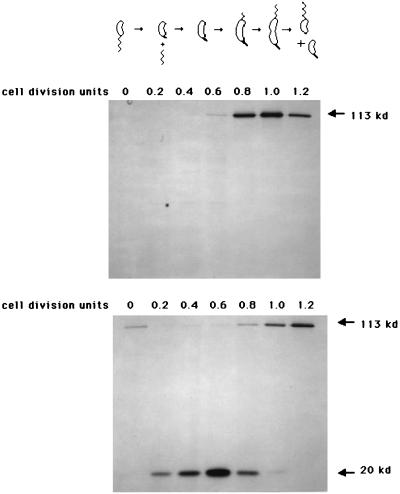
One possible function of FlbT in wild-type cells may be to influence the temporal expression of flagellin genes. For example, FlbT activity may exist to shut off flagellin synthesis following the completion of flagellar assembly. To test this hypothesis, the temporal expression of fljK::lacZ was assayed in wild-type and flbT mutant backgrounds. Cell cycle expression of fljK was assayed by synchronizing a culture containing an integrated fljK::lacZ protein fusion. The isolated swarmer cells were suspended in fresh medium and allowed to progress through the cell cycle. At various times thereafter, proteins were pulse-labeled with Tran35S-label, and radioactive β-galactosidase was immunoprecipitated from the cell extracts and subjected to PAGE. In wild-type cells, the fljK::lacZ fusion was expressed under temporal control. As previously shown (49, 79), expression was observed in swarmer cells (0 cell division units) and was turned off in stalked cells (0.45 cell division unit) (Fig. 7). Later in the cell cycle, when the cells reached the predivisional stage, expression of the fljK::lacZ fusion returned and continued until cell division occurred. In a flbT mutant strain the fljK::lacZ fusion had an altered pattern of expression. A full-length, labeled FljK::LacZ fusion was synthesized at the same period in the cell cycle as observed in wild-type cells. In contrast to the case for wild-type cells, in newly formed stalked cells and early predivisional cells of the flbT mutant strain, a smaller polypeptide of approximately 20 kDa was synthesized. Since a monoclonal antibody was employed in these experiments, the most likely source of this polypeptide is β-galactosidase. The best interpretation of this observation is that the fljK::lacZ fusion is expressed throughout the C. crescentus cell cycle but that protein which is expressed in stalked and early predivisional cells is subject to proteolysis. In this case, proteolysis is dependent on the fusion of fljK to lacZ, since fusions that express full-length lacZ at this time of the cell cycle, such as gyrB- or recF-lacZ transcription fusions, do not exhibit proteolysis of β-galactosidase (67). Therefore, we hypothesize that since fljK is not transcribed in stalked cells, the extended fljK mRNA stability observed in the flbT mutant is responsible for fljK::lacZ expression in stalked cells.
FIG. 7.
Effect of a flbT mutation on the temporal expression of a fljK::lacZ translation fusion. The temporal expression of β-galactosidase was assayed with either C. crescentus NA1000 or SC276 cells containing a fljK::lacZ translation reporter fusion. Isolated swarmer cells were suspended in fresh M2 medium and were permitted to progress through the cell cycle. At various times during the cell cycle (0, 30, 60, 90, 120, 150, and 180 min), an aliquot was removed and proteins were labeled with Tran35S-label for 10 min. Labeled protein was immunoprecipitated with a monoclonal anti-β-galactosidase antibody and subjected to SDS-PAGE as described in Materials and Methods. The gel was dried and exposed to X-ray film. (Top) fljK::lacZ expression in wild-type strain NA1000. The drawing above the fluorogram shows the cell types present at each time point, as determined by light microscopy. Labeled β-galactosidase is indicated by an arrow. (Bottom) fljK::lacZ expression in a flbT mutant strain, SC276. A low-molecular-mass (approximately 20-kDa) proteolytically generated fragment of β-galactosidase is also indicated by an arrow.
DISCUSSION
Flagellar biogenesis in C. crescentus is characterized by cell cycle assembly of a polar flagellar structure. The assembly of the polar flagellum is a complex process that requires over 50 gene products (17) and is regulated both by progression through the cell cycle and by the assembly of the structure itself. In this report, we have identified flbT as a key player in coupling flagellar assembly to the expression of flagellin. The gene, encoding FlbT, maps to a region that contains a cluster of late-acting flagellar genes (70), including those encoding proteins involved in the posttranslational modification of flagellin (44) as well as three flagellin genes, fljL, fljK, and fljJ. Mutations in flbT have been shown to have pleiotropic effects, including impaired motility, defects in the temporally regulated loss of the flagellum from stalked cells, and a marked increase in flagellin protein synthesis (70, 73). The last property suggests that flbT might function in the negative regulation of flagellin expression. The data presented here indicate that FlbT functions to inhibit flagellin expression in the absence of an intermediate flagellar structure into which flagellin monomers can be incorporated. In support of this view, mutations in flbT restore the expression of the fljK flagellin gene in the absence of a flagellar hook structure (i.e., in a flgE mutant strain). Although the precise nature of the flbT mutation in strain SC276 is not known, we have found, using anti-FlbT antibodies, that this strain produces no detectable FlbT protein (4). This suggests that the restoration of flagellin expression observed in flgE flbT double mutants is a consequence of a loss of FlbT and is probably not attributable to a gain-of-function allele. This is supported by the observation that deletions in flbT can also restore fljK::lacZ expression in a flgE mutant.
Immunoblot analysis showed that the flbT mutation could restore flagellin synthesis in all class II and class III flagellar mutant strains tested, suggesting that flbT is a general regulator that couples assembly to flagellin gene expression. Surprisingly, a mutation in flbT could also restore flagellin expression in rpoN, flbD, and flbE mutant strains, which are deficient in essential trans-acting factors. Recent analysis has revealed that the β cluster of flagellins, which contains three copies of 25-kDa flagellin genes, is not regulated by rpoN, flbD, or flbE (18), suggesting that the flagellin present in these mutants derives from this cluster. Interestingly, although the flagellins in the β cluster differ in their transcriptional regulation, they apparently have a common mechanism of regulation in response to flagellum assembly.
The negative regulation of flagellin expression in class III mutant strains is most likely mediated through a posttranscriptional mechanism. This conclusion is supported by the observation that although the fljK gene is transcribed in mutants that do not assemble a hook structure, there is little, if any, flagellin protein present. Deletion analysis of the fljK transcript showed that the expression of fljK was reduced in a hook mutant if the entire coding region was absent. Deletion of the 5′ untranslated sequences resulted in a marked decrease of fljK expression. Even though expression was decreased in a hook mutant, this fusion exhibited a relatively modest increase in activity in a FlbT-hook double mutant. Based on these data, we hypothesize that FlbT exerts its primary effect through acting on the 5′ untranslated leader of the fljK transcript. This is consistent with previous experiments demonstrating that sequences within the 5′ untranslated region of the fljK transcript were important for regulation by flagellar assembly (2). The absence of hook assembly presumably results in an increased turnover of fljK mRNA. Furthermore, mutations in flbT apparently decrease this high rate of mRNA turnover. From the experiments presented here, it is impossible to distinguish whether the apparent increase in mRNA turnover in class III mutants is a direct consequence of message instability in these strains or is brought about by an inhibition of translation, which as a secondary result leads to unstable mRNA. For example, it has been suggested that in some cases in bacteria, message stability is directly proportional to the formation of a translational complex (32, 35, 55). In the absence of ribosome binding, RNase E sites are exposed, causing rapid degradation of message (66). We envision two possible mechanisms whereby the flbT gene product promotes the turnover of flagellin mRNA in class III flagellar mutants. Previous experiments (47), as well as those reported here, have demonstrated that fljK mRNA has an unusually long half-life for a bacterial message. One plausible possibility is that a trans-acting factor(s) stabilizes fljK mRNA, and FlbT functions to antagonize its activity. Alternatively, FlbT may act directly to either destabilize fljK mRNA or prevent translation. Either model is consistent with the observation that mRNA sequences upstream of the translation initiation codon are required for FlbT to exert its maximal effect.
This mechanism of posttranscriptional repression of flagellin gene expression contrasts with an analogous regulation of flagellin gene expression in S. typhimurium. In S. typhimurium, the flagellin and chemotaxis genes are not expressed in mutant strains that have defects in flagellar assembly (41, 43). These genes are transcribed by ς28-containing RNA polymerase. Negative regulation is accomplished through an anti-sigma factor encoded by flgM (24, 25, 59). In the absence of flagellar assembly, the intracellular levels of the flgM gene product rise and the protein binds to the ς28 subunit of RNA polymerase, inhibiting its activity. Once a functional hook structure is assembled, FlgM is exported from the cell via the nascent flagellar structure, thereby relieving repression (34). In C. crescentus, as well, the transcription of class III genes (e.g., those encoding the basal body rods, outer rings, and hook) is repressed in the absence of the assembly of a flagellar structure encoded by class II gene products (Fig. 1). A mutation in a single uncharacterized gene, called bfa, can restore transcription of class III genes in the absence of flagellar assembly.
The regulation of flagellin gene expression by flbT represents another control point in flagellar biogenesis in C. crescentus, which is absent in enteric bacteria. What is the logic in possessing two different mechanisms to regulate flagellar biogenesis in response to assembly? The transcription of both class III and class IV flagellar genes is positively regulated by the FlbD transcription factor (7, 8, 55, 79, 81). FlbD is a member of the two-component response regulators (64) and is phosphorylated in response to a cell cycle cue (79), possibly an early stage in cell division (78). Once cells have progressed past this critical cell cycle step, they are competent to transcribe both class III and class IV flagellar genes. We speculate that it is at this stage when FlbT inhibits the expression of the class IV flagellin genes until a hook structure is assembled. With analogy to regulation in enteric bacteria, FlbT repression would then be inactivated, and any newly transcribed flagellin mRNA would be expressed. This model would ensure that flagellin is not synthesized until the correct stage in flagellar assembly is completed.
The flbT gene product may have an additional regulatory role later in the cell cycle, following cell division. As noted above, flagellin mRNA possesses an unusually long half-life. We have previously shown that fljK is transcribed in the swarmer compartment of the predivisional cell (31, 78, 79). The transcription of fljK ceases abruptly upon the completion of cell division; however, the mRNA remains within the progeny swarmer cell and is continually translated (46). Therefore, swarmer compartment-specific transcription of fljK results in generation of a supply of fljK mRNA for the nascent swarmer progeny cell. This mRNA is translated into flagellin protein, which is assembled in the flagellar filament. Thus, the requirement for continued filament growth in the progeny swarmer cells is fulfilled both by the swarmer compartment-specific transcription of fljK and by the unusual stability of flagellin mRNA. We hypothesize that FlbT plays a critical role in shutting off fljK translation in swarmer cells once the demand for new flagellin monomers is satisfied. This idea is supported by the observation that fljK is synthesized at relatively high levels in swarmer cells possessing mutant flbT and continues to be translated after the swarmer cells have differentiated into stalked cells. Therefore, FlbT, and presumably the assembly of the flagellum, contributes to the temporal expression of fljK. This result is similar to what occurs in bfa mutant cells, where the cessation of transcription of class III promoters during the cell cycle is significantly delayed (45). This suggests that the function of these two gene products in wild-type cells is to repress the synthesis of flagellar genes following the completion of a specific stage of flagellar assembly. Negative regulatory pathways that couple flagellar assembly to gene expression in other organisms, such as those present in enteric bacteria, may serve a similar purpose, perhaps functioning to regulate the number of flagella synthesized by each cell.
As noted above, one consequence of a mutation in flbT is the expression of fljK at an inappropriate time in the cell cycle. In this mutant strain, the FljK::LacZ fusion protein is degraded when swarmer cells differentiate into stalked cells. This is evidenced by the appearance of a small (approximately 20-kDa) polypeptide that reacts with the monoclonal anti-β-galactosidase antibody employed in this experiment. Proteolysis of β-galactosidase is apparently dependent on the presence of FljK amino acid sequence at the amino terminus. We have shown previously that β-galactosidase is not subject to proteolysis at this stage of the cell cycle (66). Therefore, C. crescentus possesses a developmentally regulated protease that can degrade intracellular flagellin specifically in stalked cells. One hallmark of the C. crescentus developmental program is the timed degradation of proteins, often in a cell-type-specific fashion. For example, the transcription factor CtrA (15), the FliF protein (MS ring) (37), and the methyl-accepting chemotaxis receptor (1) are all degraded specifically in stalked cells. One documented protease which is involved in this process is the highly conserved ClpXP protease. The ClpXP protease degrades CtrA, an essential response regulator that controls early flagellar gene transcription as well as the initiation of DNA replication (36). The ClpXP protease recognizes disordered C-terminal sequences, which possess some degree of conservation, as its substrates. Therefore, ClpXP is probably not involved in the degradation of FljK::LacZ, since the amino terminus of FljK appears to be the determinant for proteolysis. This indicates that C. crescentus probably possesses at least one additional developmentally regulated protease whose activity is restricted to the stalked cell type. In summary, C. crescentus utilizes a diverse array of regulatory mechanisms, such as cell cycle- and cell-type-regulated transcriptional activation, flagellar assembly-regulated transcriptional and posttranscriptional repression, and cell-type-specific proteolysis, in order to regulate flagellar biogenesis. The simultaneous operation of these pathways ensures that flagellum synthesis is tightly coupled to the formation of a differentiated daughter cell.
ACKNOWLEDGMENTS
We are grateful to Charles Boyd, G. Craig Draper, Jenny England, and Rachel Muir for critical reading of the manuscript.
E.K.M. was supported by Public Health Service predoctoral fellowship GM07104. This work was supported by Public Health Service grants GM48417 to J.W.G. and GM50547 to B.E. from the National Institutes of Health.
REFERENCES
- 1.Alley M R K, Maddock J, Shapiro L. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- 2.Anderson D K, Newton A. Posttranscriptional regulation of Caulobacter flagellin genes by a late flagellum assembly checkpoint. J Bacteriol. 1997;179:2281–2288. doi: 10.1128/jb.179.7.2281-2288.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson D K, Ohta N, Wu J, Newton A. Regulation of the Caulobacter crescentus rpoN gene and function of the purified sigma 54 in flagellar gene transcription. Mol Gen Genet. 1995;246:697–706. doi: 10.1007/BF00290715. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, P. E., and J. W. Gober. Unpublished data.
- 5.Ausubel F M, Brent R, Kingston R E, Moore D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989. [Google Scholar]
- 6.Bellofatto V, Shapiro L, Hodgson D. Generation of a Tn5 promoter probe and its use in the study of gene expression in Caulobacter crescentus. Proc Natl Acad Sci USA. 1984;81:1035–1039. doi: 10.1073/pnas.81.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson A K, Ramakrishnan G, Ohta N, Feng J, Ninfa A J, Newton A. The Caulobacter crescentus FlbD protein acts at ftr sequence elements both to activate and to repress transcription of cell cycle-regulated flagellar genes. Proc Natl Acad Sci USA. 1994;91:4989–4993. doi: 10.1073/pnas.91.11.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson A K, Wu J, Newton A. The role of FlbD in regulation of flagellar gene transcription in Caulobacter crescentus. Res Microbiol. 1994;145:420–430. doi: 10.1016/0923-2508(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 9.Brun Y, Marczynski G, Shapiro L. The expression of asymmetry during cell differentiation. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 10.Brun Y V, Shapiro L. A temporally controlled sigma-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 1992;6:2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- 11.Champer R, Dingwall A, Shapiro L. Cascade regulation of Caulobacter flagellar and chemotaxis genes. J Mol Biol. 1987;194:71–80. doi: 10.1016/0022-2836(87)90716-9. [DOI] [PubMed] [Google Scholar]
- 12.Chen L-S, Mullin D, Newton A. Identification, nucleotide sequence and control of developmentally regulated promoters in the hook operon region of Caulobacter crescentus. Proc Natl Acad Sci USA. 1986;83:2860–2864. doi: 10.1073/pnas.83.9.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingwall A, Zhuang W Y, Quon K, Shapiro L. Expression of an early gene in the flagellar regulatory hierarchy is sensitive to an interruption in DNA replication. J Bacteriol. 1992;174:1760–1768. doi: 10.1128/jb.174.6.1760-1768.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ditta G, Stanfield D, Corbin D, Helinski D R. Broad host range cloning system for gram-negative bacteria: construction of a gene bank for Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domian I J, Quon K C, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 16.Driks A, Schoenlein P V, DeRosier D J, Shapiro L, Ely B. A Caulobacter gene involved in polar morphogenesis. J Bacteriol. 1990;172:2113–2123. doi: 10.1128/jb.172.4.2113-2123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ely B, Ely T W. Use of pulsed field gel electrophoresis and transposon mutagenesis to estimate the minimal number of genes required for motility in Caulobacter crescentus. Genetics. 1989;123:649–654. doi: 10.1093/genetics/123.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ely B, Leclerc G. The Caulobacter crescentus fljMNO flagellin genes. GenBank accession no. AF040268. 1998. [Google Scholar]
- 19.Ely B, Johnson R C. Generalized transduction in Caulobacter crescentus. Genetics. 1977;87:391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ely B, Croft R H, Gerardot C J. Genetic mapping of genes required for motility in Caulobacter crescentus. Genetics. 1984;108:523–532. doi: 10.1093/genetics/108.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gill P R, Agabian N. A comparative structural analysis of the flagellin monomers of Caulobacter crescentus indicates that these proteins are encoded by two genes. J Bacteriol. 1982;150:925–933. doi: 10.1128/jb.150.2.925-933.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gill P R, Agabian N. The nucleotide sequence of the Mr = 28,500 flagellin gene of Caulobacter crescentus. J Biol Chem. 1983;258:7395–7401. [PubMed] [Google Scholar]
- 24.Gillen K L, Hughes K T. Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J Bacteriol. 1991;173:2301–2310. doi: 10.1128/jb.173.7.2301-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillen K L, Hughes K T. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J Bacteriol. 1991;173:6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gober J W, Marques M. Regulation of cellular differentiation in Caulobacter crescentus. Microbiol Rev. 1995;59:31–47. doi: 10.1128/mr.59.1.31-47.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gober J W, Shapiro L. Integration host factor is required for the activation of developmentally regulated genes in Caulobacter. Genes Dev. 1990;4:1494–1504. doi: 10.1101/gad.4.9.1494. [DOI] [PubMed] [Google Scholar]
- 28.Gober J W, Shapiro L. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol Biol Cell. 1992;3:913–926. doi: 10.1091/mbc.3.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gober J W, Boyd C H, Jarvis M, Mangan E K, Rizzo M F, Wingrove J A. Temporal and spatial regulation of fliP, an early flagellar gene of Caulobacter crescentus that is required for motility and normal cell division. J Bacteriol. 1995;177:3656–3667. doi: 10.1128/jb.177.13.3656-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gober J W, Xu H, Dingwall A, Shapiro L. Identification of cis and trans elements involved in the timed control of a Caulobacter flagellar gene. J Mol Biol. 1991;217:247–257. doi: 10.1016/0022-2836(91)90539-i. [DOI] [PubMed] [Google Scholar]
- 31.Gober J W, Champer R, Reuter S, Shapiro L. Expression of positional information during cell differentiation in Caulobacter. Cell. 1991;64:381–391. doi: 10.1016/0092-8674(91)90646-g. [DOI] [PubMed] [Google Scholar]
- 32.Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 33.Gomes S L, Shapiro L. Differential expression and positioning of chemotaxis methylation proteins in Caulobacter. J Mol Biol. 1984;178:551–568. doi: 10.1016/0022-2836(84)90238-9. [DOI] [PubMed] [Google Scholar]
- 34.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 35.Iost I, Dreyfus M. The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 1995;14:3252–3261. doi: 10.1002/j.1460-2075.1995.tb07328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenal U, Shapiro L. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 1996;15:2393–2406. [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson R C, Ely B. Isolation of spontaneously derived mutants of C. crescentus. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson R C, Ely B. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J Bacteriol. 1979;137:627–634. doi: 10.1128/jb.137.1.627-634.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khambaty F M, Ely B. Molecular genetics of the flgI region and its role in flagellum biosynthesis in Caulobacter crescentus. J Bacteriol. 1992;174:4101–4109. doi: 10.1128/jb.174.12.4101-4109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986;168:1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kunkel T A, Roberts J D. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 43.Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leclerc G, Wang S P, Ely B. A new class of Caulobacter crescentus flagellar genes. J Bacteriol. 1998;180:5010–5019. doi: 10.1128/jb.180.19.5010-5019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangan E, Bartamian M, Gober J W. A mutation that uncouples flagellum assembly from transcription alters the temporal pattern of flagellar gene expression in Caulobacter crescentus. J Bacteriol. 1995;177:3176–3184. doi: 10.1128/jb.177.11.3176-3184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marques M V, Gober J W. Activation of a temporally regulated Caulobacter promoter by upstream and downstream sequence elements. Mol Microbiol. 1995;16:279–289. doi: 10.1111/j.1365-2958.1995.tb02300.x. [DOI] [PubMed] [Google Scholar]
- 47.Milhausen M, Agabian N. Caulobacter flagellin mRNA segregates asymmetrically at cell division. Nature. 1983;302:630–632. doi: 10.1038/302630a0. [DOI] [PubMed] [Google Scholar]
- 48.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- 49.Minnich S A, Newton A. Promoter mapping and cell regulation of flagellin gene transcription in Caulobacter crescentus. Proc Natl Acad Sci USA. 1987;84:1142–1146. doi: 10.1073/pnas.84.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohr C D, MacKichan J K, Shapiro L. A membrane-associated protein, FliX, is required for an early step in Caulobacter flagellar assembly. J Bacteriol. 1998;180:2175–2185. doi: 10.1128/jb.180.8.2175-2185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mullin D, Minnich S, Chen L S, Newton A. A set of positively regulated flagellar gene promoters in Caulobacter crescentus with sequence homology to the nif gene promoters of Klebsiella pneumoniae. J Mol Biol. 1987;195:939–943. doi: 10.1016/0022-2836(87)90497-9. [DOI] [PubMed] [Google Scholar]
- 52.Mullin D A, Newton A. Ntr-like promoters and upstream regulatory sequence ftr are required for transcription of a developmentally regulated Caulobacter crescentus flagellar gene. J Bacteriol. 1989;171:3218–3227. doi: 10.1128/jb.171.6.3218-3227.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullin D A, Newton A. A ς54 promoter and downstream sequence elements ftr2 and ftr3 are required for regulated expression of divergent transcription units flaN and flbG in Caulobacter crescentus. J Bacteriol. 1993;175:2067–2076. doi: 10.1128/jb.175.7.2067-2076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullin D A, Van Way S M, Blankenship C A, Mullin A H. FlbD has a DNA-binding activity near its carboxy terminus that recognizes ftr sequences involved in positive and negative regulation of flagellar gene transcription in Caulobacter crescentus. J Bacteriol. 1994;176:5971–5981. doi: 10.1128/jb.176.19.5971-5981.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagai H, Harumi Y, Takashi Y. Interplay of two cis-acting mRNA regions in translational control of ς32 synthesis during the heat shock response of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:10515–10519. doi: 10.1073/pnas.88.23.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newton A, Ohta N, Ramakrishnan G, Mullin D, Raymond G. Genetic switching in the flagellar gene hierarchy of Caulobacter requires negative as well as positive regulation of transcription. Proc Natl Acad Sci USA. 1989;86:6651–6655. doi: 10.1073/pnas.86.17.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohta N, Swanson E, Ely B, Newton A. Physical mapping and complementation analysis of transposon Tn5 mutations in Caulobacter crescentus: organization of transcriptional units in the hook gene cluster. J Bacteriol. 1984;158:897–904. doi: 10.1128/jb.158.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohta N, Chen L-S, Swenson E, Newton A. Transcriptional regulation of a periodically controlled flagellar gene operon in Caulobacter crescentus. J Mol Biol. 1985;186:107–115. doi: 10.1016/0022-2836(85)90261-x. [DOI] [PubMed] [Google Scholar]
- 59.Ohta N, Chen L-S, Mullin D A, Newton A. Timing of flagellar gene expression in the Caulobacter cell cycle is determined by a transcriptional cascade of positive regulatory genes. J Bacteriol. 1991;173:1514–1522. doi: 10.1128/jb.173.4.1514-1522.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Onishi K, Kutsukake K, Suzuki H, Iino T. A novel transcriptional regulatory mechanism in the flagellar regulon of Salmonella typhimurium: an anti-sigma factor inhibits the activity of the flagellum-specific sigma factor, ςF. Mol Microbiol. 1992;6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- 61.Poindexter J S. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quon K C, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 63.Ramakrishnan G, Newton A. FlbD of Caulobacter crescentus is a homologue of the NtrC (NRI) protein and activates ς54-dependent flagellar gene promoters. Proc Natl Acad Sci USA. 1990;87:2369–2373. doi: 10.1073/pnas.87.6.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramakrishnan G, Zhao J-L, Newton A. The cell cycle-regulated flagellar gene flbF of Caulobacter crescentus is homologous to a virulence locus (lcrD) of Yersinia pestis. J Bacteriol. 1991;173:7283–7292. doi: 10.1128/jb.173.22.7283-7292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramakrishnan G, Zhao J L, Newton A. Multiple structural proteins are required for both transcriptional activation and negative autoregulation of Caulobacter crescentus flagellar genes. J Bacteriol. 1994;176:7587–7600. doi: 10.1128/jb.176.24.7587-7600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rapaport L R, Mackie G A. Influence of translational efficiency on the stability of the mRNA for ribosomal protein S20 in Escherichia coli. J Bacteriol. 1994;176:992–998. doi: 10.1128/jb.176.4.992-998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rizzo M F, Shapiro L, Gober J. Asymmetric expression of the gyrase B gene from the replication-competent chromosome in the Caulobacter predivisional cell. J Bacteriol. 1993;175:6970–6981. doi: 10.1128/jb.175.21.6970-6981.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanders L A, Van Way S, Mullin D A. Characterization of the Caulobacter crescentus flbF promoter and identification of the inferred flbF product as a homolog of the LcrD protein from a Yersinia enterocolitica virulence plasmid. J Bacteriol. 1992;174:857–866. doi: 10.1128/jb.174.3.857-866.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schoenlein P V, Ely B. Characterization of strains containing mutations in the contiguous flaF, flaT, or flbA-flaG transcription unit and identification of a novel Fla phenotype in Caulobacter crescentus. J Bacteriol. 1989;171:1554–1561. doi: 10.1128/jb.171.3.1554-1561.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schoenlein P V, Gallman L S, Ely B. Organization of the flaFG gene cluster and identification of two additional genes involved in flagellum biogenesis in Caulobacter crescentus. J Bacteriol. 1989;171:1544–1553. doi: 10.1128/jb.171.3.1544-1553.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schoenlein P V, Gallman L S, Winkler M E, Ely B. Nucleotide sequence of the Caulobacter crescentus flaF and flbT genes and an analysis of codon usage in organisms with G+T-rich genomes. Gene. 1990;93:17–25. doi: 10.1016/0378-1119(90)90130-j. [DOI] [PubMed] [Google Scholar]
- 73.Schoenlein P V, Lui J, Gallman L, Ely B. The Caulobacter crescentus flaFG region regulates synthesis and assembly of flagellin proteins encoded by two genetically unlinked gene clusters. J Bacteriol. 1992;174:6046–6053. doi: 10.1128/jb.174.19.6046-6053.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simon R, Piefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;1:784–790. [Google Scholar]
- 75.Stephens C, Mohr C, Boyd C, Maddock J, Gober J, Shapiro L. Identification of the fliI and fliJ components of the Caulobacter flagellar type III protein secretion system. J Bacteriol. 1997;179:5355–5365. doi: 10.1128/jb.179.17.5355-5365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stephens C M, Shapiro L. An unusual promoter controls cell-cycle regulation and dependence on DNA replication of the Caulobacter fliLM early flagellar operon. Mol Microbiol. 1993;9:1169–1179. doi: 10.1111/j.1365-2958.1993.tb01246.x. [DOI] [PubMed] [Google Scholar]
- 77.Wang S P, Sharma P L, Schoenlein P V, Ely B. A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc Natl Acad Sci USA. 1993;90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wingrove J A, Gober J W. Identification of an asymmetrically localized sensor histidine kinase responsible for temporally and spatially regulated transcription. Science. 1996;274:597–601. doi: 10.1126/science.274.5287.597. [DOI] [PubMed] [Google Scholar]
- 79.Wingrove J A, Mangan E K, Gober J W. Spatial and temporal phosphorylation of a transcriptional activator regulates pole-specific gene expression in Caulobacter. Genes Dev. 1993;7:1979–1992. doi: 10.1101/gad.7.10.1979. [DOI] [PubMed] [Google Scholar]
- 80.Wu J, Newton A. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol Microbiol. 1997;24:233–239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]
- 81.Wu J, Benson A K, Newton A. Global regulation of a ς54-dependent flagellar gene family in Caulobacter crescentus by the transcriptional activator FlbD. J Bacteriol. 1995;177:3241–3450. doi: 10.1128/jb.177.11.3241-3250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu H, Dingwall A, Shapiro L. Negative transcriptional regulation in the Caulobacter flagellar hierarchy. Proc Natl Acad Sci USA. 1989;86:6656–6660. doi: 10.1073/pnas.86.17.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu J, Shapiro L. Early Caulobacter crescentus genes fliL and fliM are required for flagellar gene expression and normal cell division. J Bacteriol. 1992;174:3327–3338. doi: 10.1128/jb.174.10.3327-3338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhuang W Y, Shapiro L. Caulobacter FliQ and FliR membrane proteins, required for flagellar biogenesis and cell division, belong to a family of virulence factor export proteins. J Bacteriol. 1995;177:343–356. doi: 10.1128/jb.177.2.343-356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]