ABSTRACT
Colorectal cancer (CRC) is the third most common malignant tumor worldwide. The incidence and mortality rates of CRC have been increasing in China, possibly due to economic development, lifestyle, and dietary changes. Evidence suggests that gut microbiota plays an essential role in the tumorigenesis of CRC. Gut dysbiosis, specific pathogenic microbes, metabolites, virulence factors, and microbial carcinogenic mechanisms contribute to the initiation and progression of CRC. Gut microbiota biomarkers have potential translational applications in CRC screening and early diagnosis. Gut microbiota-related interventions could improve anti-tumor therapy’s efficacy and severe intestinal toxic effects. Chinese researchers have made many achievements in the relationship between gut microbiota and CRC, although some challenges remain. This review summarizes the current evidence from China on the role of gut microbiota in CRC, mainly including the gut microbiota characteristics, especially Fusobacterium nucleatum and Parvimonas micra, which have been identified to be enriched in CRC patients; microbial pathogens such as F. nucleatum and enterotoxigenic Bacteroides fragilis, and P. micra, which Chinese scientists have extensively studied; diagnostic biomarkers especially F. nucleatum; therapeutic effects, including microecological agents represented by certain Lactobacillus strains, fecal microbiota transplantation, and traditional Chinese medicines such as Berberine and Curcumin. More efforts should be focused on exploring the underlying mechanisms of microbial pathogenesis of CRC and providing novel gut microbiota-related therapeutic and preventive strategies.
KEYWORDS: Gut microbiota, colorectal cancer, microbial carcinogenesis, microbial biomarkers, anti-tumor therapy, microbiota interventions
Introduction
Colorectal cancer (CRC) threatens the health and causes severe social burdens, accounting for about 10% of all new cancer cases.1 Data from the National Cancer Center of China in 2022 showed that CRC ranked fourth in incidence, fifth in mortality among male patients, third in incidence, and fourth in mortality among female patients.2 Europe, Oceania, and northern America have the highest CRC incidence and mortality rate globally. However, recent studies showed that the incidence and mortality of CRC in some regions decreased but increased in China.3 This trend might be influenced by adopting Western dietary and lifestyle patterns and the accompanying alterations in the gut microbiota.4 Improving early diagnosis and treatment and reducing the disease burden of CRC is a significant public health problem. The gut microbiota plays an essential role in CRC occurrence and progression. This review focally addresses Chinese studies on CRC and gut microbiota to describe the characteristics of the intestinal microbiota associated with CRC, the possible microbial mechanisms, the gut microbiota-related biomarkers for screening, early diagnosis, and risk warnings. It also addresses the role of the intestinal microbiota in chemoradiotherapy and immunotherapy and the application of intestinal microbiota interventions with Chinese characteristics, including microecological agents, fecal microbiota transplantation (FMT), and traditional Chinese medicine (TCM).
CRC-associated gut microbiota profiles
Intestinal bacterial microbiota and CRC
There are characteristic changes in gut microbiota in various CRC stages, including colorectal adenoma (CRA) and early and advanced CRC.5–8 The CRC cohort-specific noises, like ethnic, geographical, and genetic heterogeneity, might distort the structure of gut microbiota dysbiosis and lead to inconsistent conclusions.5 Searching for a potential core co-set of microorganisms from different cohorts that might be carcinogenic is of great interest to CRC studies.6 Therefore, current studies focused on the gut bacterial microbiota related to CRC across the discovery and validation cohorts based on different ethnic and countries. Unlike CRC patients in Austria, Germany, and France, the gut bacterial composition of CRC patients in China has a unique characteristic with a lower proportion of Firmicutes and a higher proportion of Verrucomicrobia.9 Generally, the characteristics of the intestinal bacterial microbiota of CRC in recent Chinese studies are detailed in Table 1. These findings suggest significant differences in the intestinal bacterial microbiota of Chinese CRC patients with respect to different populations,10–14 disease progression15–19 and prognosis.20,21 In the southern Chinese population, the proportion of intestinal Fusobacteria is higher than in Western populations, and CRC in this population may be linked to F. varium and other Fusobacteria species in addition to Fusobacterium nucleatum.22 Several studies from China have integrated the gut metagenomic data of CRC patients among Chinese and other ethnic cohorts and found many bacterial genera, including Fusobacterium, Parvimonas, Peptostreptococcus, Porphyromonas, Gemella, Prevotella, Solobacterium, are associated with CRC. CRC-related bacterial species include F. nucleatum, Escherichia coli, Bacteroides fragilis, P. micra, P. stomatis, P. ascharolytica, P. somerae, P. intermedia, P. anaerobius, Clostridium symbiosum, S. moorei, and others.5, 23–25 The abundance of F. nucleatum changes in the CRA stage and increases in intramucosal carcinoma and advanced CRC; the relative abundance of Atopobium parvulum and Actinomyces odontolyticus increases significantly multiple CRA and intramucosal cancer stages.26 The incidence of early-onset CRC has been increasing globally, which could display adverse clinical and histopathological features. The characteristic changes of gut microbiota associated with early-onset CRC are unclear. A few studies on the Chinese population showed that Flavonifractor plautii, Actinomyces, and Schaalia cardiffensis were the critical microbes in early-onset CRC patients.27,28 Moreover, the diversity of the intestinal bacterial microbiota in left-sided colon cancer is higher than in right-sided colon cancer, and the relative abundance of B. vulgatus and C. perfringens is higher.29
Table 1.
Characteristics of the intestinal bacterial microbiota of CRC in recent Chinese studies.
| Authors | Population | Sample collection | Microbial diversity | Significant enrichment | Significant losses |
|---|---|---|---|---|---|
| Dai et al.5 | CRC vs. controls | Feces | Decrease in the Hong Kong cohort | Bacteroides fragilis, Fusobacterium nucleatum, Porphyromonas asaccharolytica, Parvimonas micra, Prevotella intermedia, and Alistipes finegoldii | |
| Xie et al.10 | CRC vs. controls | Feces | Clostridium symbiosum and F. nucleatum | ||
| Zhang et al.11 | CRC vs. controls | Feces | No statistical differences | Peptostreptococcus stomatis, P. micra, Gemella morbillorum, P. asaccharolytica, S. moorei, F. nucleatum, Ruminococcus torques, Campylobacter rectus, Clostridium scindens, and Caproicibacterium lactatifermentans | Eubacterium eligens, Eubacterium hadrum, Eubacterium hallII, Eubacterium desmolans, Blautia faecis, Roseburia faecis, Ruminococcus lactaris, and Streptococcus salivarius |
| Yu et al.12 | CRC vs. controls | Feces | Decrease | P. micra, Solobacterium moorei, and F. nucleatum | |
| Li et al.13 | CRC vs. HC | Feces | B. fragilis | Faecalibacterium prausnitzii, Bifidobacterium adolescent, Clostridium clostridioforme, Blautia producta and R. faecis | |
| Chen et al.14 | CRC vs. HC | Feces | The number of viable colonies of Escherichia coli and Enterococcus faecalis | The number of living colonies of Campylobacter, Bifidobacterium, and Lactobacillus | |
| Yuan et al.15 | CRC vs. HC | Feces | Decrease | Bifidobacterium, Bacteroides, and Megasphaera | Collinsella and Faecalibacterium |
| Chang et al.16 | CRC vs. HC | Feces | No statistical differences | G. morbillorum and Shigella sonnei | Streptococcus gallolyticus |
| Du et al.17 | CRC vs. HC | Feces | Decrease | Bacteroidota, Fusobacteriota, and Campylobacter | Firmicutes |
| Coker et al.18 | CRC vs. HC | Feces | P. stomatis, F. nucleatum, P. micra, Peptostreptococcus anaerobius, and B. fragilis | Eubacterium ventriosum, Roseburia interinalis and Roseburia inulivorans | |
| Yeoh et al.19 | CRC vs. non-CRC | Feces | Fusobacterium gonidiaformans and F. nucleatum | ||
| Yang et al.20 | CRC vs. healthy family members | Feces | Roseburia inulinivorans, Clostridium ramosum, Porphyromonas gingivalis, F. nucleatum, and G. morbillorum | ||
| Li et al.21 | CRC | Tumor mucosa vs normal mucosa | No statistical differences | Fusobacterium, Gemella, Campylobacter, and Streptococcus | Rhodococcus, Blautia, and Dorea |
| Liu et al.7 | CRC | Tumor tissues vs. adjacent normal tissues | No statistical differences | Peptostreptococcus and Fusobacterium | |
| CRC vs. CRA | Tumor tissues | Decrease | Fusobacterium, Bacteroides, Parvimonas, and Prevotella | ||
| Feng et al.8 | CRC vs. advanced CRA or HC | Feces | No statistical differences | A. putredinis, Lachnospiraceae bacterium and E. coli | Streptococcus thermophilus |
| Hua et al.22 | CRC vs. CRA vs. HC | Feces | No statistical differences | Acidaminococcus exhibited a decreased relative abundance from HC to CRA and CRC | |
| Gao et al.23 | CRC vs. CRA vs. HC | Feces | Firmicutes exhibited a decreased relative abundance from HC to CRA and CRC | ||
| Wu et al.24 | CRC vs. CRA | Feces | No statistical differences | F. nucleatum, P. asaccharolytica, P. stomatis, P. micra, and Escherichia-Shigella sp. | Blautia obeum, Butyricicoccus faecihominis, and Dorea longicatena |
| Jin et al.25 | CRC vs. CRC postoperative | Feces | No statistical differences | Gemella, Howardella, and Parascardovia | Tyzzerella 3 and Lawsonella |
| Huang et al.26 | CRC vs. HC | Feces | Increase | Negativicutes and Clostridia | Clostridia and Negativicutes |
| CRC cases after chemotherapy vs. CRC | No statistical differences | Verrucomicrobia and Akkermansiaceae | |||
| Huo et al.27 | CRC recurrence vs. non-CRC recurrence | Tumor tissues | No statistical differences | Firmicutes, Bacteroidetes, and Fusobacteria | |
| Off-tumor tissues | Firmicutes and Bacteroidetes | ||||
| Nakatsu et al.28 | early-stage CRC vs. late-stage CRC | Lesion-adjacent mucosae | No statistical differences | Fusobacterium, Parvimonas, Gemella, and Leptotrichia | Bacteroides, Blautia, Faecalibacterium prausnitzii, Collinsella aerofaciens and Alistipes putredinis |
| Xu et al.29 | young-onset CRC vs. old-onset CRC | Tumor tissues | Decrease | Schaalia cardiffensis, Diaphorobacter, Actinomyces, Actinomycetaceae, and Actinomycetales | Acidobacteriota and Pyramidobacter piscolens |
| Yang et al.30 | young-onset CRC vs. old-onset CRC | Feces | Increase | Fusobacterium, Flavonifractor, and Odoribacter | Streptococcus, Fusobacterium, and Gemella |
| Zhong et al.31 | Left-sided colon cancer vs. right-sided colon cancer | Tumor tissue and feces | Increase | Clostridium perfringens, Bacteroides coprocola DSM 17,136, Collinsella aerofaciens, and S. gallolyticus subsp. Macedonicus in patients from Harbin | Bifidobacterium dentium and Ruminococcus sp. 15975 in patients from Harbin |
| Bacteroides vulgatus in patients from Xiamen | B. fragilis and S. gallolyticus subsp. Macedonicus in patients from Xiamen |
CRC: colorectal cancer; CRA: colorectal adenoma; HC: healthy controls.
Gut mycobiota and viral microbiota in CRC
The enteric mycobiota is an essential component of the intestinal microbiota that is relatively unexplored.30 High-throughput sequencing and bioinformatics techniques demonstrated the ecologic association of gut mycobiota with the pathogenesis of human diseases.31 Recent studies identified dysbiosis of intestinal fungal microbiota in CRC.32–34 The ratio of Basidiomycota/Ascomycota and the abundance of Malassezia are increased, Saccharomyces and Pneumocystis are decreased, six genera are enriched, and the abundances of 38 species are changed.33 S. cerevisiae is a probiotic absent in the intestine of CRC patients; previous studies showed that S. cerevisiae delays CRC in APCmin/+ mice.33,35 A meta-analysis of seven datasets found that the abundance of six fungi, including Aspergillus rambelli, was increased in CRC patients, and subsequent in vitro and in vivo experiments showed that A. rambelli promotes the growth of CRC cells in mice.32 That study found that intestinal fungal-fungal and fungal-bacterial interactions are significantly enhanced in CRC and positively correlated with the course of CRC.32 The SYK/CARD9 signaling pathway protects against CRC by restricting the gut mycobiota-mediated expansion of myeloid-derived suppressor cells.36 Studies are needed to clarify the mechanism of intestinal mycobiome participating in the carcinogenesis of CRC.
The enteric virome, an essential component and regulator of gut microflora, affects the intestinal microbiota’s structure and abundance, potentially impacting CRC occurrence, progression, and outcomes by altering bacterial-host communities.37–39 The abundance and diversity of gut viral microbiota such as Siphoviridae, Myoviridae, and Podoviridae increase significantly, and Herelleviridae is significantly depleted in CRC patients.37 Intestinal viral dysregulation is associated with early and late stages of CRC, and viruses such as Betabaculovirus, Punalikevirus, and Mulikevirus were associated with clinical outcomes.38 A meta-analysis of metagenomic data showed that bacteriophages of Porphyromonas, Fusobacterium, and Hungatella were enriched in CRC patients.40 Another study found that F. nucleatum, Peptacetobacter hiranonis, and P. micra bacteriophages are abundant in CRC patients.41 Remodeling the bacteriophage structure might have potential therapeutic value in CRC prevention and treatment. M13 bacteriophage binds to F. nucleatum and transforms into M13@Ag, improving the tumor immune microenvironment and inhibiting the proliferation of immunosuppressive myeloid-derived suppressor cells; it acts with the immunosuppressive agent PD-L1 or chemotherapeutic agents to prolong the survival of tumor-bearing mice significantly.42 Intestinal bacteriophages have potential applications in screening CRC-related biomarkers and treating drug resistance associated with anti-tumor therapy. The interactions of gut viral microbiota with bacterial microflora and human host and their carcinogenic and anti-tumor effects in CRC need further investigation.
Intestinal microbial metabolites associated with CRC
The dietary pattern of the Chinese has been plant-based (high in cereal and fiber) in the past few decades. However, the consumption of sugar, fat, and animal-source foods has been increasing among the Chinese population in recent years, which may be associated with an increased incidence of CRC.43 Gut microbiota could directly interact with dietary compounds like dietary fiber and produce some metabolites such as short-chain fatty acids (SCFAs) and bile acids (BAs). The change in dietary patterns might alter the gut microbiota and its metabolites. Then, the disrupted gut microbiota-related metabolites contribute to CRC carcinogenesis.44
SCFAs
SCFAs, including butyrate, propionate, and acetate, are metabolites generated from the gut microbial fermentation of insoluble dietary fiber and build connections among dietary patterns, gut microbiota, and intestinal function. Many bacteria, such as Clostridium and Bifidobacterium, could transform dietary fiber into SCFAs.45 SCFAs serve as energy substrates of colonic epithelial cells and maintain intestinal homeostasis through several vital biological processes. For example, SCFAs could regulate the metabolism of colonic epithelial cells, enhance gut barrier function, and modulate the immune response of the intestine, and the activation of G protein-coupled receptors and the inhibition of histone deacetylase activity may be the potential mechanisms.46
A high-fiber diet could prevent and reduce the occurrence of CRC, which was associated with the increased abundance of SCFAs-producing bacteria and the raised level of SCFAs in the intestine.46 Moreover, a low-fiber diet results in a lack of butyrate-producing bacteria and a lower level of butyrate in the gut observed in CRA and CRC patients. However, the interaction among SCFAs, the gut microbiota, and CRC is complex. SCFAs could antagonize the proliferation, accelerate the apoptosis of CRC cells, and suppress the inflammation-related CRC carcinogenesis pathways.46 SCFAs reduce the burden of carcinogens such as BAs by activating drug-metabolizing enzymes and inhibiting the degradation of primary BAs to secondary BAs.47 SCFAs also serve as a tumor suppressor to modulate gene expression through epigenetic effects.48 In addition, the dysregulation of SCFAs glucose metabolism could be initiated through the intestinal dysbiosis of CRC, and the interaction between SCFAs transporters and glycolysis might correlate with the initiation and progression of CRC.49
Gut microbiota-derived SCFAs can be potentially applied in CRC prevention and treatment. Supplementation of dietary fiber or postbiotics containing SCFAs might play an essential role in CRC prevention;50 the appropriate formulation and amounts and the efficacy evaluation should be further investigated. Furthermore, several preclinical studies showed that SCFAs-producing bacteria and butyrate could enhance the efficacy of chemotherapy drugs such as oxaliplatin and irinotecan and improve the response to immunotherapy;51 and then SCFAs could alleviate the adverse effect caused by chemotherapeutic adverse effects such as colitis. Studies based on animal models showed the potential value of intestinal microbiota-derived SCFAs in CRC.52 In-depth investigations are needed before the clinical transformation and application of SCFAs in CRC prevention and treatment.
BAs
Individuals who consume high-fat diets have a higher incidence of CRC. Western diets, including high-fat, red, and processed meat, increase the level of fecal secondary BAs, mostly deoxycholic acid and lithocholic acid. The secondary BAs metabolized by gut microbiota are carcinogenetic factors in CRC development.53 Clostridium and Eubacterium mediate BA-related 7α-dehydroxylation and sulfidation. Elevated levels of secondary BAs exert detrimental tumor-promoting effects via mechanisms such as colonic epithelial barrier function injury, oxidative damage to DNA, inflammation, activation of NFκB signaling pathway, and increased cell proliferation.54 Studies also showed that cholecystectomy might be associated with CRC, which may be due to the disruption of BAs secretion and intestinal microbiota imbalance.55 BAs modulate the intestinal microbiota, which regulates the BA pool; the disruption of BA-gut microbiota crosstalk contributes to CRC development. However, the synthesis, transport, and metabolism of BAs and the mechanisms of BA-gut microbiota interactions involved in carcinogenesis remain unclear and should be further investigated. Moreover, the modulation of gut microbiota, BAs, and BA-related receptor signaling pathways (such as farnesoid X receptor and G protein-coupled bile acid receptor 1) might be novel therapeutic targets and research frontiers for CRC.56
Hydrogen sulfide (H2S)
H2S is generated from the degradation of food-derived nutrients in the intestine by sulfate-reducing bacteria (SRB) such as Desulfovibrio spp. H2S promotes CRC occurrence and development by damaging intestinal epithelial cells, inducing free radical release, DNA damage, and colonic mucosal inflammation, inhibiting butyrate oxidation, cytochrome oxidase, and DNA methylation57. SRB-related intestinal microbial sulfur metabolism is a potential trigger of CRC. A high-fat and high-protein diet can promote the growth of SRB, metabolizing a large amount of genotoxic H2S related to CRC progression.58 There are elevated intestinal H2S levels in CRC patients, and endogenous and microbial H2S produced by cysteine at CRC sites show an upward trend.59
The microbial pathogenesis of CRC
CRC carcinogenesis involves genetic and environmental factors. Genetic mutations, epigenetic changes, inflammation, immune regulation, and metabolic and hormonal disorders contribute to CRC. The main pathway in colorectal carcinogenesis is the “adenoma-carcinoma sequence,” which describes that most CRC cases are sporadic and progress slowly from normal to dysplastic epithelium to carcinoma throughout years.60 Besides, the gut microbiota is the essential environmental factor contributing to the initiation, progression, metastasis, and CRC outcomes. The intestinal microbiota of CRC patients promotes colorectal dysplasia and tumor formation in a germ-free mouse model, providing evidence for the gut microbiota’s involvement in CRC progression.61 According to available studies, the gut microbiota-related carcinogenic mechanisms of CRC involve an imbalance of intestinal microflora, invasion and colonization of pathogenic microbes, and impaired intestinal barrier function induced by microbial metabolites and virulence factors. The tumor microenvironment is altered by inducing the secretion of pro-inflammatory cytokines and chronic inflammation surrounding intestinal epithelial cells, activating the Wnt/β-catenin and NF-κB pro-inflammatory signaling pathways and exacerbating intestinal epithelial cell DNA damage, ultimately initiating CRC progression (summarized in Figure 1).
Figure 1.
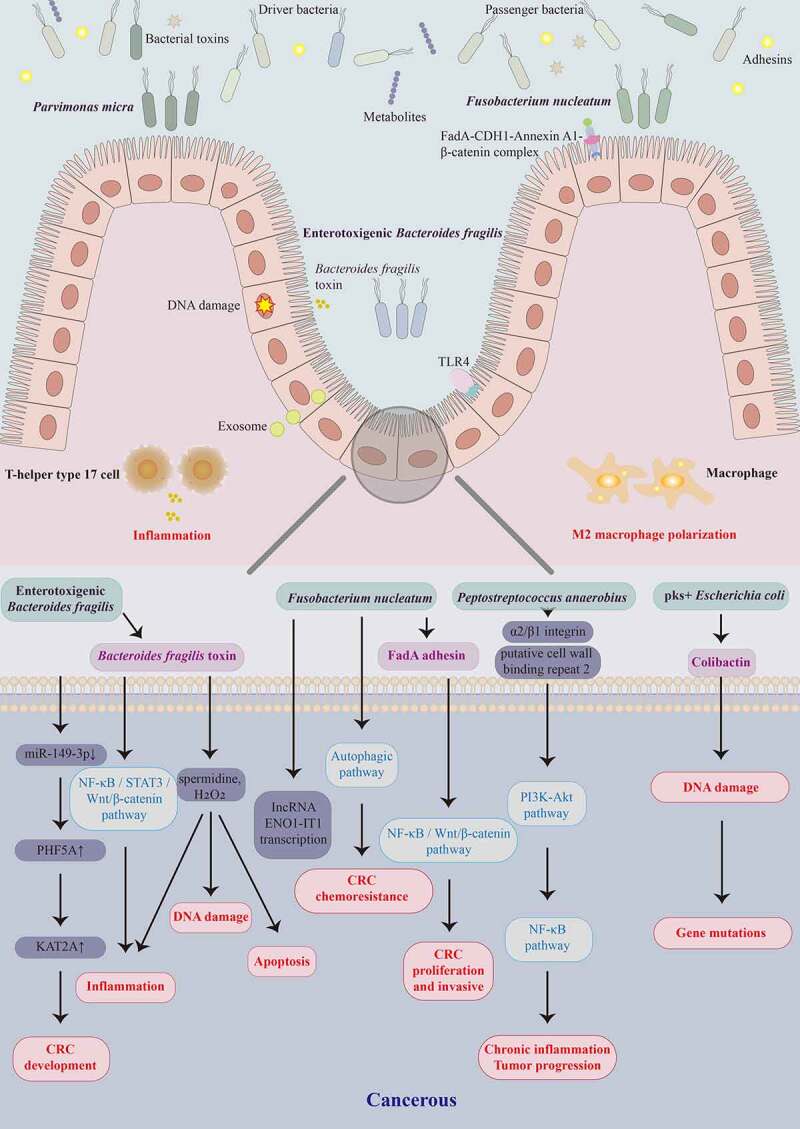
Schematic representation of gut microbiota and their metabolites involved in colorectal carcinogenesis at the cellular and molecular levels. Gut bacteria such as Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis, Peptostreptococcus anaerobius, pks+ Escherichia coli, and Parvimonas micra, and their virulence factors FadA, B. fragilis toxin, and colibactin contribute to the colorectal cancer (CRC) development by activating different pathways that trigger DNA damage, intestinal inflammation, macrophage polarization, and apoptosis.
There are two models (“alpha-bug” and “driver-passenger”) of intestinal microbial carcinogenesis related to CRC.62 The alpha-bug model holds Helicobacter pylori in gastric cancer and hepatitis B virus in primary liver cancer stimulates the host to produce inflammatory and immune responses, proto-oncogene activation, and tumor suppressor gene inhibition.62 Enterotoxigenic B. fragilis (ETBF) -related enterotoxin compromises intestinal epithelial permeability, activation of several tumor-initiating relevant Wnt/β-catenin and STAT3 pathways, and immune responses from persistent Th17 activation, leading to CRC progression.6 The abundance of these so-called promoter bacterial microbes significantly increases in cancer or para-cancerous tissues; however, they appear to decrease during CRC progression. The alpha-bug model does not explain the increased abundance of undifferentiated gut microflora in the early stage of CRC progression. Therefore, a driver-passenger model was proposed to explain the complex changes in the intestinal microbiota associated with CRC.63 For example, E. coli and ETBF would be driver bacteria that induce DNA damage in intestinal epithelial cells and trigger inflammation to initiate carcinogenesis. These organisms form a unique tumor microenvironment conducive to increasing opportunistic pathogens called passenger bacteria. These passenger bacteria, such as Streptococcus bovis, S. gallolyticus, Roseburia, and Fusobacterium, might participate in cancer promotion and even competitively inhibit driver bacteria growth. On the other hand, the traditional “adenoma-carcinoma sequence” is the primary pathway for CRC occurrence and development, and the other pathway of colorectal serrated lesions;60 the former involves APC, KRAS, BRAF, TP53, SMAD4, and other gene mutations, while the latter involves CpG island methylation phenotype, MLH1, MGMT, and MSI.64
Research has demonstrated that specific “oncomicrobes” promote CRC initiation and progression, with F. nucleatum, ETBF, pks+ E. coli, and P. micra being extensively studied.6 F. nucleatum activates the TLR4/KEAP1/NRF2 axis and the NF-κB signaling pathway, accompanied by inducing M2 macrophage polarization and macrophage infiltration in the tumor microenvironment to promote tumor growth.65 In light of this evidence, a “two-hit” model has been proposed with pro-carcinogenic features that may initiate CRC development, in which somatic mutations are the first hit and F. nucleatum is the second, exacerbating CRC development after benign cells become cancerous.66 Several Chinese research teams contribute to elucidating the gut microbiota-mediated pathogenesis of CRC. Fang et al. from Shanghai found that F. nucleatum could activate IncRNA ENO1-IT1 transcription to promote CRC initiation and development and activates the autophagic pathway to promote CRC chemoresistance,67,68 while enterotoxigenic B. fragilis promotes Th17 cell differentiation through downregulation of miR-149-3p. The team proposed for the first time that gut microbiota and host co-metabolism regulate the gut immune microenvironment. The gut microbiota promotes colorectal carcinogenesis by mediating the host urea cycle and disrupting intestinal immune homeostasis.69 Furthermore, Yu et al. from Hong Kong focused on the microbial-mediated pathways in CRC. P. micra induces tumors in mice by inducing colorectal cell proliferation and modulating the Th17 immune response,70 while P. anaerobius promotes colorectal tumorigenesis by binding to integrin α2/β1 and activating NF-κB signaling pathway.71
Additionally, many studies focused on the characteristics of the intestinal microbial virulence factors associated with CRC. Gut dysbiosis is carcinogenic through intestinal microbial-associated virulence factors that induce inflammation or immunosuppression. The genetic toxicity of intestinal microbial-associated virulence factors leads to host DNA damage, also involved in CRC initiation and development. The unique FadA adhesin secreted by F. nucleatum binds to E-cadherin (CDH1) and forms a FadA-CDH1-Annexin A1-β-catenin complex that activates Wnt/β-catenin signaling to promote initiation and progression of CRC.72,73 F. nucleatum adheres to E-cadherin on the surface of CRC cells and activates the Wnt/β-catenin and NF-κB inflammatory pathways, increasing the proliferation, and invasive activities of CRC cells, promoting metastasis.74 The Fap2 protein of F. nucleatum directly interacts with TIGIT, leading to the bacterium-dependent tumor immune evasion mechanism.75 ETBF, a subtype of B. fragilis, secretes B. fragilis toxin (BFT) to activate the NF-κB, STAT3, and Wnt/β-catenin signaling pathways in colonic epithelium cells, triggering inflammation responses and causing excessive cell proliferation; in this way, ETBF promotes carcinogenesis in CRC.76 BFT produces spermidine and H2O2 as by-products of polyamine catabolism, leading to apoptosis, inflammation, and DNA damage.77 Colibactin produced by pks+ E. coli strain induces DNA double-strand breaks in host cells through its deoxyribonuclease activity, activating DNA damage signaling cascades and increasing the frequency of gene mutations associated with CRC development.78 Nevertheless, the microbial-associated pathogenesis of CRC and CRA remains to be elucidated. Notably, the research into the pathogenesis of CRC is not limited to several novel pathogenic microbes and their virulence factors to promote CRC initiation and development but continues to advance and deepen.
Gut microbiota-related biomarkers for the screening and early CRC diagnosis
Although the incidence rate of CRC in China is lower than that in Europe and northern America, the number of CRC patients is the largest globally for the large population.3 Most CRC patients are in the middle or late stage of diagnosis and lose the opportunity for surgery, leading to poor outcomes and high mortality rates. The overall burden for CRC diagnosis and treatment remains severe globally. The recent China guideline for the screening and early detection of CRC has recommended that individuals with a high risk of CRC should undergo screening at 40 years of age, and the optimal age for starting CRC screening is 45 years old.79 Colonoscopy is regarded as the gold standard for CRC screening, which could reduce the incidence and mortality of CRC.80 However, as an invasive procedure, colonoscopy is challenging as a large-scale screening method.81 The coverage and participation rate were also lower in China than in America, which limited the effectiveness of nationwide CRC screening.3 In addition, fecal occult blood testing, fecal immunochemical test (FIT), and blood DNA methylation biomarkers are the recommended noninvasive screening methods, which are superior to colonoscopy regarding clinical compliance and cost-effectiveness; nevertheless, they still have limited sensitivity for advanced CRA and CRC.82 Longitudinal studies involving various stages of CRC and cross-sectional studies of different CRC cohorts suggested that the intestinal microbiota is specific in CRC and its precancerous lesions.5,24,83 This finding provides direct evidence for the potential role of microbial biomarkers in CRC screening and early detection. With the development of multi-omics and bioinformatics artificial intelligence algorithms, innovating noninvasive intestinal microbiota-related biomarkers have become an essential research field in CRC screening and early diagnosis. The intestinal microbiota-related biomarkers for the screening and early diagnosis of CRC in recent Chinese studies are presented in Table 2.
Table 2.
Gut microbiota-related biomarkers for the screening and early diagnosis of CRC in recent Chinese studies.
| Authors | Cohort | Gut microbiota-related biomarkers | AUC |
|---|---|---|---|
| Bacterial-related biomarkers | |||
| Gao et al.23 | CRC vs. CRA | the combined panel of 12 species (Parvimonas unclassified, Fusobacterium nucleatum, Roseburia intestinalis, Gardnerella vaginalis, Peptostreptococcus anaerobius, Peptostreptococcus stomatis, Lactobacillus iners, Gemella morbillorum, Porphyromonas asaccharolytica, Faecalibacterium prausnitzii, Atopobium vaginae, and Parvimonas micra) and three metabolites | 0.994 |
| Coker et al.18 | CRC vs. NC | 11 metabolite biomarkers and six bacterial species (F. nucleatum, P. anaerobius, P. micra, Roseburia inulinivorans, Eikenella corrodens, and Xanthomonas perforans) | 0.9417 |
| Yang et al.20 | CRC vs. NC | gene 8,122,329 (unknown function from Coprobacillus) | 0.930 |
| Chen et al.85 | CRC vs. CRA | Eight gut microbiome-associated serum metabolites | 0.92 |
| Wu et al.24 | CRC vs. CRA | 24 differential biomarkers (Streptococcus thermophilus TH1435, P. micra, Bacteroides dorei, Clostridium scindens, Erysipelatoclostridium ramosum, Blautia sp., Eubacterium coprostanoligenes group sp., Lachnospira pectinoschiza, Ruminococcaceae UCG-002 sp., Ruminococcus bromii, Porphyromonas sp. HMSC077F02, Porphyromonas sp. 2007b, Eubacterium ruminantium, Tyzzerella 3 sp., Hungatella hathewayi WAL-18680, Blautia faecis, Bacteroides nordii, Lachnospiraceae UCG-010 sp., Eubacterium ventriosum group sp., Streptococcus infantarius, R. intestinalis, Merdibacter massiliensis, Ruminococcus gnavus group sp., Roseburia hominis A2–183) with age and body mass index | 0.89 |
| Xie et al.10 | CRC vs. NC | Combination of Clostridium symbiosum and F. nucleatum abundance with FIT and CEA test | 0.876 |
| Hua et al.22 | CRC vs. CRA | Combination of the top ten species (Butyricimonas synergistica, Agrobacterium larrymoorei, Bacteroides plebeius, Lachnospiraceae bacterium feline oral taxon 001, C. scindens, Prevotella heparinolytica, bacterium LD2013, Streptococcus mutans, L. bacterium 19gly4, and Eubacterium hallii) | 0.8554 |
| Yu et al.12 | CRC vs. NC | Two gene biomarkers (from F. nucleatum and P. micra) | 0.84 |
| Dai et al.5 | CRC vs. NC | 7 CRC-enriched bacterial biomarkers (B. fragilis, F. nucleatum, P. asaccharolytica, P. micra, Prevotella intermedia, Alistipes finegoldii, and Thermanaerovibrio acidaminovorans) | 0.80 |
| Liang et al.86 | CRC vs. NC | A simple linear combination of four bacteria (F. nucleatum + Clostridium hathewayi + undefined species (labeled as m7) - Bacteroides clarus) | 0.886 |
| Fungal-related biomarkers | |||
| Coker et al.36 | CRC vs. NC | Fourteen fungal biomarkers (Aspergillus flavus, Kwoniella mangrovensis, Pseudogymnoascus sp. VKM F-4518, Debaryomyces fabryi, Aspergillus sydowii, Moniliophthora perniciosa, Kwoniella heavenensis, Aspergillus ochraceoroseus, Talaromyces islandicus, Malassezia globosa, Pseudogymnoascus sp. VKM F-4520, Aspergillus rambelli, Pneumocystis murina, and Nosemia apis) | 0.93 |
| Gao et al.37 | CRC vs. NC | Thirteen fungal biomarkers (Phanerochaete chrysosporium, A. flavus, Entomophthora muscae, A. rambelli, fungal sp ARF18, Metschnikowia cubensis, Fusarium pininemorale, Spraguea lophii, Candida versatilis, Lachancea waltii, Taxomyces andreanae, Exophiala mesophila, and Brettanomyces anomalus) | 0.757 |
| Viral-related biomarkers | |||
| Nakatsu et al.41 | CRC vs. NC | Twenty-two viral biomarkers (Orthobunyavirus, Inovirus, Tunalikevirus, L5likevirus, Phikzlikevirus, Betabaculovirus, Sp6likevirus, Sfi21dtunalikevirus, Punalikevirus, Lambdalikevirus, C2likevirus, Mulikevirus, Twortlikevirus, γ-Sphaerolipovirus, Circovirus, Spounalikevirus, Cytomegalovirus, Epsilon15likevirus, N15likevirus, Phikmvlikevirus, Cyprinivirus, and Lymphocryptovirus) | 0.802 |
| Multi-kingdom biomarkers | |||
| Lin et al.35 | CRC vs. NC | Five fungi (Aspergillus kawachii, Rhizophagus clarus, Baudoinia panamericana, A. rambelli, Trichoderma atroviride) and nine bacteria (P. micra, F. nucfeatum, G. morbillorum, Porphyromonas asaccharolytica, Dialister pneumosintes, Anaerostipes hadrus, Romboutsia ilealis, Brachyspira pilosicoli, and Streptococcus safivarius) | 0.9002 |
| Liu et al.9 | CRC vs. NC | 11 bacterial (F. nucleatum, P. micra, G. morbillorum, Ruminococcus bicirculans, R. intestinalis, Pseudobutyrivibrio xylanivorans, Streptococcus anginosus, and Eubacterium eligens), four fungal (A. rambelli, Sistotremastrum suecicum, T. islandicus, and Aspergillus niger) and one archaeal (Pyrobaculum arsenaticum) biomarkers | 0.83 |
AUC: area under the receiver-operating characteristics curve; CRC: colorectal cancer; CRA: colorectal adenoma; NC: normal controls; rpoB: RNA polymerase subunit β; FIT: fecal immunochemical test; CEA: carcinoembryonic antigen.
Bacterial biomarkers
Several recent studies have focused on gut microbial-related biomarkers for noninvasive diagnosis of CRC and its precancerous lesions,24,83–85, with significant studies by Chinese scientists. F. nucleatum is the leading candidate bacterial biomarker for early diagnosis, risk, and outcomes prediction.84,85 F. nucleatum abundance in patients with CRC and its precancerous lesions was significantly higher than in the control group, and the level of F. nucleatum was more elevated in cancer and para-cancerous tissues in CRC.86 Changes in the ratio of F. nucleatum to Faecalibacterium prausnitzii and Bifidobacterium in stool samples from CRC patients might identify early CRC.84 A Chinese study found that detecting F. nucleatum DNA in oral saliva was superior to traditional tumor markers such as CEA and CA-199 in diagnosing CRC, and F. nucleatum DNA level was related to overall survival and was an independent factor for predicting CRC outcomes.87 Subsequently, quantitative PCR analysis in another Chinese cohort identified F. nucleatum-related butyryl-CoA dehydrogenase and P. micra-related rpoB as discriminators of CRC patients with an area under the ROC curve (AUC) of 0.84; these genes were enriched in patients with early CRC, suggesting that bacteria-related gene markers have the potential for early CRC diagnosis.24 Compared with the culture and identification of specific microbial strains, the quantitative analysis of CRC-specific bacterial genes might improve the sensitivity and specificity of CRC screening and early diagnosis, reducing the cost and presenting higher clinical operability. For the bacterial gene marker m3 based on F. nucleatum, Lachnochlostridium sp., and C. hathewayi, the sensitivity and specificity for distinguishing CRA were superior to F. nucleatum or the FIT method. The m3 gene combined with F. nucleatum, C. hathewayi, B. clarus, and FIT had a higher diagnostic ability for CRC.88 Furthermore, Fang et al. found that FIT combined with quantitative PCR detection of fecal F. nucleatum DNA improved the sensitivity and specificity in diagnosing and predicting CRC.89 F. nucleatum IgA and IgG antibody-based assays combined with traditional tumor markers such as CEA and CA-199 are also used for CRC screening.90 Combining gut microbiota-related biomarkers with conventional approaches such as FIT to improve the efficiency of screening and early diagnosis for CRC is essential to the clinical translational application of gut microbiota.88,89
In addition to F. nucleatum, other intestinal bacterial biomarkers are used for screening for CRA and early CRC. Some studies analyzed the intestinal metagenomic characteristics of CRA, early to advanced CRC at different stages, and revealed characteristic changes of the intestinal microbiota at the precancerous stage of CRC.26 Fang et al. validated a set of gut microbiota-based diagnostic models developed to diagnose laterally spreading tumors, the primary precursor lesions of CRC, with an AUC of 0.92.91 Besides, Yu et al. identified four CRC-enriched bacterial-related genes in Chinese exploratory and Danish validation cohorts and further validated them in French and Australian populations with AUCs of 0.72 and 0.77, respectively.24 A.parvulum and A.odontolyticus were only significantly increased in multiple CRA and intramucosal carcinoma, suggesting that there are specific microbial biomarkers for early screening of CRC precancerous lesions.26 A random forest classifier was constructed with 11 bacterial phylotypes to distinguish CRA from the control group. A classifier with 26 bacterial phylotypes was constructed to distinguish CRC from CRA with an AUC of 0.89; this classifier was validated in two independent cohorts with AUCs of 0.78 and 0.84, respectively, and these findings were confirmed in a new cohort using real-time quantitative PCR.83 In addition to screening and early diagnosis of CRC, Wei et al. also validated a set of gut microbiota-based diagnostic models to distinguish the newly developed adenomas from the healthy subjects, which may help prevent cancer recurrence in postoperative patients.92
In recent years, multi-omics research showed the potential of intestinal microbial metabolites in CRC screening and early diagnosis.43,93,94 One study developed an intestinal microbiome-related eight serum metabolites panel to predict CRC and CRA with an AUC of 0.92 in the validation cohort, which was significantly better than tumor markers such as CEA, showing the potential application value of serum metabolites associated with intestinal microbiota in identifying CRC and CRA.93 Through fecal metagenomic and metabolomic analysis of CRA and CRC, at different stages from CRA to CRC, Yu et al. found that norvaline and myristic acid showed an increasing trend, and 20 CRC-enriched metabolites distinguished CRC from CRA and healthy control; further integration of microbial metagenomic data showed that the ability to distinguish CRC from CRA and healthy control was improved with AUC of 0.94 and 0.92 respectively.44 Moreover, a few studies from China tried to find the gut microbial biomarkers for early-onset CRC.27,28,94 One study showed that multi-omics signatures of early-onset CRC might be associated with enriched F. plauti and increased tryptophan, BAs, and choline metabolism. They constructed a microbiome-derived predictive model based on metagenomic, metabolomic, and KEGG orthology gene markers to detect early-onset CRC.94 Gut microbial-associated biomarkers have shown significant advantages in the noninvasive diagnosis of CRC, although further validation studies are needed before these biomarkers can be used in clinical practice.
Viral and fungal biomarkers
Several studies focused on the gut virome- and mycobiome-associated biomarkers for CRC.33,38,39 The virome biomarker, including 22 viral metagenomic taxa, distinguished CRC patients from control subjects across discovery and validation cohorts (AUC = 0.8).38 Moreover, significant changes in intestinal mycobiome were associated with CRC, with an increased abundance of Malassezia and decreased Saccharomycetes and pneumocystis. Fourteen fungal microbes distinguished CRC with an AUC of 0.93.33 Combining fungal and bacterial biomarkers might be more potent for diagnosing CRC; combining A. rambelli and F. nucleatum improved diagnostic accuracy by 1.4%–10.6%.32
The specificity and sensitivity of biomarkers identified from the intestinal microbiota are superior to existing clinical methods for CRC screening and early diagnosis. The gut bacterial, fungal, and viral biomarkers for CRC have the potential for clinical translational application; nevertheless, further investigation is needed. Integrating and optimizing intestinal microbiota-related biomarker panels with traditional clinical methods can improve CRC screening and diagnosis levels.
Gut microbiota and anti-tumor therapy in CRC
Intestinal microbiota dysbiosis and specific pathogenic microbes have carcinogenic and pro-tumorigenic effects. Substantial evidence shows that the gut microbiota is associated with the efficacy or adverse effects of anti-tumor treatments such as chemoradiotherapy and immunotherapy.95 The gut microbiota is a potential biomarker to predict anti-tumor responses and adverse reactions. The gut microbiota regulates the efficacy of anti-tumor therapy and clinical CRC outcomes.
Chemotherapy
The intestinal microbiota influences the efficacy of chemotherapy, drug resistance, and intestinal-associated toxicity. The gut microbiota mediates the anti-tumor effects of chemotherapeutic drugs (e.g., 5-FU, gemcitabine, and platinum) through various mechanisms, including translocation, immunomodulation, metabolism, enzymatic degradation, reduced diversity, and ecological variation.96 The gut microbiota is related to the efficacy of chemotherapy for CRC.97,98 Patients with CRC who respond to chemotherapy have a relatively high abundance of Sutterella and Roseburia; a high abundance of Fusobacterium indicates non-response and poor clinical outcomes.99,100 Moreover, the gut microbiota might be involved in chemotherapy resistance. F. nucleatum is enriched in tumor tissues of recurrent CRC patients after chemotherapy, and the abundance of F. nucleatum negatively correlates with the CRC outcomes. F. nucleatum induces drug resistance to platinum and 5-FU in CRC by affecting tumor cell autophagy.68 A study showed that metronidazole reduced the amount of F. nucleatum and shrank the tumor body, suggesting the application of anti-F. nucleatum before chemotherapy in F. nucleatum-positive CRC patients might enhance chemotherapeutic efficacy.101 F. nucleatum is emerging as a microbial biomarker for predicting chemotherapeutic efficacy, drug resistance, and outcomes and is becoming a therapeutic target for CRC. The composition and function of the gut microbiota are altered by cytotoxic chemotherapeutic agents, further affecting microbial-associated drug metabolism.102 The extent of tumor regression in tumor-bearing mice treated with platinum-based chemotherapy is significantly lower after antibiotic treatment, and germ-free mice do not respond to platinum treatment.103 Platinum induces reactive oxygen species (ROS) aggregation, the primary anti-tumor mechanism of platinum. However, no apparent ROS aggregation was observed in germ-free mice or tumor-bearing mice models treated with antibiotics, suggesting that the intestinal microbiota is involved in platinum-induced DNA damage and tumor cell apoptosis.104 The intestinal microbiota assists platinum in activating tumor-related inflammatory cells to generate ROS, enhancing the role of platinum in killing tumor cells.103 The influence of intestinal microbial-related metabolites on chemotherapy should not be ignored. For example, butyric acid enhances the anti-tumor cytotoxic effect of CD8+T cells in vitro and promotes the anti-tumor effect of platinum-based chemotherapeutic drugs.51 Supplementation of probiotics alleviates 5-FU and platinum-related intestinal injuries and adverse reactions.105 Lactobacillus supplementation during chemotherapy in CRC patients treated with 5-FU reduces mortality and improves gastrointestinal symptoms such as diarrhea.106 Furthermore, the gut microbiota modulates irinotecan’s metabolism and adverse effects. Irinotecan is a prodrug of SN-38 used to treat CRC. The incidence of irinotecan-related grade 3–4 diarrhea is 20%-40%, which affects its anti-tumor efficacy. SN-38 is glucuronidated to SN-38 G by liver enzymes and excreted to the intestine, where intestinal bacterial β-glucuronidase hydrolyzes SN-38 G to SN-38, resulting in intestinal mucosal injury and diarrhea.96,107 Probiotic supplementation improves diarrhea and intestinal mucosal damage associated with irinotecan treatment.108 The development of intestinal microbiota biomarkers for predicting the efficacy of CRC chemotherapy and microbial interventions for improving the chemotherapeutic efficacy and adverse reactions need to be further studied.
Radiotherapy
The intestinal microbiota might play a specific role in CRC radiotherapy, influencing radiotherapy’s efficacy and participating in adverse intestinal events such as mucositis and radiation enteritis. Radiotherapy causes significant variation in the structure, diversity, and abundance of the intestinal microbiota, characterized by decreased abundance of Firmicutes and an increased abundance of Proteus, Ackermann, Fusobacteria, and Bacteroides.102 The abundance of pathogenic microbes in advanced CRC patients after radiotherapy is significantly reduced, while the abundance of beneficial microbes such as Lactobacillus and Streptococcus is increased. The increase of beneficial bacteria may be related to the efficacy of radiotherapy. Bacterial taxa such as Thermi and Sphingomonadaceae identify patients with advanced rectal cancer who have achieved complete pathological response and non-pathological complete response after neoadjuvant chemoradiotherapy.109 Oral microbiota alterations influence the gut bacterial composition within CRC tumors, and oral bacteria such as F. nucleatum migrate to the CRC locus and impair radiotherapy’s therapeutic efficacy and outcomes.110
On the other hand, exposure to high-energy radiation causes intestinal mucosal barrier damage and intestinal crypt cell apoptosis, leading to intestinal dysbiosis and increasing the risk of radiation enteritis.102 After radiotherapy, the high abundance of Clostridia, Rothia, and Koalas might be related to intestinal epithelium inflammatory reactions and barrier dysfunction, which lead to radiation enteritis.111 Regulating intestinal microbiota by supplementing probiotics and FMT might effectively prevent and treat radiation enteritis.112 Bifidobacterium, Acidophilus, Streptococcus, and L. casei alleviate the severity of radiation enteritis.113 The intestinal microbiota is expected to be an interventional target for radiation enteritis.
Immunotherapy
Immunotherapy has been used in hematologic malignancies and immune-sensitive tumors. Immune checkpoint inhibitors (ICIs) have been explored in treating CRC in a subset of CRC patients with MSI-H phenotype.114 For metastatic patients with specific subtypes such as MSI-H or DNA mismatch repair deficiency, immunotherapy can achieve a more sustained therapeutic response,115 and combined use of ICIs prolongs survival in patients with advanced refractory CRC.116 Improving the curative effect and reducing intestinal toxicity is a critical problem in CRC immunotherapy.
Cancer immunotherapy involves the gut microbiota, which plays an essential role in the complex interactions among tumors, the immune system, and ICIs.117 The intestinal microbiota is associated with the efficacy of CRC immunotherapy. ICIs did not inhibit CRC progression in germ-free mice, and the antibiotic treatment weakened the anti-tumor effect of ICIs in mice with normal intestinal microflora.117 The effectiveness of ICIs depends on intestinal microbes such as A. muciniphila, B. fragilis, Bifidobacterium sp., Eubacterium limosum, Faecalibacterium sp., Prevotella sp., and Alistipes shahii. A preclinical study showed that A. muciniphila and Prevotella sp. might play an essential role in maintaining the efficacy of ICIs.117 The abundance and positive detection of the Fusobacterium in non-responders were significantly higher than in responders when combining regofinil with treprizumab to treat refractory and advanced CRC.100 B. fragilis induces dendritic cells to mature and helper T lymphocytes to produce an immune response, then enhance the efficacy of ICIs. Bifidobacterium enhances the killing effect of CD8+T cells on tumors by changing the activity of dendritic cells. The gut microbiota is indispensable for the immune response of immunotherapy, which promotes host immunity, improves the host immune microenvironment, and affects the antigen presentation process, thereby changing the outcome of immunotherapy.118
Intestinal microbiota dysbiosis is involved in ICIs-associated colitis.119 Increases in Bacteroidetes are associated with resistance to ICIs-associated colitis; polyamine transport and vitamin B synthesis pathways are associated with colitis predisposition.120 Intestinal microbiota interventions improve metabolic pathways and immunotherapy’s efficacy.121 A set of 11 commensal bacterial strains enhance the therapeutic efficacy of ICIs in syngeneic tumor models.122 Understanding the mechanisms of the intestinal microbiota in promoting ICIs efficacy or drug resistance, exploring microbiota-related biomarkers, enhancing ICIs efficacy, and treating ICIs-related colitis through microecological interventions are expected to revolutionize tumor immunotherapy.
Gut microbiota modulation in the prevention and treatment of CRC
Studies have shown that CRC could be prevented by the intestinal microbiota modulation such as diet interventions, probiotics supplement and antibiotics administration. Moreover, there is mounting evidence that several microecological agents, FMT, and TCM could influence the therapeutic effects and clinical outcomes of CRC by modulating the gut microbiota. Therefore, we discuss the promising gut microbiota modulators that are expected to be applied in the prevention and treatment of CRC (Figure 2).
Figure 2.
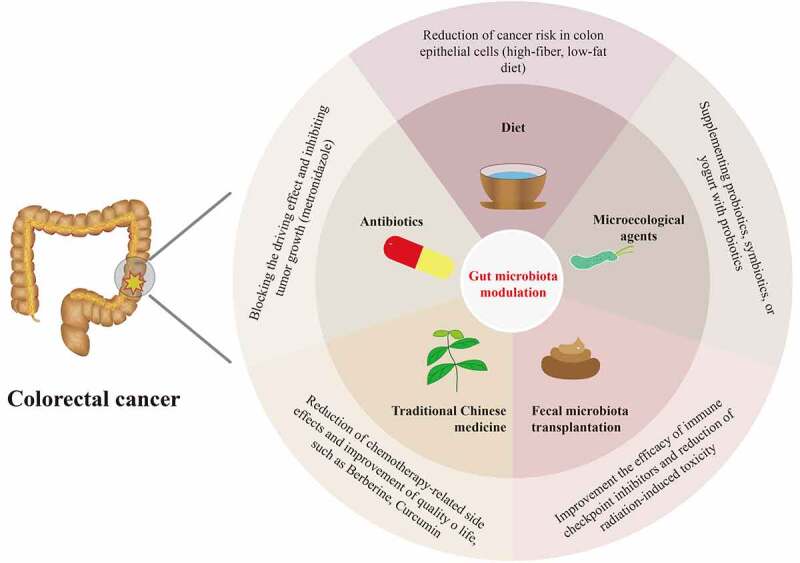
Overview of candidates for gut microbiota modulation for the prevention and treatment of colorectal cancer, including diet modulations, microecological agents, fecal microbiota transplantation, antibiotics, and traditional Chinese medicine.
Diet modulations
Chinese medicine theory holds that “medical and edible food and medicine are from the same root,” and an old Chinese proverb says, “Diseases enter by the mouth.” Diet plays a central role in maintaining intestinal microecological homeostasis, and dietary factors influence CRC. Excessive intake of red meat and processed meat products can increase the risk of CRC, while a high-fiber diet helps prevent CRC.50 A study showed that the abundance of carbohydrate fermentation and butyrate-producing microorganisms in a native African population was higher; in contrast, more microorganisms related to BAs metabolism were found in African Americans.123 These differences are related to different dietary structures. The intestinal flora structure of African Americans changed significantly after taking the high-fiber, low-fat diet. CRC-related molecular markers were reduced in tissues, suggesting that the high-fiber, low-fat diet inhibits harmful intestinal bacteria, improves the abundance of beneficial bacteria, increases butyrate production and glycolysis capacity, reduces the synthesis of secondary BAs, and reduces the risk of colon epithelial cell carcinogenesis.59 Dietary factors affect CRC occurrence and progression by affecting intestinal microbiota and metabolites. Dietary interventions such as functional food supplements containing probiotics or other ingredients maintain the stability of the intestinal mucus layer and the homeostasis of gut microbiota-host interactions.124
Microecological agents
The role of microecological agents such as probiotics, prebiotics, synbiotics, and even postbiotics in CRC is gradually being recognized. Probiotic products are subject to significant differences in strains, dosage forms, study populations, evaluation standards, and other factors at home and abroad, and the clinical application of probiotic preparations in China has its unique characteristics. The probiotics approved by the National Health Commission of China for use in humans mainly include Lactobacillus, Bifidobacterium, Enterococcus, Streptococcus, Bacillus, Clostridium, and Yeast at the genus levels.125
Microbiota modulators present a new vision for the prevention and the anti-tumor treatment of CRC. Evidence has suggested that high consumption of yogurt with probiotics moderately reduces the risk of CRA and CRC.126,127 Notably, both L. casei Zhang and L. rhamnosus Probio-M9 probiotic strains from China reduced the development of colon tumors in mice by improving intestinal microbial dysbiosis.128,129 Probiotics inhibit the development of CRC by increasing the abundance of probiotics, inhibiting various pathogenic bacteria, producing SCFAs, down-regulating the expression of pro-inflammatory factors, modulating the immune system, and promoting apoptosis.130 Supplementing probiotics, symbiotics, or yogurt with probiotics for those CRC patients receiving chemoradiotherapy or surgery could lower the complications, such as chemotherapy-related diarrhea/colitis, radiation enteritis, and postoperative adverse events.131,132 Preclinical studies showed that Enterobacteriaceae, Bifidobacterium, and Akkermansia muciniphila could improve the efficacy of CRC immunotherapy.133 Besides, L. rhamnosus Probio-M9 enhances the efficacy and responsiveness of anti-PD-1 immunotherapy, which may be a potential candidate for microecological agents.134 In the future, developing novel microecological agents in adjuvant anti-CRC therapy deserves further anticipation.
FMT
The oldest description of FMT is from Chinese medicine. “Prescription Collection of Fifty-Two Diseases” recorded the use of “golden juice” to treat infectious diseases as early as the Western Zhou Dynasty of China (about the tenth century BC). Dr. Ge Hong, in the Eastern Jin Dynasty of China (about 1700 years ago), edited the “Prescription Collection of Emergency” and called the FMT preparation “yellow soup,” which was used to treat food poisoning and severe diarrhea.135 FMT is a critical way of modulating the imbalance of gut microbiota by transplanting functional fecal microflora of healthy individuals into the gastrointestinal tract of patients. FMT is primarily used for treating refractory or recurrent C. difficile infections, with a more than 90% cure rate. In addition, there are prospects for therapeutic application in inflammatory bowel disease, functional intestinal disorders, metabolic diseases, and rheumatoid immune diseases.135–137
FMT has become a research hotspot associated with cancer therapy, especially for enhancing the efficacy of ICIs, improving ICIs efficacy, drug resistance, and intestinal toxicity.138,139 Data regarding FMT in CRC are somewhat limited; however, a preclinical study showed that a mixture of 11 commensal bacteria enhanced the activity of ICIs and inhibited tumor growth in a mouse model of CRC.122 A study showed that FMT improved the drug resistance of melanoma patients to ICIs and restored the immunotherapeutic response, possibly related to the activation of T cell activity in tumors.122,138 Currently, many clinical studies are being performed to evaluate FMT for improving the efficacy and treatment sensitivity of ICIs. In addition, FMT modulates the intestinal toxicity of chemoradiotherapy and immunotherapy, including severe chemotherapy-related diarrhea or colitis, radiation enteritis, and ICI-related colitis.112,140 FMT has been used to treat refractory ICIs-associated colitis, and complete remission has been reported after single or multiple FMT.141 Preclinical studies showed that FMT alleviates severe diarrhea and intestinal mucositis associated with 5-FU and platinum in CRC mice.140 FMT improved diarrhea, abdominal pain, hematochezia, and other symptoms associated with radiation enteritis,142 suggesting the value of FMT in reducing radiation-induced toxicity and improving radiotherapy outcomes.112 Nevertheless, in-depth studies are needed before applying FMT to cancer therapy; definitions are needed for indications, donor screening, sample quality control, transplantation timing, and safety of FMT for CRC prevention and treatment.
Antibiotics
Antibiotics have direct bactericidal or bacteriostatic effects on intestinal microflora; however, the relationship between antibacterial drugs and CRC is controversial. A large case-control study showed an association between antimicrobial agents and CRC.143 Theoretically, antibiotics might block its drive effect and inhibit tumor growth for F. nucleatum, E. coli sp., and other CRC-related driver bacteria. Studies found that F. nucleatum in primary lesions and liver metastases of CRC; the number of F. nucleatum in tumors decreased, and tumor growth slowed after treatment with metronidazole, suggesting that antimicrobials may have potential therapeutic value for F. nucleatum infection-related CRC.101 The influence of antibiotics on the gut microbiota and its role in CRC needs to be further studied.
TCM
TCM has a long history of application in preventing and treating CRC as a predominant source of natural medicines and herbal products. In China, TCM has excellent potential to combat colorectal carcinogenesis while reducing the side effects associated with chemotherapy and improving the quality of life of CRC patients. TCM could inhibit CRC development by regulating CRC-related signaling pathways, such as PI3K/Akt, NF-κB, MAPK, and Wnt/β-catenin.144
Accumulating evidence suggests the fundamental role of the gut microbiota in the prevention and treatment of CRC through TCM, with the most frequently studied TCM ingredients being Berberine and Curcumin. A randomized double-blind controlled trial has demonstrated that Berberine effectively reduces the risk of CRA recurrence, suggesting that it is a promising agent for preventing and treating CRC.145 The specific role of gut microbiota in Berberine’s inhibition of CRC tumorigenesis has been elucidated in a set of studies.146,147 A recent investigation of the fecal microbiota of the CRC mouse model using 16S rRNA profiling identified a significant decrease in microbial richness without loss of diversity and an increase in the relative abundance of beneficial bacteria after Berberine treatment, accompanied by inhibition of several inflammatory and oncogenic pathways, such as NF-κB pathway.147 Preclinical studies have shown that Berberine inhibits CRC initiation and progression by regulating the intestinal microbiota and modulating the tumor microenvironment.146 Besides, Curcumin is identified as a well-tolerated chemotherapeutic adjuvant to oxaliplatin chemotherapy in patients with colorectal liver metastases. It mediates cell cycle arrest and ameliorates Treg-associated inflammation by regulating gut microbiota to exert anti-tumor effects.148 However, there is a lack of research data on the interactions between TCM and intestinal microbiota in CRC. Future research on the role of TCM in CRC prevention and treatment from the perspective of gut microbiota deserves in-depth study.
Conclusion
In the past decades, the morbidity and mortality of CRC in China have increased, transitioning to developed countries such as Europe and the United States. A large population, unique genetics of multiple ethnic groups, variable environments, lifestyles, and dietary and gut microbiota structures are characteristics of China. Gut microbiota dysbiosis is associated with CRC progression. Some specific microbes contribute to CRC carcinogenesis, including F. nucleatum, pks+ E. coli, ETBF, and P. micra. However, the role of fungi and viruses in CRC needs further investigation. The microbial pathogenesis of CRC involves gut microbiota dysbiosis, invasion and colonization of pathogens, microbial-related metabolites, and virulence, which induce pro-inflammatory signaling pathways and exacerbate intestinal epithelial DNA damage, ultimately initiating CRC progression. However, high-quality in vitro and in vivo validation and in-depth evaluation are needed to clarify the gut microbiota-related mechanisms in the carcinogenesis of CRC. Early diagnosis and prediction by microbial biomarkers are promising research areas with significant clinical translational value. Integrating and optimizing intestinal microbiota-related biomarker panels with traditional clinical methods to improve CRC screening and diagnosis levels is promising and worth digging into. Gut microbiota interventions, including microecological agents and FMT, hold potential clinical applications to enhance drug efficacy, improve drug resistance, and reduce the severe intestinal toxicity associated with anti-tumor therapy of CRC. Moreover, TCM plays anti-tumor roles in CRC as an adjuvant treatment scheme in China. However, in-depth studies combined with the intestinal microbiota are still needed. Today, international cooperation is essential for searching for gut microbiota-related carcinogenic pathways, biomarkers, and interventional approaches to CRC.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Authors contribution
ZKW and YSY were responsible for the idea, outlined, and prepared the initial and final drafts of the manuscript. ZKW, WYD, and NNZ drafted the manuscript. JYF and YSY revised the manuscript. All authors approved the final draft submitted.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F.. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. A Cancer J Clin. 2021;71(3):209–24. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Xia C, Dong X, Li H, Cao M, Sun D, He S, Yang F, Yan X, Zhang S, Li N, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J. 2022;135(5):584–590. doi: 10.1097/CM9.0000000000002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li N, Lu B, Luo C, Cai J, Lu M, Zhang Y, Chen H, Dai M. Incidence, mortality, survival, risk factor and screening of colorectal cancer: a comparison among China, Europe, and northern America. Cancer Lett. 2021;522:255–268. doi: 10.1016/j.canlet.2021.09.034. [DOI] [PubMed] [Google Scholar]
- 4.Zheng X, Hur J, Nguyen LH, Liu J, Song M, Wu K, Smith-Warner SA, Ogino S, Willett WC, Chan AT, et al. Comprehensive assessment of diet quality and risk of precursors of early-onset colorectal cancer. J Natl Cancer Inst. 2021;113(5):543–552. doi: 10.1093/jnci/djaa164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, Chan FKL, Kristiansen K, Sung JJY, Wong SH, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6(1):70. doi: 10.1186/s40168-018-0451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 7.Liu W, Zhang X, Xu H, Li S, Lau HC, Chen Q, Zhang B, Zhao L, Chen H, Sung JJ, et al. Microbial community heterogeneity within colorectal neoplasia and its correlation with colorectal carcinogenesis. Gastroenterol. 2021;160(7):2395–2408. doi: 10.1053/j.gastro.2021.02.020. [DOI] [PubMed] [Google Scholar]
- 8.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat Commun. 2015;6(1):6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 9.Liu NN, Jiao N, Tan JC, Wang Z, Wu D, Wang AJ, Chen J, Tao L, Zhou C, Fang W, et al. Multi-kingdom microbiota analyses identify bacterial–fungal interactions and biomarkers of colorectal cancer across cohorts. Nature Microbiol. 2022;7(2):238–250. doi: 10.1038/s41564-021-01030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie YH, Gao QY, Cai GX, Sun XM, Sun XM, Zou TH, Chen HM, Yu SY, Qiu YW, Gu WQ, et al. Fecal clostridium symbiosum for noninvasive detection of early and advanced colorectal cancer: test and validation studies. EBioMed. 2017;25:32–40. doi: 10.1016/j.ebiom.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Yu X, Yu E, Wang N, Cai Q, Shuai Q, Yan F, Jiang L, Wang H, Liu J, et al. Changes in gut microbiota and plasma inflammatory factors across the stages of colorectal tumorigenesis: a case-control study. BMC Microbiol. 2018;18(1):92. doi: 10.1186/s12866-018-1232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 13.Li N, Bai C, Zhao L, Ge Y, Li X. Characterization of the fecal microbiota in gastrointestinal cancer patients and healthy people. Clin Transl Oncol. 2022;24(6):1134–1147. doi: 10.1007/s12094-021-02754-y. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, Xu L, Wu T, Li J, Hua L. Analysis of abnormal intestinal flora on risk of intestinal cancer and effect of heparin on formation of bacterial biofilm. Bioengineered. 2022;13(1):894–904. doi: 10.1080/21655979.2021.2014388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan D, Tao Y, Wang H, Wang J, Cao Y, Cao W, Pan S, Yu Z. A comprehensive analysis of the microbiota composition and host driver gene mutations in colorectal cancer. Invest New Drugs. 2022;40(5):884–894. doi: 10.1007/s10637-022-01263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang H, Mishra R, Cen C, Tang Y, Ma C, Wasti S, Wang Y, Ou Q, Chen K, Zhang J. Metagenomic analyses expand bacterial and functional profiling biomarkers for colorectal cancer in a Hainan Cohort, China. Curr Microbiol. 2021;78(2):705–712. doi: 10.1007/s00284-020-02299-3. [DOI] [PubMed] [Google Scholar]
- 17.Du X, Li Q, Tang Z, Yan L, Zhang L, Zheng Q, Zeng X, Chen G, Yue H, Li J, et al. Alterations of the gut microbiome and fecal metabolome in colorectal cancer: Implication of intestinal metabolism for tumorigenesis. Front Physiol. 2022;13:854545. doi: 10.3389/fphys.2022.854545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coker OO, Liu C, Wu WKK, Wong SH, Jia W, Sung JJY, Yu J. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. 2022;10(1):35. doi: 10.1186/s40168-021-01208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeoh YK, Chen Z, Wong MCS, Hui M, Yu J, Ng SC, Sung JJY, Chan FKL, Chan PKS. Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut. 2020;69(11):1998–2007. doi: 10.1136/gutjnl-2019-319635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Li D, Yang Z, Dai W, Feng X, Liu Y, Jiang Y, Li P, Li Y, Tang B, et al. Establishing high-accuracy biomarkers for colorectal cancer by comparing fecal microbiomes in patients with healthy families. Gut Microbes. 2020;11(4):918–929. doi: 10.1080/19490976.2020.1712986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Cao H, Fei B, Gao Q, Yi W, Han W, Bao C, Xu J, Zhao W, Zhang F. Gut microbiota signatures in tumor, para-cancerous, normal mucosa, and feces in colorectal cancer patients. Front Cell Dev Biol. 2022;10:916961. doi: 10.3389/fcell.2022.916961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua H, Sun Y, He X, Chen Y, Teng L, Lu C. Intestinal microbiota in colorectal adenoma-carcinoma sequence. Front Med. 2022;9:888340. doi: 10.3389/fmed.2022.888340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao R, Wu C, Zhu Y, Kong C, Zhu Y, Gao Y, Zhang X, Yang R, Zhong H, Xiong X, et al. Integrated analysis of colorectal cancer reveals cross-cohort gut microbial signatures and associated serum metabolites. Gastroenterology. 2022;163(4):1024–37.e9. doi: 10.1053/j.gastro.2022.06.069. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, Jiao N, Zhu R, Zhang Y, Wu D, Wang AJ, Fang S, Tao L, Li Y, Cheng S, et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat Commun. 2021;12(1):3063. doi: 10.1038/s41467-021-23265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Y, Liu Y, Zhao L, Zhao F, Feng J, Li S, Chen H, Sun J, Zhu B, Geng R, et al. Gut microbiota in patients after surgical treatment for colorectal cancer. Environ Microbiol. 2019;21(2):772–783. doi: 10.1111/1462-2920.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang R, He K, Duan X, Xiao J, Wang H, Xiang G. Changes of intestinal microflora in colorectal cancer patients after surgical resection and chemotherapy. Comput Math Methods Med. 2022;2022:1940846. doi: 10.1155/2022/1940846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Huo RX, Wang YJ, Hou SB, Wang W, Zhang CZ, Wan XH. Gut mucosal microbiota profiles linked to colorectal cancer recurrence. World J Gastroenterol. 2022;28(18):1946–1964. doi: 10.3748/wjg.v28.i18.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Z, Lv Z, Chen F, Zhang Y, Xu Z, Huo J, Liu W, Yu S, Tuersun A, Zhao J, et al. Dysbiosis of human tumor microbiome and aberrant residence of Actinomyces in tumor-associated fibroblasts in young-onset colorectal cancer. Front Immunol. 2022;13:1008975. doi: 10.3389/fimmu.2022.1008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Du L, Shi D, Kong C, Liu J, Liu G, Li X, Ma Y. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat Commun. 2021;12(1):6757. doi: 10.1038/s41467-021-27112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong M, Xiong Y, Ye Z, Zhao J, Zhong L, Liu Y, Zhu Y, Tian L, Qiu X, Hong X. Microbial community profiling distinguishes left-sided and right-sided colon cancer. Front Cell Infect Microbiol. 2020;10:498502. doi: 10.3389/fcimb.2020.498502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 33.Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol. 2017;17(10):635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastro Hepat. 2019;16(6):331–345. doi: 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- 35.Lin Y, Lau HC, Liu Y, Kang X, Wang Y, Ting NL, Kwong TN, Han J, Liu W, Liu C, et al. Altered mycobiota signatures and enriched pathogenic aspergillus rambelli are associated with colorectal cancer based on multicohort fecal metagenomic analyses. Gastroenterology. 2022;163(4):908–921. doi: 10.1053/j.gastro.2022.06.038. [DOI] [PubMed] [Google Scholar]
- 36.Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, Chan FKL, Sung JJY, Yu J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2019;68(4):654–662. doi: 10.1136/gutjnl-2018-317178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao R, Xia K, Wu M, Zhong H, Sun J, Zhu Y, Huang L, Wu X, Yin L, Yang R, et al. Alterations of gut mycobiota profiles in adenoma and colorectal cancer. Front Cell Infect Microbiol. 2022;12:839435. doi: 10.3389/fcimb.2022.839435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, Fruehauf J, Goldsmith JD, Xu H, Katchar KK, Koon HW, Zhao D, Kokkotou EG, Pothoulakis C, Kelly CP. Saccharomyces boulardii inhibits EGF receptor signaling and intestinal tumor growth in Apc(min) mice. Gastroenterology. 2009;137(3):914–923. doi: 10.1053/j.gastro.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T, Fan C, Yao A, Xu X, Zheng G, You Y, Jiang C, Zhao X, Hou Y, Hung MC, et al. The adaptor protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity. 2018;49(3):504–14.e4. doi: 10.1016/j.immuni.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo W, Michail S, Sun F. Metagenomic analyses of multiple gut datasets revealed the association of phage signatures in colorectal cancer. Frontiers Cellular Infect Microbiol. 2022;12:918010. doi: 10.3389/fcimb.2022.918010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, Li X, Szeto CH, Sugimura N, Lam TY, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 2018;155(2):529–41.e5. doi: 10.1053/j.gastro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 42.Gao R, Zhu Y, Kong C, Xia K, Li H, Zhu Y, Zhang X, Liu Y, Zhong H, Yang R, et al. Alterations, interactions, and diagnostic potential of gut bacteria and viruses in colorectal cancer. Front Cell Infect Microbiol. 2021;11:657867. doi: 10.3389/fcimb.2021.657867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F, Li S, Guo R, Song F, Zhang Y, Wang X, Huo X, Lv Q, Ullah H, Wang G, et al. Meta-analysis of fecal viromes demonstrates high diagnostic potential of the gut viral signatures for colorectal cancer and adenoma risk assessment. J Adv Res. 2022;49:103–114. doi: 10.1016/j.jare.2022.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen S, Huo D, Ma C, Jiang S, Zhang J, Xu ZZ. Expanding the colorectal cancer biomarkers based on the human gut phageome. Microbiol Spectr. 2021;9(3):e0009021. doi: 10.1128/Spectrum.00090-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong X, Pan P, Zheng DW, Bao P, Zeng X, Zhang XZ. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci Adv. 2020;6(20):eaba1590. doi: 10.1126/sciadv.aba1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Wei H, Zhou Y, Szeto CH, Li C, Lin Y, Coker OO, Lau HCH, Chan AWH, Sung JJY, et al. High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterology. 2022;162(1):135–49.e2. doi: 10.1053/j.gastro.2021.08.041. [DOI] [PubMed] [Google Scholar]
- 47.Chen D, Jin D, Huang S, Wu J, Xu M, Liu T, Dong W, Liu X, Wang S, Zhong W, et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020;469:456–467. doi: 10.1016/j.canlet.2019.11.019. [DOI] [PubMed] [Google Scholar]
- 48.Hou H, Chen D, Zhang K, Zhang W, Liu T, Wang S, Dai X, Wang B, Zhong W, Cao H. Gut microbiota-derived short-chain fatty acids and colorectal cancer: Ready for clinical translation? Cancer Lett. 2022;526:225–235. doi: 10.1016/j.canlet.2021.11.027. [DOI] [PubMed] [Google Scholar]
- 49.Keane JM, Joyce SA, Gahan CGM, Hyland NP, Houston A. Microbial metabolites as molecular mediators of host-microbe symbiosis in colorectal cancer. Results Probl Cell Differ. 2020;69:581–603. doi: 10.1007/978-3-030-51849-3_22. [DOI] [PubMed] [Google Scholar]
- 50.Ryu TY, Kim K, Son MY, Min JK, Kim J, Han TS, Kim DS, Cho HS. Downregulation of PRMT1, a histone arginine methyltransferase, by sodium propionate induces cell apoptosis in colon cancer. Oncol Rep. 2019;41(3):1691–1699. doi: 10.3892/or.2018.6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang G, Yu Y, Wang YZ, Wang JJ, Guan R, Sun Y, Shi F, Gao J, Fu XL. Role of SCFAs in gut microbiome and glycolysis for colorectal cancer therapy. J Cell Physiol. 2019;234(10):17023–17049. doi: 10.1002/jcp.28436. [DOI] [PubMed] [Google Scholar]
- 52.Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158(2):322–340. doi: 10.1053/j.gastro.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He Y, Fu L, Li Y, Wang W, Gong M, Zhang J, Dong X, Huang J, Wang Q, Mackay CR, et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metab. 2021;33(5):988–1000.e7. doi: 10.1016/j.cmet.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 54.Ma X, Zhou Z, Zhang X, Fan M, Hong Y, Feng Y, Dong Q, Diao H, Wang G. Sodium butyrate modulates gut microbiota and immune response in colorectal cancer liver metastatic mice. Cell Biol Toxicol. 2020;36(5):509–515. doi: 10.1007/s10565-020-09518-4. [DOI] [PubMed] [Google Scholar]
- 55.Ridlon JM, Wolf PG, Gaskins HR. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes. 2016;7(3):201–215. doi: 10.1080/19490976.2016.1150414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Májer F, Sharma R, Mullins C, Keogh L, Phipps S, Duggan S, Kelleher D, Keely S, Long A, Radics G, et al. New highly toxic bile acids derived from deoxycholic acid, chenodeoxycholic acid and lithocholic acid. Bioorg Med Chem. 2014;22(1):256–268. doi: 10.1016/j.bmc.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 57.Ma Y, Qu R, Zhang Y, Jiang C, Zhang Z, Fu W. Progress in the study of colorectal cancer caused by altered gut microbiota after cholecystectomy. Front Endocrinol (Lausanne). 2022;13:815999. doi: 10.3389/fendo.2022.815999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai J, Sun L, Gonzalez FJ. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe. 2022;30(3):289–300. doi: 10.1016/j.chom.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 2007;5(5):455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 60.Guo FF, Yu TC, Hong J, Fang JY. Emerging roles of hydrogen sulfide in inflammatory and neoplastic colonic diseases. Front Physiol. 2016;7:156. doi: 10.3389/fphys.2016.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 63.Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, Xiao X, Kwong TNY, Tsoi H, Wu WKK, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017;153(6):1621–33.e6. doi: 10.1053/j.gastro.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 64.Yu YN, Fang JY. Gut microbiota and colorectal cancer. Gastrointestinal Tumors. 2015;2(1):26–32. doi: 10.1159/000380892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10(8):575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 66.Li J, Ma X, Chakravarti D, Shalapour S, DePinho RA. Genetic and biological hallmarks of colorectal cancer. Genes Dev. 2021;35(11–12):787–820. doi: 10.1101/gad.348226.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kong C, Yan X, Zhu Y, Zhu H, Luo Y, Liu P, Ferrandon S, Kalady MF, Gao R, He J, et al. Fusobacterium nucleatum promotes the development of colorectal cancer by activating a cytochrome P450/epoxyoctadecenoic acid axis via TLR4/Keap1/NRF2 signaling. Cancer Res. 2021;81(17):4485–4498. doi: 10.1158/0008-5472.Can-21-0453. [DOI] [PubMed] [Google Scholar]
- 68.Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D, Dalerba P, Wang TC, Han YW. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep. 2019;20(4). doi: 10.15252/embr.201847638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang X, Xie Y, Yan T, Yu T, Sun T, et al. F. Nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2021;70(11):2123–2137. doi: 10.1136/gutjnl-2020-322780. [DOI] [PubMed] [Google Scholar]
- 70.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–63.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen H, Tong T, Lu SY, Ji L, Xuan B, Zhao G, Yan Y, Song L, Zhao L, Xie Y, et al. Urea cycle activation triggered by host-microbiota maladaptation driving colorectal tumorigenesis. Cell Metab. 2023;35(4):651–666.e7. doi: 10.1016/j.cmet.2023.03.003. [DOI] [PubMed] [Google Scholar]
- 72.Zhao L, Zhang X, Zhou Y, Fu K, Lau HC, Chun TW, Cheung AH, Coker OO, Wei H, Wu WK, et al. Parvimonas micra promotes colorectal tumorigenesis and is associated with prognosis of colorectal cancer patients. Oncogene. 2022;41(36):4200–4210. doi: 10.1038/s41388-022-02395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, Coker OO, Chan AWH, Chan FKL, Sung JJY, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nature Microbiol. 2019;4(12):2319–2330. doi: 10.1038/s41564-019-0541-3. [DOI] [PubMed] [Google Scholar]
- 74.Chen S, Su T, Zhang Y, Lee A, He J, Ge Q, Wang L, Si J, Zhuo W, Wang L. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes. 2020;11(3):511–525. doi: 10.1080/19490976.2019.1695494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu C, Fan L, Lin Y, Shen W, Qi Y, Zhang Y, Chen Z, Wang L, Long Y, Hou T, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. 2021;13(1):1980347. doi: 10.1080/19490976.2021.1980347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sánchez-Alcoholado L, Ramos-Molina B, Otero A, Laborda-Illanes A, Ordóñez R, Medina JA, Gómez-Millán J, Queipo-Ortuño MI. The role of the gut microbiome in colorectal cancer development and therapy response. Cancers Basel. 2020;12(6):1406. doi: 10.3390/cancers12061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe. 2018;23(2):203–14.e5. doi: 10.1016/j.chom.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, Hacker-Prietz A, Rabizadeh S, Woster PM, Sears CL, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108(37):15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135(4):1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.CollectiveName:National Cancer Center CEGotDoCGftSED, Early Treatment of Colorectal C . [China guideline for the screening, early detection and early treatment of colorectal cancer (2020, Beijing)]. Zhonghua Zhong Liu Za Zhi. 2021;43(1):16–38. doi: 10.3760/cma.j.cn112152-20210105-00010. [DOI] [PubMed] [Google Scholar]
- 82.Zhang J, Cheng Z, Ma Y, He C, Lu Y, Zhao Y, Chang X, Zhang Y, Bai Y, Cheng N. Effectiveness of screening modalities in colorectal cancer: a network meta-analysis. Clin Colorectal Cancer. 2017;16(4):252–263. doi: 10.1016/j.clcc.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 83.Chen H, Li N, Ren J, Feng X, Lyu Z, Wei L, Li X, Guo L, Zheng Z, Zou S, et al. Participation and yield of a population-based colorectal cancer screening programme in China. Gut. 2019;68(8):1450–1457. doi: 10.1136/gutjnl-2018-317124. [DOI] [PubMed] [Google Scholar]
- 84.Zou J, Xiao Z, Wu Y, Yang J, Cui N. Noninvasive fecal testing for colorectal cancer. Clin Chim Acta. 2022;524:123–131. doi: 10.1016/j.cca.2021.10.030. [DOI] [PubMed] [Google Scholar]
- 85.Chen F, Dai X, Zhou CC, Li KX, Zhang YJ, Lou XY, Zhu YM, Sun YL, Peng BX, Cui W. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut. 2022;71(7):1315–1325. doi: 10.1136/gutjnl-2020-323476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, Brim H, Ashktorab H, Ng SC, Ng SSM, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 2017;23(8):2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 87.Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H, Xue Y, Wu Y, Wang Y, Liu W, et al. A simple and novel fecal biomarker for colorectal cancer: Ratio of Fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clin Chem. 2018;64(9):1327–1337. doi: 10.1373/clinchem.2018.289728. [DOI] [PubMed] [Google Scholar]
- 88.Wang N, Fang JY. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 2022;31(2):159–172. doi: 10.1016/j.tim.2022.08.010. [DOI] [PubMed] [Google Scholar]
- 89.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang X, Zhang Y, Gui X, Zhang Y, Zhang Z, Chen W, Zhang X, Wang Y, Zhang M, Shang Z, et al. Salivary Fusobacterium nucleatum serves as a potential biomarker for colorectal cancer. iScience. 2022;25(5):104203. doi: 10.1016/j.isci.2022.104203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liang JQ, Li T, Nakatsu G, Chen YX, Yau TO, Chu E, Wong S, Szeto CH, Ng SC, Chan FKL, et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut. 2020;69(7):1248–1257. doi: 10.1136/gutjnl-2019-318532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang HF, Li LF, Guo SH, Zeng QY, Ning F, Liu WL, Zhang G. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci Rep. 2016;6:33440. doi: 10.1038/srep33440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shen X, Li J, Li J, Zhang Y, Li X, Cui Y, Gao Q, Chen X, Chen Y, Fang JY. Fecal enterotoxigenic bacteroides fragilis-peptostreptococcus stomatis-parvimonas micra biomarker for noninvasive diagnosis and prognosis of colorectal laterally spreading tumor. Front Oncol. 2021;11:661048. doi: 10.3389/fonc.2021.661048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kong C, Liang L, Liu G, Du L, Yang Y, Liu J, Shi D, Li X, Ma Y. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut. 2022;72(6):1129–1142. doi: 10.1136/gutjnl-2022-327156. [DOI] [PubMed] [Google Scholar]
- 95.Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Sci. 2018;359(6382):1366–1370. doi: 10.1126/science.aar6918. [DOI] [PubMed] [Google Scholar]
- 96.Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastro Hepat. 2017;14(6):356–365. doi: 10.1038/nrgastro.2017.20. [DOI] [PubMed] [Google Scholar]
- 97.Deng X, Li Z, Li G, Li B, Jin X, Lyu G. Comparison of microbiota in patients treated by surgery or chemotherapy by 16S rRNA sequencing reveals potential biomarkers for colorectal cancer therapy. Front Microbiol. 2018;9:1607. doi: 10.3389/fmicb.2018.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yi Y, Shen L, Shi W, Xia F, Zhang H, Wang Y, Zhang J, Wang Y, Sun X, Zhang Z, et al. Gut microbiome components predict response to neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer: A prospective, longitudinal study. Clin Cancer Res. 2021;27(5):1329–1340. doi: 10.1158/1078-0432.CCR-20-3445. [DOI] [PubMed] [Google Scholar]
- 99.Li N, Bai C, Zhao L, Sun Z, Ge Y, Li X. The relationship between gut microbiome features and chemotherapy response in gastrointestinal cancer. Front Oncol. 2021;11:781697. doi: 10.3389/fonc.2021.781697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang F, He MM, Yao YC, Zhao X, Wang ZQ, Jin Y, Luo HY, Li JB, Wang FH, Qiu MZ, et al. Regorafenib plus toripalimab in patients with metastatic colorectal cancer: a phase Ib/II clinical trial and gut microbiome analysis. Cell Rep Med. 2021;2(9):100383. doi: 10.1016/j.xcrm.2021.100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science (New York, NY). 2017;358(6369):1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Al-Qadami G, Van Sebille Y, Le H, Bowen J. Gut microbiota: implications for radiotherapy response and radiotherapy-induced mucositis. Expert Rev Gastroenterol Hepatol. 2019;13(5):485–496. doi: 10.1080/17474124.2019.1595586. [DOI] [PubMed] [Google Scholar]
- 103.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science (New York, NY). 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18(1). doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang CW, Liu CY, Lee HC, Huang YH, Li LH, Chiau JC, Wang TE, Chu CH, Shih SC, Tsai TH, et al. Lactobacillus casei variety rhamnosus probiotic preventively attenuates 5-Fluorouracil/Oxaliplatin-induced intestinal injury in a syngeneic colorectal cancer model. Front Microbiol. 2018;9:983. doi: 10.3389/fmicb.2018.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Osterlund P, Ruotsalainen T, Korpela R, Saxelin M, Ollus A, Valta P, Kouri M, Elomaa I, Joensuu H. Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: a randomised study. Br J Cancer. 2007;97(8):1028–1034. doi: 10.1038/sj.bjc.6603990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science (New York, NY). 2010;330(6005):831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mego M, Chovanec J, Vochyanova-Andrezalova I, Konkolovsky P, Mikulova M, Reckova M, Miskovska V, Bystricky B, Beniak J, Medvecova L, et al. Prevention of irinotecan induced diarrhea by probiotics: A randomized double blind, placebo controlled pilot study. Complement Ther Med. 2015;23(3):356–362. doi: 10.1016/j.ctim.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 109.Fan Q, Shang F, Chen C, Zhou H, Fan J, Yang M, Nie X, Liu L, Cai K, Liu H. Microbial characteristics of locally advanced rectal cancer patients after neoadjuvant chemoradiation therapy according to pathologic response. Cancer Manag Res. 2021;13:2655–2667. doi: 10.2147/CMAR.S294936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dong J, Li Y, Xiao H, Zhang S, Wang B, Wang H, Li Y, Fan S, Cui M. Oral microbiota affects the efficacy and prognosis of radiotherapy for colorectal cancer in mouse models. Cell Rep. 2021;37(4):109886. doi: 10.1016/j.celrep.2021.109886. [DOI] [PubMed] [Google Scholar]
- 111.Wang Z, Wang Q, Wang X, Zhu L, Chen J, Zhang B, Chen Y, Yuan Z. Gut microbial dysbiosis is associated with development and progression of radiation enteritis during pelvic radiotherapy. J Cell Mol Med. 2019;23(5):3747–3756. doi: 10.1111/jcmm.14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cui M, Xiao H, Li Y, Zhou L, Zhao S, Luo D, Zheng Q, Dong J, Zhao Y, Zhang X, et al. Faecal microbiota transplantation protects against radiation-induced toxicity. EMBO Mol Med. 2017;9(4):448–461. doi: 10.15252/emmm.201606932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Crawford PA, Gordon JI. Microbial regulation of intestinal radiosensitivity. Proc Natl Acad Sci U S A. 2005;102(37):13254–13259. doi: 10.1073/pnas.0504830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.André T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383(23):2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 115.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R, Hill A, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 116.Chen EX, Jonker DJ, Loree JM, Kennecke HF, Berry SR, Couture F, Ahmad CE, Goffin JR, Kavan P, Harb M, et al. Effect of combined immune checkpoint inhibition vs best supportive care alone in patients with advanced colorectal cancer: The Canadian cancer trials group CO.26 study. JAMA Oncol. 2020;6(6):831–838. doi: 10.1001/jamaoncol.2020.0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu X, Lv J, Guo F, Li J, Jia Y, Jiang D, Wang N, Zhang C, Kong L, Liu Y, et al. Gut microbiome influences the efficacy of PD-1 antibody immunotherapy on MSS-Type colorectal cancer via metabolic pathway. Front Microbiol. 2020;11:814. doi: 10.3389/fmicb.2020.00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (New York, NY). 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (New York, NY). 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, No D, Gobourne A, Littmann E, Huttenhower C, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. doi: 10.1038/ncomms10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang J, Zheng X, Kang W, Hao H, Mao Y, Zhang H, Chen Y, Tan Y, He Y, Zhao W, et al. Metagenomic and metabolomic analyses reveal synergistic effects of fecal microbiota transplantation and anti-PD-1 therapy on treating colorectal cancer. Front Immunol. 2022;13:874922. doi: 10.3389/fimmu.2022.874922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565(7741):600–605. doi: 10.1038/s41586-019-0878-z. [DOI] [PubMed] [Google Scholar]
- 123.Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EMM, Sanders ME, Shamir R, Swann JR, Szajewska H, et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastro Hepat. 2021;18(9):649–667. doi: 10.1038/s41575-021-00440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oh NS, Lee JY, Kim YT, Kim SH, Lee JH. Cancer-protective effect of a synbiotic combination between Lactobacillus gasseri 505 and a Cudrania tricuspidata leaf extract on colitis-associated colorectal cancer. Gut Microbes. 2020;12(1):1785803. doi: 10.1080/19490976.2020.1785803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Microecology CSo . Chinese expert consensus on the clinical application of microecological agents. Chin J Microecol. 2020;32(8):953–965. [Google Scholar]
- 126.Kim CE, Yoon LS, Michels KB, Tranfield W, Jacobs JP, May FP. The impact of prebiotic, probiotic, and synbiotic supplements and yogurt consumption on the risk of colorectal neoplasia among adults: A systematic review. Nutrients. 2022;14(22). doi: 10.3390/nu14224937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pala V, Sieri S, Berrino F, Vineis P, Sacerdote C, Palli D, Masala G, Panico S, Mattiello A, Tumino R, et al. Yogurt consumption and risk of colorectal cancer in the Italian European prospective investigation into cancer and nutrition cohort. Int J Cancer. 2011;129(11):2712–2719. doi: 10.1002/ijc.26193. [DOI] [PubMed] [Google Scholar]
- 128.Zhang Y, Ma C, Zhao J, Xu H, Hou Q, Zhang H. Lactobacillus casei Zhang and vitamin K2 prevent intestinal tumorigenesis in mice via adiponectin-elevated different signaling pathways. Oncotarget. 2017;8(15):24719–24727. doi: 10.18632/oncotarget.15791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu H, Hiraishi K, Kurahara LH, Nakano-Narusawa Y, Li X, Hu Y, Matsuda Y, Zhang H, Hirano K. Inhibitory effects of breast milk-derived lactobacillus rhamnosus Probio-M9 on colitis-associated carcinogenesis by restoration of the gut microbiota in a mouse model. Nutrients. 2021;13(4). doi: 10.3390/nu13041143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xie X, He Y, Li H, Yu D, Na L, Sun T, Zhang D, Shi X, Xia Y, Jiang T, et al. Effects of prebiotics on immunologic indicators and intestinal microbiota structure in perioperative colorectal cancer patients. Nutrition. 2019;61:132–142. doi: 10.1016/j.nut.2018.10.038. [DOI] [PubMed] [Google Scholar]
- 131.Chen Y, Qi A, Teng D, Li S, Yan Y, Hu S, Du X. Probiotics and synbiotics for preventing postoperative infectious complications in colorectal cancer patients: a systematic review and meta-analysis. Tech Coloproctol. 2022;26(6):425–436. doi: 10.1007/s10151-022-02585-1. [DOI] [PubMed] [Google Scholar]
- 132.Zhang JW, Du P, Gao J, Yang BR, Fang WJ, Ying CM. Preoperative probiotics decrease postoperative infectious complications of colorectal cancer. Am J Med Sci. 2012;343(3):199–205. doi: 10.1097/MAJ.0b013e31823aace6. [DOI] [PubMed] [Google Scholar]
- 133.Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15(6):382–396. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 134.Gao G, Ma T, Zhang T, Jin H, Li Y, Kwok LY, Zhang H, Sun Z. Adjunctive probiotic lactobacillus rhamnosus Probio-M9 administration enhances the effect of Anti-PD-1 antitumor therapy via restoring antibiotic-disrupted gut microbiota. Front Immunol. 2021;12:772532. doi: 10.3389/fimmu.2021.772532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shi YC, Yang YS. Fecal microbiota transplantation: Current status and challenges in China. JGH open: an open access journal of gastroenterology and hepatology. 2018;2(4):114–116. doi: 10.1002/jgh3.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ren R, Gao X, Shi Y, Li J, Peng L, Sun G, Wang Z, Yan B, Zhi J, Yang Y. Long-term efficacy of low-intensity single donor fecal microbiota transplantation in ulcerative colitis and outcome-specific gut bacteria. Front Microbiol. 2021;12:742255. doi: 10.3389/fmicb.2021.742255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao HJ, Luo X, Shi YC, Li JF, Pan F, Ren RR, Peng LH, Shi XY, Yang G, Wang J, et al. The efficacy of fecal microbiota transplantation for children with Tourette syndrome: A preliminary study. Front Psychiatry. 2020;11:554441. doi: 10.3389/fpsyt.2020.554441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, Deblasio RN, Menna C, Ding Q, Pagliano O, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science (New York, NY). 2021;371(6529):595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kaźmierczak-Siedlecka K, Daca A, Fic M, van de Wetering T, Folwarski M, Makarewicz W. Therapeutic methods of gut microbiota modification in colorectal cancer management - fecal microbiota transplantation, prebiotics, probiotics, and synbiotics. Gut Microbes. 2020;11(6):1518–1530. doi: 10.1080/19490976.2020.1764309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chang CW, Lee HC, Li LH, Chiang Chiau JS, Wang TE, Chuang WH, Chen MJ, Wang HY, Shih SC, Liu CY, et al. Fecal microbiota transplantation prevents intestinal injury, upregulation of toll-like receptors, and 5-Fluorouracil/Oxaliplatin-induced toxicity in colorectal cancer. Int J Mol Sci. 2020;21(2). doi: 10.3390/ijms21020386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, Jiang ZD, Abu-Sbeih H, Sanchez CA, Chang CC, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med. 2018;24(12):1804–1808. doi: 10.1038/s41591-018-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jian Y, Zhang D, Liu M, Wang Y, Xu ZX. The impact of gut microbiota on radiation-induced enteritis. Frontiers Cellular Infect Microbiol. 2021;11:586392. doi: 10.3389/fcimb.2021.586392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang J, Haines C, Watson AJM, Hart AR, Platt MJ, Pardoll DM, Cosgrove SE, Gebo KA, Sears CL. Oral antibiotic use and risk of colorectal cancer in the United Kingdom, 1989-2012: a matched case-control study. Gut. 2019;68(11):1971–1978. doi: 10.1136/gutjnl-2019-318593. [DOI] [PubMed] [Google Scholar]
- 144.Chen JF, Wu SW, Shi ZM, Hu B. Traditional Chinese medicine for colorectal cancer treatment: potential targets and mechanisms of action. Chin Med. 2023;18(1):14. doi: 10.1186/s13020-023-00719-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen YX, Gao QY, Zou TH, Wang BM, Liu SD, Sheng JQ, Ren JL, Zou XP, Liu ZJ, Song YY, et al. Berberine versus placebo for the prevention of recurrence of colorectal adenoma: a multicentre, double-blinded, randomised controlled study. Lancet Gastroenterol Hepatol. 2020;5(3):267–275. doi: 10.1016/s2468-1253(19)30409-1. [DOI] [PubMed] [Google Scholar]
- 146.Yu YN, Yu TC, Zhao HJ, Sun TT, Chen HM, Chen HY, An HF, Weng YR, Yu J, Li M, et al. Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget. 2015;6(31):32013–32026. doi: 10.18632/oncotarget.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen H, Ye C, Cai B, Zhang F, Wang X, Zhang J, Zhang Z, Guo Y, Yao Q. Berberine inhibits intestinal carcinogenesis by suppressing intestinal pro-inflammatory genes and oncogenic factors through modulating gut microbiota. BMC Cancer. 2022;22(1):566. doi: 10.1186/s12885-022-09635-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Weng W, Goel A. Curcumin and colorectal cancer: An update and current perspective on this natural medicine. Semin Cancer Biol. 2022;80:73–86. doi: 10.1016/j.semcancer.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]


