Abstract
A small open reading frame from the Escherichia coli chromosome, bcrCEC, encodes a homologue to the BcrC subunit of the bacitracin permease from Bacillus licheniformis. We show that disruption of the chromosomal bcrCEC gene causes bacitracin sensitivity and, conversely, that BcrCEC confers bacitracin resistance when expressed from a multicopy plasmid.
Bacitracin is an antibiotic produced by certain species of Bacillus as a mixture of related cyclic polypeptides. Its primary mode of action is interference with bacterial cell wall synthesis through inhibition of dephosphorylation of the peptidoglycan carrier C55-isoprenyl pyrophosphate (IPP) (14). In Bacillus licheniformis, an active ABC-type efflux system comprised of three proteins, BcrA, BcrB, and BcrC, has been shown to be responsible for the resistance of the cells to bacitracin (10). Being a potent antimicrobial drug, bacitracin is used clinically in combination with other antibiotics. However, despite its clinical importance, little is known about the mechanisms by which bacteria other than B. licheniformis acquire resistance to this antibiotic. Only indirect mechanisms of bacitracin resistance, involving IPP, have been described for Escherichia coli and other gram-negative bacteria (4, 11).
Recently, we have isolated an E. coli genomic clone (P2) encoding a multidrug resistance protein (MdfA) with a broad spectrum of drug specificity (6) (also termed Cmr [9]). The DNA sequence of E. coli between mdfA and deoR near min 19 revealed an open reading frame of 198 codons (orf1, termed here bcrCEC). In agreement with the work of Blattner et al. (3), bcrCEC encodes a putative hydrophobic protein (termed here BcrCEC) that exhibits approximately 30% identity to one of the integral membrane subunits of the B. licheniformis bacitracin resistance complex (BcrC) (10). Accordingly, the hydrophobicity profiles of BcrC from B. licheniformis and its homologue from E. coli are nearly identical along most of their primary sequences (data not shown). This suggests a common function. In addition, no significant sequence matches were found with other known proteins, whereas BcrCEC showed approximately 80% identity to a putative permease from Salmonella typhimurium, described as a lipoprotein similar to bacitracin permease from B. licheniformis (15). Here we report that BcrCEC modulates bacitracin resistance in E. coli.
In order to characterize the product of the bcrCEC gene, plasmid pT-bcrCEC was constructed by deleting sequences from the P2 plasmid, leaving intact bcrCEC and its own promoter and also an appropriately oriented T7 promoter (Fig. 1). We then proceeded to verify that the bcrCEC gene product is a transmembrane protein, as expected from its structure. To get a detectable amount of protein, the plasmid pT-bcrCEC was first transformed into E. coli BL21(DE3), and the gene was expressed from the T7 promoter in the presence of [35S]methionine. Membrane fractions were then prepared as described elsewhere (17), and proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A radiolabeled protein with the expected molecular mass for BcrCEC (23 kDa) was observed in the sample from membrane fractions of isopropyl-β-d-thiogalactopyranoside-induced cells, thus confirming the anticipated membrane localization (data not shown).
FIG. 1.
Gene arrangements in the plasmids P2 and pT-bcrCEC. (Upper panel) Organization of the chromosomal E. coli DNA clone P2. (Lower panel) DNA fragment in plasmid pT-bcrCEC.
The putative involvement of BcrCEC in bacitracin resistance was studied by chromosomal bcrCEC gene disruption and by mild overexpression of BcrCEC from pT-bcrCEC. Disruption of the chromosomal bcrCEC was achieved by insertion of a kanamycin resistance cassette from plasmid pUC-4K into the unique BsaBI site present in bcrCEC (Fig. 1). The new plasmid pT-bcrCEC::kan was linearized, and the bcrCEC::kan allele was transferred to the chromosome of JC7623 (recBC sbcB) by homologous recombination. The replacement of bcrCEC by bcrCEC::kan in Kanr Amps transformants was verified by PCR analysis. The bcrCEC::kan mutation was transferred to E. coli UT5600 by P1 transduction (1, 16). In order to test the effect of mild overexpression of BcrCEC, wild-type and mutant cells were transformed by pT-bcrCEC or pT-bcrCEC::kan. Resistance to bacitracin was tested by plating diluted samples of overnight cultures on solid Luria-Bertani medium containing various concentrations of bacitracin (Fig. 2).
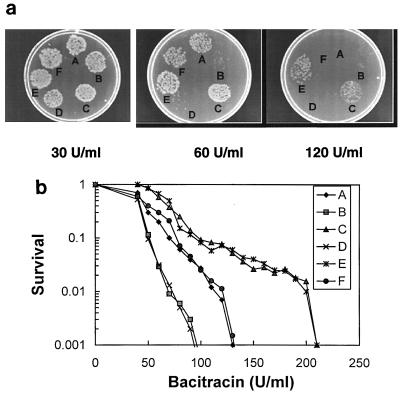
FIG. 2.
Bacitracin resistance of UT5600 and its derivatives. Samples (5 μl) of a 104 dilution of overnight cultures of E. coli UT5600 and derivatives were spotted onto solid Luria-Bertani medium containing indicated concentrations of bacitracin and incubated at 37°C overnight. (a) Photographs. (b) Relative numbers of developed colonies on the various spots (averages of five experiments). A, bcrCEC+; B, bcrCEC::kan; C, bcrCEC::kan/pT-bcrCEC; D, bcrCEC::kan/pT-bcrCEC::kan; E, bcrCEC+/pT-bcrCEC; F, bcrCEC+/pT-bcrCEC::kan.
The results show that cells harboring the chromosomal bcrCEC disrupted gene were two times more sensitive to bacitracin than were wild-type cells (Fig. 2a; MICs of 60 and 120 U/ml, respectively). Furthermore, it was possible to complement this increased sensitivity by transformation with pT-bcrCEC but not by transformation with pT-bcrCEC::kan. This fact indicates that the bacitracin resistance phenotype is specifically related to the status of the bcrCEC gene. The same magnitude of bacitracin sensitization was observed when the bcrCEC::kan allele was transferred into a galU derivative of UT5600, UTL2 (2, 5), 100 times more sensitive to bacitracin because of an altered cell envelope (data not shown). This result suggests that the resistance mediated by BcrCEC is likely to be independent of an intact outer membrane. We note that resistance to other drugs such as vancomycin, chloramphenicol, and tetraphenylphosphorium was not affected in all cases (data not shown).
Overexpression of BcrCEC from its own promoter on a multicopy plasmid allowed the cells to resist up to 200 U of bacitracin per ml (Fig. 2a; drug concentration enabling 1% survival), independently of the carried chromosomal allele, either bcrCEC+ or bcrCEC::kan, thus increasing the resistance to bacitracin by a factor of two or three, respectively. Similar effects of increased resistance were observed when BcrCEC was overexpressed in the bcrCEC::kan galU derivative (data not shown).
The modest magnitude of the effect of overexpression of BcrCEC may be explained in various ways. For example, since it shows identity with a subunit of the bacitracin pump from B. licheniformis, we may expect the transmembrane BcrCEC protein to be part of a bacitracin ABC transporter and therefore to interact with BcrA and BcrB analogs. The low concentration of these proteins, chromosomally encoded but not overexpressed in our assay, would be responsible for the low effect. However, an extensive search for such molecules by sequence comparisons on the complete chromosome has revealed no obvious BcrB homologues in E. coli. Alternatively, BcrCEC could interact heterologously with another ABC system, contributing to substrate recognition and hijacking the energy-utilizing subunit for active export (12). Another possibility, although unlikely, is that BcrCEC acts alone as a monomer or homooligomer, with a low rate of transport. A prediction of the pump model is that the modest increase in the bacitracin extrusion rate would be unnoticeable in the galU permeable mutant, being masked by the high entry rate of bacitracin. However, the UTL2 mutant showed the same response as the UT5600 wild type, which makes this transport model less likely. Another model would postulate a partial protection of the bacitracin target, IPP, by BcrCEC protein, altering the structure of the pyrophosphate end or facilitating the access of pyrophosphatase to its substrate in the presence of bacitracin. Lastly, we cannot exclude the possibility that the observed effects are only secondary consequences of BcrCEC protein action on the cell metabolism.
In summary, the identity between BcrCEC and a subunit from the B. licheniformis bacitracin pump suggested to us a possible involvement of this protein in E. coli resistance to bacitracin, as indeed was verified in our study. The nature of this modulation of antibiotic resistance remains, however, unknown.
Nucleotide sequence accession number.
The bcrCEC GenBank-EMBL-DDBJ accession no. is U00096, f198.
REFERENCES
- 1.Bailone A, Sommer S, Knezevic J, Devoret R. Substitution of UmuD′ for UmuD does not affect SOS mutagenesis. Biochimie. 1991;73:471–478. doi: 10.1016/0300-9084(91)90114-g. [DOI] [PubMed] [Google Scholar]
- 2.Beja O, Bibi E. Functional expression of mouse Mdr1 in an outer membrane permeability mutant of Escherichia coli. Proc Natl Acad Sci USA. 1996;93:5969–5974. doi: 10.1073/pnas.93.12.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Cain B D, Norton P J, Eubanks W, Nick H S, Allen C M. Amplification of the bacA gene confers bacitracin resistance to Escherichia coli. J Bacteriol. 1993;175:3784–3789. doi: 10.1128/jb.175.12.3784-3789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edgar R, Bibi E. A single membrane-embedded negative charge is critical for recognizing positively charged drugs by the Escherichia coli multidrug resistance protein MdfA. EMBO J. 1999;18:822–832. doi: 10.1093/emboj/18.4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinary broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horii Z, Clark A J. Genetic analysis of the RecF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973;80:327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 8.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 9.Nilsen I W, Bakke I, Vader A, Olsvik O, El-Gewely M R. Isolation of cmr, a novel Escherichia coli chloramphenicol resistance gene encoding a putative efflux pump. J Bacteriol. 1996;178:3188–3193. doi: 10.1128/jb.178.11.3188-3193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Podlesek Z, Comino A, Herzog-Velikonja B, Zgur-Bertok D, Komel R, Grabnar M. Bacillus licheniformis bacitracin resistance ABC transporter: relationship to mammalian multidrug resistance. Mol Microbiol. 1995;16:969–976. doi: 10.1111/j.1365-2958.1995.tb02322.x. [DOI] [PubMed] [Google Scholar]
- 11.Pollock T J, Thorne L, Yamazaki M, Mikolajczac M J, Armentrout R W. Mechanism of bacitracin resistance in gram-negative bacteria that synthesize exopolysaccharides. J Bacteriol. 1994;176:93–98. doi: 10.1128/jb.176.20.6229-6237.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross J I, Eady E A, Cove J H, Baumberg S. Identification of a chromosomally encoded ABC-transport system with which the staphylococcal erythromycin exporter MsrA may interact. Gene. 1995;153:93–98. doi: 10.1016/0378-1119(94)00833-e. [DOI] [PubMed] [Google Scholar]
- 13.Studier F W, Moffatt B A. Use of the bacteriophage T7 RNA polymerase to direct selective high expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 14.Toscano W A, Storm D R. Bacitracin. In: Tipper D J, editor. Antibiotic inhibitors of bacterial cell wall synthesis. Elmsford, N.Y: Pergamon Press, Inc.; 1987. pp. 101–113. [Google Scholar]
- 15.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 16.Winans S C, Elledge S J, Krueger J H, Walker G C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985;161:1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zelazny A, Bibi E. Biogenesis and topology of integral membrane proteins: characterization of LacY-Cat hybrids. Biochemistry. 1996;35:10872–10878. doi: 10.1021/bi960815d. [DOI] [PubMed] [Google Scholar]