Abstract
Fungal infections are responsible for significant morbidity and mortality. Resistance to the limited number of agents in the antifungal armamentarium among pathogenic fungi represents a growing public health threat. Particularly concerning is the emerging fungal pathogen Candida auris that frequently exhibits resistance to the triazole class of antifungals and amphotericin B, and for which isolates resistant to all of the major antifungal classes have been reported. In this brief review, we provide an overview of what is currently known about the molecular and genetic basis for antifungal resistance in this fungal pathogen.
Introduction
First reported in 2009 after being isolated from otorrheal specimens in Japan, Candida auris rapidly became a healthcare-associated and multidrug-resistant pathogen of global concern [1,2••]. By 2013, virtually simultaneous outbreaks of antifungal-resistant C. auris causing invasive candidiasis had occurred in South Asia, South Africa, and South America, and C. auris has now been identified in more than 45 countries across six continents, including more than 1700 confirmed clinical cases of C. auris infections in the United States [3]. Further contributing to the clinical significance of this organism are its proclivity to colonize both environmental surfaces and patients, challenges associated with reliable identification in the clinical microbiology laboratory, and the markedly decreased susceptibility to currently available antifungal agents observed in clinical isolates [4,5]. Collectively, these characteristics combined led to C. auris being one of only five pathogens, and the single fungal pathogen, to be categorized as an urgent threat to public health by the Centers for Disease Control and Prevention (CDC) in the U.S. Antibiotic Resistance Threat report [6].
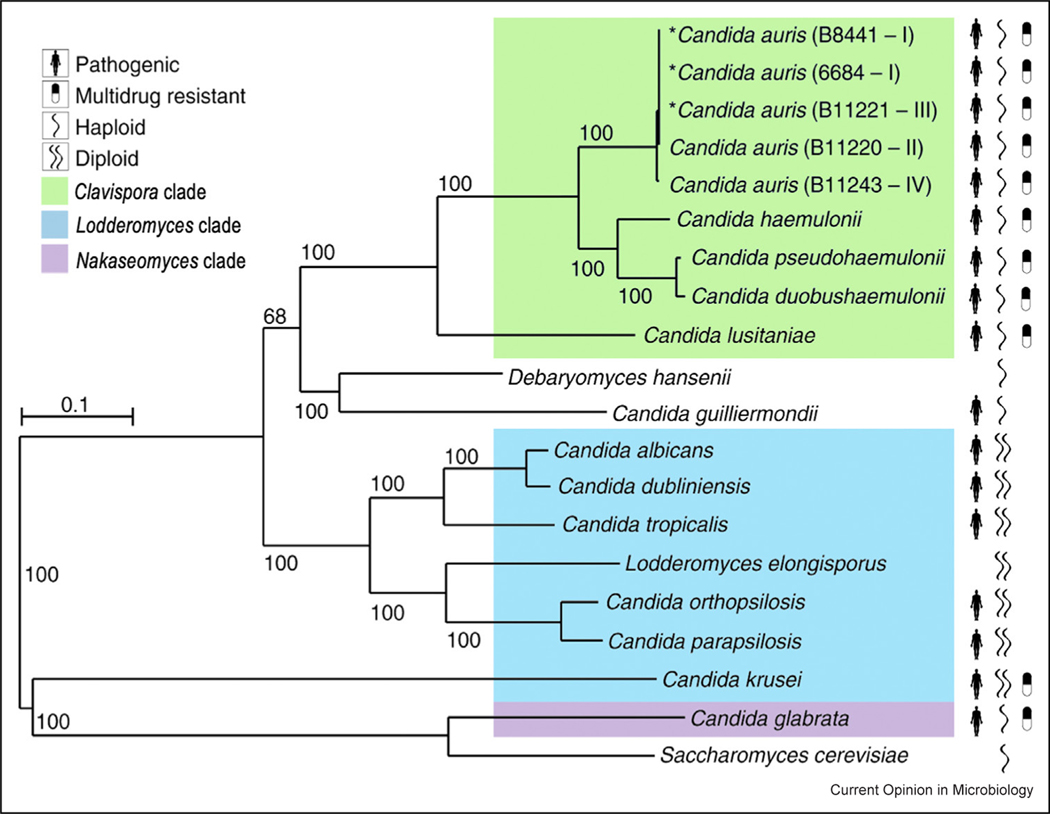
In this brief review, we provide an overview of the molecular and genetic basis of antifungal resistance in this emerging multidrug-resistant pathogen. The species of the Clavispora genus, commonly also referred to as Candida, including the pathogens C. auris, C. haemulonii, and C. lusitaniae, are evolutionarily divergent from the Lodderomyces group species of Candida, which includes C. albicans and the other commonly encountered pathogenic species of Candida (Figure 1) [7••]. In contrast to the Lodderomyces group, the pathogens of the Clavispora genus exhibit haploid genomes, and multidrug resistance is much more commonly observed among clinical isolates [2••,7••,8]. The C. auris genome is predicted to contain 5421–5546 protein-coding genes, and to date, five distinct genetic clades within the species C. auris have been identified. While there is low genetic divergence within the clades, there is high divergence between the clades and notable gene-content differences, including the loss of subtelomeric proteins linked to the cell surface in Clade II and the unique presence of a gene cluster involved in L-rhamnose assimilation in Clade III [9,10]. While isolates from all five clades have been shown to cause invasive infections, isolates of Clades I, III, and IV are more commonly associated with outbreaks of invasive infections and are more often found to be multidrug-resistant, with genes linked to resistance conserved across all clades. Conversely, isolates of Clade II are most often isolated from patients with ear infections and isolates from Clade V have only rarely been reported as causing infections. Using a Bayesian molecular clock analysis, it has recently been shown that the identified major genetic clades diversified within the past 360 years. However, the multidrug-resistant subgroups from all genetic classes of C. auris emerged much more recently (within the past 40 years), contemporaneously with the introduction of the most widely used modern antifungals [11].
Figure 1.
Phylogenomic changes in C. auris and related species. Maximum likelihood phylogeny using 1570 core genes based on 1000 replicates, across 20 annotated genome assemblies, including C. auris and related Candida species. Branch lengths indicate the mean number of changes per site. Drug-resistant species are observed in three clades, Clavispora (including C. auris), Lodderomyces, and Nakaseomyces.
Adapted from Ref. [7••] under the terms of CC BY license.
Only three classes of antifungal agents are useful for the treatment of severe Candida infections: the polyenes (amphotericin B), the triazoles (fluconazole, voriconazole, isavuconazole, itraconazole, and posaconazole), and the echinocandins (caspofungin, micafungin, and anidulafungin). Amphotericin B is only available for intravenous administration and its use is further limited by significant toxicity. Somewhat less-toxic lipid-based formulations of amphotericin B exist but are limited by their high cost. The triazole antifungals, such as fluconazole, are much less toxic, inexpensive, and available for both oral and intravenous administration. The echinocandins, which have emerged as frontline therapy for invasive candidiasis, are likewise less toxic but are only available for intravenous administration [12]. Of great concern, resistance to echinocandins can emerge while on treatment and these resistant isolates have spread in hospitals causing local outbreaks [13]. Importantly, there are few new antifungal classes on the near horizon. Ibrexafungerp represents the only novel class of antifungals, the triterpenoids, to be introduced in nearly 30 years. While this agent may prove to have utility in the treatment of severe Candida infections, including those caused by C. auris, clinical experience with ibrexafungerp remains limited and it is currently only indicated as an orally administered treatment for vulvovaginal candidiasis [14]. Moreover, only a handful of other prospective antifungals are currently in phase II/III or early development, some of which may eventually have utility for treating Candida infections, including those due to C. auris [15]. However, the clinical efficacy of these agents has not been established and they are likely years away from coming to market.
Both epidemiological data and clinical experience for the treatment of C. auris infections are limited and thus epidemiological cutoff values for antifungal susceptibility and true clinical breakpoints cannot yet be established. However, the CDC has proposed tentative breakpoints based on existing clinical and susceptibility data for C. auris in an effort to provide guidance to clinicians [16]. Using these tentative breakpoints, nearly 50% of clinical isolates are considered to be resistant to amphotericin B, 5% are resistant to echinocandins, and approximately 90% are resistant to fluconazole. Multidrug resistance, or resistance to two or more different classes of antifungals, is observed in > 30% of clinical isolates and resistance to all three of these antifungal classes has recently been reported [13].
Resistance to fluconazole
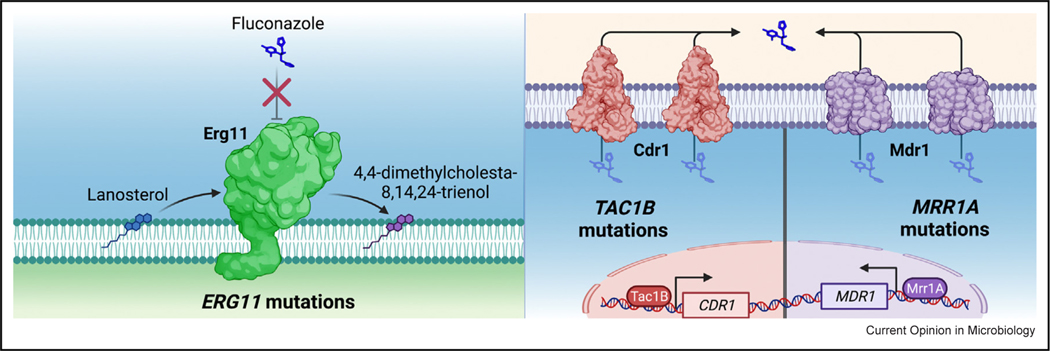
Fluconazole and other triazole antifungals act by competitively inhibiting sterol demethylase, a key enzyme of the ergosterol-biosynthesis pathway in fungi. As most C. auris clinical isolates are resistant to fluconazole, its utility against this pathogen is greatly limited. One mechanism of fluconazole resistance repeatedly identified in C. auris is mutation of the gene encoding the sterol-demethylase enzyme (ERG11). Three mutations in particular have been frequently observed in resistant clinical isolates. These encode the amino acid substitutions VF125AL (commonly referred to as F126L), Y132F, and K143R (Figure 2a). Importantly, associations between these mutations and specific genetic clades of C. auris have been observed (2). Only isolates from Clade III have been found to possess the ERG11 mutation encoding the VF125AL substitution, while the K143R-encoding mutation has only been reported in Clade-I (subclade c) isolates and a small number of individual isolates from Clade II and Clade IV [11,17]. The ERG11 mutation encoding the Y132F substitution however has been found in a large number of isolates from both Clade I (subclade b) and Clade IV. Mutations in the sterol-demethylase gene encoding the Y132F and K143R substitutions have been shown to directly contribute to triazole resistance in other Candida species, including Candida albicans. However, fluconazole-susceptible C. auris clinical isolates carrying these ERG11 mutations have been described previously and isolates with these ERG11 mutations can vary widely in MIC [18•,19•]. Nevertheless, the vast majority of these isolates are found to have MIC above the CDC-proposed resistance breakpoint of 32 mg/L.
Figure 2.
Mutations in C. auris ERG11, TAC1B, and MRR1A contribute to clinical fluconazole resistance.
The specific contribution of the most common ERG11 mutations to fluconazole resistance in C. auris was recently assessed using Cas9 gene editing to introduce mutations encoding the VF125AL, Y132F, and K143R substitutions into C. auris clinical isolate AR0387 that exhibits a fluconazole MIC of 1 mg/L [20•]. These mutations resulted in a fluconazole MIC between 8 and 16 mg/L (8–16-fold change) (Table 1). Of note however, none of these mutations alone resulted in clinical resistance, as defined by the CDC-proposed breakpoint of ≥32 mg/L, when expressed in this clinical-isolate background. In a complementary experiment, the mutation leading to the K143R substitution that is observed in resistant clinical isolate AR0390 (fluconazole MIC of 256 mg/L) was also corrected to the wild-type sequence. This change reduced the fluconazole MIC to 32 mg/L, indicating that, while this ERG11 mutation contributes to resistance in this isolate, it does not account for it in its entirety. One factor that may amplify the effects of mutations in ERG11 is an increase in ERG11 copy number as a result of aneuploidy. In fact, multiple fluconazole-resistant Clade-III clinical isolates (all harboring the VF125AL-encoding mutation) and one fluconazole-resistant Clade-I isolate (harboring the Y132F-encoding mutation) have been reported to carry two and even three copies of these fluconazole-resistance-conferring ERG11 alleles [11].
Table 1.
Characterized mechanisms of triazole resistance and their associated impact on fluconazole MIC.
| Triazole-resistance mechanism | Amino acid substitutions | Impact on fluconazole MIC if known |
|---|---|---|
| ERG11 mutations | – | – |
| Predominant | VF125AL, Y132F, K143R | 8–16-fold increase |
| Less-commonly reported | L43H, Q357K, F444L, G459S, I466M, Y501H | Fourfold increase |
| TAC1B mutations | – | – |
| Predominant | A640V, A657V, F862_N866del | 16-fold increase |
| Less-commonly reported | A15T, S192N, S195C, F214S, K247E, R495G, A583S, P595L/H, S611P, A651T, M653V | Fourfold increase |
| MRR1A mutations | – | – |
| Predominant | N647T | Fourfold increase |
Mutations in bold have been characterized and impact on fluconazole MIC is shown in the adjacent column.
Additional, less common mutations in C. auris ERG11 have also been reported, particularly among isolates from Clade IV, and some of these have been investigated experimentally. Introduction of the mutation encoding the rarely observed F444L substitution into a Clade-IV isolate was found to increase fluconazole MIC fourfold [21]. Conversely, the heterologous expression of C. auris ERG11 alleles, including either of the mutations encoding the I466M and Y501H substitutions in S. cerevisiae, was not found to impact fluconazole or voriconazole MIC [22]. The ERG11 mutations encoding the L43H and Q357K substitutions, which to date have only been reported in single Clade-II isolates obtained from patient ear samples, have yet to be studied and thus their impact on fluconazole resistance remains unclear [17].
In addition to mutations in ERG11, another common contributor to clinical triazole resistance among multiple species of Candida is increased expression of effluxpump-encoding genes [23]. This is typified by C. albicans where both the ATP-binding cassette (ABC)-type efflux-pump-encoding genes CDR1 and CDR2 and the major facilitator superfamily (MFS) transporter gene MDR1 contribute to fluconazole resistance. Moreover, in C. glabrata, nearly all clinical triazole resistance is attributable to overexpression of the (ABC)-type transporter genes CgCDR1, CgPDH1, and CgSNQ2 [24]. The C. auris genome includes many efflux-pump-encoding genes of both the ABC and MFS classes. Fluconazole-resistant clinical isolates of C. auris have been shown to exhibit elevated expression of multiple efflux-pump-encoding genes, and the increased expression of the C. auris ABC-type efflux-pump-encoding gene CDR1 has been shown to substantially contribute to clinical triazole resistance [25,26]. In fact, deletion of CDR1 reduced fluconazole MIC from 256 to 4 mg/L (64-fold reduction) and restored clinical fluconazole susceptibility in the resistant Clade-I clinical isolate AR0390 [25].
Overexpression of genes encoding drug transporters among resistant isolates of other Candida species is predominantly due to activating mutations in genes encoding zinc-cluster transcription factors (ZCF) such as TAC1 and MRR1 in C. albicans and PDR1 in C. glabrata. The C. auris genome contains 68 genes predicted to encode ZCF, including multiple genes with a high degree of homology to C. albicans TAC1 and MRR1. Deletion of one of two C. auris homologs of C. albicans TAC1, called TAC1B, was shown to reduce both fluconazole and voriconazole MIC in fluconazole-resistant clinical isolates from both Clade III and Clade IV [27]. Furthermore, a study of experimentally evolving fluconazole resistance in a Clade-I C. auris clinical isolate observed multiple independently evolved lineages to overexpress CDR1 and have acquired TAC1B mutations (leading to the amino acid substitutions F214S or R495G) after just a single passage in media containing fluconazole. A second passage in fluconazole was observed to further increase fluconazole resistance, possibly as a result of an identified increased copy number of the TAC1B gene [19•]. Similar mutations in TAC1B and TAC1B copy number variations were also observed to arise during in vitro evolution experiments conducted with clinical isolates of both Clade I and II ([28], X).
The clinical significance of mutations in TAC1B was further highlighted by an analysis of the genome sequences of 304 globally distributed C. auris clinical isolates representing each of four of the major genetic clades [19•]. In this collection, mutations in TAC1B were, after ERG11, most frequently associated with fluconazole resistance. Among the mutations identified in this study, three TAC1B mutations, encoding A640V, A657V, and F862_N866del, were commonly found in the most highly resistant isolates from Clades Ic, Ib, and IV, respectively. Introduction of the most common of these (the mutation leading to the A640V substitution) into a fluconazole-susceptible isolate (AR0387, Clade I) increased the fluconazole MIC from 1 to 8 mg/L, whereas correction of this mutation to the wild-type allele in a resistant clinical isolate (AR0390, Clade I) reduced the MIC from 256 to 16 mg/L [19•]. Additionally, a subsequent study demonstrated another TAC1B mutation (encoding S611P) to confer a fourfold increase in fluconazole resistance among four Clade-IV clinical isolates [21]. Altogether, these findings indicate that Tac1B is a major determinant of fluconazole resistance, likely through the regulation of the ABC transporter gene CDR1 and possibly other transporters (Figure 2b).
While the contribution of CDR1 overexpression to clinical fluconazole resistance has been established in isolates from multiple genetic clades, the role of MFS transporters and their transcriptional regulators in fluconazole resistance may be unique to C. auris isolates from Clade III. Nearly all fluconazole-resistant clinical isolates from this clade are found to have a single mutation (resulting in the N647T substitution) in the C. auris ZCF-encoding gene with the highest sequence similarity to C. albicans MRR1, named MRR1A [29]. Furthermore, Clade-III isolates with this mutation in MRR1A, have been observed to express the C. auris MFS-type transporter gene, MDR1, at levels substantially greater than observed in fluconazole-resistant clinical isolates of C. auris from other genetic clades [29]. While deletion of MRR1A in a fluconazole-resistant clinical isolate from Clade IV (wild-type MRR1A sequence) had no impact on fluconazole or voriconazole susceptibility, deletion of MRR1A in an isolate from Clade III, which carries the N647T mutation in MRR1A, resulted in a modest reduction in both fluconazole and voriconazole MIC [27]. Moreover, introduction of the Clade-III-associated N647T mutation in MRR1A into a susceptible Clade-IV clinical isolate was recently found to increase fluconazole and voriconazole MIC by fourfold [30]•. Taken together, the findings of these studies clearly demonstrate both that clinical fluconazole resistance in C. auris is likely to be the result of a combination of multiple resistance mechanisms and mutational stacking in single isolates, and that there are likely important differences in the mechanisms contributing to fluconazole resistance among clinical isolates from different genetic clades.
Resistance to the echinocandins
The echinocandins act by inhibiting the synthesis of a major constituent of the fungal cell wall, 1,3-beta-glucan. In C. albicans and Candida glabrata, mutations in FKS1 (also called GSC1 in C. albicans), leading to amino acid substitutions in the Fks subunit of glucan synthase, have been shown to be the major cause of resistance to the echinocandins. These substitutions occur in two conserved ‘hot spot’ regions (amino acid regions 637–654 and 1345–1365 in C. albicans) and significantly decrease the sensitivity of glucan synthase to inhibition by the echinocandins. Such substitutions are associated with poor drug response in pharmacodynamic studies in animal-infection models and are associated with clinical failures. Approximately 5% of C. auris clinical isolates are found to be resistant to the echinocandins, and in many cases, these resistant isolates are observed to emerge on therapy [2••,11,31•,32]. To date, three different mutations, leading to substitutions in the first hot spot at the amino acid position S639 (S639F, S639P, and S639Y), have most commonly been observed in echinocandin-resistant isolates of C. aruis (Table 2) [18•,33–35]. None however have been directly tested for their effects on echinocandin susceptibility. Additional mutations in both hot-spot region one (encoding D642Y, S639T, F635L/Y, and F635del substitutions) and two (encoding the R1354S/H substitutions) have also been reported less frequently [31•,36–39]. Notably, a number of these less common mutations have been observed in echinocandin-susceptible isolates exhibiting only mild elevations in echinocandin MIC, further demonstrating the importance of characterization of the direct impact each of these mutations has in C. auris.
Table 2.
Mutations in C. auris FKS1 associated with decreased susceptibility to the echinocandins.
| FKS1 mutations | Amino acid substitutions |
|---|---|
| Commonly reported | – |
| Hot spot 1 | S639F/P/Y |
| Less-commonly reported | – |
| Hot spot 1 | F635el, F635L/Y, S639T, D642Y |
| Hot spot 2 |
Resistance to amphotericin B
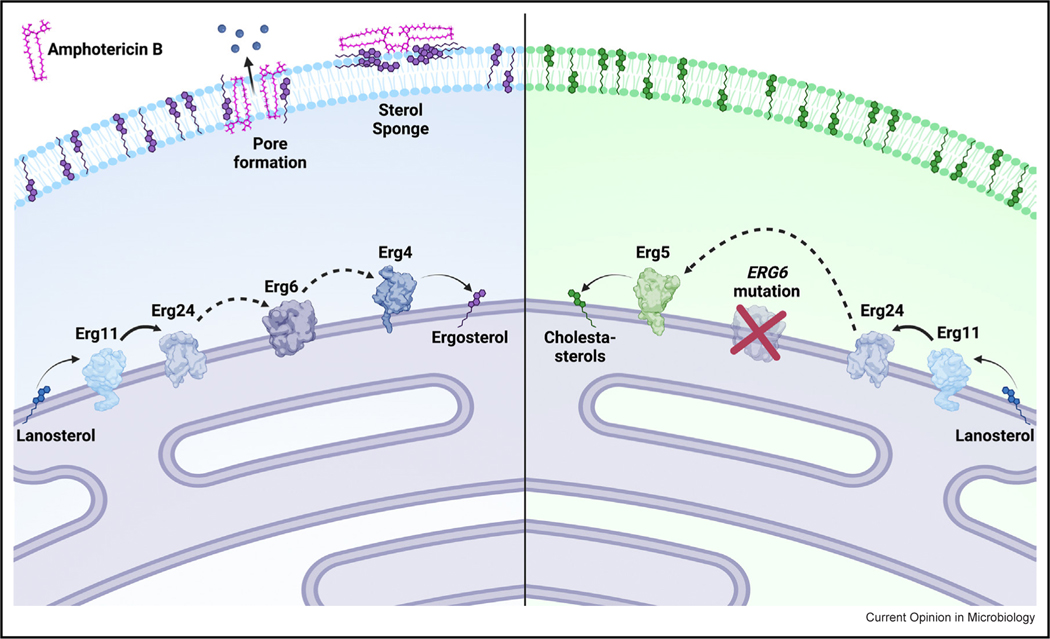
Amphotericin B exerts antifungal activity by directly binding to the predominant fungal membrane sterol, ergosterol, leading to the formation of pores in the cell membrane and/or sequestration of ergosterols (sterol sponging) (Figure 3a). In Candida species, mutations in ergosterol-biosynthesis genes ERG2, ERG3, ERG5, ERG6, and ERG11 reduce susceptibility to amphotericin B, likely as a result of reduced ergosterol in the cell membrane of these isolates [40–42]. Generally, clinical resistance to amphotericin B is remarkably rare among fungi. Presumably, this is because mutations that affect this critical membrane sterol are often associated with significant fitness costs, and amphotericin B typically exerts fungicidal activity. However, while amphotericin-B resistance is uncommon among clinically common species of Candida, multiple species of the Clavispora genus are known to frequently exhibit reduced susceptibility to this agent, including C. auris. Using tentative breakpoints proposed by the CDC, approximately 50% of all C. auris isolates are amphotericin-B-resistant. However, the mechanism(s) by which C. auris becomes resistant to amphotericin B are poorly understood.
Figure 3.
Mutations in C. auris ERG6 confer high-level amphotericin-B resistance through diversion of sterol biosynthesis toward cholesta-type sterols.
In a recently characterized series of four isogenic C. auris isolates from a single patient, resistance to amphotericin B was observed to emerge on therapy. The first isolate in this series exhibited an amphotericin-B MIC of 0.75 mg/L, the second and third > 32 mg/L, and the fourth 0.75 mg/L (by E-test). Notably, the fourth isolate also exhibited resistance to caspofungin (likely due to a mutation in FKS1), and all four isolates were highly resistant to fluconazole (likely due to mutations in both ERG11 and TAC1B), making all but the first isolate in the series multidrug-resistant (resistant to agents from at least two classes). Sequencing of the C. auris sterol-methyltransferase gene, ERG6, revealed the two isolates with elevated amphotericin-B MICs to harbor an indel-type mutation, which resulted in a frameshift and subsequent early stop codon. Introducing this mutation into the ERG6 allele of the original susceptible clinical isolate conferred amphotericin B resistance, while restoration of the wild-type ERG6 allele in the two resistant isolates completely restored amphotericin B susceptibility. Interestingly, the final amphotericin-B-susceptible clinical isolate was found to have acquired a second concomitant indel mutation in ERG6, which restored the native reading frame, resulting in loss of the early stop codon, possibly selected for to mitigate the diminished fitness observed among the two amphotericin-B-resistant isolates. Sterol profiling showed accumulation of cholestatype sterols and an absence of ergosterol (the target of amphotericin B) in all ERG6 mutants consistent with loss of sterol-methyltransferase (Erg6) activity (Figure 3b). Thus, mutations in ERG6 were demonstrated to be the first of likely multiple mechanisms of amphotericin-B resistance operative in clinical isolates of C. auris [38••]. Additionally, in vitro evolution experiments have generated amphotericin-B-resistant strains harboring nonsense mutations in both ERG11 and ERG3 (23), supporting that mutations in other ergosterol-biosynthesis genes may also be involved in clinical resistance as has been observed in other species of Candida. However, as many amphotericin-B-resistant isolates of C. auris exhibit lower levels of resistance (MIC 2–4 mg/L), and the vast majority are observed to lack mutations in ergosterol-biosynthesis genes, it appears likely that other mechanisms of amphotericin-B resistance remain to be identified.
Conclusion
Candida auris represents a unique threat among fungal pathogens owing to its rapid emergence in independent regions across the globe, its ability to colonize patients and persist on healthcare-associated surfaces, and its proclivity to achieve multidrug resistance. Indeed, the great breadth of antifungal resistance observed in C. auris makes it particularly challenging, leaving few and sometimes no reliable therapeutic options for treating infections due to this organism. While important mechanisms of resistance have been uncovered for the triazoles, echinocandins, and amphotericin B, there remains substantial resistance that cannot be explained by what has been learned to date. Examining differences between C. auris and related yeast species may be useful for understanding the unusual emergence and spread of this pathogen and its propensity to be resistant to one or more antifungal agents. Comparing and contrasting C. auris with other often antifungal-resistant Clavispora species, as well as comparisons within specific clades of C. auris, will not only aid the identification of individual resistance determinants, but also will further shed light on the evolutionary trajectory of resistance within populations of this species across the world. Moreover, continued advances in the genetic manipulation of this organism will facilitate validation of resistance mechanisms that will ultimately lead to better diagnostics, novel therapeutics, and strategies to reclaim the utility of the triazole, echinocandin, and polyene classes of antifungals against resistant C. auris infections.
Acknowledgements
J.M.R. is supported by the Society of Infectious Diseases Pharmacists Young Investigator Research Award and a St. Jude Children’s Research Hospital Children’s Infection Defense Center Grant. C.A.C. is supported by the National Institutes of Health NIAID award U19AI110818 to the Broad Institute and is a CIFAR fellow in the Fungal Kingdom Program. P.D.R. is supported by St. Jude Children’s Research Hospital and National Institutes of Health NIAID R01 Grants R01 AI058145, R01 AI131620, and R01 AI143197.
Given his role as Guest Editor, David Rogers had no involvement in the peer-review of this article and has no access to information regarding its peer-review. Full responsibility for the editorial process for this article was delegated to Richard Lee.
Footnotes
Conflict of interest statement
The authors report no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H: Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 2009, 53:41–44. [DOI] [PubMed] [Google Scholar]
- 2. Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, et al. : Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 2017, 64:134–140. •• Original description of clinical outbreaks of C. auris and the emergence of distinct genetic clades.
- 3.Anonymous: Tracking Candida auris; 2019. 〈https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html〉.
- 4.Lockhart SR: Candida auris and multidrug resistance: defining the new normal. Fungal Genet Biol 2019, 131:103243. [DOI] [PubMed] [Google Scholar]
- 5.Caceres DH, Forsberg K, Welsh RM, Sexton DJ, Lockhart SR, Jackson BR, Chiller T: Candida auris: a review of recommendations for detection and control in healthcare settings. J Fungi (4) 2019, 5:111, 10.3390/jof5040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anonymous: CDC’s Antibiotic Resistance Threats in the United States; 2019. 〈https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf〉.
- 7. Munoz JF, Gade L, Chow NA, Loparev VN, Juieng P, Berkow EL, Farrer RA, Litvintseva AP, Cuomo CA: Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun 2018, 9:5346. •• Describes the first high quality genome assemblies for all four of the major genetic clades of C. auris and the genetic relatedness of C. auris to species of Candida and Clavispora genuses.
- 8.Gade L, Munoz JF, Sheth M, Wagner D, Berkow EL, Forsberg K, Jackson BR, Ramos-Castro R, Escandon P, Dolande M, et al. : Understanding the emergence of multidrug-resistant Candida: using whole-genome sequencing to describe the population structure of Candida haemulonii species complex. Front Genet 2020, 11:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz JF, Welsh RM, Shea T, Batra D, Gade L, Howard D, Rowe LA, Meis JF, Litvintseva AP, Cuomo CA: Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics (1) 2021, 218:iyab029, 10.1093/genetics/iyab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambaraghassi G, Dufresne PJ, Dufresne SF, Vallieres E, Munoz JF, Cuomo CA, Berkow EL, Lockhart SR, Luong ML: Identification of Candida auris by use of the updated Vitek 2 yeast identification system, Version 8.01: a multilaboratory evaluation study. J Clin Microbiol (11) 2019, 57:e00884–19, 10.1128/JCM.00884-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow NA, Munoz JF, Gade L, Berkow EL, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, et al. : Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis RE, Rogers PD: Invasive fungal infections. In Pharmacotherapy Principles and Practice. Edited by Chisholm-Burns MA, Wells BG, Schwinghammer TL, Malone PM, Kolesar JM, Rotschafer JC, DiPiro JT:1229–1245.
- 13.Lyman M, Forsberg K, Reuben J, Dang T, Free R, Seagle EE, Sexton DJ, Soda E, Jones H, Hawkins D, et al. : Notes from the field: transmission of pan-resistant and echinocandin-resistant Candida auris in health care facilities — Texas and the District of Columbia, January-April 2021. MMWR Morb Mortal Wkly Rep 2021, 70:1022–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobel R, Nyirjesy P, Ghannoum MA, Delchev DA, Azie NE, Angulo D, Harriott IA, Borroto-Esoda K, Sobel JD: Efficacy and safety of oral ibrexafungerp for the treatment of acute vulvovaginal candidiasis: a global phase 3, randomised, placebo-controlled superiority study (VANISH 306). BJOG 2022, 129:412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamoth F, Lewis RE, Kontoyiannis DP: Investigational antifungal agents for invasive mycoses: a clinical perspective. Clin Infect Dis (3) 2022, 75:534–544, 10.1093/cid/ciab1070. [DOI] [PubMed] [Google Scholar]
- 16.Anonymous: Candida auris: Antifungal Susceptibility Testing and interpretation. Centers for Disease Control and Prevention, Atlanta, GA; 2020. 〈https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html〉. [Google Scholar]
- 17.Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, Lee SY, Chun S, Lee JH, Choi HJ, et al. : Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol (4) 2019, 57:e01624–18, 10.1128/JCM.01624-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, et al. : A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 2018, 73:891–899. • First characterization of ERG11 and FKS1 mutations among a large collection of C. auris clinical isolates.
- 19. Rybak JM, Munoz JF, Barker KS, Parker JE, Esquivel BD, Berkow EL, Lockhart SR, Gade L, Palmer GE, White TC, et al. : Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio (3) 2020, 11:e00365–20, 10.1128/mBio.00365-20. • First characterization of the predominance of mutations in C. auris TAC1B among a global collection of C. aruis, and assessment of the direct impact of mutations in C. auris TAC1B on triazole susceptibility.
- 20. Rybak JM, Sharma C, Doorley LA, Barker KS, Palmer GE, Rogers PD: Delineation of the direct contribution of Candida auris ERG11 mutations to clinical triazole resistance. Microbiol Spectr 2021, 9:e0158521. • First characterization of the impact of the most common mutations in C. auris ERG11 on fluconazole resistance, and determination that these mutations alone are likely insufficient to confer clinical triazole resistance.
- 21.Li J, Coste AT, Liechti M, Bachmann D, Sanglard D, Lamoth F: Novel ERG11 and TAC1b mutations associated with azole resistance in Candida auris. Antimicrob Agents Chemother (2) 2021, 65:e02663–20, 10.1128/AAC.02663-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Healey KR, Kordalewska M, Jimenez Ortigosa C, Singh A, Berrio I, Chowdhary A, Perlin DS: Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother (10) 2018, 62:e01427–18, 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD: Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front Microbiol 2016, 7:2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whaley SG, Zhang Q, Caudle KE, Rogers PD: Relative contribution of the ABC transporters Cdr1, Pdh1, and Snq2 to azole resistance in Candida glabrata. Antimicrob Agents Chemother (10) 2018, 62:e01070–18, 10.1128/AAC.01070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rybak JM, Doorley LA, Nishimoto AT, Barker KS, Palmer GE, Rogers PD: Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob Agents Chemother (4) 2019, 63:e00057–19, 10.1128/AAC.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SH, Iyer KR, Pardeshi L, Munoz JF, Robbins N, Cuomo CA, Wong KH, Cowen LE: Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. mBio (1) 2019, 10, 10.1128/mBio.02529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayr EM, Ramirez-Zavala B, Kruger I, Morschhauser J, Zinc A: A zinc cluster transcription factor contributes to the intrinsic fluconazole resistance of Candida auris. mSphere (2) 2020, 5:e00279–20, 10.1128/mSphere.00279-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carolus H, Pierson S, Munoz JF, Subotic A, Cruz RB, Cuomo CA, Van Dijck P: Genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug resistance. mBio (2) 2021, 12:e03333–20, 10.1128/mBio.03333-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer KR, Camara K, Daniel-Ivad M, Trilles R, Pimentel-Elardo SM, Fossen JL, Marchillo K, Liu Z, Singh S, Munoz JF, et al. : An oxindole efflux inhibitor potentiates azoles and impairs virulence in the fungal pathogen Candida auris. Nat Commun 2020, 11:6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J, Coste AT, Bachmann D, Sanglard D, Lamoth F: Deciphering the Mrr1/Mdr1 pathway in azole resistance of Candida auris. Antimicrob Agents Chemother 2022, 66:e0006722. • First demonstration of the impact of mutations in C. auris MRR1A on fluconazole susceptibilities.
- 31. Al-Obaid I, Asadzadeh M, Ahmad S, Alobaid K, Alfouzan W, Bafna R, Emara M, Joseph L: Fatal breakthrough Candidemia in an immunocompromised patient in Kuwait due to Candida auris exhibiting reduced susceptibility to echinocandins and carrying a novel mutation in Hotspot-1 of FKS1. J Fungi (3) 2022, 8:267, 10.3390/jof8030267. • Detailed description of the emergence of parallel lineages of C. auris with reduced echinocandin susceptibility in a single patient and discussion of the various known FKS1 mutations associated with echinocandin resistance.
- 32.Biagi MJ, Wiederhold NP, Gibas C, Wickes BL, Lozano V, Bleasdale SC, Danziger L: Development of high-level echinocandin resistance in a patient with recurrent Candida auris Candidemia secondary to chronic Candiduria. Open Forum Infect Dis 2019, 6:ofz262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kordalewska M, Lee A, Park S, Berrio I, Chowdhary A, Zhao Y, Perlin DS: Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob Agents Chemother (6) 2018, 62:e00238–18, 10.1128/AAC.00238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berkow EL, Lockhart SR: Activity of CD101, a long-acting echinocandin, against clinical isolates of Candida auris. Diagn Microbiol Infect Dis 2018, 90:196–197. [DOI] [PubMed] [Google Scholar]
- 35.Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, Fisher MC, Schelenz S: Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect 2018, 7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alfouzan W, Ahmad S, Dhar R, Asadzadeh M, Almerdasi N, Abdo NM, Joseph L, de Groot T, Alali WQ, Khan Z, et al. : Molecular epidemiology of Candida auris outbreak in a major secondary-care hospital in Kuwait. J Fungi (4) 2020, 6:307, 10.3390/jof6040307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma D, Paul RA, Rudramurthy SM, Kashyap N, Bhattacharya S, Soman R, Shankarnarayan SA, Chavan D, Singh S, Das P, et al. : Impact of FKS1 genotype on echinocandin in vitro susceptibility in Candida auris and in vivo response in a murine model of infection. Antimicrob Agents Chemother 2022, 66:e0165221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rybak JM, Barker KS, Munoz JF, Parker JE, Ahmad S, Mokaddas E, Abdullah A, Elhagracy RS, Kelly SL, Cuomo CA, et al. : In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin Microbiol Infect (6) 2021, 28:838–843, 10.1016/j.cmi.2021.11.024. •• Identification and characterization of the first known mechanism of amphotericin B resistance in C. auris.
- 39.Asadzadeh M, Mokaddas E, Ahmad S, Abdullah AA, de Groot T, Meis JF, Shetty SA: Molecular characterisation of Candida auris isolates from immunocompromised patients in a tertiary-care hospital in Kuwait reveals a novel mutation in FKS1 conferring reduced susceptibility to echinocandins. Mycoses 2022, 65:331–343. [DOI] [PubMed] [Google Scholar]
- 40.Geber A, Hitchcock CA, Swartz JE, Pullen FS, Marsden KE, KwonChung KJ, Bennett JE: Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother 1995, 39:2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hull CM, Bader O, Parker JE, Weig M, Gross U, Warrilow AG, Kelly DE, Kelly SL: Two clinical isolates of Candida glabrata exhibiting reduced sensitivity to amphotericin B both harbor mutations in ERG2. Antimicrob Agents Chemother 2012, 56:6417–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Kelly DE, Kelly SL: A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14alpha-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob Agents Chemother 2010, 54:3578–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]