Abstract
Deletion and mutational analysis of the promoter P-dapA from Corynebacterium glutamicum was performed to identify regions and particular nucleotides important for its function. An extended −10 region and a stretch of six T’s at positions −55 to −50 were found to be the most important elements in the promoter function. The results of mutational analysis of P-dapA are consistent with the conclusions of statistical computer-aided analysis of 44 C. glutamicum promoter sequences.
Use of Corynebacterium glutamicum in industrial production of amino acids incited genetic studies on amino acid biosynthesis pathways and on central metabolism in this gram-positive species (14). However, efficient utilization of gene cloning techniques and construction of overproducing strains with precisely tuned levels of expression of individual biosynthetic genes are still impeded by the lack of data on transcription initiation signals in C. glutamicum and on elements regulating their activity.
Computer-aided analysis of C. glutamicum promoters.
Recently, we isolated a number of C. glutamicum promoters and mapped the respective transcription start (TS) sites. Using the programs PROBAB and PROMSCAN, we analyzed 33 C. glutamicum promoters and defined the consensus DNA sequence of promoters of vegetative genes in C. glutamicum (10). Now we have used the same programs to analyze a set of 44 C. glutamicum promoter sequences. The result was essentially the same as that of the previous analysis, with some minor deviations. In the −35 region, the bases in hexamer TTGGCA were rather evenly conserved (48 to 55%). In the −10 region, the bases of the hexamer T1A2(T/G)3A4A5T6 were conserved to the following levels: T6, 91%; T1, 80%; A2, 73%; A4, 52%; and A5, 48%. At the nonconserved third position, T and G occurred with the same frequency (32%). Outside the −10 hexamer, the G at the second base upstream and the G at the second base downstream were less highly conserved (55 and 43%, respectively). The main motifs, TTGGCA and GNTANAATNG, were substantially less conserved than the canonical hexamers TTGACA and TATAAT of Escherichia coli (1, 3) and Bacillus subtilis (4). A low level of conservation of bases was conspicuous particularly in the −35 region, which was hardly recognizable in many of the analyzed promoters. This finding suggested that the −35 region plays a less important role in the function of C. glutamicum promoters.
Until now, no particular C. glutamicum promoter has been analyzed in detail. We have chosen the promoter of the dapA gene (P-dapA) for this purpose. The dapA gene codes for dihydrodipicolinate synthase, the enzyme catalyzing the first reaction in the lysine-specific biosynthetic pathway.
Cloning of P-dapA.
In P-dapA, the putative −10 hexamer TAACCT shares 3 bases with the consensus sequence of the C. glutamicum −10 region, TANAAT, while the −35 hexamer is not apparent in the common −35 region (10). The sequence TTAACC (−39 to −34), with 3 bases matching those found in the C. glutamicum consensus sequence might function as the −35 hexamer; however, the spacing difference of 19 bases makes it less probable. We constructed a transcriptional fusion of P-dapA, carried on the 400-bp BamHI-KpnI fragment, to the cat reporter gene in the multicopy promoter-probe vector pET2 (17) replicating in C. glutamicum. To avoid possible effects of varying the plasmid copy number, the promoter fragment was also cloned in the integrative promoter-probe vector pRIM2 (17) for assay in a single-copy system. The vector pRIM2 was transferred into C. glutamicum by the conjugation technique described by Schäfer et al. (15).
Activity of P-dapA.
Activity of the wild-type (WT) promoter and of its deletion and mutation derivatives was evaluated by chloramphenicol acetyltransferase (CAT) assay done by the method of Shaw (16). One unit of enzyme activity was defined as 1 μmol of chloramphenicol acetylated per minute.
Activity of the promoter cloned into pET2 and into pRIM2 (and integrated in the chromosome) was measured in CGXII minimal medium (6) and in Casitone-yeast extract complete medium (11). Activity of P-dapA in minimal medium was about 85% of the activity found in complete medium in both multicopy and single-copy systems (0.11 and 0.007 U · mg of protein−1, respectively). Results of measurements of the promoter activity during growth in Casitone-yeast extract medium showed little deviation during the lag, mid-log, and stationary phases (0.14, 0.13, and 0.16 U · mg of protein−1, respectively). These results suggested that the promoter is not regulated or that the regulation plays a minor role. All further measurements were done during the mid-log phase of growth in complete medium.
Deletion analysis of P-dapA.
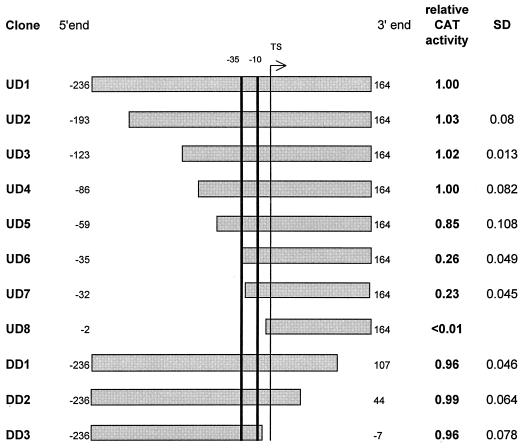
Deletion analysis of the 400-bp fragment (Fig. 1) carrying P-dapA was performed to define the extent of sequence necessary for promoter function. PCR was used to create fragments of various lengths, with appropriate restriction sites attached to the ends for cloning. The truncated fragments were cloned in pET2 and tested for their promoter activity (Fig. 1). Deletions at the 5′ end as far as position −86 did not affect P-dapA strength. Deletions downstream of the TS, including a deletion up to position −7 that encompassed the TS (clone DD3), also resulted in little or no effect on promoter activity. A remarkable reduction of activity (to 26%) was caused by the deletion of nucleotides between positions −59 and −35 (UD6). Deletion of 3 more bases covering the possible −35 region resulted in a negligible reduction of activity. A further deletion that removed the −10 region (UD8) abolished promoter activity completely. We conclude from these results that (i) the region necessary for full P-dapA activity is not longer than 80 bp (−86 to −7), (ii) the −10 element is essential for P-dapA activity, and (iii) the 22-bp sequence upstream of position −35 is necessary for full activity of the promoter P-dapA.
FIG. 1.
Deletion mapping of the promoter P-dapA. The horizontal shaded boxes represent the relative lengths of DNA fragments carrying the promoter studied. The 5′ and 3′ ends of the fragments relative to the TS are shown. The thick vertical bars mark the positions of potential −35 and −10 regions of the promoter. The vertical bar with an arrow on the top indicates the position of TS. PCR-generated fragments were cloned into pET2 to construct cat transcriptional fusions for determination of promoter activity. CAT activity is expressed relative to that measured with full-length fragment (UD1). Standard deviations (SD) from three independent measurements are shown.
Mutations in the −59 to −35 region.
To identify which bases upstream of position −35 were essential for promoter activity, 1- to 4-base changes were introduced by oligonucleotide-directed mutagenesis (5) into the sequence between positions −59 and −35. These substitutions were done in the sequences potentially important in promoter function (Fig. 2), as follows: in the AT-rich region at the 5′ end of the −35 region (clone ME77), in the stretch of four G’s (clones ME77 and MH1) forming part of a 6-base inverted repeat with the sequence AACCCC within the −35 region, and in the stretch of six T’s (clones P4 and R2) which might induce an intrinsic bend in a DNA strand. The TAA→GGG change at positions −38 to −36 in P-dapA mutant ME77 resulted in a level of activity about 70% of that of the WT promoter. This relatively small reduction of activity caused by the replacement of bases forming part of a potential −35 region (TTAACC or AACCCC with a spacing difference of 19 or 17 bp, respectively) reflected the probable minor role of the −35 region in promoter function. Disruption of the inverted repeat (Fig. 2) by a substitution of AAA for GGG at positions −46 to −44 (clone MH1) had also a small negative effect (reducing the level of activity to 80% of the WT level of activity). In contrast, both single-base (−53T→A) and double-base (−53,−52TT→CC) substitutions in the six-T tract reduced the promoter activity to about 20% (promoters P4 and R2). Removing this run of six consecutive T’s was therefore probably mainly responsible for the marked decrease of activity of the clone with a deletion at the 5′ end at position −35. We hypothesize that the T tract induces a DNA bend (12) or serves as an UP element (13) in P-dapA.
FIG. 2.
Effects of mutations in the −59 to −35 region of P-dapA on promoter activity. The arrows mark the inverted repeat. Replacement bases are indicated in bold face. Transcriptional activity of the mutated fragments was evaluated in the vector pET2. TS marks the transcription start (underlined A). CAT activity is expressed relative to that measured with the WT promoter. Standard deviations (SD) from three independent measurements are shown.
Mutations in the −10 region.
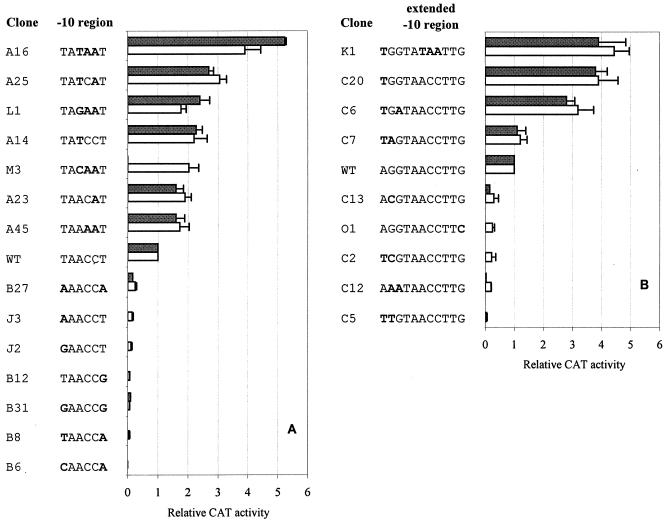
All transcriptional fusions of the promoter mutations to the cat reporter were borne by the plasmid pET2. Most of them were also integrated into the chromosome with pRIM2, and promoter strength was evaluated in both systems. To prove that the sequence TAACCT from positions −14 to −9 functions as the −10 hexamer, −14T and/or −9T, occupying the most conserved positions, were replaced with other bases. In all these cases (clones B27, J3, J2, B12, B31, B8, and B6), activity of the promoter was nearly eliminated (Fig. 3A). This result confirmed that these T’s are essential for promoter function and that the sequence TAACCT is really the −10 hexamer of P-dapA. All substitutions of one, two, or three bases increasing the homology with the consensus sequence TANAAT gave rise to promoters stronger than the original one (Fig. 3A). We made all possible changes at the third position of the hexamer (which is not conserved according to the statistical analysis of C. glutamicum promoters) combined with substitutions of AA for CC at positions −11 and −10. Among them, the sequence TAAAAT (mutant A45) yielded the lowest levels of activity (1.5- and 1.7-fold times the WT promoter activity level in the single-copy and multicopy systems, respectively), while the hexamer TATAAT (A16) represented the strongest sequence variation (with 5.2- and 3.9-fold times the activity of the WT promoter). The hexamers TAGAAT (L1) and TACAAT (M3) formed promoters of intermediate strength (with 1.8- to 2.3-fold times the WT promoter activity).
FIG. 3.
Effects of mutations in the −10 region of P-dapA on promoter activity. Replacement bases are indicated in boldface. CAT activity measured by using the transcriptional fusion to the cat reporter gene in the multicopy vector pET2 is shown by open bars; activity measured after integration of the promoter fragment with the vector pRIM2 into the C. glutamicum chromosome is shown by solid bars. CAT activity is expressed relative to that measured with the WT promoter. Standard deviations (SD) from three independent measurements are indicated with error bars. (A) Mutations within the −10 hexamer; (B) mutations outside of the −10 hexamer.
From the level of base conservation (55%), it follows that the G at the second base upstream of the −10 hexamer (−16 in P-dapA) may also play a significant role in the functioning of C. glutamicum promoters. In agreement with this assumption, replacement of −16G in P-dapA with any other base (clones C13, C2, C12, and C5) led to a dramatic drop in activity (Fig. 3B). We showed recently that a G→C transversion at this position in the promoter of the InCg integron resulted in a 5-fold decrease in the promoter activity in C. glutamicum (9). In conclusion, both statistical and experimental evidence show clearly that the G at the second position upstream of the −10 hexamer is an important base in C. glutamicum promoters.
In some gram-positive bacteria the conserved TGN motif upstream of the −10 hexamer was found to participate in promoter function (2, 4), thus forming an extended −10 region. In contrast, a T just upstream of the G is not conserved in C. glutamicum. Nevertheless, the substitution of T for A at position −17, creating the TG motif in P-dapA, resulted in a fourfold increase in promoter activity (clone C20) (Fig. 3B). In the double mutant C7, with a strength comparable to the WT P-dapA, the −17A→T change seems to compensate for the negative effect of the substitution of A for G at position −16. However, in two other poor promoters, C2 and C5, the substitution of T for A at position −17 could not compensate for the deleterious mutations −16G→C and −16G→T. This result confirms the greater importance of G in the TG motif in corynebacteria. The TG dinucleotide in the −16 region was also found to be essential for the functioning of some E. coli promoters (7), though it is not conserved in this species (3). The role of an extended −10 region in promoter function was found to be particularly important for the promoters missing the −35 motif (8). Within 12 C. glutamicum promoters lacking a discernible −35 hexamer, the G at the second base upstream of the −10 hexamer appears in 10 cases (83%); however, the TG dimer appears in only 4 promoters (33%). We assume that the presence of G or TG in the −16 region may compensate for a weak −35 hexamer in these promoters. When the most efficient up mutations in the extended −10 region were combined (TGGTATAAT), the promoter created (K1) had a strength similar to that of promoters A16 and C20 (Fig. 3B).
In addition to the G at the second base upstream of the −10 hexamer, the G at the second base downstream of this hexamer was found to be weakly conserved (43%) in C. glutamicum promoter sequences. A single transversion, −7G→C, in P-dapA mutant O1 had a strong negative effect, giving rise to the promoter with only 14% of the activity of the native promoter (Fig. 3B). These results indicate that the G at the second base downstream of the −10 hexamer may also form a part of the extended −10 element, specific to C. glutamicum.
Conclusion.
Mutational analysis of the dapA promoter supports the results of computer analysis of the set of C. glutamicum promoters aimed at defining the consensus sequence of C. glutamicum promoters of vegetative genes. We suggest that the extended −10 region is more important for transcription initiation than the −35 region in many C. glutamicum promoters. Mutagenesis study of other promoters, in particular those dependent on the −35 region, is necessary to establish the final C. glutamicum promoter consensus sequence.
Acknowledgments
We thank L. Eggeling (Jülich) for helpful discussions and A. Šroglová for excellent technical assistance.
The work was supported by grant 021/41325652/930 from Forschungszentrum Jülich, Germany, by grant 204/97/0528 from the Grant Agency of the Czech Republic, and by NATO Linkage Grant HTECH.LG 940257.
REFERENCES
- 1.Galas D J, Eggert M, Waterman M S. Rigorous pattern-recognition methods for DNA sequences. J Mol Biol. 1985;186:117–128. doi: 10.1016/0022-2836(85)90262-1. [DOI] [PubMed] [Google Scholar]
- 2.Graves M C, Rabinowitz J C. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. Evidence for “extended” promoter elements in gram-positive organisms. J Biol Chem. 1986;261:11409–11415. [PubMed] [Google Scholar]
- 3.Harley C B, Reynolds R P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helmann J D. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 6.Keilhauer C, Eggeling L, Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 8.Kumar A, Malloch R A, Fujita N, Smillie D A, Ishihama A, Hayward R S. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 9.Nešvera J, Hochmannová J, Pátek M. An integron of class 1 is present on the plasmid pCG4 from Gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol Lett. 1998;169:391–395. doi: 10.1111/j.1574-6968.1998.tb13345.x. [DOI] [PubMed] [Google Scholar]
- 10.Pátek M, Eikmanns B J, Pátek J, Sahm H. Promoters from Corynebacterium glutamicum: cloning, molecular analysis and search for a consensus motif. Microbiology. 1996;142:1297–1309. doi: 10.1099/13500872-142-5-1297. [DOI] [PubMed] [Google Scholar]
- 11.Pátek M, Nešvera J, Hochmannová J, Štokrová J. Transfection of Brevibacterium flavum with bacteriophage BFB10 DNA. Folia Microbiol. 1988;33:247–254. [Google Scholar]
- 12.Perez-Martin J, de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu Rev Microbiol. 1997;51:593–628. doi: 10.1146/annurev.micro.51.1.593. [DOI] [PubMed] [Google Scholar]
- 13.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 14.Sahm H, Eggeling L, Eikmanns B, Krämer R. Metabolic design in amino acid-producing bacterium Corynebacterium glutamicum. FEMS Microbiol Rev. 1995;16:243–252. [Google Scholar]
- 15.Schäfer A, Kalinowski J, Simon R, Seep-Feldhaus A H, Pühler A. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J Bacteriol. 1990;172:1663–1666. doi: 10.1128/jb.172.3.1663-1666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw W V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 17.Vašicová P, Abrhámová Z, Nešvera J, Pátek M, Sahm H, Eikmanns B. Integrative and autonomously replicating vectors for analysis of promoters in Corynebacterium glutamicum. Biotechnol Tech. 1998;12:743–746. [Google Scholar]