Abstract
Background
Strong surveillance systems with wide geographic coverage are needed to detect and respond to reports of antimalarial drug resistance on the African continent. We aimed to assess the utility and feasibility of using blood-fed mosquitos (xenomonitoring) to conduct rapid surveillance of molecular markers associated with resistance in human populations.
Methods
We conducted three cross-sectional surveys in two rainy seasons and the interim dry season in southwest Burkina Faso between Oct 10, 2018, and Sept 17, 2019. We collected human blood samples and blood-fed mosquitos residing in household clusters across seven village sectors. Samples were assessed for Plasmodium falciparum with ultrasensitive quantitative PCR, genotyped for two markers of reduced drug susceptibility, pfmdr1 256A>T (Asn86Tyr) and pfcrt 227A>C (Lys76Thr), and sequenced for four markers of clonality. We assessed statistical equivalence using a 10% margin of equivalence.
Findings
We identified 551 infections in 1483 human blood samples (mean multiplicity of infection [MOI] 1·94, SD 1·47) and 346 infections in 2151 mosquito blood meals (mean MOI 2·2, SD 1·67). The frequency of pfmdr1 Asn86Tyr was 4% in survey 1, 2% in survey 2, and 12% in survey 3 in human samples, and 3% in survey 1, 0% in survey 2, and 8% in survey 3 in mosquito blood meals, and inter-host frequencies were statistically equivalent in surveys 1 and 2 (p<0·0001) but not Survey 3 (p=0·062) within a tolerability of 0·10. The frequency of pfcrt Lys76Thr was 16% in survey 1, 55% in survey 2, and 11% in survey 3 in humans and 40% in survey 1, 72% in survey 2, and 13% in survey 3 in mosquitos, and inter-host frequencies were equivalent in survey 3 only (p=0·032) within a tolerability of 0·10. In simulations, multiple but not preferential feeding behaviour in mosquitos reduced the accuracy of frequency estimates between hosts, particularly for markers circulating at higher frequencies.
Interpretation
Molecular markers in mosquito blood meals and in humans exhibited similar temporal trends but frequencies were not statistically equivalent in all scenarios. More work is needed to determine empirical and pragmatic thresholds of difference. Xenomonitoring might be an efficient tool to provide rapid information on emerging antimalarial resistance in regions with insufficient surveillance.
Funding
National Institute of Allergy and Infectious Diseases.
Introduction
Drug resistance continues to pose major challenges for malaria control. Artemisinin-based combination therapies (ACTs) are the only approved drugs to treat malaria in sub-Saharan Africa, a region that accounts for approximately 95% of global malaria cases and deaths.1 Artemether–lumefantrine and artesunate–amodiaquine are the most commonly used ACTs in sub-Saharan Africa, but the genotypic landscape of resistance has changed dramatically since their introduction in 2004.2,3 Parasites with mutations conferring reduced partner drug susceptibility have become widespread throughout sub-Saharan Africa, and parasites with reduced in-vivo artemisinin susceptibility were reported for the first time in sub-Saharan Africa in Rwanda and Uganda in the past 2 years.2–5 Surveillance systems with broad geographic and temporal coverage are urgently needed to monitor the geospatial spread of resistance and rapidly inform treatment and prophylaxis policies.
Surveillance of antimalarial drug resistance relies on three complementary approaches: in-vivo treatment efficacy studies, in-vitro and ex-vivo parasite sensitivity assays, and the detection of molecular markers in infected human subjects—ie, mutations or copy number variations within the parasite genome associated with reduced sensitivity to antimalarial drugs.6 Although treatment efficacy studies remain the gold standard for demonstrating clinical failure, and in-vitro drug assays can assess drug efficacy on local parasite strains and the impact of mutations on parasite growth and development, these methods are extremely labour-intensive and costly.3 Meanwhile, the assessment of validated markers requires less infrastructure and can provide data on drug resistance more rapidly than other methods.6
Two genes, P falciparum chloroquine resistance transporter (pfcrt) and multidrug resistance gene 1 (pfmdr1), are often targeted for molecular surveillance, because they mediate susceptibility to multiple drugs, including amodiaquine and lumefantrine, partner drugs of the most commonly used ACTs in sub-Saharan Africa.3,7 Several modelling studies have shown that preexisting partner drug resistance facilitates the emergence and spread of artemisinin resistance, although the pathway to multidrug resistance suggests that it is not necessary for emergence, and might differ for each ACT.8,9 Yet despite the importance of molecular surveillance, such efforts remain spatially and temporally limited in sub-Saharan Africa, failing to reach large swaths of the sub-continent.2,3
Xenomonitoring has been cited as a potentially more cost-effective, feasible, and acceptable method to conduct surveillance than traditional approaches relying on invasive human blood sampling.10,11 Mosquito blood meals can be used to monitor locations and patient populations often excluded from standard surveillance efforts, including places with limited infrastructure for humanbased studies and asymptomatic individuals, allowing for more geographically widespread and representative assessments of resistance landscapes.11,12 Samples can further be used to investigate multiple aspects of Plasmodium epidemiology and biology, including transmission dynamics of specific genotypes and concomitant rates of other Plasmodium species, vector composition, insecticide resistance, and other indices.
Although previous studies have collected mosquitos to assess the proportion and transmission potential of drug resistance-associated markers (appendix 2 p 10), sample sizes and methods do not explicitly inform on whether and how xenomonitoring could be used for surveillance. We therefore aimed to assess whether the frequency of two molecular markers conferring reduced ACT partner drug susceptibility (pfcrt 227A>C [Lys76Thr] and pfmdr1 256A>T [Asn86Tyr], henceforth referred to as pfcrt Lys76Thr and pfmdr1 Asn86Tyr) found in mosquito blood meals would be statistically equivalent to the proportion of the same markers in humans surveyed simultaneously in time and space. We also aimed to determine whether mosquito collection would be feasible within existing entomological infrastructure and acceptable to community members and the effects of mosquito feeding behaviors and multiclonal infections common in high transmission settings.
Methods
Study design and household selection
We conducted three cross-sectional surveys between Oct 10, 2018, and Sept 17, 2019 (comprising two rainy seasons and the interim dry season), in Bama, Burkina Faso, a region in the southwest of the country, to compare the frequency of markers of reduced antimalarial susceptibility in humans and mosquito blood meals (appendix 2 p 11). Bama has a population of 24 000, is divided into seven village sectors, and is known for its rice cultivation. Malaria transmission is seasonal, peaking in the rainy season (May–October), and highly endemic, with a entomological inoculation rate of up to 2·35 infectious bites per day in the rainy season.13 The principal malaria vector is Anopheles gambiae sensu latu with Anopheles funestus present at lower proportions.13 At the time of the study, the national treatment policy for uncomplicated Plasmodium falciparum malaria included both artemether–lumefantrine and artesunate–amodiaquine as first-line options, with artemether– lumefantrine generally favoured. Seasonal malaria chemoprevention with sulfadoxine–pyrimethamine and amodiaquine was administered monthly to children under five years old during the rainy seasons.1
We used a two-stage cluster sampling design (appendix 2 p 2) to enrol concessions, which are semienclosed residential areas that are generally comprised of extended families with multiple sleeping houses for nuclear families. Criteria for inclusion included self-reported residency in selected concessions and age older than 3 months. Criteria for exclusion included any known bleeding disorder. Symptomatic status was not assessed. Written consent was obtained for eligible adults and parents or guardians of eligible children. The study was approved by the Yale University Human Investigations Committee, the Comité d’Ethique Institutionnel pour la Recherche de la Santé, and local authorities in Bama.
Household sampling
Dried blood spots from individuals residing within selected concessions and households were obtained using capillary finger pricks and stored on 903 Protein Saver and FTA filter paper cards (Whatman, Maidstone, UK). Indoor-resting mosquitos were collected on the same day as human dried blood spot collection for each concession, between 0500 h and 1000 h AM, with vacuum aspiration using battery-powered Insecta-Zooka field aspirators (BioQuip, Compton, CA, USA).12 Abdominal contents of ten or fewer blood-fed Anopheline mosquitos from each household (appendix 2 p 2) were pressed onto 96-well format CloneSaver cards (Whatman).12 Seasonal malaria chemoprevention status was verbally confirmed by caregivers and randomly verified using record cards.
DNA extraction and P falciparum detection
DNA was extracted from two hole punches of human and mosquito dried blood spots using 3-mm Integra Miltex disposable biopsy punches and the Mag-Bind Blood & Tissue DNA HDQ kit (Omega Bio-Tek, Norcross, GA, USA) with the KingFisher Flex Magnetic Particle Processor (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer protocols.12 Samples containing P falciparum DNA were detected by quantitative PCR (qPCR) targeting the var acidic terminal sequence (varATS; appendix 2 p 2). Primer and probe sequences and thermocycling conditions for qPCR and all assays mentioned hereafter are presented in appendix 2 (pp 11–12).
Multiplicity of infection genotyping
Amplicon-based sequencing was performed to determine the multiplicity of infection (MOI) in a subset of P falciparum-positive samples (appendix 2 p 2). We sequenced four markers of P falciparum diversity: conserved Plasmodium membrane protein (cpmp), circumsporozoite surface protein (csp), merozoite surface protein 7 (msp7), and conserved Plasmodium protein (cpp).14 Sequencing libraries were generated, sequenced, and analysed according to previously published protocols (appendix 2 pp 2–3). MOI was estimated as the maximum number of unique haplotypes identified at any marker for each sample.
Antimalarial drug resistance genotyping
Loci pfcrt Lys76Thr and pfmdr1 Asn86Tyr associated with susceptibility to multiple ACT and non-ACT antimalarials (and therefore commonly assessed in molecular surveillance efforts) were selected for proof-of-concept genotyping.3,7 Mutation genotyping was conducted using high resolution melting with a LightCycler 96 Instrument (Roche, Basel, Switzerland; appendix 2 p 3).15 Instrument software was used to visualise normalised melting probe peaks. A selection of inconclusive and results-discordant samples were sent for Sanger sequencing (Keck Oligonucleotide Synthesis, New Haven, CT, USA) followed by manual inspection with Geneious version 2021.1.
Statistical analysis
A-priori power and sample size considerations were based on demonstrating that the proportions of mutations were significantly equivalent in humans and blood-fed mosquitos at each timepoint, assuming a margin of inferiority of 10%, with a 2-tailed alpha of 0·05 and 80% power. We estimated both the proportion and frequency of molecular markers, because monoclonal infections are common in our study population (appendix 2 p 4). In brief, proportion estimates were calculated as the number of mixed, mutant, and wild-type infections in a given subset, and frequency estimates were calculated using maximum likelihood models that incorporated MOI.16 Equivalence testing with 10% equivalence margins using two one-sided t tests was used to compare the proportion and frequencies of molecular markers in humans and mosquitos.17
We determined the prevalence of infection by fitting a logistic model in a generalised estimating equations framework to account for clustering of infections by concession and village sector (appendix 2 p 3). We evaluated the correlation between the number of mixed and mutant genotypes between groups (hosts, ages) at multiple spatial and cluster scales using binomial logistic regression and controlling for season, survey, and number of malaria-positive mosquitos captured per household or concession.
We also conducted simulations to assess two facets of mosquito feeding behaviour, preferential biting and multiple feeding. In brief, we generated household and concession data where molecular marker x was binomially distributed at varying true proportions among monoclonal P falciparum-infected individuals, and then mosquitos fed either preferentially or multiple times on randomly selected individuals. We estimated the proportion of x in humans and mosquitos by averaging across households and over 1000 iterations (appendix 2 p 4). Sensitivity analyses were also conducted to assess equivalence margins and the impact of MOI on human-mosquito equivalence results. All analyses were done in R version 4.1.0 (packages deailed in the appendix 2 [p 3]).
Role of the funding source
The funder of the study played no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
We collected 1483 dried blood spots from humans and 2151 dried blood spots from blood-fed Anopheles mosquitoes from all three surveys spanning a 1-year period (table 1). Concessions were comprised of a mean of 6·8 households (SD 2·9), 8·6 adults (4·4), 3·6 children younger than 5 years (4·1), and 5·7 children aged 5–18 years (4·0). 900 (61%) participants were female. Self-reported insecticide-treated bed net usage was 95% (145 of 153 concessions), with concessions having a mean of 8·2 insecticide-treated bed nets (SD 6·7) or 2·5 people per insecticide-treated bed net (SD 1·7). A mean of 1·3 people per concession (7·2%, SD 1·8) had reported treatment for symptomatic malaria in the last 30 days. All children younger than 5 years received monthly prophylactic sulfadoxine–pyrimethamine and amodiaquine during the rainy season. 152 (99%) of 153 adults surveyed expressed comfort with mosquito aspirations (appendix 2 p 6).
Table 1:
Survey samples
| Survey 1 | Survey 2 | Survey 3 | |
|---|---|---|---|
|
| |||
| Dates | Oct 10–15, 2018 | April 1–8, 2019 | Sept 12–17, 2019 |
| Season | Late rainy season | Late dry season | Peak rainy season |
| Sampling design* | Household-based | Concession-based | Concession-based |
| Village sectors sampled, n | 7 | 7 | 7 |
| Concessions sampled, n | 102 | 51 | 51 |
| Sleeping households sampled, n | 102 | 255 | 270 |
| Participants, n | 452 | 388 | 643 |
| Mean per concession (SD) | 4·5 (2·3) | 7·6 (5·3) | 12·6 (7·2) |
| Participants aged <5 years, n/N (%) | 56/334 (17%) | 94/385 (24%) | 112/634 (18%) |
| Participants aged 5–18 years, n/N (%) | 134/334 (40%) | 109/385 (28%) | 233/634 (37%) |
| Participants aged >18 years, n/N (%) | 144/334 (43%) | 182/385 (47%) | 289/634 (46%) |
| Mean age (SD) | 20 (19) | 19 (18) | 20 (18) |
| Female participants, n/N (%) | 276/448 (62%) | 239/387 (62%) | 385/640 (60%) |
| Male participants, n/N (%) | 172/448 (38%) | 148/387 (38%) | 255/640 (40%) |
| Blood-fed mosquitos | 1020 | 371 | 760 |
| Mean per household (SD) | 10·0 (0·0) | 1·5 (2·2) | 3·7 (2·8) |
| Mean per concession (SD) | 10·0 (0·0)† | 7·3 (6·8) | 14·9 (8·3) |
| Anopheles gambiae complex, proportion of mosquitoes captured. | NC | 0·97 | 0·99 |
NC=not collected.
Survey 1 used household-based sampling, in which one household was randomly selected per concession, whereas surveys 2 and 3 used concession-based sampling, where all households were sampled within a selected concession.
We set a maximum of ten blood-fed mosquitos collected per household. In Survey 1, in which only one household in selected concessions was sampled, this maximum was reached for all households
Overall, we identified 551 (37·2%) of 1483 individuals infected with P falciparum, with point prevalence estimates of 53·5% (95% CI 48·3–58·7) in survey 1, 36·9% (31·3–42·7) in survey 2, and 25·8% (22·0–30·0) in survey 3 when clustering by concession (appendix 2 p 13). Stratified by 5-year age increments, individuals aged 10–20 years had the highest infection rates, and children younger than 5 years had the lowest rates in all surveys (appendix 2 p 17). In mosquitos, we identified 346 (16·1%) of 2151 infected blood meals or midguts, with point prevalence estimates of 18·7% (95% CI 15·0–22·9) in survey 1, 14·8% (11·7–18·7) in survey 2, and 13·2% (9·4–18·1) in survey 3 when clustering by concession. Similar estimates for humans and mosquitos were reported when clustering by village sector but displayed wider CIs (table 2). varATS Ct values were significantly lower in humans with P falciparum infection than in mosquito blood meals and were higher for both hosts in the dry season (appendix 2 p 5).
Table 2:
Plasmodium falciparum infections in humans and mosquitos
| Survey 1 | Survey 2 | Survey 3 | |
|---|---|---|---|
|
| |||
| Humans | |||
| Number infected | 242 | 143 | 166 |
| Mean Ct (SD) | 33·3 (5·2) | 37·9 (3·9) | 37·6 (3·8) |
| Mean MOI (SD, max); n | 2·86 (1·73, 7); 22 | 2·86 (1·99, 7); 14 | 2·00 (1·34, 5); 21 |
| Prevalence (95% CI) by concession cluster | 53·5% (48·3–58·7) | 36·9% (31·3–42·7) | 25·8% (22·0–30·0) |
| Prevalence (95% CI) by village sector cluster | 52·7% (44·3–61·0) | 37·4% (26·9–49·3) | 26·3% (21·9–31·2) |
| Mosquitoes | |||
| Number infected | 191 | 55 | 100 |
| Mean Ct (SD) | 38·6 (4·1) | 39·0 (2·9) | 37·1 (4·6) |
| Mean MOI (SD, max); n | 3·94 (2·52, 9); 16 | 2·25 (1·34, 6); 16 | 2·50 (1·38, 6); 24 |
| Prevalence (95% CI) by concession cluster | 18·7% (15·0–22·9) | 14·8% (11·7–18·7) | 13·2% (9·4–18·1) |
| Prevalence (95% CI) by village sector cluster | 22·2% (14·8–31·9) | 14·8% (11·6–18·8) | 14·1% (8·0–23·6) |
Ct=cycle threshold for qPCR detection of P falciparum targeting the var acidic terminal sequence. MOI=multiplicity of infection, calculated as the maximum number of unique haplotypes at any of four markers for each sample and averaged across all subgroups.
We sequenced P falciparum-positive human blood samples (survey 1 n=22, survey 2 n=14, survey 3 n=21) and mosquito blood meals (survey 1 n=16, survey 2 n=16, survey 3 n=24) and observed 95 unique haplotypes for the cpmp marker, 74 for cpp, 45 for csp, and 35 for msp7 (appendix 2 p 19). In humans, mean MOI was 2·86 (SD 1·73) in survey 1, 2·86 (1·99) in survey 2, and 2·15 (1·34) in survey 3 (table 2); changes over time were not significant. Aggregated across all surveys, children younger than 10 years had higher mean MOI (3·1, SD 1·85) than those aged 10 years or older (2·13, SD 1·6; p=0·048), but it did not significantly differ for each survey (appendix 2 p 13). In mosquitos, mean MOI was 3·94 (SD 2·52) in survey 1, 2·25 (1·34) in survey 2, and 2·50 (1·38) in survey 3 (table 2); MOI was significantly higher in survey 1 than in survey 2 (p=0·027) or survey 3 (p=0·048). MOI in humans and mosquitos was not significantly different for any survey (p=0·15 for survey 1, p=0·35 for survey 2, p=0·23 for survey 3).
The proportion of mutant pfmdr1 Asn86Tyr infections remained at 5% or less of genotyped samples throughout the study in both humans and mosquitos (table 3). Compared to the baseline survey, the proportion of mixed pfmdr1 Asn86Tyr infections decreased in the dry season then increased in the following rainy season in both hosts. Individuals younger than 18 years had significantly higher proportions of mixed infections (23 [22%] of 104) than did adults (two [5%] of 42) only in survey 3 (p=0·014; appendix 2 p 13).
Table 3:
Drug resistance-associated genotypes and frequencies for genotyped human blood samples and mosquito blood meals
| Humans | Mosquitos | Two one-sided t-tests p value | Null hypothesis significance testing p value | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| n (%) | Allele frequency* | n (%) | Allele frequency* | |||
|
| ||||||
| pfmdr1 Asn86Tyr | ||||||
| Survey 1 genotyped samples | 210 | 171 | <0·0001 | 0·60 | ||
| Wild type (Asn86) | 192 (91%) | 96% | 156 (92%) | 97% | ·· | ·· |
| Mixed (Asn86 or Tyr86) | 12 (6%) | ·· | 10 (5%) | ·· | ·· | ·· |
| Mutant (Tyr86) | 6 (3%) | 4% | 5 (3%) | 3% | ·· | ·· |
| Survey 2 genotyped samples | 119 | 52 | <0·0001 | 0·12 | ||
| Wild type (Asn86) | 114 (96%) | 98% | 52 (100%) | 100% | ·· | ·· |
| Mixed (Asn86 or Tyr86) | 2 (2%) | ·· | 0 | ·· | ·· | ·· |
| Mutant (Tyr86) | 3 (2%) | 2% | 0 | 0% | ·· | ·· |
| Survey 3 genotyped samples | 149 | 91 | 0·062 | 0·30 | ||
| Wild type (Asn86) | 116 (68%) | 88% | 78 (86%) | 92% | ·· | ·· |
| Mixed (Asn86 or Tyr86) | 25 (17%) | ·· | 10 (11%) | ·· | ·· | ·· |
| Mutant (Tyr86) | 8 (5%) | 12% | 3 (3%) | 8% | ·· | ·· |
| pfcrt Lys76Thr | ||||||
| Survey 1 genotyped samples | 200 | 167 | 0·99 | <0·0001 | ||
| Wild type (Lys76) | 146 (73%) | 84% | 67 (40%) | 60% | ·· | ·· |
| Mixed (Lys76 or Thr76) | 38 (19%) | ·· | 58 (35%) | ·· | ·· | ·· |
| Mutant (Thr76) | 16 (8%) | 16% | 42 (25%) | 40% | ·· | ·· |
| Survey 2 genotyped samples | 140 | 48 | 0·82 | 0·028 | ||
| Wild type (Lys76) | 30 (21%) | 45% | 4 (8%) | 28% | ·· | ·· |
| Mixed (Lys76 or Thr76) | 66 (48%) | ·· | 20 (42%) | ·· | ·· | ·· |
| Mutant (Thr76) | 44 (31%) | 55% | 24 (50%) | 72% | ·· | ·· |
| Survey 3 genotyped samples | 142 | 96 | 0·032 | 0·64 | ||
| Wild type (Lys76) | 118 (83%) | 89% | 74 (77%) | 87% | ·· | ·· |
| Mixed (Lys76 or Thr76) | 16 (11%) | ·· | 17 (18%) | ·· | ·· | ·· |
| Mutant (Thr76) | 8 (6%) | 11% | 5 (5%) | 13% | ·· | ·· |
Allele frequencies, which account for mixed infections, were estimated by maximising the likelihood of the observed proportion of a molecular marker in any given survey given the average multiplicity of infection for the respective marker, survey, and host population.
For marker pfcrt Lys76Thr, the proportion of mixed or mutant infections was more variable between surveys and hosts. Mixed or mutant genotypes made up 27% (54 of 200) malaria-positive human samples and 60% (100 of 167) of malaria-positive mosquitos in survey 1, 79% (110 of 140) and 92% (44 of 48) in survey 2, and 17% (24 of 142) and 23% (22 of 96) in survey 3 (table 3). The proportion of mixed and mutant pfcrt (Lys76Thr) genotypes was significantly higher in individuals younger than 18 years than in adults in survey 3 only (20 [25%] of 95 vs two [5%] of 38; 0·0080)—ie, the rainy season surveys—and there was no significant difference between age groups in survey 2 (appendix 2 p 13). Compared with survey 1, the proportion of mixed and mutant infections increased significantly in the dry season in both age groups (p<0·0001) and then decreased in survey 3 to similar proportions to the baseline (appendix 2 p 13).
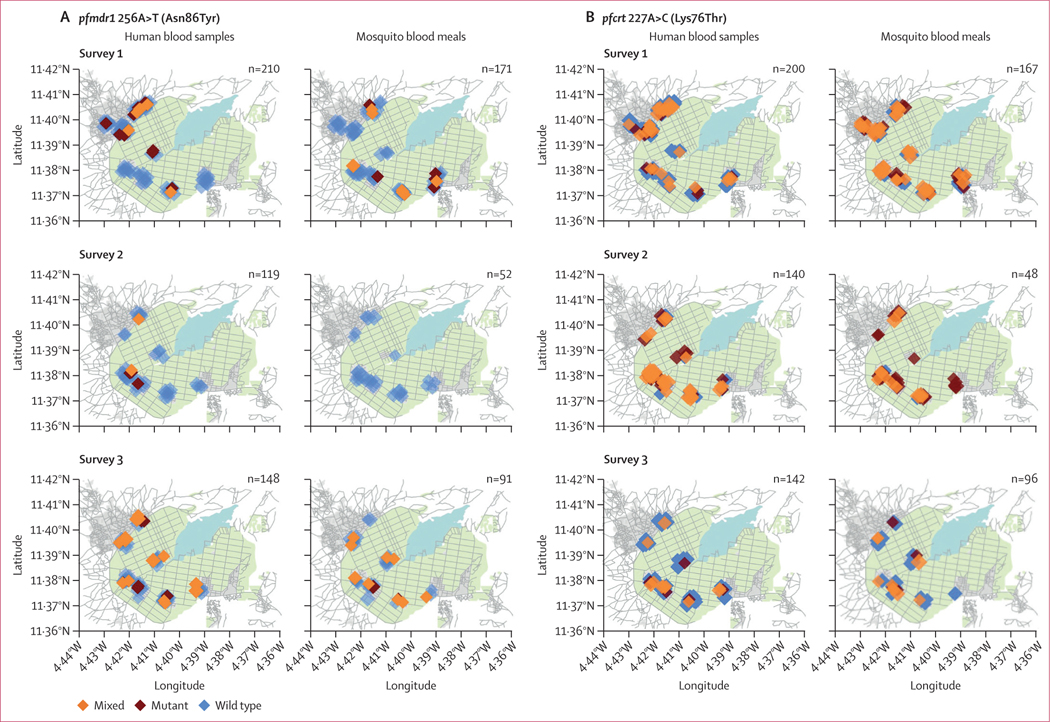
At the concession level (figure 1), the proportion of mixed or mutant genotypes in mosquitos was significantly associated with the proportion of the same in humans for pfmdr1 Asn86Tyr (odds ratio [OR] 5·72, 95% CI 0·94–34·9; p=0·042) and pfcrt Lys76Thr (OR 66·16, 95% CI 15·8–276·9; p=0·0020). Missing data are discussed in appendix 2 (p 5).
Figure 1: Spatial comparisons of allelic proportions in humans and mosquito blood meals.
(A) pfmdr1 256A>T (Asn86Tyr). (B) pfcrt 227A>C (Lys76Thr). Each diamond represents one genotyped sample, coloured according to genotype: wild type (blue), mutant (red), and mixed (orange). Background graphics include streets (lines, grey), farmland for rice cultivation (shaded, green) and lakes or marshland (shaded, blue); grey areas demarcate different village sectors. All geographic points are randomly jittered to protect anonymity. Sample sizes are shown in each panel. Maps adapted from OpenStreetMap.
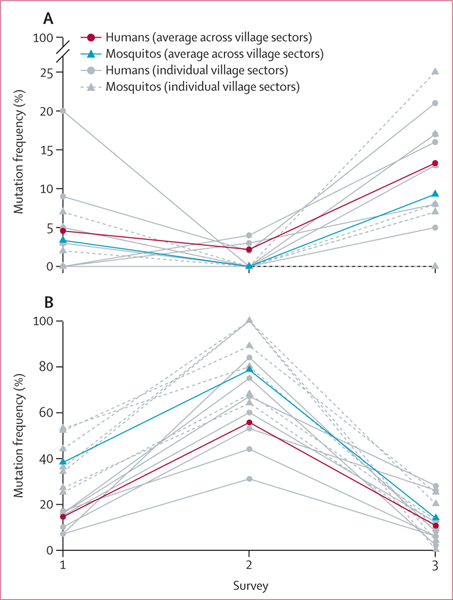
We found that the frequency of the mutation pfmdr1 Asn86Tyr in humans and mosquitos was statistically equivalent (within a tolerability of 0·10) in surveys 1 and 2 (p<0·0001) but not in survey 3 (p=0·062); however, frequencies were also not statistically different by Fisher’s exact test in survey 3 (p=0·30). The frequency of pfcrt Lys76Thr was statistically equivalent in humans and mosquitos in survey 3 only (p=0·032). The frequency of the pfcrt Lys76Thr mutation was significantly higher in mosquitos than in humans in survey 1 (p<0·0001) and suvey 2 (p=0·0010). Of the seven village sectors, we found that statistical equivalence in human and mosquito frequencies occurred in four sectors in survey 1, six in survey 2, and zero in survey 3 for pfmdr1 and in zero sectors in surveys 1 and 2 and one in survey 3 for pfcrt. Statistical difference was observed in only one village sector across all three surveys for pfmdr1 and in four sub-villages in survey 1, four in survey 2, and zero in survey 3 for pfcrt (appendix 2 p 14). Averaged over all sectors, mutation frequency over time was similar in humans and mosquitos for both markers (figure 2).
Figure 2: Mutation frequencies in hosts over time.
(A) pfmdr1 256A>T (Asn86Tyr). (B) pfcrt 227A>C (Lys76Thr). Grey lines and points show mutation frequency estimations for each of the seven village sectors in humans and mosquitos. Bold lines display weighted frequency averages across all village sectors for each survey and host.
In sensitivity analyses aimed at understanding the effect of MOI and equivalence margins on our results, we found that adjusting mean MOI in humans and mosquitos had little effect on the likelihood of statistical equivalence for surveys 1 and 2. For Survey 3, statistical equivalence was observed in 64% of all possible MOI permutations for pfmdr1 Asn86Tyr and and 65% for pfcrt Lys76Thr (appendix 2 p 20). Adjusting the equivalence margins from 0·02 to 0·25 affected the likelihood of equivalence for all surveys for pfmdr1 and for survey 3 for pfcrt (appendix 2 p 15).
In simulations to understand potential effects of vector feeding behaviors, we found that preferential biting of specific individuals did not substantially affect frequency results at the community level (appendix 2 pp 5–6, 21–22). However, consistent multiple feeding behavior (interrupted or previous) substantially increased frequency and variability of mutations in mosquitos compared with humans, because more mosquitos fed twice and because the true proportion of the mutation in humans increased to 0·50 (appendix 2 pp 5–6, 22).
Discussion
In this study, we assessed the use of blood-fed mosquitos to supplement antimalarial resistance surveillance efforts. Our study was designed to accurately capture and compare the frequency of resistance-associated molecular markers in humans and blood-fed mosquito populations sampled simultaneously in space and time, as supplement to more traditional surveillance methods. In a high transmission region of Burkina Faso across multiple seasons, we found that inter-host marker frequencies were statistically equivalent in half of possible comparisons, within a 10% margin of equivalence, and that human and mosquito frequencies exhibited similar temporal trends. Supplementing our empirical results with simulations accounting for mosquito feeding behaviors, we speculate that xenomonitoring efforts in endemic regions could more precisely reflect human populations when applied to markers circulating at lower frequencies, potentially including emergent artemisinin resistance markers.
We found that frequencies of mixed or mutant pfmdr1 Asn86Tyr genotypes in humans and mosquitos were comparable across all surveys but that mosquitos harboured higher frequencies of mixed or mutant pfcrt Lys76Thr genotypes than humans. We have multiple potential hypotheses to help explain these discrepancies. First, some proportion of blood-fed mosquitos probably fed on multiple individuals in a single feeding session and could harbour oocysts or ookinetes in midguts from previous feeds, increasing the probability of observing mixed infections in whole midgut specimens, particularly when those strains are circulating at higher proportions, as demonstrated in our simulation results.18 Some evidence suggests that mutant pfcrt Lys76Thr parasites produce higher gametocyte densities than wild-type strains, increasing their probability of ingestion by mosquitos.19,20 Further, Plasmodium undergoes genetic recombination, population bottlenecks, and changes in ploidy during mosquito life cycle stages that might confound comparisons with human populations.21
The higher frequencies of pfcrt Lys76Thr genotypes detected in mosquitos might also be associated with preferential feeding on adolescents. In our study, individuals younger than 18 years old were found to have higher frequencies of mixed or mutant pfcrt Lys76Thr infections than adults for both rainy seasons. Similarly, individuals younger than 10 years of age had a significantly higher MOI than those aged 10 years or older (with similar but non-significant trends observed in those <18 years vs those ≥18 years), supporting other work in our study region finding negative correlations between MOI and age, and increasing the possibility of observing mixed infections for markers circulating at higher frequencies.22 An additional factor that might have affected mutation prevalence in younger individuals, particularly those younger than 5 years old, is the selective pressure of seasonal malaria chemoprevention.23
The increased frequency of pfcrt Lys76Thr mutations in the dry season in both humans and mosquitos was not expected. The opposite trend is typically observed due to less drug pressure from ACTs in the dry season coupled with deleterious fitness costs associated with the mutation.19,24 However, the decline in pfcrt Lys76Thr frequencies in the subsequent rainy season suggests that, in the face of a probable population bottleneck related to a decline in malaria prevalence (appendix 2 p 5), mutant strains might have been unable to maintain a selective advantage over wild-type strains. These findings support recent meta-analytic evidence suggesting that seasonality might buffer selection of any genotype in hightransmission settings, regardless of local drug pressure.2
Our approach had notable limitations. Indoor aspirations favoured anthropophilic, endophagic mosquitos.10 Both mosquito-based and community-based human sampling are complicated by low-density and asymptomatic infections, in which assays often operate near sensitivity limits, and samples required multiple modifications (appendix 2 p 3).25 Future studies might consider pooling mosquitos by household or concession and using assays that directly estimate the proportions of markers.26 Multiclonality also affected frequency estimates; although more precise xenomonitoring might be possible in settings with less complex infections, sample sizes must be adjusted to account for lower malaria prevalence and mosquito abundance, and further work can assess feasibility in lower transmission regions. We were not able to track individuals over time, limiting possible inferences on the persistence of mutant strains between seasons. Future work could help determine evidence-based and validated thresholds for statistical equivalence that are meaningfully associated with resistance profiles, affecting study design.
Previous studies have analysed resistance-associated markers in field-collected mosquitos and a subset have compared these results to markers in infected humans (appendix 2 p 10).27–30 In Equatorial Guinea and Uganda, studies found no significant difference in the proportion of mutations in mosquitos and humans, aside from a modest significant increase in pfmdr1 Asn86Tyr in mosquito thoraxes compared to humans in Uganda.27,28 Two studies in Zambia found lower rates of pfdhfr mutations and pfcrt Lys76Thr in mosquito midguts than in humans.29,30 However, previous studies were powered to investigate fitness costs of resistant strains across the parasite life cycle and were not designed to detect statistical equivalence. Our work assessed xenomonitoring for the explicit purpose of surveillance. Differences in pfcrt between humans and mosquitos might reflect the long-term transmission potential of these genotypes and could be further investigated by assessing genotypes in salivary glands.
Our findings highlight the importance of routine surveillance across multiple seasons and years: a broader repository of molecular data would help disambiguate stochastic anomalies from longitudinal trends, elucidate the role of the mosquito in the transmission of specific markers, and help determine locations to conduct more resource-intensive treatment efficacy studies, because these are crucial to monitor clinical resistance. Xenomonitoring might offer a complementary tool to address practical challenges of surveillance, particularly to detect local markers as they are identified. In endemic regions with experienced vector-control departments, xenomonitoring presents a highly feasible and acceptable (appendix 2 p 6) approach that can be leveraged and combined with efforts to monitor vector composition and insecticide resistance, and an opportunity to promote collaboration between entomological and clinical teams. As sub-Saharan Africa begins to contend with the emergence of artemisinin resistance in multiple locations, alongside a background high rate of mutations which affect partner drug susceptibility, implementing effective surveillance strategies has taken on immediate urgency.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed and Web of Science using combinations of the terms “Plasmodium”, “drug resistance”, “mosquitos”, “vectors”, “molecular markers”, “genotypes”, “fitness”, “sampling”, “xenosurveillance”, and “xenomonitoring” for articles published between database inception and Sept 1, 2022. No language restrictions were applied to the search. We identified ten studies that assessed the presence of one or more antimalarial resistance-associated molecular markers in Plasmodium falciparum or Plasmodium vivax parasites at any developmental stage within any body part of field-caught mosquitos. Of these, four studies directly compared the proportions of molecular markers in mosquitos to those genotyped in human populations, but, to our knowledge, none directly included mosquito blood meals in these comparisons.
Added value of this study
Our study was designed and powered to assess xenomonitoring, or the use of mosquito blood meals, for the explicit purpose of surveying resistance-associated markers in human populations. We proposed that blood-fed mosquitos could be used as an accurate, efficient, and non-invasive proxy to monitor molecular markers of drug resistance in humans, and that discrepancies in molecular markers between hosts could be informative of fitness costs associated with local strains of drug-resistant or drug-susceptible malaria. We found that the frequency of molecular markers in mosquito blood meals and humans exhibited similar temporal trends but were not statistically equivalent within a 10% margin of equivalence in all scenarios. Markers in mosquitos might more precisely reflect human populations when they circulate at lower frequencies.
Implications of all the available evidence
Reports of resistance to first-line drugs, artemisinin-based combination therapies, are increasing in frequency in sub-Saharan Africa. However, the distribution of data on drug resistance remains limited in time and space. Our study validates the use of mosquito blood meals as an informative, complementary tool for molecular surveillance efforts, particularly for markers circulating at lower frequencies, including emerging markers of drug resistance.
Acknowledgments
We are indebted to all the individuals and families who participated in this study and to the larger communities of Bama and Soumousso. We also thank our extensive team of entomology and phlebotomy technicians affiliated with Institut de Recherche en Sciences de la Santé and Centre Muraz whose efforts were indispensable in sample collection. We appreciate constructive feedback from Dan Weinberger (Yale School of Public Health), Christian Nsanzabana (Swiss Tropical and Public Health Institute), and Choukri Ben Mamoun (Yale School of Medicine). This study was funded by the National Institutes of Health National Institute of Allergy and Infectious Diseases (grants R21AI135477 to SP and BDF and F31AI150168 to HYE). We received additional support from the Fogarty International Center (grant K01TW010496 to AKB and 5D43TW010540 to FAS), the Yale Institute of Biospheric Studies, and the Yale MacMillan Center.
Footnotes
Declaration of interests We declare no competing interests.
For the French translation of the abstract see Online for appendix 1
For the ACT Partner Drug Molecular Surveyor of the Worldwide Antimalarial Resistance Network see http://www.wwarn.org/molecular/surveyor/#0
For the GitHub repository see https://github.com/hannaehrlich/
Data sharing
Molecular data will be made available in ACT Partner Drug Molecular Surveyor of the Worldwide Antimalarial Resistance Network and GitHub upon publication.
References
- 1.WHO. World malaria report 2022. Geneva: World Health Organization, 2022. https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2022 (accessed Jan 20, 2023). [Google Scholar]
- 2.Ehrlich HY, Bei AK, Weinberger DM, Warren JL, Parikh S. Mapping partner drug resistance to guide antimalarial combination therapy policies in sub-Saharan Africa. Proc Natl Acad Sci USA 2021; 118: e2100685118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehrlich HY, Jones J, Parikh S. Molecular surveillance of antimalarial partner drug resistance in sub-Saharan Africa: a spatial-temporal evidence mapping study. Lancet Microbe 2020; 1: e209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uwimana A, Umulisa N, Venkatesan M, et al. Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect Dis 2021; 21: 1120–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balikagala B, Fukuda N, Ikeda M, et al. Evidence of artemisininresistant malaria in Africa. N Engl J Med 2021; 385: 1163–71. [DOI] [PubMed] [Google Scholar]
- 6.Nsanzabana C, Djalle D, Guérin PJ, Ménard D, González IJ. Tools for surveillance of anti-malarial drug resistance: an assessment of the current landscape. Malar J 2018; 17: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otienoburu SD, Suay I, Garcia S, et al. An online mapping database of molecular markers of drug resistance in Plasmodium falciparum: the ACT Partner Drug Molecular Surveyor. Malar J 2019; 18: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watson OJ, Gao B, Nguyen TD, et al. Pre-existing partner-drug resistance to artemisinin combination therapies facilitates the emergence and spread of artemisinin resistance: a consensus modelling study. Lancet Microbe 2022; 3: e701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masserey T, Lee T, Golumbeanu M, et al. The influence of biological, epidemiological, and treatment factors on the establishment and spread of drug-resistant Plasmodium falciparum. eLife 2022; 11: e77634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nascimento J, Sampaio VS, Karl S, et al. Use of anthropophilic culicid-based xenosurveillance as a proxy for Plasmodium vivax malaria burden and transmission hotspots identification. PLoS Negl Trop Dis 2018; 12: e0006909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameron MM, Ramesh A. The use of molecular xenomonitoring for surveillance of mosquito-borne diseases. Philos Trans R Soc B 2021; 376: 20190816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grubaugh ND, Sharma S, Krajacich BJ, et al. Xenosurveillance: a novel mosquito-based approach for examining the humanpathogen landscape. PLoS Negl Trop Dis 2015; 9: e0003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epopa PS, Collins CM, North A, et al. Seasonal malaria vector and transmission dynamics in western Burkina Faso. Malar J 2019; 18: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruenberg M, Lerch A, Beck H-P, Felger I. Amplicon deep sequencing improves Plasmodium falciparum genotyping in clinical trials of antimalarial drugs. Sci Rep 2019; 9: 17790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ndiaye YD, Diédhiou CK, Bei AK, et al. High resolution melting: a useful field-deployable method to measure dhfr and dhps drug resistance in both highly and lowly endemic Plasmodium populations. Malar J 2017; 16: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okell LC, Griffin JT, Roper C. Mapping sulphadoxinepyrimethamine-resistant Plasmodium falciparum malaria in infected humans and in parasite populations in Africa. Sci Rep 2017; 7: 7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker LE, Luman ET, McCauley MM, Chu SY. Assessing equivalence: an alternative to the use of difference tests for measuring disparities in vaccination coverage. Am J Epidemiol 2002; 156: 1056–61. [DOI] [PubMed] [Google Scholar]
- 18.Guelbéogo WM, Gonçalves BP, Grignard L, et al. Variation in natural exposure to anopheles mosquitoes and its effects on malaria transmission. eLife 2018; 7: e32625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ord R, Alexander N, Dunyo S, et al. Seasonal carriage of pfcrt and pfmdr1 alleles in Gambian Plasmodium falciparum imply reduced fitness of chloroquine-resistant parasites. J Infect Dis 2007; 196: 1613–19. [DOI] [PubMed] [Google Scholar]
- 20.Ecker A, Lakshmanan V, Sinnis P, Coppens I, Fidock DA. Evidence that mutant PfCRT facilitates the transmission to mosquitoes of chloroquine-treated Plasmodium gametocytes. J Infect Dis 2011; 203: 228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee AH, Symington LS, Fidock DA. DNA repair mechanisms and their biological roles in the malaria parasite Plasmodium falciparum. Microbiol Mol Biol Rev 2014; 78: 469–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sondo P, Derra K, Rouamba T, et al. Determinants of Plasmodium falciparum multiplicity of infection and genetic diversity in Burkina Faso. Parasites Vectors 2020; 13: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Somé AF, Zongo I, Compaoré Y-D, et al. Selection of drug resistance-mediating Plasmodium falciparum genetic polymorphisms by seasonal malaria chemoprevention in Burkina Faso. Antimicrob Agents Chemother 2014; 58: 3660–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babiker HA, Satti G, Ferguson H, Bayoumi R, Walliker D. Drug resistant Plasmodium falciparum in an area of seasonal transmission. Acta Tropica 2005; 94: 260–68. [DOI] [PubMed] [Google Scholar]
- 25.Mzilahowa T, McCall PJ, Hastings IM. “Sexual” population structure and genetics of the malaria agent P falciparum. PLoS One 2007; 2: e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith-Aguasca R, Gupta H, Uberegui E, et al. Mosquitoes as a feasible sentinel group for anti-malarial resistance surveillance by next generation sequencing of Plasmodium falciparum. Malar J 2019; 18: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendes C, Salgueiro P, Gonzalez V, et al. Genetic diversity and signatures of selection of drug resistance in Plasmodium populations from both human and mosquito hosts in continental Equatorial Guinea. Malar J 2013; 12: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conrad MD, Mota D, Musiime A, et al. Comparative prevalence of Plasmodium falciparum resistance-associated genetic polymorphisms in parasites infecting humans and mosquitoes in Uganda. Am J Trop Med Hyg 2017; 97: 1576–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mharakurwa S, Kumwenda T, Mkulama MA, et al. Malaria antifolate resistance with contrasting Plasmodium falciparum dihydrofolate reductase (DHFR) polymorphisms in humans and Anopheles mosquitoes. Proc Natl Acad Sci USA 2011; 108: 18796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mharakurwa S, Sialumano M, Liu K, Scott A, Thuma P. Selection for chloroquine-sensitive Plasmodium falciparum by wild Anopheles arabiensis in southern Zambia. Malar J 2013; 12: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Molecular data will be made available in ACT Partner Drug Molecular Surveyor of the Worldwide Antimalarial Resistance Network and GitHub upon publication.