Abstract
Purpose
There is great promise in use of machine learning (ML) for the diagnosis, prognosis, and treatment of various medical conditions in ophthalmology and beyond. Applications of ML for ocular neoplasms are in early development and this review synthesizes the current state of ML in ocular oncology.
Methods
We queried PubMed and Web of Science and evaluated 804 publications, excluding nonhuman studies. Metrics on ML algorithm performance were collected and the Prediction model study Risk Of Bias ASsessment Tool was used to evaluate bias. We report the results of 63 unique studies.
Results
Research regarding ML applications to intraocular cancers has leveraged multiple algorithms and data sources. Convolutional neural networks (CNNs) were one of the most commonly used ML algorithms and most work has focused on uveal melanoma and retinoblastoma. The majority of ML models discussed here were developed for diagnosis and prognosis. Algorithms for diagnosis primarily leveraged imaging (e.g., optical coherence tomography) as inputs, whereas those for prognosis leveraged combinations of gene expression, tumor characteristics, and patient demographics.
Conclusions
ML has the potential to improve the management of intraocular cancers. Published ML models perform well, but were occasionally limited by small sample sizes owing to the low prevalence of intraocular cancers. This could be overcome with synthetic data enhancement and low-shot ML techniques. CNNs can be integrated into existing diagnostic workflows, while non-neural networks perform well in determining prognosis.
Keywords: ocular oncology, machine learning, artificial intelligence, uveal melanoma, retinoblastoma
Multiple malignancy types can affect the eyes or periorbita. In adults, the most common primary intraocular cancer is uveal melanoma (UM), whereas retinoblastoma (Rb) is the most common in children. Intraocular cancers are particularly insidious, because they can be asymptomatic in the early phases and, if not treated aggressively, can threaten vision and life.1 Mortality can reach 60% in some instances,2,3 with significant risk for metastatic disease in UM.1,4 Although progress has been made on improving the management of some of these cancers such as Rb, there has been little improvement in the treatment and prognoses of others such as UM.5
There is a continuing need to improve the accuracy and usability of tools used for the diagnosis, prognostication, and treatment of intraocular cancers.6,7 Machine learning (ML) could play a key role in developing such technologies and can integrate into existing ophthalmologic workflow (e.g., imaging analysis and electronic medical records). ML in health care is growing by 40% per year and could cut more than $100B in annual health care costs over the next 5 years by assisting with administrative workflow, image analysis, treatment planning, and patient monitoring.8 Ophthalmology has embraced the use of ML, particularly in the screening and diagnosis of diabetic retinopathy, AMD, and glaucoma.9 To date, the US Food and Drug Administration (FDA) has approved six ML-enabled devices for such applications.10 There is significant interest in applying ML to improve outcomes in patients with ocular malignancies,11 although validated tools in this area are lagging behind other ophthalmic applications. The purpose of this review was to analyze the current state of science and review the research that has directly assessed the use of ML in the diagnosis, prognosis, and treatment of ocular malignancies, as well as to explore the overarching trends in ML approaches to ocular oncologic conditions.
Methods
A literature search was performed in June 2023 using PubMed and Web of Science with the compound search term and exclusion process seen in Supplemental Figure S1. Two reviewers (ASC and TMH) independently assessed inclusion criteria for all papers and two separate reviewers (NKA and YES) confirmed inclusion. For each paper, two reviewers (ASC and TMH) collected data including the type(s) of intraocular cancer studied, data source (e.g., human, database), number of data points, and data modality (e.g., eye images, tumor samples). Each study was classified based on stage of clinical workflow (e.g., diagnosis, prognosis, treatment) to which its results applied. The studies were analyzed overall by data modalities, sample size, and clinical condition studied. Timeline and citation analyses were performed, using citation data gathered from Google Scholar. Three reviewers (ASC, TMH, and CCC) assessed risk of bias using the Prediction model study Risk Of Bias ASsessment Tool (PROBAST), which is a validated system for assessing risk of bias across the four domains of participants, predictors, outcomes, and analysis. Citation data were gathered from Google Scholar. Further subanalyses based on primary clinical diagnosis studied were performed and are reported by clinical focus, including UM, Rb, and combinations of other ocular malignancies.
Literature Analysis
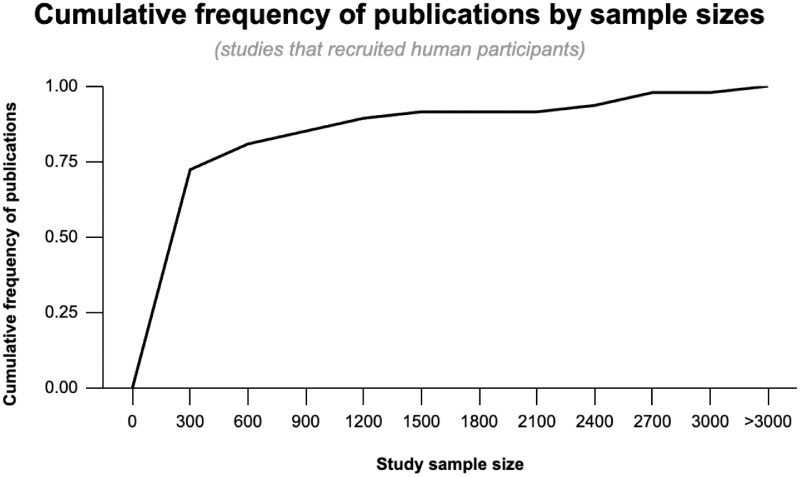
Of the 804 publications (published between 2002 and 2023), 63 met the inclusion and exclusion criteria with 629 studies excluded based on review of abstracts and 112 based on full article text review. The 63 studies included in this analysis had a wide range of sample sizes and relied on varied data modalities (e.g., human recruitment, image databases). Median sample size was 153 (interquartile range, 78–426). The smallest studies, primarily focused on methods development, had 1 participant,12,13 and the largest analyzed 52,982 images.14 For studies that recruited human participants (n = 47 [75%]), 72% recruited between 1 and 300 participants (Fig. 1). In total, 51% of papers studied UM, 25% Rb, and 24% other ocular cancers.
Figure 1.
Cumulative frequency of publications by study size. Seventy-two percent of studies that exclusively recruited human participants had a sample size of less than 300.
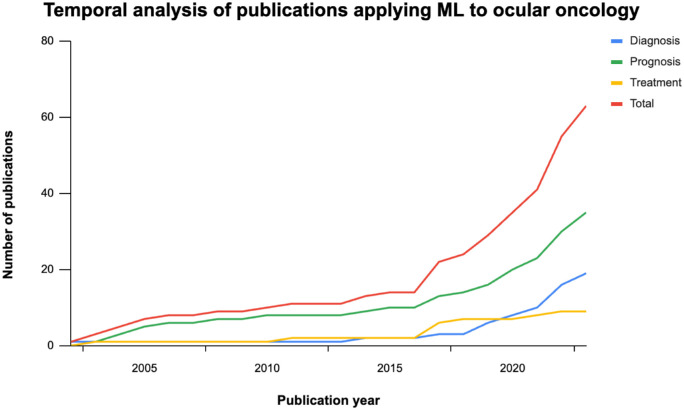
A timeline analysis revealed a slow increase in publications from 2000 to 2015, with a rapid increase since. Notably, studies related to treatment appear to have plateaued (Fig. 2). The studies analyzed here have been cited 1126 times with a median citation count of 4 (interquartile range, 1–19).
Figure 2.
Temporal analysis of publications. There has been an increase in publications related to ML in intraocular oncology starting in 2016. Publications related to cancer prognosis are the most popular. More work is needed related to treatment planning.
Summary of Clinically Important ML Techniques
ML algorithms have three primary purposes: (1) learn from data, (2) perform tasks given new data, and (3) improve with experience.15 Numerous ML algorithms have been developed over the years and these algorithms can be categorized broadly into neural networks (NNs) and non-NNs.
NNs consist of sequential layers of nodes that are interconnected with each other. This structure enables the discovery of complex, nonlinear relationships between input variables that allows for many applications, including data classification. The clinical use of NNs is varied and includes early detection of liver fibrosis,16 analysis of complex electrocardiograms,17 monitoring of Parkinson disease,18 and diagnosing glaucoma.19 Convolutional NNs (CNNs) are a specific type of NN that are used for image analysis. Clinically, CNNs have been used to classify lung cancer20 and differentiate types of infectious keratitis.21
Non-NN algorithms encompass a large number of ML techniques that include logistic regression, decision trees, and support vector machines (SVM). SVMs are commonly used in medicine as they are capable of multidimensional classification (i.e., binary classification with a large number of input variables). SVMs accomplish this by using a kernel function that maps original inputs to higher dimensional space, creating a better separation between categories within the data. SVMs have been used in ophthalmology to detect subclinical keratoconus using topography data.22
Many ML algorithms were used in the papers analyzed herein. Regardless of the specific algorithm or implementation, an important requirement for ML model development is the performance of complex, nonlinear tasks to a level that meets or exceeds human performance.
Bias Assessment
A PROBAST analysis of the 63 papers analyzed here revealed that all papers were either at low or medium risk of bias as per the 21 PROBAST questions (Supplemental File 1). An analysis of participant selection revealed a low risk of bias; the vast majority of studies used appropriate data sources and clearly defined inclusion and exclusion criteria. Risk of bias in predictor variable selection was particularly low in studies that leveraged CNNs, because there is minimal user involvement in defining model inputs. The papers also had a low to medium risk of bias in outcome definitions, because the majority of papers had clearly defined model end points and systematic methods for evaluating these end points. Potentials for risk of bias were most difficult to evaluate for the analysis category, because many papers did not include information regarding specific steps of data processing and analysis. Additionally, some of the questions in the analysis category (e.g., Do predictors and their assigned weights in the final model correspond to the results from the reported multivariable analysis?) do not necessarily apply to certain ML algorithms such as CNNs. Regardless, there need to be more standardized guidelines in the ML space regarding the reporting of model development, data processing, and final analysis.
ML Studies in UM
UM arises from melanocytes in the iris, ciliary body, or choroid23 and is the most common primary intraocular malignancy worldwide.24 Current challenges in the management of UM include improving accuracy of early diagnosis and developing reliable markers for prognostication (e.g., metastatic risk).
Diagnosis
Diagnosing intraocular cancers can be challenging, with some methods for diagnosing anterior segment tumors having an error rate approaching 40%.25 Most ML models developed to diagnose intraocular malignancies, including UM, use ocular images (e.g., external eye pictures taken with digital cameras, magnetic resonance imaging [MRI]) as inputs.
Oyedotun et al.26 developed a CNN to detect iris nevi from The Eye Cancer Foundation's pictures, achieving an accuracy of 94% in binary diagnosis (i.e., iris nevus present or not) (Table 1). Other image-based diagnostic methods using CNN, artificial neural networks, and radial basis function networks have achieved similar results.27,28 Although iris nevi are not cancerous, 8% can transform into melanoma over 15 years,29 making proper diagnosis important. This study highlights the usefulness of CNNs in augmenting the confidence of visual diagnosis by ophthalmologists. Other groups have taken a different approach and have not used CNNs for image analysis. Su et al.30 found that multilayer perceptron performed the best in diagnosing UM when trained on a combination of features extracted from T2-weighted and contrast-enhanced T1 weighted MRIs. Ultrasound images, which are commonly obtained when managing ocular cancers, combined with patient demographics were also used to train a model that diagnosed UM with an accuracy of 93%.31
Table 1.
Studies That Applied ML for Management of UM
| Author | Year | Type(s) of Cancer | Data Source | No. of Data Points | Data Modality | ML Algorithm(s) Used | Key Takeaways | Conflicts of Interest | Funding Sources |
|---|---|---|---|---|---|---|---|---|---|
| Diagnosis | |||||||||
| Jegelevicius et al. | 2002 | UM | Human | 89 | Patient and tumor characteristics | Decision tree | Decision tree achieved 93.3% accuracy in classifying tumor versus not tumor | Not listed | Not listed |
| Oyedotun et al. | 2017 | Iris nevus | Database | 500 | Eye images | CNN, Deep belief network | CNN and DBN achieved approximately 94% recognition rates in determining nevus affected versus unaffected | Not listed | Not listed |
| Song et al. | 2019 | UM | Human | 84 | Patient serum | Logistic regression | Multivariate logistic regression identified a panel of heat shock protein 27 and osteopontin that discriminated between UM and healthy controls with an AUC of 0.98 | None | National Cancer Institute |
| Su et al. | 2020 | UM | Human | 245 | MRI: T1, T2-weighted, and postcontrast-enhanced T1WI | Logistic regression, multilayer perceptron, and SVM | Multilayer perceptron classifier obtained the best discriminant effect in classifying UM versus non-UM | None | Beijing Municipal Administration of Hospitals’ Ascent Plan; Beijing Municipal Administration of Hospitals’ Clinical Medicine Development of Special Funding Support; High Level Health Technical Personnel of Bureau of Health in Beijing; Beijing Key Laboratory of Intraocular Tumor Diagnosis and Treatment |
| Zabor et al. | 2022 | Choroidal melanoma | Human | 363 | Patient and tumor characteristics | Lasso LR | The model had an AUC of 0.86 when differentiating choroidal nevus and melanoma. The most important variables in making this prediction included subretinal fluid, height, distance to optic disc, and orange pigment | A.D.S: Editor-in-Chief of Ocular Oncology and Pathology. Aura Biosciences (stock options), IsoAid LLC (consultancy), Immunocore (consultancy), Isoaid (consultancy), and Eckert and Zeigler (consultancy) | Unrestricted grant from Research to Prevent Blindness to the Cole Eye Institute |
| Santos-Bustos et al. | 2022 | UM | Human | 183 | Eye images | CNN with transfer learning | The model was able to achieve sensitivity, precision and accuracy, of 99%, 98%, and 99%, respectively in diagnosing UM | None | None listed |
| Olaniyi et al. | 2023 | UM | Human | 380 | Eye images | ANN; radial basis function network | ANN and radial basis function network identified melanoma with recognition rates of 92.31% and 94.70% | None | None listed |
| Prognosis | |||||||||
| Damato et al. | 2003 | UM | Human | 2543 | Patient and tumor characteristics | ANN | RMS error for survival prediction by the ANN was 3.8 years compared with 4.3 years by the clinical expert | None | Not listed |
| Kaiserman et al. | 2004 | UM | Human | 231 | Patient and tumor characteristics | ANN | 3–layers ANN had a diagnostic precision of 84% in predicting 5-year survival | None | None |
| Kaiserman et al. | 2005 | Choroidal melanoma | Human | 153 | Patient and tumor characteristics | ANN | ANN with 1 hidden layer of 16 neurons had 84% forecasting accuracy in predicting 5-year mortality compared with the human expert forecasting ability of <70% | None | Not listed |
| Ehlers et al. | 2005 | UM | Human | 74 tumor samples | Gene expression profiles | SVM | TRAM1 and NBS1 ha the largest SVM discriminant in determining cytologic severity | Not listed | NIH, a Research to Prevent Blindness, Inc. Physician-Scientist Award, a Macula Society Research Award, the National Eye Institute and Research to Prevent Blindness, Inc. |
| Kaiserman et al. | 2006 | Choroidal melanoma | Human | 165 | Tumor characteristics | ANN | ANN did not perform better than the KI index in evaluating prognosis | None | Not listed |
| Damato et al. | 2008 | Choroidal melanoma | Human | 2655 | Patient and tumor characteristics | CHENN | All-cause survival curves generated by CHENN matched those produced with Kaplan-Meier analysis (Kolmogorov-Smirnov, P < 0.05). Estimated melanoma-related mortality was lower with CHENN in older patients because the model accounted for competing risks | Not listed | EU Network of Excellence; National Commissioning Group, Department of Health of England, London |
| Onken et al. | 2010 | UM | Human | 609 tumor samples | Gene expression profiles | SVM | SVM trained on expression assay of 15 genes identified tumors with high metastatic risk | Harbour and Washington University may receive income based on a license of related technology by the University to Castle Biosciences, Inc. | National Cancer Institute, Barnes-Jewish Hospital Foundation, Kling Family Foundation, Tumori Foundation, Horncrest Foundation, a Research to Prevent Blindness David F. Weeks Professorship, Research to Prevent Blindness, Inc. Unrestricted grant, and the National Institutes of Health Vision Core grant |
| Harbour et al. | 2014 | UM | N/A* | – | Gene expression profiles | SVM | Expression assay of 15 genes enabled accurate predictions of metastasis risk | Not listed | Not listed |
| Augsburger et al. | 2015 | Primary choroidal and ciliochoroidal melanomas | Human | 80 | RNA microarray | SVM | Two-site sampling of tumors may be advisable to lessen the probability of underestimating risk of metastasis and death | None | Research to Prevent Blindness, Inc; Quest for Vision Fund; National Eye Institute; National Cancer Institute; Barnes-Jewish Hospital Foundation; Kling Family Foundation; Tumori Foundation; Horncrest Foundation; Research to Prevent Blindness |
| Plasseraud et al. | 2017 | UM | Human | 131 tumor samples | Gene expression profiles | SVM | SVM trained on expression assay of 15 genes has reliable and replicable results | All authors are employees and option holders at Castle Biosciences, Inc. | Study was sponsored by Castle Biosciences, Inc. |
| Vaquero-Garcia et al. | 2017 | UM | Human | 1127 | Patient and tumor characteristics | Logistic regression, decision trees, random forest | Model that used clinical and tumor characteristics in addition to chromosome 3 information had the highest AUC of 85% in predicting metastases at 48 months | None | National Institute on Aging; Penn Institute for Biomedical Informatics; National Cancer Institute |
| Eason et al. | 2018 | UM | Database | 121 patients | Gene expression profiles | K-means clustering | K-means clustering was integral to an overall clustering pipeline that analyzed gene expression in UM to reveal profiles associated with metastases and poorer outcomes | Author AS has ownership interest as an inventor for the patent “Colorectal cancer classification with different prognosis and personalized therapeutic responses” | Institute of Cancer Research; Engineering and Physical Sciences Research Council; The Royal Marsden NHS Foundation Trust; The Institute of Cancer Research, London |
| Serghiou et al. | 2019 | Choroidal melanoma | Human | 1022 | Patient and tumor characteristics | Elastic Net, Bayesian generative learning, gradient tree boosting and deep NNs | Different models could predict final visual acuity and need for enucleation | None | National Eye Institute; Research to Prevent Blindness Inc; That Man May See Inc |
| Sun et al. | 2019 | UM | Human | 47 | Pathology slides | CNN | CNN had an overall diagnostic accuracy of 97.1% and AUC of 0.99 in predicting BAP1 expression in individual high-resolution patches | None | Karolinska Institutet; Stockholm County Council; Beijing University of Posts and Telecommunications |
| Zhang et al. | 2020 | UM | Human | 184 | H&E–stained sections | CNN | CNN had an AUC of 0.9 in predicting nuclear BAP1 expression in UM using H&E–stained slides. This task is difficult for human pathologists and this software could be useful for laboratories without access to high quality immunohistochemistry facilities or genetic testing | None | Not listed |
| Liu et al. | 2020 | UM | Human | 20 | H&E stained sections | CNN | CNN achieved 75% accuracy in determining gene expression class from H&E stains | J.F.A.: Consultant - DORC International B.V., Allergan, Inc., Bayer, Mallinckrodt; Financial support - Topcon; Royalties - Springer SBM LLC. Z.M.C.: Financial support - Castle Biosciences, Inc., Immunocore, LLC | Emerson Collective Cancer Research Fund; Research to Prevent Blindness; Radiological Society of North America |
| Zhang et al. | 2020 | UM | TCGA database | 80 | mRNA profile | Regression | Immune cell-based signature was significantly associated with overall survival | None | Not listed |
| Hou et al. | 2021 | UM | Human/TCGA | 138 (80 TCGA, 50 human) | DNA methylation data | Likelihood-based boosting | ML-based discovery of DNA methylation-driven signature had strong prognostic value in determining survival | None | Scientific Research Foundation for Talents of Wenzhou Medical University |
| Zhang et al. | 2021 | UM | Human | 3017 | Iris images | Random forest, CNN | CNN showed no significant correlation between UM incidence and iris color | KZ was employed by SenseTime Group, an artificial intelligence technology company | Capital Health Research and Development of Special; Science & Technology Project of Beijing Municipal Science & Technology Commission; Beijing Municipal Administration of Hospitals’ Ascent Plan |
| Chi et al. | 2022 | UM | Database | 108 | Microarray | SVM | An SVM-based algorithm was able to identify genes related to sphingolipid metabolism that could help predict survival | None | Luzhou Science and Technology Department Applied Basic Research program;Sichuan Province Science and Technology Department of foreign (border) high-end talent introduction project |
| Liu et al. | 2022 | UM | Human | 89 | H&E–stained FNA samples | Attention-based NN | The model predicted tumor gene expression profiles with an AUC of 0.94, accuracy, sensitivity, and specificity of approximately 92% | T.Y.A.L.: Grant from Emerson Collective Cancer Research Fund; Pending patent: MH2# 0184.0097-PRO. Z.C.: Grants from Emerson Collective Cancer Research Fund; Pending patent: MH2# 0184.0097-PRO | Emerson Collective Cancer Research Fund and Research to Prevent Blindness Unrestricted Grant to Wilmer Eye Institute |
| Wang et al. | 2022 | UM | Database | 92 | Microarray | LR | High ROBO1 and low FMN1 expression were associated with longer metastasis-free survival | None | None |
| Lv et al. | 2022 | UM | Database | 171 | Microarray | Lasso regression | The model-based epithelial-mesenchymal transition score had an AUC between 0.75 and 0.85 in predicting poor prognosis | None | National Key R&D Program of China; National Natural Science Foundation of China |
| Luo et al. | 2022 | UM | Human | 454 | Patient and tumor characteristics | Random forest | AUC was 0.71 in predicting death using data from one post-brachytherapy follow-up. AUC increased to 0.88 when using data from three follow-ups. AUC for predicting metastasis was 0.73 using data from one follow-up and 0.85 with data from three follow-ups | KZ: Employed by InferVision health care Science and Technology Limited Company | National Natural Science Foundation of China; Beijing Natural Science Foundation; Scientific Research Common Program of Beijing Municipal Commission of Education; Beijing Municipal Administration of Hospital's Youth Programme; Beijing Dongcheng District Outstanding Talents Cultivating Plan; the Capital Health Research and Development of Special; Science & Technology Project of Beijing Municipal Science & Technology Commission |
| Lv et al. | 2022 | UM | Database | 171 | RNA sequencing – | ConsesnsusClusterPlus (unsupervised clustering) | There were three patient subtypes identified and those with high tumor-infiltrated immune cells scores generally had a better prognosis | None | None listed |
| Donizy et al. | 2022 | UM | Human | 164 | Tumor characteristics | Random forest; survival gradient boosting | AUC for random forest and survival gradient boosting were 0.67 and 0.78 for overall survival and 0.66 and 0.78 for progression-free survival | None | Polish Ministry of Education and Science |
| Meng et al. | 2023 | UM | Database | 143 | Microarray | Unsupervised clustering (unspecified algorithm) | Two clusters based on metastasis-associated gene expression had different survival rates, with specific pathways (e.g., JAK/STAT, mTOR) displaying higher activity in the high-risk cluster | None | None listed |
| Geng et al. | 2023 | UM | Database | 143 | Microarray | Unsupervised clustering (unspecified algorithm) | Clustering identified a biomarker based on expression of PI3K/AKT/mTOR pathway-related genes that could be used for prognostication in UM | None | Natural Science Foundation of Hebei Province and S&T Program of Hebei |
| Liu et al. | 2023 | UM | Database | 108 | RNA sequencing | LASSO | Glycosylation-related genes served significant prognostic value in determining survival in UM. Higher gene activity was correlated with poorer outcomes | None | None listed |
| Li et al. | 2023 | UM | Database | 259 | RNA sequencing | LASSO | LASSO regression identified a set of immune-related genes whose expression helped predict prognosis for patients with UM | None | National Natural Science Foundation of China; China Postdoctoral Science Foundation; Henan Postdoctoral Foundation; Fund of Key Laboratory of Modern Teaching Technology; Ministry of Education P.R. China; Shannxi Province Key Science and Natural Project |
| Treatment | |||||||||
| Bolis et al. | 2017 | UM | TCGA database | 10,080 | RNA-sequencing data | Penalized linear regression and nonlinear regression models such as ensemble (random Forest) and kernel-based methods (SVM) | UM had the highest predicted sensitivity to all-trans retinoic acid, suggesting that all-trans retinoic acid should be tested as a possible therapeutic for metastatic disease | None | Associazione Italiana per la Ricerca contro il Cancro; Fondazione Italo Monzino |
| Cavusculu et al. | 2017 | UM | Human | 480 | Eye videos | Linear discriminant analysis | Linear discriminant analysis has no classification error in identifying when patients' eyes are closed. This model could be used during radiotherapy to immediately halt radiation when the eye is closed and prevent beams directed to healthy regions of the eye | Not listed | Not listed |
CHENN, conditional hazard estimating neural network; H&E, hematoxylin and eosin; mTOR, mammalian target of rapamycin; RMS, root mean square; TCGA, The Cancer Genome Atlas.
The majority of studies were related to diagnosis or prognosis and used varied ML algorithms and data sources. *Methods development.
Song et al.32 developed a logistic regression model that used serum biomarkers rather than ocular images for early diagnosis of UM primary tumor and metastasis. The model identified a two-marker panel of heat shock protein 27 and osteopontin with an area under the receiver operator curve (AUC) of 0.98 when differentiating between UM and control. A single panel, including only melanoma inhibitory activity protein, had an AUC of 0.78 in differentiating between disease-free survivors and those with metastases.32 Significant research is being conducted to investigate the role of serum biomarkers in the management of UM.33 The clinical utility of this work with respect to ML models could be optimized further by integrating biomarkers with clinical imaging data, as Zabor et al.34 have shown with their model, which is able to differentiate between UM and choroidal nevus with an AUC of 0.86 using patient and tumor characteristics.
Prognosis
Differential gene expression is important in the development and evolution of many cancers, including UM. Genetic data have been leveraged to understand the prognosis of this cancer. Early work in this area was conducted by Harbour et al.,35 who used an SVM to differentiate high- and low-risk UM, and found that NBS1 expression correlated with survival. Further work by the Harbour group and affiliates has focused on integrating their SVM model into point of care genetic analysis assays that are currently being used as a commercial assay to predict metastatic risk.36–38 DNA methylation and gene expression data have also identified genes with prognostic value for UM for both overall survival39–45 and metastatic risk.46–48 In addition, gene expression analyses have shown that the presence of specific immune cells (e.g., CD8+ T cells) is associated with overall survival in UM. Combining this immunologic information with clinical (e.g., patient age) and pathological (e.g., tumor stage) data achieved an AUC of approximately 0.8 in predicting survival.49 Other investigators have shown the promise of assessing gene expression from CNN-based image processing of cytopathologic slides.50
Clinical data on patient demographics and tumor characteristics have been effective in training ML models to determine prognosis for intraocular cancers. NNs have been the most common algorithm used for this purpose and have leveraged demographic information such as patient age and sex and various tumor features such as size, location, mitotic rate, and chromosomal abnormalities. Damato et al.51 trained a NN using data from 2543 patients with UM to predict time to metastatic death using clinical and histopathologic features. The survival prediction error by the NN was approximately 3.8 years compared with approximately 4.3 years by a clinical expert.51 A study by Taktak et al.52 reported similar results. Pe'er et al.53 developed a NN to estimate 5-year survival in patients with choroidal melanoma, achieving an accuracy of 84% compared with the less than 70% accuracy by achieved by clinicians. Prognostic ML algorithms have been compared with Kaplan-Meier analyses and found to have a similar performance, although ML methods were superior in estimating survival of older patients. In these models, tumor diameter along with histological and cytogenetic features such as monosomy 3 were most important in predicting survival.54 Donizy et al.55 built prognostication models and similarly found that features such as BAP1 expression, nucleoli size, and mitotic rate helped predict progression-free survival with an AUC of 0.78.
ML models for determining prognosis have gone beyond predicting survival. Serghiou et al.56 developed multiple ML models to predict visual outcomes after proton beam radiotherapy for choroidal melanoma. Post-treatment visual acuity was best predicted by factors including tumor thickness, radiation received by the macula, and total radiation received by the overall globe volume. Need for enucleation was predicted with an AUC of 0.8 and tumor features such as thickness and stage were the most important for this prediction.56 Luo et al.57 developed a similar model, but for predicting metastasis and death after brachytherapy and achieved a maximum AUC of 0.85. Many groups have focused on predicting metastatic risk using pathology images from enucleation and local resection samples. Zhang et al.58 developed a CNN that used hematoxylin and eosin–stained slides, without other specialized stains, to assess nuclear BAP1 expression, which is a surrogate for metastatic prognostic factors such as monosomy 3 and BAP1 mutation. The CNN achieved an AUC of 0.93,58 which is especially impressive given that identifying nuclear BAP expression based solely on hematoxylin and eosin staining is difficult, even for experienced pathologists. In contrast, a CNN developed by Sun et al.59 to assess BAP1 expression from slides specifically stained for BAP1 achieved an AUC of 0.99. Other investigators have developed CNN-based methods for gene expression profiling and achieved an AUC of 0.94.60 Vaquero-Garcia et al.61 used a non–image-based approach to predict metastases, relying on tumor features such as location and chromosomal copy number to train a model that predicted 48-month metastatic risk with an accuracy of 85%. Not all ML models developed to predict metastasis have been successful. Kaiserman et al.62 developed a NN to predict conversion of choroidal nevi to melanomas using features such as nevus thickness, base diameter, and reflectivity, but the model did not predict malignant transformation with greater accuracy compared with existing metrics.
Treatment
Research applying ML to optimize UM treatment is in its infancy. Bolis et al.63 used RNA sequencing data from The Cancer Genome Atlas to predict tumor sensitivity to all-trans retinoic acid and found that UM had the highest predicted sensitivity. Because metastatic UM lacks many treatment options, all-trans retinoic acid–based therapeutics could be explored.63
ML Studies in Rb
Rb is the most common primary intraocular cancer in children and is lethal if untreated.64,65 However, treatment advances have led to near 100% survival in developed countries, with the possibility of eye salvage in many cases.66 Current work in the management of Rb has focused on developing vision-sparing treatments and novel diagnosis techniques for use in resource-poor areas.
Diagnosis
The initial diagnosis of Rb is heralded commonly by leukocoria or white pupil reflex. In 2019, Munson et al.14 designed the ComputeR-Assisted Detector of Leukocoria (CRADLE) to assist parents in augmenting clinical leukocoria screening (Table 2). CRADLE is a CNN-based mobile application that analyzes pictures stored on mobile devices and provides alerts if leukocoria is detected. The CNN was tested using 52,982 facial photographs of 40 different children with unilateral Rb (n = 8), bilateral Rb (n = 7), Coat's disease, cataract, amblyopia, and hyperopia (n = 5), or no ocular disorder (n = 20). The testing data comprised pictures of children in everyday settings (e.g., eating dinner, playing). CRADLE's sensitivity was 90% for diagnosing children 2 years or younger and the algorithm enabled leukocoria detection from photographs taken 1.3 years before clinical diagnosis. Applications like this, especially if incorporated into everyday devices such as mobile phones, can enable low-cost, widescale leukocoria screening and enable earlier intervention and improved visual outcomes with this cancer.14 Further validation of this concept was provided by Bernard et al. whose CNN-based algorithm loaded onto Android smartphones achieved an AUC of 0.93 in identifying leukocoria in pediatric clinics in Ethiopia.67
Table 2.
Studies That Applied ML for Management of Rb
| Author | Year | Type(s) of Cancer | Data Source | Number of Data Points | Data Modality | ML Algorithm(s) Used | Key Takeaways | Conflicts of Interest | Funding Sources |
|---|---|---|---|---|---|---|---|---|---|
| Diagnosis | |||||||||
| Rivas-Perea et al. | 2014 | Rb | Human | 72 images (144 eyes) | Eye images | ANN, discriminant analysis (linear and quadratic), SVM | Combination of SVM, linear discriminant analysis/quadratic discriminant analysis, and SVM in a softmax classification achieved a 92% accuracy rate in detecting leukocoria from eye images | None | National Council for Science and Technology (Mexico); Baylor University |
| Munson et al. | 2019 | Rb | Human | 52982 pictures from 40 children | Pictures | CNN | A smartphone application could detect leukocoria in photographs taken an average of 1.3 years before diagnosis | BFS, GH, and RH are the inventors of the CRADLE/White Eye Detector Application | The National Science Foundation; Robert A. Welch Foundation |
| Rao et al. | 2020 | Rb | Human | 124 images | Eye images | CNN, SVM, ANN | CNN had an accuracy of 98.6% in detecting leukocoria | Not listed | Not listed |
| Bernard et al. | 2022 | Rb | Human | 1457 | Eye images | CNN | The EyeScreen software was able to identify leukocoria with an AUC of 0.93, sensitivity of 87%, and specificity of 73% | C.N.: Board of directors – World Association of Eye Hospitals; Board trustee – American Society of Ophthalmic Plastic and Reconstructive Surgery Foundation H.D.: Advisory Board – Castle Bioscience; Financial support – Aura Bioscience | University of Michigan Medical School; the Ravitz Foundation; the Richard N. and Marilyn K. Witham Professorship; and an anonymous foundation |
| Kaliki et al. | 2023 | Rb | Human | 771 | Eye images | CNN | CNN was effective in detecting Rb, more so when given all images of an eye versus an individual image | None | The Operation Eyesight Universal Institute for Eye Cancer (SK) and Hyderabad Eye Research Foundation (SK), Hyderabad, India |
| Kumar et al. | 2023 | Rb | Human | 278 | MRI, computed tomography scan, fundus images | CNN | Two different CNNs were trained (AlexNet and ResNet50) to identify Rb. ResNet50 achieved an accuracy of 93.16%, which is higher than other models reported in the literature | None | None listed |
| Prognosis | |||||||||
| Ciller et al. | 2017 | Rb | Human | 1186 images from 28 children | Fundus images | CNN | CNN performed well in automatic segmentation and worked in <1 second compared with manual delineation taking 1–2 minutes | None | None |
| Alvarez-Suarez et al. | 2020 | Rb | Human | 8 patients | Gene expression profiles | Clustering | Supervised and unsupervised clustering identified commonly and heterogeneously expressed genes, including 4 possible therapeutic targets | None | Instituto Mexicano del Seguro Social; National Council for Science and Technology (Mexico); National Institutes of Health |
| Liu et al. | 2021 | Rb | Human | 66 | Aqueous humor samples | Regression, logistic regression, decision tree | Ridge regression had an AUC between 0.81 and 0.91 in differentiating between early and advanced disease | None | Ministry of Science and Technology of China; National Natural Science Foundation of China; Shanghai Institutions of Higher Learning; Shanghai Rising-Star Program; Shanghai Municipal Health Commission; National Research Center for Translational Medicine·Shanghai; Shanghai Municipal Science and Technology Major Project |
| Im et al. | 2023 | Rb | Human | 76 | Aqueous humor liquid biopsy | Logistic regression | Logistic regression–based analysis of nucleic acid concentration helped to predict Rb disease burden | J.LB. and L.X.: Patent application entitled: Aqueous humor cell free DNA for diagnostic and prognostic evaluation of ophthalmic disease | National Cancer Institute; The Wright Foundation; Children's Oncology Group/ St. Baldrick's Foundation; Danhakl Family Foundation; Hyundai Hope on Wheels; Childhood Eye Cancer Trust; Children's Cancer Research Fund; The Berle & Lucy Adams Chair in Cancer Research; The Larry and Celia Moh Foundation; The Institute for Families Inc; Children's Hospital Los Angeles; The Knights Templar Eye Foundation; Las Madrinas Endowment in Experimental Therapeutics for Ophthalmology; Research to Prevent Blindness Career Development Award; National Eye Institute |
| Treatment | |||||||||
| Strijbis et al. | 2021 | Rb | Human | 60 | MRI: T1-, T2-weighted, and fast imaging employing steady-state acquisition | CNN | CNN can obtain accurate ocular structure and tumor segmentations in Rb | None | Cancer Center Amsterdam; Swiss National Science Foundation |
| Hung et al. | 2011 | Rb | Human | 1 | MRI | ANN | ANN can help to optimize the performance of algorithms to segment tumors in MRIs | Not listed | Not listed |
| Lin et al. | 2003 | Rb | Human | 1 | MRI | ANN | Fuzzy logic combined with ANN can help with tumor segmentation from MRI | Not listed | National Science Council of Taiwan |
| Ciller et al. | 2017 | Rb | Human | 16 | MRI scans | Random forest and CNN | CNN and random forest perform well in segmenting tumors from MRI | None | Swiss Cancer League; Swiss National Science Foundation; Hasler Stiftung; University of Lausanne; University of Geneva; Centre Hospitalier Universitaire Vaudois; Hôpitaux Universitaires de Genève; University of Bern (UniBe); Leenaards and the Jeantet Foundations |
| Han et al. | 2018 | Rb | Human | 62 | Gene expression profiles | SVM | SVM had an AUC of 0.79 using differentially expressed genes to classify between Rb and normal | None | None |
| Kakkassery et al. | 2022 | Rb | Cell line | N/A | Rb cell lines | Multiple (e.g., LASSO, elastic nets, random forests, extreme gradient boosting) | Pathways related to retinoid metabolism and transport and sphingolipid biosynthesis could play a role in etoposide resistance seen in Rb | T.G.: Member of the BigOmics Analytics Advisory Board | KinderAugenKrebsStiftung; Karl and Charlotte Spohn Stiftung; Ad Infinitum Foundation |
ANN, artificial neuralnetwork.
Most studies were related to treatment and primarily used MRI images to train ML models.
Other groups have used CNNs, SVMs, and NNs to identify leukocoria from facial images taken in nonclinical settings. CNN outperformed the other two ML algorithms, achieving an accuracy of 98.6% and sensitivity of 97.6%. However, the CNN specificity of 63.8% was the lowest of all the algorithms.68 To address low specificity, studies have designed ensemble models that combine NNs with other algorithms to achieve promising performance in leukocoria detection, with a specificity of 89%.69 Notably, all three of the aforementioned models use nonclinical digital images (e.g., image of child at birthday party or playground) as their inputs. This increases opportunities for early diagnosis in areas without ophthalmic care, nonophthalmic clinical settings, and at home. Further work, with higher quality eye images, has been conducted and achieved a specificity of 85% with a sensitivity of 99%.70 In contrast, some efforts have been made to design support systems that assist physicians in better diagnosing Rb in the clinical setting. Kumar et al.71 created CNN-based models to MRI and computed tomography scans that could identify Rb with an accuracy of 93.16%, which is higher than many other models in the literature.
Prognosis
Gene expression analysis has been useful in determining the prognosis of Rb. Alvarez-Suarez et al.72 leveraged supervised and unsupervised clustering to identify gene expression patterns associated with Rb, with some genes predicting unilateral versus bilateral disease. Building off initial work conducted by Berry et al.,73,74 Liu et al.75 analyzed metabolic activity from aqueous humor samples to stage Rb, achieving an AUC of 0.9 and accuracy of 80%. Similar methods could be used to assess disease progression more quantitatively. For example, logistic regression-based analysis of nucleic acid content from aqueous humor samples has been used to better quantify disease burden at diagnosis and during treatment.76
Treatment
Multiple treatment approaches exist for Rb, ranging from focal therapy alone or in combination with systemic chemotherapy, intra-arterial chemosurgery, and enucleation. These approaches vary in their technical requirements and costs, which affect their availability and use in the United States and elsewhere in the world. ML approaches may be helpful for remote diagnosis and tumor stratification for treatment recommendation. Important for ML programs is accurate tumor segmentation in imaging (e.g., MRI, ultrasound examination), because both tumor size and depth of infiltration can impact treatment approach and complexity. Ciller et al.77 developed a CNN that segments Rb tumors using fundus images, thereby decreasing physician-to-physician variability with manual segmentation and simplifying long-term tracking of tumors. A combination random forest and CNN model, also developed by Ciller et al.,78 showed strong performance in segmenting Rb tumors when using a new set of features that combined information about tumor shape and position. In 2021, Strijbis et al.79 developed a CNN to segment Rb from T1- and T2-weighted MRI. The CNN had high correlation with expert manual segmentation in assessing eye and tumor volume and tumor spatial location.79 Several similar models have been developed to assist physicians with segmentation and treatment planning,12,13 but these methods have yet to be tested with larger samples.
Finally, ML has been used to identify potential therapeutic targets for Rb. Han et al.80 used SVM to analyze gene expression in 62 Rb samples from enucleated eyes and demonstrated effective differentiation between Rb and controls based on expression of seven genes. These genes may warrant further investigation for targeting in Rb treatment.80 Similar ML-based genetic analyses have revealed mechanisms for chemotherapy resistance in Rb including pathways related to retinoid metabolism and sphingolipid synthesis.81
ML Studies in Other Ocular Cancers
There are other classes of ocular cancers that involve the orbit and surrounding structures (e.g., conjunctival melanoma, orbital teratoma).82,83 These cancers can be especially difficult to diagnose and manage given their low incidence. Therefore, ML-enabled clinical decision support systems could assist physicians to improve patient outcomes.
Diagnosis
Developing ML models for rare diseases is challenging given the relative lack of training and testing data (Table 3). One solution is to use synthetic data augmentation. In 2021, Yoo et al.84 trained a CNN to diagnose conditions of the conjunctiva (e.g., melanoma, pterygium), some of which have incidences as low as 0.3 per 1,000,000. Given the small set of images that existed to train the CNN, the group used data augmentation to enhance the size and variety of the training data. This augmentation involved image processing techniques including changing image quality through adding noise or flipping images about a vertical axis. More advanced augmentation used generative NNs to synthesize new images with representative features from existing images in the dataset. With this augmentation, the model achieved an accuracy of 97% in the detection of conjunctival melanoma.84 Other CNN-based algorithms have been successful in diagnosing ocular adnexal lymphoma,85 eyelid basal cell,86 and general eyelid tumors.87
Table 3.
Studies That Applied ML for Management of Other Ocular Cancers (i.e., Non-Rb, and Noniris/UM)
| Author | Year | Type(s) of Cancer | Data Source | No. of Data Points | Data Modality | ML Algorithm(s) Used | Key Takeaways | Conflicts of Interest | Funding Sources |
|---|---|---|---|---|---|---|---|---|---|
| Diagnosis | |||||||||
| Habibalahi et al. | 2019 | Ocular surface squamous neoplasia | Human | 18 | Auto fluorescence multispectral images of biopsy specimens | K-nearest neighbor, SVM | Multispectral autofluorescence imaging can be used in combination with ML to detect the boundary of OSSN | None | Personal Eyes Pty Ltd; ARC Centre of Excellence for Nanoscale Biophotonics; Australian research council; iMQRES scholarship |
| Yoo et al. | 2021 | Conjunctival melanoma | Images from Google and smartphone images from synthetic eye models | 398 | Eye images | 5 different CNN architectures | The algorithm achieved an accuracy of 87.5% in 4 class classification (conjunctival melanoma, conjunctival nevus, and melanosis, pterygium, and normal conjunctiva) and 97.2% for binary classification (conjunctival melanoma or not) | Ik Hee Ryu and Jin Kuk Kim are executives of VISUWORKS, Inc., which is a Korean artificial intelligence startup providing medical ML solutions | Aerospace Medical Center of Korea Air Force, South Korea |
| Hou et al. | 2021 | Adnexal lymphoma | Human | 56 | MRI | SVM with linear kernel | SVM with contrast-enhanced MRI was significantly better than the radiology resident in differentiating between ocular adnexal lymphoma and idiopathic orbital inflammation | None | National Natural Science Foundation of China; China Postdoctoral Science Foundation; Shaanxi Key R&D Plan; Xi'an Science and Technology Plan Project; Scientific Research Foundation of Xi'an Fourth Hospital |
| Xie et al. | 2022 | Ocular adnexal lymphoma | Human | 89 | Patient and tumor characteristics | CNN | The model trained on a combination of clinical and imaging data had an AUC of 0.95 in differentiating between ocular adnexal lymphoma and idiopathic orbital inflammation | None | National Natural Science Foundation of China; Shaanxi Key R&D Plan; Shaanxi International Science and Technology Cooperation Program; China Postdoctoral Science Foundation; Xi'an Science and Technology Plan |
| Luo et al. | 2022 | Eyelid basal cell and sebaceous carcinoma | Human | 296 | H&E–stained sections | CNN | CNN achieved an accuracy of 0.983 in diagnosing eyelid basal cell carcinoma and sebaceous carcinoma, compared with accuracies of 0.644, 0.729, and 0.831 for pathologists | XG is a technical consultant of Jinan Guoke Medical Engineering and Technology Development Co | National Natural Science Foundation of China; Science and Technology Commission of Shanghai; Innovative research team of high-level local universities in Shanghai; Key Research and Development Program of Shandong Province; Key Research and Development Program of Jiangsu Province; Jiangsu Province Engineering Research Center of Diagnosis and Treatment of Children's Malignant Tumour; Shandong Province Natural Science Foundation |
| Hui et al. | 2022 | Eyelid tumors | Human | 345 | Eye images | CNN | Eight CNNs were trained and achieved a maximum AUC of 0.889. The deep-learning systems performed comparably with senior ophthalmologists | None | National Natural Science Foundation of China; The Special Fund of the Pediatric Medical Coordinated Development Center of Beijing Hospitals Authority |
| Prognosis | |||||||||
| Taktak et al. | 2004 | Intraocular melanoma | Human | 2331 | Tumor characteristics | ANN | RMS error was 3.7 years for the ANN and 4.3 years for the clinical expert for 15 years survival | Not listed | Not listed |
| Treatment | |||||||||
| Tan et al. | 2017 | Periocular basal cell carcinoma | Human | 156 | Patient and tumor characteristics | Naive Bayes and decision trees | A 3-variable decision tree was able to predict the operative complexity | None | None |
ANN, artificial neural network; H&E, hematoxylin and eosin.
Most studies focused on diagnosis, though the literature is sparse in this field.
Non-CNN algorithms have been applied successfully to diagnose rarer classes of ocular cancers. Hou et al.88 found that an SVM trained using MRI could differentiate between ocular adnexal lymphoma and idiopathic orbital inflammation with an AUC of 0.8, a significant performance improvement compared with a radiology resident. Finally, Habibalahi et al.89 showed that k-nearest neighbor and SVM could differentiate between normal tissue and ocular surface squamous neoplasia using fluorescence biopsy histopathologic images. The outputs from algorithms like this could be used in the operating room to classify cancer margins better.
Treatment
There has been little work investigating use of ML in treating rarer classes of ocular cancers. Tan et al.90 built a decision tree–based model that assessed the complexity of reconstruction after periocular basal cell excision, achieving an AUC of 0.85 with only three predictive variables: (1) preoperative assessment of complexity, (2) surgical delays (e.g., <75 or >75 days), and tumor size (e.g., <14 mm or >14 mm).
Discussion
ML is a powerful tool and is rapidly increasing in popularity for clinical applications. Ophthalmology lends itself well to ML-based technologies, given the relative ease of acquiring disease-related data and images (e.g., primary fundus photographs, optical coherence tomography images, and corneal topography). In fact, numerous ML algorithms have already been applied to various subspecialties within ophthalmology, but ocular oncology has been relatively underexplored in this regard. This review highlights 63 publications demonstrating the current state of science in using ML to assist in the diagnosis, prognosis, and treatment of ocular cancers. Most studies focused on developing ML algorithms for UM or Rb, but rarer forms of ocular cancers are also represented. These ML algorithms have been trained using a variety of data sources, including imaging (e.g., MRI, ultrasound examination), gene expression arrays, and demographic data, which demonstrates the breadth of information that can be leveraged to develop the models. Although NNs were the most popular algorithm used, non-NN algorithms have also been developed successfully for applications to ocular malignancies.
Analyses of studies that recruited human participants revealed that 72% had fewer than 300 participants. This is important because training ML models is a data-intensive process.91 In general, the more varied the data that are available to train an ML model, the more accurate and generalizable its outputs will be.92 Smaller sample sizes could also contribute to variation in algorithm-to-algorithm performance. In ocular oncology, small sample sizes are related to the low incidence and prevalence of the cancers being studied, which can limit ML model performance. Methods to work around this issue include leveraging synthetic data enhancement with imaging data using techniques similar to those seen in Santos-Bustos et al.28 and Olaniyi et al.,27 collaborating with ocular oncology centers of excellence that have large in-house data repositories, and adopting “low-shot” ML algorithms that can be trained with relatively small training datasets.93 Regardless of sample size, the studies analyzed in this review demonstrated strong performance.
Bias in ML model design and development is important to evaluate in assessing a model's real-world applicability. Analysis of the funding sources and author conflicts of interest of the studies evaluated here revealed that most studies were funded by nonprofit or government agencies. Although some authors declared conflicts of interest with private artificial intelligence or pharmaceutical companies, none seemed to interact directly with the authors’ works assessed herein. Continued reporting of funding sources and conflicts of interest will be crucial to maintain the independence of the studies contributing to this growing field.
Core aspects of ML model development such as algorithm selection (e.g., NN versus decision tree) and parameter identification (e.g., learning rates for gradient descent) require significant trial and error.94–97 In reality, multiple ML algorithms could achieve acceptable performance on a task given the same data.98,99 However, certain ML algorithms have specific advantages for applications in ophthalmology. CNNs were commonly leveraged in the studies presented here; they provide robust, deep learning without the need for feature engineering since clinical images themselves (e.g., fundus photographs) are used as model inputs. As stated elsewhere in this article, image data also enable synthetic data enhancement to augment small datasets. One disadvantage of CNNs is their black box nature, which can make it difficult to understand how exactly the models are making decisions. To increase end-user interpretability, researchers could leverage explainability techniques, such as saliency maps, which visually highlight key image features that an algorithm used to make a decision.100 These saliency maps also work to increase physician confidence in model outputs. Non-NN algorithms often allow for more mechanistic insight and interpretability and generally require less training data compared with NNs. Common non-NN algorithms discussed here include decision trees, that were used by Jegelevicius et al.,31 Serghiou et al.,56 and Tan et al.90 Although decision trees can be effective, they tend to overfit training data, thereby limiting their generalizability. In those cases, groups can use techniques such as boosting or bagging to decrease bias and variance, respectively. Bagged decision trees can be particularly useful to mitigate overfitting, which can result from analyzing small datasets.
Fifty of the 63 studies analyzed here were published after 2014. A breakdown of the publications by category revealed that papers related to prognosis and diagnosis were the primary drivers of this recent increase. There remains significant work to be done. More tools are needed to help predict metastases, especially for UM. This goal can be accomplished through developing monitoring blood assays or creating tools that identify proteins or genetic expression associated with metastatic spread. Metastatic UM is nearly universally fatal,101,102 so predictive tools have great potential to improve patient outcomes. There are also opportunities to better leverage ML to identify therapeutic gene targets or genes that predispose individuals to developing specific intraocular cancers.
The ultimate goal for integrating ML into day-to-day clinical workflows is to develop FDA-approved solutions. There is great opportunity to build on existing work to achieve this goal. To date, the FDA has approved 521 ML-enabled devices, with six for use in ophthalmology, mostly for the detection of diabetic retinopathy.10 With an increasing focus on ML in health care, the FDA has created new protocols to better assist researchers in developing ML solutions and navigating the FDA approval process.103 The plan published by the FDA to improve evaluation of ML-enabled technologies includes outlining good ML practices for researchers to follow, creating guidelines for algorithm transparency, supporting intramural and extramural research on ML algorithm evaluation and improvement, and establishing more robust guidelines pertaining to real-world data collection and postapproval monitoring.
Conclusions
There is great promise in developing ML approaches to improve management of patients with intraocular cancers across the workflow of diagnosis, prognosis, and treatment. Further work is needed to continue validating accurate and easy-to-use solutions that better integrate into existing clinical workflows, with a specific focus on creating tools to help diagnose and treat intraocular malignancies. Work toward this goal will hopefully improve future outcomes for patients with intraocular cancers.
Supplementary Material
Acknowledgments
Author Contributions: ASC and YS devised the study. ASC, TMH, CCC, and NKB performed the literature search and conducted the analyses. ASC and TMH wrote the manuscript with support from YS. All authors read and approved the final version of the manuscript.
Disclosure: A.S. Chandrabhatla, None; T.M. Horgan, None; C.C. Cotton, None; N.K. Ambati, None; Y.E. Shildkrot, Castle Biosciences (C), Genentech/Roche (E)
References
- 1. Kaliki S, Shields CL.. Uveal melanoma: relatively rare but deadly cancer. Eye (London). 2017; 31(2): 241–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Papakostas TD, Lane AM, Morrison M, Gragoudas ES, Kim IK.. Long-term outcomes after proton beam irradiation in patients with large choroidal melanomas. JAMA Ophthalmol. 2017; 135(11): 1191–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seider MI, Mruthyunjaya P.. Molecular prognostics for uveal melanoma. Retina. 2018; 38(2): 211–219. [DOI] [PubMed] [Google Scholar]
- 4. Maheshwari A, Finger PT.. Cancers of the eye. Cancer Metastasis Rev. 2018; 37(4): 677–690. [DOI] [PubMed] [Google Scholar]
- 5. Valasapalli S, Guddati AK.. Nation-wide trends in incidence-based mortality of patients with ocular melanoma in USA: 2000 to 2018. Int J Gen Med. 2021; 14: 4171–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bianciotto C, Shields CL, Guzman JM, et al.. Assessment of anterior segment tumors with ultrasound biomicroscopy versus anterior segment optical coherence tomography in 200 cases. Ophthalmology. 2011; 118(7): 1297–1302. [DOI] [PubMed] [Google Scholar]
- 7. Damato B. Progress in the management of patients with uveal melanoma. The 2012 Ashton Lecture. Eye (Lond). 2012; 26(9): 1157–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bohr A, Memarzadeh K.. The rise of artificial intelligence in healthcare applications. Artificial Intelligence in Healthcare. Published online 2020: 25–60. [Google Scholar]
- 9. Balyen L, Peto T.. Promising artificial intelligence-machine learning-deep learning algorithms in ophthalmology. Asia Pac J Ophthalmol (Phila). 2019; 8(3): 264–272. [DOI] [PubMed] [Google Scholar]
- 10. FDA. Artificial intelligence and machine learning (AI/ML)-enabled medical devices. Published online September 22, 2021. Available at: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices. Accessed January 15, 2022.
- 11. Shields CL, Lally SE, Dalvin LA, et al.. White Paper on ophthalmic imaging for choroidal nevus identification and transformation into melanoma. Transl Vis Sci Technol. 2021; 10(2): 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hung WL, Chen DH, Yang MS.. Suppressed fuzzy-soft learning vector quantization for MRI segmentation. Artif Intell Med. 2011; 52(1): 33–43. [DOI] [PubMed] [Google Scholar]
- 13. Lin KCR, Yang MS, Liu HC, Lirng JF, Wang PN.. Generalized Kohonen's competitive learning algorithms for ophthalmological MR image segmentation. Magn Reson Imaging. 2003; 21(8): 863–870. [DOI] [PubMed] [Google Scholar]
- 14. Munson MC, Plewman DL, Baumer KM, et al.. Autonomous early detection of eye disease in childhood photographs. Sci Adv. 2019; 5(10): eaax6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Y, Chen PHC, Krause J, Peng L.. How to read articles that use machine learning: users’ guides to the medical literature. JAMA. 2019; 322(18): 1806–1816. [DOI] [PubMed] [Google Scholar]
- 16. Sarvestany SS, Kwong JC, Azhie A, et al.. Development and validation of an ensemble machine learning framework for detection of all-cause advanced hepatic fibrosis: a retrospective cohort study. Lancet Digital Health. 2022; 4(3): e188–e199. [DOI] [PubMed] [Google Scholar]
- 17. Bos JM, Attia ZI, Albert DE, Noseworthy PA, Friedman PA, Ackerman MJ.. Use of artificial intelligence and deep neural networks in evaluation of patients with electrocardiographically concealed long QT syndrome from the surface 12-lead electrocardiogram. JAMA Cardiol. 2021; 6(5): 532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chandrabhatla AS, Pomeraniec IJ, Ksendzovsky A.. Co-evolution of machine learning and digital technologies to improve monitoring of Parkinson's disease motor symptoms. npj Digit Med. 2022; 5(1): 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thompson AC, Jammal AA, Berchuck SI, Mariottoni EB, Medeiros FA.. Assessment of a segmentation-free deep learning algorithm for diagnosing glaucoma from optical coherence tomography scans. JAMA Ophthalmology. 2020; 138(4): 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Page AL, Ballot E, Truntzer C, et al.. Using a convolutional neural network for classification of squamous and non-squamous non-small cell lung cancer based on diagnostic histopathology HES images. Sci Rep. 2021; 11(1): 23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Redd TK, Prajna NV, Srinivasan M, et al.. Image-based differentiation of bacterial and fungal keratitis using deep convolutional neural networks. Ophthalmology Science. 2022; 2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arbelaez MC, Versaci F, Vestri G, Barboni P, Savini G.. Use of a support vector machine for keratoconus and subclinical keratoconus detection by topographic and tomographic data. Ophthalmology. 2012; 119(11): 2231–2238. [DOI] [PubMed] [Google Scholar]
- 23. Bilmin K, Synoradzki KJ, Czarnecka AM, et al.. New perspectives for eye-sparing treatment strategies in primary uveal melanoma. Cancers (Basel). 2021; 14(1): 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grisanti S, Tura A. Uveal Melanoma. In: Scott JF, Gerstenblith MR, eds. Noncutaneous Melanoma. Singapore: Codon Publications; 2018. Available at: http://www.ncbi.nlm.nih.gov/books/NBK506991/. Accessed April 18, 2022. [Google Scholar]
- 25. Char DH. Uveal melanoma: differential diagnosis. In: Damato B, Singh AD, eds. Clinical Ophthalmic Oncology: Uveal Tumors. Berlin: Springer; 2014: 99–111, doi: 10.1007/978-3-642-54255-8_8 [DOI] [Google Scholar]
- 26. Oyedotun O, Khashman A.. Iris nevus diagnosis: convolutional neural network and deep belief network. Turk J Elec Eng & Comp Sci. Published online 2017:10, doi: 10.3906/elk-1507-190 [DOI] [Google Scholar]
- 27. Olaniyi EO, Komolafe TE, Oyedotun OK, Oyemakinde TT, Abdelaziz M, Khashman A.. Eye melanoma diagnosis system using statistical texture feature extraction and soft computing techniques. J Biomed Phys Eng. 2023; 13(1): 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santos-Bustos DF, Nguyen BM, Espitia HE.. Towards automated eye cancer classification via VGG and ResNet networks using transfer learning. Engineering Science and Technology, an International Journal. 2022; 35: 101214, doi: 10.1016/j.jestch.2022.101214 [DOI] [Google Scholar]
- 29. Shields CL, Kaliki S, Hutchinson A, et al.. Iris nevus growth into melanoma: analysis of 1611 consecutive eyes: the ABCDEF guide. Ophthalmology. 2013; 120(4): 766–772. [DOI] [PubMed] [Google Scholar]
- 30. Su Y, Xu X, Zuo P, et al.. Value of MR-based radiomics in differentiating uveal melanoma from other intraocular masses in adults. Eur J Radiol. 2020; 131: 109268. [DOI] [PubMed] [Google Scholar]
- 31. Jegelevicius D, Paunksnis A, Barzdžiukas V.. Application of data mining technique for diagnosis of posterior uveal melanoma. Informatica. 2002; 13(4): 455–464. [Google Scholar]
- 32. Song J, Merbs SL, Sokoll LJ, Chan DW, Zhang Z.. A multiplex immunoassay of serum biomarkers for the detection of uveal melanoma. Clin Proteomics. 2019; 16: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bande Rodríguez MF, Fernandez Marta B, Lago Baameiro N, et al.. Blood biomarkers of uveal melanoma: current perspectives. Clin Ophthalmol. 2020; 14: 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zabor EC, Raval V, Luo S, Pelayes DE, Singh AD.. A prediction model to discriminate small choroidal melanoma from choroidal nevus. Ocul Oncol Pathol. 2022; 8(1): 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ehlers JP, Harbour JW.. NBS1 expression as a prognostic marker in uveal melanoma. Clin Cancer Res. 2005; 11(5): 1849–1853. [DOI] [PubMed] [Google Scholar]
- 36. Harbour JW. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol Biol. 2014; 1102: 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Onken MD, Worley LA, Tuscan MD, Harbour JW.. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Molec Diagn. 2010; 12(4): 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Plasseraud KM, Wilkinson JK, Oelschlager KM, et al.. Gene expression profiling in uveal melanoma: technical reliability and correlation of molecular class with pathologic characteristics. Diagn Pathol. 2017; 12(1): 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hou P, Bao S, Fan D, et al.. Machine learning-based integrative analysis of methylome and transcriptome identifies novel prognostic DNA methylation signature in uveal melanoma. Brief Bioinform. 2021; 22(4): bbaa371. [DOI] [PubMed] [Google Scholar]
- 40. Chi H, Peng G, Yang J, et al.. Machine learning to construct sphingolipid metabolism genes signature to characterize the immune landscape and prognosis of patients with uveal melanoma. Front Endocrinol (Lausanne). 2022; 13: 1056310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lv Y, He L, Jin M, Sun W, Tan G, Liu Z. EMT-related gene signature predicts the prognosis in uveal melanoma patients. J Oncol. 2022; 2022: 5436988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lv X, Ding M, Liu Y.. Landscape of infiltrated immune cell characterization in uveal melanoma to improve immune checkpoint blockade therapy. Front Immunol. 2022; 13: 848455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Geng Y, Geng Y, Liu X, et al.. PI3K/AKT/mTOR pathway-derived risk score exhibits correlation with immune infiltration in uveal melanoma patients. Front Oncol. 2023; 13: 1167930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu J, Zhang P, Yang F, et al.. Integrating single-cell analysis and machine learning to create glycosylation-based gene signature for prognostic prediction of uveal melanoma. Front Endocrinol (Lausanne). 2023; 14: 1163046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li X, Kang J, Yue J, et al.. Identification and validation of immunogenic cell death-related score in uveal melanoma to improve prediction of prognosis and response to immunotherapy. Aging (Albany NY). 2023; 15(9): 3442–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eason K, Nyamundanda G, Sadanandam A.. polyClustR: defining communities of reconciled cancer subtypes with biological and prognostic significance. BMC Bioinform. 2018; 19(1): 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang T, Wang Z, Yang J, Chen Y, Min H.. Screening and identification of key biomarkers in metastatic uveal melanoma: evidence from a bioinformatic analysis. J Clin Med. 2022; 11(23): 7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meng S, Zhu T, Fan Z, et al.. Integrated single-cell and transcriptome sequencing analyses develops a metastasis-based risk score system for prognosis and immunotherapy response in uveal melanoma. Front Pharmacol. 2023; 14: 1138452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Z, Ni Y, Chen G, Wei Y, Peng M, Zhang S.. Construction of immune-related risk signature for uveal melanoma. Artif Cells Nanomed Biotechnol. 2020; 48(1): 912–919. [DOI] [PubMed] [Google Scholar]
- 50. Liu TYA, Zhu H, Chen H, et al.. Gene expression profile prediction in uveal melanoma using deep learning: a pilot study for the development of an alternative survival prediction tool. Ophthalmol Retina. 2020; 4(12): 1213–1215. [DOI] [PubMed] [Google Scholar]
- 51. Damato BE, Fisher AC, Taktak AF.. Prediction of metastatic death from uveal melanoma using a Bayesian artificial neural network. Invest Ophthalmol Vis Sci. 2003; 44(13): 2159. [Google Scholar]
- 52. Taktak AFG, Fisher AC, Damato BE.. Modelling survival after treatment of intraocular melanoma using artificial neural networks and Bayes theorem. Phys Med Biol. 2004; 49(1): 87–98. [DOI] [PubMed] [Google Scholar]
- 53. Kaiserman I, Rosner M, Pe'er J. Forecasting the prognosis of choroidal melanoma with an artificial neural network. Ophthalmology. 2005; 112(9): 1608. [DOI] [PubMed] [Google Scholar]
- 54. Damato B, Eleuteri A, Fisher AC, Coupland SE, Taktak AFG.. Artificial neural networks estimating survival probability after treatment of choroidal melanoma. Ophthalmology. 2008; 115(9): 1598–1607. [DOI] [PubMed] [Google Scholar]
- 55. Donizy P, Krzyzinski M, Markiewicz A, et al.. Machine learning models demonstrate that clinicopathologic variables are comparable to gene expression prognostic signature in predicting survival in uveal melanoma. Eur J Cancer. 2022; 174: 251–260. [DOI] [PubMed] [Google Scholar]
- 56. Serghiou S, Damato BE, Afshar AR.. Use of machine learning for prediction of ocular conservation and visual outcomes after proton beam radiotherapy for choroidal melanoma. Invest Ophthalmol Vis Sci. 2019; 60(9): 962. [Google Scholar]
- 57. Luo J, Chen Y, Yang Y, et al.. Prognosis prediction of uveal melanoma after plaque brachytherapy based on ultrasound with machine learning. Front Med (Lausanne). 2021; 8: 777142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang H, Kalirai H, Acha-Sagredo A, Yang X, Zheng Y, Coupland SE.. Piloting a deep learning model for predicting nuclear BAP1 immunohistochemical expression of uveal melanoma from hematoxylin-and-eosin sections. Transl Vis Sci Technol. 2020; 9(2): 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun M, Zhou W, Qi X, et al.. Prediction of BAP1 expression in uveal melanoma using densely-connected deep classification networks. Cancers (Basel). 2019; 11(10): 1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu TYA, Chen H, Gomez C, Correa ZM, Unberath M.. Direct gene expression profile prediction for uveal melanoma from digital cytopathology images via deep learning and salient image region identification. Ophthalmol Sci. 2023; 3(1): 100240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vaquero-Garcia J, Lalonde E, Ewens KG, et al.. PRiMeUM: a model for predicting risk of metastasis in uveal melanoma. Invest Ophthalmol Vis Sci. 2017; 58(10): 4096–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kaiserman I, Kaiserman N, Pe'er J. Long term ultrasonic follow up of choroidal naevi and their transformation to melanomas. Br J Ophthalmol. 2006; 90(8): 994–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bolis M, Garattini E, Paroni G, et al.. Network-guided modeling allows tumor-type independent prediction of sensitivity to all-trans-retinoic acid. Ann Oncol. 2017; 28(3): 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ancona-Lezama D, Dalvin LA, Shields CL.. Modern treatment of retinoblastoma: A 2020 review. Indian J Ophthalmol. 2020; 68(11): 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Global Retinoblastoma Study Group. Global retinoblastoma presentation and analysis by national income level. JAMA Oncol. 2020; 6(5): 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fabian ID, Onadim Z, Karaa E, et al.. The management of retinoblastoma. Oncogene. 2018; 37(12): 1551–1560. [DOI] [PubMed] [Google Scholar]
- 67. Bernard A, Xia SZ, Saleh S, et al.. EyeScreen: development and potential of a novel machine learning application to detect leukocoria. Ophthalmol Sci. 2022; 2(3): 100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Subrahmanyeswara Rao B. Accurate leukocoria predictor based on deep VGG-net CNN technique. IET Image Processing. 2020; 14(10): 2241–2248. [Google Scholar]
- 69. Rivas-Perea P, Baker E, Hamerly G, Shaw BF.. Detection of leukocoria using a soft fusion of expert classifiers under non-clinical settings. BMC Ophthalmol. 2014; 14: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. YS, Vempuluru VS, Ghose N, Patil G, Viriyala R, Dhara KK. Artificial intelligence and machine learning in ocular oncology: retinoblastoma. Indian J Ophthalmol. 2023; 71(2): 424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kumar P, Suganthi D, Valarmathi K, et al.. A multi-thresholding-based discriminative neural classifier for detection of retinoblastoma using CNN models. Biomed Res Int. 2023; 2023: 5803661. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72. Alvarez-Suarez DE, Tovar H, Hernández-Lemus E, et al.. Discovery of a transcriptomic core of genes shared in 8 primary retinoblastoma with a novel detection score analysis. J Cancer Res Clin Oncol. 2020; 146(8): 2029–2040. [DOI] [PubMed] [Google Scholar]
- 73. Berry JL, Xu L, Kooi I, et al.. Genomic cfDNA analysis of aqueous humor in retinoblastoma predicts eye salvage: the surrogate tumor biopsy for retinoblastoma. Mol Cancer Res. 2018; 16(11): 1701–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Berry JL, Xu L, Murphree AL, et al.. Potential of aqueous humor as a surrogate tumor biopsy for retinoblastoma. JAMA Ophthalmol. 2017; 135(11): 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu W, Luo Y, Dai J, et al.. Monitoring retinoblastoma by machine learning of aqueous humor metabolic fingerprinting. Small Methods. 2022; 6(1): e2101220. [DOI] [PubMed] [Google Scholar]
- 76. Im DH, Pike S, Reid MW, et al.. A multicenter analysis of nucleic acid quantification using aqueous humor liquid biopsy in retinoblastoma. Ophthalmol Sci. 2023; 3(3): 100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ciller C, Ivo S., De Zanet S, Apostolopoulos S, et al.. Automatic segmentation of retinoblastoma in fundus image photography using convolutional neural networks. Invest Ophthalmol Vis Sci. 2017; 58(8): 3332. [Google Scholar]
- 78. Ciller C, De Zanet S, Kamnitsas K, et al.. Multi-channel MRI segmentation of eye structures and tumors using patient-specific features. PLoS One. 2017; 12(3): e0173900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Strijbis VIJ, de Bloeme CM, Jansen RW, et al.. Multi-view convolutional neural networks for automated ocular structure and tumor segmentation in retinoblastoma. Sci Rep. 2021; 11(1): 14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Han L, Cheng MH, Zhang M, Cheng K.. Identifying key genes in retinoblastoma by comparing classifications of several kinds of significant genes. J Cancer Res Ther. 2018; 14(Suppl): S22–S27. [DOI] [PubMed] [Google Scholar]
- 81. Kakkassery V, Gemoll T, Kraemer MM, et al.. Protein profiling of WERI-RB1 and etoposide-resistant WERI-ETOR reveals new insights into topoisomerase inhibitor resistance in retinoblastoma. Int J Mol Sci. 2022; 23(7): 4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Brouwer NJ, Verdijk RM, Heegaard S, Marinkovic M, Esmaeli B, Jager MJ.. Conjunctival melanoma: new insights in tumour genetics and immunology, leading to new therapeutic options. Prog Retin Eye Res. 2022; 86: 100971. [DOI] [PubMed] [Google Scholar]
- 83. Kisser U, Heichel J, Glien A.. Rare diseases of the orbit. Laryngorhinootologie. 2021; 100(Suppl 1): S1–S79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yoo TK, Choi JY, Kim HK, Ryu IH, Kim JK.. Adopting low-shot deep learning for the detection of conjunctival melanoma using ocular surface images. Comput Methods Programs Biomed. 2021; 205: 106086. [DOI] [PubMed] [Google Scholar]
- 85. Xie X, Yang L, Zhao F, et al.. A deep learning model combining multimodal radiomics, clinical and imaging features for differentiating ocular adnexal lymphoma from idiopathic orbital inflammation. Eur Radiol. 2022; 32(10): 6922–6932. [DOI] [PubMed] [Google Scholar]
- 86. Luo Y, Zhang J, Yang Y, et al.. Deep learning-based fully automated differential diagnosis of eyelid basal cell and sebaceous carcinoma using whole slide images. Quant Imaging Med Surg. 2022; 12(8): 4166–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hui S, Dong L, Zhang K, et al.. Noninvasive identification of Benign and malignant eyelid tumors using clinical images via deep learning system. J Big Data. 2022; 9(1): 84. [Google Scholar]
- 88. Hou Y, Xie X, Chen J, et al.. Bag-of-features-based radiomics for differentiation of ocular adnexal lymphoma and idiopathic orbital inflammation from contrast-enhanced MRI. Eur Radiol. 2021; 31(1): 24–33. [DOI] [PubMed] [Google Scholar]
- 89. Habibalahi A, Bala C, Allende A, Anwer AG, Goldys EM.. Novel automated non invasive detection of ocular surface squamous neoplasia using multispectral autofluorescence imaging. Ocul Surf. 2019; 17(3): 540–550. [DOI] [PubMed] [Google Scholar]
- 90. Tan E, Lin F, Sheck L, Salmon P, Ng S.. A practical decision-tree model to predict complexity of reconstructive surgery after periocular basal cell carcinoma excision. J Eur Acad Dermatol Venereol. 2017; 31(4): 717–723. [DOI] [PubMed] [Google Scholar]
- 91. Ozaydin B, Berner ES, Cimino JJ.. Appropriate use of machine learning in healthcare. Intell Based Med. 2021; 5: 100041. [Google Scholar]
- 92. Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G.. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med. 2018; 178(11): 1544–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Burlina P, Paul W, Mathew P, Joshi N, Pacheco KD, Bressler NM.. Low-shot deep learning of diabetic retinopathy with potential applications to address artificial intelligence bias in retinal diagnostics and rare ophthalmic diseases. JAMA Ophthalmol. 2020; 138(10): 1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chicco D. Ten quick tips for machine learning in computational biology. BioData Mining. 2017; 10(1): 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Raschka S. Model evaluation, model selection, and algorithm selection in machine learning. arXiv:181112808 [cs, stat]. Published online November 10, 2020, http://arxiv.org/abs/1811.12808. Accessed October 18, 2021.
- 96. Ali S, Smith KA.. On learning algorithm selection for classification. Appl Soft Comput. 2006; 6(2): 119–138. [Google Scholar]
- 97. Kotthoff L, Gent IP, Miguel I. An evaluation of machine learning in algorithm selection for search problems. AI Communications. 2012; 25(3): 257–270. [Google Scholar]
- 98. Lee I, Shin YJ.. Machine learning for enterprises: applications, algorithm selection, and challenges. Business Horizons. 2020; 63(2): 157–170. [Google Scholar]
- 99. Awan SE, Bennamoun M, Sohel F, Sanfilippo FM, Dwivedi G.. Machine learning-based prediction of heart failure readmission or death: implications of choosing the right model and the right metrics. ESC Heart Fail. 2019; 6(2): 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhang Y, Weng Y, Lund J.. Applications of explainable artificial intelligence in diagnosis and surgery. Diagnostics (Basel). 2022; 12(2): 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lane AM, Kim IK, Gragoudas ES.. Survival rates in patients after treatment for metastasis from uveal melanoma. JAMA Ophthalmology. 2018; 136(9): 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Woodman SE. Metastatic uveal melanoma: biology and emerging treatments. Cancer J. 2012; 18(2): 148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Artificial intelligence/machine learning (AI/ML)-based software as a medical device (SaMD) action plan. Published online January 2021. Available at: https://www.fda.gov/media/145022/download.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.