Abstract
The ophD gene, encoding a permease for phthalate transport, was cloned from Burkholderia cepacia ATCC 17616. Expression of the gene in Escherichia coli results in the ability to transport phthalate rapidly into the cell. Uptake inhibition experiments show that 4-hydroxyphthalate, 4-chlorophthalate, 4-methylphthalate, and cinchomeronate compete for the phthalate permease. An ophD knockout mutant of 17616 grows slightly more slowly on phthalate but is still able to take up phthalate at rates equivalent to that of the wild-type strain. This means that 17616 must have a second phthalate-inducible phthalate uptake system.
Phthalate degradation by Burkholderia cepacia DBO1 (ATCC 29424) has been well studied at both the genetic (3, 4) and biochemical levels (1, 6, 10, 18). The four genes encoding the pathway enzymes are arranged in at least three operons (3), and a fourth operon codes for a quinolinate phosphoribosyl transferase that enhances the ability of DBO1 to grow on phthalate (4). Analysis of the nucleotide sequence identified a gene (ophD) encoding a nonfunctional putative permease containing a frameshift mutation (3). Since DBO1 is able to transport phthalate into the cell at wild-type levels even when this frameshifted permease gene is deleted (3), there must be an as-yet-unidentified mechanism for transporting phthalate into the cell. B. cepacia ATCC 17616 is also able to grow on phthalate and shows a restriction fragment length polymorphism pattern identical to that of DBO1 when probed with the cloned DBO1 genes for phthalate degradation. The present work was performed to investigate the ability of 17616 to transport phthalate into the cell.
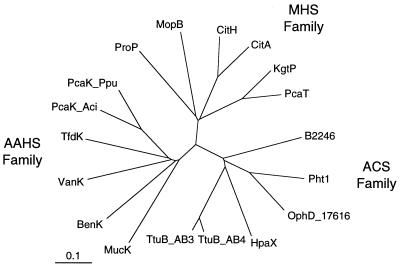
The frameshifted putative permease in DBO1 synthesizes two polypeptides, designated Orf1 and OphD (3). The region encoding these two proteins was PCR amplified from both DBO1 and 17616 by using the primers ophD-N (5′-GGCATATGGCACATTCAACGTTGCACTCCG-3′) and ophD-C (5′-CCCTGCAGTGTCACGCGCCGGATCGCTGCG-3′) according to standard procedures (3). The ophD-N primer contains the existing translation initiation codon ATG within a new NdeI site, and the ophD-C primer contains the existing stop codon TGA followed by a new PstI site. The PCR products were cloned into the pCRII-TOPO plasmid (Invitrogen, Carlsbad, Calif.), and the sequences of several clones were examined. The nucleotide sequence of this region from 17616 (GenBank accession no. AF152094) is only 1 base different from the DBO1 sequence (3): it has an additional A at position 301, numbering from the ATG initiation codon. The 17616 nucleotide sequence with the additional base thus codes for a “normal” permease rather than the two polypeptides seen in DBO1 (3). Comparison of the OphD of 17616 to other proteins reveals that it belongs to the major facilitator superfamily (MFS) of transport proteins (16). It has 12 membrane-spanning α-helices (11, 23), typical of members of MFSs. The OphD of 17616 is most closely related (Fig. 1) to permeases for transport of acidic compounds in the anion:cation symporter (ACS) family (16) such as the putative phthalate transporter Pht1 from Pseudomonas putida NMH102-2 (57.7% identity and 67.1% similarity), putative tartrate transporters (TtuB) from Agrobacterium vitis AB3 (39% identity and 50.9% similarity) and AB4 (42.2% identity and 53.6% similarity), and the putative p-hydroxyphenylacetate permease HpaX from Escherichia coli (36.0% identity and 47.2% similarity). The OphD of 17616 shows little similarity to members of the well-studied aromatic acid:H+ symporter (AAHS) family (16) such as the P. putida p-hydroxybenzoate and protocatechuate transporter PcaK and the Acinetobacter sp. benzoate transporter BenK (Fig. 1). MopB, a 4-methylphthalate-specific permease from B. cepacia Pc701, is able to transport both 4-methylphthalate and phthalate into the cell (19). However, despite the fact that it transports phthalate, MopB shows low similarity to the OphD of 17616 and falls into the metabolite:H+ symporter (MHS) family (16).
FIG. 1.
Dendrogram showing the relationship of the 17616 ophD gene product to selected transport proteins. Permeases involved in aromatic acid transport fall into three families as defined by Pao et al. (16): the ACS family, the AAHS family, and the MHS family. The ACS family includes the phthalate transporter OphD from B. cepacia 17616 (this work), the putative phthalate transporter Pht1 from P. putida NMH102-2 (15), the putative tartrate transporters (TtuB) from A. vitis AB3 (20) and AB4 (7), the putative p-hydroxyphenylacetate permease HpaX from E. coli (17), and the putative transporter B2246 from E. coli (2). The MHS family includes the 4-methylphthalate transporter MopB from B. cepacia Pc701 (19), the citrate transporters CitH from Klebsiella pneumoniae (24) and CitA from Salmonella typhimurium (22), the α-ketoglutarate transporter KgtP from E. coli (21), the proline/betaine transporter ProP from E. coli (8), and the dicarboxylate transporter PcaT from P. putida PRS2000 (GenBank accession no. U48776). The AAHS family includes the p-hydroxybenzoate and protocatechuate transporter PcaK from P. putida (14), the putative aromatic acid transporter PcaK from Acinetobacter sp. strain ADP1 (12), the benzoate transporter BenK from Acinetobacter sp. strain ADP1 (5), the putative vanillate transporter VanK from Acinetobacter sp. strain ADP1 (GenBank accession no. AF009672), the muconate transporter MucK from Acinetobacter sp. strain ADP1 (25), and the 2,4-dichlorophenoxyacetate transporter TfdK from Ralstonia eutropha (13). The amino acid sequences were aligned with the Pileup program of the Genetics Computer Group package (9), the alignment was confirmed by visual inspection, and the phylogenetic tree was calculated with the PAUP program by using the minimal-distance method.
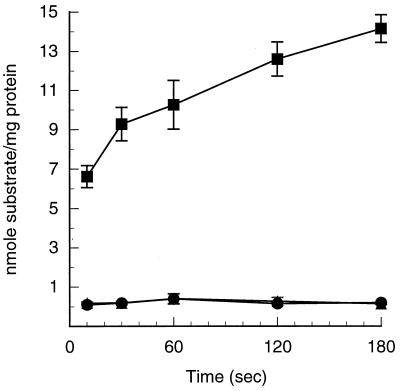
The cloned 1.3-kb PCR products containing orf1/ophDDBO1 and ophD17616 were moved separately into the expression vector pALTER-Ex1 (Promega, Madison, Wis.) by using the added NdeI and PstI cutting sites. E. coli JM109 carrying either clone (designated pGJZ1351 and pGJZ1352, respectively) was cultured in Luria broth containing 15 μg of tetracycline/ml until the cell optical density at 600 nm reached 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was then added to induce gene expression. The cells were collected 1.5 h after induction and washed twice with 50 mM phosphate buffer. Phthalate transport assays were performed as described earlier (3) except that 100 μl of cells rather than 300 μl was added to the assay mixture. JM109 expressing the OphD of 17616 rapidly transported phthalate into the cell, while JM109 expressing Orf1 and OphD of DBO1 or carrying the vector pALTER-Ex1 showed no accumulation of phthalate (Fig. 2). This data demonstrates that the frameshift in Orf1 and OphD of DBO1 results in loss of the ability of this protein to transport phthalate and proves that the OphD of 17616 is functional for transporting phthalate into the cells.
FIG. 2.
Phthalate uptake by IPTG-induced E. coli JM109 containing the cloned 17616 ophD (■), the cloned DBO1 orf1/ophD (●), or the vector pALTER-Ex1 (▴). Error bars, standard deviation from three independent experiments.
A number of compounds were tested for the ability to inhibit transport of radiolabeled phthalate by JM109 expressing the OphD of 17616 (Table 1). In a control experiment, a 20-fold excess of unlabeled phthalate inhibited the rate of transport of radiolabeled phthalate by 91%. Phthalate substituted at the 4 position with either a hydroxyl, a methyl, or a chloro group inhibited phthalate transport significantly when present in a 20-fold excess (86, 66, and 32%, respectively). Phthalate substituted with large polar substituent groups at the 4 position (4-sulfophthalate and 4-nitrophthalate) inhibited phthalate transport less than 10%. It is surprising that the structurally similar compound quinolinate (2,3-pyridinedicarboxylic acid) showed no detectable effect on phthalate transport, while cinchomeronate (3,4-pyridinedicarboxylic acid) inhibited phthalate transport 27%. This may be due to a disrupting effect of a nitrogen atom in the aromatic ring near the carboxyl groups. The structurally similar compounds o-chlorobenzoate and salicylate did not inhibit phthalate transport, probably because two carboxyl groups are required for substrate binding. The fact that compounds with two carboxyl groups, such as tartrate and maleic acid, did not inhibit phthalate transport implies that the aromatic ring structure is important for substrate recognition.
TABLE 1.
Substrate inhibition of phthalate uptake by E. coli JM109 expressing ophD from B. cepacia 17616
| Competing substratea | % Inhibition of phthalate uptakeb |
|---|---|
| Phthalate | 91 ± 1 |
| 4-Hydroxyphthalate | 86 ± 2 |
| 4-Chlorophthalate | 66 ± 4 |
| 4-Methylphthalate | 32 ± 4 |
| Cinchomeronate | 27 ± 6 |
The concentrations of phthalate and competing substrate were 50 μM and 1 mM, respectively.
The following compounds inhibited phthalate uptake less than 10%: 4-nitrophthalate, 4-sulfophthalate, 3-nitrophthalate, quinolinate, o-chlorobenzoate, salicylate, benzoate, p-hydroxybenzoate, p-hydroxyphenylacetate, maleic acid, and tartrate. Values are averages from three to six experiments ± standard deviations.
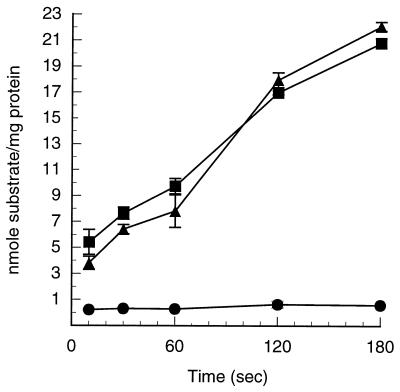
It was previously shown that DBO1 has an inducible phthalate transporter and that deletion of orf1 and ophD had no effect on either growth or the phthalate transport rate (3). It is thus possible that 17616 has a second phthalate-inducible transport system. A 17616 ophD knockout mutant was constructed by gene replacement with a kanamycin resistance gene cassette by following the same procedure as that used to make the analogous mutant of DBO1 (3). One strain resulting from a double crossover, designated 17616-CZ1, was saved for analysis. The mutant strain grows slightly more slowly on minimal medium (26) with 10 mM phthalate than does the wild type (doubling time, 113 ± 4 min versus 97 ± 1 min). Transport assays with 17616 grown on phthalate showed a rapid accumulation of phthalate at the rate of 5.6 nmol/min/mg of protein (Fig. 3). In contrast, 17616 grown on 10 mM p-hydroxybenzoate did not transport phthalate at any measurable rate (Fig. 3), demonstrating that the phthalate transport ability of 17616, like that of DBO1, is inducible. Interestingly, the knockout mutant 17616-CZ1 took up phthalate at the same rate as the wild-type strain, 17616 (Fig. 3). The data indicates that although 17616 has a functional phthalate transporter encoded by ophD, a second phthalate transport system exists. That this transport system is specific for phthalate is shown by the fact that 17616 grown on p-hydroxybenzoate did not show any ability to take up phthalate in the transport assay. It is entirely probable that, despite the fact that 17616 and DBO1 were independently isolated on different coasts of the United States, they have a common ancestor with two phthalate transporters and that DBO1 for some reason gained a frameshift mutation in ophD, resulting in a nonfunctional protein product.
FIG. 3.
Phthalate uptake by B. cepacia 17616 (wild type) following growth on phthalate (■) or p-hydroxybenzoate (●) and by 17616-CZ1 (ophD knockout mutant) following growth on phthalate (▴). Error bars, standard deviation from three independent experiments.
Nucleotide sequence accession number.
The nucleotide sequence of ophD from B. cepacia ATCC 17616 has been deposited in the GenBank database under accession no. AF152094.
Acknowledgments
This material is based on work supported by the National Science Foundation under grants CHE-9810248 and MCB-9257750.
Footnotes
Center for Environmental BioInorganic Chemistry (CEBIC) publication 4.
REFERENCES
- 1.Batie C J, LaHaie E, Ballou D P. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J Biol Chem. 1987;262:1510–1518. [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Chang H K, Zylstra G J. Novel organization of the genes for phthalate degradation from Burkholderia cepacia DBO1. J Bacteriol. 1998;180:6529–6537. doi: 10.1128/jb.180.24.6529-6537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang H K, Zylstra G J. Role of quinolinate phosphoribosyl transferase in degradation of phthalate by Burkholderia cepacia DBO1. J Bacteriol. 1999;181:3069–3075. doi: 10.1128/jb.181.10.3069-3075.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier L S, Nichols N N, Neidle E L. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J Bacteriol. 1997;179:5943–5946. doi: 10.1128/jb.179.18.5943-5946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correll C C, Batie C J, Ballou D P, Ludwig M L. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe–2S] Science. 1992;258:1604–1610. doi: 10.1126/science.1280857. [DOI] [PubMed] [Google Scholar]
- 7.Crouzet P, Otten L. Sequence and mutational analysis of a tartrate utilization operon from Agrobacterium vitis. J Bacteriol. 1995;177:6518–6526. doi: 10.1128/jb.177.22.6518-6526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culham D E, Lasby B, Marangoni A G, Milner J L, Steer B A, van Nues R W, Wood J M. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J Mol Biol. 1993;229:268–276. doi: 10.1006/jmbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gassner G T, Ballou D P, Landrum G A, Whittaker J W. Magnetic circular dichroism studies on the mononuclear ferrous active site of phthalate dioxygenase from Pseudomonas cepacia show a change of ligation state on substrate binding. Biochemistry. 1993;32:4820–4825. doi: 10.1021/bi00069a017. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann K, Stoffel W. TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 12.Kowalchuk G A, Hartnett G B, Benson A, Houghton J E, Ngai K L, Ornston L N. Contrasting patterns of evolutionary divergence within the Acinetobacter calcoaceticus pca operon. Gene. 1994;146:23–30. doi: 10.1016/0378-1119(94)90829-x. [DOI] [PubMed] [Google Scholar]
- 13.Leveau J H, Zehnder A J, van der Meer J R. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4) J Bacteriol. 1998;180:2237–2243. doi: 10.1128/jb.180.8.2237-2243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols N N, Harwood C S. PcaK, a high-affinity permease for the aromatic compound 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J Bacteriol. 1997;179:5056–5061. doi: 10.1128/jb.179.16.5056-5061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nomura Y, Nakagawa M, Ogawa N, Harashima S, Oshima Y. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J Ferment Bioeng. 1992;74:333–344. [Google Scholar]
- 16.Pao S S, Paulsen I T, Saier M H J. Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prieto M A, Diaz E, Garcia J L. Molecular characterization of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli W: engineering a mobile aromatic degradative cluster. J Bacteriol. 1996;178:111–120. doi: 10.1128/jb.178.1.111-120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pujar B G, Ribbons D W. Phthalate metabolism in Pseudomonas fluorescens PHK: purification and properties of 4,5-dihydroxyphthalate decarboxylase. Appl Environ Microbiol. 1985;49:374–376. doi: 10.1128/aem.49.2.374-376.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saint C P, Romas P. 4-Methylphthalate catabolism in Burkholderia (Pseudomonas) cepacia Pc701: a gene encoding a phthalate-specific permease forms part of a novel gene cluster. Microbiology. 1996;142:2407–2418. doi: 10.1099/00221287-142-9-2407. [DOI] [PubMed] [Google Scholar]
- 20.Salomone J Y, Crouzet P, De Ruffray P, Otten L. Characterization and distribution of tartrate utilization genes in the grapevine pathogen Agrobacterium vitis. Mol Plant-Microbe Interact. 1996;9:401–408. doi: 10.1094/mpmi-9-0401. [DOI] [PubMed] [Google Scholar]
- 21.Seol W, Shatkin A J. Escherichia coli kgtP encodes an alpha-ketoglutarate transporter. Proc Natl Acad Sci USA. 1991;88:3802–3806. doi: 10.1073/pnas.88.9.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimamoto T, Izawa H, Daimon H, Ishiguro N, Shinagawa M, Sakano Y, Tsuda M, Tsuchiya T. Cloning and nucleotide sequence of the gene (citA) encoding a citrate carrier from Salmonella typhimurium. J Biochem (Tokyo) 1991;110:22–28. doi: 10.1093/oxfordjournals.jbchem.a123537. [DOI] [PubMed] [Google Scholar]
- 23.TMpred—Prediction of Transmembrane Regions and Orientation. [Online.] European Molecular Biology network. http://www.ch.embnet.org/software/TMPRED_form.html. [17 May 1999, last date accessed.]
- 24.van der Rest M E, Schwarz E, Oesterhelt D, Konings W N. DNA sequence of a citrate carrier of Klebsiella pneumoniae. Eur J Biochem. 1990;189:401–407. doi: 10.1111/j.1432-1033.1990.tb15502.x. [DOI] [PubMed] [Google Scholar]
- 25.Williams P A, Shaw L E. mucK, a gene in Acinetobacter calcoaceticus ADP1 (BD413), encodes the ability to grow on exogenous cis,cis-muconate as the sole carbon source. J Bacteriol. 1997;179:5935–5942. doi: 10.1128/jb.179.18.5935-5942.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zylstra G J, Olsen R H, Ballou D P. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase genes. J Bacteriol. 1989;171:5907–5914. doi: 10.1128/jb.171.11.5907-5914.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]