Abstract
We report here the characterization of the catalytic component (ISPNAR) of a new naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. The genes encoding the two subunits of ISPNAR are not homologous to their previously characterized counterparts in Pseudomonas. The deduced amino acid sequences have only 33 and 29% identity with the corresponding subunits in Pseudomonas putida NCIB 9816-4, for which the tertiary structure has been reported.
Members of the genus Rhodococcus are found in many environmental niches and have a remarkable ability to metabolize a wide variety of xenobiotic compounds (6, 25). Naphthalene is released into the environment as coal tar and coal tar products such as creosote (17), and bacteria which degrade naphthalene are widely distributed in nature (3). The first step in the catabolism of naphthalene by bacteria has been elucidated for Pseudomonas spp. and involves the oxidation of naphthalene to (+)-cis-(1R,2S)-dihydroxy-1,2-dihydronaphthalene (naphthalene cis-dihydrodiol), which is catalyzed by naphthalene 1,2-dioxygenase (NDO) (EC 1.14.12.12) (11). To date only one NDO has been well characterized, namely, that of Pseudomonas putida NCIB 9816-4. In this sense, therefore, NDO is a paradigm for further studies of this important group of enzymes.
The Pseudomonas NDO is a member of a family of over 40 multicomponent, non-heme iron oxygenases (2). Initially, an iron-sulfur flavoprotein transfers electrons from NAD(P)H to a putative Rieske [2Fe-2S] iron sulfur center in a ferredoxin protein. The electrons are then transferred to the catalytic component of the enzyme to facilitate the addition of dioxygen to the substrate, naphthalene. The catalytic component of the enzyme, ISPNAP, consists of two nonidentical subunits (α and β) needed for the reaction. Recently, the three-dimensional structure has been elucidated by X-ray crystallography and shown to be a 3α and 3β structure (12). The role of the β subunit has not been determined; however, the α subunit is directly involved in catalysis. Electrons are passed to a Rieske [2Fe-2S] center of one α subunit and then to a mononuclear iron at the active site in an adjacent α subunit. The apparent presence of a Rieske [2Fe-2S] center and a mononuclear iron moiety at the active site is typical of such dioxygenases (2). The catabolism of naphthalene by Rhodococcus strain B4 to the metabolites salicylic acid and gentisic acid has been reported (7). Many putative NDO sequences are available in gene data banks; however, most are not based on biochemical confirmation of their induced expression and function. Also, they are all very closely homologous at the DNA level to either the Pseudomonas NDO (23) or the 2,4-dinitrotoluene dioxygenase from Burkholderia sp. strain DNT (24).
We have previously reported that Rhodococcus strain NCIMB12038 degrades 1-naphthol to salicylic acid and gentisic acid (15). This strain has been shown also to catabolize naphthalene and to express NDO enzyme activity in cell extracts, with naphthalene cis-dihydrodiol confirmed as the product (1). We report here the purification and characteristics of a new naphthalene-induced NDO ISPNAR from Rhodococcus sp. strain NCIMB12038 and the complete nucleotide sequence of the genes encoding the α and β subunits.
Characterization of the Rhodococcus NDO.
The conditions used to grow Rhodococcus sp. strain NCIMB12038 were as described by Allen et al. (1). The NDO assay used was based on that of Ensley et al. (5), where [14C]naphthalene is converted to a relatively nonvolatile product, cis-1,2-dihydroxy-1,2-dihydro[14C]naphthalene (naphthalene cis-dihydrodiol) (1). An optimum NADH concentration of 2 mM was reported for NDO activity in P. putida NCIB 9816-4 (5). However, our results suggest that with the Rhodococcus sp. enzyme, the optimum concentration is much higher; we assayed NDO in the presence of 5.5 mM NADH.
The effect of other coenzymes on NDO activity was determined, and NADPH was found to be usable as an alternative electron donor to NADH. When flavin adenine dinucleotide (FAD) or flavin mononucleotide was added, enzyme activity also increased. A similar effect was observed with P. putida NCIB 9816-4 (5), and the data suggest that a flavoprotein is also required for NDO activity in Rhodococcus sp. Subsequently, 5 μM FAD was added routinely to NDO assays.
For purification of the protein required for NDO activity, which was subsequently designated ISPNAR, naphthalene-grown cells were resuspended in 50 mM TEG buffer (50 mM Tris-HCl [pH 7.8] containing 10% [vol/vol] ethanol, 10% [vol/vol] glycerol, and 0.5 mM dithiothreitol) (4), disrupted with a French pressure cell, centrifuged (40,000 × g for 1 h), and applied to a Blue Sepharose (heparin-Sepharose CL-6B and Pharmacia FPLC system) affinity column eluted with TEG buffer containing 1 mM NAD (8). The nonbinding NDO active fraction was concentrated (100-kDa ultrafiltration membrane; Vivascience), applied to an anion-exchange column (Pharmacia Q-Sepharose LKB), and eluted with a continuous potassium chloride salt gradient (0 to 1 M). Two fractions were needed to restore enzyme activity: fraction A (yellow) eluted between 0.41 and 0.43 M KCl, and fraction B (brown) eluted between 0.44 and 0.46 M KCl. Both fractions were concentrated by using a 5-kDa ultrafiltration membrane (Vivascience). Fraction B was further purified by gel filtration chromatography (Pharmacia Superose 12 HR) and contained a protein that had NDO activity and which could be reconstituted in the presence of fraction A. The data show that 8.2-fold purification of the protein was achieved with 53% recovery of enzyme activity (Table 1). Surprisingly, NDO activity was present in the first fraction eluted from the Blue Sepharose column without added NAD+. This observation was unexpected, as it was shown previously that one of the components of the NDO complex from P. putida NCIB 9816-4 bound to this type of affinity column (8).
TABLE 1.
Purification of the ISPNAR protein from Rhodococcus sp. strain NCIMB12038
| Purification stagea | Protein (mg) | Activity (U)b | Sp act (U · mg−1) | Recovery (%) |
|---|---|---|---|---|
| (i) Cell extract | 198 | 144.7 | 0.731 | 100 |
| (ii) Blue Sepharose column | 137 | 113.8 | 0.831 | 79 |
| (iii) Q-Sepharose column | 23 | 119.6 | 5.225c | 82 |
| (iv) Superose 12 column | 13 | 77.4 | 5.971 | 53 |
See text for details.
One unit represents production of 1.0 nmol of cis-dihydrodiol · min−1.
Fractions A and B were both necessary for enzyme activity. When either fraction was assayed alone, coenzyme-dependent specific activity was reduced to approximately 1 and 14%, respectively, of the combined activity. In enzyme assays used for stages iii and iv, 0.3 mg of fraction A and 0.1 mg of fraction B or ISPNAR were combined in 200 μl of assay mixture.
Although the fold purification may seem low, the results also indicate that the recovered ISPNAR protein constituted 6.6% of the total protein in the cell extract. This should be compared with P. putida NCIB 9816-4, where the analogous ISPNAP protein constituted only 0.9% of the total cell-free protein (4). However, too much emphasis should not be placed on the importance of specific activity measurements in this case. The specific activity of multicomponent enzymes such as NDO is dependent on protein concentration and the relative amounts of the different enzyme components in the assay mixture (i.e., that of the purified ISPNAR protein and the components present in fraction A, as indicated in Table 1). This was confirmed when specific activity was measured while varying the amount of fraction A in the assay but with a constant amount of purified ISPNAR protein (data not shown). These factors explain why there is an apparent increase in the recovery of enzyme activity between steps ii and iii of the purification procedure (Table 1): the components of the dioxygenase enzyme complex were separated at this point.
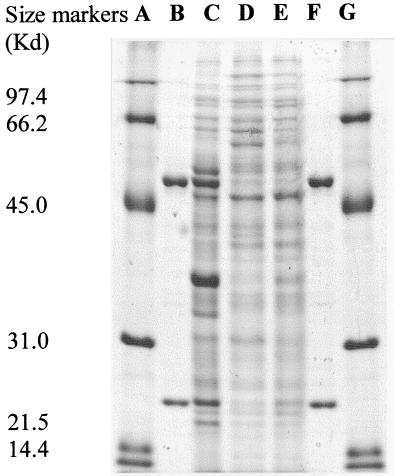
Polyacrylamide gel electrophoresis (PAGE) of the active ISPNAR protein under nondenaturing conditions (10% [wt/vol] acrylamide gel) revealed the presence of a single band that stained for protein (not shown). PAGE (sodium dodecyl sulfate [SDS] and native gels) was performed by published procedures (14). The molecular mass of the ISPNAR protein was estimated to be 155 kDa from the retention time of the protein on the Superose 12 column used in the purification protocol. Analysis of the purified protein by SDS-PAGE revealed that it was comprised of two subunits, of 55 and 23 kDa, as indicated in Fig. 1. The larger protein is here designated the α subunit, and the smaller is designated the β subunit. The data suggest that in solution, the ISPNAR complex has an α2β2 configuration. Although in similar experiments the P. putida NCIB 9861-4 ISPNAP α and β proteins appeared to form a dimeric α2β2 structure (4), the three-dimensional structure indicated a more plausible α3β3 configuration (12). Hence, the α3β3 subunit configuration should also be considered for the ISPNAR enzyme.
FIG. 1.
Expression of NDO ISPNAR α and β subunits in protein extracts from Rhodococcus sp. strain NCIMB12038 grown on naphthalene (C), salicylate (D), and pyruvate (E). Purified ISPNAR α and β subunits are also shown (B and F). Size markers are low-molecular-weight standards (A and G) (Bio-Rad).
The N-terminal sequences of the ISPNAR α and β subunits (sequence data were obtained by Brian Dunbar at the Protein Sequencing Facility, University of Aberdeen, Aberdeen, United Kingdom) were found to be Met Leu Ser Asn Glu Leu Arg Gln Thr Leu Gln Lys Gly Leu His Asp Val Asn Ser Asp Trp Thr Val Pro Ala and Met Asn Thr Gln X Val Ser Asp Thr Thr Val Arg Glu Ile Thr Glu Trp Leu Tyr Met Glu Ala Glu Leu, respectively. These sequences facilitated analysis of the genes encoding the ISPNAR subunits (see below).
In a previous report (1), it was noted that NDO activity is expressed by strain NCIMB12038 during growth on naphthalene as the sole carbon source but not when grown on salicylate or pyruvate. We analyzed cell extracts of NCIMB12038 when it was grown on these substrates as the sole carbon source and by SDS-PAGE compared the proteins expressed under the various conditions with the purified ISPNAR α and β subunits (Fig. 1). The results show clearly that proteins of the same molecular weight as the purified components are expressed in naphthalene-grown cells only. This evidence strongly supports the proposal that the purified enzyme α and β subunits are directly associated with naphthalene metabolism and expression of NDO activity.
In order to further confirm the identity of the ISPNAR proteins as the catalytic components of NDO, we incubated the purified protein (combined α and β subunits) with radioactive [14C]naphthalene in a substrate binding experiment. The bound [14C]naphthalene was then separated from unbound substrate by gel filtration chromatography. A similar method was used to show that the ISPNAP component from P. putida 9816-4 binds naphthalene (4). Here, a solution of [14C]naphthalene (5.6 μl of 10 mM dimethylformamide [8.1 μCi/mmol]) was added to 100 μl of purified ISPNAR enzyme solution (0.69 mg) in Tris buffer (50 mM, pH 7.5). After incubation for 20 min at room temperature, the mixture was applied to a Sephadex G-25 gel filtration column, and fractions (1 ml) were eluted with the same buffer. The collected fractions were then assayed for radioactivity (Beckman Rackbeta 1217 scintillation counter, Amersham BCS scintillation fluid) and protein concentration (BCA assay; Pierce Inc., Rockford, Ill.). By analysis of the protein and radioactivity present in fractions collected from the gel filtration column, we were able to show that only the ISPNAR protein bound to the [14C]naphthalene. In a control experiment, the ISPNAR protein was replaced with an equal amount of bovine serum albumin and naphthalene binding was not detected.
We attempted to determine if the purified ISPNAR protein was also necessary for conversion of naphthalene to naphthalene cis-dihydrodiol. The enzyme assay was performed with the purified protein, as shown in Table 1. The reaction was terminated after 5 min by diluting the assay mixture (200 μl) with methanol (800 μl), and 30 ml of this mixture was analyzed by thin-layer chromatography (0.25 mM silica gel plates with a mobile phase comprising 8% methanol in dichloromethane). It was noted after autoradiography that only one radioactive metabolite was present, and it had the same Rf (0.5) as an authentic standard of naphthalene cis-dihydrodiol. The autoradiography procedures are described elsewhere (1). In control experiments, the assay was repeated with both the purified ISPNAR protein and the fraction A protein alone. In these cases, no radioactive naphthalene cis-dihydrodiol was detected. This confirmed that at least two proteins (one being ISPNAR and present in fraction B and the other in fraction A) were necessary for naphthalene cis-dihydrodiol formation.
Biochemical evidence that ISPNAR is a Rieske-type iron-sulfur protein.
A typical feature of the catalytic component of dioxygenase enzymes is that they are iron-sulfur proteins with Rieske-type [2Fe-2S] centers (9). We therefore sought to determine the iron and sulfur content of the purified ISPNAR protein and found that 2.4 g-atoms of iron (± 0.2 [n = 3]) and 2.1 g-atoms of sulfur (± 0.9 [n = 3]) were present per αβ unit of protein. Published methods were used to determine the iron (8) and sulfur (22) contents of the purified proteins, and the sulfur assay was calibrated with ferredoxin from Clostridium pasteurinum (Sigma).
The UV spectrum of the oxidized ISPNAR protein was also obtained in 50 mM Tris buffer (pH 7.5) by using a Beckman DU 640B spectrophotometer (4). Absorbance maxima of 334 nm (ɛ = 23.9 mM−1 · cm−1), 462 nm (ɛ = 14.1 mM−1 · cm−1), and 566 nm (ɛ = 7.5 mM−1 · cm−1 [shoulder]) were characteristic of a Rieske-type [2Fe-2S] iron sulfur center being present in the protein (2). These absorbance maxima are also identical to those observed with the ISPNAP protein from P. putida NCIB 9816-4 (4). This conclusion was further supported by the fact that when the protein was reduced with sodium dithionite, the absorbance maxima at 462 and 566 nm disappeared and a new maximum was observed at 519 nm. To obtain the reduced UV spectrum of the ISPNAR protein, the method of Latimer et al. (16) was used. The enzyme (170 μg of protein in 50 μl) was added to a 1 M solution of oxygen-free sodium dithionite (2.5 ml in 50 mM Tris buffer, pH 7.5) in a sealed quartz cuvette. The mixture was then purged with nitrogen before spectroscopy. A similar observation was noted for the purified ISPNAP protein when it was reduced enzymatically (4). When air was purged through the reduced ISPNAR mixture, the oxidized spectrum was partially restored. Overall, these observations indicate that ISPNAR is a redox protein, with shifts in absorbance maxima of this type being also observed for other Rieske-type [2Fe-2S] oxygenases and flavoproteins (10, 16, 20).
Nucleotide sequence analysis of the narAa and narAb genes.
N-terminal sequences of the ISPNAR α and β subunits were used to design primers for analysis of the NDO (narA) locus. It was assumed that the order of the genes encoding these subunits in Rhodococcus is the same as was shown for Pseudomonas species. The likely codon usage preference of Rhodococcus (assuming a high G+C content) was taken into account for the design of three degenerate primers: F200 (forward primer, amino acid residues 16 to 20 of the ISPNAR α subunit), 5′-GACGTSAACWSSGACTGGAC-3′; F201 (forward primer, amino acid residues 4 to 9 of the ISPNAR α subunit), 5′-AACGAGCTSCGSCAGAC-3′; R202 (reverse primer, amino acid residues 24 to 18 of the ISPNAR β subunit), 5′-TCSGCCTCCATGTASAGCCA-3′. Total DNA from the Rhodococcus strain was isolated as described by Kulakova et al. (13), and amplification of Rhodococcus sequences was done with Taq+ DNA polymerase (Stratagene). Reactions were carried out in volumes of 25 μl with deoxynucleoside triphosphates at 200 μM concentrations and primers at 0.6 μM each. The following temperature profile was used: denaturation at 95°C for 3 min, followed by 35 cycles of 95°C for 40 s, 55°C for 30 s, and 72°C for 1 min. Both pairs of primers (F200-R202 and F201-R202) yielded fragments of about 1.5 kbp, analysis of which revealed that they contained sequences corresponding to the N-terminal amino acid sequences of the larger, α subunit and the smaller, β subunit of NDO. Nucleotide sequences upstream and downstream of this region were obtained by the inverse PCR approach. For this, total DNA was digested with either the MluI, XhoI, or SacII restriction enzyme. Then, the preparations were ligated with T4 DNA ligase and used in PCRs with different sets of divergent primers (conditions as above except that the primer concentration was 0.15 μM).
A sequence of 2,619 bp was thus obtained with the Taq Dye Deoxy Terminator Cycle Sequencing Kit and an automatic sequencer (Applied Biosystems model 373A). This sequence contained two open reading frames that we have designated narAa, corresponding to the α subunit, and narAb, corresponding to the β subunit. The narAa gene (bp 510 to 1922) encodes a putative protein of 470 amino acids, in which a sequence of 25 N-terminal amino acids is identical to that of the NCIMB12038 NDO ISPNAR α subunit iron-sulfur protein. Similarly, the narAb gene (bp 1926 to 2444) encodes a putative protein of 172 amino acids; 23 N-terminal amino acids were identical to that of the NCIMB12038 NDO ISPNAR β subunit iron-sulfur protein (note that residue 5 was not determined with certainty in the N-terminal sequence). Both genes are preceded by putative ribosome binding sites (21).
Database searches (with the FASTA and BLAST programs [19] and the EMBL and GenBank databases) and homology analysis revealed that the putative products of the narAa and narAb genes have some similarity (31 to 39% amino acid identity) with the α and β subunits of a number of aromatic-ring-hydroxylating dioxygenases (results not presented). The identity of the deduced amino acid sequences of the narAa and narAb genes with the N-terminal sequences of the α and β subunits of the Rhodococcus NDO and the similarity of these amino acid sequences to different aromatic-ring-hydroxylating dioxygenases suggest that the sequenced genes indeed encode the corresponding subunits of NDO.
Alignment of the α subunits of the Rhodococcus sp. strain NCIMB12038 NDO and the P. putida 9816-4 NDO is shown in Fig. 2. It is worth noting that the β subunits of these enzymes show a slightly lower level of overall similarity than the corresponding α subunits (results not presented). There is little known to date about the catalytic role of the β subunit, which makes it difficult to discuss the significance of the conserved amino acid sequences in these proteins.
FIG. 2.
Amino acid sequence alignment of the ISPNAR α subunits of naphthalene dioxygenases of P. putida 9816-4 (NahAc) and Rhodococcus sp. strain NCIMB12038 (NarAa). Sequences were aligned by the CLUSTALW program, and manual corrections were introduced to permit alignment of the TVFPN sequences.
Despite significant differences in the amino acid sequences of the α subunits, they show conservation of key catalytic residues, as summarized in Table 2. In general, there appears to be more conservation of the amino acid residues within the catalytic regions than in the other regions putatively associated with ferredoxin binding and substrate specificity. It is worth bearing in mind that the sequence of amino acids in the putative ferredoxin protein binding site region of the Pseudomonas ISPNAP α subunit is very different from that in the analogous region of the Rhodococcus ISPNAR α subunit. In addition to these observations, we note the surprising conservation of a five-amino-acid sequence located in a region of the ISPNAR α subunit which has coordinates close to the substrate cleft, as suggested in the model of Kauppi et al. (12).
TABLE 2.
Conservation of key amino acids in α subunits of NDOsa
| Rieske center
|
Active site
|
Putative ferredoxin binding region
|
Electron transfer (Rieske active site)
|
Region of unknown function
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| NahAc | NarAa | NahAc | NarAa | NahAc | NarAa | NahAc | NarAa | NahAc | NarAa |
| C81 | C88 | N201 | N209 | K97 | S104 | W106 | W113 | T299 | T297 |
| C101 | C108 | H208 | H216 | G98 | H105 | V117 | V124 | V300 | V298 |
| H83 | H90 | H213 | H221 | V100 | R107 | R84 | R91 | F301 | F299 |
| H104 | H111 | D205b | D213 | Q115 | V122 | E200 | D208 | P302 | P300 |
| D362 | D372 | S116 | G123 | N303 | N301 | ||||
| P118c | P125c | ||||||||
| W211 | M219 | ||||||||
In summary, we have isolated a new naphthalene dioxygenase whose catalytic component has a greatly divergent amino acid sequence compared to dioxygenases with similar biochemical properties. Our belief that the ISPNAR protein is the catalytic component of the naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038 is based on a number of biochemical observations. The purified protein binds naphthalene and is necessary for its subsequent conversion to naphthalene cis-dihydrodiol. Like the ISPNAP protein from Pseudomonas, ISPNAR is also an iron-sulfur protein, with good evidence for the presence of a Rieske [2Fe-2S] center. The ISPNAR protein is comprised of α and β subunits with similar size and configuration (α2β2) to ISPNAP and is clearly expressed only during growth on naphthalene (and not during growth on pyruvate or salicylate) in Rhodococcus sp. However, the low overall similarity of the Rhodococcus and Pseudomonas NDOs raises an important question concerning the evolutionary origins of these enzymes. They may have diverged from the same ancestral dioxygenase, or they may be the result of convergent evolution of different proteins. If the latter is the case, then the close similarity of the catalytic regions may be due to functional demands rather than actual conservation of amino acid residues.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been assigned GenBank accession no. AF082663.
Acknowledgments
We are grateful to D. T. Gibson for helpful comments.
REFERENCES
- 1.Allen C C R, Boyd D R, Larkin M J, Reid K A, Sharma N D, Wilson K. Metabolism of naphthalene, 1-naphthol, indene, and indole by Rhodococcus sp. strain NCIMB 12038. Appl Environ Microbiol. 1997;63:151–155. doi: 10.1128/aem.63.1.151-155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler C S, Mason J R. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv Microb Physiol. 1997;38:47–84. doi: 10.1016/s0065-2911(08)60155-1. [DOI] [PubMed] [Google Scholar]
- 3.Cerniglia C E. Biodegradation of polycyclic aeromatic hydrocarbons. Biodegradation. 1992;3:351–368. [Google Scholar]
- 4.Ensley B D, Gibson D T. Naphthalene dioxygenase: purification and properties of a terminal oxygenase component. J Bacteriol. 1983;155:505–511. doi: 10.1128/jb.155.2.505-511.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ensley B D, Gibson D T, Laborde A L. Oxidation of naphthalene by a multi-component enzyme system from Pseudomonas sp. strain NCIB 9816. J Bacteriol. 1982;149:948–954. doi: 10.1128/jb.149.3.948-954.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finnerty W R. The biology and genetics of the genus Rhodococcus. Annu Rev Microbiol. 1992;46:193–218. doi: 10.1146/annurev.mi.46.100192.001205. [DOI] [PubMed] [Google Scholar]
- 7.Grund E, Denecke B, Eichenlaub R. Naphthalene degradation via salicylate and gentisate by Rhodococcus sp. strain B4. Appl Environ Microbiol. 1992;58:1874–1877. doi: 10.1128/aem.58.6.1874-1877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haigler B E, Gibson D T. Purification and properties of NADH-ferredoxinNAP reductase, a component of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Bacteriol. 1990;172:457–464. doi: 10.1128/jb.172.1.457-464.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harayama S, Kok M, Niedle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki T, Isogai Y, Iizuka T, Oshima T. Sulredoxin: a novel iron-sulfur protein of the thermoacidophilic archaeon Sulfolobus sp. strain 7 with a Rieske-type [2Fe-2S] center. J Bacteriol. 1995;177:2576–2582. doi: 10.1128/jb.177.9.2576-2582.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeffrey A M, Yeh H J C, Jerina D M, Patel T R, Davey J F, Gibson D T. Initial reactions in the oxidation of naphthalene by Pseudomonas putida. Biochemistry. 1975;14:575–584. doi: 10.1021/bi00674a018. [DOI] [PubMed] [Google Scholar]
- 12.Kauppi B, Lee K, Carrendano E, Parales R E, Gibson D T, Eklund H, Ramaswamy S. Structure of an aromatic-ring-hydroxylating dioxygenase—naphthalene 1,2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 13.Kulakova A N, Stafford T M, Larkin M J, Kulakov L A. Plasmid pRTL1 controlling 1-chloroalkane degradation by Rhodococcus rhodochrous NCIMB13064. Plasmid. 1995;33:208–217. doi: 10.1006/plas.1995.1022. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Larkin M J, Day M J. The metabolism of carbaryl by three isolates, Pseudomonas spp (NCIB12042 and 12043) and Rhodococcus sp (NCIB12038), from garden soil. J Appl Bacteriol. 1986;60:233–242. doi: 10.1111/j.1365-2672.1986.tb01078.x. [DOI] [PubMed] [Google Scholar]
- 16.Latimer M T, Painter M H, Ferry J G. Characterization of an iron-sulfur flavoprotein from Methanosarina thermophila. J Biol Chem. 1996;271:24023–24028. doi: 10.1074/jbc.271.39.24023. [DOI] [PubMed] [Google Scholar]
- 17.Mueller J G, Chapman P J, Pritchard P H. Creosote-contaminated sites—their potential for bioremediation. Environ Sci Technol. 1989;23:1197–1201. [Google Scholar]
- 18.Parales R E, Parales J G, Gibson D T. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. J Bacteriol. 1999;181:1831–1837. doi: 10.1128/jb.181.6.1831-1837.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pikus J D, Studts J M, Adim C, Kauffmann K E, Munck E, Steffan R J, McClay K, Fox B G. Recombinant toluene-4-monooxygenase: catalytic and Mössbauer studies of the purified diiron and Rieske components of a four-protein complex. Biochemistry. 1996;35:9106–9119. doi: 10.1021/bi960456m. [DOI] [PubMed] [Google Scholar]
- 21.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosomal binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel L M. A direct microdetermination for sulfide. Anal Biochem. 1965;11:126–132. doi: 10.1016/0003-2697(65)90051-5. [DOI] [PubMed] [Google Scholar]
- 23.Simon M J, Osslund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W C, Cruden D L, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 24.Suen W C, Haigler B E, Spain J C. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J Bacteriol. 1996;178:4926–4934. doi: 10.1128/jb.178.16.4926-4934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warhurst A M, Fewson C A. Biotransformations catalyzed by the genus Rhodococcus. Crit Rev Biotechnol. 1994;14:29–73. doi: 10.3109/07388559409079833. [DOI] [PubMed] [Google Scholar]